Abstract
Introduction
Covid-19 infection was declared a global pandemic by WHO on March 11, 2020. GRP78 protein is known to be involved in the intrusion of numerous viruses. Our current study tries to provide some insight into the variation of GRP78 protein levels in patients with Covid-19 (−) pneumonia, Covid-19 (+) pneumonia, and CT negative Covid-19 infection in comparison to the normal population through a larger number of cases.
Materials and methods
42 patients who have Covid-19 (−) pneumonia; 72 patients who have Covid-19 infection (30 pneumonia,42 CT negative patients) and 30 patient who have no known diseases (control group) have included in the study after the clinical and radiological evaluation. Serum GRP78 levels of the subjects were measured through a commercially available enzyme-linked immunosorbent assay (ELISA) kit.
Results
The GRP78 level was found to be significantly higher in the Covid-19 infection group than both Covid-19 (−) pneumonia and control group (p = 0.031 and p = 0.0001, respectively).No significant difference was evident between Covid-19 (−) pneumonia, Covid-19 (+) pneumonia and CT negative Covid 19 infection groups with respect to GRP78 levels (p = 0.09). In addition, the GRP78 levels were significantly higher in the Covid-19 (−) pneumonia group than the control group (p = 0.0001).
Conclusion
This prospective case-control study reveals that the serum GRP78 levels significantly increased during Covid-19 infection in comparison to both the Covid-19 (−) pneumonia and the control group. As the association between SARS-CoV-2 virus and GRP78 protein is revealed more clearly, this association may come to the fore as a therapeutic target.
Keywords: Endoplasmic reticulum stress, GRP78, Covid-19 infection, Pneumonia
1. Introduction
Caused by a new type of coronavirus (SARS-CoV-2) that emerged in China in late 2019 and then were imported throughout the world, Covid-19 infection was declared a global pandemic by WHO on March 11, 2020 [1]. Covid-19 infection may manifest itself in severe clinical conditions, ranging from viral upper respiratory tract infection to pneumonia, sepsis, septic shock, and even acute respiratory distress syndrome (ARDS), in symptomatic patients [2].
Spike proteins play a major role in the infectivity of coronaviruses. Binding of the Virus Spike protein (S) to the cell surface receptor triggers coronavirus infection. SARS-CoV-2 virus, which leads to the emergence of Covid-19 infection, is an enveloped and single-stranded RNA virus that penetrates the host cell through the receptor-mediated endocytosis mechanism. The SARS-CoV-2 spike protein is a signal peptide with S1 and S2 domains and especially the S1 domain determines the tropism of coronaviruses [[3], [4], [5], [6]].
While S1 subunit of the SARS-CoV-2 spike (S) glycoprotein allows it to bind to the host ACE2 receptor, the S2 subunit is implicated in the fusion of the virus to the cell [4,7,8]. The virus internalization to the host cell occurs with the contribution of cellular proteases, such as cathepsins, transmembrane protease serine 2 (TMPRSS2) and human airway trypsin-like protease (HAT), as well as ACE-2 [9].
Viral glycoproteins are the main triggers of ER stress in the cell and bring about unfolded protein accumulation in the ER lumen, activating the UPR signaling pathway [10]. With the activated UPR pathway, the synthesis of GRP78 and other chaperone proteins increases, and protein kinase R-like ER kinase (PERK) protein and eIF2αSer51 protein are activated by autophosphorylation [11]. Subsequently, inflammation and apoptosis pathways are stimulated [12].
GRP78 protein is known to be involved in the intrusion of numerous viruses, such as Bat Coronavirus, Ebola Virus, MERS-CoV, Dengue Virus, Japanese Encephalitis Virus, Influenza Virus, and Zika Virus, into the host cell [13]. Previous research has demonstrated that MERS-CoV virus binds to GRP78 protein as well as dipeptidyl peptidase 4 (DPP4) protein for internalization to the host cell [13]. The fact that GRP78 protein expression is increased in SARS-CoV infection has also been documented in earlier studies [14].
The association between the SARS-CoV-2 virus and the GRP78 protein has been investigated in several studies, and the utility of this association has been proposed as a potential therapeutic target [[15], [16], [17]]. As molecular docking prediction studies reveal, if the virus is to enter the target cell expressing cell-surface GRP78, binding seems more convenient between Region IV of the SARS-CoV-2 spike protein and the GRP78 substrate binding domain (SBD). In our previous works, we reported that a marked increase emerges in serum GRP78 level in SARS-CoV-2 pneumonia, and that serum GRP78 mRNA levels rise substantially in Covid-19 pneumonia [16,17].
Within this context, our current study tries to provide some insight into the variation of GRP78 protein levels in patients with Covid-19 (−) pneumonia, Covid-19 (+) pneumonia, and CT negative Covid-19 infection in comparison to the normal population through a larger number of cases.
2. Material and methods
2.1. Study type
The present study is a prospective case-control study, and the required approval was obtained from the Ethics Committee of Pamukkale University prior to the study (60116787-020/26598 numbered).
2.1.1. Study population
The present study included the patients who were admitted between April 2020 and June 2020 to Covid-19 pandemic outpatient clinic of the emergency department (ED) with symptoms of upper respiratory tract infection and pneumonia, were asymptomatic, were established to be Covid-19 PCR (+) during contact tracing, and presented to the ED for further examination and treatment. After the required information concerning the study was provided both to the patient group and to the healthy control group, the written consent forms were obtained from all the subjects who agreed to participate in the study. The healthy volunteers with no known chronic or acute disease or drug use as well as no recent history of infection were included study as the control group. Once these subjects were assessed in accordance with the inclusion and exclusion criteria, they were divided into three groups as the Covid-19 (−) pneumonia group, the Covid-19 infection group, and the healthy control group.
2.1.2. Covid-19 (−) pneumonia group
This cohort consisted of the patients (a) who presented to the Covid-19 outpatient policlinic of the ED with pneumonia symptoms, (b) whose CT imagings were not compatible with Covid-19 pneumonia in accordance with the Radiological Society of North America Expert Consensus (RSNAEC) criteria [18], (c) whose nasopharyngeal swab samples taken in the ED were negative for PCR, and (d) who gave their informed consent to participate in the study.
2.1.3. Covid-19 infection group
This cohort included the patients (a) who presented to the Covid-19 outpatient policlinic of the ED with pneumonia symptoms, (b) whose CT imagings were compatible with Covid-19 pneumonia in accordance with the RSNAEC criteria and whose PCR tests were positive, (c) whose Covid-19 PCR tests were positive as a result of contact tracing, and (d) who presented to the ED for further examination.
2.1.4. Healthy group (control group)
This cohort involved the volunteers (a) who had no known acute, subacute or chronic disease history, (b) who did not suffer from any infection in the last fortnight, (c) who were not on a particular medication, (d) who presented to the ED with reasons other than infectious complaints, and (e) who gave their written consent to participate in the study.
The exclusion criteria consisted of diagnosis of kidney and liver failure, acute pulmonary embolism, chronic inflammatory disease history (rheumatological disease, autoimmune disease), pregnancy, presence of any cancer diagnosis, chronic obstructive pulmonary disease, asthma disease, and history of cerebrovascular disease. In addition, the patients whose CT imagings were compatible with Covid-19 pneumonia but whose PCR tests were negative were also excluded from the study.
2.2. CT evaluation and clinical evaluation
2.2.1. CT evaluation
Chest CT performed at the time of admission of the patients to the ED was assessed under the criteria of the Radiological Society of North America Expert Consensus by an emergency physician who followed up the patient clinically. The pneumonia cases were classified in line with these criteria and recorded in the clinical classification dataset [18].
2.2.2. Clinical evaluation
The clinical assessment of the subjects was performed in accordance with Covid-19 diagnosis and treatment guidelines of Ministry of Health [19]. As this guide was updated, the patient management algorithm was also edited. The Pneumonia Severity Index and CURB-65 scores of the subjects were calculated as suggested in the literature and then recorded in the dataset [20,21].
2.3. Blood samples and laboratory parameters
Complete blood count, C-reactive protein (CRP), creatinine, urea, d-dimer, and ferritin parameters, which are routinely checked during admission to the ED, were recorded in the dataset. For GRP78 level measurement, after 3 mL blood sample was taken into a dry tube and centrifuged at 4000 rpm for 10 min, its serum section was separated, and the GRP78 level was analyzed by the Enzyme-Linked Immunosorbent Assay (ELISA) method. In the control group, on the other hand, after 3 mL of blood was drawn into a dry tube, and another 3 mL of blood was placed into an EDTA tube, the GRP78 level was analyzed through the same methods in the same laboratory. The laboratory parameters of the blood samples requested from the patients in the ED for examination were recorded in the dataset.
2.4. Measurement of GRP78 levels
Serum GRP78 levels of the subjects were measured through a commercially available enzyme-linked immunosorbent assay (ELISA) kit (Human Glucose Regulated Protein 78 (GRP78) ELISA Kit, Sun Long, SL2048Hu, China), as per the manufacturer's protocol. The detection rate of this kit is 16 pg/mL.
2.5. Data analysis
As a result of the power analysis made in line with the presumptions since a similarly-organized reference study did not exist, at least 84 people (min. 28 for each cohort) were needed to achieve 90% power at 95% confidence interval, assuming that the projected effect size would be medium-high (f = 0.4). All the dataset obtained from the study were analyzed through SPSS package program. The continuous variables were presented as mean ± standard deviation and median (IQR), whereas the categorical variables were provided as numbers and percentages. A Kolmogorov-Smirnov test was run to calculate the parametric distribution of the continuous data. When the parametric test assumptions were not met, Kruskal-Wallis variance analysisis and Mann-Whitney U test were performed for the comparison of independent group differences. In addition, the relationships between continuous variables were analyzed using Spearman correlation analysis, chi-square test was used for analyzing categorical variables. The significance level was defined as p < 0.05 for all the analyses.
3. Results
A total of 846 patients presented to the Covid-19 outpatient clinic of the ED between April 2020 and June 2020. 67 of these patients were diagnosed with Covid-19 pneumonia and 65 patients with Covid-19 infection (other than pneumonia) and then hospitalized in the service.
52 subjects whose CT imagings scanned in the pandemic outpatient clinic were compatible with bronchopneumonia were established to be Covid-19 PCR-negative as a result of the nasal swab samples taken in the ED. Out of these 52 subjects, 10 were excluded due to the exclusion criteria, and 42 patients were eventually included in the Covid-19 (−) pneumonia group.
37 subjects hospitalized for Covid-19 pneumonia after being assessed (in CT and clinical terms) in the pandemic outpatient clinic were excluded from the study under the exclusion criteria. Of the 65 subjects diagnosed with Covid-19 negative as a result of their CT imaging, 16 were excluded as they could not be informed about the study due to the congestion in the ED, while 7 subjects were excluded since they had one of the exclusion criteria. A total of 72 patients (30 Covid-19 pneumonia; 42 CT-negative Covid-19 infection) were included in the Covid-19 infection group.
A statistical power analysis was run, based on the results we obtained in the study. The effect size found for the difference of serum GRP78 concentrations between the three groups was f = 0.6, considered to be medium-high using Cohen's criteria. Accordingly, the power obtained in the study for this effect size corresponded to 96.1% at 95% confidence level.
The Covid-19 (−) pneumonia and Covid-19 infection cohorts were compared to the healthy age- and sex-matched control group (p = 0.354 and p = 0.051, respectively). The median duration of symptoms in the Covid-19 (−) pneumonia and Covid-19 infection groups were 3 days (2–7) and 3.5 days (2–6), respectively, and the symptom durations of both cohorts were similar before admission to the ED (p = 0.867). However, fever values tended to be higher in the Covid-19 (−) pneumonia group (p = 0.037), while the sPO2 median value was lower in the same group (p = 0.006). On the other hand, no significant difference was evident between the systolic and diastolic pressure median values of the Covid-19 (−) pneumonia and Covid 19 infection groups (p = 0.709 and p = 0.601, respectively) (Table 1 ).
Table 1.
Clinical and demographical datas of the groups.
| Control group (N = 30) |
Covid-19 (−) pneumonia (N = 42) |
Covid-19 infection (N = 72) |
p-Value | ||
|---|---|---|---|---|---|
| Gender N (%) |
Male | 12 (40%) | 24 (57.1%) | 34 (50%) | 0.358 |
| Female | 18 (60%) | 18 (42.9%) | 34 (50%) | ||
| Control group (N = 30) |
Covid-19 (−) pneumonia (N = 42) |
Covid-19 infection (N = 72) |
p-Value | ||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
||
| Age | 45.86 ± 11.44 | 45.5 (38–55.25) |
51.83 ± 14.82 | 55.5 (40.25–63) |
44.98 ± 18.91 | 40 (31–58) |
⁎0.051 |
| Symptom onset (day) | 4.78 ± 4.09 | 3 (2–7) |
4.94 ± 5.03 | 3.5 (2–6) |
⁎0.867 | ||
| Fever (°C) | 36.58 ± 0.24 | 36.6 (36.6–36.75) |
37.1 ± 0.84 | 36.9 (36.57–37.62) |
36.8 ± 0.64 | 36.7 (36.5–37.07) |
⁎0.037 |
| sPO2 | 96.2 ± 2.17 | 96 (95–98) |
93.04 ± 7.23 | 95 (93–96) |
95.63 ± 4.09 | 97 (94.25–98) |
⁎0.006 |
| Systolic blood pressure (mm/Hg) | 129.6 ± 20.48 | 120 (112–150) |
125.1 ± 23.61 | 120 (110–140) |
126.1 ± 15.56 | 120 (115.25–140) |
⁎0.709 |
| Diastolic blood pressure (mm/Hg) | 78.65 ± 8.66 | 80 (70–85) |
76.14 ± 12.54 | 78 (70–80) |
76.39 ± 9.39 | 80 (70–80) |
⁎0.601 |
p values are derived from Kruskal Wallis test.
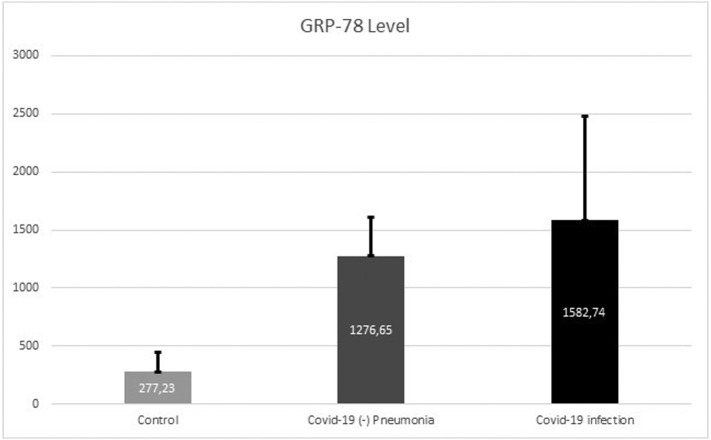
The median value for serum GRP78 level was 1429.14 pg/mL (1167.43–1710.3) in the Covid-19 infection group, 1283.13 pg/mL (1018.05–1536.62) in the Covid-19 (−) pneumonia group, and 258.5 pg/mL (153.5–353.75) in the control group. The GRP78 level was found to be significantly higher in the Covid-19 infection group than both Covid-19 (−) pneumonia and control group (p = 0.031 and p = 0.0001, respectively). In addition, the GRP78 levels were significantly higher in the Covid-19 (−) pneumonia group than the control group (p = 0.0001) (Table 2 ) (Fig. 1 ).
Table 2.
Serum GRP-78 levels of the groups.
| Control group (N = 30) |
Covid-19 (−) pneumonia (N = 42) |
Covid-19 infection (N = 72) |
p-Value | ||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
||
| GRP-78 (pg/mL) | 277.23 ± 167.5 | 258.5 (153.5–353.75) |
1276.65 ± 331.65 | 1283.13 (1018.05–1536.62) |
1582.74 ± 900 | 1429.14 (1167.43–1710.3) |
⁎p = 0.0001 ⁎⁎p = 0.031 ⁎⁎⁎p = 0.0001 |
GRP78, Glucose Regulated Protein.
p-Value derived from Kruskal-Wallis test and refers to the comparison between covid-19 infection, broncopneumonia and control group.
p-Value is derived from Kruskal-Wallis test, and refers to the comparison between Covid-19 (−) pneumonia and Covid-19 infection groups.
p-Value is derived from Kruskal-Wallis test and refers to the comparison between Control and Covid-19 (−) pneumonia groups.
Fig. 1.
Serum GRP-78 levels of the groups.
WBC count, neutrophil count, and serum CRP level were observed to be lower in Covid-19 infection group than the Covid-19 (−) pneumonia group (p = 0.002; p = 0.0001; p = 0.0001). However, no significant difference was noted between these groups in terms of serum urea, creatinine, d-dimer, ferritin and hsTnT levels (p = 0.101; p = 0.163; p = 0.128; p = 0.143 and p = 0.118, respectively) (Table 3 ).
Table 3.
Laboratory parameters of the groups.
| Control group (N = 30) |
Covid-19 (−) pneumonia (N = 42) |
Covid-19 infection (N = 72) |
p Value | ||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
||
| WBC (K/μL) | 7.89 ± 2.18 | 7.59 (6.29–9.48) |
10.38 ± 5.39 | 9.11 (6.95–13.01) |
7.71 ± 4.08 | 6.46 (4.78–9.34) |
1p = 0.0003 2p = 0.002 3p = 0.354 |
| Hemoglobin (g/dl) | 13.8 ± 1.6 | 13.55 (12.74–14.97) |
13.43 ± 2.39 | 13.95 (11.67–15.2) |
14.1 ± 2.16 | 14.3 (12.69–15.7) |
1p = 0.41 |
| Neutrophil count (K/μL) | 8.12 ± 5.26 | 6.47 (4.53–10.76) |
5.07 ± 3.59 | 3.64 (2.86–6.51) |
4p = 0.0001 | ||
| Lymphocyte count (K/μL) | 1.55 ± 0.75 | 1.36 (1.07–1.97) |
1.93 ± 0.97 | 1.82 (1.3–2.54) |
4p = 0.035 | ||
| Platelet count (K/μL) | 264.6 ± 59.69 | 264 (222–292.25) |
242.09 ± 78.68 | 234 (169–303.5) |
239.76 ± 70.6 | 239.5 (192.5–290.25) |
1p = 0.293 |
| C-reactive protein (mg/L) | 5.98 ± 17 | 1.75 (1.05–3.72) |
75.01 ± 90.43 | 38.8 (14.37–105.93) |
28.91 ± 57.05 | 3.37 (1.34–29.82) |
1p = 0.0001 2p = 0.0001 3p = 0.0001 |
| Urea (mg/dl) | 19.18 ± 5.45 | 18.2 (15.4–22.4) |
40.76 ± 26.52 | 31 (23–46.25) |
30.92 ± 19.59 | 26.5 (20.25–33.75) |
1p = 0.0001 |
| Creatinine (mg/dl) | 0.78 ± 0.13 | 0.79 (0.66–0.91) |
1.1 ± 1.39 | 0.85 (0.7–1.05) |
0.91 ± 0.36 | 0.86 (0.64–1.03) |
1p = 0.163 |
| D-dimer (ng/mL) | 419.72 ± 403.98 | 272 (148.5–536.5) |
429.49 ± 590.2 | 198 (82.5–491) |
4p = 0.128 | ||
| Ferritin (μg/L) | 367.2 ± 516.3 | 194 (44–352.2) |
213.26 ± 289.8 | 127.9 (35.2–237.5) |
4p = 0.143 | ||
| hsTnT (μg/L) | 6.93 ± 5.47 | 5 (4–7.25) |
96.27 ± 434.3 | 10.85 (3–28.22) |
9.23 ± 13.63 | 5 (3–9.11) |
1p = 0.118 |
WBC, white blood cell; hsTnT, high sensitive troponin T.
p-Value derived from Kruskal-Wallis test and refers to the comparison between covid-19 infection, Covid-19 (−) pneumonia and control group.
p-Value is derived from Kruskal-Wallis test, and refers to the comparison between Covid-19 (−) pneumonia and Covid-19 infection subgroups.
p-Value is derived from Kruskal-Wallis test and refers to the comparison between Control and Covid-19 (−) pneumonia subgroups.
p-Value is derived from Mann Whitney U test and refers to the comparison between Covid-19 (−) pneumonia and Covid-19 infection subgroups.
When the Covid-19 infection group was divided into subgroups as Covid-19 (+) pneumonia and CT negative pneumonia, no significant difference was evident between Covid-19 (−) pneumonia, Covid-19 (+) pneumonia and CT negative Covid 19 infection groups with respect to GRP78 levels (p = 0.09). However, serum GRP78 levels were significantly higher in these subgroups in comparison to the control group (p = 0.0001). The median value of PSI and CURB-65 scores of the subjects diagnosed with pneumonia remained close to each other in Covid-19 (−) and (+) pneumonia groups (p = 0.803 and p = 0.942 respectively) (Table 4 ).
Table 4.
Clinical and demographical datas of the groups.
| Covid-19 (−) pneumonia (N = 42) |
Covid-19 pneumonia (N = 30) |
CT negative Covid-19 infection (N = 42) |
p-Value | ||||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
Mean ± SD | Median (IQR) |
||
| GRP-78 | 1276.65 ± 331.65 | 1283.13 (1018,05–1536.62) |
1462.13 ± 489 | 1474.86 (1002–1784.71) |
1662.18 ± 1088.6 | 1420.7 (1269.9–1594.5) |
1p = 0.09 2p = 0.517 3p = 0.656 |
| CURB-65 score | 1.29 ± 1 | 1 (1–2) | 1.32 ± 0.98 | 1 (0.5–2) | 2p = 0.942 | ||
| PSI score | 79.17 ± 42.52 | 80.5 (37.75–123.5) | 75.72 ± 37.96 | 68 (45.75–111.75) | 2p = 0.803 | ||
| Symptom duration (Day) |
4.78 ± 4.09 | 3 (2–7) | 6 ± 6.03 | 4 (2.5–6.5) | 4.03 ± 3.93 | 2 (2–5) | 2p = 0.177 |
GRP78, Glucose Regulated Protein; PSI, pneumonia severity index, CT, computed tomography.
p-Value derived from Kruskal-Wallis test and refers to the comparison between both subgroups.
p-Value is derived from Mann Whitney U test, and refers to the comparison between Covid-19 (−) pneumonia and Covid-19 pneumonia subgroups.
p-Value is derived from Mann Whitney U test and refers to the comparison between CT Negative Covid 19 Infection and Covid19 pneumonia subgroups.
As far as the relationship of serum GRP78 levels with laboratory and clinical parameters is concerned, a significant and weak-positive correlation was established between serum CRP level and serum GRP78 levels (r = 0.223 and p = 0.008). In a similar vein, a significant and weak-positive correlation was noted between serum urea levels and GRP78 levels (r = 0.31 and p = 0.0001) (Table 5 ).
Table 5.
Correlations between GRP-78 level and laboratory and clinical parameters.
| GRP-78 | ||
|---|---|---|
| CURB-65 score | Rho | 0,012 |
| p value | 0,921 | |
| PSI score | Rho | 0,039 |
| p value | 0,730 | |
| Fever | Rho | −0,016 |
| p value | 0,868 | |
| sPO2 | Rho | −0,017 |
| p value | 0,857 | |
| Systolic blood pressure | Rho | −0,066 |
| p value | 0,493 | |
| Diastolic blood pressure | Rho | 0,043 |
| p value | 0,652 | |
| WBC count | Rho | 0,065 |
| p value | 0,446 | |
| Hemoglobin | Rho | 0,006 |
| p value | 0,947 | |
| Netrophil count | Rho | 0,017 |
| p value | 0,857 | |
| Lymphocyte count | Rho | 0,0001 |
| p value | 0,999 | |
| Platelet | Rho | −0,056 |
| p value | 0,508 | |
| C-reactive protein | Rho | 0,223 |
| p value | 0,008 | |
| Urea | Rho | 0,310 |
| p value | 0,0001 | |
| Creatinine | Rho | 0,080 |
| p value | 0,346 | |
| D-dimer | Rho | −0,012 |
| p value | 0,906 | |
| Ferritin | Rho | −0,060 |
| p value | 0,582 | |
| hsTnT | Rho | −0,010 |
| p value | 0,915 | |
| CT severity score | Rho | −0,095 |
| p value | 0,439 | |
| Symptom duration | Rho | −0,148 |
| p value | ,150 | |
p and rho values are derived from Spearman Correlations. GRP-78, Glucose Regulated Protein; PSI, pneumonia severity index; WBC, white blood cell; hsTnT, high sensitive troponin T; CT, computed tomography.
4. Discussion
Although a range of options is currently available in the treatment of Covid-19 infection, efforts towards new therapeutic targets are underway [22]. The effectiveness of multiple mechanisms and drugs against Covid-19 infection is tested through repurposing studies, and researchers attempt to come up with potential promising drugs [23].
GRP78, one of the ER stress markers previously studied in conditions such as heart failure [24], cardiac arrest [25], hepatitis infection [26], is known to play a key role, especially in coronavirus infections. Previous research has documented that, when ER stress occurs in viral infections, GRP78 is overexpressed and translocated from the ER to the cell membrane, and that it is also involved in the internalization of numerous viruses, such as Bat Coronavirus, MERS-CoV, Ebola Virus, Dengue Virus, Japanese Encephalitis Virus, Influenza Virus, and Zika Virus, to the host cell. In addition, it is well-established that GRP78 expression tends to increase in SARS-CoV infection [13,14]. Versteeg et al. conclude that MHV and SARS-CoV infections induce ER stress [10].
A recent molecular docking study has revealed that the spike protein of the SARS-CoV-2 virus has a tight binding affinity with the Region IV of substrate binding domain (SBD β) and the GRP78 protein, suggesting that this association might be considered as one of the therapeutic targets. This study also concludes that the SARS-CoV-2 virus is associated with the GRP78 protein during its internalization to the host cell, as in other coronaviruses [15]. In our previous trials with smaller populations, we demonstrated that the GRP78 protein levels and mRNA levels were also increased in the serum during Covid-19 infection [16,17]. Likewise, Palmeira et al. established GRP78 mRNA levels to be significantly higher in patients with SARS-CoV-2 pneumonia than the control group and the SARS-CoV-2 (−) pneumonia group. In addition, a docking study on GRP78 inhibitors and GRP78 protein was conducted within this study, based on the relationship between the SARS-CoV-2 spike protein and GRP78 protein, and it was further noted that Imatinib, a BCR-ABL inhibitor, turned out to present the highest affinity [17]. Similarly, a study on cell culture conducted by Dyall et al. identifies Imatinib as a promising antiviral agent in inhibiting MERS-CoV and SARS-CoV viruses [27].
In this study carried out with a larger population, we have corroborated the prediction that GRP78 levels increase in Covid-19 infection. Indeed, the fact that we identified the serum GRP78 level to be higher in the Covid-19 infection group than both the control and Covid-19 (−) pneumonia group points to the association between Covid-19 infection and GRP78 protein. Besides, the fact that the GRP78 levels were approximately 5 times higher in Covid-19 sufferers than the healthy control group is particularly noteworthy in shedding light on the ER stress that occurs during Covid-19 infection.
Endothelial dysfunction is known to emerge in the lung tissue in conditions, such as pneumonia and ARDS [28]. It is also known that HSP70 proteins are released into the blood as a result of endothelial dysfunction [29]. The GRP78 protein, also a member of the HSP70 protein family, was found higher in the Covid-19 (−) pneumonia group than the control group in our study, leading us to conclude that the increased serum GRP78 levels may stem from endothelial dysfunction in the lung tissue. The lack of significant difference between the GRP78 levels of the Covid-19 (−) pneumonia and Covid-19 (+) pneumonia groups might be attributed to the similar levels of PSI and CURB-65 scores, which indicate the clinical conditions of the patients in the groups.
The higher serum GRP78 levels in the CT negative Covid-19 infection subgroup than the control group indicates that the GRP78 levels are likely to rise substantially even without pneumonia caused by Covid-19 infection. This suggests that the SARS-CoV-2 virus tends to increase ER stress before ending up with pneumonia as well as leading to an increase in the expression of the GRP78 protein, which facilitates internalization of the virus to the host cell. Further to this, the significant positive correlation, albeit at a weak degree, between CRP and GRP78 levels in our study seems to support the association between the GRP78 protein and the presence of infection. Although this study alone may not suffice to arrive at a definite decision on this issue, more comprehensive information will be obtained through future studies to be conducted on cell culture and at the tissue level. A recent study has stated that GRP78 chaperone protein stands out as an important target in coronavirus infections [30]. Another study has stated importance of GRP-78 protein in cross vaccination against Covid-19 infection [31]. Our study revealed the clinical importance of the GRP-78 protein, which was previously emphasized for Covid-19 infection.
A range of limitations may have restricted the generalizability of our present results. For one thing, not taking the lung tissue samples from the Covid-19 patients as well as not studying the GRP78 levels from the tissue samples can be considered as important limitations, but we preferred to study the samples from peripheral blood due to the risk of transmission of the disease. Additionally, literature falls short of addressing the time duration when GRP78 levels increase in in-vivo settings.
5. Conclusion
This prospective case-control study reveals that the serum GRP78 levels significantly increased during Covid-19 infection in comparison to both the Covid-19 (−) pneumonia and the control group. We hypothesize that the significantly higher rate of GRP78 also in the CT negative Covid-19 infection cohort than the control group might increase ER stress even before SARS-CoV-2 virus induces pneumonia and may bring about an increase in the expression of GRP78 protein, which mediates its entry in the host cell. We are also of the opinion that these assumptions need to be validated with further studies on cell culture, as in the studies on MERS-CoV and SARS-CoV viruses, to obtain more comprehensive information. As the association between SARS-CoV-2 virus and GRP78 protein is revealed more clearly, this association may come to the fore as a therapeutic target.
Acknowledgment
The authors declare that they have no conflicts of interests.
There is no funding statement for this study.
References
- 1.CDC CDC; 2019. Novel coronavirus, Wuhan, China. https://www.cdc.gov/coronavirus/2019-ncov/about/index.html Available at.
- 2.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls [Internet] StatPearls Publishing; Treasure Island (FL): 2020 Jan. Features, evaluation, and treatment of coronavirus (COVID-19) [updated 2020 Aug 10]https://www.ncbi.nlm.nih.gov/books/NBK554776/ Available from: [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;3:1–9. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada Y., Liu X.B., Fang S.G., Tay F.P., Liu D.X. Acquisition of cell-cell fusion activity by amino acid substitutions in spike protein determines the infectivity of a coronavirus in cultured cells. PLoS One. 2009;4:e6130. doi: 10.1371/journal.pone.0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shereen M.A., Khan S., Kazmi A., et al. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;16(24):91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Versteeg G.A., van de Nes P.S., Bredenbeek P.J., Spaan W.J.M. The coronavirus spike protein induces endoplasmic reticulum stress and Upregulation of intracellular chemokine mRNA concentrations. J. Virol. 2007;81:10981–10990. doi: 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Kwak D., Lu Z., et al. Endoplasmic reticulum stress sensor protein kinase R-like endoplasmic reticulum kinase (PERK) protects against pressure overload-induced heart failure and lung remodeling. Hypertension. 2014;64:738–744. doi: 10.1161/HYPERTENSIONAHA.114.03811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang K. Endoplasmic reticulum stress response and transcriptional reprogramming. Front. Genet. 2015;5:460. doi: 10.3389/fgene.2014.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu H., Chan C.M., Zhang X., Wang Y., Yuan S., Zhou J., et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018;293(30):11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan C.P., Siu K.L., Chin K.T., Yuen K.Y., Zheng B., Jin D.Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006;80(18):9279–9287. doi: 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., et al. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Inf. Secur. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Köseler A., Sabirli R., Gören T., et al. Endoplasmic reticulum stress markers in SARS-COV-2 infection and pneumonia: case-control study. In Vivo. 2020;34:1645–1650. doi: 10.21873/invivo.11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmeira A., Sousa E., Köseler A., et al. Preliminary virtual screening studies to identify GRP78 inhibitors which may interfere with SARS-CoV-2 infection. Pharmaceuticals (Basel) 2020;13:132. doi: 10.3390/ph13060132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson S., Kay F.U., Abbara S., et al. Radiological Society of North America Expert Consensus Statement on Reporting Chest CT Findings Related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiol. Cardiothoracic Imaging. 2020;2:2. doi: 10.1148/ryct.2020200152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turkish Ministry of Health Covid-19 diasnosis and treatment guideline. https://covid19bilgi.saglik.gov.tr/depo/rehberler/covid-19-rehberi/COVID-19_REHBERI_ERISKIN_HASTA_TEDAVISI.pdf Accessed at.
- 20.Lim W.S., van der Eerden M.M., Laing R., et al. Defining CAP severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–382. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioachimescu O.C., Ioachimescu A.G., Iannini P.B. Severity scoring in community-acquired pneumonia caused by Streptococcus pneumoniae: a 5-year experience. Int. J. Antimicrob. Agents. 2004;24:485–490. doi: 10.1016/j.ijantimicag.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 22.COVID-19 therapeutics tracker. https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-therapeutics-tracker Accessed at.
- 23.Ulm J.W., Nelson S.F. COVID-19 drug repurposing: summary statistics on current clinical trials and promising untested candidates. Transbound. Emerg. Dis. 2020;3 doi: 10.1111/tbed.13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabirli R., Koseler A., Mansur N., et al. Predictive value of endoplasmic reticulum stress markers in low ejection fractional heart failure. In Vivo. 2019;33:1581–1592. doi: 10.21873/invivo.11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ardic S., Yilmaz S., Demir S., et al. Endoplasmic reticulum stress markers are of no value in predicting cardiopulmonary resuscitation success and survival in out-of hospital cardiac arrest: a nested case-control study. Turk. J. Emerg. Med. 2019;19:58–63. doi: 10.1016/j.tjem.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirrik S., Çetinkol Y., Altunçekiç Yildirim A., Çalgin M.K., Noyan T. Circulating glucose-regulated protein 78 levels in patients with chronic hepatitis B infection. Viral. Hepat. J. 2018;24:85–89. [Google Scholar]
- 27.Dyall J., Coleman C.M., Hart B.J., et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller-Redetzky H.C., Suttorp N., Witzenrath M. Dynamics of pulmonary endothelial barrier function in acute inflammation: mechanisms and therapeutic perspectives. Cell Tissue Res. 2014;355(3):657–673. doi: 10.1007/s00441-014-1821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barabutis N. Unfolded protein response in acute respiratory distress syndrome. Lung. 2019;197:827–828. doi: 10.1007/s00408-019-00279-4. [DOI] [PubMed] [Google Scholar]
- 30.Elfiky A.A., Baghdady A.M., Ali S.A., et al. GRP78 targeting: hitting two birds with a stone. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elfiky A.A., Ibrahim I.M., Ismail A.M., Elshemey W.M. A possible role for GRP78 in cross vaccination against COVID-19 [published online ahead of print, 2020 Sep 10] J. Inf. Secur. 2020;30598-3:S0163–S4453. doi: 10.1016/j.jinf.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]