Abstract
The emergence of Coronavirus Disease 2019 (COVID-19) in early December 2019 has caused immense damage to health and global well-being. Currently, there are approximately five million confirmed cases and the novel virus is still spreading rapidly all over the world. Many hospitals across the globe are not yet equipped with an adequate amount of testing kits and the manual Reverse Transcription-Polymerase Chain Reaction (RT-PCR) test is time-consuming and troublesome. It is hence very important to design an automated and early diagnosis system which can provide fast decision and greatly reduce the diagnosis error. The chest X-ray images along with emerging Artificial Intelligence (AI) methodologies, in particular Deep Learning (DL) algorithms have recently become a worthy choice for early COVID-19 screening. This paper proposes a DL assisted automated method using X-ray images for early diagnosis of COVID-19 infection. We evaluate the effectiveness of eight pre-trained Convolutional Neural Network (CNN) models such as AlexNet, VGG-16, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50 and Inception-V3 for classification of COVID-19 from normal cases. Also, comparative analyses have been made among these models by considering several important factors such as batch size, learning rate, number of epochs, and type of optimizers with an aim to find the best suited model. The models have been validated on publicly available chest X-ray images and the best performance is obtained by ResNet-34 with an accuracy of 98.33%. This study will be useful for researchers to think for the design of more effective CNN based models for early COVID-19 detection.
Keywords: COVID-19, SARS-CoV-2, Optimization algorithms, Convolutional Neural Networks, Chest X-ray
Highlights
-
•
A deep learning assisted automated method is proposed for COVID-19 diagnosis.
-
•
A comprehensive study among eight pre-trained CNN models is performed.
-
•
The impact of several hyper-parameters have been analyzed.
-
•
The best performing model is obtained and compared with the state-of-the-art methods.
-
•
The model could assist radiologists for accurate and stable COVID-19 diagnosis.
1. Introduction
The outbreak of Coronavirus Disease 2019 (COVID-19) has put the globe under tremendous pressure since early December 2019. To date, more than five million people across the globe have been infected, and there are approximately three lakhs confirmed death cases as reported by the World Health Organization (WHO) [1]. It is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and the common symptoms of COVID-19 include fever, myalgia, dry cough, headache, sore throat, and chest pain [2], [3] and therefore, it is considered as a respiratory disease. It can take around 14 days to show complete symptoms in the infected person. Currently, there is no explicit treatment or drug available to heal this disease. However, the utmost common practice used in the diagnosis of COVID-19 is called Reverse Transcription-Polymerase Chain Reaction (RT-PCR) [4], [5]. Recently, it has been found that medical imaging techniques such as X-ray and Computed Tomography (CT) play a crucial role in testing COVID-19 cases [4], [6], [7], [8]. Since the virus generally causes infection in the lungs, the chest radiography images (chest X-ray or CT images) have been widely considered [9] and the interpretation of these images is manually performed by radiologists to find some visual indicators for COVID-19 infection. These visual indicators can be served as an alternative method for the rapid screening of infected patients.
The conventional diagnosis process has become relatively faster but still causes a high risk for medical staff. Moreover, it is costly and there exists a limited number of diagnostic test kits. On the other hand, medical imaging techniques (e.g., X-ray and CT) based screening are relatively safe, faster, and easily accessible. Compared to CT imaging, X-ray imaging has been extensively used for COVID-19 screening as it requires less imaging time, lower cost, and X-ray scanners are widely available even in rural areas [7], [10]. However, the visual inspection of X-ray images by radiologists at a larger scale is time-consuming, cumbersome, and may lead to inaccurate diagnosis due to lack of prior knowledge about the virus-infected regions. Thus, there is a strong need for design of automated methods to obtain a faster and accurate COVID-19 diagnosis. The recent automated methods used contemporary Artificial Intelligence (AI) technologies (mostly Deep Learning (DL) algorithms) to enhance the power of X-ray imaging and aimed to reduce the workload of radiologists [4]. The DL models, in particular, Convolutional Neural Networks (CNN) have shown to be effective than traditional AI methods and have been widely used for analyzing several medical images [11], [12], [13], [14], [15]. Recently, CNN has been successfully applied to detect pneumonia in chest X-ray images [16], [17], [18], [19].
Recently, studies have been conducted using DL models related to the diagnosis of COVID-19 through X-ray images. For instance, Ozturk et al. [20] developed a DL network termed as DarkCovidNet based on X-ray images for automated COVID-19 diagnosis. The model achieved a higher accuracy of 87.02% and 98.08% for multi-class (COVID-19, normal, and pneumonia) and two-class (COVID-19 and normal) cases. Hemdan et al. [21] developed a COVIDX-Net model considering X-ray images. Seven different CNN models have been used to train the COVIDX-Net model and the model is validated on 50 X-ray images (25 normal and 25 COVID-19 cases). Wang and Wong [22] designed a deep CNN model referred to as COVID-Net for COVID-19 detection which obtained a testing accuracy of 92.6%. Apostolopoulos et al. [23] evaluated a set of existing CNN models for classification of COVID-19 cases and obtained the highest testing accuracy of 93.48% and 98.75% for three and binary classes. Narin et al. [24] yielded testing accuracy of 98% on a dataset of 100 images (50 normal and 50 COVID-19 cases) using the ResNet50 model. Sethy and Behera [25] obtained the features from different pre-trained CNN architectures using chest X-ray images. The ResNet50 coupled with Support Vector Machine (SVM) classifier achieved the highest accuracy of 95.38% using 50 samples (25 normal and 25 COVID-19 cases). Ucar and Korkmaz [26] suggested a COVIDiagnosis-Net model using SqueezeNet and Bayesian optimizer to achieve a testing accuracy of 98.3% over three-class classification cases. Toğaçar et al. [27] designed an automated method for classification of COVID-19 cases from normal and pneumonia cases using two DL models (MobileNetV2 and SqueezeNet) and SVM classifier. In their study, the original dataset was reconstructed using Fuzzy Color and Stacking techniques prior to the application of DL models. The features obtained by the DL models were further processed using Social Mimic Optimization (SMO) algorithm to derive efficient features and finally SVM was applied on the combined feature set. Recently, Farooq and Hafeez [28] built a ResNet baed CNN model named as COVID-ResNet for classification of COVID-19 and three other cases (normal, bacterial pneumonia and viral pneumonia). They obtained an accuracy of 96.23% over a publicly available dataset (i.e., COVIDx); however, only 68 COVID-19 samples were considered in this study. The major challenge lies while applying DL models is to collect an adequate number of samples with proper annotations for effective training. The literature reveals that the earlier models are validated using a fewer number of samples and the data in most cases are imbalanced. The pre-trained models adopted for the classification of COVID-19 cases are not fully investigated empirically. Furthermore, a comprehensive comparative study among the pre-trained CNN models has not yet reported. Hence, there exists a scope to perform an empirical study of different pre-trained CNN models to obtain the best performing model that can be used as an alternative tool for the rapid diagnosis of COVID-19.
In this study, we proposed a DL empowered automated COVID-19 screening method using chest X-ray images. Eight most successful pre-trained CNN models namely VGG-16, AlexNet, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50 and Inception-V3 have been taken into consideration based on the concept of Transfer Learning (TL). A comparative analysis has been made among all these models by considering several factors such as batch size, learning rate, number of epochs, misclassification rate, and type of optimization techniques, and finally, the best performing model has been obtained and compared with the state-of-the-art methods. The models have been validated using a larger number of chest X-ray images collected from covid-chestxray-dataset [29] and ChestX-ray8 [30] datasets. To mitigate the data scarcity and data imbalance problem, a multi-scale offline augmentation technique followed by data balancing and normalization has been adopted. A comparison of the proposed model with other state-of-the-art methods has been made in the context of COVID-19 class sensitivity. The suggested model for COVID-19 diagnosis is easy to implement, follows an end to end architecture without the need for manual feature engineering and could assist radiologists for accurate and stable diagnosis of COVID-19 infection.
The major contributions of this proposed research can be outlined as follows:
-
•
The effectiveness of the eight most efficient pre-trained deep CNN models, namely, VGG-16, Inception-V3, ResNet-34, MobileNet-V2, AlexNet, GoogleNet, ResNet-50, and SqueezeNet have been compared comprehensively. The impact of several hyper- parameters such as learning rate, batch size, number of epochs, and optimization techniques have been studied. Eventually, the best model is derived that will be useful for the researchers to design a more efficient CNN based solution for early detection of COVID-19 infection.
-
•
The data in the publicly available datasets are limited and imbalanced. To overcome this, we performed multi-operation data augmentation while balancing the samples for both COVID-19 and normal classes.
The remainder of the paper is structured as follows. Section 2 gives a description of the samples used for validation of different CNN models and describes the proposed automated model for COVID-19 detection. The results and comparisons are presented in Section 3. The concluding remarks are outlined in Section 4.
2. Materials and method
In this section, we present a detailed description of the proposed methodology designed for COVID-19 detection and the data used for the validation of the proposed model.
2.1. Dataset
To validate the proposed method, the chest X-ray images have been collected from two different sources. The dataset (covid-chestxray-dataset [29]) prepared by Cohen JP has been used to collect the COVID-19 X-ray images that considers images from various open sources and has been updated regularly. While chest X-ray images of normal category have been collected from a GitHub repository1 which contains 500 images selected from ChestX-ray8 dataset [30] In our experiment, 203 COVID-19 frontal-view chest X-ray images have been selected from covid-chestxray-dataset. To deal with the data imbalance problem, the same 203 frontal-view chest X-ray images of healthy lungs have been randomly selected from [30]. The example of chest X-ray images from both normal and COVID-19 classes are depicted in Fig. 1. Since the information about the actual data split was not provided in the datasets, 70% of the data was used for training and remaining 30% for testing which resulted in 286 images (143 COVID-19 and 143 normal cases) in the training set and 120 images (60 COVID-19 and 60 normal cases) in the test set.
Fig. 1.
Samples of frontal-view chest X-ray images from the dataset: (a) COVID-19 case and (b) normal case.
2.2. Proposed methodology
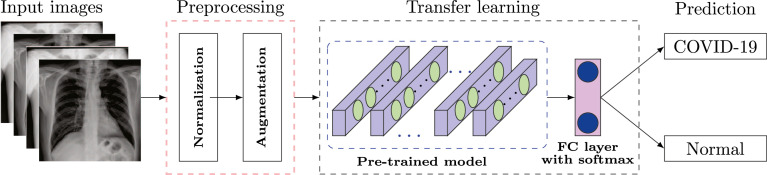
The proposed model for automated diagnosis of COVID-19 cases is illustrated in Fig. 2. The model aims to classify a given chest X-ray image into normal or COVID-19 category which includes two vital stages: preprocessing (normalization and augmentation) and classification using pre-trained CNN architectures. The description of each stage is detailed in the subsequent sections.
Fig. 2.
Overview of the proposed automated COVID-19 diagnosis method using frontal-view chest X-ray images.
2.2.1. Preprocessing
This section gives a detailed description of the methods used at the preprocessing stage.
Normalization Normalization of data is an essential step and is generally used to maintain numerical stability in the CNN architectures. With normalization, a CNN model is likely to learn faster and the gradient descent is more likely to be stable [31]. Therefore, in this study, the pixel values of the input images have been normalized in between the range 0–1. The images used in the considered datasets are gray-scale images and the rescaling was achieved by multiplying 1/255 with the pixel values.
Data Augmentation The CNN models require a vast amount of data for effective training and have shown to perform better on bigger datasets [11], [13]. However, the available training X-ray images in the considered dataset is very less (i.e., 286 X-ray images). This has been a major concern while performing analysis of medical images using DL algorithms since it is hard to collect medical data. To handle this problem, data augmentation technique has been widely employed which helps in expanding the number of images using a set of transformations while preserving class labels. Augmentation also increases variability in the images and serves as a regularizer at the dataset level [32]. The techniques adopted in this study for augmenting the training images are illustrated in Fig. 3.
Fig. 3.
Illustration of different data augmentation techniques used in this study.
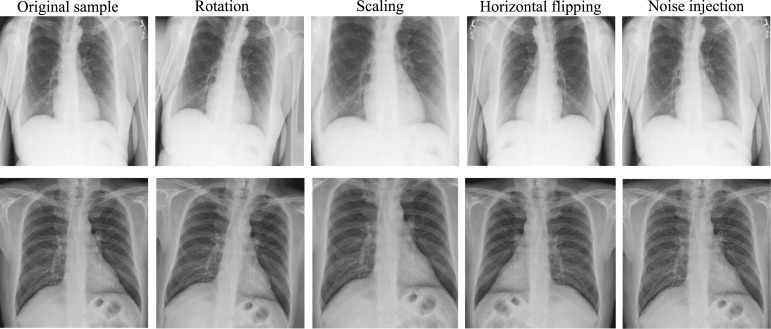
The images were augmented using the following techniques: (1) images were rotated by the angles of 5 degrees clockwise, (2) images were scaled by a measure of 15%, (3) images were performed horizontal flipping, and (4) images were added Gaussian noise with a zero mean and variance 0.25. It is worth noting that all these techniques have been applied over the training samples and the example results of each technique are depicted in Fig. 4. Finally, a larger training set containing 1430 images was obtained which is 5 times more than the original training images.
Fig. 4.
Sample results of data augmentation.
2.2.2. COVID-19 prediction using pre-trained CNN models
The CNN models have been proved to obtain superior results in a wide range of medical image processing applications. However, training these models from scratch would be difficult for prediction of COVID-19 cases due to the limited availability of X-ray samples. The application of pre-trained models using the concept of Transfer Learning (TL) can be useful in such circumstances. In TL, the knowledge gained by a DL model trained from a large dataset is used to solve a related task with a comparatively smaller dataset. This helps in eliminating the need for a large dataset and longer learning time as required by DL methods that are trained from scratch [11], [32], [33]. In this study, eight pre-trained models such as AlexNet [34], VGG-16 [35], GoogleNet [36], MobileNet-V2 [37], SqueezeNet [38], ResNet-34 [39], ResNet-50 [39], and Inception-V3 [40] have been used for classification of COVID-19 from normal cases. These networks have been achieved dramatic success in a wide range of computer vision and medical image analysis problems and hence were chosen in this study to distinguish COVID-19 infection from normal cases. It is worth noting here that these models were originally trained on a large-scale labeled dataset called ImageNet [34] and later fine-tuned over the chest X-ray images. The last layer in these models has been removed and a new Fully Connected (FC) layer is inserted with an output size of two that represents two different classes (normal and COVID-19). In these resulted models, only the final FC layer is trained, whereas other layers are initialized with pre-trained weights. The hyperparameters play a key role for tuning these DL models and were kept constant to derive a fair comparison. The details of the parameter settings are discussed in Section 3.1.
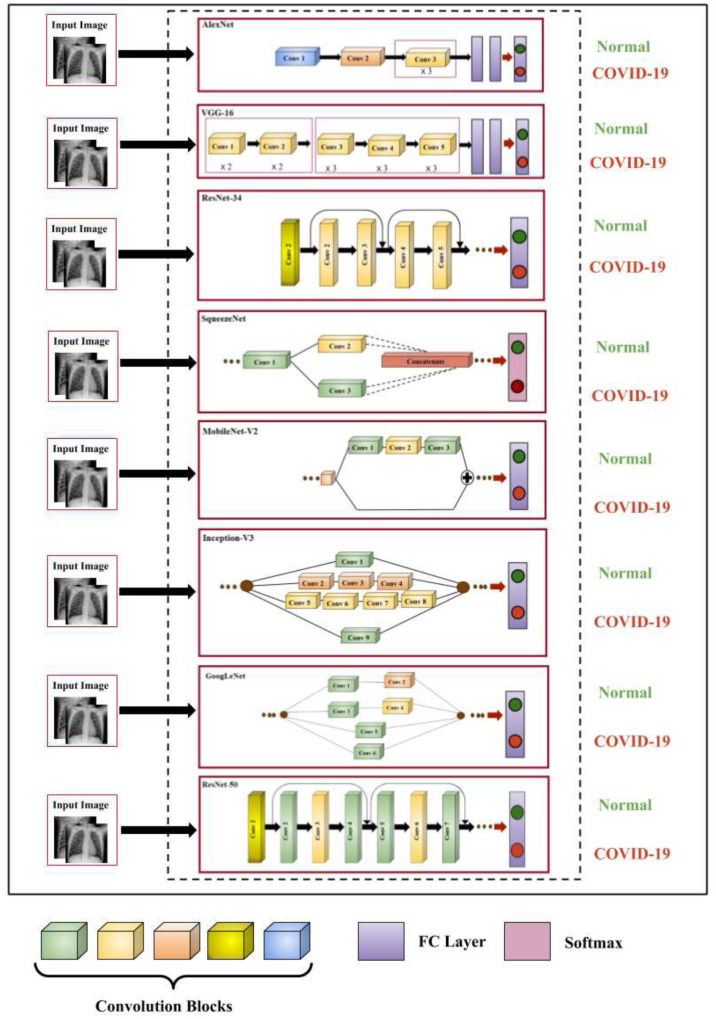
The architectural overview of the pre-trained CNN models is tabulated in Table 1 and the major components of each network are illustrated in Fig. 5. AlexNet comprises of five convolutional layers and three FC layers and was trained over 1.2 million images from 1000 categories [34]. ReLU activation is used in this network. The first and second FC layers have 4096 neurons, whereas the final FC layer has 1000 neurons. In VGG-16 network, there is an increased number of layers i.e., 13 convolutional layers and three FC layers [35]. The filters in this network are restricted to 3 × 3 with stride and padding of 1. The model was trained over a million images of 1000 categories. SqueezeNet executes better performance than AlexNet with comparatively fewer parameters [38]. It has one convolution layer at the beginning and the end and has eight fire modules in between.
Table 1.
Architectural descriptions of the pre-trained CNN models used in this study.
| Model | Layers | Parameters (in million) | Input layer size | Output layer size |
|---|---|---|---|---|
| AlexNet | 8 | 60 | (224,224,3) | (2,1) |
| VGG-16 | 16 | 138 | (224,224,3) | (2,1) |
| GoogleNet | 22 | 5 | (224,224,3) | (2,1) |
| MobileNet-V2 | 53 | 3.4 | (224,224,3) | (2,1) |
| SqueezeNet | 18 | 1.25 | (224,224,3) | (2,1) |
| ResNet-34 | 34 | 21.8 | (224,224,3) | (2,1) |
| ResNet-50 | 50 | 25.6 | (224,224,3) | (2,1) |
| Inception-V3 | 42 | 24 | (299,299,3) | (2,1) |
Fig. 5.
Illustration of the major components of eight pre-trained CNN models. Convolution blocks of different colors indicate filters of different size. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
ResNet-34 is a deep residual network and is designed based on the concept of residual learning [39]. It consists of one standalone convolution layer and 16 residual bocks followed by one FC layer. This network mainly overcomes the degradation problem by introducing residual connections. ResNet-50 is another variant of ResNet that consists of same number of residual block as with ResNet-34, but, the structure of residual block is different [39]. ResNet-50 model replaces each two layer residual block of ResNet-34 with a three layer bottleneck block that uses 1 × 1 convolutions. GoogleNet is a 22 layer deep architecture with 5 million parameters which is considerably than the parameters used in AlexNet and VGG models [36]. The inception module is the basic building block of this model that helps in processing the data using multiple filters in parallel. Inception-V3 is a variant of Inception-V2 and avoids representational bottlenecks [40]. It has more effective computations by using factorization techniques and achieves a low error rate as opposed to its earlier variants. This network contains 42 layers with an input image size of 299 × 299. MobileNet-V2 is designed based on the ideas of MobileNet-V1 that utilize depth wise separable convolution as efficient building blocks. But, this version introduces a new layer module that is the inverted residual with linear bottleneck [37]. It is a small and cost-effective architecture that helps in achieving high performance with limited resources. It has 19 residual bottleneck layers. The main purpose of this research is to determine the best performing DL model for COVID-19 screening that can drive forward researchers for development of more effective AI based solutions.
3. Experiments and results
This section presents the results obtained from several experiments. We performed a comprehensive experimental analysis for prediction of COVID-19 from X-ray images using eight pre-trained CNN models namely, AlexNet, VGG-16, GoogleNet, MobileNet-V2, Squeezenet, ResNet-34, ResNet-50 and Inception-V3. We analyzed the impact of several hyperparameters associated with these models and carried out a comparative analysis among eight CNN models. Finally, the best performing model is obtained. We also compared the results with recent state-of-the-art approaches.
3.1. Experimental setup and performance metrics used
The CNN models were evaluated using chest X-ray samples collected from covid-chestxray-dataset [29] and ChestX-ray8 dataset [30]. The details of the data splitting used in our study with and without augmentation are shown in Table 2. The augmented samples have been used for training the model. After performing augmentation on training images, 1430 X-ray images have been obtained which were further divided into a train and validation set using a splitting ratio of 70% and 30% and thereby, resulting in 1002 training images and 428 validation images as shown in Table 2. The validation set has been used to prevent the model from overfitting and obtain an optimal model.
Table 2.
Details of data splitting with and without augmentation.
| Class | Original dataset |
Augmented dataset |
|||
|---|---|---|---|---|---|
| Train | Test | Train | Validation | Test | |
| COVID-19 | 143 | 60 | 501 | 214 | 60 |
| Normal | 143 | 60 | 501 | 214 | 60 |
| Total | 286 | 120 | 1002 | 428 | 120 |
The input X-ray images were initially resized to 224 × 224 while using AlexNet, VGG-16, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50, and to 299 × 299 while using Inception-V3. We implemented our algorithms using PyTorch toolbox. For TL, the batch size was set as 32. Each model was trained for 50 epochs. The batch size and number of epochs have been determined empirically. The training was performed using Adam optimizer and the learning rate has been empirically decided. The performance of each model was evaluated based on different metrics like F1-score, Specificity (Spe), Sensitivity (Sen), Precision (Pre), Accuracy (Acc), and Area Under the ROC Curve (AUC). These metrics were computed by different parameters of the confusion matrix such as True Positive (TP), False Positive (FP), True Negative (TN) and False Negative (FN) [41], [42]. The metrics are defined as follows:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
In this study, COVID-19 infections and normal cases were considered as positive and negative cases respectively. Therefore, and indicate the accurately predicted normal and COVID-19 cases respectively, whereas and indicate the inaccurately predicted normal and COVID-19 cases respectively.
3.2. Results
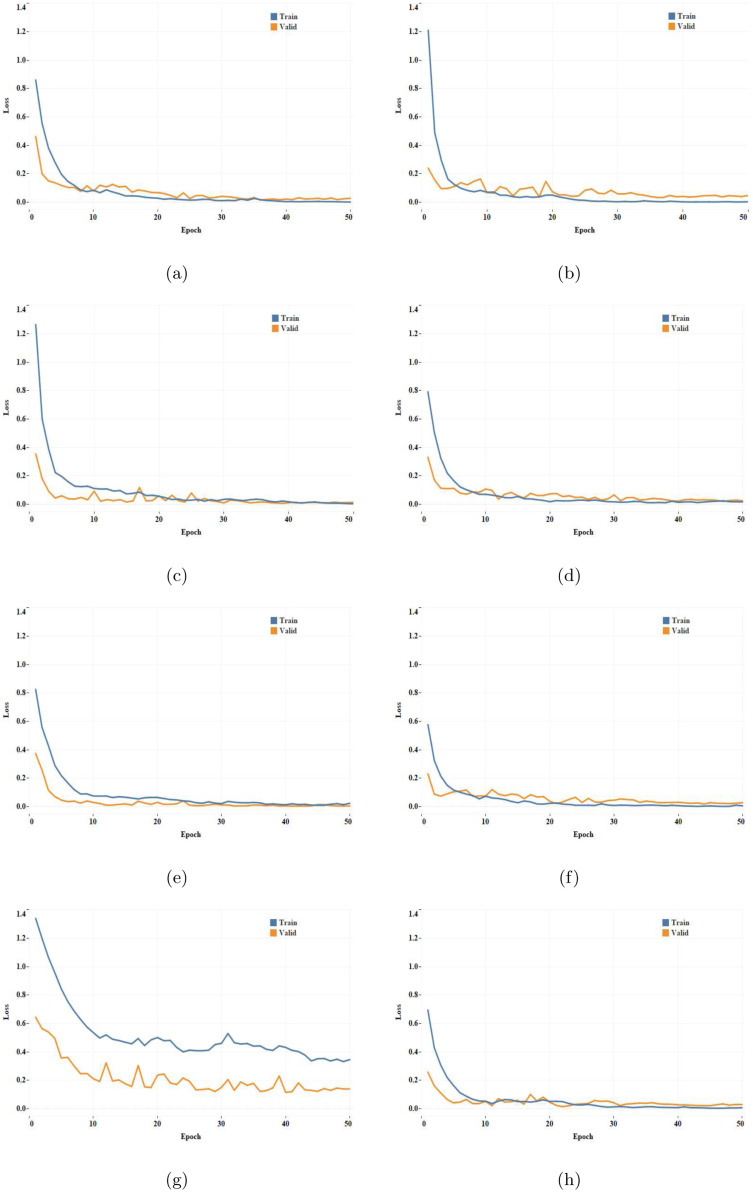
The training performance in terms of training loss, validation loss and validation accuracy obtained by different networks at different epochs are listed in Table 3. Fig. 6 illustrates the training and validation loss across different iterations for all networks.
Table 3.
Trainings performance of all the proposed models.
| Model | Epoch | Train loss | Valid loss | Valid accuracy (%) |
|---|---|---|---|---|
| AlexNet | 1 | 0.8252 | 0.3738 | 84.81 |
| … | … | … | ||
| 49 | 0.0143 | 0.0053 | 98.73 | |
| 50 | 0.0232 | 0.0043 | 99.05 | |
| VGG-16 | 1 | 0.8113 | 0.3149 | 84.57 |
| … | … | … | ||
| 49 | 0.1759 | 0.1859 | 96.30 | |
| 50 | 0.1724 | 0.1886 | 96.15 | |
| GoogleNet | 1 | 0.4749 | 0.2396 | 81.35 |
| … | … | … | ||
| 49 | 0.0020 | 0.0396 | 98.27 | |
| 50 | 0.0028 | 0.0477 | 98.62 | |
| MobileNet-V2 | 1 | 0.5772 | 0.2308 | 90.65 |
| … | … | … | ||
| 49 | 0.0102 | 0.0243 | 97.92 | |
| 50 | 0.0057 | 0.0270 | 97.23 | |
| SqueezeNet | 1 | 0.6943 | 0.2576 | 90.89 |
| … | … | … | ||
| 49 | 0.0057 | 0.0298 | 97.37 | |
| 50 | 0.0071 | 0.0289 | 97.65 | |
| ResNet-34 | 1 | 0.8606 | 0.4616 | 77.80 |
| … | … | … | ||
| 49 | 0.0019 | 0.0245 | 98.68 | |
| 50 | 0.0015 | 0.0270 | 98.13 | |
| ResNet-50 | 1 | 0.5923 | 0.3542 | 83.84 |
| … | … | … | ||
| 49 | 0.0050 | 0.0126 | 98.71 | |
| 50 | 0.0035 | 0.0137 | 98.78 | |
| Inception-V3 | 1 | 1.3397 | 0.6440 | 69.83 |
| … | … | … | ||
| 49 | 0.3319 | 0.1395 | 94.97 | |
| 50 | 0.3460 | 0.1393 | 95.13 | |
Fig. 6.
Loss convergence plot obtained for different CNN architectures: (a) ResNet-34, (b) ResNet-50, (c) GoogleNet, (d) VGG-16, (e) AlexNet, (f) MobileNet-V2, (g) Inception-V3, and (h) SqueezeNet.
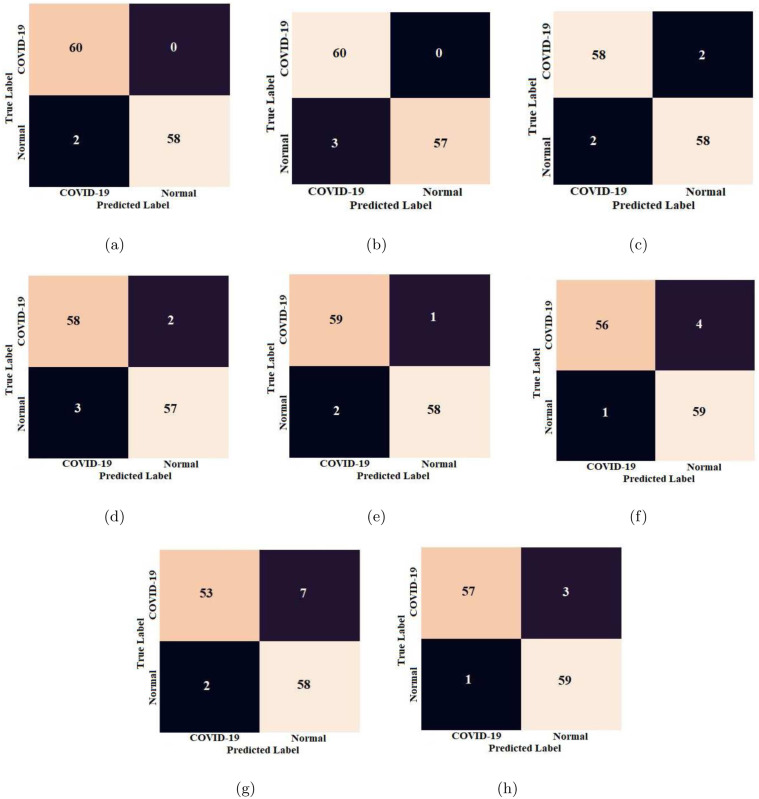
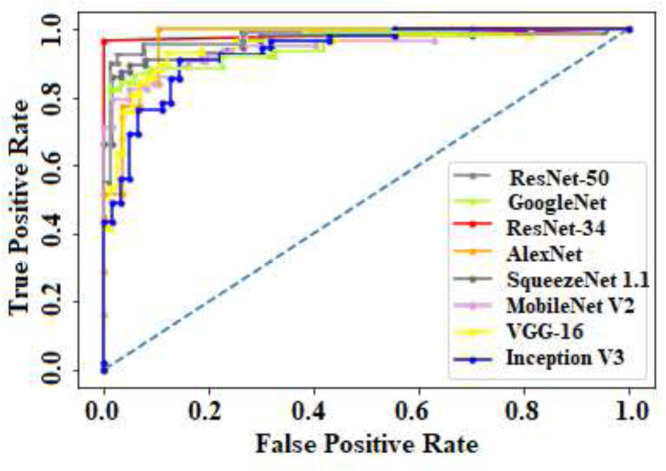
We presented the confusion matrices of all eight CNN models on the test data in Fig. 7. It can be observed that our proposed method with ResNet-34 and ResNet-50 could able to classify all COVID-19 infection cases accurately. The ROC curves of all models are depicted in Fig. 8.
Fig. 7.
Confusion matrices obtained for different CNN models: (a) ResNet-34, (b) ResNet-50, (c) GoogleNet, (d) VGG-16, (e) AlexNet, (f) MobileNet-V2, (g) Inception-V3, and (h) SqueezeNet.
Fig. 8.
ROC curves of eight different CNN models used in this study.
The detailed classification results obtained from all networks are compared in terms of various metrics and are tabulated in Table 4. It can be seen that the ResNet-34 model achieved the highest performance with a precision of 96.77%, specificity of 96.67%, F1-score of 0.9836, accuracy of 98.33%, and AUC of 0.9836. Also, it is noticed that the obtained sensitivity for COVID-19 class is remarkably higher (i.e., 100%) for both ResNet models. AlexNet network was found to be the second-best performer for COVID-19 prediction which obtained a precision of 96.72%, sensitivity of 98.33%, specificity of 96.67%, F1-score of 0.9752, accuracy of 97.50%, and AUC of 0.9642.
Table 4.
Classification results comparison of all eight CNN models.
| Model | Pre (%) | Sen (%) | Spe (%) | F1-score | Acc (%) | AUC |
|---|---|---|---|---|---|---|
| ResNet-34 | 96.77 | 100.00 | 96.67 | 0.9836 | 98.33 | 0.9836 |
| ResNet-50 | 95.24 | 100.00 | 95.00 | 0.9756 | 97.50 | 0.9731 |
| GoogleNet | 96.67 | 96.67 | 96.67 | 0.9667 | 96.67 | 0.9696 |
| VGG-16 | 95.08 | 96.67 | 95.00 | 0.9587 | 95.83 | 0.9487 |
| AlexNet | 96.72 | 98.33 | 96.67 | 0.9752 | 97.50 | 0.9642 |
| MobileNet-V2 | 98.24 | 93.33 | 98.33 | 0.9573 | 95.83 | 0.9506 |
| Inception-V3 | 96.36 | 88.33 | 96.67 | 0.9217 | 92.50 | 0.9342 |
| SqueezeNet | 98.27 | 95.00 | 98.33 | 0.9661 | 96.67 | 0.9705 |
3.2.1. Results comparison with different optimization methods
Adam optimizer [43] has been chosen for performing the training of all networks. To evaluate the effectiveness, its results were compared with of other efficient optimization methods such as SGD [44], Adadelta [45], and RMSProp [46]. Table 5 lists the classification results of different optimizers for two different and best performing CNN models such as ResNet-34 and AlexNet. The results have been computed over the test set. It can be seen that Adam optimizer outperforms other competitive optimization methods.
Table 5.
Classification performance (in %) comparison among different optimizers.
| Model | Optimizer | Pre (%) | Sen (%) | Spe (%) | F1-score | Acc (%) |
|---|---|---|---|---|---|---|
| ResNet-34 | SGD | 95.00 | 100.00 | 95.24 | 0.9740 | 97.50 |
| Adadelta | 95.00 | 93.44 | 94.92 | 0.9420 | 94.17 | |
| RMSProp | 96.67 | 98.31 | 96.72 | 0.9748 | 97.50 | |
| Adam | 96.77 | 100.00 | 96.67 | 0.9836 | 98.33 | |
| AlexNet | SGD | 98.33 | 93.65 | 98.25 | 0.9593 | 95.83 |
| Adadelta | 91.67 | 93.22 | 91.80 | 0.9240 | 92.50 | |
| RMSProp | 95.00 | 98.28 | 95.16 | 0.9661 | 96.67 | |
| Adam | 96.72 | 98.33 | 96.67 | 0.9752 | 97.50 | |
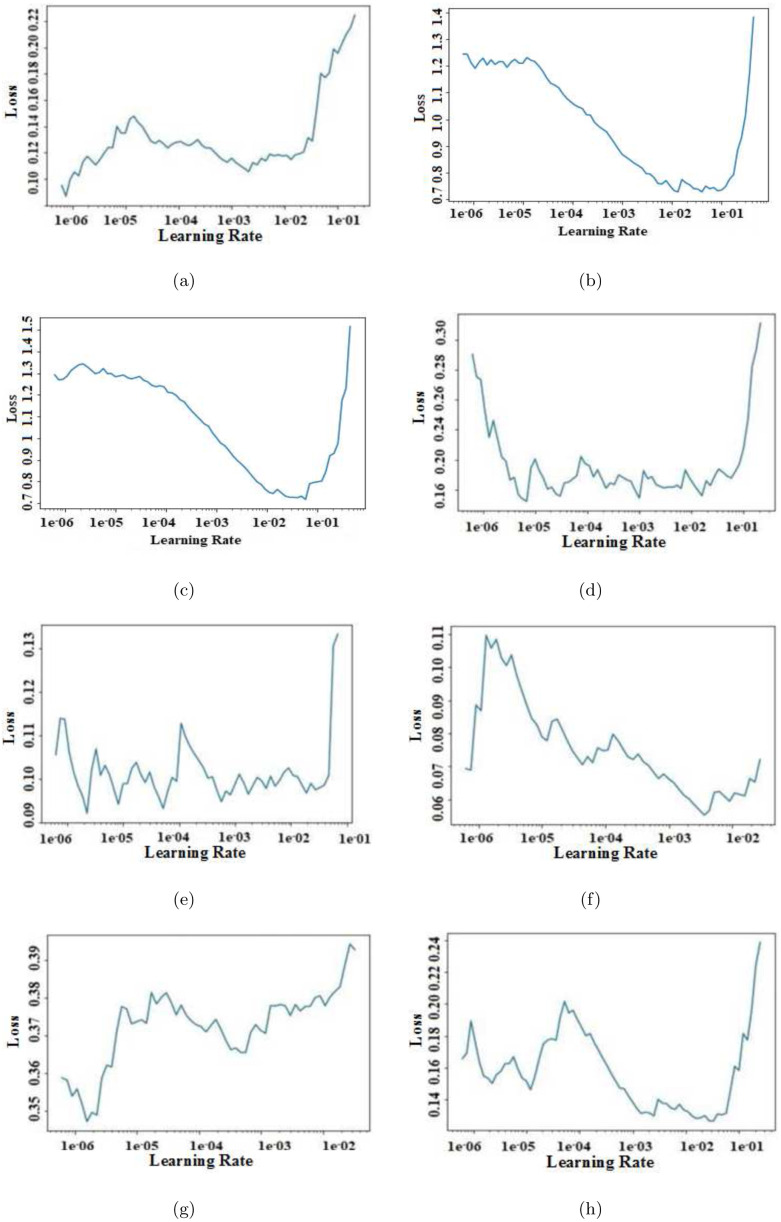
3.2.2. Optimal learning rate selection
To determine the optimal learning rate for all networks, a graph has been plotted between different possible learning rates and the validation loss as shown in Fig. 9. The best learning rate of a model was chosen based on the minimum loss.
Fig. 9.
Plot between learning rate and loss obtained for all models: (a) ResNet-34, (b) ResNet-50, (c) GoogleNet, (d) VGG-16, (e) AlexNet, (f) MobileNet-V2, (g) Inception-V3, and (h) SqueezeNet.
3.2.3. Results comparison with different batch sizes
Batch size is one of the most crucial hyper-parameters for tuning the DL models. In this experiment, the impact of batch size on testing accuracy is studied. Table 6 shows the test accuracy of all networks when trained using different batch sizes such as 8, 16 and 32. It can be seen that a higher and stable testing performance is obtained for all network models with batch size 32 and hence, a batch size of 32 has been chosen in the study.
Table 6.
Testing accuracy (in %) obtained by the CNN models trained with different batch sizes.
| Model | Batch size |
||
|---|---|---|---|
| 8 | 16 | 32 | |
| ResNet-34 | 98.33 | 98.33 | 98.33 |
| ResNet-50 | 96.67 | 97.50 | 97.50 |
| GoogleNet | 95.83 | 95.83 | 96.67 |
| VGG-16 | 96.67 | 95.83 | 95.83 |
| AlexNet | 96.67 | 96.67 | 97.50 |
| MobileNet-V2 | 95.00 | 96.67 | 95.83 |
| Inception-V3 | 92.50 | 91.67 | 92.50 |
| SqueezeNet | 96.67 | 95.83 | 96.67 |
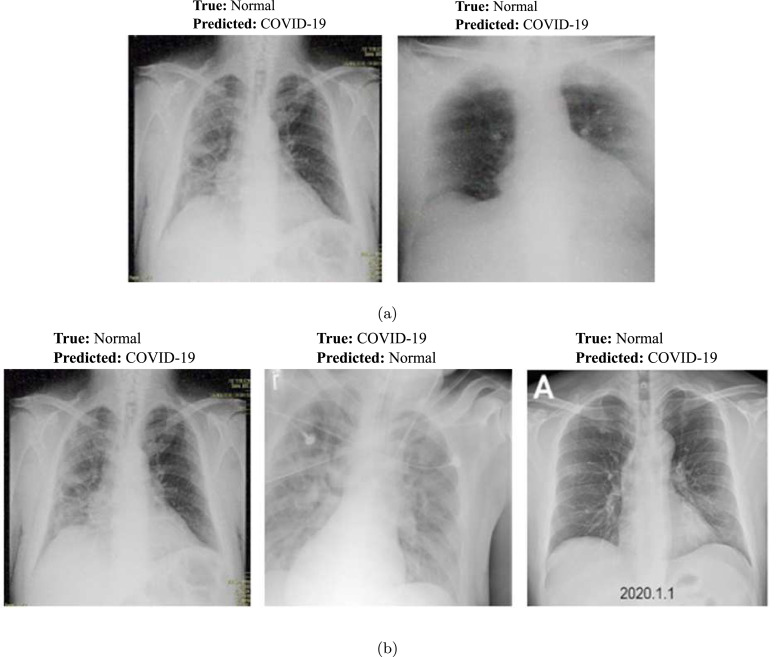
3.2.4. Misclassification results analysis
The misclassification samples predicted by two best performing CNN models: ResNet-34 and AlexNet are depicted in Fig. 10. The misclassification was possibly occurred due to the similar imaging features between the normal and COVID-19 infection cases.
Fig. 10.
Illustration of misclassification results obtained by two best networks: (a) ResNet-34 and (b) AlexNet.
Table 8.
COVID-19 class performance comparison with the state-of-the-art approaches.
| Reference | COVID-19 class sensitivity (%) |
|---|---|
| Hemdan et al. [21] | 100.00 |
| Narin et al. [24] | 96.00 |
| Sethy and Behera [25] | NR |
| Toğaçar et al. [27] | 99.32 |
| Wang and Wong [22] | 87.10 |
| Ucar and Korkmaz [26] | 100.00 |
| Farooq and Hafeez [28] | 100.00 |
| Ozturk et al. [20] | 90.65 |
| Proposed method | 100.00 |
NR: Not reported
3.3. Comparison with state-of-the-art methods
The results achieved by the best CNN model are compared with the recently proposed DL methods for automated COVID-19 diagnosis using chest X-ray images in Table 7. It can be observed that the proposed method achieved higher performance than the other existing schemes. Compared to most of the studies (Hemdan et al. [21], Narin et al. [24] and Sethy and Behera [25]), the proposed study considered a fairly large number of chest X-ray samples to validate the CNN models. On the other hand, Toğaçar et al. [27], Ozturk et al. [20], Wang and Wong [22], Ucar and Korkmaz [26] and Farooq and Hafeez [28] used comparatively larger datasets to validate their models, but these datasets led to the class imbalance problem and accommodated less number of COVID-19 samples in general. But, the dataset used in the proposed study has equal class distribution for normal and COVID-19 infection cases. In [26], the class imbalance problem was resolved using offline augmentation technique. Also, it can be noticed that binary classification was performed in most of the studies; however, multi-class (mostly three or four) classification was performed in [20], [22], [26], [27] and [28]. Only the study in [28] considered four classes such as COVID-19, normal, bacterial pneumonia, and viral pneumonia and an accuracy of 96.23% was achieved using a model called COVID-ResNet. However, the major weakness of this model is that it was validated on very less number of COVID-19 samples (i.e., 68). To summarize, the multi-class COVID-19 classification task is more vital and challenging due to the similar manifestation of COVID-19 with different types of pneumonia. Therefore, the performance in these cases is comparatively lower and is yet to be improved. For example, the accuracies obtained in [22] and [20] are 92.60% and 87.02% respectively.
Table 7.
Comparison with state-of-the-art deep learning COVID-19 detection approaches using chest X-ray images.
| Reference | Method | Classes | Number of X-ray samples | Acc (%) |
|---|---|---|---|---|
| Hemdan et al. [21] | COVIDX-Net | COVID-19 | 50 | 90.00 |
| Normal | C: 25 and N: 25 | |||
| Narin et al. [24] | ResNet-50 | COVID-19 | 100 | 98.00 |
| Normal | C: 50 and N: 50 | |||
| Sethy and Behera [25] | ResNet-50 and SVM | COVID-19 | 50 | 95.38 |
| Normal | C: 25 and N: 25 | |||
| Toğaçar et al. [27] | SqueezeNet and MobileNetV2 | COVID-19 | 458 | 98.25 |
| SMO and SVM | Normal | C: 295, N: 65 and P: 98 | ||
| Pneumonia | ||||
| Wang and Wong [22] | COVID-Net | COVID-19 | 13800 | 92.60 |
| Normal | C: 183, N: – and P: – | |||
| Pneumonia | ||||
| Ucar and Korkmaz [26] | Bayes-SqueezeNet | COVID-19 | 5949 | 98.30 |
| Normal | C: 76, N: 1583 and P: 4290 | |||
| Pneumonia | ||||
| Farooq and Hafeez [28] | COVID-ResNet | COVID-19 | 5941 | 96.23 |
| Normal | C: 68, N: –, BP: –, VP: – | |||
| Bacterial pneumonia | ||||
| Viral pneumonia | ||||
| Ozturk et al. [20] | DarkCovidNet | COVID-19 | 625 | 98.08 |
| Normal | C: 125 and N: 500 | |||
| COVID-19 | 1125 | 87.02 | ||
| Normal | C: 125, N: 500 and P: 500 | |||
| Pneumonia | ||||
| Proposed method | ResNet-34 | COVID-19 | 406 | 98.33 |
| Normal | C: 203 and N: 203 |
C: COVID-19, N: Normal, P: Pneumonia, BP: Bacterial pneumonia, VP: Viral pneumonia
Furthermore, a performance comparison has been made between the proposed scheme and existing methods in the context of COVID-19 class sensitivity and the results are given in Table 8. Wang and Wong [22] achieved the lowest COVID-19 class sensitivity of 87.10%. It can be seen that the proposed method achieved 100% COVID-19 class sensitivity. Also, Hemdan et al. [21], Ucar and Korkmaz [26], and Farooq and Hafeez [28] obtained similar COVID-19 class sensitivity (i.e., 100%). However, the actual test set used in these studies contained comparatively less number of COVID-19 samples.
3.4. Discussion
The study of chest X-ray images for accurate prediction of COVID-19 infection has been attracting remarkable attention since the release of a dataset developed by Cohen [29]. Thereafter, several attempts have been made to develop a reliable diagnosis model using DL techniques. The concept of TL has been extensively used with CNN based networks. However, most of the earlier methods were evaluated using a limited amount of data. Further, in some cases, the data are imbalanced.
In this study, we have comprehensively evaluated the effectiveness of the eight most effective CNN models such as AlexNet, VGG-16, MobileNet-V2, Squeezenet, ResNet-34, and Inception-V3 for prediction of COVID-19 infections in X-ray images. Extensive experiments were conducted on a comparatively large dataset by considering several factors to determine the best performing model for automated COVID-19 screening. The COVID-19 X-ray images and normal X-ray images were taken from two sources [29] and [30] respectively. To handle the data imbalance issue, the same amount of data was chosen for both the groups. The experimental results and the detailed comparative analysis among all methods demonstrated the superiority of ResNet models. We achieved accuracy and sensitivity of 98.33% and 100.00% with ResNet-34 network. The model is cost-effective and can aid the radiologists to verify their decisions. The major purpose of this research to take faster decisions for quarantine of patients that can ultimately help in reducing the spread of COVID-19 infection. The major weakness of the proposed study is that it is validated using a limited amount of COVID-19 samples. To date, there exists no large dataset publicly available as this is an ongoing and new pandemic. But, in the future, we plan to validate our approach using large datasets.
4. Conclusion
In this study, a DL based automated method was proposed for efficient classification of COVID-19 infection cases from normal cases using chest X-ray images. Several pre-trained CNN architectures using the concept of TL were studied by considering several crucial factors and their results over a set of publicly available X-ray samples were compared. The results indicated that ResNet-34 outperformed other competitive networks with an accuracy of 98.33% and can hence be regarded as a potential model for prediction of COVID-19 infection. The model can be utilized by the radiologists to verify their screening and thereby, reducing their workload. This study also paves the way for further development of effective deep CNN models (using residual connections) for a more accurate diagnosis of COVID-19 infection. The suggested DL model is developed to earn significant performance for binary classification (COVID-19 versus normal) and a limited number of studies have been proposed to date for multi-class classification (COVID versus pneumonia versus normal). Hence, in future studies, the effectiveness of the proposed model will be verified for multi-class classification problem. Further, we plan to explore the use of optimization algorithms along with the DL models used in this study to design a more reliable model.
CRediT authorship contribution statement
Soumya Ranjan Nayak: Formal analysis, Resources, Validation, Writing - original draft. Deepak Ranjan Nayak: Conceptualization, Methodology, Software, Writing - review & editing, Visualization, Supervision. Utkarsh Sinha: Data curation, Investigation, Software, Writing - original draft. Vaibhav Arora: Data curation, Investigation, Software, Writing - original draft. Ram Bilas Pachori: Investigation, Writing - review & editing, Supervision, Validation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
References
- 1.WHO . 2020. Coronavirus disease 2019 (COVID-19) situation report – 127. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200526-covid-19-sitrep-127.pdf?sfvrsn=7b6655ab_8. [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elasnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J. Biomol. Struct. Dyn. 2020:1–22. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 5.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong D., Tang Z., Wang S., Hui H., Gong L., Lu Y., Xue Z., Liao H., Chen F., Yang F. The role of imaging in the detection and management of COVID-19: A review. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 7.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 8.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on COVID-19: An update—radiology scientific expert panel. Radiology. 2020;296(2):1–2. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338. [Google Scholar]
- 11.Hoo-Chang S., Roth H.R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging. 2016;35(5):1285. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A., Van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 13.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 14.Nayak D.R., Dash R., Majhi B., Pachori R.B., Zhang Y. A deep stacked random vector functional link network autoencoder for diagnosis of brain abnormalities and breast cancer. Biomed. Signal Process. Control. 2020;58 [Google Scholar]
- 15.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.X. Gu, L. Pan, H. Liang, R. Yang, Classification of bacterial and viral childhood pneumonia using deep learning in chest radiography, in: Proceedings of the 3rd International Conference on Multimedia and Image Processing, 2018, pp. 88–93.
- 17.Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P., Moreira C., Damaševičius R., de Albuquerque V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl. Sci. 2020;10(2):559. [Google Scholar]
- 18.Lakhani P., Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 19.Rajpurkar P., Irvin J., Ball R.L., Zhu K., Yang B., Mehta H., Duan T., Ding D., Bagul A., Langlotz C.P. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15(11) doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. COVIDX-Net: A framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 22.Wang L., Wong A. 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethy P.K., Behera S.K. 2020. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. [DOI] [Google Scholar]
- 26.Ucar F., Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-Ray images. Med. Hypotheses. 2020 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farooq M., Hafeez A. 2020. COVID-ResNet: A deep learning framework for screening of COVID19 from radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 29.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. https://github.com/ieee8023/covid-chestxray-dataset. [Google Scholar]
- 30.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2097–2106.
- 31.Swati Z.N.K., Zhao Q., Kabir M., Ali F., Ali Z., Ahmed S., Lu J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019;75:34–46. doi: 10.1016/j.compmedimag.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Nayak D.R., Dash R., Majhi B. Automated diagnosis of multi-class brain abnormalities using MRI images: A deep convolutional neural network based method. Pattern Recognit. Lett. 2020;138:385–391. [Google Scholar]
- 33.M. Oquab, L. Bottou, I. Laptev, J. Sivic, Learning and transferring mid-level image representations using convolutional neural networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2014, pp. 1717–1724.
- 34.Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 35.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 36.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going deeper with convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 1–9.
- 37.M. Sandler, A. Howard, M. Zhu, A. Zhmoginov, L.-C. Chen, MobileNetV2: Inverted residuals and linear bottlenecks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 4510–4520.
- 38.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. 2016. SqueezeNet: Alexnet-level accuracy with 50x fewer parameters and 0.5 MB model size. arXiv preprint arXiv:1602.07360. [Google Scholar]
- 39.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778.
- 40.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, Rethinking the inception architecture for computer vision, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, 2818–2826.
- 41.Toğaçar M., Ergen B., Cömert Z. BrainMRNet: Brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med. Hypotheses. 2020;134 doi: 10.1016/j.mehy.2019.109531. [DOI] [PubMed] [Google Scholar]
- 42.Toğaçar M., Özkurt K.B., Ergen B., Cömert Z. BreastNet: A novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Physica A. 2020;545 [Google Scholar]
- 43.Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- 44.I. Sutskever, J. Martens, G. Dahl, G. Hinton, On the importance of initialization and momentum in deep learning, in: International Conference on Machine Learning, 2013, pp. 1139–1147.
- 45.Zeiler M.D. 2012. Adadelta: an adaptive learning rate method. arXiv preprint arXiv:1212.5701. [Google Scholar]
- 46.Hinton G., Srivastava N., Swersky K. Neural Networks for Machine Learning. 2012. Lecture 6a overview of mini-batch gradient descent course. [Google Scholar]