SUMMARY
Multicellular development depends on generating and precisely positioning distinct cell types within tissues. During leaf development, pores in the epidermis called stomata are spaced at least one cell apart for optimal gas exchange. This pattern is primarily driven by iterative asymmetric cell divisions (ACDs) in stomatal progenitors, which generate most of the cells in the tissue. A plasma membrane-associated polarity crescent defined by BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) and BREVIS RADIX family (BRXf) proteins is required for asymmetric divisions and proper stomatal pattern, but the cellular mechanisms that orient ACDs remain unclear. Here, utilizing long-term, quantitative time-lapse microscopy, we identified two oppositely oriented nuclear migrations that precede and succeed ACD during epidermal patterning. The pre- and post-division migrations are dependent on microtubules and actin, respectively, and the polarity crescent is the unifying landmark that is both necessary and sufficient to orient both nuclear migrations. We identified a specific and essential role for MYOXI-I in controlling post-ACD nuclear migration. Loss of MYOXI-I decreases stomatal density, owing to an inability to accurately orient a specific subset of ACDs. Taken together, our analyses revealed successive and polarity-driven nuclear migrations that regulate ACD orientation in the Arabidopsis stomatal lineage.
Keywords: Asymmetric division, stomata, nuclear migration, cytoskeleton, leaf development, tissue patterning, MYOXI-I, Arabidopsis
Graphical Abstract
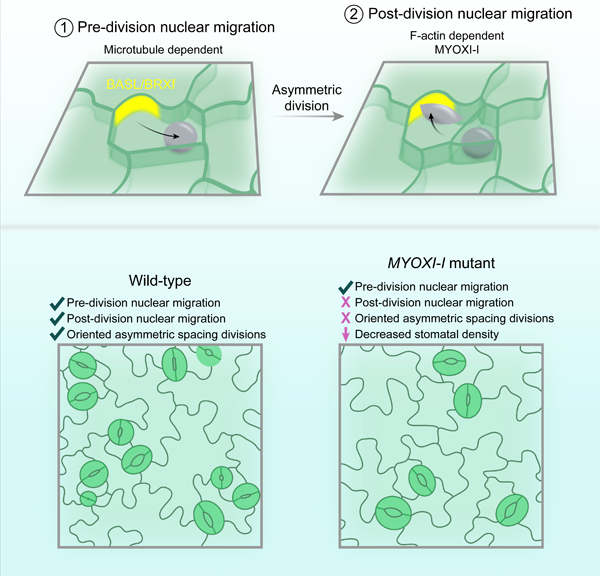
eTOC Blurb
How are asymmetric divisions oriented in developing plant tissues? Muroyama et al. find that oppositely oriented nuclear migrations flank asymmetric divisions in the Arabidopsis stomatal lineage. Despite having distinct cytoskeletal requirements, both migrations use BASL as a spatial landmark and are collectively required for division orientation.
INTRODUCTION
Plant development frequently relies on asymmetric cell divisions (ACDs) that establish differential daughter cell size, placement, and fate [1–3]. In most cases, ACDs are executed in a highly predictable manner and, importantly, with stereotyped orientations relative to a cellular niche or a tissue axis. However, developmental patterns can also originate from a more flexible approach, exemplified by the Arabidopsis leaf epidermis, where iterative ACDs are dynamically re-oriented in the absence of a fixed niche.
Stomata, two-celled valves that mediate gas exchange in the leaf, are patterned by successive ACDs in progenitors that generate daughters of unequal size and cellular fate: a smaller meristemoid and a larger stomatal lineage ground cell (SLGC) (Figure 1A). Each meristemoid will generate the two guard cells that create the stomatal pore, while SLGCs eventually mature into lobed pavement cells (PCs). In Arabidopsis, meristemoids and SLGCs can perform additional ACDs, termed amplifying and spacing divisions, respectively, to modulate stomatal density and distribution [4]. All ACDs, regardless of cell type, are oriented to prevent meristemoids from being generated adjacent to existing meristemoids, guard mother cells (GMCs), or stomata [5, 6]. Transcriptional [7], hormonal [8, 9], and environmental [10, 11] inputs can modulate the balance of successive ACDs, but the intracellular mechanisms that orient divisions in response to these varied cues remain poorly defined.
Figure 1. Successive nuclear migrations bookend ACD in the stomatal lineage.
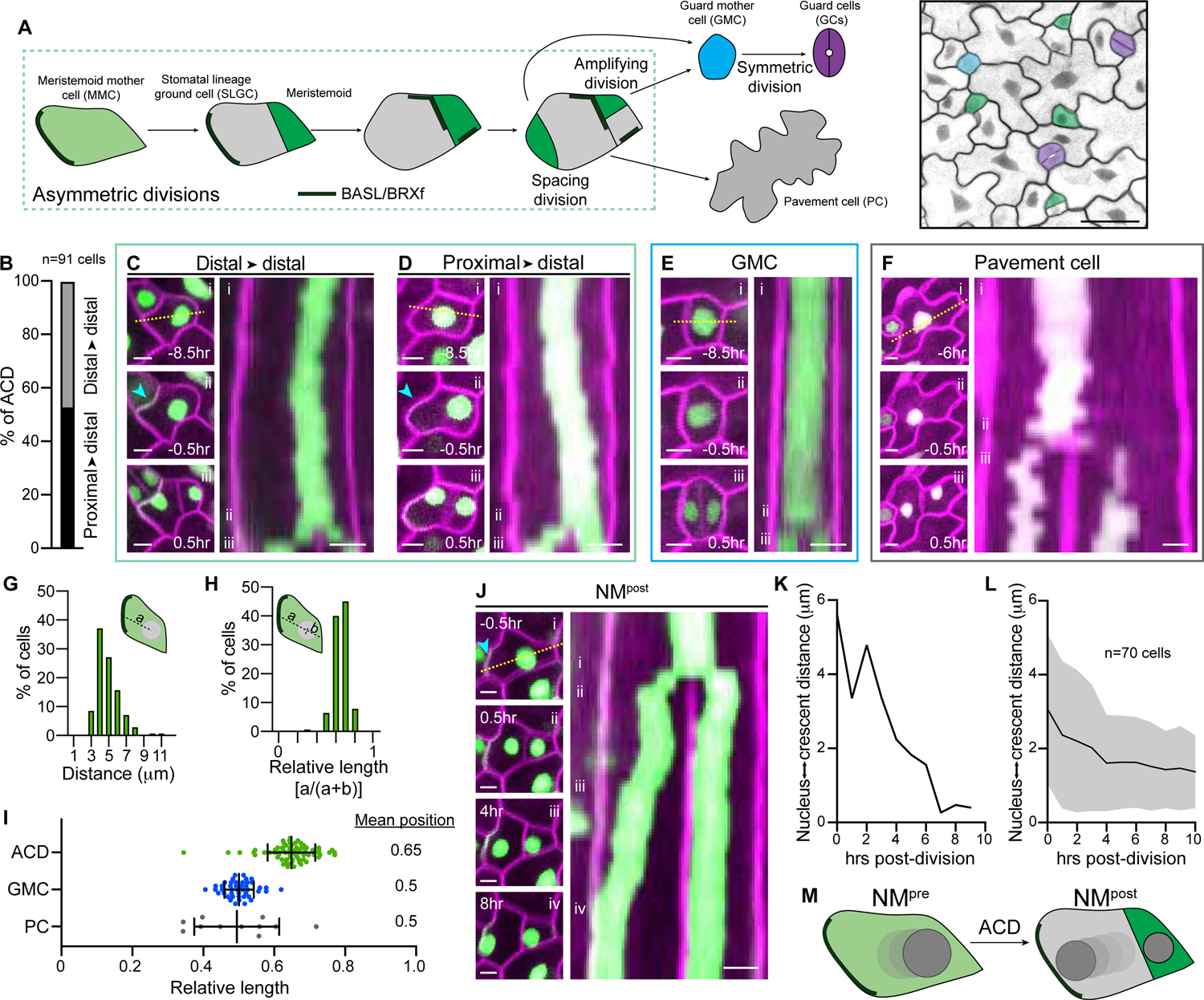
A. Schematic of the Arabidopsis stomatal lineage divisions, cell types, and polarity crescents (left). Epidermis of a 3dpg cotyledon (right) from the R2D2 BRXL2::BRXL2-YFP ML1::mCherry-RCI2A line, pseudo-colored to indicate cell types. Scale bar-25μm.
B. Percent of asymmetrically dividing cells with nuclei initially positioned proximal or distal to polarizing BRXL2. n-91 cells.
C–D. Examples of two classes of nuclear positioning before ACD in R2D2 BRXL2::BRXL2-YFP ML1::mCherry-RCI2A cotyledons. Stills (left) illustrate asymmetrically dividing cells (i) 8.5hr before division, (ii) 0.5hr before division, and (iii) 0.5hr post-ACD. Kymograph (right) generated from the dotted line in (i). The cyan arrowhead indicates the polarity crescent. Scale bars-5μm.
E. Stills (left) and kymograph (right) showing the nuclear position during a representative GMC division (same format as in Figure 1C). Scale bars-50μm.
F. Stills (left) and kymograph (right) showing the nuclear position during a representative PC division (i) 6hr before division, (ii) 0.5hr before division, and (iii) 0.5hr post-ACD. Scale bars-5μm.
G–H. Distribution of the distances from the pre-ACD nuclear center to the polarity crescent, plotted as absolute distance from the distal wall (G) or as a relative distance along the cell axis orthogonal to the division plane (H). n-140 cells.
I. Relative nuclear positions before division in the indicated cell types. The ACD data is derived from those plotted in Figures 1G and 1H (see Methods for details). ACD-70 cells, GMC-44 cells, PC-11 cells. Data are represented as mean±standard deviation.
J. Stills (left) and kymograph (right) of NMpost in an SLGC. Stills show the cell (i) 0.5hr before ACD, (ii) 0.5hr after ACD, (iii) 4hr after ACD, and (iv) 8hr after ACD. Kymograph (right) generated from the dotted line in (i). The cyan arrowhead indicates the polarity crescent. Scale bars-5μm.
K. SLGC nucleus-to-crescent distance during NMpost, quantified from the cell shown in Figure 1J.
L. Average SLGC nucleus-to-crescent distance during NMpost. Data are represented as mean±standard deviation.
M. Schematic showing the two, oppositely oriented nuclear movements, NMpre and NMpost, that flank ACD. Nuclear color deepens in later timepoints.
Previous work developed a theoretical “polarity switching” model that provides a conceptual framework to understand how reorientation of sequential ACDs establishes the final stomatal pattern [12]. At its core, this model depends on the dynamic behavior of a cortical polarity crescent. Constituents of the crescent include BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) [13] and BREVIS RADIX family (BRXf) [14] proteins that polarize at the plasma membrane several hours before ACD; subsequently, only the SLGC inherits the polarity crescent, where BASL scaffolds a MAP kinase signaling cascade to suppress meristemoid identity [15, 16]. The behavior of the BASL/BRXf crescent is consistent with the polarity switching model, but how it defines successive division planes is completely unknown.
Here, we report that the BASL/BRXf polarity crescent is required to initiate and orient two nuclear migrations during Arabidopsis stomatal patterning. The first migration depends on microtubules and positions the nucleus away from the polarity crescent to ensure that the meristemoid is generated distal to cortical BASL/BRXf. Following cytokinesis, a second, actin-dependent nuclear migration reorients the nucleus toward the BASL/BRXf crescent, thereby reversing the cellular polarity in one of the two daughters. We identified MYOXI-I as a myosin required for the second migration, and analysis of the stomatal phenotypes of myoxi-i suggest that post-division nuclear migration is required to reorient and prime successive ACDs to enforce the one-cell spacing rule. Our data fit a model where back-to-back nuclear migrations are employed in the leaf epidermis to enable repeated rounds of asymmetric and oriented divisions that reinforce distinct cellular identities and proper tissue patterning.
RESULTS
Nuclear migration precedes asymmetric cell division in the stomatal lineage
To identify the mechanisms that orient ACDs in the stomatal lineage, we performed time-lapse analyses of developing cotyledons triply labeled with reporters for the epidermal plasma membrane (ML1::mCherry-RCI2A), nuclei (R2D2) [17], and BRXL2 (BRXL2::BRXL2-YFP) (Figure 1A, right). Because the z-dimensions of nuclei and cells of interest were uniform at the developmental stages we tracked (Figure S1A), we considered ACD parameters in two dimensions.
Before ACD, nuclei were largely distal to the polarity crescent (Figures 1B and 1I), leading us to hypothesize that nuclear positioning may underlie division orientation (Video S1). Based on our data, we classified asymmetrically dividing cells broadly into two categories. In approximately half of progenitors, the BRXL2 domain polarized at the plasma membrane away from the offset nucleus. In these cases, the nucleus maintained its distal position until mitotic onset (Figures 1B and 1C). In the other progenitors, where BRXL2 polarized proximal to the initial nuclear position, we observed directed nuclear migration away from the polarity crescent (Figures 1B and 1D). We refer to this nuclear migration as NMpre. Imaging of NMpre in plants harboring a fluorescent reporter for S-phase (HTR2::CDT1a-RFP) [18] and quantification of migration times revealed that NMpre is likely initiated at the S/G2 transition (Figures S1B and S1C).
No directional migrations preceded the symmetric GMC divisions that generate the guard cell pair (Figures 1E and 1I, Video S1). Similarly, rare pavement cell divisions were preceded by stochastic, randomly oriented nuclear movement without persistent directionality (Figures 1F and 1I, Video S1). As NMpre preceded both amplifying and spacing divisions, we conclude that NMpre is intrinsic and specific to the ACD program in the stomatal lineage (Figures S1D–S1F).
Other well-characterized pre-ACD migrations in Arabidopsis involve cells with highly stereotyped shapes and sizes (e.g., in lateral root primordia or embryos) [2, 3]. In contrast, asymmetrically dividing stomatal lineage cells had highly variable morphologies and sizes (>8-fold difference in cell areas) (Figure S1G). While nuclear migrations that transit a consistent absolute distance would be sufficient to generate robust asymmetry in uniformly sized cells, a similar mechanism seemed at odds with the heterogenous nature of stomatal progenitors. Indeed, absolute distance from the polarity crescent was an inaccurate predictor of final nuclear position before mitotic onset. Re-mapping the data as relative distances along the longest cell axis indicated that NMpre likely integrates cell size information to reliably establish distal nuclear asymmetry in stomatal progenitors (Figures 1G and 1H).
A second nuclear migration follows ACD in SLGCs
Our time-lapse analyses revealed a second, unexpected nuclear migration in SLGCs that started after cytokinesis (hereafter termed NMpost) and moved the nucleus toward the BRXL2 crescent (Figures 1J–1L, Video S2). NMpost progressed until either (1) the nucleus reached the BRXL2 crescent or (2) BRXL2 signal disappeared. NMpost lengths were variable (3.3±2.8 hours, mean±standard deviation, n=50 cells), reflecting heterogeneity in both the SLGC size and the distance from the division plane to the polarity crescent (Figures S1H and S1I). Upon reaching the BRXL2 crescent, nuclear movements were largely suppressed until the disappearance of the crescent triggered resumption of stochastic nuclear motion (Figure S1J, Video S2). Therefore, asymmetric divisions are flanked by two oppositely oriented nuclear migrations in the Arabidopsis stomatal lineage: NMpre orients the nucleus away from the polarity crescent before ACD and NMpost reorients the SLGC nucleus toward the polarity site post-division (Figure 1M).
Nuclear migration is dominated by cell-autonomous polarity signals
Signaling between neighboring cells is essential to enforce proper stomatal spacing, in part through regulation of ACD orientation. The direction of NMpre could be controlled by these non-cell autonomous cues or be cell-autonomously directed by BASL/BRXf location. To distinguish between these two possibilities, we quantified pre-ACD nuclear position in the TOO MANY MOUTHS (TMM) mutant, which is unable to perceive secreted signaling peptides, resulting in frequently misplaced polarity crescents [6, 13, 19]. Before ACD, nuclei in measured tmm cells were always distal to the polarity crescent, regardless of its intracellular position (Figures S2A and S2B). Importantly, we identified a subset of tmm cells where BASL was inappropriately polarized away from existing stomata. Nuclei migrated toward the stomata in these cases, indicating that the polarity crescent position overrides potential direct intercellular signaling from neighboring stomata. Based on these data, we conclude that cell-intrinsic cues direct NMpre and that any intercellular communication that impacts nuclear position does so through the crescent.
The BASL/BRXf crescent is the spatial landmark that orients NMpre and NMpost
Our analysis of NMpre and NMpost in wild-type and tmm cells revealed a striking correlation between the directions of both nuclear movements and BRXL2 location. Membrane-localized polarity complexes often orient metazoan asymmetric divisions [20]. While mutations in, or overexpression of, polarity proteins can affect division patterns in Arabidopsis [21, 22], whether the intracellular positions of these polarized complexes directly orient divisions remains unknown. To test whether the BASL/BRXf crescent is the spatial landmark required for NMpre and NMpost, we first monitored nuclear movement in basl cotyledons (basl-2 35S::PIP2A-RFP ML1::H2B-YFP) (Figure 2A). Without BASL, NMpre and NMpost were both lost (Figures 2B, 2C, 2E and 2F, Video S3). Instead, nuclear position in basl was dominated by random, stochastic motion before and after ACD, demonstrating that polarity crescent formation is a prerequisite for both nuclear migrations.
Figure 2. NMpre and NMpost are oriented by the polarity crescent.
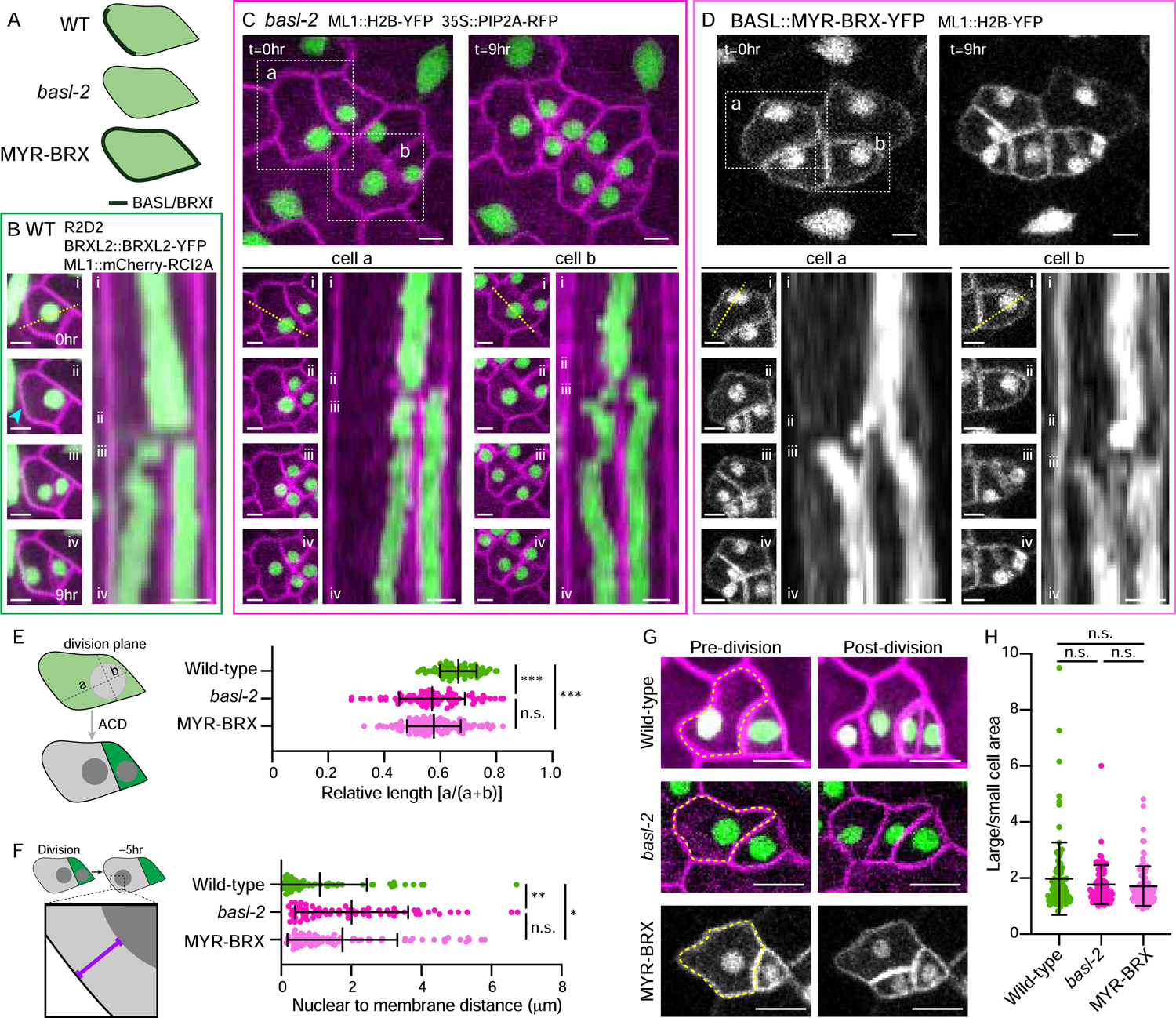
A. Schematic of the genotypes used and corresponding polarity protein localizations.
B. Stills (left) and kymograph (right) showing NMpre and NMpost over nine hours in wild-type cells. Stills correspond to the indicated time frames in the kymograph showing the (i) initial, (ii) pre-mitotic, (iii) post-division, and (iv) final nuclear positions. Kymograph (right) generated from the dotted line in (i). The cyan arrowhead indicates the polarity crescent. Scale bars-5μm.
C–D. Lower panels as in Figure 2B, with top panels showing broader fields at the start (t=0hr) and end (t=9hr), for basl-2 (C) and MYR-BRX (D). Two representative cells for each genotype are boxed. Scale bars-5μm.
E. Schematic illustrating the quantification method (left). Quantification of the nuclear position pre-ACD relative to the membrane opposite the division plane in the indicated genotypes (right). Wild-type-70 cells, basl-2-90 cells, MYR-BRX-128 cells. Data are represented as mean±standard deviation. n.s.-not significant, *** - p<0.001.
F. Schematic illustrating the quantification method (left). Quantification of the nuclear position relative to the membrane opposite the division plane five hours post-division in the indicated genotypes (right). Wild-type-64 cells, basl-2-69 cells, MYR-BRX-80 cells. Data are represented as mean±standard deviation. n.s.-not significant, * - p<0.05, ** - p<0.01.
G. Representative images of cells immediately before (left) and after (right) ACD in wild-type, basl-2, and MYR-BRX. Yellow dotted lines indicate the dividing cells. Scale bars-10μm.
H. Daughter cell size ratios in the indicated genotypes. Wild-type-99 cells, basl-2-83 cells, MYR-BRX-96 cells. Data are represented as mean±standard deviation. n.s.-not significant,
To test whether the polarity crescent position also controls the directions of NMpre and NMpost, we ectopically localized the polarity proteins along the plasma membrane through forced BRX myristoylation [14] and monitored nuclear movements (Figures 2A, 2B, and 2D–2F, Video S3). In MYR-BRX (BASL::MYR-BRX-YFP ML1::H2B-YFP), nuclei initiated NMpre (see cell a, frames i and ii in Figure 2D) but were not properly oriented. As a result, final nuclear position before mitotic onset was more variable and randomized in MYR-BRX than in wild-type. During wild-type NMpost, nuclei typically move to the plasma membrane opposite the division plane, where the BASL/BRXf crescent is often polarized. When the polarity domain was ectopically spread in MYR-BRX, the SLGC nucleus would move to a random cell wall during NMpost (Figures 2D and 2F). In extreme cases, nuclei moved toward the newly formed cell wall in MYR-BRX (see cell b in Figure 2D). These data demonstrate that the location of the BASL/BRXf polarity crescent is the essential spatial signal that orients nuclear migrations before and after ACD in the stomatal lineage.
Ectopic BASL is sufficient to drive nuclear repositioning
A rigorous test of whether cortical polarity is sufficient to guide nuclear position is to ectopically induce polarity crescents in naïve tissues. Using a stabilized version of BASL shown previously to generate polar crescents in post-mitotic cells (35S::GFP-BASL-IC) [13], we assayed nuclear position in the hypocotyl epidermis following forced polarization. Epidermal cells in the hypocotyl are non-mitotic, so we reasoned that ectopic BASL crescents would induce a migration analogous to NMpost. Indeed, while nuclei in control cells were centered, nuclei in 35S::GFP-BASL IC-expressing hypocotyls were more likely to be (1) offset from the cell center and (2) proximal to the ectopic crescents (Figures S2C–E). Similar results were found in pavement cells with ectopic BASL crescents (Figures S2F–H). We conclude, therefore, that cortical BASL can recruit more broadly expressed cellular machinery to position nuclei, even outside the stomatal lineage.
Nuclear migrations are required for proper daughter cell placement
In other plant tissues, pre-ACD nuclear migrations enable the production of daughters of differing sizes and fates and also direct the relative placement of these cells [23, 24]. Surprisingly, our time-lapse analyses of basl and MYR-BRX cotyledons revealed that unequally sized daughter cells could still be generated without appropriately oriented nuclear migrations (Figures 2G and 2H). The random nuclear movements in basl and the incorrectly oriented nuclear migrations in MYR-BRX create roughly equivalent outcomes on daughter cell sizes; in both cases, nuclei are frequently offset from the cell center pre-ACD but in an inappropriate place relative to neighbor cells of specific identities (Figure 2G). Combining the de-centering of nuclei, the irregular cell shapes of stomatal progenitors and the minimal-wall rule that broadly governs division plane placement in plant cells [25–27] provides a parsimonious explanation for how differentially sized daughter cells are created from stomatal progenitor divisions, even without directed nuclear migrations (Figure 2H). However, without NMpre and NMpost, tissue-wide spacing of meristemoids is incorrect owing to an inability to orient ACDs relative to neighbor cells (Figure 2G). Therefore, we propose that the stomatal clusters found in basl arise from an inability to orient divisions without NMpre and NMpost, coupled with BASL’s post-division function suppressing division potential [16, 28].
NMpre and NMpost require distinct cytoskeletal pathways
We hypothesized that the opposite orientations of NMpre and NMpost could provide an elegant means to reverse cellular polarity using a fixed polarity landmark. To test this, we sought a way to specifically disrupt the more enigmatic NMpost. We began by investigating the cytoskeletal requirements for NMpre and NMpost by assaying nuclear movement in cotyledons treated with DMSO (control), 1μM latrunculin B (latB), or 10μM oryzalin. LatB and oryzalin treatments efficiently depolymerized F-actin and microtubules, respectively, in the epidermis and had their expected impacts on nuclear movement in PCs (Figures S3A–S3D). Because F-actin is essential for pre-ACD nuclear migration in other developmental contexts, including subsidiary cell recruitment in Zea mays [1] and lateral root initiation in Arabidopsis [29], we were surprised that NMpre proceeded normally in latB-treated stomatal progenitors (Figures 3A, 3B and 3D). Oryzalin treatment, however, abrogated NMpre, indicating that pre-divisional migration is microtubule dependent. These results demonstrate that NMpre is mechanistically distinct from other pre-division migrations previously described in Arabidopsis.
Figure 3. NMpre and NMpost require distinct cytoskeletal pathways.
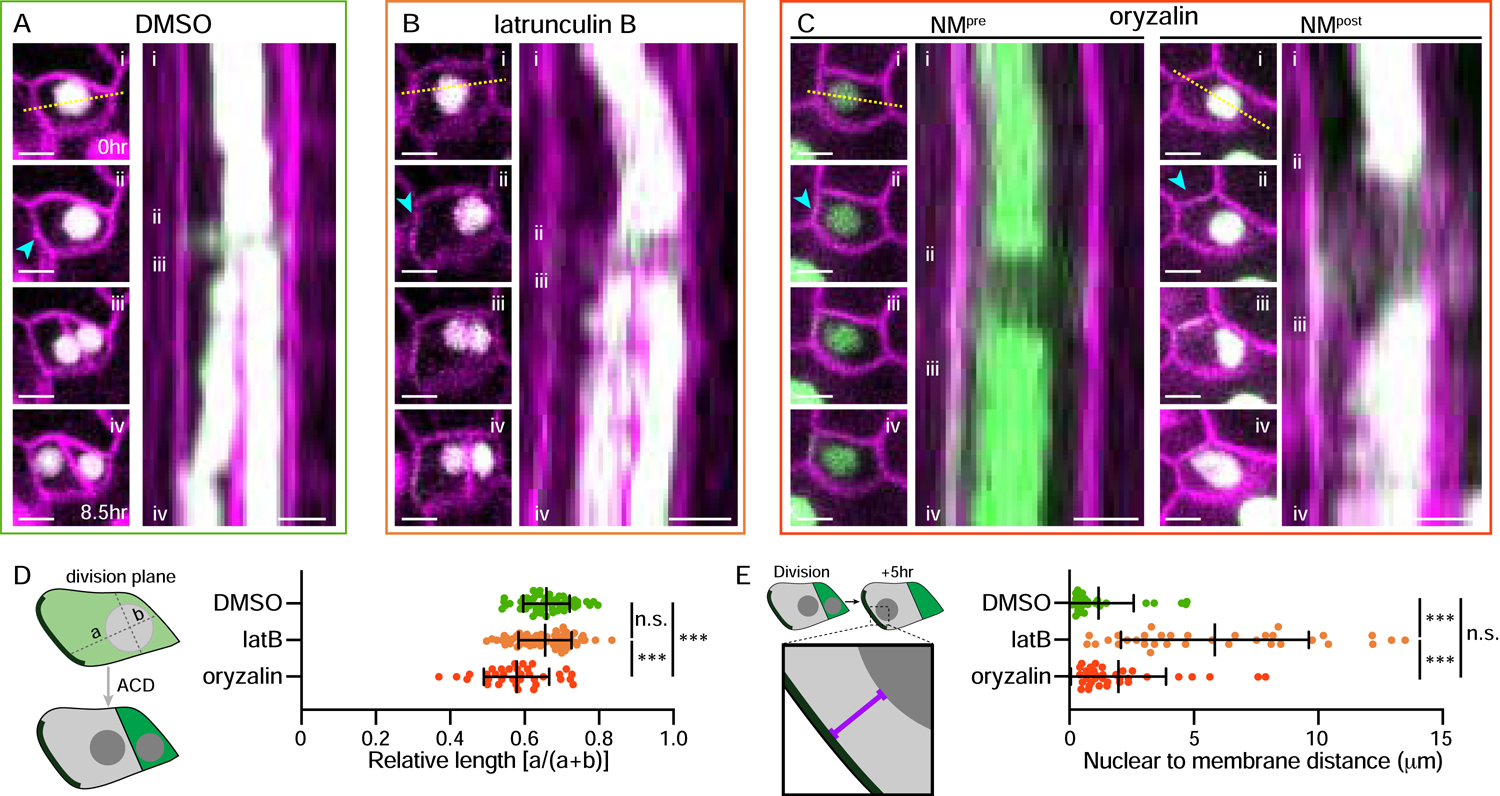
A–C. Stills (left) and kymographs (right) showing nuclear movement over 8.5hr in cells treated with (A) DMSO, (B) 1μM latrunculin B and (C) 10μM oryzalin. Stills correspond to the indicated time frames in the kymograph showing the (i) initial, (ii) pre-mitotic, (iii) post-division, and (iv) final nuclear positions. Kymograph (right) generated from the dotted line in (i). Two examples are provided for oryzalin treatment (C) to more clearly highlight NMpre and NMpost. Note the longer time between (ii) and (iii) in (C) due to the requirement for microtubules for mitosis. The cyan arrowheads indicate the polarity crescents. Scale bars-5μm.
D. Schematic illustrating the quantification method (left). Quantification of the nuclear position pre-ACD in the indicated treatments, oriented relative to the polarity crescent (right). DMSO-54 cells, latB-77 cells, oryzalin-35 cells. Data are represented as mean±standard deviation. n.s.-not significant, *** - p<0.001.
E. Schematic illustrating the quantification method (left). Quantification of the nuclear distance to the polarity crescent in SLGCs five hours after division (right). DMSO-29 cells, latB-34 cells, oryzalin-35 cells. Data are represented as mean±standard deviation. n.s.-not significant, *** - p<0.001.
See also Figure S3.
As cell divisions failed in oryzalin-treated cotyledons, we could not assay NMpost in SLGCs; however, the single nucleus that reformed following an extended mitosis in oryzalin-treated cells migrated toward the polarity crescent (Figures 3C and 3E), indicating that NMpost does not rely on microtubules. In contrast, NMpost completely failed in latB-treated cotyledons; without F-actin, SLGC nuclei remained near the division plane for the duration of our imaging (Figures 3B and 3E). NMpost disruption was not an indirect consequence of latB-mediated alternations to the polarity crescent, as BRXL2 was inherited by and maintained in SLGCs normally. We confirmed that associations between the nuclear envelope and reporters for microtubules and F-actin could be observed during NMpre and NMpost, respectively (Figures S3E and S3F). Therefore, the two ACD-associated nuclear migrations in the Arabidopsis stomatal lineage utilize separate cytoskeletal pathways; NMpre is microtubule dependent while NMpost requires F-actin.
MYOXI-I regulates NMpost
As NMpost required F-actin, we reasoned that it would also depend on myosin. The myosin XI family regulates various aspects of intracellular organization [30]. We noted that SLGC nuclei adopted an elongated (spindle-shaped) morphology during NMpost (Figure S4A), and MYOXI-I (At4g33200) was previously shown to 1) associate with the nuclear envelope and 2) control the spindle shape of pavement cell nuclei [31]. Therefore, we introduced plasma membrane (ML1::mCherry-RCI2A), nuclear (ML1::H2B-YFP), and polarity (BRXL2::BRXL2-YFP) reporters into the previously characterized myoxi-i3 and myoxi-i4 mutants [31] and performed time-lapse imaging of developing cotyledons. Pavement cell nuclei were rounder in myoxi-i lines, as previously reported, and PC stochastic movement was suppressed, although not as strongly as upon latB treatment (Figures S4B–S4E). As both myoxi-i3 and myoxi-i4 yielded identical results, we report the cellular phenotypes of myoxi-i4. NMpost was largely lost in myoxi-i4; SLGC nuclei failed to migrate toward the polarity crescent and remained near the division plane (Figures 4A–C). As expected, NMpre still initiated in myoxi-i4 (Figure S4F), so we conclude that MYOXI-I is specifically required for NMpost during stomatal lineage ACD.
Figure 4. MYOXI-I regulates NMpost and stomatal patterning.
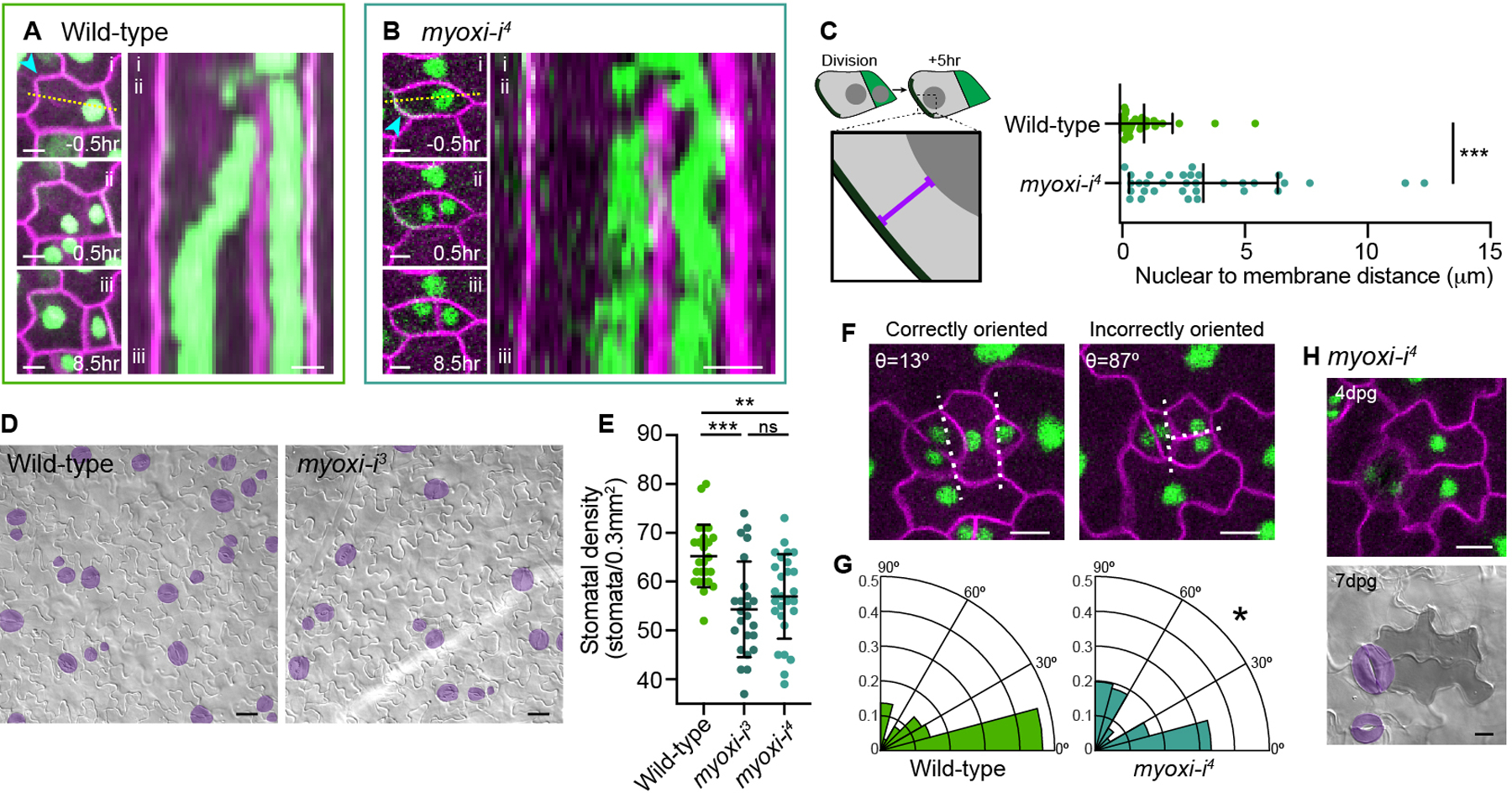
A. Stills (left) and kymograph (right) of NMpost in a wild-type SLGC. Stills show the cell (i) 0.5hr before ACD, (ii) 0.5hr after ACD and (iii) 8hr after ACD. Kymograph (right) generated from the dotted line in (i). The cyan arrowhead indicates the polarity crescent. Scale bars-5μm.
B. Stills (left) and kymograph (right) of NMpost in myoxi-i4, formatted as in Figure 4A. The cyan arrowhead indicates the polarity crescent. Scale bars-5μm.
C. Schematic illustrating the quantification method (left). Quantification of the nuclear distance to the polarity crescent in SLGCs five hours after division (right). Wild-type-31 cells, myoxi-i4-33 cells. Data are represented as mean±standard deviation. n.s.-not significant, *** - p<0.001.
D. Representative images of 7dpg wild-type and myoxi-i3 cotyledons. Stomata are pseudo-colored purple. Scale bars-25μm.
E. Quantification of abaxial stomatal density at 7dpg in wild-type and myoxi-i cotyledons. Wild-type-25 seedlings, myoxi-i3-24 seedlings, myoxi-i4-27 seedlings. Data are represented as mean±standard deviation. n.s.-not significant, ** - p<0.01, *** - p<0.001.
F. Representative images of correctly and incorrectly oriented spacing divisions, both from myoxi-i4. Dotted lines indicate the vectors used to calculate the spacing division angle (θ). Scale bars-10μm.
G. Quantification of spacing division angles in wild-type and myoxi-i4. Wild-type-88 cells, myoxi-i4-81 cells. Kolmogorov-Smirnov test-*- p<0.05.
H. Representative images of the abnormal cellular lobing phenotype at 4dpg (top) and 7dpg (below) following incorrect spacing divisions in myoxi-i4. Scale bars-10μm.
See also Figure S4.
NMpost is necessary to orient spacing divisions
Having identified that myoxi-i specifically lacked NMpost, we assessed the developmental functions of post-ACD nuclear migration. Cotyledons from 7 days post germination (dpg) myoxi-i3 and myoxi-i4 seedlings had lower stomatal densities than wild-type (Figures 4D and 4E). Closer examination revealed that the lower stomatal density in myoxi-i resulted from a specific decrease in the number of stomata formed through asymmetric spacing divisions. At 4dpg, when spacing divisions frequently occur, we noted numerous misoriented spacing divisions in myoxi-i4, and the quantification of division angles confirmed that significantly more spacing divisions were orthogonal to existing stomata in myoxi-i4 than in wild-type (Figures 4F and 4G).
Misoriented spacing divisions in myoxi-i generated two daughters that began to lobe, suggesting that they had both acquired SLGC fate (Figure 4H). At 7dpg, cells created by these misoriented divisions were observable as lobed (pavement cell-like) cell pairs (Figure 4H, bottom). The correctly oriented spacing divisions in myoxi-i4 generated the typical meristemoid/SLGC pair, indicating that disruption of MYOXI-I does not generally equalize cell identities after ACD. In summary, MYOXI-I is required for post-division nuclear migration, spacing division orientation, and stomatal patterning.
DISCUSSION
We defined two nuclear migrations that bookend each ACD in the Arabidopsis stomatal lineage. Nuclear migrations preceding ACD are prevalent during plant development, and our work adds the Arabidopsis stomatal lineage to the compendium of cell types that utilize this mechanism. While several examples of polarity domains that guide pre-ACD nuclear migration have been described in other plant species [1, 23, 32], cortical BASL/BRXf’s necessity and sufficiency for nuclear migrations and its role in coordinating the spatial positioning of successive ACDs are new. The use of time-lapse imaging to capture ACDs was critical, as it suggests that BASL/BRXf orient asymmetric divisions, rather than strictly define their physical asymmetry. We propose that future models of BASL functionality be expanded beyond kinase scaffolding to include nuclear positioning as an important parameter.
While it is tempting to speculate that conserved principles translate cortical polarity to pre-division nuclear migration across species, our studies of NMpre highlight several key differences, including the cytoskeletal elements involved and the cell cycle state [33], that argue against universality in nuclear positioning mechanisms. Critically, like the conserved PAR proteins in animals, previously identified plant polarity domains act as “attractive” cues to orient nuclear migration; in contrast, BASL/BRXf “repels” the nucleus before division. Why might stomatal formation depend on a fundamentally different strategy for pre-ACD nuclear orientation? One possible explanation lies in our identification of post-ACD nuclear migration in SLGCs. NMpost reverses nuclear position by “attracting” nuclei to the BASL/BRXf crescent in an actin-dependent manner. This suggests NMpost may share more mechanistic commonalities with pre-ACD nuclear migration in other cell types than does NMpre. Orienting two opposed nuclear migrations using a single polarity crescent is an elegant way to reorient cellular polarity in a cell cycle-dependent manner; as BASL switches from a “repulsive” to an “attractive” landmark at the end of mitosis, it can reverse cellular polarity in SLGCs while remaining fixed in place.
A key finding from our study is that disruption of MYOXI-I specifically perturbs post-division nuclear migration in the Arabidopsis stomatal lineage. We have shown that a specific subset of ACDs, spacing divisions, are more frequently misoriented in myoxi-i, ultimately leading to the production of fewer stomata. Based on our data and previous localization studies of MYOXI-I [31, 34, 35], we believe that the most parsimonious explanation for the stomatal patterning defect is that failure to properly execute NMpost in SLGCs increases the likelihood of misorienting subsequent spacing divisions in those cells. Other members of the myosin XI family have been implicated in division orientation in plants [36], but likely do so through direct effects on the mitotic machinery and/or cell wall guidance. We have not observed broad division defects that would suggest a similar role for MYOXI-I, and no studies, to our knowledge, have reported co-localization of MYOXI-I with the newly forming cell plate. However, whether other, unknown cellular effects of MYOXI-I disruption, such as the failure to segregate fate determinants, contribute to the patterning phenotype will require further exploration.
Polarity switching was a provocative “first principles” model that could describe how stomatal progenitors can create their own cellular neighborhoods in the dicot leaf [12], but it did not provide a specific mechanism by which polarity crescent location and division orientation could be coupled. Our data provide a plausible mechanistic basis for the polarity switching model and suggest that post-ACD nuclear migration may be a unique adaptation to accommodate the flexible nature of ACD and neighbor cell spacing in the dicot stomatal lineage.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Dominique Bergmann (dbergmann@stanford.edu).
Materials Availability
There are no restrictions on materials generated for this manuscript.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
All Arabidopsis thaliana lines reported in this study are in the Columbia (Col-0) background. Transgenic reporter lines were generated by agrobacterium-mediated transformation. Seeds were sterilized in 20% bleach with 0.1% Tween-20 for 10 minutes, washed 3x in sterile dH20, and plated on ½ Murashige and Skoog (MS) plates supplemented with 0.5% sucrose. Plates were stratified in the dark at 4ºC for 2 days and then grown vertically at 22°C under long day conditions (16hr light/8hr dark).
Method Details
Plant material
All reported lines are in the A. thaliana Col-0 background. The ML1::mCherry-RCI2A BRXL2::BRXL2-YFP R2D2 (RPS5A::DII-n3xVenus RPS5A::mDII-ntdTomato) [17] line was generated by crossing. HTR2::CDT1a-RFP ML1::YFP-RCI2A was generated by introducing HTR2::CDT1a-RFP [18] into the ML1::YFP-RCI2A [37] background by agrobacterium-mediated transformation. 35S::GFP-BASL-IC lines were created using the construct reported in [13]. basl-2 (WiscDsLox264F02) [13], tmm-1 E1728 BASL::GFP-BASL [13], and BASL::MYR-BRX-YFP [14] lines were previously reported. BASL::MYR-BRX-YFP and basl-2 35S::PIP2A-RFP were transformed with ML1::H2B-YFP [37] (kind gift from Dr. Adrienne Roeder) to generate BASL::MYR-BRX-YFP ML1::H2B-YFP and basl-2 35S::PIP2A-RFP ML1::H2B-YFP, respectively. 35S::YFP-ABD2-YFP and UBQ10::mCherry-ABD2-mCherry lines (kind gift from Dr. Elison Blancaflor) were previously reported [38]. BASL::YFP-BASL TMM::mCherry-TUA5 lines were generated by introducing TMM::mCherry-TUA5 R4pGWB601 into BASL::YFP-BASL [14] by agrobacterium-mediated transformation. The 35S::mCherry-TUA5 line was previously reported [39].
myoxi-i alleles are referenced in keeping with the numbering used in [31]; myoxi-i3 (SALK_092026) refers to kaku1–3 and myoxi-i4 (SALK_082443) refers to kaku1–4. Genotyping of myoxi-i3 and myoxi-i4 was performed using LBb1.3/myoxi-i3 F/myoxi-i3 R and LBb1.3/myoxi-i4 F/myoxi-i4 R, respectively (see Key Resources Table for the primer sequences). myoxi-i3 and myoxi-i4 reporter lines were generated by agrobacterium-mediated transformation with ML1::mCherry-RCI2A, BRXL2::BRXL2-YFP, and ML1::H2B-YFP reporters.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Agrobacterium tumefaciens GV3101 | N/A | N/A |
| Escherichia coli TOP10 | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Propidium iodide | Life Technologies/Thermo Fisher Scientific | P3566 |
| Oryzalin | Sigma-Aldrich | 36182 |
| Latrunculin B | Sigma-Aldrich | L5288–1MG |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis: BASL::YFP-BASL ML1::mCherry-RCI2A | [14] | N/A |
| Arabidopsis: tmm-1 E1728 BASL::GFP-BASL | [13] | N/A |
| Arabidopsis: basl-2 ML1::H2B-YFP 35S::PIP2A-RFP | This manuscript | N/A |
| Arabidopsis: BASL::MYR-BRX-YFP ML1::H2B-YFP | This manuscript | N/A |
| Arabidopsis: 35S::mCherry-TUA5 | [39] | N/A |
| Arabidopsis: 35S::YFP-ABD2-YFP | [38] | N/A |
| Arabidopsis: UBQ10::mCherry-ABD2-mCherry | [38] | N/A |
| Arabidopsis: BASL::YFP-BASL TMM::mCherry-TUA5 | This manuscript | N/A |
| Arabidopsis: BASL::CFP-BASL 35S::YFP-ABD2-YFP | This manuscript | N/A |
| Arabidopsis: myoxi-i3 | Arabidopsis Biological Resource Center | SALK_092026 |
| Arabidopsis: myoxi-i4 | Arabidopsis Biological Resource Center | SALK_082443 |
| Arabidopsis: myoxi-i4 BRXL2::BRXL2-YFP ML1::H2B-YFP ML1::mCherry-RCI2A | This manuscript | N/A |
| Arabidopsis: ML1::mCherry-RCI2A ML1::H2B-YFP RPS5A::DII-n3xVenus RPS5A::mDII-ntdTomato | This manuscript | N/A |
| Arabidopsis: HTR2::CDT1a-RFP ML1::YFP-RCI2A | This manuscript | N/A |
| Oligonucleotides | ||
| Primer: myoxi-i3 F-ATGATTTTCGCTATTGCATGG | This manuscript | N/A |
| Primer: myoxi-i3 R-GCTTCATTAGCAACCTGCAAG | This manuscript | N/A |
| Primer: myoxi-i4 F-TTCTGCAATTTCAATTCAGGC | This manuscript | N/A |
| Primer: myoxi-i4 R-CAGCAGACTTCTCCTTCATGG | This manuscript | N/A |
| Primer: TUA5 cDNA F-CACCATGAGGGAAATTATTAGCATTC | This manuscript | N/A |
| Primer: TUA5 cDNA R-TCAATAGTCTTCACCTTCATC | This manuscript | N/A |
| Primer: H2B cDNA F-CACCATGGCGAAGGCAGATAAGAAAC | This manuscript | N/A |
| Primer H2B cDNA R-AGAACTCGTAAACTTCGTAACC | This manuscript | N/A |
| Primer: LBb1.3-ATTTTGCCGATTTCGGAAC | [43] | N/A |
| Recombinant DNA | ||
| Plasmid: R4pGWB601 | [37] | RIKEN BRC: pdi00133 |
| Plasmid: R4pGWB640 | [40] | RIKEN BRC: pdi00145 |
| Plasmid: TMMpro pDONR P4-P1R | [19] | N/A |
| Plasmid: ML1pro pDONR P4-P1R | [41] | N/A |
| Plasmid: TUA5 cDNA pENTR/D-TOPO | This manuscript | N/A |
| Plasmid: H2B cDNA pENTR/D-TOPO | This manuscript | N/A |
| Plasmid: TMM::mCherry-TUA5 R4pGWB601 | This manuscript | N/A |
| Plasmid:: ML1::H2B-YFP | [37] | N/A |
| Plasmid: HTR2::CDT1a-RFP | [18] | N/A |
| Plasmid: 35S::GFP-BASL-IC | [13] | N/A |
| Plasmid:: ML1::H2B-YFP R4pGWB640 | This manuscript | N/A |
| Plasmid: BRXL2::BRXL2-YFP pHGY | [14] | N/A |
| Plasmid: ML1::mCherry-RCI2A R4pGWB401 | [44] | N/A |
| Plasmid: pENTR/D-TOPO | Invitrogen | K240020 |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPad Software | https://graphpad.com/scientific-software/prism/ |
| FIJI | NIH | https://imagej.net/Fiji |
Cloning
Cloning was done using Gateway technology (Invitrogen). To generate TMM::mCherry-TUA5 R4pGWB601, the TUA5 (At5g19780) sequence was amplified from A. thaliana cDNA using TUA5 cDNA F and TUA5 cDNA R (see Key Resources Table for the primer sequences) and subcloned into pENTR/D-TOPO (Invitrogen). The mCherry sequence was inserted at the N-terminus of TUA5 using NotI sites to create mCherry-TUA5 in pENTR/D-TOPO. TMM::mCherry-TUA5 was generated by Gateway cloning, combining mCherrry-TUA5 pENTR/D-TOPO, the TMM promoter in pDONR P4-P1R (contains 540bp upstream of the TMM gene) [19], and R4pGWB601 [40]. To clone ML1::H2B-YFP, the H2B (At5g22880) sequence was amplified from A. thaliana cDNA using H2B cDNA F and H2B cDNA R (see Key Resources Table for the primer sequences) and subcloned into pENTR/D-TOPO. ML1::H2B-YFP was generated by Gateway cloning combining H2B pENTR/D-TOPO, the ML1 promoter in pDONR P4-P1R (contains 3385bp upstream of the ATML1 gene) [41], and R4pGWB640 [40]. All plasmids generated for this study were confirmed by sequencing.
Microscopy and imaging
For time-lapse experiments, 3dpg and 4dpg seedlings were imaged in a custom flow chamber maintained in a ¼ MS solution supplemented with 0.75% sucrose [41]. The flow chamber was connected to a peristaltic pump, set to a rate of 2ml/hr to maintain a constant flow of fresh media. All time-lapse experiments were performed on a Leica SP5 confocal microscope with HyD detectors using 25x NA0.95 and 40x NA1.1 water objectives. Our time-lapse acquisition setup does not allow a paired design, so different genotypes and treatments were necessarily acquired in different imaging sessions. Time-lapse intervals ranged from 10–30 minutes depending on the experiment. All figures were generated from datasets with 30 minute intervals, with the exception of the basl-2 time-lapse shown in Figure 2, which came from a dataset with 20 minute intervals. Images of the cytoskeletal association with the nuclear envelope were acquired on a spinning disk confocal microscope with a Leica DMI6000 stand, Photometrics Evolve EMCCD camera, and 100x oil NA1.4 objective. The phenotypic analysis of 7dpg control, myoxi-i-3 and myoxi-i-4 was performed on cleared tissue samples using a Leica DM6B microscope with 20x NA0.7 and 40x NA0.85 air objectives. To prepare the cleared samples for phenotyping, cotyledons were placed in 7:1 ethanol:acetic acid for at least 24 hours. Samples were rinsed in dH2O and mounted on slides in Hoyer’s medium. Phenotypic analysis was performed on the abaxial sides of the cotyledons, and consistent regions were used for the stomatal density quantification to control for regional differences in stomatal density within the cotyledon. For the images in Figure S1A, seedlings were incubated in a 0.1mg/ml propidium iodide solution for 5 minutes before imaging. For Figure S2, seedlings were incubated in a 0.1mg/ml propidium iodide solution for >30 minutes to allow visualization of both the cell outlines and the nuclei.
Drug treatments
To assay the efficacy of the cytoskeletal inhibitors, respective reporter lines were incubated with DMSO, 1μM latrunculin B (Sigma-Aldrich) or 10μM oryzalin (Sigma-Aldrich), in ¼ MS + 0.75% sucrose, and images were acquired using a Leica SP5 confocal microscope after 2 hours. For time-lapse experiments using cytoskeletal inhibitors, the MS solution was supplemented with DMSO, 1μM latrunculin B, or 10μM oryzalin prior to seedling loading and all flow-through contained the same drug concentration. During the analysis, cell behaviors were only quantified after cellular indicators of drug penetrance were apparent: for latB, cessation of pavement cell nuclear movement was the benchmark, and for oryzalin mitotic failure was the benchmark.
Image quantification
For all time-lapse analyses, we began by registering the images using the Correct 3D Drift FIJI plugin [42] to account for whole organ growth. Additional registration on cropped regions of interest (ROIs) was performed as necessary to correct smaller drifts in localized areas. For calculating pre-ACD nuclear position (for Figure 1, 2, 3, 4, S1), the frame immediately prior to nuclear envelope breakdown was used to calculate the nuclear position along the longest axis orthogonal to the division plane. Nuclear position was determined based on the nuclear center in these measurements. Notably, nuclear positions in tmm-1 (Figure S1) were derived from single images where we inferred that cells with cortical and nuclear BASL signal were cells poised to undergo ACD. Note that data for NMpre in wild-type used in Figure 1, S1, and 2 were derived from 140 cells observed in 5 independent experiments. We pooled the cells for initial characterization of NMpre (Figure 1G, 1H, and S1F). These data were randomly split into two groups of 70 cells: one group was used for Figure 1I and one that was re-quantified according to the method shown in Figure 2E (left) and used for comparison with the polarity mutants. To quantify pre-ACD nuclear position in the polarity mutants, which lack a single polarity crescent to define the SLGC and use as an internal landmark, the larger daughter generated from the ACD was oriented to the left, as the SLGC is oriented in wild-type (Figure 2E). The wild-type samples were treated the same way for this quantification.
For the nuclear tracking in SLGCs during NMpost (Figure 1K and 1L, Figure S1H), nuclei were first segmented and xy coordinates defining the nuclear outline for each frame in the time-lapse movie were collected. Next, the polarity crescent position was manually defined in each frame of the time-lapse movie, and the shortest distances from the crescent to the nuclear periphery were calculated to define the nuclear to crescent distance over time. The nuclear position in SLGCs five hours post-division was calculated from time-lapse movies as the distance from the nuclear periphery to the polarity crescent. To quantify post-ACD nuclear position for the polarity mutants, the larger daughter cell generated from the ACD was oriented to the left, as the SLGC is oriented in wild-type. Without the polarity crescent to use as an internal reference, the nuclear position relative to the cell membrane distal to the division plane was calculated. For the quantification shown in Figure 2F, the wild-type cells were quantified in the same way as the polarity mutants to make them directly comparable.
Daughter cell size ratios in wild-type, basl-2, and MYR-BRX (Figure 2H) were quantified from time-lapse movies where individual ACDs could be tracked. Cell outlines were generated from frames acquired no more than 30 minutes after completion of cytokinesis to ensure that daughter cell size ratios accurately reflected the sizes generated by the division plane with minimal contribution from cellular growth.
To quantify the nuclear position relative to the polarity crescent in 35S::GFP-BASL-IC-expressing hypocotyls (Figure S2E), nuclei were first manually segmented, and the xy coordinates of the nuclear centers were used for downstream measurements. The cell outline was segmented and classified into “crescent” and “non-crescent” coordinates based on the cortical GFP-BASL-IC signal. The distances from the nuclear center to the “crescent” and “non-crescent” domains were calculated and averaged to obtain “crescent avg” and “non-crescent avg,” respectively. Crescent distance is therefore defined as . A value <1 indicates that the nucleus is closer, on average, to the crescent than to the rest of the cortex, while a value >1 indicates the converse. Control cells lack a polarity cell to use as a reference point and have non-randomly positioned nuclei. Therefore, to calculate the crescent distance score in control cells, cropped ROIs of individual cells were first randomly rotated. A “crescent” sized cortical domain (~1/6 of the cell perimeter) was manually defined on the same side of the cell across all rotated images. For control randomized (1) in Figure S2E, the “crescent” domain was always defined at the bottom of the rotated cell, and for control randomized (2) in Figure S2E, the “crescent” domain was always defined at the top of the rotated cells. Therefore, the data shown for the control cells in Figure S2E originate from the same pool of rotated cells, with opposing crescent regions. As an additional control, the 35S::GFP-BASL-IC cells were subjected to the same randomization regime of random rotation and consistent “crescent” definition. In this case, the real position of the GFP-BASL-IC crescent was disregarded. The results of this randomization are shown in the 35S::GFP-BASL-IC randomized column in Figure S2E.
Stochastic pavement cell nuclear movements were calculated from time-lapse movies. Nuclei were segmented, and the coordinates of the nuclear center were used to calculate the distance traveled per 30 minutes. The median value from the time-lapse movie was used to define the nuclear movement of an individual pavement cell.
Quantification of spacing division angles in wild-type and myoxi-i4 was performed in images acquired from 4dpg cotyledons. Only spacing divisions adjacent to stomata were used for these analyses. To consistently define the spacing division angle, one line was drawn through the two cell wall intersections where the guard cell meets the recently divided SLGC. A second line was drawn along the newly formed cell wall. The angle (θ) between these two lines was defined as the spacing division angle.
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8. Student’s t-tests were used to compare two samples. For comparison of three or more samples, one-way ANOVAs were used to test for significant interactions. When significant, Tukey’s post-hoc tests were used for pairwise comparisons. For measuring the distribution of spacing division angles, the Kolmogorov-Smirnov test was used. p<0.05 was used as the significance cut-off. For all graphs, n.s.-not significant, * - p<0.05, ** - p<0.01, *** - p<0.001. All graphs show mean±standard deviation for each group.
Supplementary Material
Video S1. Pre-division nuclear movement during stomatal lineage divisions. Related to Figure 1 and Figure S1.
Examples of cell type-specific pre-division nuclear behaviors in various stomatal lineage cells in the epidermis of 3dpg cotyledons. Time-lapse movies were acquired from BRXL2::BRXL2-YFP ML1::mCherry-RCI2A R2D2.
Video S2. Post-division nuclear movement in SLGCs. Related to Figure 1, Figure S1, and Figure S4.
Examples of post-division nuclear behavior in SLGCs. Time-lapse movies were acquired from BRXL2::BRXL2-YFP ML1::mCherry-RCI2A R2D2.
Video S3. Divisions in polarity mutants. Related to Figure 2.
Examples of divisions in stomatal progenitors in wild-type (BRXL2::BRXL2-YFP ML1::mCherry-RCI2A R2D2), basl-2 (basl-2 ML1::H2B-YFP 35S::PIP2A-RFP), and MYR-BRX (BASL::MYR-BRX-YFP ML1::H2B-YFP).
Highlights.
Nuclear migrations flank asymmetric divisions in the Arabidopsis stomatal lineage
A BASL/BRXf polarity crescent orients both nuclear migrations
MYOXI-I is required for nuclear migration after asymmetric division
Loss of directed nuclear migrations correlates with division orientation defects
ACKNOWLEDGEMENTS
We thank Martin Bringmann for reagent generation. We thank members of the Bergmann lab for critical reading and constructive insights during manuscript preparation. A.M. is supported by a postdoctoral fellowship from the National Institutes of Health (F32 GM133102–01). Y.G. is supported by funds from Stanford University and the Howard Hughes Medical Institute. D.C.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Cartwright HN, Humphries JA, and Smith LG (2009). PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323, 649–651. [DOI] [PubMed] [Google Scholar]
- 2.De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol 20, 1697–1706. [DOI] [PubMed] [Google Scholar]
- 3.Kimata Y, Higaki T, Kawashima T, Kurihara D, Sato Y, Yamada T, Hasezawa S, Berger F, Higashiyama T, and Ueda M (2016). Cytoskeleton dynamics control the first asymmetric cell division in Arabidopsis zygote. Proc. Natl. Acad. Sci. USA 113, 14157–14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau OS, and Bergmann DC (2012). Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139, 3683–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hara K, Kajita R, Torii KU, Bergmann DC, and Kakimoto T (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21, 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler M, Nadeau J, and Sack FD (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacAlister CA, Ohashi-Ito K, and Bergmann DC (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540. [DOI] [PubMed] [Google Scholar]
- 8.Kim TW, Michniewicz M, Bergmann DC, and Wang ZY (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vatén A, Soyars CL, Tarr PT, Nimchuk ZL, and Bergmann DC (2018). Modulation of Asymmetric Division Diversity through Cytokinin and SPEECHLESS Regulatory Interactions in the Arabidopsis Stomatal Lineage. Dev. Cell 47, 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau OS, Song Z, Zhou Z, Davies KA, Chang J, Yang X, Wang S, Lucyshyn D, Tay IHZ, Wigge PA, et al. (2018). Direct Control of SPEECHLESS by PIF4 in the High-Temperature Response of Stomatal Development. Curr. Biol 28, 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang CY, Lian HL, Wang FF, Huang JR, and Yang HQ (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson S, Barbier de Reuille P, Chan J, Bergmann D, Prusinkiewicz P, and Coen E (2011). Generation of spatial patterns through cell polarity switching. Science 333, 1436–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong J, MacAlister CA, and Bergmann DC (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe MH, Dong J, Weimer AK, and Bergmann DC (2019). A Plant-Specific Polarity Module Establishes Cell Fate Asymmetry in the Arabidopsis Stomatal Lineage. bioRxiv, 614636.
- 15.Zhang Y, Guo X, and Dong J (2016). Phosphorylation of the Polarity Protein BASL Differentiates Asymmetric Cell Fate through MAPKs and SPCH. Curr. Biol 26, 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Wang P, Shao W, Zhu JK, and Dong J (2015). The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, and Weijers D (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin K, Ueda M, Takagi H, Kajihara T, Sugamata Aki S, Nobusawa T, Umeda-Hara C, and Umeda M (2014). A dual-color marker system for in vivo visualization of cell cycle progression in Arabidopsis. Plant J 80, 541–552. [DOI] [PubMed] [Google Scholar]
- 19.Nadeau JA, and Sack FD (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. [DOI] [PubMed] [Google Scholar]
- 20.Venkei ZG, and Yamashita YM (2018). Emerging mechanisms of asymmetric stem cell division. J. Cell Biol 217, 3785–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campos R, Goff J, Rodriguez-Furlan C, and Van Norman JM (2020). The Arabidopsis Receptor Kinase IRK Is Polarized and Represses Specific Cell Divisions in Roots. Dev. Cell 52, 183–195. [DOI] [PubMed] [Google Scholar]
- 22.van Dop M, Fiedler M, Mutte S, de Keijzer J, Olijslager L, Albrecht C, Liao CY, Janson ME, Bienz M, and Weijers D (2020). DIX Domain Polymerization Drives Assembly of Plant Cell Polarity Complexes. Cell 180, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi P, and Goshima G (2020). Rho of Plants GTPases and Cytoskeletal Elements Control Nuclear Positioning and Asymmetric Cell Division during Physcomitrella patens Branching. Curr. Biol 30, 2860–2868. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida S, Barbier de Reuille P, Lane B, Bassel GW, Prusinkiewicz P, Smith RS, and Weijers D (2014). Genetic control of plant development by overriding a geometric division rule. Dev. Cell 29, 75–87. [DOI] [PubMed] [Google Scholar]
- 25.Besson S, and Dumais J (2011). Universal rule for the symmetric division of plant cells. Proc. Natl. Acad. Sci. USA 108, 6294–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez P, Allsman LA, Brakke KA, Hoyt C, Hayes J, Liang H, Neher W, Rui Y, Roberts AM, Moradifam A, et al. (2018). Predicting Division Planes of Three-Dimensional Cells by Soap-Film Minimization. Plant Cell 30, 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moukhtar J, Trubuil A, Belcram K, Legland D, Khadir Z, Urbain A, Palauqui JC, and Andrey P (2019). Cell geometry determines symmetric and asymmetric division plane selection in Arabidopsis early embryos. PLoS Comput. Biol. 15, e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houbaert A, Zhang C, Tiwari M, Wang K, de Marcos Serrano A, Savatin DV, Urs MJ, Zhiponova MK, Gudesblat GE, Vanhoutte I, et al. (2018). POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563, 574–578. [DOI] [PubMed] [Google Scholar]
- 29.Vilches Barro A, Stöckle D, Thellmann M, Ruiz-Duarte P, Bald L, Louveaux M, von Born P, Denninger P, Goh T, Fukaki H, et al. (2019). Cytoskeleton Dynamics Are Necessary for Early Events of Lateral Root Initiation in Arabidopsis. Curr. Biol 29, 2443–2454. [DOI] [PubMed] [Google Scholar]
- 30.Duan Z, and Tominaga M (2018). Actin-myosin XI: an intracellular control network in plants. Biochem. Biophys. Res. Commun 506, 403–408. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, and Hara-Nishimura I (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol 23, 1776–1781. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Facette M, Humphries JA, Shen Z, Park Y, Sutimantanapi D, Sylvester AW, Briggs SP, and Smith LG (2012). Identification of PAN2 by quantitative proteomics as a leucine-rich repeat-receptor-like kinase acting upstream of PAN1 to polarize cell division in maize. Plant Cell 24, 4577–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennard JL, and Cleary AL (1997). Pre-mitotic nuclear migration in subsidiary mother cells of Tradescantia occurs in G1 of the cell cycle and requires F-actin. Cell Motil. Cytoskel 36, 55–67. [DOI] [PubMed] [Google Scholar]
- 34.Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, and Sparkes IA (2009). A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol 150, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haraguchi T, Tominaga M, Nakano A, Yamamoto K, and Ito K (2016). Myosin XI-I is Mechanically and Enzymatically Unique Among Class-XI Myosins in Arabidopsis. Plant Cell Physiol 57, 1732–1743. [DOI] [PubMed] [Google Scholar]
- 36.Abu-Abied M, Belausov E, Hagay S, Peremyslov V, Dolja V, and Sadot E (2018). Myosin XI-K is involved in root organogenesis, polar auxin transport, and cell division. J. Exp. Bot 69, 2869–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roeder AH, Chickarmane V, Cunha A, Obara B, Manjunath BS, and Meyerowitz EM (2010). Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol 8, e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyachok J, Sparks JA, Liao F, Wang YS, and Blancaflor EB (2014). Fluorescent protein-based reporters of the actin cytoskeleton in living plant cells: fluorophore variant, actin binding domain, and promoter considerations. Cytoskeleton 71, 311–327. [DOI] [PubMed] [Google Scholar]
- 39.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, and Ehrhardt DW (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol 11, 797–806. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, et al. (2010). Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotech. Bioch 74, 1315–1319. [DOI] [PubMed] [Google Scholar]
- 41.Davies KA, and Bergmann DC (2014). Functional specialization of stomatal bHLHs through modification of DNA-binding and phosphoregulation potential. Proc. Natl. Acad. Sci. USA 111, 15585–15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parslow A, Cardona A, and Bryson-Richardson RJ (2014). Sample drift correction following 4D confocal time-lapse imaging. J. Vis. Exp 86, 51086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Malley RC, Barragan CC, and Ecker JR (2015). A user’s guide to the Arabidopsis T-DNA insertion mutant collections. Methods Mol. Biol 1284, 323–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bringmann M, and Bergmann DC (2017). Tissue-wide Mechanical Forces Influence the Polarity of Stomatal Stem Cells in Arabidopsis. Curr. Biol 27, 877–883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Pre-division nuclear movement during stomatal lineage divisions. Related to Figure 1 and Figure S1.
Examples of cell type-specific pre-division nuclear behaviors in various stomatal lineage cells in the epidermis of 3dpg cotyledons. Time-lapse movies were acquired from BRXL2::BRXL2-YFP ML1::mCherry-RCI2A R2D2.
Video S2. Post-division nuclear movement in SLGCs. Related to Figure 1, Figure S1, and Figure S4.
Examples of post-division nuclear behavior in SLGCs. Time-lapse movies were acquired from BRXL2::BRXL2-YFP ML1::mCherry-RCI2A R2D2.
Video S3. Divisions in polarity mutants. Related to Figure 2.
Examples of divisions in stomatal progenitors in wild-type (BRXL2::BRXL2-YFP ML1::mCherry-RCI2A R2D2), basl-2 (basl-2 ML1::H2B-YFP 35S::PIP2A-RFP), and MYR-BRX (BASL::MYR-BRX-YFP ML1::H2B-YFP).
Data Availability Statement
This study did not generate any unique datasets or code.


