Abstract
Background
Microglia, the primary immune cells of the brain, are implicated in alcohol use disorder (AUD). However, it is not known if microglial activation contributes to the transition from alcohol use to AUD or is a consequence of alcohol intake.
Methods
We investigated the role of microglia in a mouse model of alcohol dependence using a colony stimulating factor 1 receptor inhibitor (PLX5622) to deplete microglia and a chronic intermittent ethanol vapor two-bottle choice drinking procedure. Additionally, we examined anxiety-like behavior during withdrawal. We then analyzed synaptic neuroadaptations in the central nucleus of the amygdala (CeA) and gene expression changes in the medial prefrontal cortex (mPFC) and CeA from the same animals used for behavioral studies.
Results
PLX5622 prevented escalations in voluntary alcohol intake and decreased anxiety-like behavior associated with alcohol dependence. PLX5622 also reversed expression changes in inflammatory-related genes and glutamatergic and GABAergic genes in the mPFC and CeA. At the cellular level in these animals, microglia depletion reduced inhibitory GABAA and excitatory glutamate receptor-mediated synaptic transmission in the CeA, supporting the hypothesis that microglia regulate dependence-induced changes in neuronal function.
Conclusions
Our multifaceted approach is the first to link microglia to the molecular, cellular and behavioral changes associated with the development of alcohol dependence, suggesting that microglia may also be critical for the development and progression of AUD.
Keywords: Microglia, alcohol, PLX5622, ethanol dependence, transcriptome, electrophysiology
Introduction
Microglia (MG), the resident macrophages of the brain, play critical roles in development and homeostasis, as well as in injury and disease. MG are present in mouse brain after embryonic day 9 (1) and respond to localized signals that drive changes in transcription, morphology and proliferation (2,3). Loss of homeostatic control during disease leads to re-expression of developmental pathways and eventually to a disease phenotype (3). This dysregulation produces abnormal synaptic engulfment (4), loss of blood-brain-barrier integrity (5), peripheral immune cell recruitment (5) and chronic immune activation (6).
Gene expression profiling in human brain has linked immune activation with alcohol use disorder (AUD) (7–10), supporting a role for brain MG in the addiction process. A critical part of AUD pathology involves aberrant immune/inflammatory pathways in both the brain and periphery (9). Immune and inflammatory genes also regulate alcohol drinking and related behaviors in rodent models (11). MG express and secrete immune molecules that regulate neuronal transmission important in alcohol behaviors (12,13). Binge-like alcohol exposure primes and activates MG, causing heightened proliferation, morphological changes and immune responses in postmortem brains of individuals with AUD and in brains from ethanol-exposed rodents (14,15). MG depletion, however, blunts neuroinflammatory responses after binge ethanol withdrawal (16). It is not known if MG are contributing to pathogenesis and AUD progression or, rather, are activated as a consequence of disease pathology.
We tested the hypothesis that MG participate in the development of alcohol dependence in mice using a combination of behavior, histology, RNA-sequencing (RNA-seq) and electrophysiology. We examined the medial prefrontal cortex (mPFC) and central nucleus of the amygdala (CeA), key brain regions in the neurocircuitry of AUD. We used an established mouse drinking model of alcohol dependence (chronic intermittent ethanol vapor two-bottle choice, CIE-2BC) that mimics phenotypes in AUD (13,15), and PLX5622, a colony stimulating factor 1 receptor inhibitor (CSFR1) to deplete MG (17). CIE-2BC increased MG proliferation in the mPFC, and MG depletion prevented dependence-induced escalation of drinking, decreased withdrawal-associated anxiety behavior and reversed changes in immune, glutamatergic and GABAergic genes in the mPFC and CeA. MG depletion also decreased CeA glutamatergic and GABAergic synaptic transmission. This is the first multidisciplinary study in the same animals causally linking MG to molecular, cellular and behavioral changes associated with alcohol dependence.
Materials & Methods
All procedures were approved by The Scripps Research Institute Institutional Animal Care and Use Committee and conforms to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. A detailed description of experimental approaches and statistical analyses can be found in Supplemental Information.
Chronic intermittent ethanol two-bottle choice procedure
The CIE-2BC procedure was used as described (18,19). We initially used a cohort of 12 male C57BL/6J mice (cohort 1) to examine MG proliferation and tested three groups: 1) mice that exhibit escalating alcohol intake after CIE vapor, i.e., alcohol dependent (Dep); 2) control mice with the same voluntary drinking history that were only exposed to air vapor and do not show escalations in alcohol intake, i.e., non-dependent (Non-Dep); and 3) alcohol-naïve control mice (Naïve). Blood alcohol levels were measured bi-weekly on the third or fourth day of ethanol vapor exposure (Figure S1b).
MG depletion
A second cohort of 48 male C57BL/6J mice was treated with either chow containing PLX5622 to deplete MG or with control chow. Mice were started on the MG depletion diet four weeks before the CIE-2BC procedure and remained on PLX5622 chow throughout the study (Figure S1).
Anxiety-like behavioral assay
The novelty suppressed feeding test (NSFT) was performed as described (20). Four days following their last 2BC drinking session, mice were food deprived (24h) and tested for latency to eat a piece of regular chow in a novel arena and then latency to eat a piece of regular chow in their home cage (Figure S1a).
Immunohistochemistry (IHC)
Animals used for IHC (and for RNA isolation and electrophysiology) were exposed to a final 16h-bout of ethanol vapor and euthanized within 2h of removal from the chamber. Mice (n=23) were anesthetized, transcardially perfused and brains were removed and fixed in Z-Fix, then flash frozen and sectioned (35μm) on a cryostat and cryoprotected. IHC was performed as described (19,21). Free-floating sections were stained for IBA1 (Ionized Calcium Binding Adaptor Molecule 1) and TMEM119 (Transmembrane Protein 119) in the mPFC and CeA. Sections were imaged and analyzed as described (19,21).
RNA isolation and sequencing
Cohort 2 mice were exposed to a final 16h-bout of ethanol vapor and mPFC (n=34) and CeA (n=19) were dissected. Total RNA was isolated as described (22,23) and submitted to the Genomic Sequencing and Analysis Facility at the University of Texas at Austin for RNA-seq. Raw and processed data are available on Gene Expression Omnibus (GEO; GSE142571).
Electrophysiology
Mice (n=5 animals/group) were used within 2h of removal from the vapor chambers. Ex vivo brain slices were prepared as described (19,24). Using whole-cell patch-clamp, we recorded pharmacologically isolated spontaneous GABAA receptor-mediated inhibitory postsynaptic currents (sIPSCs) or glutamate receptor-mediated excitatory postsynaptic currents (sEPSCs) from neurons in the medial subdivision of CeA. Intrinsic membrane properties and excitability were assessed in current-clamp mode using a protocol comprised of hyperpolarizing and depolarizing steps.
Drugs
AP-5 (NMDA receptor antagonist), bicuculline (GABAA receptor antagonist), CGP 55845A (GABAB receptor antagonist) and DNQX (AMPA and kainate receptor antagonist) were purchased from Tocris (Bristol, UK). Drugs were dissolved in aCSF and applied by bath perfusion. PLX5622 (Plexxikon Inc.; Berkeley, CA) was formulated at a dose of 1200 ppm in RMH1800 chow (Research Diets; New Brunswick, NJ). Control chow (RMH 1800) was also provided by Research Diets.
Statistics
Data are presented as mean ± standard error of the mean (SEM) values. Data were analyzed using Prism 8.01 (GraphPad, San Diego, CA) or R programming (P<0.05 was the criterion for statistical significance, and n represents the number of mice, cells or images). IHC data were analyzed by one-way ANOVA with Bonferroni post hoc tests (Fig 1) or by two-way ANOVA. We used three-way ANOVA with repeated measures to examine Time*Diet*CIE interactions for 2BC alcohol intake. Planned orthogonal contrasts analyzed effects of CIE and MG depletion on anxiety-related behavior and escalation in voluntary alcohol intake (25). Mann-Whitney U-Tests were additionally used for anxiety-related behavior. Differential gene expression analysis was performed using a multiple factor design with interactions, except for planned orthogonal contrasts in the CeA between MG depletion/Dep and Control Diet/Dep groups. Genes with a nominal significance threshold of P<0.05 were considered significant. This threshold was selected to balance type-1 and type-2 error rates when performing pathway and network analyses. Electrophysiological data were analyzed using two-way ANOVA with Sidak’s multiple comparisons post hoc tests.
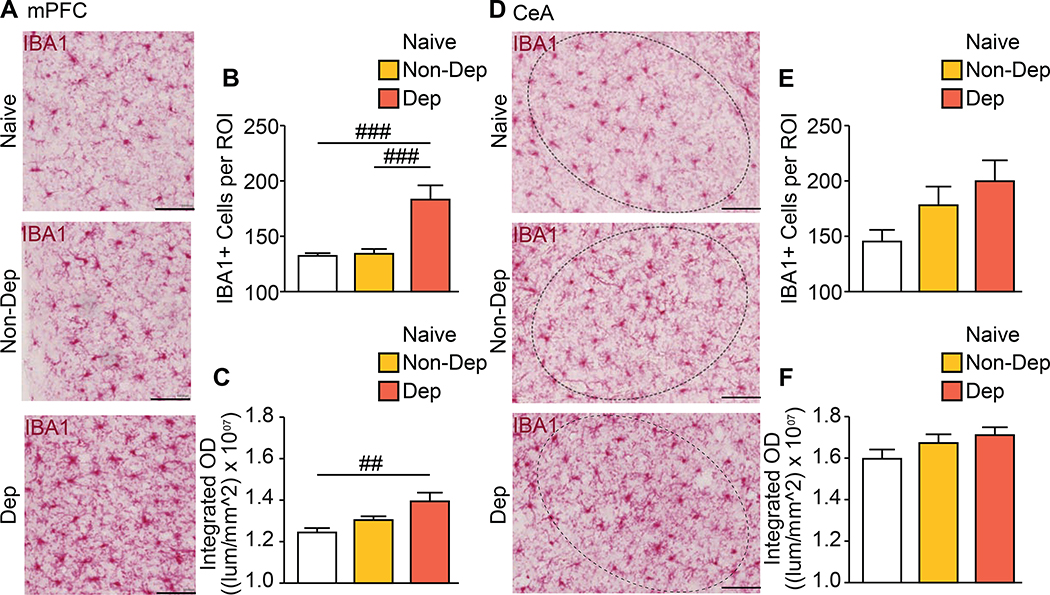
Figure 1. Increased number of MG in mPFC of dependent mice.
(a) Representative images of Ionized Calcium Binding Adaptor Molecule 1 (IBA1)-labeled (red) MG in the medial prefrontal cortex (mPFC) of Naïve, non-dependent (Non-Dep) and alcohol-dependent (Dep) mice. Summary graph of (b) the number of IBA1+ cells and (c) integrated optical density (OD) present in the mPFC. (d) Representative images of IBA1-labeled (red) MG in the central nucleus of the amygdala (CeA) of Naïve, non-dependent (Non-Dep) and dependent (Dep) mice. Circled region of interest (ROI) indicates the CeA. Summary graphs of (e) the number of IBA1+ cells and (f) integrated OD present in the CeA. Scale bar = 100 μm. Data are presented as mean±SEM values; ## P<0.01; ### P<0.001.
Results
MG proliferation in mPFC in dependent mice
We first determined if the CIE-2BC procedure induces brain region-specific proliferation of MG, as observed in AUD (15). We compared alcohol-dependent (Dep) mice exhibiting escalations in voluntary alcohol intake, non-dependent (Non-Dep) mice that do not show escalations and ethanol-naïve controls (Naïve). We used IBA1 as a general MG marker to determine changes in the number of MG-like cells in the CeA and mPFC (26). There was an increased number of cells expressing IBA1 in the mPFC of Dep mice (one-way ANOVA: F(2,85)=14.14; P<0.0001) compared with Non-Dep (Padj<0.0001) and Naïve (Padj<0.0001) mice. Likewise, the density of IBA1 expression increased in Dep mice (ANOVA: F(2,85)=5.87; P<0.01) compared with Naïve (Padj=0.0031), but not compared with Non-Dep mice (Figure 1a-c). In the CeA, however, there were no group differences in the number of cells expressing IBA1 or expression density, although a trend toward increased IBA1-expressing cells in Dep mice was observed (P=0.069) (Figure 1d-f, Figure S1). These results support strong MG proliferation in cortical areas similar to humans with AUD (15).
MG depletion blunts escalation of alcohol intake and anxiety-like behavior in dependent mice
Based on the robust MG proliferation in Dep mice, we tested the hypothesis that proliferation is associated with alcohol dependence using four groups of male mice: 1) MG intact, non-dependent (Control Diet/Non-Dep); 2) MG depleted, non-dependent (MG Depletion/Non-Dep); 3) MG intact, dependent (Control Diet/Dep); and 4) MG depleted, dependent (MG Depletion/Dep). Timelines for CIE-2BC and behavioral testing are in Figure S2a. MG depletion in the CeA and mPFC was verified using IBA1 and TMEM119 (Figures S3–4).
There was no escalation of alcohol drinking in MG Depletion/Dep mice (orthogonal contrast 1, Control Diet/Dep vs. MG Depletion/Dep + Control Diet/Non-Dep + MG Depletion/Non-Dep: t(3,44)=2.63; P=0.012, Figure 2a). There was also no difference in MG-depletion/Dep vs. Non-Dep mice (orthogonal contrast 2: MG Depletion/Dep vs. Control Diet/Non-Dep + MG Depletion/Non-Dep mice t(3,44)=0.006; P=0.99, Figure 2a). MG depletion also did not alter non-dependent drinking (orthogonal contrast 3: Control Diet/Non-Dep vs. MG Depletion/Non-Dep t(3,44)=0.17; P=0.86, Figure 2a). These findings support the hypothesis that MG depletion impedes dependence-induced escalation in drinking.
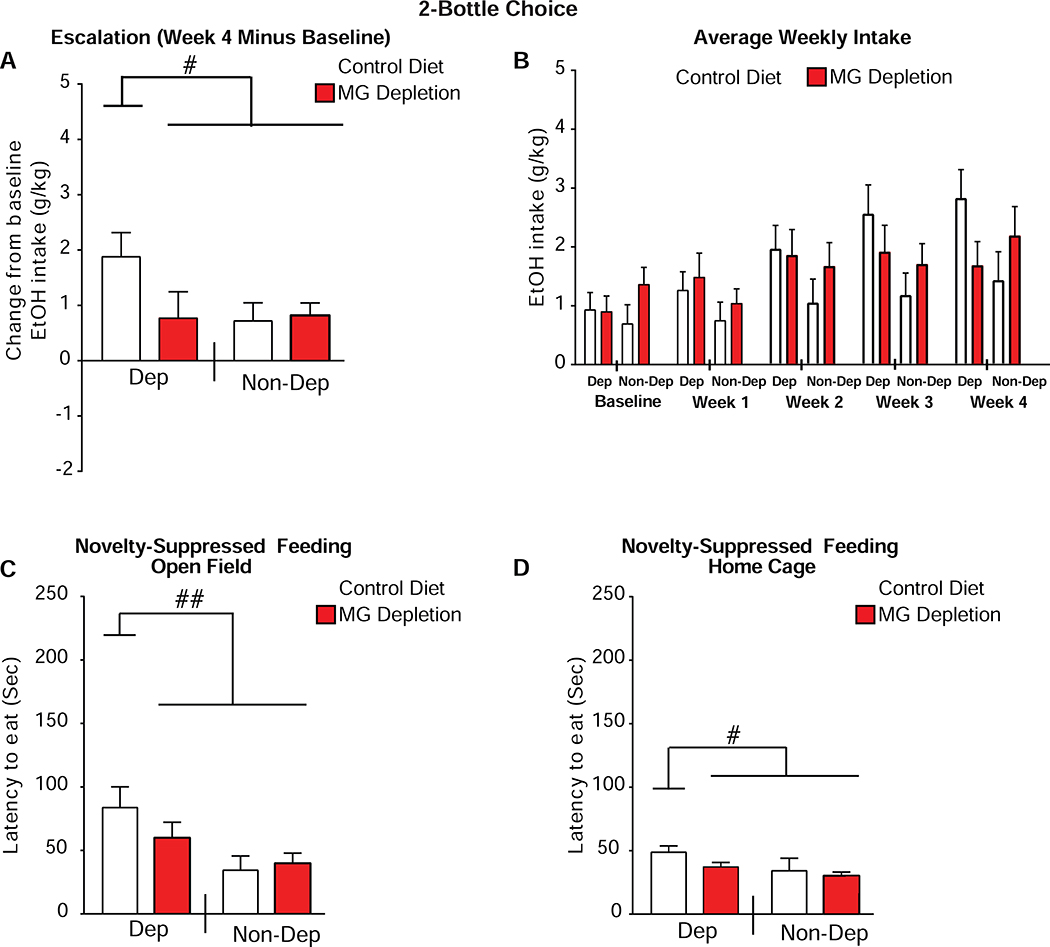
Figure 2. MG depletion prevents dependence-induced escalation in alcohol intake.
(a) Change in average weekly two bottle choice (2BC) ethanol (EtOH) intake (escalation) between week 4 and baseline. (b) Average weekly 2BC intake after each week of chronic intermittent ethanol (CIE) treatment. (c) Novelty-suppressed feeding test (NSFT) and latency to eat in the open field. (d) NSFT and latency to eat in the home cage. n=12 mice per group, except for the Control Diet/Non-Dep (non-dependent) group in the NSFT (n=9). Data are presented as mean±SEM values; # P<0.05, ## P<0.01, planned orthogonal contrasts.
There was a trend toward a within-subjects interaction, indicating that both MG depletion and alcohol dependence influenced escalation over 4 weeks of 2BC drinking (Time*Diet*CIE interaction, two-way ANOVA: F(2,99)=2.70; P=0.063, Greenhouse-Geisser correction, Figure 2b). Blood alcohol levels during vapor exposure did not differ between Control Diet/Dep and MG Depletion/Dep mice or Control Diet/Non-Dep and MG Depletion/Non-Dep mice (Figure S2b). Although MG Depletion/Non-Dep mice initially weighed about 4.67% more than the other groups (Diet*Alcohol, F(1,44)=4.31; P=0.044), by the end of the experiment the interaction was absent (F(1,44)=0.23; P=0.66).
We examined the effect of MG depletion on anxiety-related behavior using NSFT. Control Diet/Dep mice exhibited a significant protraction in latency to eat in the open field compared with the other groups in the open field (t(3,41)=2.84; P=0.006, Figure 2c) and the home cage (t(3,41)=2.69; P=0.01, Figure 2d). For orthogonal contrasts examining latency in the open field, MG Depletion/Dep mice did not differ from the pooled Non-Dep groups (Control Diet/Non-Dep + MG Depletion/Non-Dep, t(3,41)=1.61; P=0.115), nor did the Non-Dep groups differ from one another t(3,41)=−1.55; P=0.128). MG Depletion/Dep mice did not show an increased latency to eat in this novel environment, suggesting that MG depletion reduces anxiety-like behavior in abstinent mice. While latencies to eat were significantly greater in the Control Diet/Dep mice under both conditions, the effect was larger in the open field (Mann-Whitney U-Tests: Control Diet/Dep mice (P=0.003), all other P’s≥0.117). Therefore, MG depletion prevented both dependence-induced escalation of drinking and ameliorated anxiety-related behavior, suggesting that MG regulate behaviors associated with the transition from voluntary alcohol drinking to dependence.
MG depletion reduces expression of neuroimmune and glutamatergic genes in mPFC of dependent mice
Given increased MG proliferation in the mPFC of alcohol-dependent mice, we profiled gene expression in this region from the same mice that underwent the behavioral assays. We note that gene expression and microglial phenotype vary substantially when alcohol is present, relative to acute or protracted alcohol withdrawal (16,27,28). Our results represent expression data within 2h of the last ethanol exposure.
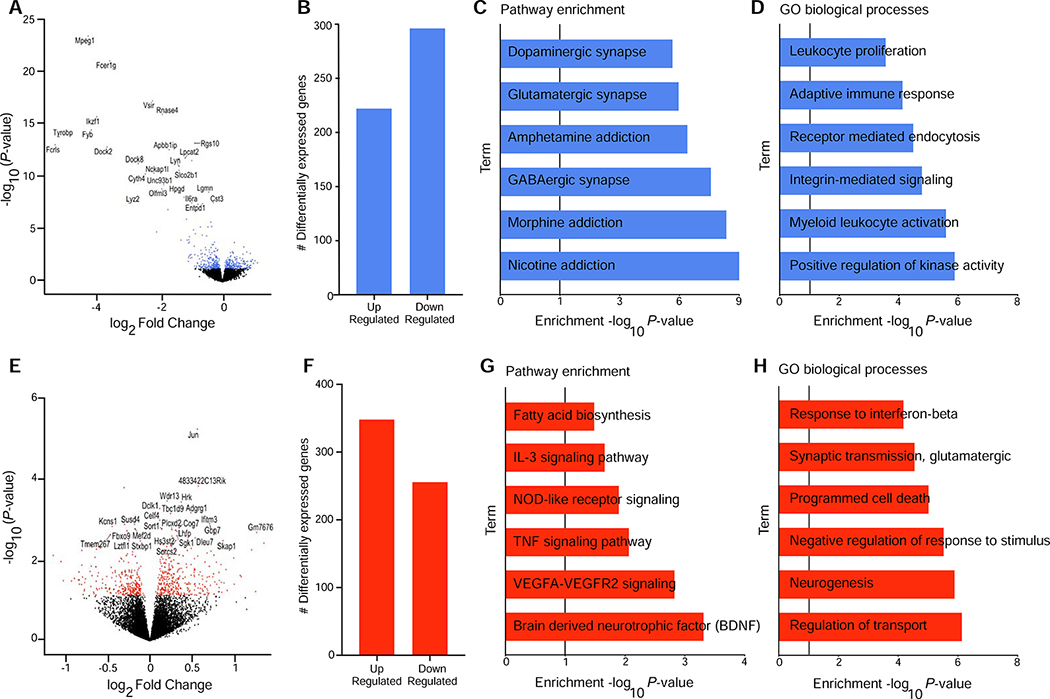
There were 511 and 588 differentially expressed genes (P<0.05) following MG depletion (Control Diet vs. MG Depletion) or alcohol dependence (Non-Dep vs. Dep mice), respectively (Figure 3a,b,e,f). After MG depletion, enrichment analysis supported strong downregulation of gene ontologies (GO) such as myeloid leukocyte activation (GO:0002274), immune response (GO:0006955) (Figure 3d) and glutamatergic and GABAergic synaptic pathways (Figure 3c). After alcohol dependence, there was upregulation of type 1 interferon signaling (GO:0060337), glutamatergic transmission (GO:0035249) and the Brain Derived Neurotrophic Factor (BDNF) pathway (Figure 3g,h). For the interaction between MG depletion and dependence, 270 genes were differentially expressed showing enrichment in anion and cofactor transport (GO:0051186, GO:0006820) (Figure S5). Table S1 shows the complete list of differentially expressed genes.
Figure 3. mPFC transcriptome is altered in response to MG depletion and alcohol dependence.
(a) Volcano plot showing log2 Fold Change against log10 P-value for the main effect of microglia (MG) Depletion. Differentially expressed genes (DEG) (P<0.05) are shown as blue dots. The top (DEG) are labeled. (b) Number of upregulated and downregulated DEG in the medial prefrontal cortex (mPFC) (P<0.05) after MG depletion. (c, d) Pathway enrichment (Webgestalt) and gene ontology (GO) biological process (Webgestalt) analyses, respectively for genes identified after MG depletion. (e) Volcano plot showing log2 Fold Change against log10 P-value for the main effect of alcohol dependence. DEG (P<0.05) are shown as red dots. The top DEG are labeled. (f) Number of upregulated and downregulated DEG in the mPFC (P<0.05) after alcohol dependence. (g, h) Pathway enrichment (Enrichr) and gene ontology (GO) biological process (Webgestalt) analyses, respectively for genes identified after alcohol dependence.
We next performed a systems-level analysis of mPFC gene expression using weighted gene co-expression network analysis (WGCNA) to identify shared molecular targets of MG depletion and alcohol dependence. WGCNA identifies gene expression variation associated with AUD-related traits and other complex disorders (29). Module eigengenes for treatment, alcohol intake and escalation of drinking were identified to examine the biological relationships with co-expression networks (Figure S6).
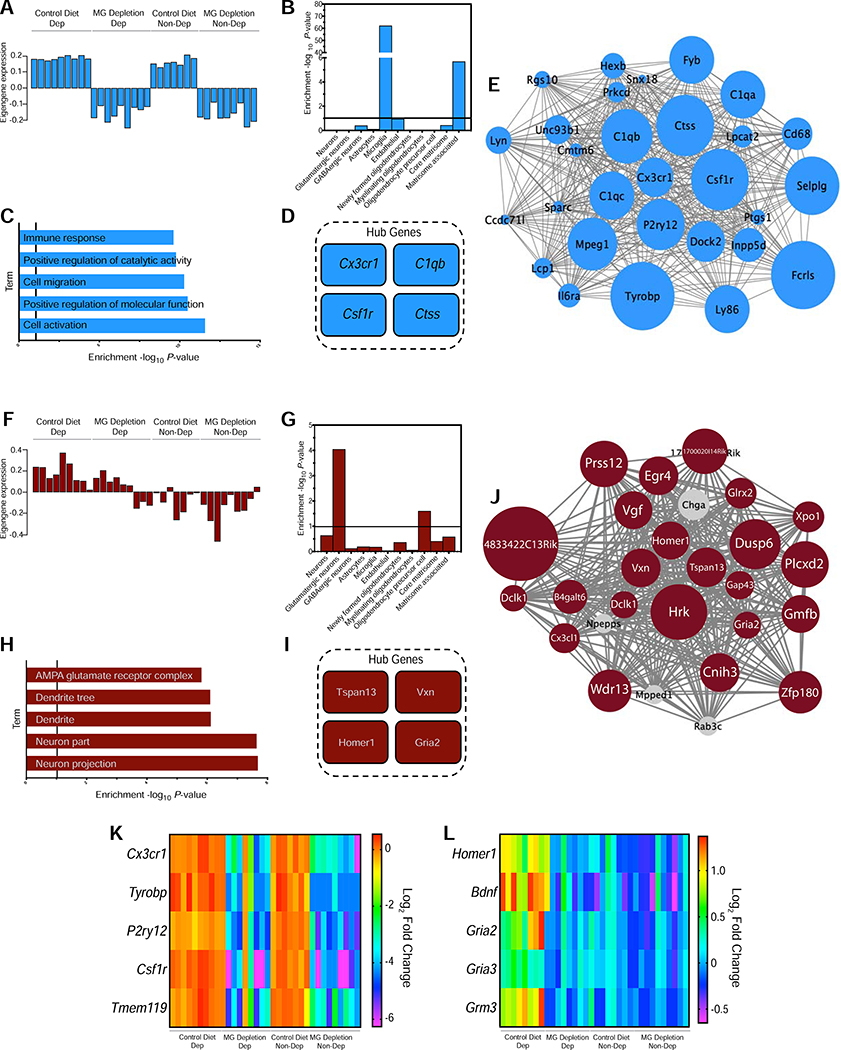
Module 10 (M10) revealed eigengenes that were highly correlated with MG Depletion (r=−0.59, P=3×10−04) (Table S2 cerulean, Figure S6). Co-expressed genes (288 genes) highly enriched in MG markers were downregulated in both MG Depletion/Non-Dep and MG Depletion/Dep mice, suggesting that MG depletion prevents the upregulation of MG neuroimmune responses even after alcohol dependence (Figure 4a,b). M10 included genes enriched in immune response (GO:0006955) and genes that regulate alcohol consumption (B2m, Beta-2 Microglobulin; Ctss, Cathepsin S; Il6, Interleukin 6; Il10, Interleukin 10) (11) (Figure 4c). There was no upregulation of inflammation-related genes, suggesting that MG depletion does not create a compensatory immune response, consistent with previous studies (30). After MG depletion, MG markers were the highest-ranking differentially expressed hub genes: Cx3cr1 (C-X3-C Motif Chemokine Receptor 1), C1qb (Complement C1q B Chain), Ctss, and Csf1r (Figure 4d,e). Figure 4k is a representative heatmap of select MG markers. The M10 depletion signature indicates that MG depletion prevents neuroimmune activation in alcohol-dependent animals.
Figure 4. Gene co-expression modules in the mPFC associated with MG depletion and alcohol dependence.
(a, f) Corresponding module eigengene values (y-axis) across samples (x-axis) in the medial prefrontal cortex. (b, g) Enrichment (y-axis) for the cell-type specific genes (x-axis) belonging to the co-expression module. (c, h) Relevant gene ontology (GO) categories enriched in the M10 and M14 modules, respectively. (d, i) The genes most strongly correlated with the module eigengene value are shown in the Hub Gene inset. (e,j) Visualization of the M10 and M14 modules, respectively. The top 30 connections are shown for each module. Genes that are significantly differentially expressed are shown in the module color, and genes that are in grey are not differentially expressed. Size of the circle represents magnitude of log2 Fold Change. (k) Heatmap displaying expression levels of select microglia (MG) marker-related differentially expressed genes from M10 across treatment groups. Color indicates expression level (regularized log transformed gene counts). (l) Heatmap displaying levels of select glutamatergic-related differentially expressed genes from M14 across treatment groups. Color indicates expression level (regularized log transformed gene counts).
When comparing Non-Dep vs. Dep mice, we identified co-expressed genes strongly correlated with alcohol dependence (M14, r=0.63, P=9×10−05, 126 genes) (Table S2 darkred, Figure S6). Genes were upregulated in Control Diet/Dep mice, but were unaltered in MG Depletion/Dep mice (Figure 4f). M14 was enriched in glutamatergic neuronal genes and synaptic processes, particularly at glutamatergic synapses (Figure 4g,h), consistent with findings in frontal cortex from individuals with AUD (29). Additionally, hub genes involved in glutamatergic signaling (Homer1, Homer Scaffold Protein 1 and Gria2, Glutamate Ionotropic Receptor AMPA Type Subunit 2) have been associated with AUD (Figure 4i,j) (31,32). A representative heatmap of select genes is shown in Figure 4l. These results suggest that MG depletion downregulates neuroimmune- and MG-related genes, and MG depletion may prevent the upregulation of glutamatergic-related genes in the mPFC that occurs in alcohol dependence.
MG depletion prevents coordinated changes in neuroimmune response, and in glutamatergic and GABAergic gene expression in CeA and mPFC
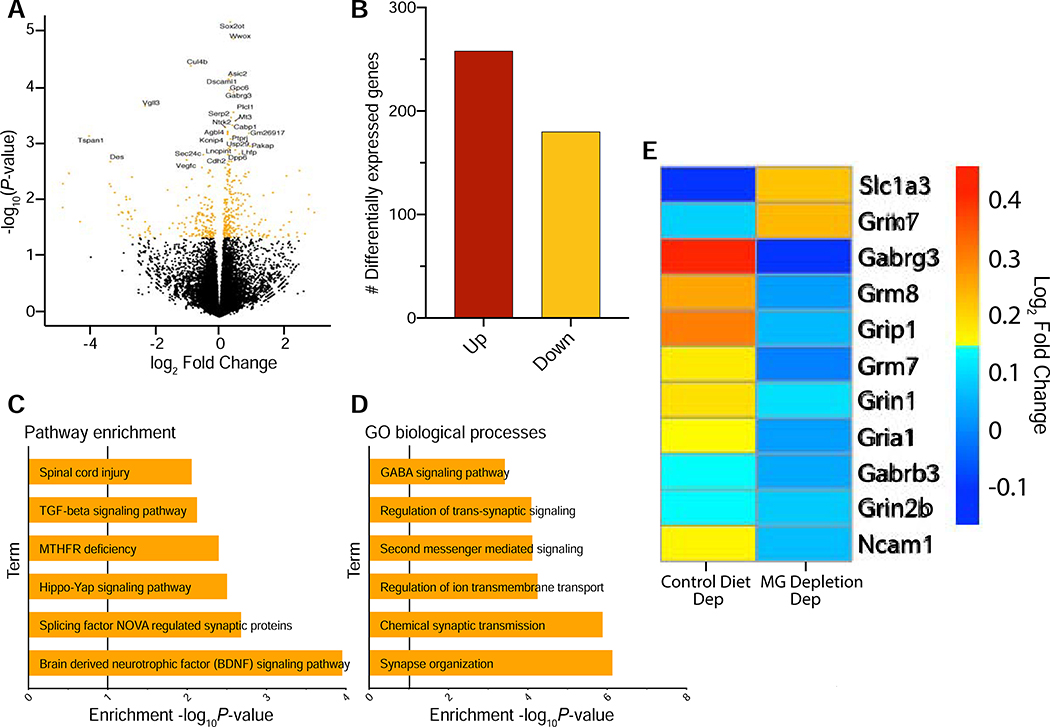
The CeA is implicated in the escalation of alcohol drinking in dependent animals and in withdrawal-associated anxiety behaviors across species (33–36). For Non-Dep vs. Dep mice, 439 CeA genes were differentially expressed (Figure 5a, Table S3), the majority of which were upregulated (Figure 5b). Enrichment analysis indicated that these genes were involved in synaptic transmission (GO:0004890, GO:0099177, GO:0043269; GO:0007268; GO:0050808) and BDNF pathways (Figure 5c,d). Several of the synaptic genes involved glutamatergic and GABAergic signaling, including Slc1a3 (Solute Carrier Family 1 Member 3), Grm7 (Glutamate Metabotropic Receptor 7), Grin1 (Glutamate Ionotropic Receptor NMDA Type Subunit 1), Grin2b, Grip1 (Glutamate Receptor Interacting Protein 1) and Gabrg3 (Gamma-Aminobutyric Acid Type A Receptor Gamma 3 Subunit) (Figure 5e). However, in MG Depletion/Dep vs. Control Diet/Dep mice, upregulation of these genes was not observed (Figure 5e).
Figure 5. Differential gene expression after alcohol dependence in the CeA.
(a) Volcano plot showing log2 fold-change against log10 P-value in central nucleus of the amygdala (CeA). Differentially expressed genes (DEG) (P < 0.05) are shown as orange dots. The top DEG are labeled. (b) Number of upregulated and downregulated DEG in the CeA (P<0.05) after alcohol dependence. (c, d) Pathway enrichment and gene ontology (GO) biological process analysis for genes identified after alcohol dependence. (e) Heatmap displaying expression levels of differentially expressed glutamatergic and GABAergic genes in the CeA after alcohol dependence (Control Diet/Dep) or after alcohol dependence and microglia (MG) Depletion (MG Depletion/Dep). Color indicates expression level (regularized log transformed gene counts).
We examined similarity in CeA and mPFC expression changes after alcohol dependence and MG depletion using a rank rank hypergeometric overlap test (RRHO) to identify patterns of overlap between expression profiles (37). MG depletion produced similar gene expression changes in both regions (Figure S7). For Control Diet vs. MG Depletion, we identified a robust overlap (max −log10(P-value)=36) in genes downregulated in both regions (Figure S7a). The most significant co-downregulated genes were MG markers (Csf1r, Cx3cr1) and genes related to myeloid leukocyte activation (GO:0002274), neutrophil-mediated immunity (GO:0002446) and immune response (GO:0006955), suggesting a prominent MG depletion signature (Figure S7b,c). We also observed a small but significant overlap in expression signatures between the mPFC and CeA for the orthogonal contrasts MG Depletion/Dep vs. MG Depletion/Non-Dep mice (max −log10(P-value)=6) (Figure S8a). Co-downregulated genes between regions were related to myeloid leukocyte activation (GO:0002274), neutrophil-mediated immunity (GO:0002446) and transmitter-gated ion channel activity (GO:0022824) (Figure S8b,c). Many of the co-regulated immune genes associated with MG Depletion/Dep are implicated in alcohol consumption (Il6, B2m, Ctss, Il10), further suggesting MG depletion prevents dependence-induced neuroimmune activation and escalation of drinking (11). Many synaptic genes that are co-downregulated in MG Depletion/Dep animals also regulate glutamatergic (Grm5, Gria3, Grik1) and GABAergic (Gabrb3, Gabra3) function (Figure S8b). These coordinated changes in mPFC/CeA transcriptomes implicate key mechanisms by which MG depletion might counteract changes associated with alcohol dependence.
MG depletion reduces glutamatergic and GABAergic signaling in the CeA
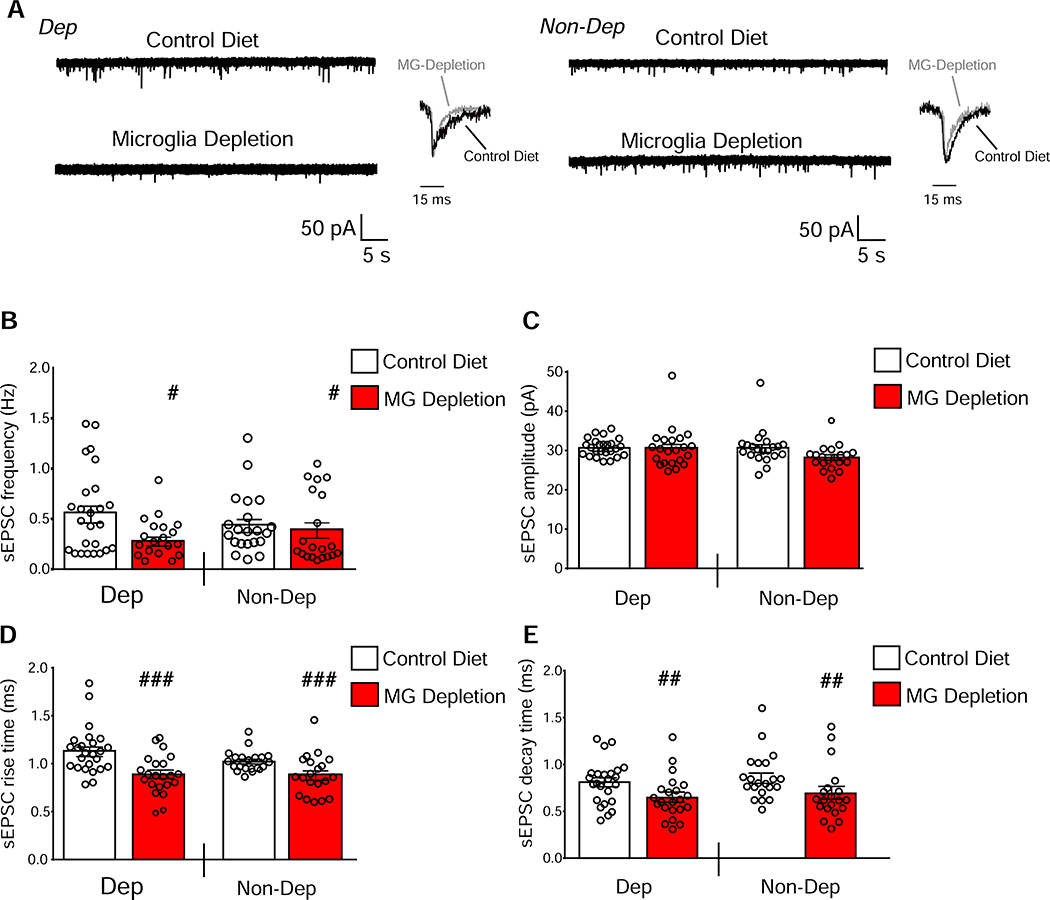
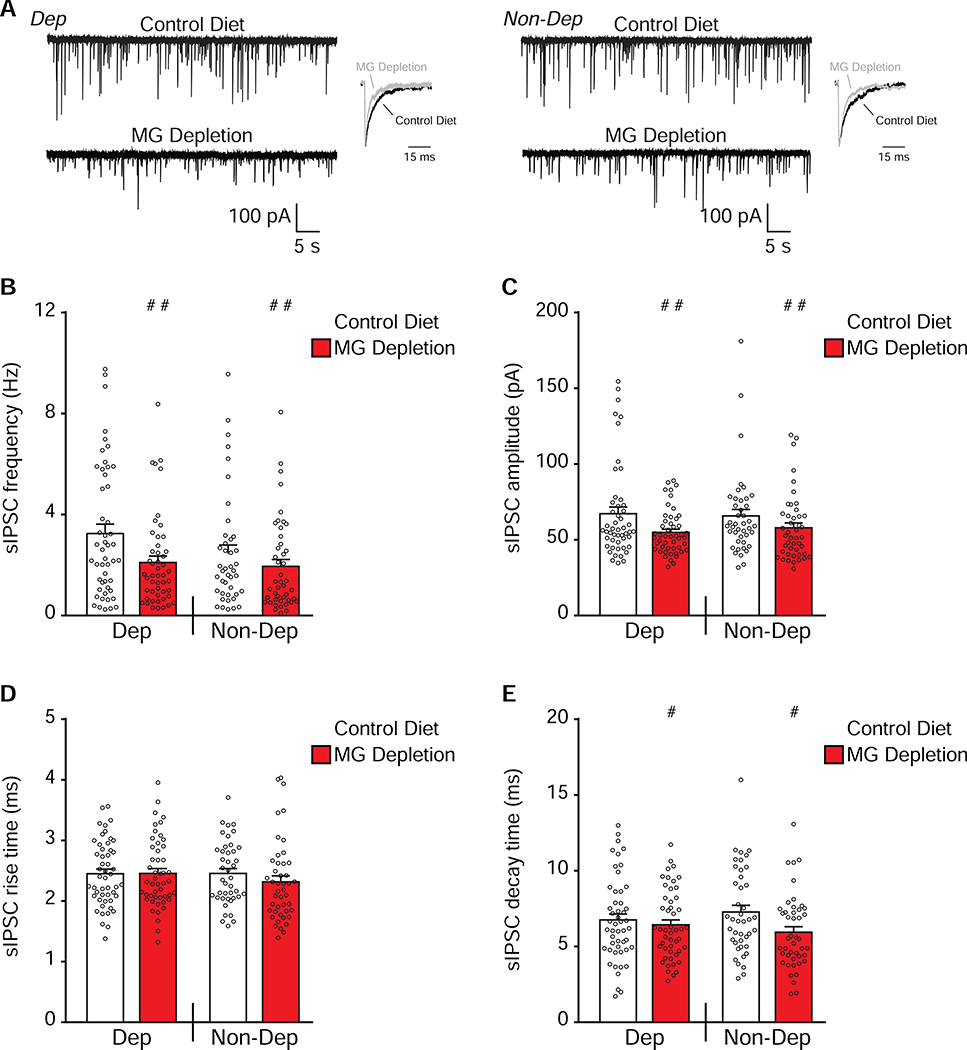
Coordinated gene expression changes in CeA GABAergic and glutamatergic synaptic function led us to study the effect of MG depletion on CeA synaptic transmission in the same mice used for behavioral and transcriptomic analyses. We recorded spontaneous excitatory and inhibitory postsynaptic currents (sE/IPSCs) and found significant effects of MG depletion on sEPSC frequencies (F(1,84)=4.97; P=0.029), rise times (F(1,84)=19.30; P<0.001) and decay times (F(1,84)=8.27; P=0.0051), but no main effects of CIE-2BC or interaction effects (Figure 6). We found significant main effects of MG depletion on sIPSC frequencies (F(1,182)=6.85; P=0.0096), amplitudes (F(1,182)=7.80; P=0.0058) and decay times (F(1,182)=4.75; P=0.031), but no main effects of CIE-2BC and no interaction effects (Figure 7). Decreases in E/IPSC frequencies are associated with reductions in neurotransmitter release, while changes in amplitudes or kinetics are associated with altered postsynaptic receptor function, spine morphology or density (38). Thus, MG depletion, despite alcohol treatment, decreases glutamatergic and GABAergic synaptic function in the CeA via a combination of pre- and post-synaptic mechanisms.
Figure 6. MG depletion reduces glutamatergic transmission in the CeA even after alcohol dependence.
(a) Representative sEPSC traces and scaled averages in central nucleus of the amygdala (CeA) neurons from Control Diet or microglia (MG) Depletion mice that were alcohol-dependent (Dep, on left) or non-dependent (Non-Dep, on right). (b-e) MG depletion significantly decreased the mean sEPSC frequency (b), rise time (d) and decay time (e) in CeA neurons of Dep and Non-Dep mice (n=22–26 cells from 5 mice/group). Data are presented as mean±SEM values. # P<0.05, ##P<0.01 or ###P<0.001 main effect of diet by two-way ANOVA.
Figure 7. MG depletion reduces GABAergic transmission in the CeA even after alcohol dependence.
(a) Representative sIPSC traces and scaled averages in the central nucleus of the amygdala (CeA) neurons from Control Diet or microglia (MG) Depletion mice that were alcohol-dependent (Dep, on left) or non-dependent (Non-Dep, on right). (b-e) MG depletion significantly decreased the mean sIPSC frequency (b), amplitude (c) and decay time (e) in CeA neurons of Dep and Non-Dep mice (n=42–50 cells from 5 mice/group). Data are presented as mean±SEM values; # P<0.05, ## P<0.01 or ### P<0.001 main effect of diet by two-way ANOVA.
We found similar passive CeA membrane properties and current-voltage relationships in all groups, whereas active membrane properties were significantly different, suggesting that MG depletion decreases the overall excitability of CeA neurons (Supplemental Information, Figure S9, Tables S4–5).
Discussion
Alcohol exposure alters MG function and expression of immune-related genes, and there is ample literature supporting neuroimmune signaling in alcohol behaviors (11). We investigated the role of MG in alcohol dependence using the well-established CIE-2BC mouse model that produces neurobiological and transcriptional changes that overlap with those found in AUD. The non-dependent and alcohol-dependent groups also allow us to study MG depletion on moderate and excessive drinking. Moreover, performing histological, transcriptomic and electrophysiological analyses in brain regions from the same cohort of animals gave us unprecedented insight into how MG depletion impacts alcohol dependence-induced neuroadaptations and behavioral changes.
Alcohol dependence produced strong proliferation of MG in the mPFC, similar to results in AUD (15). MG transcriptomes from the frontal cortex also indicate enhanced immune responses after alcohol dependence in both mice and humans, particularly upregulated interferon pathways (22,23), suggesting the importance of cellular mechanisms (2). MG depletion prevented dependence-induced escalations in alcohol consumption without altering drinking in non-dependent mice. This new finding supports the hypothesis that MG are necessary for the transition from voluntary drinking to alcohol dependence. Another corroborating study showed that MG depletion did not alter voluntary alcohol consumption but did regulate neuroimmune-induced escalation in drinking (39). Additionally, MG depletion prior to binge alcohol drinking blocked inflammatory responses to alcohol (16). These findings suggest that MG regulate drinking behaviors after sufficient immune activation such as following alcohol dependence, binge drinking or neuroimmune-induced escalation. MG depletion using PLX5622 also blocked the proinflammatory effects of nicotine withdrawal (40), further corroborating MG as neuroimmune mediators in addiction to different drugs of abuse.
One limitation of this study is that we did not examine MG depletion in female mice. Transcriptomes and inflammatory signaling differ between brain MG from male and female animals (41,42), suggesting sex-specific functional and disease states. Under homeostatic conditions in female rodents, MG show morphological markers reflective of primed and activated hyper-inflammatory state (42,43). However, under conditions of cerebral ischemia and stroke in females, MG exhibit an anti-inflammatory and neuroprotective phenotype (41,44). In AUD and rodent drinking models, females are more vulnerable than males to the inflammatory and neurotoxic effects of ethanol (45), suggesting that MG may also be key regulators of alcohol behaviors in females.
MG function and anxiety-like behavior during alcohol withdrawal
MG are implicated in anxiety-related disorders, but their role in alcohol withdrawal-related anxiety is unknown (46). MG depletion reduced anxiety-like behavior in alcohol-dependent, but not non-dependent mice. These findings establish a link between MG function and anxiety in abstinent animals. MG function also underlies withdrawal and dependence phenotypes associated with other drugs of abuse (40,47). For example, inhibition of MG activation restored the rewarding effects of cocaine in opioid-dependent animals (47). During nicotine withdrawal, MG depletion prevented anxiogenic behaviors (40). These findings, together with immune responses, provide corroborating evidence that MG have distinct cellular functions in behavioral responses to drugs of abuse.
MG depletion reduces expression of inflammatory- and immune-related genes in alcohol dependent animals
Alcohol alters immune responses in the brain and neuroimmune signaling modulates drinking behavior (11). Our findings suggest that alcohol dependence initiates a neuroinflammatory response involving type I interferons in the mPFC. Similar results were found in astrocyte and MG transcriptomes after alcohol dependence (22). Moreover, type I interferons were enriched in co-expressed genes that interacted with both alcohol consumption traits and risk genes from human GWAS studies, suggesting a connection between an individual’s risk of becoming alcohol dependent and aberrant immune function (8).
MG depletion prevents upregulation of neuroimmune responses in the mPFC and CeA in dependent mice, suggesting that MG are necessary for neuroimmune-induced escalations in drinking. Indeed, many of the differentially expressed mPFC and CeA genes have been shown to regulate alcohol consumption (B2m, Ctss, Il6) or related behaviors (Tlr4) (11). Thus, MG depletion may prevent dependence-induced escalations in alcohol consumption by blocking aberrant neuroimmune activation following chronic alcohol exposure.
MG depletion prevents upregulation of excitatory gene expression and decreases excitatory synaptic transmission
Distinct functional and gene expression changes are necessary for the transition to alcohol dependence (10,36), such as glutamatergic dysregulation (48). Gene expression and GWAS studies from individuals with AUD revealed an association of glutamatergic genes with alcohol use phenotypes and lifetime consumption (31,32). In our mouse model of dependence, we observed upregulation of several glutamatergic genes in the mPFC and CeA, with the strongest signature in mPFC. MG depletion prevented coordinated changes in both regions and reduced spontaneous CeA glutamate transmission, suggesting that MG depletion prevents alcohol-induced hyperglutamatergic states. Consistent with our study, pharmacological or genetic inhibition of glutamatergic genes like Grm5 reduced alcohol self-administration and relapse-like behavior in rodents (49,50). A study using CX3CR1CreER transgenic mice to deplete MG found decreased levels of proteins at glutamatergic synapses and decreased spontaneous glutamate release, providing additional evidence that MG regulate glutamatergic signaling (51).
MG depletion counteracts GABAergic neuroadaptations induced by alcohol dependence and may disrupt glial-neuronal communication
GABAergic transmission increases in the CeA following alcohol dependence across species (52,53). MG depletion reduced CeA GABAergic signaling and gene expression in the CeA and mPFC. Top candidate genes implicated in AUD include transcripts for GABA receptor subunits and transporters (e.g., Slc6a11), although the strength of these associations can vary depending on the AUD subpopulation studied (33,54). Our findings suggest that MG depletion reverses known neuroadaptations in the GABAergic system following alcohol dependence, thus preventing escalation of drinking.
Bidirectional MG-neuronal communication permits tight control of synaptic activity (55) that could be compromised by MG depletion, interfering with synaptic changes in alcohol dependence. MG express many neurotransmitter receptors and alter release of neuroactive molecules via a feedback loop (55). In particular, BDNF, IL1β and TNFα (Tumor Necrosis Factor Alpha) are important for MG-neuronal communication (55) and are also implicated in AUD and other neuropsychiatric disorders (56–58). In individuals with AUD, TNFα and IL1β serum levels are elevated, whereas BDNF levels are reduced (59,60). Moreover, loss of BDNF in MG recapitulates deficits observed in MG Depletion mice, such as decreased glutamatergic synaptic proteins and spine density (51). Downregulation of Bdnf in MG may thus play a role in dependence-induced changes in CeA glutamatergic transmission and gene expression. GABAergic transmission in the CeA is regulated by IL1β (12,13), and we speculate that MG depletion normalizes both GABAergic and glutamatergic function by preventing secretion of neuromodulatory cytokines and chemokines. Further investigation is necessary to characterize the role of MG-derived molecules in the neuroadaptations that underlie alcohol dependence.
Conclusions
Our multidisciplinary study is the first to show widespread effects of MG depletion, from reduced alcohol-dependence-associated behaviors to impaired synaptic function to reversal of synaptic and neuroimmune-related gene expression. These findings suggest a complex, diverse role for MG in the development of alcohol dependence.
Supplementary Material
KEY RESOURCES TABLE.
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | anti-IBA1antibody (Rabbit) | Wako Chemicals USA, Richmond, VA | 019-19741 | 1:1000 dilution |
| anti-IBA1 antibody (Goat) | Abcam, Cambridge, MA | ab5076 | 1:500 dilution | |
| anti-TMEM119 antibody (Rabbit) | Abcam, Cambridge, MA | ab209064 | 1:500 dilution | |
| Alexa Fluor 647-conjugated donkey (Goat) | Jackson Immunoresearch, West Grove, PA | 705-605-147 | 1:500 dilution | |
| Alexa Fluor 488-conjugated Donkey (Rabbit) | Jackson Immunoresearch, West Grove, PA | 711-545-152 | 1:500 dilution | |
| Hoechst 33342 | Life Technologies, Carlsbad, CA | H3570 | 1:1000 dilution | |
| Bacterial or Viral Strain | ||||
| Biological Sample | ||||
| Cell Line | ||||
| Chemical Compound or Drug | BLOXALL | Vector Laboratories, Burlingame, CA | SP-6000 | |
| ImmPRESS™-AP Anti-Rabbit IgG (alkaline phosphatase) Polymer Detection | Vector Laboratories, Burlingame, CA | MP-5401-50 | ||
| ImmPACT Vector Red Substrate Kits | Vector Laboratories, Burlingame, CA | SK-5100 | ||
| ProLong Gold Antifade Mountant | Thermo Fischer Scientific, Waltham, MA | P10144 | ||
| PLX5622 | Plexxikon Inc., Berkeley, CA | |||
| Control Chow (RMH1800) | Research Diets, New Brunswick, NJ | |||
| AP-5 | Tocris Bioscience, Bristol, UK | |||
| Bicuculline | Tocris Bioscience, Bristol, UK | |||
| CGP 55845A | Tocris Bioscience, Bristol, UK | |||
| DNQX | Tocris Bioscience, Bristol, UK | |||
| DPX | Thermo Fischer Scientific, Waltham, MA | X1525 | ||
| Commercial Assay Or Kit | MagMAX-96 Total RNA Isolation Kit | Thermo Fischer Scientific, Waltham, MA | AM1830 | |
| DNA-free Kit | Thermo Fischer Scientific, Waltham, MA | AM1906 | ||
| ERCC RNA Spike In Kit | Thermo Fischer Scientific, Waltham, MA | 4456740 | ||
| Deposited Data; Public Database | RNA-seq data deposited into GEO; GSE142571 | |||
| Genetic Reagent | ||||
| Organism/Strain | C57BL/6J male mice | |||
| Peptide, Recombinant Protein | ||||
| Recombinant DNA | ||||
| Sequence-Based Reagent | ||||
| Software; Algorithm | Image Pro Premiere | Media Cybernetics, Inc., Rockville, MD | ||
| Mini Analysis | Synaptosoft Inc., Fort Lee, NJ | |||
| NeuroExpress software (version 19.4.09) | Dr. A. Szücs | |||
| Prism 8.01 | GraphPad, San Diego, CA | |||
| Transfected Construct | ||||
| Other | Vapor Chambers | La Jolla Alcohol Research, La Jolla, CA |
Acknowledgements and Disclosures
This work was supported by the National Institutes of Health/National Institute of Alcohol Abuse and Alcoholism [U01 AA020926, U01 AA013498, P60 AA006420, AA015566, AA017477, AA027700, AA021491, P01 AA020683, AA013520, AA006399, AA025499, AA025408, T32-AA007456]. The authors would like to thank Andrey Rymar at Plexxikon Inc. for helping to secure the chow formulation and drug distribution and Jody Mayfield for editing the manuscript. The authors report no biomedical financial interests or potential conflicts of interest. This is Scripps manuscript number 29938.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. (2010): Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. (2017): An environment-dependent transcriptional network specifies human microglia identity. Science. 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, et al. (2019): Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 50:253–271 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. (2016): Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 352:712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Fonseca AC, Matias D, Garcia C, Amaral R, Geraldo LH, Freitas C, et al. (2014): The impact of microglial activation on blood-brain barrier in brain diseases. Front Cell Neurosci. 8:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salter MW, Stevens B (2017): Microglia emerge as central players in brain disease. Nat Med. 23:1018–1027. [DOI] [PubMed] [Google Scholar]
- 7.Tay TL, Bechade C, D’Andrea I, St-Pierre MK, Henry MS, Roumier A, et al. (2017): Microglia Gone Rogue: Impacts on Psychiatric Disorders across the Lifespan. Front Mol Neurosci. 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapoor M, Wang JC, Farris SP, Liu Y, McClintick J, Gupta I, et al. (2019): Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry. 9:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson EK, Grantham EK, Warden AS, Harris RA (2019): Neuroimmune signaling in alcohol use disorder. Pharmacol Biochem Behav. 177:34–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warden AS, Mayfield RD (2017): Gene expression profiling in the human alcoholic brain. Neuropharmacology. 122:161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayfield J, Arends MA, Harris RA, Blednov YA (2016): Genes and Alcohol Consumption: Studies with Mutant Mice. Int Rev Neurobiol. 126:293–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajo M, Herman MA, Varodayan FP, Oleata CS, Madamba SG, Harris RA, et al. (2015): Role of the IL-1 receptor antagonist in ethanol-induced regulation of GABAergic transmission in the central amygdala. Brain Behav Immun. 45:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel RR, Khom S, Steinman MQ, Varodayan FP, Kiosses WB, Hedges DM, et al. (2019): IL-1beta expression is increased and regulates GABA transmission following chronic ethanol in mouse central amygdala. Brain Behav Immun. 75:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall SA, Geil CR, Nixon K (2016): Prior Binge Ethanol Exposure Potentiates the Microglial Response in a Model of Alcohol-Induced Neurodegeneration. Brain Sci. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Crews FT (2008): Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp Neurol. 210:349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter TJ, Crews FT (2017): Microglial depletion alters the brain neuroimmune response to acute binge ethanol withdrawal. J Neuroinflammation. 14:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagher NN, Najafi AR, Kayala KM, Elmore MR, White TE, Medeiros R, et al. (2015): Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J Neuroinflammation. 12:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker HC, Lopez MF (2004): Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 28:1829–1838. [DOI] [PubMed] [Google Scholar]
- 19.Bajo M, Montgomery SE, Cates LN, Nadav T, Delucchi AM, Cheng K, et al. (2016): Evaluation of TLR4 Inhibitor, T5342126, in Modulation of Ethanol-Drinking Behavior in Alcohol-Dependent Mice. Alcohol Alcohol. 51:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang TY, Renoir T, Du X, Lawrence AJ, Hannan AJ (2013): Depression-related behaviours displayed by female C57BL/6J mice during abstinence from chronic ethanol consumption are rescued by wheel-running. Eur J Neurosci. 37:1803–1810. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe SA, Sidhu H, Patel RR, Kreifeldt M, D’Ambrosio SR, Contet C, et al. (2019): Molecular, Morphological, and Functional Characterization of Corticotropin-Releasing Factor Receptor 1-Expressing Neurons in the Central Nucleus of the Amygdala. eNeuro. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson EK, Blednov YA, Harris RA, Mayfield RD (2019): Glial gene networks associated with alcohol dependence. Sci Rep. 9:10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy GM, Farris SP, Blednov YA, Harris RA, Mayfield RD (2018): Microglial-specific transcriptome changes following chronic alcohol consumption. Neuropharmacology. 128:416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajo M, Varodayan FP, Madamba SG, Robert AJ, Casal LM, Oleata CS, et al. (2015): IL-1 interacts with ethanol effects on GABAergic transmission in the mouse central amygdala. Front Pharmacol. 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry CJ, McNally GP (2012): Naloxone prevents the rapid reacquisition but not acquisition of alcohol seeking. Behav Neurosci. 126:599–604. [DOI] [PubMed] [Google Scholar]
- 26.Hopperton KE, Mohammad D, Trepanier MO, Giuliano V, Bazinet RP (2018): Markers of microglia in post-mortem brain samples from patients with Alzheimer’s disease: a systematic review. Mol Psychiatry. 23:177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall SA, McClain JA, Kelso ML, Hopkins DM, Pauly JR, Nixon K (2013): Microglial activation is not equivalent to neuroinflammation in alcohol-induced neurodegeneration: The importance of microglia phenotype. Neurobiol Dis. 54:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osterndorff-Kahanek EA, Becker HC, Lopez MF, Farris SP, Tiwari GR, Nunez YO, et al. (2015): Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS One. 10:e0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012): Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 32:1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, et al. (2018): Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci. 21:530–540. [DOI] [PubMed] [Google Scholar]
- 31.Karpyak VM, Geske JR, Colby CL, Mrazek DA, Biernacka JM (2012): Genetic variability in the NMDA-dependent AMPA trafficking cascade is associated with alcohol dependence. Addict Biol. 17:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. (2011): Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A. 108:7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Augier E, Barbier E, Dulman RS, Licheri V, Augier G, Domi E, et al. (2018): A molecular mechanism for choosing alcohol over an alternative reward. Science. 360:1321–1326. [DOI] [PubMed] [Google Scholar]
- 34.Jimenez VA, Herman MA, Cuzon Carlson VC, Walter NA, Grant KA, Roberto M (2019): Synaptic adaptations in the central amygdala and hypothalamic paraventricular nucleus associated with protracted ethanol abstinence in male rhesus monkeys. Neuropsychopharmacology. 44:982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberto M, Gilpin NW, Siggins GR (2012): The central amygdala and alcohol: role of gamma-aminobutyric acid, glutamate, and neuropeptides. Cold Spring Harb Perspect Med. 2:a012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koob GF, Volkow ND (2010): Neurocircuitry of addiction. Neuropsychopharmacology. 35:217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cahill KM, Huo Z, Tseng GC, Logan RW, Seney ML (2018): Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep. 8:9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varodayan FP, Sidhu H, Kreifeldt M, Roberto M, Contet C (2018): Morphological and functional evidence of increased excitatory signaling in the prelimbic cortex during ethanol withdrawal. Neuropharmacology. 133:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warden AS, Triplett TA, Lyu A, Grantham EK, Azzam MM, DaCosta A, Mason S, Blednov YA, Ehrlich LIR, Mayfield RD, Harris RA (2020): Microglia depletion and alcohol: Transcriptome and behavioral profiles. Addict Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adeluyi A, Guerin L, Fisher ML, Galloway A, Cole RD, Chan SSL, et al. (2019): Microglia morphology and proinflammatory signaling in the nucleus accumbens during nicotine withdrawal. Sci Adv. 5:eaax7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villa A, Gelosa P, Castiglioni L, Cimino M, Rizzi N, Pepe G, et al. (2018): Sex-Specific Features of Microglia from Adult Mice. Cell Rep. 23:3501–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz JM, Sholar PW, Bilbo SD (2012): Sex differences in microglial colonization of the developing rat brain. J Neurochem. 120:948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle HH, Eidson LN, Sinkiewicz DM, Murphy AZ (2017): Sex Differences in Microglia Activity within the Periaqueductal Gray of the Rat: A Potential Mechanism Driving the Dimorphic Effects of Morphine. J Neurosci. 37:3202–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong X, Xu L, Wei L, White RE, Ouyang YB, Giffard RG (2015): IL-4 Is Required for Sex Differences in Vulnerability to Focal Ischemia in Mice. Stroke. 46:2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alfonso-Loeches S, Pascual M, Guerri C (2013): Gender differences in alcohol-induced neurotoxicity and brain damage. Toxicology. 311:27–34. [DOI] [PubMed] [Google Scholar]
- 46.Wang YL, Han QQ, Gong WQ, Pan DH, Wang LZ, Hu W, et al. (2018): Microglial activation mediates chronic mild stress-induced depressive- and anxiety-like behavior in adult rats. J Neuroinflammation. 15:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor AM, Castonguay A, Ghogha A, Vayssiere P, Pradhan AA, Xue L, et al. (2016): Neuroimmune Regulation of GABAergic Neurons Within the Ventral Tegmental Area During Withdrawal from Chronic Morphine. Neuropsychopharmacology. 41:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao PS, Bell RL, Engleman EA, Sari Y (2015): Targeting glutamate uptake to treat alcohol use disorders. Front Neurosci. 9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Besheer J, Faccidomo S, Grondin JJ, Hodge CW (2008): Regulation of motivation to self-administer ethanol by mGluR5 in alcohol-preferring (P) rats. Alcohol Clin Exp Res. 32:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blednov YA, Harris RA (2008): Metabotropic glutamate receptor 5 (mGluR5) regulation of ethanol sedation, dependence and consumption: relationship to acamprosate actions. Int J Neuropsychopharmacol. 11:775–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, et al. (2013): Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 155:1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberto M, Madamba SG, Stouffer DG, Parsons LH, Siggins GR (2004): Increased GABA release in the central amygdala of ethanol-dependent rats. J Neurosci. 24:10159–10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pleil KE, Lowery-Gionta EG, Crowley NA, Li C, Marcinkiewcz CA, Rose JH, et al. (2015): Effects of chronic ethanol exposure on neuronal function in the prefrontal cortex and extended amygdala. Neuropharmacology. 99:735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B (2008): GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 42:184–191. [DOI] [PubMed] [Google Scholar]
- 55.Posfai B, Cserep C, Orsolits B, Denes A (2019): New Insights into Microglia-Neuron Interactions: A Neuron’s Perspective. Neuroscience. 405:103–117. [DOI] [PubMed] [Google Scholar]
- 56.Logrip ML, Barak S, Warnault V, Ron D (2015): Corticostriatal BDNF and alcohol addiction. Brain Res. 1628:60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koskela M, Back S, Voikar V, Richie CT, Domanskyi A, Harvey BK, et al. (2017): Update of neurotrophic factors in neurobiology of addiction and future directions. Neurobiol Dis. 97:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crews FT, Vetreno RP (2016): Mechanisms of neuroimmune gene induction in alcoholism. Psychopharmacology (Berl). 233:1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heberlein A, Muschler M, Wilhelm J, Frieling H, Lenz B, Groschl M, et al. (2010): BDNF and GDNF serum levels in alcohol-dependent patients during withdrawal. Prog Neuropsychopharmacol Biol Psychiatry. 34:1060–1064. [DOI] [PubMed] [Google Scholar]
- 60.Achur RN, Freeman WM, Vrana KE (2010): Circulating cytokines as biomarkers of alcohol abuse and alcoholism. J Neuroimmune Pharmacol. 5:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.