Abstract
Uncontrolled growth and an enforced epithelial-mesenchymal transition (EMT) process contribute to the poor survival rate of patients with osteosarcoma (OS). Long noncoding RNAs (lncRNAs) have been reported to be involved in the development of OS. However, the significant role of lncRNA SNHG1O on regulating proliferation and the EMT process of OS cells remains unclear. In this study, quantitative real-time PCR and fluorescence in situ hybridization (FISH) results suggested that SNHG10 levels were significantly increased in OS compared with healthy tissues. In vitro experiments (including colony formation, CCK-8, wound healing, and transwell assays) and in vivo experiments indicated that downregulation of SNHG10 significantly suppressed the proliferation and invasion of OS cells. Luciferase reporter assay and RNA immunoprecipitation (RIP) assay confirmed that SNHG10 could regulate FZD3 levels through sponging microRNA 182-5p (miR-182-5p). In addition, the SNHG10/miR-182-5p/FZD3 axis could further promote the β-catenin transfer into nuclear accumulation to maintain the activation of the Wnt singling pathway. Together, our results established that SNHG10 has an important role in promoting OS growth and invasion. By sponging miR-182-5p, SNHG10 can increase FZD3 expression and further maintain the activation of Wnt/β-catenin singling pathway in OS cells.
Keywords: osteosarcoma, SNHG10, Wnt/β-catenin, lncRNA, EMT
Graphical Abstract
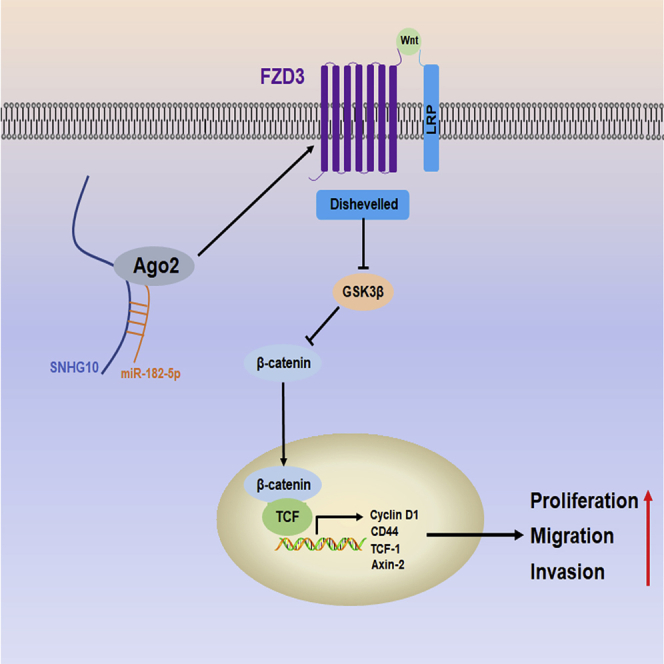
Zhu et al. demonstrate that overexpressed lncRNA SNHG10 promotes proliferation, migration, and invasion of OS cells by sponging miR-182-5p. The SNHG10/miR-182-5p axis activates the Wnt/β-catenin pathway via elevating FZD3. Targeting SNHG10 may improve the prognosis of patients with OS.
Introduction
Osteosarcoma (OS) is the most common and most malignant bone tumor. Most of OS occurs in young patients, younger than 20 years.1,2 Despite of the aggressive therapy, including surgical removal and chemotherapy with multidrugs, the 5-year overall survival rate of patients with OS remains poor (less than 30%).3,4 Therefore, treatment improvement is indispensable. The development of molecular biology and the increased scientific research about the molecular mechanisms of tumorigenesis and progression have provided a new approach to defeat tumors. Several genetic alterations have been proven to initiative and force OS,5,6 and targeting these molecules may provide new strategies for OS treatment.
Long noncoding RNAs (lncRNAs) are a newly determined class of noncoding RNAs, with a length of about 200 nucleotides.7 Increased evidence suggests that dysregulated lncRNAs expression is involved in regulating the multi-processes of OS. For example, downregulated lncRNA CEBPA-AS1 in OS was proven to promote proliferation and migration of tumor cells.8 lncRNA RP11-361F15.2 promotes OS tumorigenesis by inhibiting M2-like polarization of tumor-associated macrophages (TAMs).9 One of the most important roles of lncRNA is acting as competitive endogenous RNA (ceRNA) to competitively bind with target mRNA against microRNAs (miRNAs).10,11 Recently, lncRNA SNHGs, as well as lncRNA SNHG10, were investigated. RNA sequencing (RNA-seq) data showed that lncRNA SNHG10 is a potential prognostic biomarker for patients with lung adenocarcinoma.12 Lan et al.13 found that highly expressed SNHG10 in hepatocellular carcinoma (HCC) contributes to the malignant phenotype, and the SNHG10/DDX54/PBX3 feedback loop contributes to the growth of gastric cancer cells.14 In addition, SNHG10 suppresses non-small cell lung cancer (NSCLC) proliferation by sponging miR-543 to upregulate SIRT1.15 However, the function of SNHG10 in OS remains unclear.
In this study, we found that SNHG10 was overexpressed in OS cells and was associated with a poor prognosis. In vitro and in vivo experiments showed that SNHG10 promoted the proliferation, migration, and invasion of OS cells. In addition, we identified SNHG10 as functioning as a ceRNA by sponging miR-182-5p and thereby regulating FZD3 and the Wnt/catenin pathway.
Results
lncRNA SNHG10 Is Overexpressed in OS and Is Associated with a Poor Prognosis
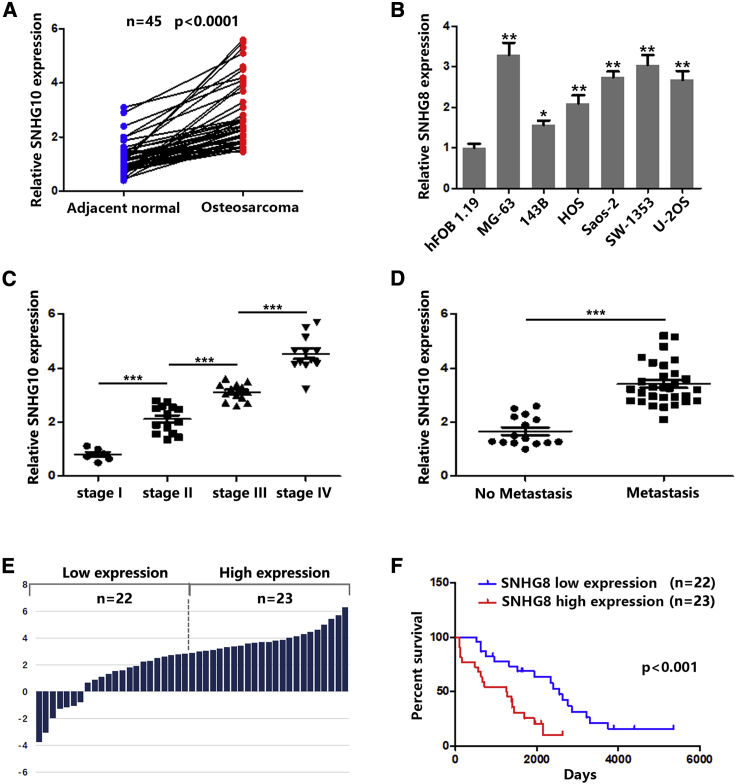
First, we examined the expression of SNHG10 in 45 paired OS tissues and adjacent tissues and found that SNHG10 was significantly upregulated in OS tissues compared with healthy tissues (Figure 1A). As shown in Figure 1B, SNHG10 was more highly expressed in OS cells than it was in healthy bronchial epithelial cells. In addition, increased SNHG10 expression level was correlated with advanced stage and lymph node metastasis in patients with OS (Figures 1C and 1D). Pearson chi-square and Fisher’s exact test results indicated that, rather than age, gender, or anatomic location, increased SNHG10 levels were correlated with larger tumor size (≥3 cm), advanced TNM stage, and lymph node metastasis (Table 1). According to the median value of the SNHG10 expression level, we divided 45 OS tissues into two groups (high and low SNHG10 levels) (Figure 1E). Kaplan-Meier survival analysis showed that higher SNHG10 indicated a poorer prognosis (Figure 1F). Together, these results suggested that the lncRNA SNHG10 could function as an oncogene, promoting OS progression.
Figure 1.
LncRNA SNHG10 Is Overexpressed in OS and Is Associated with Poor Prognosis
(A) Relative expression of SNHG10 in OS and adjacent normal samples were quantified by quantitative real-time PCR. (B) Relative expression of SNHG10 in different OS cell lines and healthy bronchial epithelial cells were quantified by quantitative real-time PCR. (C) Relative expression of SNHG10 in OS samples was analyzed according to the tumor stage. (D) Relative expression of SNHG10 in OS samples was analyzed according to metastasis status. (E) Forty-five OS samples were divided into two groups according to whether expression of SNHG10 was high or low. (F) Kaplan-Meier analysis was used to determine the association between SNHG4 high or low expression and overall survival of patients with OS. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
Table 1.
Clinical and Pathologic Characteristics of OS
| Feature | Number (n) | SNHG10 Expression |
p Value | |
|---|---|---|---|---|
| Low (n) | High (n) | |||
| Age (year) | ||||
| ≥18 | 25 | 6 | 19 | 0.681 |
| < 18 | 20 | 5 | 15 | |
| Gender | ||||
| Male | 17 | 5 | 12 | 0.635 |
| Female | 28 | 6 | 22 | |
| Tumor Size | ||||
| ≥3 | 26 | 3 | 23 | 0.024 |
| < 3 | 19 | 8 | 11 | |
| TMN Stage | ||||
| I+II | 20 | 11 | 9 | 0.011 |
| III+IV | 25 | 4 | 21 | |
| Lymph Node Metastasis | ||||
| Positive | 30 | 3 | 27 | 0.031 |
| Negative | 15 | 6 | 9 | |
| Anatomic Location | ||||
| Tibia/femur | 27 | 10 | 17 | 0.257 |
| Elsewhere | 18 | 8 | 10 | |
Knockdown of SNHG10 Restrains OS Progression In Vitro
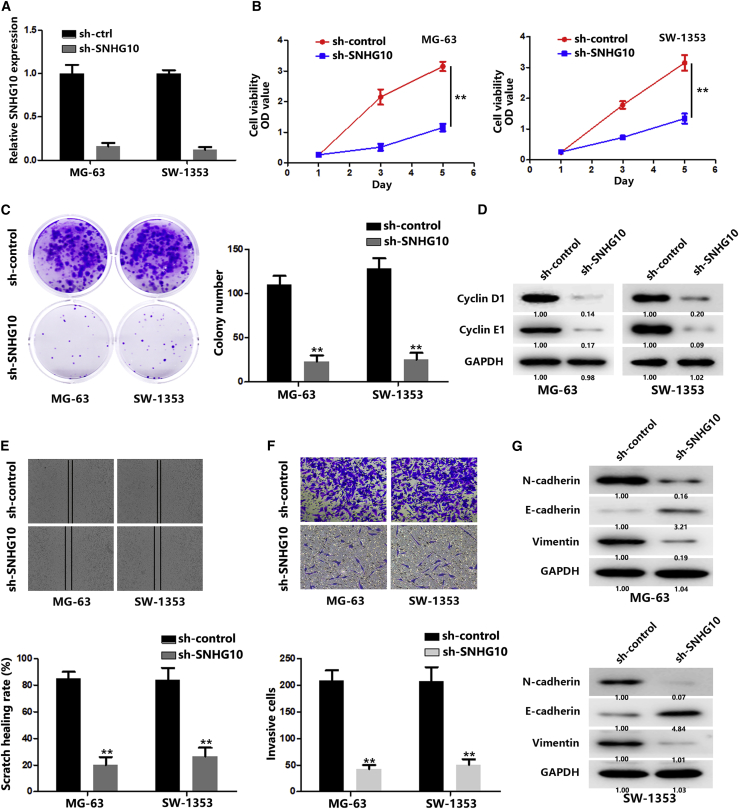
Sustaining proliferation and activating invasion are the great hallmarks of cancers.16,17 Therefore, we wondered whether overexpressed SNHG10 might act on the proliferation and invasion of OS cells. We first transfected OS cells with SNHG10-short hairpin RNA (shRNA), and the knockdown effect was confirmed by quantitative real-time PCR (Figure 2A). Then, CCK-8 and colony formation assays were performed. As shown in Figures 2B and 2C, downregulation of SNHG10 significantly decreased OS cell proliferation. In addition, cyclin D1 and E1, which have been proven to regulate cancer cell proliferation,18, 19, 20 were decreased along with SNHG10 downregulation (Figure 2D). Wound-healing and transwell assays were performed, and the results showed that downregulation of SNHG10 significantly impaired the ability of OS cells to migrate and invade (Figures 2E and 2F). In addition, increased E-cadherin and decreased N-cadherin and vimentin were observed after downregulation of SNHG10, suggesting that the epithelial-mesenchymal transition (EMT) process was suppressed (Figure 2G). Taken together, these results indicate a critical role for SNHG10 in OS proliferation and invasion.
Figure 2.
Knockdown of SNHG10 Restrains OS Progression In Vitro
(A) Relative expression of SNHG10 in OS cells was quantified by quantitative real-time PCR after transfection of sh-control (sh-ctrl) or sh-SNHG10. (B) The proliferation of transfected OS cells was evaluated with a CCK-8 assay. (C) The proliferation of transfected OS cells was evaluated with a colony formation assay. (D) The expression of cyclin D1 and E1 was quantified by western blot after transfection of sh-ctrl or sh-SNHG10. (E) The migration of transfected OS cells was evaluated with a wound healing assay. (F) The invasion of transfected OS cells was evaluated with a transwell assay. (G) The expression of E-cadherin, N-cadherin, and vimentin was quantified by western blot after transfection of sh-ctrl or sh-SNHG10. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
SNHG10 Serves as a Sponge for miR-182-5p
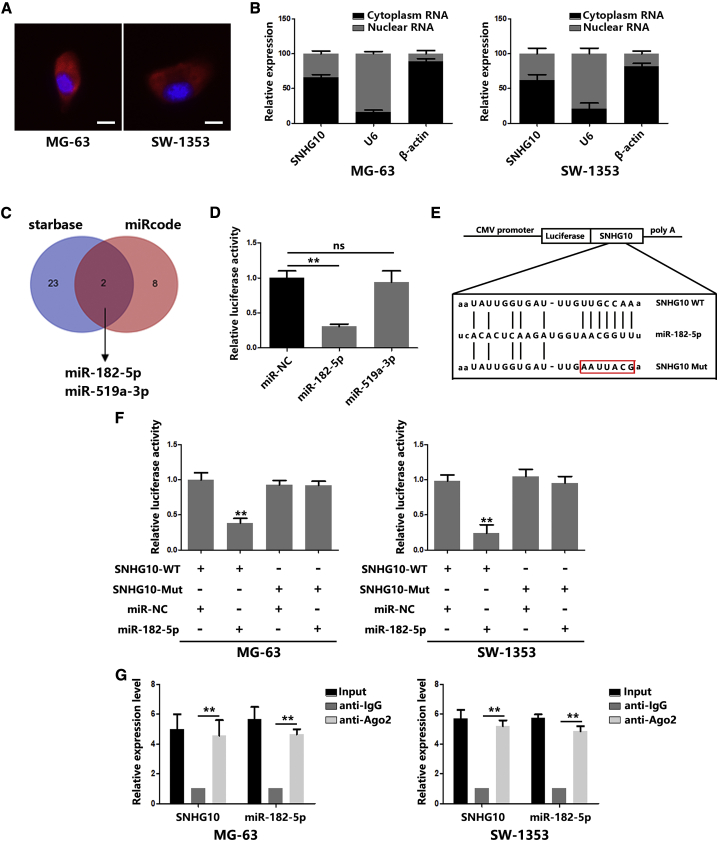
To determine the potential molecular mechanism of SNHG10 in promoting proliferation and invasion in OS cells, we first determined the distribution of SNHG10 in OS cells. Fluorescence in situ hybridization (FISH) analysis and nuclear mass separation assays were employed, and SNHG10 was found to be mostly enriched in the cytoplasm (Figures 3A and 3B). This kind of distribution gives SNHG10 the ability to be a ceRNA. Using the bioinformatic website (Database: miRcode and starBase), we found two candidate miRNAs (Figure 3C). To verify the prediction results, dual luciferase reporter assays were performed. The results indicated that miR-182-5p, instead of miR-519a-3p, significantly suppressed SNHG10-driven luciferase activity (Figure 3D). Mutations of the predicative binding sites in SNHG10 restored the decreased luciferase activity induced by miR-182-5p (Figures 3E and 3F). In addition, the results of RNA immunoprecipitation (RIP) assays confirmed the direct binding of SNHG10 and miR-182-5p with argonaute-2 (Ago2), an important component of the RNA-induced silencing complex (RISC) (Figure 3G). These data indicate that SNHG10 functions as a ceRNA by directly interacting with miR-182-5p.
Figure 3.
SNHG10 Serves as a Sponge for miR-182-5p
(A) FISH analysis indicated a subcellular location for SNHG10 in OS cells (green). Nuclei were stained with DAPI (blue). (B) Relative SNHG10 expression levels in nuclear and cytosolic fractions of the OS cells were quantified by quantitative real-time PCR. (C) A schematic drawing of the screening procedure of candidate miRNAs. (D) The luciferase reporter plasmids carrying SNHG10 were co-transfected into HEK293T cells with two miRNA-coding plasmids. (E) A schematic representation of the miR-182-5p binding sites in SNHG10 and site mutagenesis. (F) The luciferase reporter plasmid carrying wild-type (WT) or mutant (MUT) SNHG10 was co-transfected into OS cells with miR-182-5p in parallel with an empty vector. Relative luciferase activity in OS cells was determined. (G) Ago2 RIP assay analysis of the enrichment of SNHG10 and miR-182-5p pulled down from the Ago2 protein in OS cells, and the expression levels of SNHG10 and miR-182-5p was examined by quantitative real-time PCR analysis. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
SNHG10 Promotes OS Progression through miR-182-5p
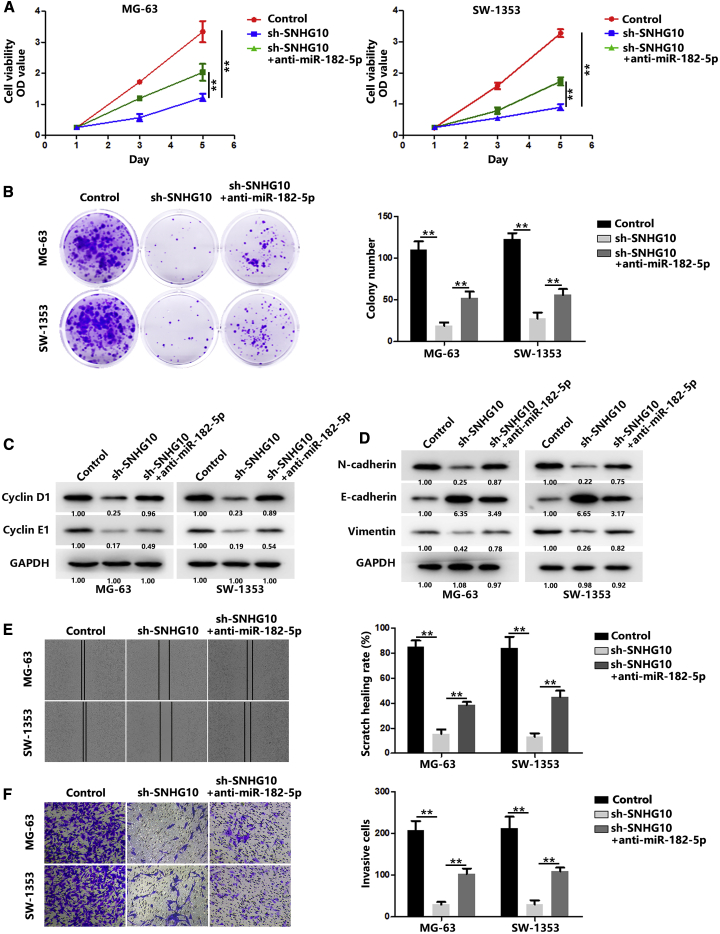
To investigate whether SNHG10 modulates proliferation and invasion through miR-182-5p, we performed rescue experiments by transfecting anti-miR-182-5p along with SNHG10 knockdown experiments. As shown in Figures 4A and 4B, the reduced proliferation caused by SNHG10 knockdown was significantly restored by anti-miR-182-5p transfection. The levels of cyclin D1 and E1 in OS cells echoed each other (Figure 4C). Similarly, inhibition of OS cell invasion and the EMT process were restored by anti-miR-182-5p transfection in SNHG10-depleting cells (Figures 4D–4F). Those results confirmed that SNHG10 functions as a ceRNA, via sponging miR-182-5p, to promote OS progression.
Figure 4.
SNHG10 Promotes OS Progression through miR-182-5p
(A) The proliferation of OS cells after transfection with sh-SNHG10 or co-transfection with a miR-182-5p mimic was evaluated with a CCK-8 assay. (B) The proliferation of OS cells after transfection with sh-SNHG10 or co-transfection with a miR-182-5p mimic was evaluated using a colony formation assay. (C) The expression of cyclin D1 and E1 in OS cells after transfection with sh-SNHG10 or co-transfection with a miR-182-5p mimic was quantified by western blot. (D) The expressions of E-cadherin, N-cadherin, and vimentin were quantified using western blot after transfection of sh-SNHG10 or co-transfection with a miR-182-5p mimic. (E) The migration of OS cells after transfection with sh-SNHG10 or co-transfection with a miR-182-5p mimic was evaluated with a wound healing assay. (F) The invasion of OS cells after transfection with sh-SNHG10 or co-transfection with a miR-182-5p mimic was evaluated with a transwell assay. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
FZD3 Is a Target of miR-182-5p
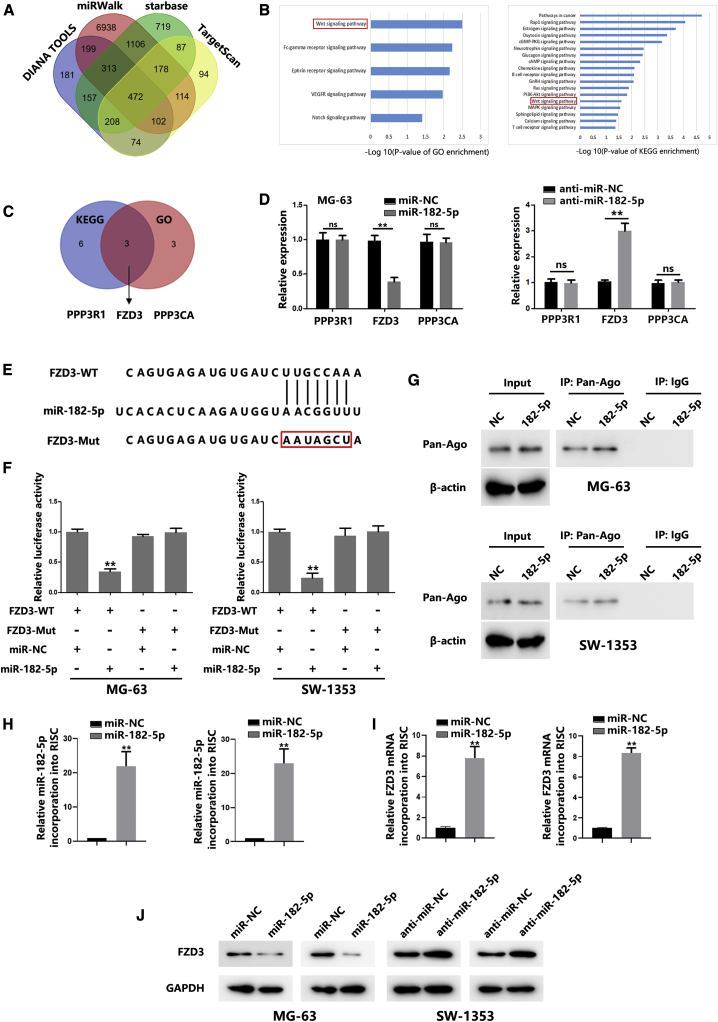
Next, we identified the putative target genes of miR-182 through four online prediction tools (Database: DIANA Tools, miRWalk, starBase, and TargetScan). As shown in Figure 5A, 472 common genes were found. Both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses suggested that these target genes are involved in the Wnt signaling pathway (Figure 5B). Furthermore, we found three common genes, which are the components of the Wnt signaling pathway from the results of GO and KEGG search (Figure 5C). Real-time PCR analysis revealed that miR-182-5p overexpression significantly inhibited FZD3 expression in OS cells (Figures 5D and S1A). In contrast, downregulation of miR-182-5p markedly increased FZD3 (Figures 5D and S1A). To further confirm that FZD3 was a direct target of miR-182-5p, a 3′ untranslated region (3′ UTR) luciferase reporter assay was performed. As shown in Figures 5E and 5F, miR-182-5p overexpression significantly inhibited the luciferase activity of the wild-type FZD3 reporter, whereas mutation of binding sites in FZD3 abrogated the repressive effect of miR-182-5p. Furthermore, miR-182-5p downregulation significantly increased luciferase activity of the wild-type FZD3 reporter, whereas the mutant FZD3 reporter was not affected (Figure S1B). In addition, RNA chromatin immunoprecipitation (RNA-ChIP) assays were performed to identify FZD3 abundance in RISC. As shown in Figures 5G–5I, miR-182-5p overexpression contributed to increased FZD3 levels in RISC. These results verified the direct interaction between miR-182-5p and FZD3. Western blot (WB) results showed that miR-182-5p overexpression significantly restrained FZD3 levels, whereas miR-182-5p inhibition significantly increased FZD3 levels in OS cells (Figure 5J). In addition to the results from cells, the level of miR-182-5p expression was found to be significantly downregulated in OS tissues compared with healthy tissues (Figure S1C). However, FZD3 mRNA was found to be significantly higher in OS tissues than it was in healthy tissues (Figure S1D). An inverse correlation between miR-182-5p and FZD3 in OS samples was observed (Figure S1E). Together, our data suggest that miR-182-5p could directly target FZD3.
Figure 5.
FZD3 Is a Target of miR-182-5p
(A) Schematic of the candidate genes of miR-182-5p using four prediction tools. (B) GO and KEGG pathway analysis of candidate targets of miR-182-5p. (C) Schematic of the common candidate genes involved in regulating the Wnt/β-catenin signaling pathway. (D) mRNA expression of PPP3R1, FZD3, and PPP3CA in OS cells transfected with miR-NC or a miR-182-5p mimic. (E) Predicted miR-182-5p target sequences in the 3′ UTRs of FZD3 genes. (F) Relative FZD3 reporter activities in OS cells co-transfected with miR-182-5p and luciferase reporters. (G) Immunoprecipitation of the Ago2/RISC (RNA-induced silencing complex) using the Pan-Ago2 antibody in OS cells overexpressing miR-NC or miR-182-5p. IgG was used as a negative control and β-actin was used as an internal control. (H) Quantitative real-time PCR analysis of miR-182-5p incorporated into RISC in OS cells overexpressing miR-182-5p compared with the levels in the control. (I) Quantitative real-time PCR of FZD3 incorporated into RISC in OS cells overexpressing miR-182-5p. (J) FZD3 expression levels in OS cells transfected with a miR-182-5p mimic or anti-miR-182-5p were quantified by western blot. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
SNHG10 Promotes OS Progression through FZD3
After identifying FZD3 as a direct target of miR-182-5p, we studied whether SNHG10 could bind competitively with FZD3. The results of Ago2 RIP assays showed that SNHG10 knockdown led to a great increase in FZD3 enrichment, and SNHG10 overexpression significantly decreased FZD3 enrichment, suggesting that SNHG10 could compete with FZD3 transcripts for an Ago2-based, miRNA-induced expression complex (Figures S2A and S2B). A luciferase reporter assay showed that SNHG10 knockdown significantly decreased the luciferase activity of the FZD3 reporter, and the results could be rescued by a miR-182-5p sponge. Moreover, the luciferase activity of the mutant FZD3 reporter was not affected. Instead, SNHG10 overexpression significantly increased the luciferase activity of the wild-type FZD3 reporter, rather than the mutant FZD3 reporter, whereas miR-182-5p abolished that effect (Figures S2C and S2D). In addition, SNHG10 knockdown seriously reduced FZD3 protein levels, whereas SNHG10 overexpression remarkably increased FZD3 levels (Figure S2E). These data indicate that SNHG10 could function as a ceRNA via sponging miR-182-5p to facilitate FZD3 expression.
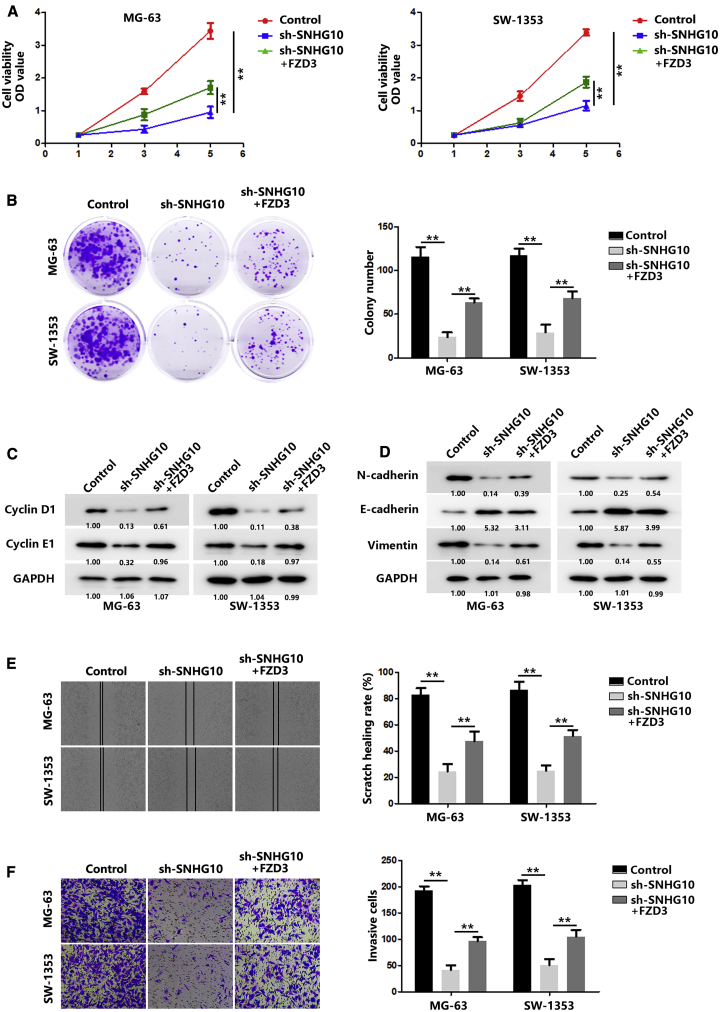
To further confirm that SNHG10 affects proliferation and invasion through FZD3, we performed a series of in vitro experiments. As shown in Figures 6A and 6B, FZD3 significantly reversed the inhibition caused by SNHG10 knockdown. The expression of cyclin D1 and E1 also exhibited similar trends (Figure 6C). Meanwhile, FZD3 restored the inhibited invasion and EMT process caused by the SNHG10 knockdown (Figures 6D–6F).
Figure 6.
SNHG10 Promotes OS Progression through FZD3
(A) The proliferation of OS cells after transfection with sh-SNHG10 or co-transfection with FZD3 was evaluated with a CCK-8 assay. (B) The proliferation of OS cells after transfection with sh-SNHG10 or co-transfection with FZD3 was evaluated with a colony-formation assay. (C) The expression of cyclin D1 and E1 in OS cells after transfection with sh-SNHG10 or co-transfection with FZD3 was quantified by western blot. (D) The expressions of E-cadherin, N-cadherin, and vimentin were quantified using western blot after transfection of sh-SNHG10 or co-transfection with FZD3. (E) The migration of OS cells after transfection with sh-SNHG10 or co-transfection with FZD3 was evaluated with a wound-healing assay. (F) The invasion of OS cells after transfection with sh-SNHG10 or co-transfection with FZD3 was evaluated with a transwell assay. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
Collectively, these data suggest that SNHG10 facilitates FZD3 expression via sponging miR-182-5p.
SNHG10 Activates the Wnt/β-Catenin Pathway to Promote OS Progression
The Wnt/β-catenin signaling pathway has been reported to be involved in controlling OS development.21 Because FZD3 is a direct target of miR-182-5p, we investigated whether miR-182-5p affected Wnt by regulating FDZ3. Overexpression of miR-182-5p significantly decreased β-catenin nuclear accumulation, whereas β-catenin was increased in the cytoplasm (Figures S3A and S3B). In addition, the mRNA levels of downstream genes regulated by the Wnt signaling pathway were downregulated after overexpressing miR-182-5p (Figure S3C). Rescue assays showed that FZD3 could increase the decreased β-catenin levels in the nucleus caused by miR-182-5p overexpression (Figures S3A–S3C). The results of T cell factor/lymphoid enhancer biding factor (TCF/LEF) reporter assays showed that miR-182-5p overexpression significantly decreased the reporter activity, and additional FZD3 elevated the activity (Figure S3D). In contrast, si-FZD3 (FZD3 siRNA) transfection displayed an effect similar to that of XAV-939 (a small molecule inhibitor) treatment: β-catenin assembly in the nucleus caused by miR-182-5p knockdown was markedly inhibited (Figure S3D). These results suggested that miR-182-5p inhibited the Wnt/β-catenin signaling pathway by directly targeting FZD3.
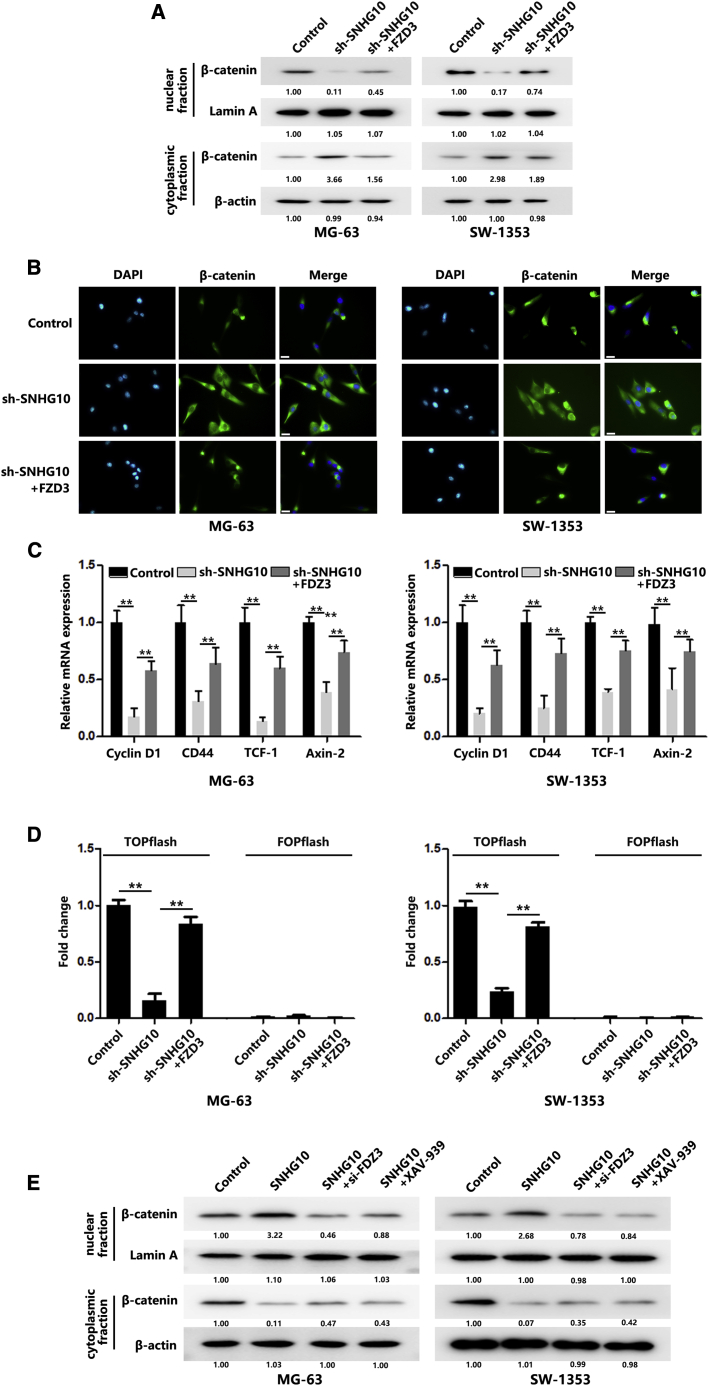
Because SNHG10 could completely bind with miR-182-5p to promote FZD3 translation, we wanted to determine whether SNHG10 could affect the Wnt/β-catenin signaling pathway through FZD3. As shown in Figures 7A and 7B, downregulation of SNHG10 significantly inhibited β-catenin assembly in the nucleus. Additional transfection of FZD3 could restore β-catenin levels in the nucleus. Quantitative real-time PCR results showed that SNHG10 knockdown could reduce mRNA levels of Wnt/β-catenin signaling targeting genes, and additional transfection of FZD3 could rescue the decrease (Figure 7C). We further performed TCF/LEF reporter assays, and the results indicated that SNHG10 downregulation significantly inhibited the reporter activity, and FZD3 overexpression could rescue the decreased activity (Figure 7D). Meanwhile, we overexpressed SNHG10 and found that nuclear β-catenin levels were significantly increased. However, additional additions of si-FZD3 or XAV-939 partially abolished the increase of nuclear β-catenin (Figure 7E). Taken together, these results indicate that Wnt/β-catenin signaling is activated by SNHG10 in OS cells.
Figure 7.
SNHG10 Activates Wnt/β-Catenin Pathway to Promote OS Progression
(A) β-catenin protein redistribution in different cellular compartments of transfected OS cells was quantified by western blot. (B) β-catenin protein redistribution in different cellular compartments of transfected OS cells was quantified using immunofluorescence staining. Scale bar, 100 μm. (C) Relative expression levels of Wnt/β-catenin pathway downstream target genes were quantified by real-time PCR. (D) Relative TCF/LEF reporter activity was measured after transfection with sh-SNHG10 or co-transfection with FZD3. (E) The β-catenin protein redistribution in different cellular compartments of transfected or XAV-939 treated OS cells was quantified using western blot. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
SNHG10 Promotes OS Tumorigenesis In Vivo
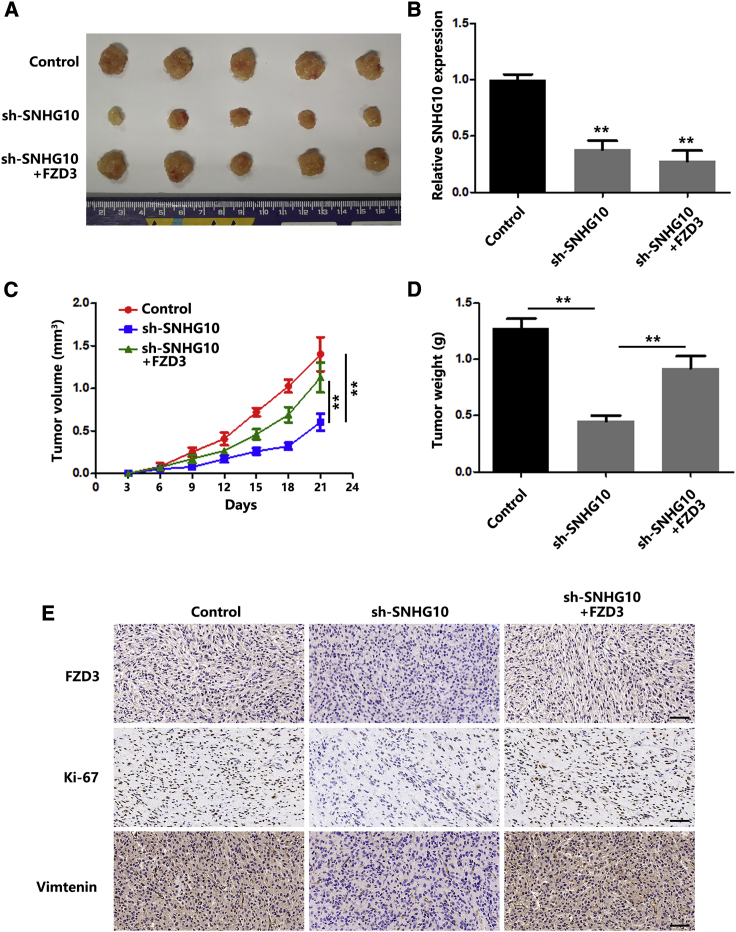
To investigate the effect of SNHG10 on OS in vivo, subcutaneous xenograft models were employed. Downregulation of SNHG10, confirmed by quantitative real-time PCR (Figure 8B), significantly inhibited tumor growth of OS compared with that of the control group. As expected, additional FZD3 overexpression repaired the tumor-growth inhibition (Figures 8A, 8C, and 8D). Ki-67 and vimentin (molecular markers for proliferation and EMT, respectively) expression levels in tumors were evaluated using immunohistochemistry (IHC). As shown in Figure 8E, lower expressions of Ki-67 and vimentin were found in sh-SNHG10 tumors compared with those of the control group. In addition, FZD3 transfection increased expression of Ki-67 and vimentin. Together, the results from the in vitro experiments were confirmed in vivo.
Figure 8.
SNHG10 Promotes OS Tumorigenesis In Vivo
(A) Subcutaneous tumors were separated and imaged at the endpoint of the experiment. Scale bar, 1 cm. (B) Relative expression of SNHG10 in subcutaneous tumor tissues was quantified by quantitative real-time PCR. (C) Tumor growth curves of different subcutaneous tumor groups are shown. (D) Tumor weights of different tumor groups are shown. (E) The expression levels of FZD3, Ki-67, and vimentin in different groups of subcutaneous tumors were evaluated by IHC. In all experiments, bars represent means ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
Discussion
Emerging evidence have proven that dysregulated lncRNAs have significant roles in modulating biological processes, such proliferation,22,23 metabolism,24 metastasis,25 and apoptosis26, in OS. lncRNA SNHGs represent a group of lncRNAs produced from a small nucleolar RNA host gene. Growing research data demonstrate that lncRNA SNHGs are involved in tumorigenesis and progression. For example, lncRNA SNHG1 acts as a ceRNA to promote OS tumorigenesis by sponging miR-326.26 Lao et al.27 showed that higher lncRNA SNHG4 predicts poor survival and recurrence in OS. Inhibition of lncRNA SNHG12 might be a new therapy to suppress OS growth.28 There have been a number of studies that have revealed the oncogenic role of SNHG10 in various tumors, such as hepatocarcinogenesis13 and lung adenocarcinoma.12 However, the expression level and function of SNHG10 in OS remains to be elucidated. Overexpressed SNHG10 in OS was determined by quantitative real-time PCR analysis of 45 paired tumor and adjacent healthy tissues. Statistical analysis results suggested that SNHG10 level was positively correlated with tumor size, TNM stage, and lymph node metastasis. Further, in vitro and in vivo experiments indicated that SNHG10 is essential for tumor growth and invasion. Additional experiments are needed to determine more about the functional role of SNHG10 in OS cells.
One of the most important molecular mechanism of lncRNA is its role as a ceRNA sponging miRNAs.29,30 Therefore, we hypothesized that SNHG10 could also function as a ceRNA. In this study, we found via FISH and nuclear mass separation assays that most of SNHG10 is distributed in the cytoplasm, and the results provided strong support for our hypothesis. Using online prediction tools, luciferase assays, and RIP assays, we confirmed that SNHG10 direct interacts with miR-182-5p. In vitro experiments showed that miR-182-5p overexpression could reverse the suppressed proliferation and invasion of OS cells caused by NSHG10 knockdown. In addition, we found that FZD3 is a direct target of miR-182-5p. In vitro experiments showed that overexpression of FZD3 could also reverse the suppressive effect caused by SNHG10 knockdown. Taken together, we found that SNHG10 could competitively regulate FZD3 through sponging miR-182-5p.
Wnt/β-catenin signaling pathway is a crucial pathway in controlling OS development.31,32 The transfer of β-catenin into the nucleus induces activation of a large number of transcription factors, further regulating the downstream signaling cascade.33 Previous studies have identified several lncRNAs that could regulate Wnt/β-catenin. For example, Cui et al found that SNHG1 overexpression contributes to non-small-cell-lung-cancer progression by activating Wnt/β-catenin.34 SNHG1 was proven to activate Wnt/β-catenin signaling in OS as well.35 In our study, using GO and KEGG pathway analyses, we found that the potential downstream genes of miR-182-5p are associated with Wnt/β-catenin. WB and immunofluorescence (IF) results showed that miR-182-5p functions as a Wnt inhibitor in OS cells, and overexpression of FZD3 impaired the effect of miR-182-5p. In addition, we found that downregulation of SNHG10 significantly suppressed the Wnt/β-catenin signaling pathway. Because SNHG10 could competitively promote FZD3 through sponging miR-182-5p, we co-transfected OS cells with sh-SNHG10 and FZD3. WB and IF results suggest that FZD3 significantly reverses the suppressive effect caused by SNHG10 knockdown. Together, these results suggest that the activation of the Wnt/β-catenin signaling pathway was supported by SNHG10/miR-182-5p/FZD3 axis in OS cells.
Materials and Methods
Clinical Samples
We used 45 paired OS and healthy tissues collected from the Department of Orthopedics, Huaihe Hospital of Henan University. This study was approved by the institutional review board and theethics committee of Henan University and written informedconsent was obtained from all patients, in accordance with the guidelines established in the Declaration of Helsinki. All samples were collected during surgery and were immediately frozen in liquid nitrogen for further experiments.
Cell Cultures
The human OS cell lines MG-63, 143B, HOS, Saos-2, SW-1353, U-20S, and hFOB 1.19 (a human osteoblast cell line) were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum and antibiotics (100 units/mL penicillin and 100 units/mL streptomycin). OS cells were maintained at 37°C with 5% CO2; hFOB 1.19 cells were maintained at 34°C with 5% CO2.
Cell Transfection
Cell transfection was performed with the Lipofectamine 2000 reagent. The sequence of anti-miRNA, miRNA mimics, siRNAs, and plasmids are listed below.
Anti-miR-182-5p: 5′-UUUGGCAAUGGUAGAACUCACACU-3′,
miR-182-5p mimics: 5′-UUUGGCAAUGGUAGAACUCACACCG-3′,
si-FZD3: 5′-CCTCCTTGTCCAAATATGTACTTCA-3′.
To obtain stable SNHG10-downregulated OS cells, we designed an empty vector and SNHG10 shRNA (5′-CCGGGGGAACTAAGAAGTGAATTCGCTCGAG GCTTAAGTGAAGAATCAAGGGTTTTTG-3′). The empty vector and the sh-SNHG10 were transfected into the OS cells with a lentivirus, then stable cells were selected with puromycin.
FISH
To identify the expression level of SNHG10 in OS and normal tissues, FISH analysis was performed as previously described.36 FISH probe was designed to target the following sequences.
SNHG10-1: 5′-ACCGGGGTTCCAGCGCTCGGGCCGTAGCCT-3′
SNHG10-2: 5′-AGGACGATGCTTGGAACGTGGTAAGTGCTCCTATTG-3′
RNA Extraction and Quantification
RNA extraction and quantitative real-time PCR tests were performed as previously described.36 Briefly, total RNA from cells and tissues were extracted with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). A PARIS kit (Thermo Fisher Scientific) was employed to extract nuclear and cytoplasmic RNA separately. cDNA was synthesized with the PrimeScript RT reagent kit. Quantitative real-time PCR analysis was performed with SYBR Green Premix Ex Taq. For miRNA detection, the stem-loop-specific primer method was used. The miRNA-specific reverse-transcription primers and qPCR primers were obtained from RiboBio (Guangzhou, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used as normalization controls for lncRNA and miRNA. The primer information is provided in the Supplemental Materials and Methods.
WB Assay
WB assays were performed as previously described.36 Briefly, protein was extracted from cells and tissues with RIPA lysis buffer (MilliporeSigma, Billerica, MA, USA) and was quantified with the BCA kit (Beyotime Biotechnology, Haimen, China). Protein samples were subjected to 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. FZD3, β-catenin, and GAPDH antibodies were used to incubate the membranes. The next day, the membranes were incubated with secondary antibodies. Lastly, bolts were achieved with a chemiluminescence detection system. The antibody information is provided in the Supplemental Materials and Methods.
Immunofluorescence Staining
Briefly, OS cells in the logarithmic growth phase were fixed with 4% paraformaldehyde for a half an hour. BSA was used as the blocking solution. After blocking, the β-catenin antibody was employed to incubate the cells overnight. The next day, the second antibody (Alexa Fluor 647) was used to incubate cells for 1 h. The nuclei were stained with DAPI.
Cell Viability and Colony Formation Assay
To evaluate OS cell proliferation, CCK-8 and colony formation assays were performed. For the CCK-8 assay, transfected OS cells were harvested and incubated with CCK-8 solution for 1 h. The optical density (OD) value was detected at 450 nm. For the colony formation assay, transfected OS cells were harvested and seeded in 6-well plates. After 2 weeks, the colonies were stained with crystal violet staining solution.
Wound-Healing Assay
A wound-healing assay was performed to assess the migration of OS cells. Briefly, cells were scratched with a 20-μL tip to make a gap. The width of the gaps was observed and measured after 48 h.
Transwell Assay
A transwell assay was performed to evaluate the ability of OS cells to invade. Briefly, transfected OS cells (1 × 105) were seeded into the upper chamber (Corning Costar, Corning, NY, USA), which were pre-coated with matrigel (Corning); 48 h later, the cells remaining in the upper chamber were removed, and cells at the bottom of the chamber were stained with crystal violet and measured.
Nuclear Mass Separation Assays
To identify the distribution of SNHG10 in OS cells, we isolated the cytoplasmic and nuclear RNA separately using PARIS kit (Thermo Fisher Scientific) following the manufacturer’s instructions.37
Dual Luciferase Reporter Assay
Dual luciferase reporter assay was performed as described previously. Briefly, to identify the interaction between SNHG10 and miR-182-5p, we used wild-type and mutant SNHG10 reporter plasmids that contained a period sequence of SNHG10 (5′-CCAGCGCGGAAUAUUGGUGAUAUUGUUGCCAAAAAAGCTTGACCCATC-3′). HEK293T cells were co-transfected with SNHG10 reporter plasmids and miR-NC or miR-182-5p; 48 h later, the dual luciferase reporter assay system was used to measure the fluorescent changes.
Similarly, to identify the interaction between FZD3 and miR-182-5p, we used wild-type and mutant FZD3 reporter plasmids that contained wild-type and mutant binding sites for miR-182-5p (GenePharma, Shanghai, China). HEK293T cells were co-transfected with FZD3 reporter plasmids and miR-NC or miR-182-5p; 48 h later, the dual luciferase reporter assay system was used to measure the fluorescent changes.
RIP Assay
To identify the role of SNHG10 as a ceRNA sponging miR-182-5p, a RIP assay was performed with Magna RNA-binding protein immunoprecipitation kit (MilliporeSigma). Briefly, lysis from OS cells was incubated with Ago2 or immunoglobulin G (IgG)-coated magnetic beads. Proteinase K was used to incubate samples, and the RNA was immunoprecipitated. After measuring the concentration and assessing the quality, SNHG10 and miR-182-5p expression was measured in purified RNA by quantitative real-time PCR.
TCF/LEF Reporter Assay
To assess activation of the Wnt signaling pathway, the TCF/LEF reporter kit (catalog no. 60500, BPS Bioscience, San Diego, CA, USA) was used according to the manufacturer’s instructions.38
Xenograft Tumor Model
Briefly, stably transfected OS cells (1 × 107) were subcutaneously injected in 15 female immune-deficient mice. Tumor volume was measured every 3 days. After euthanasia, the mice tumor weights were measured, and IHC was used to measure FZD3, Ki-67, and vimentin levels in the tumors.
Statistical Analysis
GraphPad Software (La Jolla, CA, USA) was employed for statistical analysis. All experiments were performed at least three times independently. All data are presented as means ± SD, and t test and ANOVA were used for comparisons. The Spearman rank test was used to evaluate the correlations between SNHG10 and FZD3 or miR-182-5p and FZD3. Survival analysis was performed with log-rank tests, as shown in Figure 8C. p <0.05 was considered statistically different.
Author Contributions
Y.L., S.Z., and X.W. designed the study, analyzed data, and drafted the manuscript. X.W., J.W., and G.X. participated in the manuscript preparation and performed the in vitro and in vivo experiments. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank all the members of our laboratory.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.010.
Supplemental Information
References
- 1.Tian W., Li Y., Zhang J., Li J., Gao J. Combined analysis of DNA methylation and gene expression profiles of osteosarcoma identified several prognosis signatures. Gene. 2018;650:7–14. doi: 10.1016/j.gene.2018.01.093. [DOI] [PubMed] [Google Scholar]
- 2.Berner K., Johannesen T.B., Berner A., Haugland H.K., Bjerkehagen B., Bøhler P.J., Bruland O.S. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015;54:25–33. doi: 10.3109/0284186X.2014.923934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu D., Kahen E., Cubitt C.L., McGuire J., Kreahling J., Lee J., Altiok S., Lynch C.C., Sullivan D.M., Reed D.R. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci. Rep. 2015;5:16991. doi: 10.1038/srep16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Adamopoulos C., Gargalionis A.N., Basdra E.K., Papavassiliou A.G. Deciphering signaling networks in osteosarcoma pathobiology. Exp. Biol. Med. (Maywood) 2016;241:1296–1305. doi: 10.1177/1535370216648806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kushlinskii N.E., Fridman M.V., Braga E.A. Molecular Mechanisms and microRNAs in Osteosarcoma Pathogenesis. Biochemistry (Mosc.) 2016;81:315–328. doi: 10.1134/S0006297916040027. [DOI] [PubMed] [Google Scholar]
- 7.Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia P., Gu R., Zhang W., Sun Y.F. lncRNA CEBPA-AS1 overexpression inhibits proliferation and migration and stimulates apoptosis of OS cells via notch signaling. Mol. Ther. Nucleic Acids. 2020;19:1470–1481. doi: 10.1016/j.omtn.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Yang D., Liu K., Fan L., Liang W., Xu T., Jiang W., Lu H., Jiang J., Wang C., Li G., Zhang X. LncRNA RP11-361F15.2 promotes osteosarcoma tumorigenesis by inhibiting M2-Like polarization of tumor-associated macrophages of CPEB4. Cancer Lett. 2020;473:33–49. doi: 10.1016/j.canlet.2019.12.041. [DOI] [PubMed] [Google Scholar]
- 10.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen L., Wang Q., Liu R., Chen Z., Zhang X., Zhou P., Wang Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46:717–729. doi: 10.1093/nar/gkx1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li D.S., Ainiwaer J.L., Sheyhiding I., Zhang Z., Zhang L.W. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016;20:2285–2295. [PubMed] [Google Scholar]
- 13.Lan T., Yuan K., Yan X., Xu L., Liao H., Hao X., Wang J., Liu H., Chen X., Xie K. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res. 2019;79:3220–3234. doi: 10.1158/0008-5472.CAN-18-4044. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Guo H., Zhang H. SNHG10/DDX54/PBX3 feedback loop contributes to gastric cancer cell growth. Dig. Dis. Sci. 2020 doi: 10.1007/s10620-020-06488-9. Published online July 25, 2020. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Nong L., Chen M.L., Gu X.L., Zhao W.W., Liu M.H., Cheng W.W. Long Noncoding RNA SNHG10 sponges miR-543 to upregulate tumor suppressive SIRT1 in nonsmall cell lung cancer. Cancer Biother. Radiopharm. 2020 doi: 10.1089/cbr.2019.3334. Published online April 22, 2020. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Zeng X., Wang N., Zhao W., Zhang X., Teng S., Zhang Y., Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol. Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Hainaut P., Plymoth A. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr. Opin. Oncol. 2013;25:50–51. doi: 10.1097/CCO.0b013e32835b651e. [DOI] [PubMed] [Google Scholar]
- 19.John R.R., Malathi N., Ravindran C., Anandan S. Mini review: multifaceted role played by cyclin D1 in tumor behavior. Indian J. Dent. Res. 2017;28:187–192. doi: 10.4103/ijdr.IJDR_697_16. [DOI] [PubMed] [Google Scholar]
- 20.Zucchini C., Manara M.C., Cristalli C., Carrabotta M., Greco S., Pinca R.S., Ferrari C., Landuzzi L., Pasello M., Lollini P.L. ROCK2 deprivation leads to the inhibition of tumor growth and metastatic potential in osteosarcoma cells through the modulation of YAP activity. J. Exp. Clin. Cancer Res. 2019;38:503. doi: 10.1186/s13046-019-1506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam D., Angulo P., Ponnurangam S., Dandawate P., Ramamoorthy P., Srinivasan P., Iwakuma T., Weir S.J., Chastain K., Anant S. Suppressing STAT5 signaling affects osteosarcoma growth and stemness. Cell Death Dis. 2020;11:149. doi: 10.1038/s41419-020-2335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wunder J.S., Nielsen T.O., Maki R.G., O’Sullivan B., Alman B.A. Opportunities for improving the therapeutic ratio for patients with sarcoma. Lancet Oncol. 2007;8:513–524. doi: 10.1016/S1470-2045(07)70169-9. [DOI] [PubMed] [Google Scholar]
- 23.Wei G., Zhang T., Li Z., Yu N., Xue X., Zhou D., Chen Y., Zhang L., Yao X., Ji G. USF1-mediated upregulation of lncRNA GAS6-AS2 facilitates osteosarcoma progression through miR-934/BCAT1 axis. Aging (Albany NY) 2020;12:6172–6190. doi: 10.18632/aging.103015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan L., Wu X., Yin X., Du F., Liu Y., Ding X. LncRNA CCAT2 promoted osteosarcoma cell proliferation and invasion. J. Cell. Mol. Med. 2018;22:2592–2599. doi: 10.1111/jcmm.13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang Y., Zhu X., Xu Y., Tang Q., Huang Z., Zhao Z., Lu J., Song G., Xu H., Deng C., Wang J. Energy stress-induced lncRNA HAND2-AS1 represses HIF1α-mediated energy metabolism and inhibits osteosarcoma progression. Am. J. Cancer Res. 2018;8:526–537. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Kong D. LncRNA GAS5 represses osteosarcoma cells growth and metastasis via sponging MiR-203a. Cell. Physiol. Biochem. 2018;45:844–855. doi: 10.1159/000487178. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Cao L., Wu J., Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int. J. Oncol. 2018;52:77–88. doi: 10.3892/ijo.2017.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu R., Feng F., Yu X., Liu Z., Lao L. LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma. Cell Prolif. 2018;51:e12515. doi: 10.1111/cpr.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Zheng J., Xue Y., Qu C., Chen J., Wang Z., Li Z., Zhang L., Liu Y. Inhibition of TDP43-Mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol. Ther. Nucleic Acids. 2018;10:142–158. doi: 10.1016/j.omtn.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 31.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Liu G., Wu Y., Ma J., Wu H., Xie Z., Chen S., Yang Y., Wang S., Shen P. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol. Cancer. 2019;18:150. doi: 10.1186/s12943-019-1076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kansara M., Tsang M., Kodjabachian L., Sims N.A., Trivett M.K., Ehrich M., Dobrovic A., Slavin J., Choong P.F., Simmons P.J. Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J. Clin. Invest. 2009;119:837–851. doi: 10.1172/JCI37175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui Y., Zhang F., Zhu C., Geng L., Tian T., Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Z., Jiang C., Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018;495:238–245. doi: 10.1016/j.bbrc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Yi H., Peng R., Zhang L.Y., Sun Y., Peng H.M., Liu H.D., Yu L.J., Li A.L., Zhang Y.J., Jiang W.H., Zhang Z. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017;8:e2583. doi: 10.1038/cddis.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu D., Lu C., Qu X., Li P., Chen K., Shan L., Zhu X. LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY) 2019;11:8374–8385. doi: 10.18632/aging.102325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Amerongen R. Celebrating discoveries in Wnt signaling: how one man gave wings to an entire field. Cell. 2020;181:487–491. doi: 10.1016/j.cell.2020.03.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.