The intrinsic L1 metallo- and L2 serine-β-lactamases in Stenotrophomonas maltophilia make it naturally multidrug resistant and difficult to treat. There is a need to identify novel treatment strategies for this pathogen, especially against isolates resistant to first-line agents. Aztreonam in combination with avibactam has demonstrated potential, although data on other aztreonam–β-lactamase inhibitor (BLI) combinations are lacking. Additionally, molecular mechanisms for reduced susceptibility to these combinations have not been explored.
KEYWORDS: aztreonam, avibactam, clavulanate, relebactam, vaborbactam, Stenotrophomonas maltophilia, MDR, metallo-β-lactamase, L1, L2, smeABC, metalloenzymes, multidrug resistance
ABSTRACT
The intrinsic L1 metallo- and L2 serine-β-lactamases in Stenotrophomonas maltophilia make it naturally multidrug resistant and difficult to treat. There is a need to identify novel treatment strategies for this pathogen, especially against isolates resistant to first-line agents. Aztreonam in combination with avibactam has demonstrated potential, although data on other aztreonam–β-lactamase inhibitor (BLI) combinations are lacking. Additionally, molecular mechanisms for reduced susceptibility to these combinations have not been explored. The objectives of this study were to evaluate and compare the in vitro activities and to understand the mechanisms of resistance to aztreonam in combination with avibactam, clavulanate, relebactam, and vaborbactam against S. maltophilia. A panel of 47 clinical S. maltophilia strains nonsusceptible to levofloxacin and/or trimethoprim-sulfamethoxazole were tested against each aztreonam-BLI combination via broth microdilution, and 6 isolates were then evaluated in time-kill analyses. Three isolates with various aztreonam-BLI MICs were subjected to whole-genome sequencing and quantitative reverse transcriptase PCR. Avibactam restored aztreonam susceptibility in 98% of aztreonam-resistant isolates, compared to 61, 71, and 15% with clavulanate, relebactam, and vaborbactam, respectively. The addition of avibactam to aztreonam resulted in a ≥2-log10-CFU/ml decrease at 24 h versus aztreonam alone against 5/6 isolates compared to 1/6 with clavulanate, 4/6 with relebactam, and 2/6 with vaborbactam. Molecular analyses revealed that decreased susceptibility to aztreonam-avibactam was associated with increased expression of genes encoding L1 and L2, as well as the efflux pump (smeABC). Aztreonam-avibactam is the most promising BLI-combination against multidrug-resistant S. maltophilia. Decreased susceptibility may be due to the combination of overexpressed β-lactamases and efflux pumps. Further studies evaluating this combination against S. maltophilia are warranted.
INTRODUCTION
Stenotrophomonas maltophilia is an opportunistic pathogen that is difficult to treat due in large part to its predilection for antimicrobial resistance. Among the resistance mechanisms found in S. maltophilia are two intrinsic, inducible β-lactamases, L1 and L2. L1 is an Ambler class B metallo-β-lactamase (MBL) that confers resistance to all β-lactams (including carbapenems and β-lactam/β-lactamase inhibitors [BLIs]), except aztreonam (1). L2 is an Ambler class A β-lactamase capable of hydrolyzing most β-lactams, including extended-spectrum cephalosporins and aztreonam (2, 3). This combination of β-lactamases negates first-line Gram-negative antimicrobials and necessitates the use of potentially less efficacious, more toxic non-β-lactam agents for infections due to S. maltophilia.
Among these non-β-lactam agents, trimethoprim-sulfamethoxazole (TMP-SMZ) has traditionally been regarded as the drug combination of choice for S. maltophilia infections, but increasing reports of resistance along with toxicities and a lack of robust PK/PD data for which to optimize dosing have led clinicians to seek alternate therapies. Levofloxacin and minocycline are often considered suitable alternative agents to TMP-SMZ (4–7), although each is plagued by its own shortcomings, including increasing resistance rates, adverse drug effects, drug-drug interactions, and a dearth of high-quality preclinical or clinical data to support their use against S. maltophilia (8, 9). Therefore, there is a crucial need to identify additional safe, effective agents with reliable activity against S. maltophilia.
Given aztreonam’s ability to evade MBL-mediated hydrolysis by L1, coadministration of it with a β-lactamase inhibitor that inhibits L2 can theoretically prevent aztreonam’s hydrolysis and restore its activity. Previous studies have demonstrated that among the first-generation β-lactamase inhibitors (clavulanate, sulbactam, and tazobactam), only clavulanate exhibits appreciable activity against L2 (10), but the recent development of novel β-lactamase inhibitors (avibactam, relebactam, and vaborbactam) has sparked a renewed interest in evaluating the activity of aztreonam in combination with β-lactamase inhibitors against S. maltophilia. To date, anecdotal clinical data and in vitro susceptibility studies support the activity of the aztreonam-avibactam combination against S. maltophilia (11–14), but more robust analyses, including more strains and comparisons to other novel β-lactamase inhibitors, have not been conducted. Additionally, S. maltophilia strains demonstrate significant molecular heterogeneity, and little is known about the underlying genotypic mechanisms encoding phenotypic resistance, especially against novel β-lactamase inhibitor combinations. As such, the objective of this study was to evaluate and compare the in vitro activities of aztreonam alone and in combination with avibactam, clavulanate, relebactam, and vaborbactam against multidrug-resistant (MDR) S. maltophilia via broth microdilution testing and time-kill analyses and to investigate the molecular basis for differences in phenotypic susceptibility via whole-genome sequencing (WGS) and quantitative reverse transcriptase PCR (qRT-PCR).
(Results of this study were presented in part at the 29th European Congress of Clinical Microbiology and Infectious Diseases in Amsterdam, Netherlands, as abstract no. 6092 [45].)
RESULTS
Susceptibility testing.
The MIC50, MIC90, and MIC range of each agent against all 47 isolates are summarized in Table 1. Only 18/47 (38.3%) and 21/47 (44.7%) isolates were susceptible to levofloxacin and TMP-SMZ, respectively. Although no CLSI interpretive criteria are available for the commercially available β-lactam/β-lactamase inhibitors against S. maltophilia, the MIC50 values for each agent were ≥64 mg/liter and above their respective resistance breakpoints for Enterobacteriaceae (amoxicillin-clavulanate, ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam) and Pseudomonas aeruginosa (ceftazidime-avibactam and imipenem-relebactam). Applying CLSI interpretive criteria for ceftazidime to ceftazidime-avibactam resulted in just 25.5% susceptibility for each. All but one isolate was resistant to aztreonam alone, while susceptibility to aztreonam was restored in 45/46 (97.8%) isolates following the addition of avibactam (4 mg/liter) and in 28/46 (60.8%), 33/46 (71.3%), and 7/46 (15.2%) isolates following the additions of clavulanate (2 mg/liter), relebactam (4 mg/liter), or vaborbactam (8 mg/liter), respectively. Increasing the clavulanate concentration to 4 mg/liter changed the MIC by >1 log2 dilution against only 2 (4.3%) isolates and did not affect the overall percentage susceptible (60.8%). Decreasing the concentration of vaborbactam to 4 mg/liter changed the MIC by >1 log2 dilution against 14 (30.4%) isolates and reduced the overall percentage susceptible to 6.4%.
TABLE 1.
Activity of aztreonam–β-lactamase inhibitor combinations and comparator agents against tested clinical Stenotrophomonas maltophilia isolatesa
| Agent(s) | MIC (mg/liter) |
% susceptible | ||
|---|---|---|---|---|
| 50% | 90% | Range | ||
| Aztreonam | ≥256 | ≥256 | 8 to ≥256 | 2.1 |
| Aztreonam-avibactamb | 4 | 4 | 0.5 to 16 | 97.9 |
| Aztreonam-clavulanatec | 8 | ≥256 | 1 to ≥256 | 61.7 |
| Aztreonam-clavulanated | 4 | 128 | 1 to ≥256 | 61.7 |
| Aztreonam-relebactame | 8 | 16 | 1 to 128 | 72.3 |
| Aztreonam-vaborbactamf | 32 | 128 | 2 to ≥256 | 17.0 |
| Aztreonam-vaborbactamg | 64 | ≥256 | 2 to ≥256 | 6.4 |
| Amoxicillin-clavulanate | ≥256 | ≥256 | 16 to ≥256 | |
| Ceftazidime-avibactamh | 64 | 128 | 0.125 to ≥256 | 25.5 |
| Imipenem-relebactam | ≥64 | ≥64 | 0.5 to ≥64 | |
| Levofloxacin | 8 | ≥32 | 0.25 to ≥32 | 38.3 |
| Meropenem-vaborbactam | ≥64 | ≥64 | 0.25 to ≥64 | |
| Trimethoprim-sulfamethoxazolei | 8 | ≥16 | 0.03 to ≥16 | 44.7 |
n = 47 isolates. Susceptibility interpretations of aztreonam-based regimens were based on CLSI aztreonam interpretive criteria against P. aeruginosa (34).
Avibactam tested at 4 mg/liter.
Clavulanate tested at 2 mg/liter.
Clavulanate tested at 4 mg/liter.
Relebactam tested at 4 mg/liter.
Vaborbactam tested at 8 mg/liter.
Vaborbactam tested at 4 mg/liter.
Susceptibility interpretation based on CLSI ceftazidime interpretative criteria against S. maltophilia (34).
Reflects the MIC of the trimethoprim component only.
The MIC50, MIC90, and MIC range of each agent against all 47 isolates stratified across infection type, acquisition setting, and geographic location are displayed in Table 2. Although isolates obtained from patients with pneumonia, in the hospital setting, and from outside the United States tended to be less susceptible overall, there were no statistically significant differences in the MIC distributions.
TABLE 2.
Activity of aztreonam–β-lactamase inhibitor combinations and comparator agents against tested clinical Stenotrophomonas maltophilia isolates stratified according to infection type, acquisition setting, and geographic locationa
| Parameter and agent(s) | MIC (mg/liter) |
% susceptible | MIC (mg/liter) |
% susceptible | ||||
|---|---|---|---|---|---|---|---|---|
| 50% | 90% | Range | 50% | 90% | Range | |||
| Infection type | Pneumonia (n = 36) | Nonpneumonia (n = 11) | ||||||
| Aztreonam | ≥256 | ≥256 | 64 to ≥256 | 0 | ≥256 | ≥256 | 8 to ≥256 | 9.1 |
| Aztreonam-avibactam | 2 | 4 | 0.5 to 16 | 97.2 | 4 | 4 | 1 to 8 | 100 |
| Aztreonam-clavulanate | 8 | ≥256 | 2 to ≥256 | 61.1 | 4 | ≥256 | 1 to ≥256 | 63.6 |
| Aztreonam-relebactam | 8 | 16 | 1 to 128 | 69.4 | 4 | 16 | 1 to 32 | 81.8 |
| Aztreonam-vaborbactam | 32 | 128 | 4 to ≥256 | 13.9 | 32 | 64 | 2 to ≥256 | 27.3 |
| Amoxicillin-clavulanate | ≥256 | ≥256 | 16 to ≥256 | ≥256 | ≥256 | 64 to ≥256 | ||
| Ceftazidime-avibactamb | 64 | 128 | 0.125 to ≥256 | 22.2 | 32 | 128 | 1 to ≥256 | 36.4 |
| Imipenem-relebactam | ≥64 | ≥64 | 0.5 to ≥64 | ≥64 | ≥64 | ≥64 to ≥64 | ||
| Levofloxacin | 4 | ≥32 | 0.25 to ≥32 | 36.1 | 4 | 16 | 0.5 to ≥32 | 45.5 |
| Meropenem-vaborbactam | ≥64 | ≥64 | 0.25 to ≥64 | ≥64 | ≥64 | 16 to ≥64 | ||
| Trimethoprim-sulfamethoxazolec | 8 | ≥16 | 0.03 to ≥16 | 44.4 | 8 | ≥16 | 0.125 to ≥16 | 45.5 |
| Acquisition setting | Community (n = 21) | Nosocomial (n = 18) | ||||||
| Aztreonam | ≥256 | ≥256 | 8 to ≥256 | 4.8 | ≥256 | ≥256 | 64 to ≥256 | 0 |
| Aztreonam-avibactam | 2 | 8 | 1 to 16 | 95.2 | 2 | 4 | 0.5 to 8 | 100 |
| Aztreonam-clavulanate | 8 | ≥256 | 1 to ≥256 | 61.9 | 8 | ≥256 | 2 to ≥256 | 61.1 |
| Aztreonam-relebactam | 8 | 16 | 1 to 128 | 76.2 | 8 | 32 | 2 to 128 | 72.2 |
| Aztreonam-vaborbactam | 64 | ≥256 | 2 to ≥256 | 23.8 | 32 | ≥256 | 8 to ≥256 | 11.1 |
| Amoxicillin-clavulanate | ≥256 | ≥256 | 16 to ≥256 | ≥256 | ≥256 | ≥256 to ≥256 | ||
| Ceftazidime-avibactamb | 64 | ≥256 | 0.125 to ≥256 | 38.1 | 64 | 128 | 1 to ≥256 | 16.7 |
| Imipenem-relebactam | ≥64 | ≥64 | 0.5 to ≥64 | ≥64 | ≥64 | 32 to ≥64 | ||
| Levofloxacin | 4 | 8 | 0.5 to ≥32 | 47.6 | 8 | ≥32 | 0.25 to ≥32 | 27.8 |
| Meropenem-vaborbactam | ≥64 | ≥64 | 0.25 to ≥64 | ≥64 | ≥64 | 16 to ≥64 | ||
| Trimethoprim-sulfamethoxazolec | 8 | ≥16 | 0.125 to ≥16 | 47.6 | 8 | ≥16 | 0.125 to 8 | 44.4 |
| Location | U.S. (n = 21) | Non-U.S. (n = 26) | ||||||
| Aztreonam | ≥256 | ≥256 | 8 to ≥256 | 4.8 | ≥256 | ≥256 | 64 to ≥256 | 0 |
| Aztreonam-avibactam | 2 | 4 | 0.5 to 8 | 100 | 2 | 8 | 0.5 to 16 | 96.2 |
| Aztreonam-clavulanate | 8 | ≥256 | 1 to ≥256 | 66.7 | 8 | ≥256 | 2 to ≥256 | 57.7 |
| Aztreonam-relebactam | 8 | 16 | 1 to 32 | 66.7 | 8 | 32 | 2 to 128 | 77.0 |
| Aztreonam-vaborbactam | 32 | 128 | 2 to ≥256 | 28.6 | 32 | ≥256 | 8 to ≥256 | 7.7 |
| Amoxicillin-clavulanate | ≥256 | ≥256 | 16 to ≥256 | ≥256 | ≥256 | ≥256 to ≥256 | ||
| Ceftazidime-avibactamb | 64 | 128 | 0.125 to ≥256 | 28.6 | 64 | ≥256 | 1 to ≥256 | 23.1 |
| Imipenem-relebactam | ≥64 | ≥64 | 0.5 to ≥64 | ≥64 | ≥64 | 32 to ≥64 | ||
| Levofloxacin | 4 | ≥32 | 0.25 to ≥32 | 47.6 | 8 | ≥32 | 0.5 to ≥32 | 30.8 |
| Meropenem-vaborbactam | ≥64 | ≥64 | 0.25 to ≥64 | ≥64 | ≥64 | 32 to ≥64 | ||
| Trimethoprim-sulfamethoxazolec | 8 | ≥16 | 0.03 to ≥16 | 42.9 | 8 | ≥16 | 0.25 to ≥16 | 46.2 |
No statistically significant differences in MIC distribution were present for any agent based on infection type, acquisition setting, or location using the Mann-Whitney U test. Susceptibility interpretations of aztreonam-based regimens were based on CLSI aztreonam interpretive criteria against P. aeruginosa (34).
Susceptibility interpretation based on CLSI ceftazidime interpretative criteria against S. maltophilia (34).
Reflects the MIC of the trimethoprim component only.
Time-kill experiments.
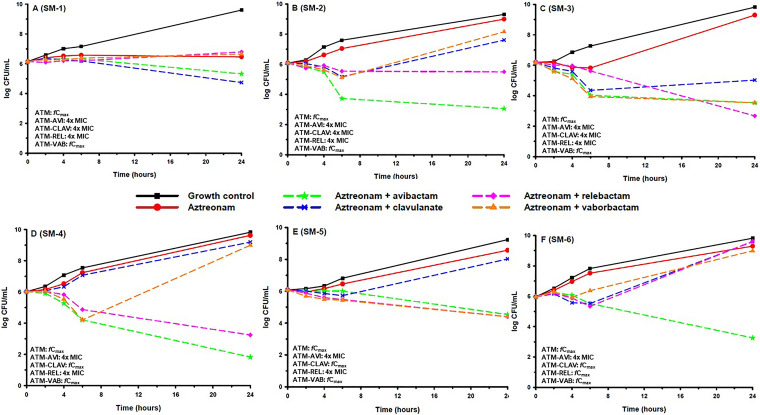
Table 3 displays MIC values of each agent against the 6 aztreonam-resistant isolates (SM-1 to -6) selected for time-kill experiments. The aztreonam-avibactam MICs of these isolates spanned each 2-fold dilution from 0.5 to 16 mg/liter, while MICs of aztreonam-clavulanate, aztreonam-relebactam, and aztreonam-vaborbactam ranged from 2 to >128, 4 to 128, and 8 to >128 mg/liter, respectively. Results from time-kill experiments with aztreonam alone and in combination with each β-lactamase inhibitor at the highest concentration tested are displayed in Fig. 1. Aztreonam alone failed to demonstrate bactericidal activity against any isolate. When combined with avibactam, a ≥2-log10-CFU/ml decrease at 24 h versus aztreonam alone was observed against 5/6 (83.3%) isolates, and bactericidal activity was restored against 3/6 (50%) isolates. None of the combinations were bactericidal against SM-1. Against SM-2 and SM-6 (Fig. 1B and F), aztreonam-avibactam was the only combination to demonstrate bactericidal activity. Aztreonam-clavulanate resulted in a ≥2-log10-CFU/ml decrease at 24 h versus aztreonam alone against 1/6 (16.7%) isolates and was not bactericidal against any (0%) isolate. Aztreonam-relebactam resulted in a ≥2-log10-CFU/ml decrease at 24 h versus aztreonam alone against 4/6 (66.7%) isolates and was bactericidal against 2/6 (33.3%) isolates. Aztreonam-vaborbactam resulted in a ≥2-log10-CFU/ml decrease at 24 h versus aztreonam alone against 2/6 (33.3%) isolates and was not bactericidal against any (0%) isolate.
TABLE 3.
MICs of tested agents against 6 S. maltophilia isolates included in time-kill experimentsa
| Isolate | MIC (mg/liter) ofb
: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ATM | ATM-AVI | ATM-CLAV | ATM-REL | ATM-VAB | AMOX-CLAV | CAZ-AVI | IMI-REL | LFX | MER-VAB | TMP-SMZc | |
| SM-1d | ≥256 | 0.5 | 2 | 4 | 8 | ≥256 | 32 | ≥64 | >16 | 32 | 8 |
| SM-2 | ≥256 | 4 | 8 | 16 | 128 | ≥256 | 64 | ≥64 | 2 | ≥64 | ≥16 |
| SM-3 | ≥256 | 1 | 4 | 4 | 64 | ≥256 | 128 | ≥64 | 16 | ≥64 | 8 |
| SM-4 | ≥256 | 8 | ≥256 | 16 | ≥256 | ≥256 | 64 | ≥64 | 1 | 32 | ≥16 |
| SM-5d | ≥256 | 2 | ≥256 | 4 | 64 | ≥256 | ≥256 | ≥64 | 8 | 32 | 8 |
| SM-6d | ≥256 | 16 | 128 | 128 | ≥256 | ≥256 | ≥256 | ≥64 | 4 | ≥64 | 0.5 |
AVI was tested at 4 mg/liter, CLAV was tested at 2 mg/liter, REL was tested at 4 mg/liter, and VAB was tested at 8 mg/liter.
ATM, aztreonam; AVI, avibactam; CLAV, clavulanate; REL, relebactam; VAB, vaborbactam; AMOX, amoxicillin; CAZ, ceftazidime; IMI, imipenem; LFX, levofloxacin; MER, meropenem; TMP-SMZ, trimethoprim-sulfamethoxazole.
Reflects the MIC of the trimethoprim component.
Subjected to whole-genome sequencing and quantitative reverse transcriptase PCR analysis.
FIG 1.
Mean log10 CFU/ml versus time profile for aztreonam(ATM) alone and in combination with avibactam (AVI), clavulanate (CLAV), relebactam (REL), or vaborbactam (VAB) against six S. maltophilia strains (A to F). Curves represent average concentrations for triplicate experiments.
WGS and analysis.
Multilocus sequence type (MLST) analysis revealed that SM-1 and SM-5 both belonged to sequence type 233 (ST233), whereas isolate SM-6 was assigned to the novel type ST440. All 3 isolates harbored the same resistance genes; however, there were differences in the sequences of those genes. In general, the evaluated resistance genes in isolates SM-1 and SM-5 (aztreonam-avibactam MICs of 0.5 and 2 mg/liter, respectively) showed greater similarity to each other than to SM-6 (aztreonam-avibactam MIC of 16 mg/liter) (Table 3). For blaL1, isolates SM-1, SM-5, and SM-6 showed 86.6, 86.5, and 86.1% sequence identities, respectively, to S. maltophilia 1275 blaL1a. All 3 isolates contained D152N and N169S substitutions in the α3-β7 loop of L1. Isolates SM-1 and SM-5 also had G161D substitutions in the α3-β7 loop and G233Y and P235A substitutions in the β12-α5 loop of L1. Conversely, none of the isolates had substitutions in the L2 active site pocket or the SDN loop or at L103. Isolates SM-1 and SM-5 had L165N, E168D, L169V, S171L, and A173V substitutions in the Ω loop of L2 whereas no Ω loop substitutions in L2 were observed in SM-6.
The smeABC, smeDEF, smeIJK, smeOP, smeR, smeT, smeVWX, and smeYZ genes for isolates SM-1, SM-5, and SM-6 all had ≥92.2% identity to those in the K279a reference strain and did not have any frameshift mutations or premature stop codons. However, isolates SM-1 and SM-5 both had frameshift mutations in smeS that led to a premature stop codon, whereas isolate SM-6 was 99.4% identical to the K279a smeS reference gene and did not possess a frameshift mutation. The lysR, mltD1, ampD, rpoE, and soxR genes in SM-1, SM-5, and SM-6 showed ≥91.6% identity to K279a, and there were no frameshift mutations or premature stop codons identified.
qRT-PCR.
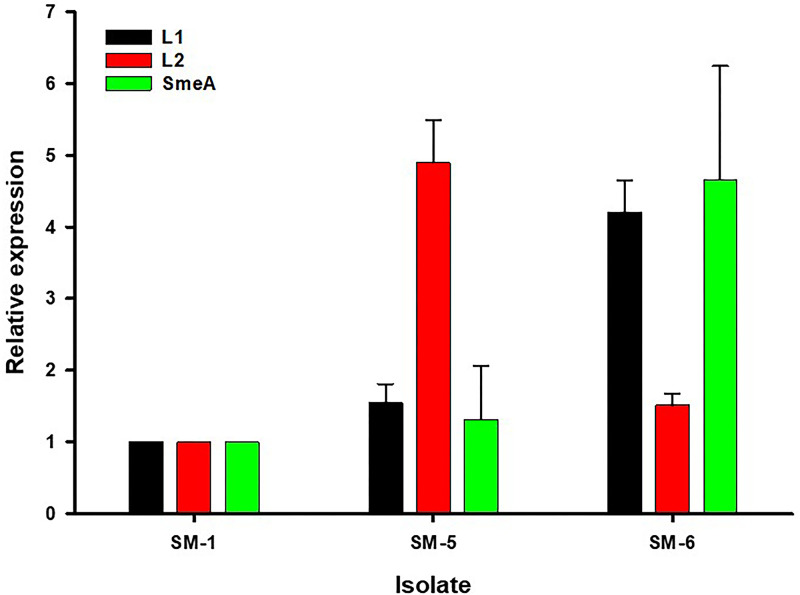
Uninduced qRT-PCR was utilized to examine the transcription levels of the intrinsic L1 and L2 β-lactamase- and SmeA-encoding genes to further elucidate the underlying mechanisms for differences observed in phenotypic susceptibilities between SM-1, SM-5, and SM-6 (Fig. 2). Mean ± standard deviation (SD) expression levels of genes encoding L1, L2, and SmeA, respectively, in SM-5 (aztreonam-avibactam MIC of 2 mg/liter) relative to SM-1 (aztreonam-avibactam MIC of 0.5 mg/liter) were 1.54 ± 0.26 (P = 0.004), 4.89 ± 0.60 (P < 0.001), and 1.31 ± 0.75 (P = 0.352). Mean ± SD expression levels of L1-, L2-, and SmeA-encoding genes, respectively, in SM-6 (aztreonam-avibactam MIC of 16 mg/liter) relative to SM-1 were 4.20 ± 0.45 (P < 0.001), 1.51 ± 0.17 (P = 0.001), and 4.66 ± 1.58 (P = 0.002). Relative to each other, the expression of genes encoding L1 and SmeA in SM-6 was significantly higher than that of SM-5, while expression of the gene encoding L2 was significantly higher in SM-5 than in SM-6.
FIG 2.
Expression of genes encoding L1, L2, and SmeA by isolates SM-1 (aztreonam-avibactam MIC of 0.5 mg/liter), SM-5 (aztreonam-avibactam MIC of 2 mg/liter), and SM-6 (aztreonam-avibactam MIC of 16 mg/liter) determined by qRT-PCR. Expression levels were normalized to 16S rRNA expression via the ΔΔCT method and are displayed relative to SM-1 as the reference. Bars represent mean ± SD values from three independent experiments in triplicate.
DISCUSSION
The recent development of novel β-lactamase inhibitors has helped stave off the postantibiotic era by providing clinicians with safe and effective treatments for infections due to multidrug-resistant Gram-negative pathogens, including carbapenem-resistant strains of Enterobacteriaceae and P. aeruginosa. However, none of the recently approved β-lactam/β-lactamase inhibitors display reliable activity against S. maltophilia due to the lack of activity of all commercially available β-lactamase inhibitors against MBLs, such as the L1 β-lactamase found intrinsically in S. maltophilia.
In the present study, the abilities of avibactam, clavulanate, relebactam, and vaborbactam to reduce the MIC and restore the bactericidal activity of aztreonam were compared head-to-head against 47 clinical S. maltophilia isolates resistant to one or both current first-line treatment options. Results of susceptibility testing demonstrated that avibactam reduced aztreonam MICs to the greatest degree and restored susceptibility in the highest number of isolates, followed by relebactam, clavulanate, and then vaborbactam. Accordingly, in time-kill experiments, aztreonam-avibactam was the most reliably bactericidal combination, while aztreonam-vaborbactam was the least.
Despite the fact that we intentionally enriched our sample with levofloxacin- and/or TMP-SMZ-resistant isolates, our results are consistent with those of Mojica et al., who have demonstrated that the addition of avibactam restores the activity of aztreonam in 82 to 97% of aztreonam-resistant S. maltophilia isolates (12, 14). Clavulanate restored aztreonam activity in fewer isolates than avibactam, which may in part be explained by the propensity for clavulanate, but not avibactam, to induce expression of L1 (1). Although previous studies have demonstrated higher rates of susceptibility to aztreonam-clavulanate than demonstrated in our work, these studies utilized a fixed 2:1 ratio of aztreonam to clavulanate, which necessitates the use of concentrations of clavulanate far above those that can be achieved in vivo even after intravenous (i.v.) dosing (15–19). Analogous to our results, a recent study of the reference K279a strain of S. maltophilia demonstrated that the addition of relebactam decreased the aztreonam MIC from 256 mg/liter to 8 mg/liter, while the addition of avibactam decreased the MIC to 2 mg/liter. This phenotypic change in MIC was reflective of their respective inhibitory concentration (IC50) value against L2, which was more than 30-fold lower for avibactam compared to relebactam (20). To the best of our knowledge, the activity of aztreonam plus relebactam (with or without imipenem) against any other MBL-producing organism has not been previously reported. Aztreonam plus vaborbactam (with or without meropenem) has previously been reported to be bactericidal against aztreonam-resistant MBL-producing Escherichia coli and Klebsiella pneumoniae, but the results of the current study suggest the activity of this combination may be attenuated against S. maltophilia (21, 22), likely secondary to the specificity and affinity of vaborbactam for the Ambler class A serine KPC enzyme (23, 24).
Clinical isolates of S. maltophilia are genetically diverse, and specific mutations in genes encoding L1, L2, and other efflux pumps and two-component regulators that confer phenotypic resistance are poorly understood, especially against modern antimicrobial agents (7, 25). Understanding the molecular mechanisms that confer nonsusceptible phenotypes to promising new therapeutic strategies such as aztreonam–β-lactamase inhibitor combinations is essential to anticipate resistance and guide optimal clinical use. A recent study of 130 clinical S. maltophilia strains demonstrated that they belonged to 90 different STs, only 27 of which were previously known, and displayed numerous novel allelic variations in blaL1 and blaL2 (14). Although several strains in this collection demonstrated MICs of aztreonam/ceftazidime-avibactam of ≥4 mg/liter, the genotypes of these strains were not specifically explored in relation to exquisitely susceptible strains. To our knowledge, our work represents the first attempt to elucidate the molecular mechanisms responsible for reduced susceptibility to aztreonam-avibactam against S. maltophilia. Previous studies have demonstrated that 4-amino-acid insertions in PBP3 are responsible for decreased aztreonam-avibactam susceptibility among strains of E. coli (26, 27). Our WGS analyses of S. maltophilia did not reveal any insertions in PBP3, and the PBP3 sequence of the K279a reference strain and SM-6 differed by only one amino acid (and that amino acid was identical in SM-1 and SM-5). Through WGS we demonstrated differences in STs and numerous substitutions in blaL1 and blaL2 across all 3 strains tested, which may be important for enzyme specificity and may play a role in reduced β-lactam susceptibility (14). Importantly, isolate SM-6 (aztreonam-avibactam MIC of 16 mg/liter) demonstrated an intact smeS, whereas SM-1 and SM-5 both had frameshift mutations leading to premature stop codons in this gene. The two-component regulator smeSR has been shown to upregulate efflux by smeABC and to lead to decreased β-lactam susceptibility (28). Our qRT-PCR results confirm the overexpression of the gene encoding SmeA in SM-6, as predicted by the intact smeS gene found in this isolate. Interestingly, increases in expression of the blaL1 and blaL2 genes were also noted in SM-6 and SM-5, respectively. Though these differences in gene expression are well correlated with the differences in aztreonam-avibactam MICs between the three S. maltophilia isolates, the underlying genetic causes of the gene expression differences remain undefined. Comparison of the blaL2 promoter regions and ampR transcriptional regulator genes in SM-5 and SM-1, which have been shown to cause changes in blaL2 expression, did not reveal any differences in those genes between these two isolates (29). There was no clear mutation in the genes we examined that may explain the elevated expression of the gene encoding L1 in SM-6. Future dedicated studies further examining the specific genotypic-phenotypic relationships among S. maltophilia against aztreonam-avibactam are warranted.
Strengths of our study include the use of a global collection of clinical isolates with resistance to levofloxacin and/or TMP-SMZ, the ability to directly compare the β-lactamase inhibitors by testing them with the same β-lactam agent, and the use of WGS and qRT-PCR to explain differences in phenotypic susceptibility against the most clinically promising combination regimen, aztreonam-avibactam. Limitations of this study include the inherently static nature of time-kill experiments and exclusion of the backbone β-lactams (i.e., amoxicillin, ceftazidime, imipenem, and meropenem) in time-kill experiments. However, previous data generated by our group suggest that the activity of aztreonam plus β-lactam/β-lactamase inhibitor combinations against MBL producers is primarily driven by the interaction between aztreonam and the β-lactamase inhibitor (21, 22). Finally, although confirmation of whether the increased mRNA expression observed via qRT-PCR analysis resulted in increased translation was outside the scope of this work, previous proteomic investigations of L1 and L2 have observed strong correlations between increased gene expression and subsequent protein production leading to phenotypic resistance (30, 31).
In summary, the results of our study suggest that avibactam most reliably restores the activity of aztreonam against MDR S. maltophilia, followed by relebactam, clavulanate, and vaborbactam. Although decreased susceptibility to aztreonam-avibactam remains rare, it may be due in part to the combination of overexpressed intrinsic β-lactamases and efflux pumps. Until the fixed combination of aztreonam-avibactam is available in the clinical arena, aztreonam with ceftazidime-avibactam may be the preferred combination for S. maltophilia infections, especially isolates resistant to levofloxacin and/or TMP-SMZ or for patients who are intolerant. Additional studies evaluating these aztreonam-based combinations in more complex in vitro and in vivo models capable of simulating humanized PK and alternate microbial environments such as biofilms are warranted.
MATERIALS AND METHODS
Bacteria and susceptibility testing.
A panel of 47 clinical S. maltophilia isolates nonsusceptible to levofloxacin and/or TMP-SMZ collected through the SENTRY Antimicrobial Surveillance Program from 2008 to 2018 were included in all experiments (32). Species identification was confirmed at JMI Laboratories (North Liberty, IA) by standard biochemical tests and via matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Billerica, MA). Among those with available data, isolates were acquired in either the community (n = 21) or nosocomial (n = 18) setting and were cultured from patients with the following types of infection: pneumonia (n = 36), bacteremia (n = 4), skin/soft tissue infection (n = 4), urinary tract infection (n = 2), and intra-abdominal infection (n = 1). Isolates were primarily collected from sites in North America (n = 21) and Europe (n = 15), followed by Asia (n = 4), Australia (n = 3), South America (n = 3), and Africa (n = 1). Isolates were maintained at −80°C in cation-adjusted Mueller-Hinton broth (CAMHB) with 20% glycerol and were subcultured twice on tryptic soy agar plates with 5% sheep blood prior to use.
Analytical-grade amoxicillin, avibactam, aztreonam, ceftazidime, clavulanate, imipenem, levofloxacin, meropenem, sulfamethoxazole, trimethoprim (Sigma-Aldrich, St. Louis, MO), relebactam, and vaborbactam (MedChemExpress, Monmouth Junction, NJ) were commercially obtained. Stock solutions of each agent were freshly prepared as single-use aliquots at the beginning of each week and kept frozen at −80°C. MICs were determined in triplicate by reference broth microdilution (BMD) at a standard inoculum according to Clinical and Laboratory Standards Institute (CLSI) guidelines using the same 0.5 McFarland suspension (33). Concentrations of the tested β-lactamase inhibitors with aztreonam were fixed at 4 mg/liter (avibactam and relebactam) and 8 mg/liter (vaborbactam) according to CLSI guidelines (33), while clavulanate was fixed at 2 mg/liter according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (34). Additionally, BMD MICs were performed with all β-lactamase inhibitor concentrations fixed at 4 mg/liter with aztreonam to allow for direct comparison. MIC values are reported as the MIC50, MIC90, and MIC range. Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 1705, and Pseudomonas aeruginosa ATCC 27853 were included as quality control organisms. Susceptibility interpretations were based on CLSI interpretative criteria for S. maltophilia against ceftazidime (±avibactam), levofloxacin, and TMP-SMZ (33). CLSI interpretative criteria against P. aeruginosa were used for aztreonam and aztreonam–β-lactamase inhibitor combinations. MIC distributions and susceptibilities were compared across three strata: infection type (pneumonia versus nonpneumonia), acquisition setting (community versus nosocomial), and geographic isolation (United States versus non-United States) via Mann-Whitney U test. A two-tailed P value of ≤0.05 was considered statistically significant. Statistical analyses were performed using SPSS version 26 (SPSS, Inc., Chicago, IL).
Time-kill experiments.
Time-kill experiments were performed as previously described (35) on a subset of six isolates selected to provide a range of MIC values for aztreonam–β-lactamase inhibitor combinations. Aztreonam was tested alone at a concentration of 112 mg/liter, corresponding to the free maximum concentration (fCmax) in plasma following a 2-g dose (36, 37). In combination experiments, aztreonam was tested at 1/4, 1/2, 1, 2, and 4× the aztreonam–β-lactamase inhibitor MIC, unless any of these concentrations exceeded the fCmax of aztreonam, in which case the fCmax was used. The concentrations of avibactam (4 mg/liter), clavulanate (2 mg/liter), relebactam (4 mg/liter), and vaborbactam (8 mg/liter) were fixed in all experiments.
WGS and analysis.
Three isolates tested in time-kill analyses (SM-1, SM-5, and SM-6) underwent WGS to identify antimicrobial resistance mechanisms associated with the various phenotypic susceptibilities observed against aztreonam-avibactam, as it was the most active of the aztreonam–β-lactamase inhibitor combinations tested. Genomic DNA was extracted using the QIAmp and HT DNA kit (Qiagen, Hilden, Germany), and the library was prepared using the Nextera XT library prep kit for Illumina. Paired-end genome sequencing was performed on an Illumina MiSeq (Illumina, San Diego, CA) 2- by 150-bp configuration (Genewiz, Inc., South Plainfield, NJ). Adapter sequences were trimmed and low-quality bases were removed using BBDuk 37.64. De novo genome assembly was performed using SPAdes 3.10 (38).
Multilocus sequence typing (MLST) was performed by comparing the de novo sequence assembly to sequence in the PubMLST database (https://pubmlst.org/smaltophilia/). Antimicrobial resistance genes were initially identified via BLAST searching the de novo assembly against the ResFinder 3.1 (39) and CARD-RGI (40) databases. Additionally, the sequences were evaluated for the presence of mutations that have previously been shown to confer antibiotic resistance in S. maltophilia by aligning them to reference genes using the Clustal Omega algorithm. The sequences of genes encoding L1 and L2 were compared to S. maltophilia 1275 blaL1a (accession no. X75074) and blaL2a (accession no. Y08562) reference genes (41). The sequences of the efflux pump genes and two-component regulator genes smeABC, smeDEF, smeIJK, smeOP, smeRS, smeT, smeVWX, and smeYZ were compared to those of S. maltophilia reference strain K279a (accession no. AM743169) (42). S. maltophilia K279a also acted as a reference strain for identification of mutations in lysR, mltD1, ampD, rpoE, and soxR.
qRT-PCR.
Cultures from the same 3 isolates subjected to WGS (SM-1, SM-5, and SM-6) were grown to log phase, and total RNA was isolated with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions and treated with RNase-free DNase I (Thermo Fisher Scientific, Waltham, MA) to remove the remaining DNA. First-strand cDNA was generated with 2 μg of total RNA using random primers and a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific, Waltham, MA). Uninduced quantitative real-time PCR was performed three times in triplicate with PowerUP SYBR green master mix (Thermo Fisher Scientific, Waltham, MA) in a StepOnePlus real-time PCR system with StepOne software (v2.0; Applied Biosystems, Foster City, CA), according to the manufacturer’s protocols. The mRNA expression levels of assayed genes (blaL1, blaL2, and smeA) were normalized to endogenous 16S rRNA levels and are reported relative to that of the most susceptible isolate (SM-1). As upregulation of the efflux pump smeABC has been shown specifically to lead to decreased β-lactam susceptibility (28), the expression of the first gene of the operon (smeA) was analyzed as a measure of smeABC expression (28, 43). The primers used for qRT-PCR are listed in Table S1 in the supplemental material. Relative expression was calculated using the threshold cycle (ΔΔCT) method (44) and compared via Student's t test and one-way analysis of variance (ANOVA). A two-tailed P value of ≤0.05 was considered statistically significant. Statistical analyses were performed using SPSS version 26 (SPSS, Inc., Chicago, IL).
Data availability.
This Whole Genome Shotgun project has been deposited in GenBank under BioProject no. PRJNA606341 with accession no. JAAIKL000000000 (SM-1), JAAIKN000000000 (SM-5), and JAAIKM000000000 (SM-6).
Supplementary Material
ACKNOWLEDGMENTS
There was no external financial support for this work. Z.P.B. was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, under grant KL2TR002002. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
E.W. serves on the speaker’s bureau for Melinta Therapeutics, Astellas Pharma, and Allergan Plc and on the advisory board for GenMark Diagnostics and Shionogi. All other authors certify no potential conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Calvopina K, Hinchliffe P, Brem J, Heesom KJ, Johnson S, Cain R, Lohans CT, Fishwick CWG, Schofield CJ, Spencer J, Avison MB. 2017. Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates. Mol Microbiol 106:492–504. doi: 10.1111/mmi.13831. [DOI] [PubMed] [Google Scholar]
- 2.Walsh TR, MacGowan AP, Bennett PM. 1997. Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 41:1460–1464. doi: 10.1128/AAC.41.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartwright SJ, Waley SG. 1984. Purification of beta-lactamases by affinity chromatography on phenylboronic acid-agarose. Biochem J 221:505–512. doi: 10.1042/bj2210505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. 2007. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis 13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gales AC, Jones RN, Forward KR, Linares J, Sader HS, Verhoef J. 2001. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997–1999). Clin Infect Dis 32(Suppl 2):S104–S113. doi: 10.1086/320183. [DOI] [PubMed] [Google Scholar]
- 6.Ko J-H, Kang C-I, Cornejo-Juárez P, Yeh K-M, Wang C-H, Cho SY, Gözel MG, Kim S-H, Hsueh P-R, Sekiya N, Matsumura Y, Lee D-G, Cho S-Y, Shiratori S, Kim Y-J, Chung DR, Peck KR. 2019. Fluoroquinolones versus trimethoprim-sulfamethoxazole for the treatment of Stenotrophomonas maltophilia infections: a systematic review and meta-analysis. Clin Microbiol Infect 25:546–554. doi: 10.1016/j.cmi.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 7.Chang YT, Lin CY, Chen YH, Hsueh PR. 2015. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol 6:893. doi: 10.3389/fmicb.2015.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flamm RK, Castanheira M, Streit JM, Jones RN. 2016. Minocycline activity tested against Acinetobacter baumannii complex, Stenotrophomonas maltophilia, and Burkholderia cepacia species complex isolates from a global surveillance program (2013). Diagn Microbiol Infect Dis 85:352–355. doi: 10.1016/j.diagmicrobio.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Farrell DJ, Sader HS, Jones RN. 2010. Antimicrobial susceptibilities of a worldwide collection of Stenotrophomonas maltophilia isolates tested against tigecycline and agents commonly used for S. maltophilia infections. Antimicrob Agents Chemother 54:2735–2737. doi: 10.1128/AAC.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecso-Bornet M, Bergogne-Berezin E. 1997. Susceptibility of 100 strains of Stenotrophomonas maltophilia to three beta-lactams and five beta-lactam-beta-lactamase inhibitor combinations. J Antimicrob Chemother 40:717–720. doi: 10.1093/jac/40.5.717. [DOI] [PubMed] [Google Scholar]
- 11.Mojica MF, Ouellette CP, Leber A, Becknell MB, Ardura MI, Perez F, Shimamura M, Bonomo RA, Aitken SL, Shelburne SA. 2016. Successful treatment of bloodstream infection due to metallo-beta-lactamase-producing Stenotrophomonas maltophilia in a renal transplant patient. Antimicrob Agents Chemother 60:5130–5134. doi: 10.1128/AAC.00264-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mojica MF, Papp-Wallace KM, Taracila MA, Barnes MD, Rutter JD, Jacobs MR, LiPuma JJ, Walsh TJ, Vila AJ, Bonomo RA. 2017. Avibactam restores the susceptibility of clinical isolates of Stenotrophomonas maltophilia to aztreonam. Antimicrob Agents Chemother 61:e00777-17. doi: 10.1128/AAC.00777-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emeraud C, Escaut L, Boucly A, Fortineau N, Bonnin RA, Naas T, Dortet L. 2019. Aztreonam plus clavulanate, tazobactam, or avibactam for treatment of infections caused by metallo-beta-lactamase-producing Gram-negative bacteria. Antimicrob Agents Chemother 63:e00010-19. doi: 10.1128/AAC.00010-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mojica MF, Rutter JD, Taracila M, Abriata LA, Fouts DE, Papp-Wallace KM, Walsh TJ, LiPuma JJ, Vila AJ, Bonomo RA. 2019. Population structure, molecular epidemiology, and beta-lactamase diversity among Stenotrophomonas maltophilia isolates in the United States. mBio 10:e00405-19. doi: 10.1128/mBio.00405-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Rodriguez JA, Garcia Sanchez JE, Munoz Bellido JL, Garcia GM, Garcia Sanchez E. 1991. Kinetics of antimicrobial activity of aztreonam/clavulanic acid (2:1) against Xanthomonas maltophilia. J Antimicrob Chemother 27:552–554. doi: 10.1093/jac/27.4.552. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Rodriguez JA, Garcia Sanchez JE, Garcia Garcia MI, Garcia Sanchez E, Munoz Bellido JL. 1991. Antibiotic susceptibility profile of Xanthomonas maltophilia. In vitro activity of beta-lactam/beta-lactamase inhibitor combinations. Diagn Microbiol Infect Dis 14:239–243. doi: 10.1016/0732-8893(91)90038-H. [DOI] [PubMed] [Google Scholar]
- 17.Munoz Bellido JL, Munoz Criado S, Garcia Garcia I, Alonso Manzanares MA, Gutierrez Zufiaurre MN, Garcia-Rodriguez JA. 1997. In vitro activities of beta-lactam-beta-lactamase inhibitor combinations against Stenotrophomonas maltophilia: correlation between methods for testing inhibitory activity, time-kill curves, and bactericidal activity. Antimicrob Agents Chemother 41:2612–2615. doi: 10.1128/AAC.41.12.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krueger TS, Clark EA, Nix DE. 2001. In vitro susceptibility of Stenotrophomonas maltophilia to various antimicrobial combinations. Diagn Microbiol Infect Dis 41:71–78. doi: 10.1016/s0732-8893(01)00281-4. [DOI] [PubMed] [Google Scholar]
- 19.GlaxoSmithKline UK. March 2016. Augmentin. Intravenous (amoxicillin-clavulanate). Package insert. GlaxoSmithKline UK, Middlesex, United Kingdom. http://www.accessdata.fda.gov/drugsatfda_docs/label/2014/022110s011lbl.pdf.
- 20.Tooke CL, Hinchliffe P, Lang PA, Mulholland AJ, Brem J, Schofield CJ, Spencer J. 2019. Molecular basis of class A beta-lactamase inhibition by relebactam. Antimicrob Agents Chemother 63:e00564-19. doi: 10.1128/AAC.00564-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biagi M, Wenzler E. 2018. Searching for the optimal treatment regimen for metallo-β-lactamase producing Enterobacteriaceae: aztreonam plus ceftazidime-avibactam vs. aztreonam plus meropenem-vaborbactam, abstr 2443. IDWeek 2018, San Francisco, CA. [DOI] [PMC free article] [PubMed]
- 22.Biagi M, Wu T, Lee M, Wenzler E. 2019. Evaluation of aztreonam-based combination therapies for NDM-producing Enterobacteriaceae: aztreonam plus ceftazidime-avibactam vs. aztreonam plus meropenem-vaborbactam, abstr 6204. ECCMID 2019, Amsterdam, Netherlands.
- 23.Pemberton OA, Tsivkovski R, Totrov M, Lomovskaya O, Chen Y. 2020. Structural basis and binding kinetics of vaborbactam in class A β-lactamase inhibition. Antimicrob Agents Chemother [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomovskaya O, Sun D, Rubio-Aparicio D, Nelson K, Tsivkovski R, Griffith DC, Dudley MN. 2017. Vaborbactam: spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother 61:e01443-17. doi: 10.1128/AAC.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdezate S, Vindel A, Martin-Davila P, Del Saz BS, Baquero F, Canton R. 2004. High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J Clin Microbiol 42:693–699. doi: 10.1128/jcm.42.2.693-699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alm RA, Johnstone MR, Lahiri SD. 2015. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J Antimicrob Chemother 70:1420–1428. doi: 10.1093/jac/dku568. [DOI] [PubMed] [Google Scholar]
- 27.Periasamy H, Joshi P, Palwe S, Shrivastava R, Bhagwat S, Patel M. 2020. High prevalence of Escherichia coli clinical isolates in India harbouring four amino acid inserts in PBP3 adversely impacting activity of aztreonam/avibactam. J Antimicrob Chemother 75:1650–1651. doi: 10.1093/jac/dkaa021. [DOI] [PubMed] [Google Scholar]
- 28.Li XZ, Zhang L, Poole K. 2002. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 46:333–343. doi: 10.1128/aac.46.2.333-343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil-Gil T, Martínez JL, Blanco P. 2020. Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: a review of current knowledge. Expert Rev Anti Infect Ther 18:335–347. doi: 10.1080/14787210.2020.1730178. [DOI] [PubMed] [Google Scholar]
- 30.Van Oudenhove L, De Vriendt K, Van Beeumen J, Mercuri PS, Devreese B. 2012. Differential proteomic analysis of the response of Stenotrophomonas maltophilia to imipenem. Appl Microbiol Biotechnol 95:717–733. doi: 10.1007/s00253-012-4167-0. [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Zou D, Wang X, Li X, Zhu L, Yin Z, Yang Z, Wei X, Han L, Wang Y, Shao C, Wang S, He X, Liu D, Liu F, Wang J, Huang L, Yuan J. 2012. Proteomic analysis of clinical isolate of Stenotrophomonas maltophilia with blaNDM-1, blaL1 and blaL2 β-lactamase genes under imipenem treatment. J Proteome Res 11:4024–4033. doi: 10.1021/pr300062v. [DOI] [PubMed] [Google Scholar]
- 32.Gales AC, Seifert H, Gur D, Castanheira M, Jones RN, Sader HS. 2019. Antimicrobial susceptibility of Acinetobacter calcoaceticus-Acinetobacter baumannii complex and Stenotrophomonas maltophilia clinical isolates: results from the SENTRY Antimicrobial Surveillance Program (1997–2016). Open Forum Infect Dis 6:S34–S46. doi: 10.1093/ofid/ofy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing: twenty-ninth informational supplement M100-S29. CLSI, Wayne, PA. [Google Scholar]
- 34.EUCAST. 2020. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf.
- 35.Biagi M, Wu T, Lee M, Patel S, Butler D, Wenzler E. 2019. Searching for the optimal treatment for metallo- and serine-beta-lactamase producing Enterobacteriaceae: aztreonam in combination with ceftazidime-avibactam or meropenem-vaborbactam. Antimicrob Agents Chemother 63:e01426-19. doi: 10.1128/AAC.01426-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scully BE, Swabb EA, Neu HC. 1983. Pharmacology of aztreonam after intravenous infusion. Antimicrob Agents Chemother 24:18–22. doi: 10.1128/aac.24.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swabb EA, Singhvi SM, Leitz MA, Frantz M, Sugerman A. 1983. Metabolism and pharmacokinetics of aztreonam in healthy subjects. Antimicrob Agents Chemother 24:394–400. doi: 10.1128/aac.24.3.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avison MB, Higgins CS, von Heldreich CJ, Bennett PM, Walsh TR. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:413–419. doi: 10.1128/AAC.45.2.413-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez MB, Martinez JL. 2018. Overexpression of the efflux pumps SmeVWX and SmeDEF is a major cause of resistance to co-trimoxazole in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 62:e00301-18. doi: 10.1128/AAC.00301-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Biagi M, Qasmieh S, Lamm D, Meyer K, Wu T, Wenzler E. 2019. Searching for optimal treatment regimens for Stenotrophomonas maltophilia resistant to levofloxacin and/or sulfamethoxazole-trimethoprim: aztreonam in combination with avibactam or vaborbactam, abstract 6092. ECCMID 2019, Amsterdam, Netherlands. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Whole Genome Shotgun project has been deposited in GenBank under BioProject no. PRJNA606341 with accession no. JAAIKL000000000 (SM-1), JAAIKN000000000 (SM-5), and JAAIKM000000000 (SM-6).