Abstract
Objective:
Myeloid-derived suppressor cells (MDSCs) contribute to HIV progression by impairing antiviral immunity; however, the mechanisms responsible for MDSC development during HIV infection are incompletely understood. HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) is a long noncoding RNA (lncRNA) that plays a pivotal role in regulating myeloid cell development via targeting HOXA1. The role of HOTAIRM1-HOXA1 in the differentiation and functions of MDSCs during HIV infection remains unclear.
Methods:
In this study, we measured MDSC induction and suppressive functions by flow cytometry, RT-PCR, and co-culture experiments using CD33+ myeloid cells derived from people living with HIV (PLHIV) on antiretroviral therapy (ART). We also manipulated the HOTAIRM1-HOXA1 axis in myeloid cells using knockdown and overexpression approaches.
Results:
We demonstrate that HOTAIRM1 and HOXA1 expressions are reciprocally upregulated and are responsible for increased levels of immunosuppressive molecules, such as arginase 1 (Arg1), inducible nitric oxide synthase (iNOS), signal transducer and activator of transcription 3 (STAT3), and reactive oxygen species (ROS), in CD33+ myeloid cells derived from PLHIV on ART. We found that overexpression of HOTAIRM1 or HOXA1 in CD33+ cells isolated from healthy individuals promoted the differentiation and suppressive functions of MDSCs, whereas silencing of HOTAIRM1 or HOXA1 expression in MDSCs derived from PLHIV significantly inhibited the frequency of MDSCs and expressions of the immunosuppressive molecules and reduced their immunosuppressive effects on T cells.
Conclusion:
These results indicate that the HOTAIRM1-HOXA1 axis enhances differentiation and suppressive functions of MDSCs and could be a potential therapeutic target for immunomodulation during latent HIV infection.
Keywords: HIV, HOTAIRM1, HOXA1, Immune suppression, lncRNA, MDSCs
Introduction
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of immature myeloid cells that are generated due to aberrant myelopoiesis under certain pathological conditions, such as cancer and inflammatory or infectious diseases [1–3]. MDSCs suppress immune responses by the production of inflammatory and immunosuppressive molecules, including arginase 1 (ARG1), inducible nitric oxide synthase (iNOS), reactive oxygen species (ROS), and signal transducer and activator of transcription 3 (STAT3), all of which are important mediators of innate immune responses against pathogenic infections [4–9].
While the immunosuppressive roles of MDSCs have been well-characterized in many tumor models, their roles in infectious diseases are less clear, especially during responses to retroviral infections that cause immunodeficiency. Although MDSCs can contribute to immune homeostasis after infection by limiting excessive inflammatory processes, their expansion may be at the expense of pathogen elimination and thus may lead to infection persistence or latency. Recent studies have shown that MDSC expansion plays a role in suppressing T cell functions and advancing disease progression during HIV infection [10–18]. However, the mechanisms that drive MDSC expansion and their suppressive functions during HIV infection remain unclear [19].
Long noncoding RNAs (lncRNAs) are genomic transcripts > 200-nt in length that do not encode proteins but possess regulatory functions [20–22]. Thus far, thousands of lncRNAs have been discovered, but their functions have not yet been characterized. Recent studies indicate that their expressions are species-, cell-, or disease stage-specific [20–23]. In particular, HOX antisense intergenic RNA myeloid 1 (HOTAIRM1) is a lncRNA encoded in the HOXA gene cluster [24–26] and appears to be the most prominent lncRNA upregulated during granulocyte differentiation and myeloid cell maturation [27–29]. While the regulatory effects of the HOTAIRM1-HOXA1 axis on hematopoiesis, leukemogenesis, and oncogenesis have been reported [30–34], the role of HOTAIRM1 in controlling viral infections, especially in the regulation of MDSC differentiation and functions during HIV infection, remains largely unknown.
In this study, we characterized the expression and function of lncRNAs in MDSCs from people living with HIV (PLHIV) on antiretroviral therapy (ART). We found that HOTAIRM1 is upregulated during HIV infection and drives MDSC expansion via upregulating HOXA1 expression. We demonstrated that the HOTAIRM1-HOXA1 axis plays an important role in the regulation of the immunosuppressive functions of MDSCs, revealing a novel mechanism of immune dysregulation during latent HIV infection on ART.
Methods
Subjects.
The study protocol was approved by the joint institutional review board at East Tennessee State University and James H. Quillen VA Medical Center (ETSU/VA IRB, Johnson City, TN). The study subjects were composed of two populations: 64 chronically HIV-1 infected individuals and 54 healthy subjects (HS). In this study, we only included PLHIV on ART with undetectable viremia (aviremia, HIV RNA < 20 copies/ml) and a CD4 count ranging from 220 ~ 1,250. Healthy subjects (HS) were negative for HBV, HCV, and HIV infection, and blood (whole blood or leukopaks) were obtained from the Physician’s Plasma Alliance LLC (Gray, TN). Written informed consent was obtained from all participants.
Cell isolation, culture, and flow cytometric analysis.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using Ficoll density gradients (GE Healthcare, Piscataway, NJ). CD33+ cells and CD4+ T cells were isolated from PBMCs using a CD33+ Cell or CD4+ T cell Isolation Kit (Miltenyi Biotec Inc., Auburn, CA). The cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA), 100 IU/ml penicillin, and 2 mM L-glutamine (Thermo Scientific, Logan, Utah) at 37°C and 5% CO2 atmosphere. Flow cytometry analysis of cell phenotypes and intracellular cytokines in PBMCs was carried out as described previously [18]. For pSTAT3 detection, the cells were stimulated as described previously [18], and brefeldin A was added 6 hours before intracellular staining and detection by flow cytometry. For IL-10 measurement, PBMCs derived from HS and PLHIV, or CD33+ cells isolated from PLHIV, following silencing with HOTAIRM1 or HOXA1 were stimulated with LPS (100 ng/ml) for 24 hours. For co-culture experiments, autologous CD4+ T cells were stimulated with 1 μg/ml anti-CD3 and 2 μg/ml anti-CD28 antibodies (BD Bioscience) or gp120 (1 μg/ml) for 2 days, followed by co-culturing with transfected CD33+ cells (at 1:2 ratio) for another two days, brefeldin A was added 6 hours before detection of IFN-γ production in CD4 T cells by flow cytometry. The following reagents were used: anti-CD4-FITC (Biolegend, San Diego, CA), anti-IFN-γ-PE (Biolegend), anti-CD33-PE (Biolegend), anti-CD14-APC (Biolegend), anti-HLA-DR-FITC (Biolegend), anti-CD3-APC (Biolegend), anti-Arg1-PE (Biolegend), anti-pSTAT3-PerCP/Cy5.5 (Biolegend), IL-10-PE (Biolegend), and anti-iNOS-PE (Novus biologicals, Centennial CO) along with isotype control antibodies (BD Bioscience, San Jose, CA). Levels of reactive oxygen species (ROS) in myeloid cells were measured using the H2DCFDA-based kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The stained cells were acquired on an AccuriTM C6 flow cytometer (BD Bioscience) and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). Isotype control antibodies (eBioscience, Waltham, MA) and single channel staining were used to determine the background levels of staining and adjust multicolor compensation as a gating strategy.
lncRNA array and RT-qPCR validation.
Total cellular RNA was isolated from CD33+ cells (pooled from 6 PLHIV and 6 HS) using the miRNeasy Mini kit (Qiagen, Valencia, CA). The RNA quality and quantity were analyzed using a BioPhotometer spectrophotometer UV/VIS, and RNA integrity was determined using gel electrophoresis. Expression profiling of lncRNAs was performed using the Arraystar gene array service (Arraystar, Rockville, MD). To validate the gene array results by real-time qPCR (RT-PCR), cDNA was generated from total RNA by the High-Capacity cDNA Reverse Transcription Kit (Thermo Scientific, Logan, Utah). The lncRNA and messenger RNA (mRNA) expression levels were assessed by real-time RT-PCR using the Taqman® fast advanced master mix (Thermo Scientific, Waltham, MA) and the CFX96 RT-PCR Detection System (Bio-Rad Laboratories Inc, Hercules, CA).
Cell transfection and co-culture experiments.
For HOTAIRM1 and HOXA1 knockdown, the CD33+ cells were transfected with 50 nM of SMART pools of siRNAs specific to HOTAIRM1, HOXA1, or a pool of scramble control siRNA (Dharmacon, Lafayette, CO) using the Human Monocyte Nucleofector Kit and Nucleofector II Device (Lonza, Allendale, NJ) following the manufacturer’s instructions. For HOTAIRM1 and HOXA1 overexpression, CD33+ cells were transfected with a HOTAIRM1 or HOXA1 expressing plasmid containing GFP (GenScript). The transfected cells were cultured for 2 days in IMEM medium (Lonza, Allendale, NJ) supplemented with 10% FBS. The cells were analyzed by flow cytometry or RT-qPCR. For cell co-culture, autologous CD4+ T cells were cultured in IMEM complete medium and stimulated with 1 μg/ml anti-CD3 plus 2 μg/ml anti-CD28 antibodies (BD Bioscience) or gp120 (1 ug/ml) for 2 days, followed by adding the transfected CD33+ cells (at 1:2 ratio), and the culture was incubated for another two days. IFN-γ production by CD4 T cells was determined by flow cytometry.
Statistical analysis.
The parametric data are presented as mean ± SD. Comparison between two groups was performed using paired t-test or unpaired t-test with or without Welch’s correction, based on the value of the F test. One-tail paired t-test was used to compare two-paired groups. The nonparametric data are presented as median with interquartile range and were analyzed by a one-tail Mann Whitney test. P-values < 0.05 or <0.01 were considered significant or very significant, respectively.
Results
MDSCs expand in HIV+ individuals on ART.
Phenotypically, human MDSCs express the common myeloid surface marker CD33 but lack the maturation marker HLA-DR. MDSCs are further categorized into monocytic (M-MDSCs) and granulocytic MDSCs (G-MDSCs) based on the differential expression of the CD14 or CD15 markers, respectively [1–3, 35, 36]. We have recently reported the expansion of M-MDSCs during latent HIV infection, which could inhibit T cell functions via promoting regulatory T cell (Treg) differentiation [18]. To better understand the role of MDSCs in PLHIV on ART, we further analyzed the frequencies of MDSCs in PBMCs by flow cytometry. We found that the frequencies of G-MDSCs (CD33+ HLA-DR−/low CD14−) also increased in PBMCs from PLHIV (Fig.1A).
Figure 1. HOTAIRM1 and HOXA1 are upregulated in MDSCs in HIV-infected individuals.
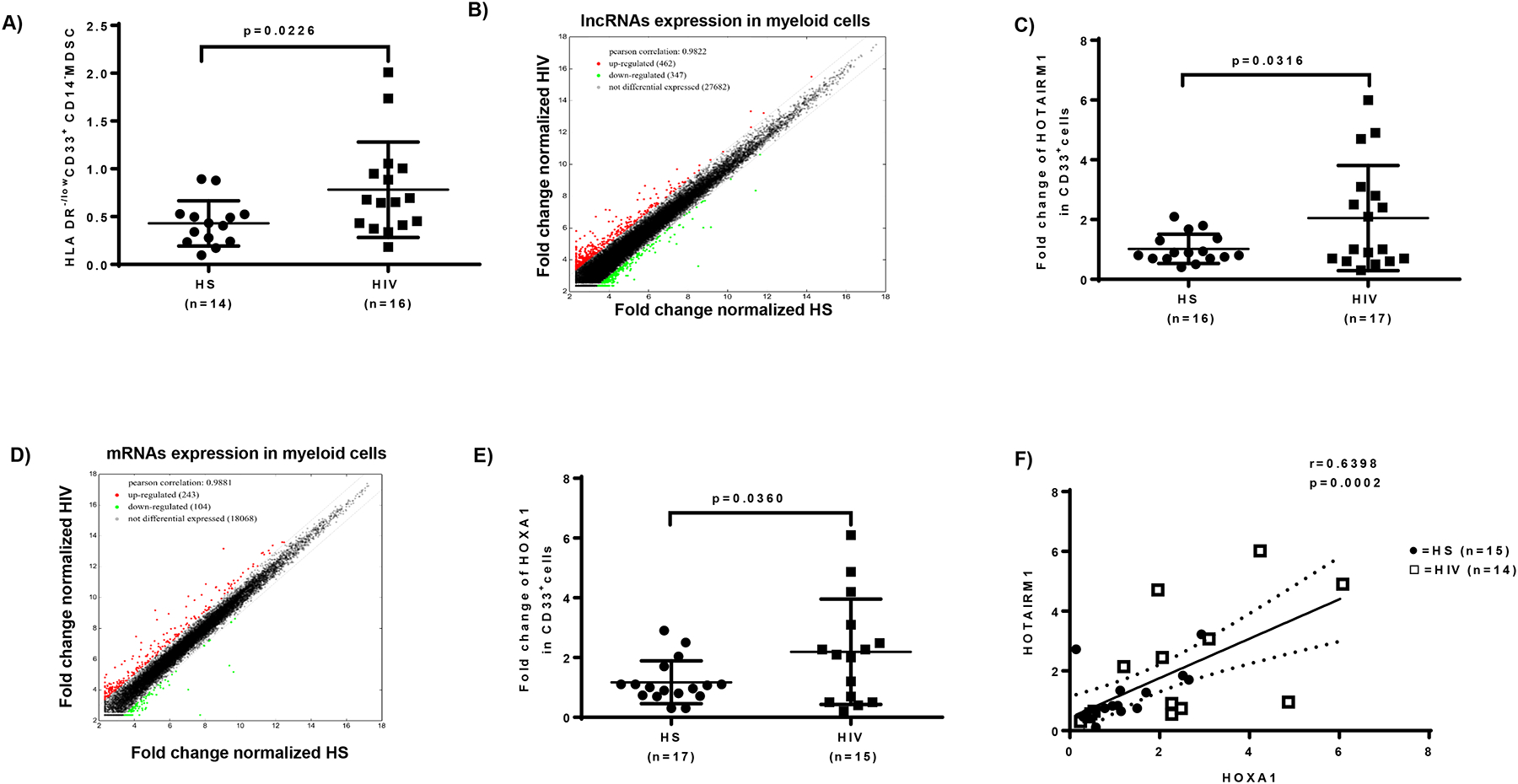
A) Expansion of G-MDSCs (HLA-DR−/lowCD33+CD14− cells) in PBMCs derived from ART-controlled, latently HIV-infected individuals compared with HS, as determined by flow cytometry. B) Scatter plot of lncRNA expression in CD33+ cells isolated from HIV-infected individuals versus HS (n=6 per group). C) HOTAIRM1 expression in CD33+ cells isolated from HIV-infected individuals versus HS, as determined by real-time PCR. D) Scatter plot of mRNA expression in CD33+ cells isolated from HIV-infected individuals versus HS (n=6 per group). E) HOXA1 expression in CD33+ cells isolated from HIV-infected individuals versus HS, as determined by real-time PCR. F) Pearson Correlation analysis of HOTAIRM1 and HOXA1 expression levels in CD33+ cells derived from the same subjects.
HOTAIRM1 and HOXA1 are upregulated in MDSCs during latent HIV infection.
To determine whether lncRNAs play a role in MDSC expansion during HIV infection, we analyzed the transcripts of lncRNAs and mRNAs in CD33+ myeloid cells isolated from ART-controlled PLHIV and HS using Arraystar gene array analysis. Among the lncRNAs analyzed (shown as a scatter plot in Fig.1B), 462 lncRNAs (red dots) were upregulated (> 2-fold), 347 lncRNAs (green dots) were downregulated, and 27,682 lncRNAs (black dots) remained unchanged in CD33+ myeloid cells from PLHIV compared to HS. Because HOTAIRM1 expression was upregulated in PLHIV, and given its critical role in granulocyte differentiation and myeloid cell maturation [27–29], we validated the upregulation of HOTAIRM1 expression by real-time PCR, which revealed a 2-fold increase in CD33+ cells derived from PLHIV (Fig.1C).
For mRNA transcripts (shown as a scatter plot in Fig.1D), 243 mRNAs (red dots) were upregulated, 104 mRNAs (green dots) were downregulated, and 18,068 mRNA transcripts remained unchanged (within 2-fold limit). Notably, the mRNA array results showed a significant upregulation of HOXA1, which is regulated by HOTAIRM1, in CD33+ cells derived from PLHIV. These results were validated by RT-PCR, which also showed a 2-fold increase in CD33+ cells from PLHIV (Fig.1E). Importantly, the levels of HOTAIRM1 positively correlated with HOXA1 expression according to the Pearson Correlation analysis (Fig.1F). Taken together, these results suggest that the expressions of HOTAIRM1 and its target gene HOXA1 are concurrently upregulated and may serve as a biomarker for MDSC expansion during HIV infection.
Elevated levels of immunosuppressive mediators in MDSCs during latent HIV infection.
MDSCs are more accurately identified by their immunosuppressive functions rather than their phenotypic markers. MDSCs suppress immune responses by producing immunosuppressive mediators, such as Arg1, iNOS, STAT3, and ROS [4–9]. Notably, Arg1 is constitutively expressed in granulocytes and represents a novel antimicrobial effector via arginine depletion in the phagolysosome [4–6]. The iNOS catalyzes the production of superoxide and free radical nitric oxide as an immune regulator [5]. STAT3 is a transcription factor and plays a pivotal role in MDSC differentiation and suppressive functions [6, 7]. ROS activate anti-oxidative pathways and induce transcriptional programs that regulate the differentiation and function of MDSCs as a part of a major mechanism to suppress T cell responses [8, 9]. To determine the mechanisms by which MDSCs exert their immunosuppressive effects during HIV infection, we measured the mRNA levels of those molecules involved in MDSC differentiation and functions. As shown in Fig.2A–C, RT-PCR analysis revealed increases in Arg1, iNOS, and STAT3 mRNA levels in CD33+ cells isolated from PLHIV compared to HS. H2DCFDA assay showed a significant increase in ROS production in CD33+ cells derived from PLHIV compared to HS. (Fig.2D). Of note, the increases in Arg1, iNOS, and STAT3 levels positively correlated with the increases in HOTAIRM1 and HOXA1 expressions in the same subjects (Fig.2E–J), suggesting that MDSCs may suppress immune responses during HIV infection by upregulating the expression of these immunosuppressive mediators through the HOTAIRM1-HOXA1 axis.
Figure 2. Upregulation of immunosuppressive molecules in MDSCs during latent HIV infection.
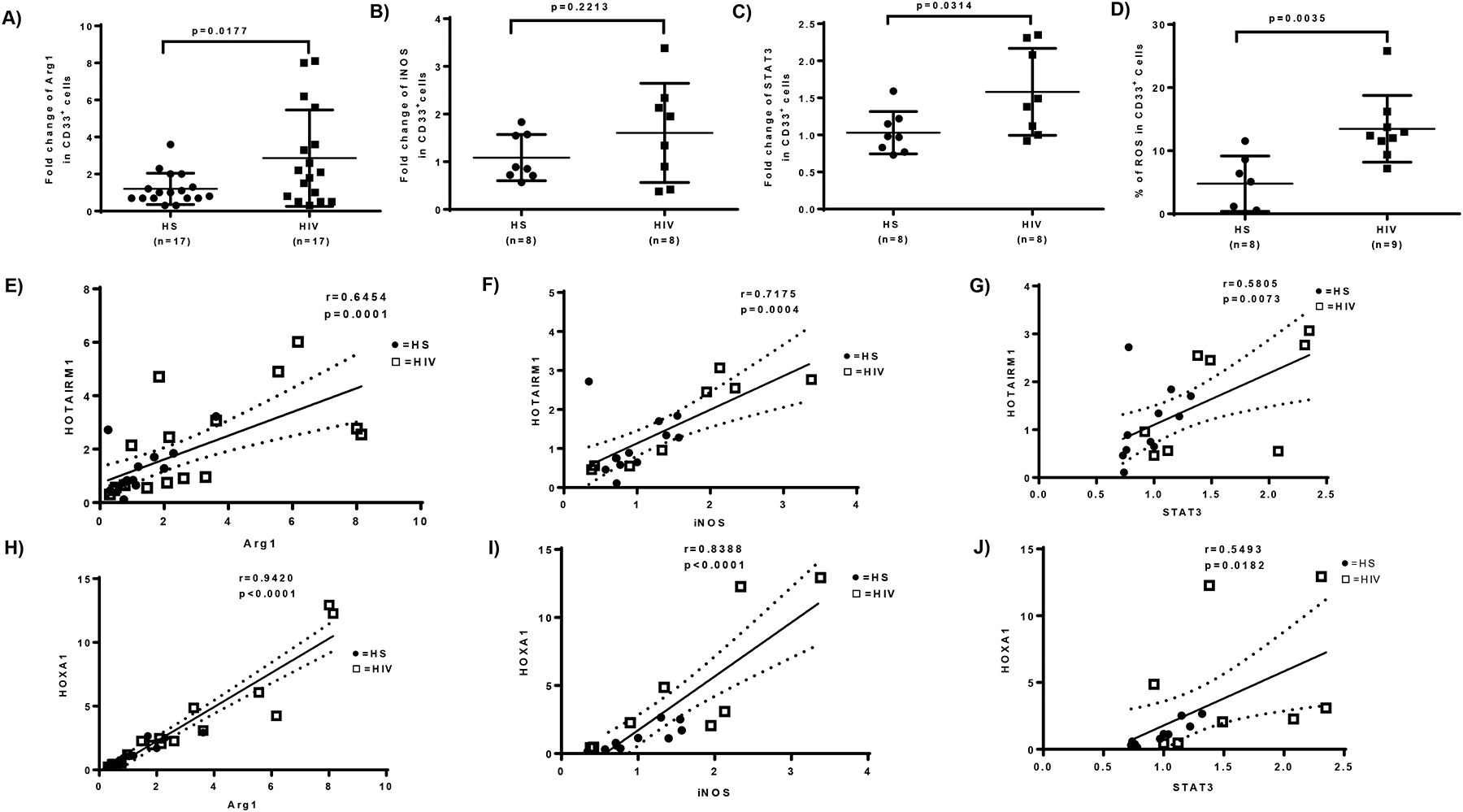
A-C) Fold change of Arg1, iNOS, and STAT3 gene expressions in CD33+ cells isolated from PLHIV and HS, analyzed by real-time PCR. D) ROS production in CD33+ cells derived from HIV-infected individuals versus HS, analyzed by the H2DCFDA assay. E-J) Relationship between HOTAIRM1 or HOXA1 and Arg1, iNOS, or STAT3 expression levels, analyzed by Pearson Correlation analysis.
HOTAIRM1 and HOXA1 regulate each other expression in MDSCs.
To elucidate the causal relationship between the expression of HOTAIRM1 and HOXA1, we transfected HS-derived CD33+ cells with HOTAIRM1- or HOXA1- expressing plasmids, followed by measuring their expression levels. As shown in Fig.3A, HOTAIRM1 mRNA levels were upregulated (~3.5-fold) in myeloid cells 2 days after HOTAIRM1 transfection. Interestingly, HOTAIRM1 expression was also upregulated (~3.5-fold) 2 days following HOXA1 transfection, suggesting that HOXA1 positively regulates HOTAIRM1 expression. While transfection of HOXA1 increased (~3.5-fold) HOXA1 levels, ectopic expression of HOTAIRM1 increased HOXA1 levels by ~4.5-fold (Fig.3B), indicating that HOTAIRM1 enhances HOXA1 expression.
Figure 3. Ectopic overexpression of HOTAIRM1 or HOXA1 in healthy CD33+ cells affects MDSC differentiation and immunosuppressive functions.
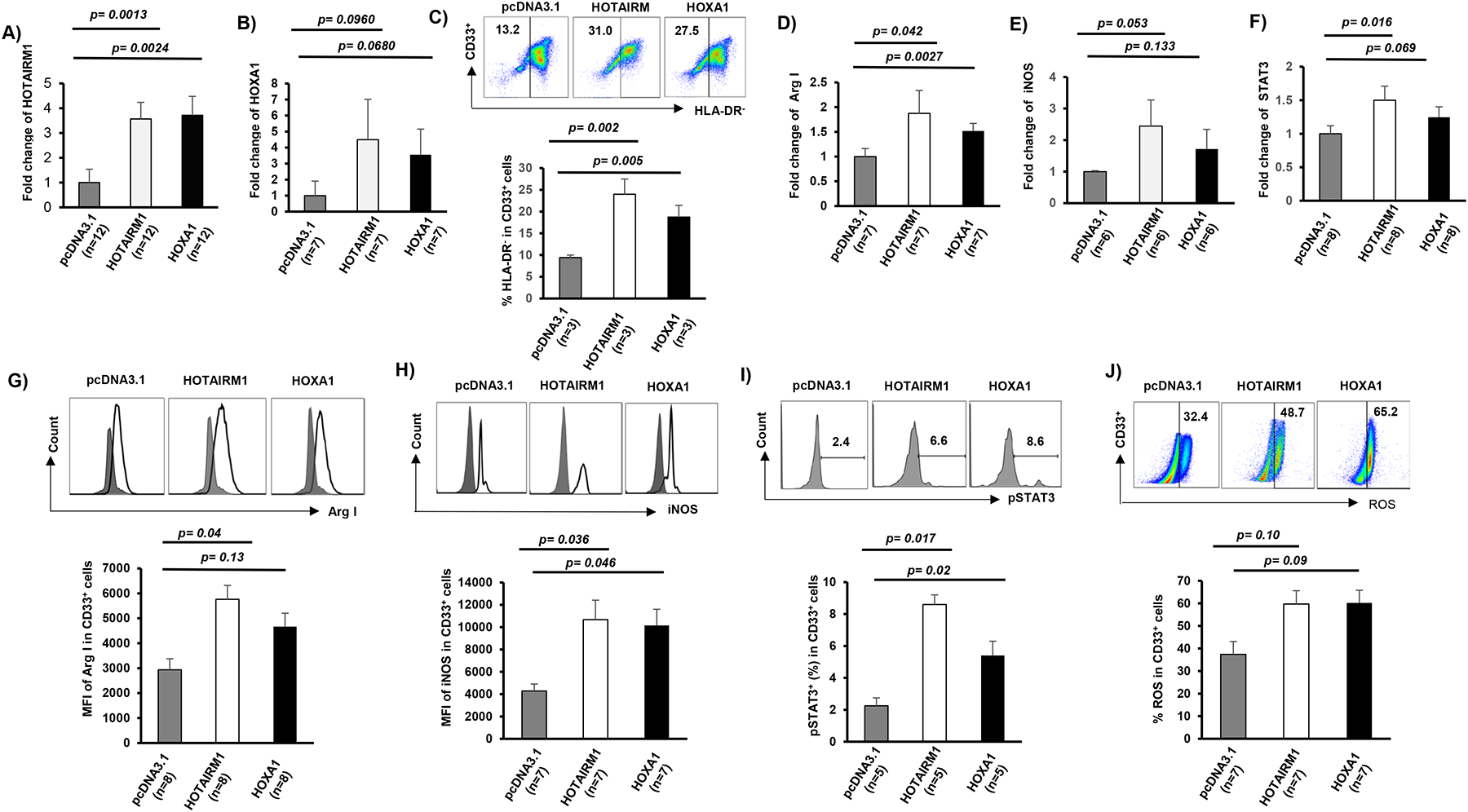
A-B) HOTAIRM1 or HOXA1 mRNA expression levels, measured by real-time PCR, in CD33+ cells transfected with pcDNA3.1 control, HOTAIRM1, or HOXA1 plasmids. C) Ectopic expression of HOTAIRM1 or HOXA1 promotes CD33+ cell differentiation into MDSCs, determined by flow cytometric analysis of higher frequencies of HLA-DR− cells. D-F) Ectopic expression of HOTAIRM1 or HOXA1 promotes Arg1, iNOS, and STAT3 mRNA expressions in CD33+ myeloid cells, determined by real-time PCR. G-J) Overexpression of HOTAIRM1 or HOXA1 in healthy CD33+ cells enhances Arg1, iNOS, and pSTAT3 expressions and ROS productions, determined by flow cytometric analysis.
We next transfected CD33+ myeloid cells derived from PLHIV with HOTAIRM1 or HOXA1 siRNAs. Compared to the control siRNA, HOTAIRM1 siRNA significantly reduced the level of HOTAIRM1 expression. Interestingly, HOXA1 siRNA also reduced the level of HOTAIRM1 expression (Fig.4A). Notably, knockdown of HOTAIRM1 or HOXA1 significantly reduced the level of HOXA1 expression in HIV-derived CD33+ myeloid cells (Fig.4B). Together, these results indicate that HOTAIRM1 and HOXA1 reciprocally regulate each other expression in CD33+ myeloid cells.
Figure 4. Silencing of HOTAIRM1 or HOXA1 expression in CD33+ cells from HIV-infected subjects abrogates MDSC differentiation and immunosuppressive functions.
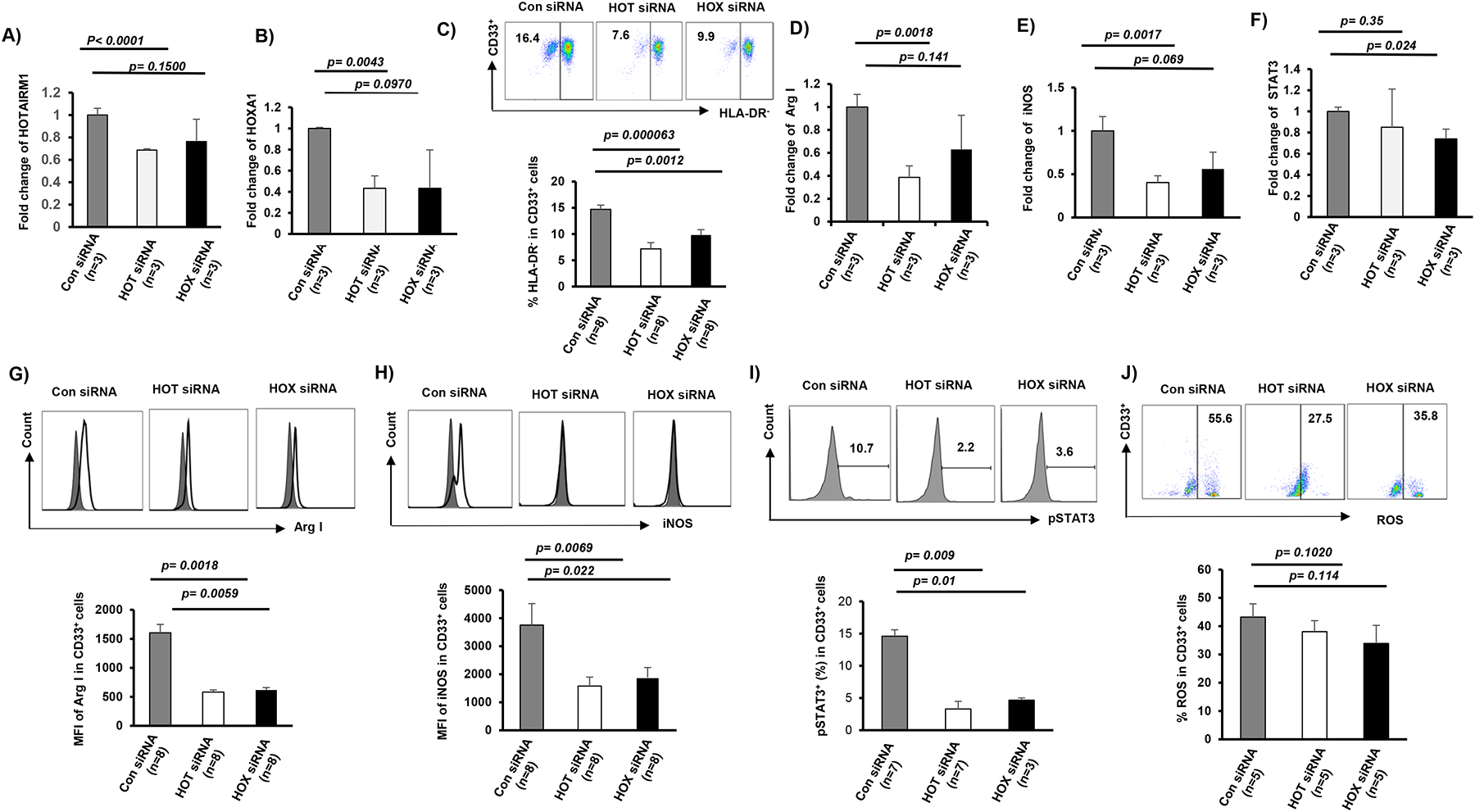
A-B) HOTAIRM1 or HOXA1 expression levels, measured by real-time PCR, in CD33+ cells transfected with control (Con) siRNA, HOTAIRM1 (HOT), or HOXA1 (HOX) siRNA. C) siRNA-mediated knockdown of HOTAIRM1 or HOXA1 expression in CD33+ cells of HIV subjects inhibits CD33+ cell differentiation into MDSCs, determined by flow cytometric analysis of lower HLA-DR− cell frequency. D-F) siRNA-mediated knockdown of HOTAIRM1 or HOXA1 expression in CD33+ cells of HIV subjects inhibits Arg1, iNOS, and STAT3 mRNA expressions, determined by real-time PCR. G-J) siRNA-mediated knockdown of HOTAIRM1 or HOXA1 expression in CD33+ cells of HIV subjects inhibits Arg1, iNOS, and pSTAT3 protein expressions and ROS production, determined by flow cytometry analysis.
Ectopic expression of HOTAIRM1 or HOXA1 promotes MDSC differentiation and suppressive functions.
Given the critical roles of HOTAIRM1 and HOXA1 in myeloid cell differentiation, we hypothesized that ectopic expressions of HOTAIRM1 or HOXA1 could enhance MDSC development and suppressive functions. To test this, we transfected CD33+ cells derived from HS with GFP-HOTAIRM1 or GFP-HOXA1 expression constructs or control plasmid (GFP-pCDNA3.1) for 2 days and then analyzed myeloid cell differentiation and the expression of Arg1, iNOS, and STAT3. Notably, transfection efficiency was about 52~62%, as determined by the rate of GFP+ in transfected cells (data not shown). Flow cytometric analysis revealed that overexpression of HOTAIRM1 or HOXA1 resulted in an immature phenotype of CD33+ differentiation, i.e., compared to control transfection, overexpression of either HOTAIRM1 or HOXA1 in CD33+ cells led to a high level of HLA-DR− expression, a feature of MDSCs (Fig.3C). In addition, the mRNA and protein levels of Arg1, iNOS, and STAT3 were increased in the HOTAIRM1- or HOXA1-transfected cells (Fig.3D–I). Correspondingly, ROS production was also increased in those cells (Fig.3J). These results suggest that the HOTAIRM1-HOXA1 axis promotes MDSC differentiation and enhances their suppressive functions.
Silencing HOTAIRM1 or HOXA1 expression reduces MDSC frequencies and suppressive functions.
Next, we asked whether knockdown of HOTAIRM1 or HOXA1 can attenuate the MDSC differentiation and immunosuppressive functions observed during HIV infection. To this end, we transfected CD33+ cells derived from PLHIV with either HOTAIRM1 or HOXA1 siRNA. Compared to the control siRNA, HOTAIRM1 siRNA or HOXA1 siRNA significantly reduced the frequency of HLA-DR− CD33+ MDSCs 2 days after transfection (Fig.4C). Transfection of HOTAIRM1 or HOXA1 siRNAs also reduced the mRNA and protein expression levels of Arg1, iNOS, and STAT3 (Fig.4D–I). ROS production was also reduced by the silencing of HOTAIRM1 or HOXA1 expression (Fig.4J). These results indicate that the HOTAIRM1-HOXA1 pathway promotes MDSC differentiation and suppressive functions.
Targeting the HOTAIRM1-HOXA1 axis in MDSCs affects T cell functions.
It is well known that MDSCs produce IL-10 as an inhibitory mediator to suppress immunity. To further elucidate the mechanisms of MDSC function, we measured IL-10 levels in MDSCs in response to LPS stimulation [18] and found that IL-10 expressions were significantly increased in both M-MDSCs and G-MDSCs in PLHIV versus HS (Fig.5A). Importantly, silencing HOTAIRM1 or HOXA1 significantly reduced the frequency of IL-10-producing myeloid cells from PLHIV (Fig.5B). We next asked whether targeting the HOTAIRM1-HOXA1 axis in myeloid cells would lead to alterations in T cell function. To address this question, we overexpressed HOTAIRM1 or HOXA1 in CD33+ cells from HS and then co-cultured them with activated autologous CD4 T cells for 2 days, followed by measuring IFN-γ production in activated T cells. As shown in Fig.5C, IFN-γ production was significantly decreased in CD4 T cells that were co-cultured with CD33+ cells overexpressing HOTAIRM1 or HOXA1. In contrast, we also silenced HOTAIRM1 or HOXA1 expression in CD33+ cells from HIV subjects and co-cultured them with autologous CD4 T cells for 2 days, followed by flow cytometric analysis. Indeed, IFN-γ production by HIV-derived CD4 T cells was restored when they were co-cultured with CD33+ cells in which HOTAIRM1 or HOXA1 expression was knocked down (Fig.5D). Because MDSCs suppressed CD4 T cell functions following activation by TCR stimulation (with anti-CD3/CD28), we asked whether HOTAIRM1-modulated MDSCs can also affect HIV-specific CD4 T cell function in response to HIV protein stimulation. Indeed, we found that HOTAIRM1- or HOXA1-silenced MDSCs derived from PLHIV also affected IFN-γ production from autologous CD4 T cells that were stimulated with an HIV protein, gp120 (Fig.5E). These results support a role for the HOTAIRM1-HOXA1 axis in promoting the MDSC immunosuppressive effects on T cell functions.
Figure 5. MDSCs suppress CD4 T cell functions.
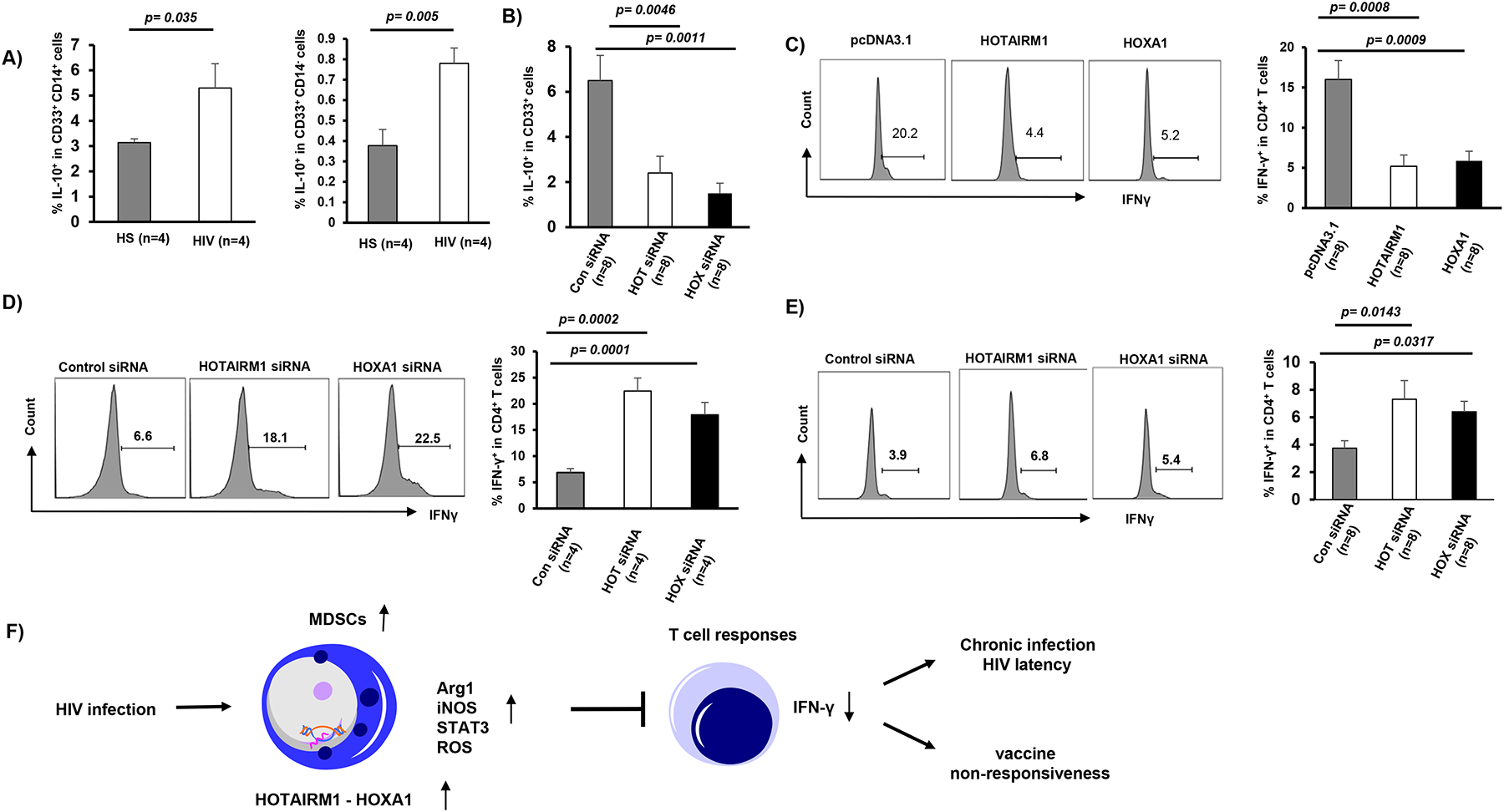
A) IL-10 expression in M-MDSCs and G-MDSCs derived from PLHIV and HS, measured by flow cytometry. B) IL-10 expression in CD33+ myeloid cells derived from PLHIV and transfected by HOTAIRM1 siRNA, HOXA1 siRNA, or control siRNA. C) IFN-γ production is suppressed in CD4 T cells following incubation with autologous HS CD33+ cells overexpressing HOTAIRM1 or HOXA1. D) IFN-γ production is restored in HIV CD4 T cells following incubation with autologous CD33+ cells silencing of HOTAIRM1 or HOXA1 expression. E) IFN-γ production is restored in HIV CD4 T cells stimulated with HIV gp120 for 2 days following incubation with autologous CD33+ cells silencing of HOTAIRM1 or HOXA1 expression. F) A model describing the role of HOTAIRM1-HOXA1 in MDSC development and the mechanism by which it suppresses host immune responses. HIV infection can induce MDSC differentiation and production of immunosuppressive molecules, such as Arg1, iNOS, pSTAT3, and ROS via induction of the HOTAIRM1-HOXA1 axis. HOTAIRM1 and HOXA1 can reciprocally regulate each other to promote MDSC differentiation and suppressive functions, which in turn inhibit T cell responses, potentially contributing to viral latency and vaccine non-responsiveness during chronic HIV infection.
Discussion
MDSCs expand and inhibit host immunity during HIV infection [10–18], but the mechanisms that regulate their expansion during HIV infection remain unclear [19]. In this study, we demonstrated that: 1) expression of lncRNA HOTAIRM1 and its target gene HOXA1 are upregulated in MDSCs that accumulate during latent HIV infection; 2) upregulation of HOTAIRM1 and HOXA1 is closely associated with the expression of immunosuppressive mediators in MDSCs; 3) overexpression of HOTAIRM1 or HOXA1 in healthy CD33+ myeloid cells promotes their differentiation into MDSCs with immunosuppressive functions, whereas silencing their expressions in CD33+ myeloid cells derived from PLHIV attenuates MDSC differentiation and immunosuppressive functions; and 4) manipulating the HOTAIRM1-HOXA1 axis in MDSCs can affect T cell functions, as demonstrated by IFN-γ production by co-culturing CD4 T cells with HOTAIRM1- or HOXA1-modulated CD33+ myeloid cells from both HS and PLHIV. Based on these findings and previous reports [10–18], we propose a model (Fig.5C) illustrating the role and mechanism by which the HOTAIRM1-HOXA1 axis promotes MDSC development to suppress the host immune responses during HIV infection. According to this model, HIV infection can induce MDSC differentiation and their production of immunosuppressive mediators, such as Arg1, iNOS, STAT3, and ROS via upregulating HOTAIRM1 and HOXA1 expressions. HOTAIRM1 and HOXA1 can regulate each other in a positive feedback manner to control MDSC development, which in turn suppresses T cell functions, thereby potentially contributing to viral persistence and vaccine non-responsiveness during latent HIV infection.
lncRNAs are key regulators of chromatin structure, affecting the epigenetic state and expression level of target genes through interactions with histone modifiers, chromatin remodeling complexes, transcriptional regulators, or the DNA methylation machinery [20–22]. In the nucleus, lncRNAs can act as a scaffold, recruiting activators or suppressors to target gene promoters, and can epigenetically regulate gene transcription by inducing histone modifications and chromatin remodeling. In the cytoplasm, lncRNAs can act as a sponge for miRNAs, which modify gene expression at the post-transcriptional level [20–22]. In the current study, we found that the lncRNA HOTAIRM1 can regulate HOXA1 expression, and vice versa, to control MDSC development during HIV infection.
Our results suggest that HOXA1 is a target for positive regulation by HOTAIRM1. HOXA1, a member of the HOXA gene cluster, is upregulated in human malignancies and acts as an oncogene. In our study, the pattern of HOTAIRM1 expression is rather similar to that of the HOXA gene, lending support to the notion that the intergenic non-coding transcription of HOX genomic regions is crucial to maintaining the active state of HOX clusters. Notably, HOX gene clusters have a specific pattern of lineage-restricted expressions, where HOXA genes are predominantly expressed in myeloid cells [37, 38]. The upregulation of some genes of the HOXA cluster has been observed in several subtypes of acute myeloid leukemia (AML) [39, 40]. Mechanistically, HOTAIRM1 contributes to three-dimensional chromatin organization changes that are required for the temporal collinear activation of HOXA genes [33]. HOTAIRM1 also contributes to the physical dissociation of chromatin loops at the cluster proximal end, which delays recruitment of the histone demethylase UTX and transcription of central HOXA genes [33]. In addition, a previous study reported that HOTAIRM1 mediates demethylation of histone proteins and reduces DNA methylation levels via epigenetic modulation of HOXA1 gene expression [32]. These studies provide examples of transcriptional control via the chromatin state and may help explain the role of HOTAIRM1 within the HOXA gene cluster.
How HOTAIRM1 and HOXA1 control each other’s expression in a mutually exclusive manner remains unclear. DNA sequence analysis of HOTAIRM1 shows poor conservation among mammals; however, it has a long CpG island that is associated with the transcription start site, as seen in almost all mammalian intergenic RNAs. Such DNA structure suggests the presence of a bi-directional promoter shared by the divergent coding and noncoding RNAs that has been proposed to facilitate the cis action of intergenic HOTAIRM1 on their genic partners [22, 23]. However, the expression of HOTAIRM1 and HOXA1 is not always synchronized, suggesting that HOTAIRM1 may also be transcribed independently from the immediately adjacent gene. For example, retinoid receptors or other transcription factors may mediate the induction of HOTAIRM1 [24]. Additional putative retinoid response elements have been predicted within the 3′ side of the HOXA cluster, including one within the CpG island embedded in the shared promoter region of HOXA1 and HOTAIRM1 [22, 23]. These studies support a positive feedback loop in the regulation of the HOXA intergenic transcript HOTAIRM1, impacting the differentiation and function of myeloid lineage cells and paving the way for further analysis of its potential regulatory function in hematopoiesis during viral infection.
Interactions between lncRNAs and mRNAs have been described previously [41, 42]. These studies showed a multilayered complexity of RNA crosstalk and competition, and that lncRNAs seem to regulate both the expression of neighboring genes and distinct genomic sequences [43]. Interestingly, the HOX genomic regions encode numerous lncRNAs, suggesting that these lncRNAs may participate in the regulation of HOX expression [24]. Specifically, HOTAIRM1 quantitatively regulates the expressions of HOXA1 and HOXA4 [27]. In addition, HOTAIRM1 regulates cell cycle progression during myeloid maturation in the NB4 human promyelocytic leukemia cells [30]. Our study clearly shows that HOTAIRM1 and HOXA1 regulate each other’s expression to control CD33+ myeloid cell differentiation into MDSCs in the setting of HIV infection. Notably, we have recently demonstrated that HCV-containing exosomes upregulate HOTAIRM1, which controls miRNA-124 expression and MDSC development [[18], and unpublished observations]. These findings are in line with a recent report showing that the HOTAIR-miR214 axis plays an essential role in the proliferation, migration, and invasion of hepatocellular carcinoma [44]. The putative cooperative role of two ncRNAs - HOTAIRM1 and miR124 - in MDSC development during HIV infection merits further investigation.
In this study, we focused on PLHIV on ART because they make up the major population in the era of ART. Due to ART-mediated control of HIV replication, a very small proportion of cells (~ one in a million cells) are infected with HIV in the PBMCs of PLHIV and latently infected cells do not express viral proteins [45]. Thus, it is unlikely that HIV infection per se causes HOTAIRM1/HOXA1 upregulation. However, we and others have shown that ART-controlled PLHIV with no viral replication can still exhibit a phenotype of immune suppression, as evidenced by MDSC expansion, suppressed T cell functions, and dampened vaccine responses [12–18]. This may be due to an early HIV-mediated suppression that persists in the latent phase, or likely a low-grade inflammation in the setting of ART-controlled, latent HIV infection. A myriad of insults may contribute to this inflammation, such as viral particles or viral RNAs/proteins released from the reservoirs, cell-secreted pro-inflammatory cytokines, HIV-enhanced gut permeability and altered gut microbiota or dysbiosis, frequent cytomegalovirus (CMV), Epstein–Barr virus (EBV), hepatitis B virus (HBV) and hepatitis C virus (HCV) coinfections, the ART regimen itself, as well as other comorbidities including malignancies, personal stresses, or social and environmental factors [46–50]. We believe that these inflammatory factors or conditions may promote HOTAIRM1/HOXA1 upregulation in the setting of latent HIV infection.
A growing list of lncRNAs have been validated as bona fide regulators of gene expression, and many studies suggest that these noncoding transcripts may serve as potential biomarkers and therapeutic targets for human diseases. To our knowledge, this is the first report showing that the HOTAIRM1-HOXA1 axis promotes MDSC development and immunosuppressive functions during latent HIV infection. Therefore, targeting this axis may provide a novel therapeutic approach for immunomodulation in conjunction with ART to protect against the immune dysregulatory effects of latent HIV infection.
Acknowledgments and disclosures:
This work was supported by National Institutes of Health grants R01AI114748 R21AI138598, and R15AG050456; VA Merit Review Awards 1I01BX002670 and 1I01BX004281; and DoD Award PR170067 (to Z.Q.Y/J.P.M). This publication is the result of work supported with resources and the use of facilities at the James H. Quillen Veterans Affairs Medical Center. The contents in this publication do not represent the views of the Department of Veterans Affairs or the United States Government. The authors declare no competing financial interests.
Footnotes
Competing interests: The authors declare no competing financial interests.
References
- 1.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol 2009; 182(8):4499–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009; 9(3):162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai J, El Gazzar M, Li GY, Moorman JP, Yao ZQ. Myeloid-derived suppressor cells: paradoxical roles in infection and immunity. J Innate Immun 2015; 7(2):116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munder M, Mollinedo F, Calafat J, Canchado J, Gil-Lamaignere C, Fuentes JM, et al. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood 2005; 105(6):2549–2556. [DOI] [PubMed] [Google Scholar]
- 5.Liu CY, Wang YM, Wang CL, Feng PH, Ko HW, Liu YH, et al. Population alterations of L-arginase- and inducible nitric oxide synthase-expressed CD11b+/CD14−/CD15+/CD33+ myeloid-derived suppressor cells and CD8+ T lymphocytes in patients with advanced-stage non-small cell lung cancer. J Cancer Res Clin Oncol 2010; 136(1):35–45. [DOI] [PubMed] [Google Scholar]
- 6.Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest 2013; 123(4):1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poschke I, Mougiakakos D, Hansson J, Masucci GV, Kiessling R. Immature immunosuppressive CD14+ HLA-DR−/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-sign. Cancer research 2010; 70(11):4335–4345. [DOI] [PubMed] [Google Scholar]
- 8.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol 2009; 182(9):5693–5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohl K, Tenbrock K. Reactive Oxygen Species as Regulators of MDSC-Mediated Immune Suppression. Front Immunol 2018; 9:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vollbrecht T, Stirner R, Tufman A, Roider J, Huber RM, Bogner JR, et al. Chronic progressive HIV-1 infection is associated with elevated levels of myeloid-derived suppressor cells. AIDS 2012; 26(12):F31–37. [DOI] [PubMed] [Google Scholar]
- 11.Macatangay BJ, Landay AL, Rinaldo CR. MDSC: a new player in HIV immunopathogenesis. AIDS (London, England) 2012; 26(12):1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin A, Cai W, Pan T, Wu K, Yang Q, Wang N, et al. Expansion of monocytic myeloid-derived suppressor cells dampens T cell function in HIV-1-seropositive individuals. J Virol 2013; 87(3):1477–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg A, Spector SA. HIV type 1 gp120-induced expansion of myeloid derived suppressor cells is dependent on interleukin 6 and suppresses immunity. J Infect Dis 2014; 209(3):441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gama L, Shirk EN, Russell JN, Carvalho KI, Li M, Queen SE, et al. Expansion of a subset of CD14highCD16negCCR2low/neg monocytes functionally similar to myeloid-derived suppressor cells during SIV and HIV infection. J Leukoc Biol 2012; 91(5):803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowers NL, Helton ES, Huijbregts RP, Goepfert PA, Heath SL, Hel Z. Immune suppression by neutrophils in HIV-1 infection: role of PD-L1/PD-1 pathway. PLoS Pathog 2014; 10(3):e1003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony D, Umbleja T, Aberg J, Kang M, Medvik K, Lederman M, et al. Lower peripheral blood CD14+ monocyte frequency and higher CD34+ progenitor cell frequency are associated with HBV vaccine induced response in HIV infected individuals. Vaccine 2011; 29(19):3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehraj V, Jenabian MA, Vyboh K, Routy JP. Immune Suppression by Myeloid Cells in HIV Infection: New Targets for Immunotherapy. Open AIDS J 2014; 8:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Zhao J, Ren JP, Wu XY, Morrison ZD, Elgazzar MA, et al. Expansion of myeloid-derived suppressor cells promotes differentiation of regulatory T cells in HIV-1+ individuals. AIDS 2016; 30(10):1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao ZQ, Moorman JP. Immune exhaustion and immune senescence: two distinct pathways for HBV vaccine failure during HCV and/or HIV infection. Archivum immunologiae et therapiae experimentalis 2013; 61(3):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends in Genetics 2017; 33(10):665–676. [DOI] [PubMed] [Google Scholar]
- 21.Morceau F, Chateauvieux S, Gaigneaux A, Dicato M, Diederich M. Long and short non-coding RNAs as regulators of hematopoietic differentiation. Int J Mol Sci 2013; 14(7):14744–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorg Chem 2019; 92:103214. [DOI] [PubMed] [Google Scholar]
- 23.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447(7146):799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129(7):1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki YT, Sano M, Kin T, Asai K, Hirose T. Coordinated expression of ncRNAs and HOX mRNAs in the human HOXA locus. Biochem Biophys Res Commun 2007; 357(3):724–730. [DOI] [PubMed] [Google Scholar]
- 26.Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA 2007; 13(2):223–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009; 113(11):2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mechanisms of development 1992; 38(3):217–227. [DOI] [PubMed] [Google Scholar]
- 29.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 2017; 24(2):212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol 2014; 11(6):777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, et al. Long Non-Coding RNA HOXA Transcript Antisense RNA Myeloid-Specific 1-HOXA1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front Immunol 2018; 9:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res 2018; 37(1):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang XQ, Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res 2017; 45(3):1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Díaz-Beyá M, Brunet S, Nomdedéu J, Pratcorona M, Cordeiro A, Gallardo D, et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015; 6(31):31613–31627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bizymi N, Bjelica S, Kittang AO, Mojsilovic S, Velegraki M, Pontikoglou C, et al. Myeloid-Derived Suppressor Cells in Hematologic Diseases: Promising Biomarkers and Treatment Targets. Hemasphere 2019; 3(1):e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pawelec G, Verschoor CP, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells: Not Only in Tumor Immunity. Front Immunol 2019; 10:1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X, Tian J, Tang X, Ma J, Wang S. Long non-coding RNAs in the regulation of myeloid cells. J Hematol Oncol 2016; 9(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong M, Goodell MA. Noncoding Regulatory RNAs in Hematopoiesis. Curr Top Dev Biol 2016; 118:245–270. [DOI] [PubMed] [Google Scholar]
- 39.De Braekeleer E, Douet-Guilbert N, Basinko A, Le Bris MJ, Morel F, De Braekeleer M. Hox gene dysregulation in acute myeloid leukemia. Future Oncol 2014; 10(3):475–495. [DOI] [PubMed] [Google Scholar]
- 40.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 2011; 118(13):3645–3656. [DOI] [PubMed] [Google Scholar]
- 41.Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene-and microRNA-expression signatures: a Cancer and Leukemia Group B study. Journal of clinical oncology 2010; 28(4):596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505(7483):344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, Shang Z, Ma Y, Ma J, Song J. HOTAIR/miR-214–3p/FLOT1 axis plays an essential role in the proliferation, migration, and invasion of hepatocellular carcinoma. Int J Clin Exp Pathol 2019; 12(1):50–63. [PMC free article] [PubMed] [Google Scholar]
- 45.Persaud D, Pierson T, Ruff C, Finzi D, Chadwick KR, Margolick JB, et al. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J Clin Invest. 2000; 105:995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leeansyah E, Cameron PU, Solomon A, Tennakoon S, Velayudham P, Gouillou M, et al. Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: a potential factor contributing to HIV-associated accelerated aging. J Infect Dis. 2013; 207:1157–65. [DOI] [PubMed] [Google Scholar]
- 47.Nasi M, De Biasi S, Gibellini L, Bianchini E, Pecorini S, Bacca V, et al. Ageing and inflammation in patients with HIV infection. Clin. Exp. Immunol 2017; 187:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai SN, Landay AL HIV and aging: role of the microbiome. Curr. Opin. HIV AIDS 2018; 13:22–27. [DOI] [PubMed] [Google Scholar]
- 49.Taddei TH, Lo Re V 3rd, Justice AC HIV, Aging, and Viral Coinfections: Taking the Long View. Curr HIV/AIDS Rep. 2016; 13:269–78. [DOI] [PubMed] [Google Scholar]
- 50.Montejano R., Stella-Ascariz N, Monge S, Bernardino JI, Pérez-Valero I, Montes ML, et al. Impact of Antiretroviral Treatment Containing Tenofovir Difumarate on the Telomere Length of Aviremic HIV-Infected Patients. J. Acquir. Immune Defic. Syndr 2017; 76:102–109. [DOI] [PubMed] [Google Scholar]


