Preclinical animal models of infection are employed to develop new agents but also to screen among molecules to rank them. There are often major differences between human pharmacokinetic (PK) profiles and those developed by animal models of infection, and these may lead to substantial differences in efficacy relative to that seen in humans. Linezolid is a repurposed agent employed to great effect for therapy of Mycobacterium tuberculosis. In this study, we used the hollow-fiber infection model (HFIM) to evaluate the impact of different pharmacokinetic profiles of mice and nonhuman primates (NHP) versus humans on bacterial cell kill as well as resistance suppression.
KEYWORDS: Mycobacterium tuberculosis, animal models, pharmacodynamics, resistance suppression
ABSTRACT
Preclinical animal models of infection are employed to develop new agents but also to screen among molecules to rank them. There are often major differences between human pharmacokinetic (PK) profiles and those developed by animal models of infection, and these may lead to substantial differences in efficacy relative to that seen in humans. Linezolid is a repurposed agent employed to great effect for therapy of Mycobacterium tuberculosis. In this study, we used the hollow-fiber infection model (HFIM) to evaluate the impact of different pharmacokinetic profiles of mice and nonhuman primates (NHP) versus humans on bacterial cell kill as well as resistance suppression. We examined both plasma and epithelial lining fluid (ELF) profiles. We examined simulated exposures equivalent to 600 mg and 900 mg daily of linezolid in humans. For both plasma and ELF exposures, the murine PK profile provided estimates of effect that were biased low relative to human and NHP PK profiles. Mathematical modeling identified a linkage between minimum concentrations (Cmin) and bacterial kill and peak concentrations (Cpeak) and resistance suppression, with the latter being supported by a prospective validation study. Finding new agents with novel mechanisms of action against M. tuberculosis is difficult. It would be a tragedy to discard a new agent because of a biased estimate of effect in a preclinical animal system. The HFIM provides a system to benchmark evaluation of new compounds in preclinical animal model systems against human PK effects (species scale-up estimates of PK), to safeguard against unwarranted rejection of promising new agents.
INTRODUCTION
New drug development for the therapy of Mycobacterium tuberculosis has lagged for multiple decades. Thankfully, we have recently seen the advent of multiple new or repurposed agents to fill this void. New agents include bedaquiline, pretomanid, and delamanid. Repurposed drugs include linezolid and moxifloxacin.
Classically, we have employed preclinical animal models to guide the choice of dose and schedule for such drugs. A real question exists as to how much information can be gleaned from such studies and whether these systems introduce bias regarding the amount of effect the drugs will have when introduced into the clinic.
Murine models are often used to predict the efficacy of antitubercular drugs because these studies are inexpensive to perform relative to human clinical trials. However, the pathology of disease in mice differs from that in humans unless the Kramnik mouse is employed (1). It is believed that tuberculosis (TB) studies in nonhuman primates (NHPs) may be better predictors of treatment outcomes in humans (2). However, few institutions have the capacity to run these trials, and they are relatively expensive to perform. Furthermore, the pharmacokinetics (PK) of drugs are often quite different in preclinical animal models relative to humans, with higher clearances and shorter half-lives.
Previously, we have shown in the hollow-fiber infection model (HFIM) that PK profiles have a significant impact on the bacterial kill and the ability of a regimen to suppress resistance emergence when we evaluated the fluoroquinolone levofloxacin for Bacillus anthracis therapy (3).
In this paper, we explore the impact of PK profile, drug dose, and site of the PK profile on the bacterial kill and ability to suppress resistance emergence using the HFIM. We examined the mouse PK profile, NHP PK profile, and human PK profile with drug exposures equivalent to doses of 600 mg and 900 mg of linezolid once daily. In addition, we created PK profiles for both plasma and epithelial lining fluid (ELF) for these systems. The organism was in log-phase growth.
RESULTS
Bacterial isolates.
The H37Rv strain of Mycobacterium tuberculosis was used for these studies. The MIC for linezolid was determined in triplicate and ranged from 0.5 to 1 mg/liter. The mutational frequency to resistance was determined twice and was 1/7.15 log10 CFU/ml and 1/7.40 log10 CFU/ml.
PK profiles in mice, NHPs, and humans.
Linezolid concentration-time curves were measured in BALB/c mice and cynomolgus macaques (Macaca fascicularis). Human concentration-time profiles were gleaned from the literature (4–6). Profiles of linezolid were developed for plasma and in ELF and are reported in Tables S1 to S4 in the supplemental material.
Protein binding.
All profiles in plasma were corrected for protein binding. ELF profiles were assumed to be unbound. Human and murine protein binding values were taken from the literature (7, 8) and were 31% and 30%, respectively. For cynomolgus macaques, protein binding was determined with linezolid concentrations ranging from 0.25 to 4.0 mg/liter in 2-fold steps. Measured binding ranged from 15.7% to 19.0%, with a mean of 16.9% and a median of 16.6%.
Antibacterial activity and resistance emergence in the HFIM.
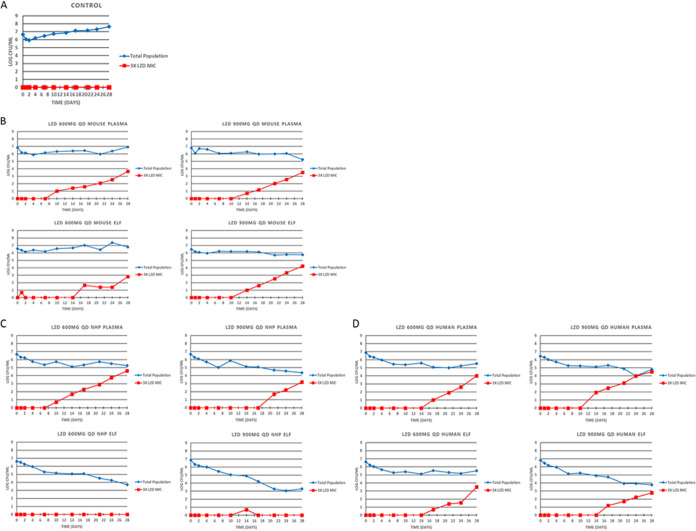
The quantitative cultures results from all the systems, doses and compartments are displayed in Fig. 1. All plasma profiles allowed resistance amplification whether at 600 mg daily or 900 mg daily, as we have demonstrated previously (9, 10). This was not seen in the NHP ELF profiles for either 600 mg daily or 900 mg daily. Our pharmacokinetic evaluations of linezolid in ELF in the NHP were surprising, in that the penetration into ELF was much greater in NHPs than in either mice or humans. The point estimate of penetration was 541% (area under the concentration-time curve for ELF [AUCELF]/AUCplasma) (see the supplemental materials). This exposure was sufficient to suppress resistance with linezolid monotherapy for a full 28 days.
FIG 1.
Impact of the differing linezolid (LZD) pharmacokinetics seen in mice, nonhuman primates (NHPs), and humans on the kill of total M. tuberculosis burden and the M. tuberculosis subpopulation with MICs that were >3× the MIC of the parent strain at simulated doses of 600 and 900 mg daily. Free drug profiles for each were simulated for plasma. For epithelial lining fluid (ELF), all drug concentrations were considered to be free drug. (A) No-treatment control; (B) murine system; (C) NHP system; (D) humans.
Modeling of drug concentrations, total M. tuberculosis burden, and lower linezolid-susceptible bacterial burden simultaneously.
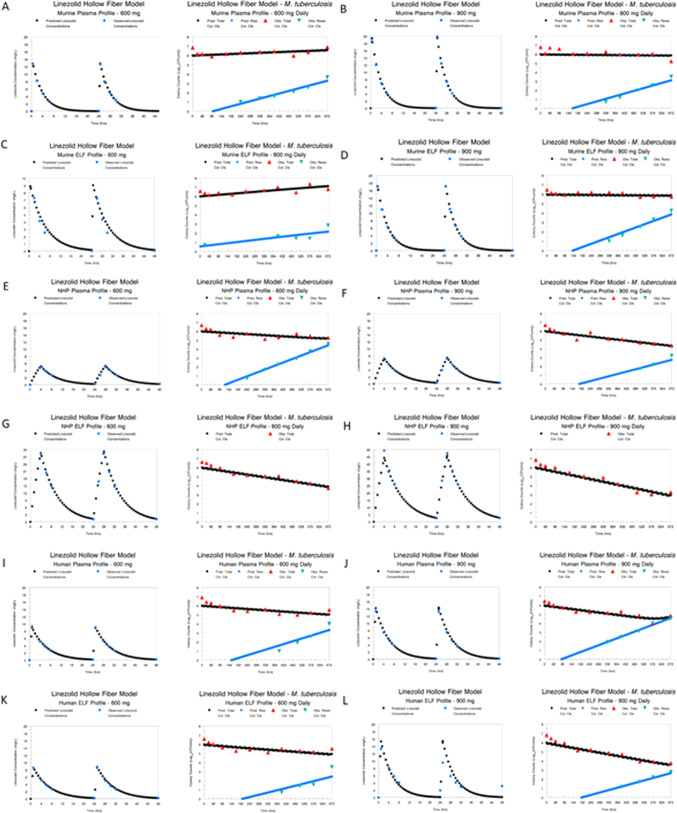
A mathematical model was fit to all the data simultaneously. Modeling was performed for the different species and different concentration-time profiles for the 600- and 900-mg daily doses independently (i.e., six different analyses: species and profile for the 600-mg and 900-mg doses). The model fits to the data are displayed for both pre-Bayesian (population) and Bayesian (individual) estimations along with measures of bias and precision in Table S5. The fit of the model to the data was acceptable for all species and profiles.
We show the Bayesian posterior predicted-observed plots by species and profile in Fig. 2. It is evident that in all cases with the exception of the NHP ELF profiles for 600 mg and 900 mg daily, there was resistance emergence. For the NHP ELF profile, there was sufficient linezolid exposure (AUC 5.41 times the plasma AUC) to counterselect amplification of less susceptible subpopulations.
FIG 2.
Modeled pharmacokinetic profile (milligrams/liter) and effect on total bacterial burden (log10 CFU/milliliter) and the effect on resistance amplification with 600- and 900-mg simulated doses of linezolid daily in a hollow-fiber infection model. (A) Murine plasma profile for 600 mg daily of linezolid and effect profile; (B) murine plasma profile for 900 mg daily of linezolid and effect profile; (C) murine ELF profile for 600 mg daily of linezolid and effect profile; (D) murine ELF profile for 900 mg daily of linezolid and effect profile; (E) NHP plasma profile for 600 mg daily of linezolid and effect profile; (F) NHP plasma profile for 900 mg daily of linezolid and effect profile; (G) NHP ELF profile for 600 mg daily of linezolid and effect profile; (H) NHP ELF profile for 900 mg daily of linezolid and effect profile; (I) human plasma profile for 600 mg daily of linezolid and effect profile; (J) human plasma profile for 900 mg daily of linezolid and effect profile; (K) human ELF profile for 600 mg daily of linezolid and effect profile; (L) human ELF profile for 900 mg daily of linezolid and effect profile.
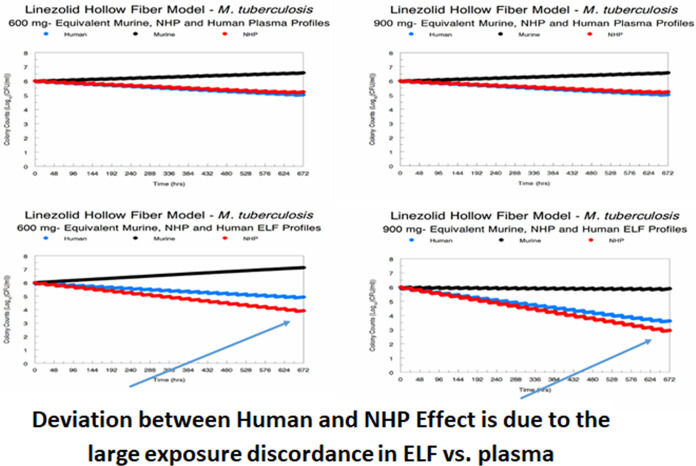
The amplification of resistant subpopulations confounds the delineation of the effect of linezolid regimens simulated in different animal species on the susceptible population. We simulated the impact of the different species and regimen profiles on the susceptible population for all three species for each dose and profile (Fig. 3). It is evident that the NHP plasma profile most closely recapitulates the human plasma effect profile and that the murine plasma effect profile is always biased to less effect. It is also plain that the NHP ELF profile is somewhat discordant from the human ELF profile, with there being greater effect in the NHP. It should be noted that this evaluation is for the susceptible population only. The discordance would have been greater if the effect on the total population, which includes the resistant population, had been evaluated (Fig. 2).There is a greater discordance for the 600-mg dose than for the 900-mg dose, which is counterintuitive. Part of the explanation is that there is a maximal kill rate (Kkill-max) that can be engendered by linezolid. The very large difference in ELF exposure relative to plasma exposure means that the 900-mg daily dose gets reasonably close to this value, whereas the 600-mg dose generates an exposure that starts off circa 33% lower, explaining the discordance. The AUC and minimum concentration (Cmin) values along with the day 28 cell kill for each species, dose, and profile are displayed in Table S6.
FIG 3.
Comparison of the effects on kill of the susceptible subpopulation of different preclinical systems and different daily doses of linezolid.
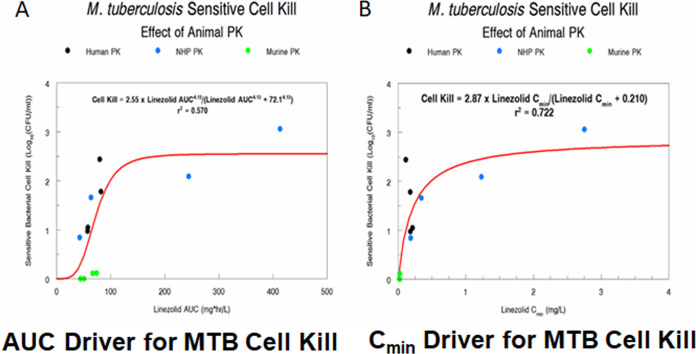
To show this graphically, we performed a sigmoid-Emax analysis with drug exposure (either AUC [Fig. 4A] or Cmin [Fig. 4B]) as the independent variable and susceptible M. tuberculosis population as the dependent variable. Net growth was accorded a bacterial kill of zero. The r2 for AUC was 0.570, and that for Cmin was 0.722. Most of the difference in the amount of variance explained is from the murine data, where the Cmin was small, but where the AUC was similar to that for humans, relative to dose. We conclude that for monotherapy with linezolid therapy, Cmin is the dynamic driver most closely linked to M. tuberculosis bacterial kill, as was shown previously (9).
FIG 4.
Sigmoid-Emax model relating exposure measures (AUC-Panel A and Cmin-Panel B) to susceptible population M. tuberculosis cell kill.
Prospective resistance suppression experiment.
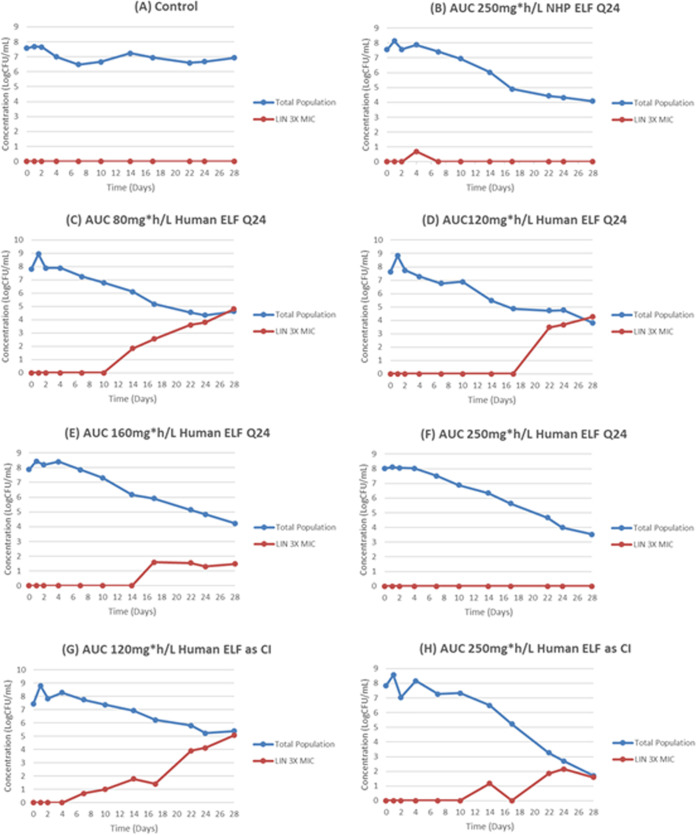
We wished to test the predictive power of the mathematical model. We performed a prospective validation experiment in the HFIM. Eight linezolid exposures were examined. Since we had previously demonstrated (Table S6) that an NHP profile exposure to linezolid of an AUC of 244 mg·h/liter suppressed resistance for 28 days, we started with this value. We then examined human profile exposures of 80, 120, 160, and 250 mg·h/liter of daily linezolid. Our mathematical model predicted that an exposure near 250 mg·h/liter would be required to suppress resistance. To examine the impact of drug profile with human pharmacokinetics, we also examined continuous-infusion exposures of linezolid producing AUC values of 120 and 250 mg·h/liter. The results are displayed in Fig. 5. The actual achieved linezolid exposures are displayed in Table S7. The NHP profile of 250 mg·h/liter suppressed resistance as it had previously. Human profiles of linezolid (ELF) of 80, 120, and 160 mg·h/liter administered daily allowed resistance emergence. The prospectively predicted exposure of 250 mg·h/liter (achieved 237 mg·h/liter) did suppress resistance for 28 days. Administering the exposure as a continuous infusion (120 and 250 mg·h/liter) allowed resistance emergence, although in the latter case it occurred after day 17. As the actual value achieved by continuous infusion was 244 mg·h/liter; this strongly implies that peak concentrations (Cpeak) are linked to resistance suppression in M. tuberculosis for linezolid.
FIG 5.
Prospective validation experiment to document the linezolid exposure required to suppress amplification of less susceptible populations. There was a no-treatment control (A), an AUC employing an NHP profile of 250 mg·h/liter (B), and human exposure profiles of 80, 120, 160, and 250 mg·h/liter administered as a daily dose (C to F); to examine the impact of human exposure profile, we generated AUCs of 120 and 250 mg·h/liter as a continuous infusion (G and H).
DISCUSSION
To rapidly develop new agents with activity against M. tuberculosis, it is critical to understand the relationship between dose/exposure and dosing frequency with bacterial cell kill, as well as suppression of amplification of less susceptible subpopulations. Preclinical systems, classically animal model systems, have been employed to elucidate such relationships. Recently, the in vitro hollow-fiber infection model (HFIM) has been employed to generate such relationships (11–16) and has been qualified as a development tool for M. tuberculosis by the European Medicines Agency (17).
We wished to evaluate the predictions of animal model systems for M. tuberculosis cell kill relative to that seen with a human pharmacokinetic profile. While there are multiple animal model systems to test M. tuberculosis therapeutics, the most common are mouse-based systems. There are multiple murine models, but we chose to evaluate the BALB/c murine model, as it is the workhorse. There are other murine models, such as the Kramnik mouse model (which uses C3HeB/FeJ mice [1]), but this differs mainly by the pathology produced. The nonhuman primate (NHP) model is often considered to be the gold standard preclinical model, as it most closely resembles humans in terms of pathology as well as TB pathogenesis. It is, however, expensive and, consequently, many experiments have small numbers of subjects.
In this experiment, we generated once-daily concentration-time profiles for the drug linezolid for plasma and ELF for humans, NHPs, and mice. The overall aim was to identify the information that was derived from the PK profiles in the murine and NHP systems, relative to the information on bacterial kill and resistance emergence seen with human PK.
Comparing the outcomes from each species for the plasma profiles, it is clear from Fig. 3 that the murine system is biased to demonstrating less effect for linezolid relative to humans, while the NHP is highly concordant. The reason is clear. We have previously demonstrated that the Cmin (trough) linezolid concentration is linked to bacterial cell kill (9) when linezolid is employed as a single agent. This is confirmed again here by our data (Fig. 4). When one examines mice versus humans for the plasma profile after once-daily dosing, the linezolid Cmin in humans are 6.9 to 7.9 times that seen in mice, even though AUC values are comparable (Table S6). The difference is driven by the clearance and half-life in mice versus humans (murine half-life = approximately 2.0 to 2.4 h, versus human half-life of 5 to 7 h). The NHP profile is quite different, with the Cmin being 0.98 to 0.52 times that seen in humans (Table S6).
The ELF profiles show differences as well. The contrast of mouse and human Cmin values shows that the human/mouse ratios were 12.5 and 5.0 for the 600- and 900-mg exposures. The profiles in ELF were substantially larger for NHPs than for humans, with Cmin ratios (human/NHP) of 0.17 and 0.041. This is due to the very large ratio of AUC values in NHPs relative to that in humans, in excess of 5.0.
A central question is whether these results are concordant with those of previous studies. Our findings demonstrated that the plasma and ELF profiles in the mouse for both 600 mg daily and 900 mg daily drove very modest effects over 28 days, maximizing at 0.115 log10 CFU/ml. In a study by Williams et al. (18), a daily dose of 150 mg/kg of body weight drove approximately a 1-log10 CFU/ml bacterial load reduction. With the pharmacokinetic profile reported in that publication, this dose would generate an AUC of 164 mg·h/liter. Our 900-mg plasma profile exposure in the mouse was 74 mg·h/liter (the largest for any murine profile). This would correlate to a daily dose of 59 mg/kg. In that study (18), the 50-mg/kg dose drove a bacterial kill of approximately 0.1 log10 CFU/ml. We may conclude that the HFIM produced results concordant with prior linezolid effect in this murine model system.
The best NHP data come from Coleman et al. (2). These authors looked at several metrics of linezolid activity in a cynomolgus macaque model. In all metrics (positron emission tomography-computed tomography [PET-CT] analysis, percentage of lesions without recoverable organisms, and reduction of lesion colony counts), linezolid demonstrated activity that was significantly better than with the no-treatment control. Consequently, the HFIM once again produced results concordant with those seen in prior evaluations.
In a human early bactericidal activity (EBA) evaluation (19), 600 mg of linezolid daily produced modest activity at day 7. Our HFIM activity at day 7 (prior to resistance emergence) for 600 mg daily was a 1.19-log10 CFU/ml bacterial load decline. In the EBA study, 600 mg of linezolid produced an average decline of approximately of 0.8 log10 CFU/ml in 10 patients over 7 days (Fig. 1 of reference 19). Again, the HFIM results were quite concordant with prior information, even for humans.
Resistance emergence confounds the evaluation of linezolid activity in the HFIM. Consequently, we calculated the activity of linezolid exposure on only the susceptible bacterial population. However, understanding the relationship between drug exposure and resistance emergence is a highly important issue. Because of the high concentrations of linezolid in the ELF in NHPs, resistance emergence was suppressed. Due to the large exposures, the likelihood of attaining these exposures without substantial toxicity is small. Nonetheless, we wished to examine the predictions of the mathematical model, and we calculated the exposure that would be likely to suppress resistance with a human pharmacokinetic profile. We generated exposures in the HFIM that ranged from 80 to 250 mg·h/liter as a daily administration with human PK parameters (and hence ELF profile). We also generated a 250-mg·h/liter exposure with an NHP profile daily, as this had completely suppressed resistance in the original experiment. Finally, we generated exposures of 120 and 250 mg·h/liter as a continuous infusion. Only the AUC ELF exposures of 250 mg·h/liter by both human and ELF profiles as daily administrations suppressed resistance. The continuous infusion of this exposure failed. These findings demonstrate the power of the mathematical modeling, as it correctly predicted prospectively an exposure that would suppress resistance emergence. It also strongly suggests that Cpeak is most closely linked to resistance suppression for linezolid for M. tuberculosis, in contradistinction to cell kill, for which Cmin is most closely linked.
In summary, we demonstrated that the difference in drug profiles generated by different pharmacokinetic parameter values in different species has a substantial impact on M. tuberculosis bacterial kill. At least for linezolid, the shorter half-life in the murine system leads to a bias in the observed degree of activity when the drug is dosed once daily. Surprisingly, when we examined the PK profile in the NHP, we found linezolid ELF exposures far in excess of the plasma exposures. The lesson here is that preclinical models need to be very carefully investigated for pharmacokinetics and the results interpreted in light of pharmacokinetic differences between humans and the preclinical animal species. To abandon a new agent because of relatively unimpressive antibacterial activity would be a horrible loss, given the magnitude of the problem with tuberculosis. It also points up the possibility to perform evaluations in the hollow-fiber infection model. This is particularly true for repurposed agents like linezolid. If human PK is not available, the experiments may still be performed. There could be two approaches. In the first, species scale-up values approximating what may be seen in humans could be employed. In the second approach, a range of empirical values for humans could be employed. We have previously demonstrated in viral systems that there are breakpoint values of PK parameters where the pharmacodynamic driver switches, so that failure can result in subpopulations of patients (20, 21). These approaches can be employed in the HFIM to safeguard against the loss of a potentially valuable new agent.
We should not throw out the murine system evaluations. Rather, we should attempt to improve the predictiveness of the system. This can be done straightforwardly. First, many laboratories only dose the mouse 5 days of the week (22–25). We have previously shown in the HFIM that this has an adverse effect (16). Changing to dosing 7 days per week is highly likely to improve the ability of murine systems to predict outcomes.
The problem shown here is that the murine PK profile can match AUC with humans, but when the pharmacodynamic driver differs from AUC, as it does with linezolid (Cmin), the faster clearance and shorter half-life in the mouse result in lower values for the pharmacodynamic index most closely linked to cell kill. To remedy this, it is critical to know the pharmacodynamic index linked to cell kill (and resistance suppression). Any algorithm for new or repurposed drug evaluation should include elucidation of the pharmacodynamic driver early on. Knowing this, the drug dosing can then be humanized. We have shown this previously (3, 26) both in the HFIM and in the murine system for Pseudomonas aeruginosa therapy. Performing animal model dose humanization for 1 to 2 days versus months of therapy is vastly different in the amount of resources required. It is likely that the murine humanization experiment could first be employed in the HFIM, and then, if desired, a partial humanization of PK profile could be attempted in the mouse. In this way, we can generate the most accurate information from our preclinical systems.
MATERIALS AND METHODS
Bacteria.
M. tuberculosis strain H37Rv (ATCC 27294) was used. Stocks of the bacteria were stored at −80°C. For each experiment, an aliquot of the bacterial stock was inoculated into filter-capped T flasks containing 7H9 Middlebrook broth that was supplemented with 0.05% Tween 80 and 10% albumin, dextrose, and catalase (ADC). The culture was incubated at 37°C and 5% CO2 on a rocker platform for 4 to 5 days to achieve log-phase growth.
Drug.
Pharmaceutical-grade linezolid was purchased from CuraScript (Orlando, FL) as a solution for injection and stored at room temperature in the dark. Prior to experimental use, linezolid was diluted to the desired concentrations in sterile deionized water.
Susceptibility testing and mutation frequency determination.
Susceptibility studies for linezolid were conducted with log-growth-phase H37Rv M. tuberculosis using the agar proportional method described by the CLSI (27) and the absolute serial dilution method on 7H10 agar–10% oleic acid, albumin, dextrose, and catalase (OADC). Briefly, a 104-CFU volume of H37Rv in log-phase growth was plated on Middlebrook 7H10 agar (Becton, Dickinson Microbiology Systems, Sparks, MD) supplemented with 10% OADC (Becton, Dickinson Microbiology Systems) containing 2-fold dilutions of linezolid. The cultures were incubated at 37°C and 5% CO2. After 4 weeks of incubation, the MICs were determined by identifying the lowest drug concentration at which there was no bacterial growth on the agar plate. For the agar proportional method, the lowest concentration of a drug that provided a 99% reduction in the bacterial density relative to the no-drug control was read as the MIC. For the absolute serial dilution method, the MIC was read as the lowest concentration of drug for which there was no growth on the agar plate.
The mutation frequency of the H37Rv strain was evaluated using methods that are described elsewhere (9). Briefly, H37Rv cultures in log-phase growth were inoculated onto plates containing Middlebrook 7H10 agar plus 10% OADC with linezolid at a concentration equivalent to 3.0 times the MIC. The mutation frequency was identified after 4 weeks of incubation at 37°C and 5% CO2.
HFIM experiments.
There were six sets of hollow-fiber infection model (HFIM) experiments. The first two were for a human pharmacokinetic profile in plasma and epithelial lining fluid (ELF) for both the 600-mg and 900-mg daily doses. The human profiles were taken from the literature (4–6) for both plasma and ELF. The second two employed a murine pharmacokinetic profile for plasma and ELF for the same doses. The profiles were measured in our laboratory (see below and Tables S1 and S2). The third two employed a nonhuman primate pharmacokinetic profile for plasma and ELF for the same doses. The profiles were measured in our laboratory (see below and Tables S3 and S4).
Sampling of the central compartment of the HFIM took place 7 or 8 times over the 28 days of the experiment. The samples were washed and then quantitatively plated onto antibiotic-free agar and antibiotic-supplemented agar to characterize the effect of each treatment regimen on the total bacterial and less susceptible bacterial populations. A volume of 200 μl was removed from the peripheral compartment and was streaked onto the zero-dilution plate. Another 100-μl volume was used to perform serial 10-fold dilutions to obtain accurate countable numbers. This was done both for antibiotic-free plates (total bacterial burden) and antibiotic-containing plates (less linezolid-susceptible organisms). For detection of mutants less susceptible to linezolid, the agar (pH 7.0) was supplemented with 10% OADC. The agar plates were read after 6 weeks of incubation at 37°C in a 5% CO2 atmosphere. Linezolid concentrations incorporated into the agar were 3.0× baseline MIC value.
Achievement of target exposure profiles.
Serial samples of media were collected 7 or 8 times from the HFIM treatment arms for assay of drug content by ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) to confirm that the targeted concentration-time profiles were achieved. The assay was previously described (9).
Plasma and bronchoalveolar lavage (BAL) fluid linezolid concentrations from murine and NHP studies were analyzed by a similar assay. Urea in mouse and NHP plasma and BAL fluid was assayed to calculate the ELF values, accounting for dilution.
The collected PK samples were stored in a –80°C freezer until quantification at the University of Florida (UF) Infectious Disease Pharmacokinetics Laboratory (IDPL). Drug concentrations were measured using validated LC-MS/MS assays on a Dionex UltiMate 3000 RS pump and a Dionex UltiMate 3000 RS autosampler (Thermo Scientific), column compartment, a TSQ Endura LC-MS/MS system, a Dell Dimension computer, and Xcalibur 2.2 SP1.48 analytical software (Thermo Scientific). The lower limit of quantification was 0.3 μg/ml. The recovery from 7H9 broth was 100%. The overall interbatch precision for quality control samples ranged from 1.34 to 3.57% for linezolid. Assays performed in human plasma, mouse plasma, 7H9, and other matrices showed no matrix effect: calibration curves could be overlaid graphically.
The urea assay has been previously described (26).
Plasma-free drug concentration was determined by ultrafiltration. Linezolid standards in saline were employed to determine nonspecific binding to the filters.
Population pharmacokinetic/pharmacodynamic mathematical model.
To determine the linezolid profile in plasma and ELF for humans, we employed two published data sets (4–6). These papers provided raw data that allowed us to model plasma and ELF data simultaneously in a population sense. For the hollow-fiber infection model data, we simultaneously modeled 3 system outputs for the analysis of the M. tuberculosis data. The system outputs were as follows: concentration of linezolid, total M. tuberculosis burden, and burden of M. tuberculosis that was less susceptible to linezolid. Population modeling was performed employing the nonparametric adaptive grid (NPAG) program of Leary et al. and Neely et al. (28, 29). Modeling choices (weighting, etc.) and goodness-of-fit evaluations were performed as previously published (9). The analyses were done for separate data sets (by species and by site [plasma versus ELF]), but each analysis included a no-treatment control and two doses of linezolid (600 mg and 900 mg) daily. Simulation was performed using the ADAPT V program of D’Argenio et al. (30) using Bayesian posterior parameter estimates.
Sigmoid-Emax effect modeling.
Identification of the parameter values was done employing the ADAPT V program of D’Argenio et al. (30). The maximum likelihood estimator was chosen for the analysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant P01AIO123036 from the NIAID.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conceptualization, G.L.D. and A.L.; methodology, G.L.D., B.D., C.A.S., S.S., S.K., M.N.N., W.M.Y., C.A.P., M.V., and A.L.; validation, G.L.D.; formal analysis, G.L.D., S.K., S.S., M.N.N., and W.M.Y.; investigation, B.D., C.A.S., C.A.P., M.V., and A.L.; writing—original data draft, G.L.D.; writing—review and editing, G.L.D., B.D., C.A.S., S.K., S.S., M.N.N., W.M.Y., M.V., C.A.P., and A.L.; supervision, G.L.D., A.L., C.A.S., S.S., M.N.N., and C.A.P.; and funding acquisition, G.L.D. and A.L.
There are no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kramnik I, Dietrich WF, Demant P, Bloom BR. 2000. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 97:8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman MT, Chen RY, Lee M, Lin PL, Dodd LE, Maiello P, Via LE, Kim Y, Marriner G, Dartois V, Scanga C, Janssen C, Wang J, Klein E, Cho SN, Barry CE III, Flynn JL. 2014. PET/CT imaging reveals a therapeutic response to oxazolidinones in macaques and humans with tuberculosis. Sci Transl Med 6:265ra167. doi: 10.1126/scitranslmed.3009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deziel MR, Heine H, Louie A, Kao M, Byrne WR, Basset J, Miller L, Bush K, Kelly M, Drusano GL. 2005. Identification of effective antimicrobial regimens for use in humans for the therapy of Bacillus anthracis infections and post-exposure prophylaxis. Antimicrob Agents Chemother 49:5099–5106. doi: 10.1128/AAC.49.12.5099-5106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boselli E, Breilh D, Caillault-Sergent A, Djabarouti S, Guillaume C, Xuereb F, Bouvet L, Rimmelé T, Saux MC, Allaouchiche B. 2012. Alveolar diffusion and pharmacokinetics of linezolid administered in continuous infusion to critically ill patients with ventilator-associated pneumonia. J Antimicrob Chemother 67:1207–1210. doi: 10.1093/jac/dks022. [DOI] [PubMed] [Google Scholar]
- 5.McGee B, Dietze R, Hadad DJ, Molino LP, Maciel EL, Boom WH, Palaci M, Johnson JL, Peloquin CA. 2009. Population pharmacokinetics of linezolid in adults with pulmonary tuberculosis. Antimicrob Agents Chemother 53:3981–3984. doi: 10.1128/AAC.01378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honeybourne D, Tobin C, Jevons G, Andrews J, Wise R. 2003. Intrapulmonary penetration of linezolid. J Antimicrob Chemother 51:1431–1434. doi: 10.1093/jac/dkg262. [DOI] [PubMed] [Google Scholar]
- 7.Slatter JG, Stalker DJ, Feenstra KL, Welshman IR, Bruss JB, Sams JP, Johnson MG, Sanders PE, Hauer MJ, Fagerness PE, Stryd RP, Peng GW, Shobe EM. 2001. Pharmacokinetics, metabolism and excretion of linezolid following an oral dose of [14C]linezolid to healthy human subjects. Drug Metab Dispos 29:1136–1145. [PubMed] [Google Scholar]
- 8.Andes D, van Ogtrop ML, Peng J, Craig WA. 2002. In vivo pharmacodynamics of a new oxazolidinone (linezolid). Antimicrob Agents Chemother 46:3484–3489. doi: 10.1128/aac.46.11.3484-3489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown AN, Drusano GL, Adams JR, Rodriquez JL, Jambunathan K, Baluya DL, Brown DL, Kwara A, Mirsalis JC, Hafner R, Louie A. 2015. Preclinical evaluations to identify optimal linezolid regimens for tuberculosis therapy. mBio 6:e01741-15. doi: 10.1128/mBio.01741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drusano GL, Neely M, Van Guilder M, Schumitzky A, Brown D, Fikes S, Peloquin C, Louie A. 2014. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS One 9:e101311. doi: 10.1371/journal.pone.0101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gumbo T, Louie A, Deziel MR, Parsons LM, Salfinger M, Drusano GL. 2004. Selection of a moxifloxacin dose that suppresses Mycobacterium tuberculosis resistance using an in vitro pharmacodynamic infection model and mathematical modeling. J Infect Dis 190:1642–1651. doi: 10.1086/424849. [DOI] [PubMed] [Google Scholar]
- 12.Gumbo T, Louie A, Liu W, Ambrose PG, Bhavnani SM, Brown D, Drusano GL. 2007. Isoniazid’s bactericidal activity ceases because of the emergence of resistance, not depletion of Mycobacterium tuberculosis in the log phase of growth. J Infect Dis 195:194–201. doi: 10.1086/510247. [DOI] [PubMed] [Google Scholar]
- 13.Gumbo T, Louie A, Liu W, Brown D, Ambrose PG, Bhavnani SM, Drusano GL. 2007. Isoniazid bactericidal activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics to predict efficacy in different ethnic populations. Antimicrob Agents Chemother 51:2329–2336. doi: 10.1128/AAC.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumbo T, Dona CS, Meek C, Leff R. 2009. Pharmacokinetics-pharmacodynamics of pyrazinamide in a novel in vitro model of tuberculosis for sterilizing effect: a paradigm for faster assessment of new antituberculosis drugs. Antimicrob Agents Chemother 53:3197–3204. doi: 10.1128/AAC.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drusano GL, Sgambati N, Eichas A, Brown DL, Kulawy R, Louie A. 2010. The combination of rifampin plus moxifloxacin is synergistic for resistance suppression, but is antagonistic for cell kill for Mycobacterium tuberculosis as determined in a hollow fiber infection model. mBio 1:e00139-10. doi: 10.1128/mBio.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drusano GL, Sgambati N, Eichas A, Brown D, Kulawy R, Louie A. 2011. Effect of administering moxifloxacin plus rifampin against Mycobacterium tuberculosis 7 of 7 days versus 5 of 7 days in an in vitro pharmacodynamic system. mBio 2:e00108-11. doi: 10.1128/mBio.00108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavaleri M, Manolis E. 2015. Hollow fiber system model for tuberculosis: the European Medicines Agency experience. Clin Infect Dis 61:S1–S4. doi: 10.1093/cid/civ484. [DOI] [PubMed] [Google Scholar]
- 18.Williams KN, Stover CK, Zhu T, Tasneen R, Tyagi S, Grosset JH, Nuermberger E. 2009. Promising antituberculosis activity of the oxazolidinone PNU 100480 relative to that of linezolid in a murine model. Antimicrob Agents Chemother 53:1314–1319. doi: 10.1128/AAC.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietze R, Hadad DJ, McGee B, Molino LP, Maciel EL, Peloquin CA, Johnson DF, Debanne SM, Eisenach K, Boom WH, Palaci M, Johnson JL. 2008. Early and extended early bactericidal activity of linezolid in pulmonary tuberculosis. Am J Respir Crit Care Med 178:1180–1185. doi: 10.1164/rccm.200806-892OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown AN, Bulitta JB, McSharry JJ, Weng Q, Adams JR, Kulawy R, Drusano GL. 2011. The effect of half-life on the pharmacodynamic index of zanamivir against influenza virus as delineated by a mathematical model. Antimicrob Agents Chemother 55:1747–1753. doi: 10.1128/AAC.01629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown AN, Adams JR, Baluya DL, Drusano GL. 2015. Pharmacokinetic determinants of virological response to raltegravir in the in vitro pharmacodynamic hollow-fiber infection model system. Antimicrob Agents Chemother 59:3771–3777. doi: 10.1128/AAC.00469-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigelow KM, Deitchman AN, Li S-Y, Barnes-Boyle K, Tyagi S, Soni H, Dooley KE, Savic R, Nuermberger EL. 29 January 2020. Pharmacodynamic correlates of linezolid activity and toxicity in murine models of tuberculosis. J Infect Dis doi: 10.1093/infdis/jiaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tasneen R, Betoudji F, Tyagi S, Li SY, Williams K, Converse PJ, Dartois V, Yang T, Mendel CM, Mdluli KE, Nuermberger EL. 2016. Contributions of oxazolidinones to the efficacy of novel regimens containing bedaquiline and pretomanid in a mouse model of tuberculosis. Antimicrob Agents Chemother 60:270–277. doi: 10.1128/AAC.01691-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Capodagli GC, Awasthi D, Shrestha R, Maharaja K, Sukheja P, Li S-G, Inoyama D, Zimmerman M, Ho Liang HP, Sarathy J, Mina M, Rasic G, Russo R, Perryman AL, Richmann T, Gupta A, Singleton E, Verma S, Husain S, Soteropoulos P, Wang Z, Morris R, Porter G, Agnihotri G, Salgame P, Ekins S, Rhee KY, Connell N, Dartois V, Neiditch MB, Freundlich JS, Alland D. 2018. Synergistic lethality of a binary inhibitor of Mycobacterium tuberculosis KasA. mBio 9:e02101-17. doi: 10.1128/mBio.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung WJ, Kornilov A, Brodsky BH, Higgins M, Sanchez T, Heifets LB, Cynamon MH, Welch J. 2008. Inhibition of M. tuberculosis in vitro in monocytes and in mice by aminomethylene pyrazinamide analogs. Tuberculosis (Edinb) 88:410–419. doi: 10.1016/j.tube.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Drusano GL, Liu W, Fikes S, Cirz R, Robbins N, Kurhanewicz S, Rodriquez J, Brown D, Baluya D, Louie A. 2014. Interaction of drug- and granulocyte-mediated killing of Pseudomonas aeruginosa in a murine pneumonia model. J Infect Dis 210:1319–1324. doi: 10.1093/infdis/jiu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing for mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed Document A24-A. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 28.Leary R, Jelliffe R, Schumitzky A, Van Guilder M. 2001. An adaptive grid non-parametric approach to pharmacokinetic and dynamic (PK/PD) models, p 389–394. Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems. IEEE Computer Society, Bethesda, MD. [Google Scholar]
- 29.Neely MN, van Guilder MG, Yamada WM, Schumitzky A, Jelliffe RW. 2012. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 34:467–476. doi: 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.