Abstract
In vitro spermatogenesis, which produces fertile spermatozoa, has been successfully performed using an organ culture method from murine tissue. Here, we provide a dataset of time-course microarray transcriptome data of in vitro cultured neonate murine testes and age-matched in vivo-derived testes. The dataset presented here is related to the article titled “Transcriptome analysis reveals inadequate spermatogenesis and immediate radical immune reactions during organ culture in vitro spermatogenesis” published in Biochemical and Biophysical Research Communications in 2020 [1]. The raw data and pre-processed data are publicly available on the GEO repository (accession number GSE147982). Furthermore, the dataset provided here includes additional metadata, detailed explanations of the experiment, results of pre-processing, analysis scripts, and lists of differentially expressed genes from in vitro culture testes and in vivo testes at each time point.
Keywords: Spermatogenesis, Organ culture, Transcriptome, Microarray
Specifications Table
| Subject | Molecular Biology |
| Specific subject area | Andrology |
| Type of data | Binary Table Figure |
| How data were acquired | Illumina MouseWG-6 v2.0 Expression beadchip |
| Data format | Raw Analyzed |
| Parameters for data collection | In vitro cultured testes for 2, 4, 6, 7, 9, and 14 days from 7 day-post-partum (dpp) mice and time-matched in vivo-derived testes of corresponding age of mice to the cultured testes (7, 9, 11, 13, 14, 16, and 21 dpp). |
| Description of data collection | Mouse testes were extracted from 7 dpp male Acr-Gfp+/+ or Acr-Gfp+/− mice (a mixture of ICR and C57BL/6) and cultured by the gas-liquid interphase culture method. The in vitro cultured testes are then collected and subjected to total RNA extraction. The total RNA from in vivo-derived testes of corresponding aged mice to the cultured testes were also extracted. The quality of the total RNA was checked and microarray analysis were performed by Illumina MouseWG-6 v2.0 Expression beadchip. |
| Data source location | RIKEN Center for Integrative Medical Sciences 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, Japan |
| Data accessibility | Repository name: Gene Expression Omnibus Data identification number: GSE147982 Direct URL to data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147982 |
| Related research article | Takeru Abe, Hajime Nishimura, Takuya Sato, Harukazu Suzuki, Takehiko Ogawa, Takahiro Suzuki, Time-course microarray transcriptome data of in vitro cultured testes and corresponding in vivo testes. Biochem Biophys Res Commun. 530 (2020) 732-738 https://doi.org/10.1016/j.bbrc.2020.06.161 |
Value of the Data
-
•
This is the first time-course transcriptome data of in vitro cultured testes, which is useful for describing the difference between in vitro cultured testes and in vivo testes.
-
•
The data benefits scientists in reproductive medicine and reproductive engineering, especially those involved in in vitro spermatogenesis.
-
•
The data can be used to identify target pathways/genes to improve in vitro spermatogenesis.
1. Data Description
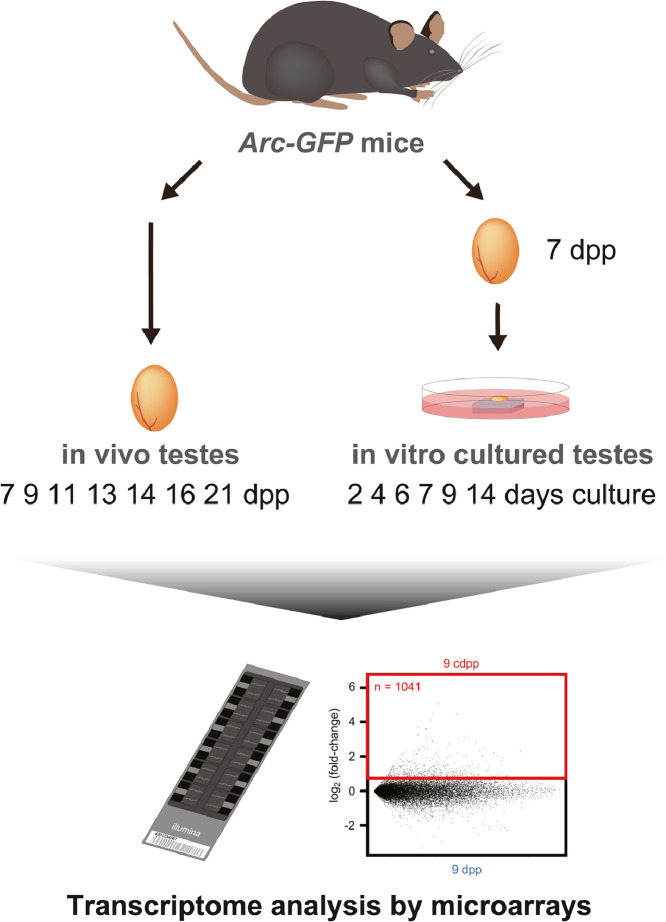
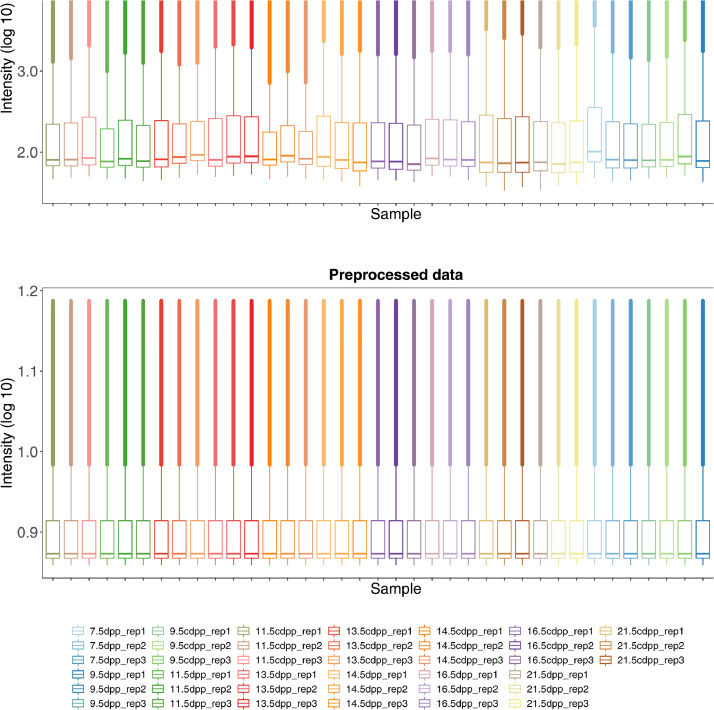
RNA was extracted from in vitro cultured testes (2, 4, 6, 7, 9, and 14 days of culture from 7 dpp mouse testes) and age-matched control in vivo-derived samples (7, 9, 11, 13, 14, 16, and 21 dpp mouse testes), followed by microarray analyses in three biological replicates. A schematic representation of the data generation is shown in Fig. 1. The raw intensity binary and text data of Illumina MouseWG-6 v2.0 Expression beadchip are provided as supplementary files (GSE147982_RAW.tar and GSE147982_non-normalized.txt.gz files, respectively) at Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE147982. The link between each GEO sample accession and the original mouse is provided in Table 1. The preprocessed data, which is subjected to background correction, variance-stabilizing transformation (VST), and quantile normalization, are also provided at GEO GSE147982. The distributions of signal intensity of raw and preprocessing data are reported in Fig. 2. Supplementary Table 1 provides the result of differential expression analysis, which is done using an empirical moderated t-statistics test with a cut-off adjusted p-value of 0.05.
Fig. 1.
Schematic representation of data generation.
Table 1.
Origin of RNA sample. The abbreviation “dpp” corresponds days post-partum (days-old) and “cddp” corresponds corresponding days post-partum, which is age-matched sample to the in vivo derived testes. Rep means biological replicate index of an experiment.
| Mouse ID | Weight (g) | GEO sample accession | ||
|---|---|---|---|---|
| 1 | 3.66 | GSM4451319 | ||
| 2 | 5.33 | GSM4451321 | ||
| 3 | 4.570 | GSM4451323 | ||
| 4 | 5.54 | GSM4451331 | ||
| 5 | 4.37 | GSM4451332 | ||
| 6 | 5.71 | GSM4451333 | ||
| 7 | 7.13 | GSM4451334 | ||
| 8 | 6.15 | GSM4451335 | ||
| 9 | 5.99 | GSM4451336 | ||
| 10 | 6.73 | GSM4451337 | ||
| 11 | 5.63 | GSM4451338 | ||
| 12 | 6.43 | GSM4451339 | ||
| 13 | 8.78 | GSM4451320 | ||
| 14 | 7.07 | GSM4451322 | ||
| 15 | 7.68 | GSM4451324 | ||
| 16 | 7.85 | GSM4451340 | ||
| 17 | 6.46 | GSM4451341 | ||
| 18 | 6.05 | GSM4451342 | ||
| 19 | 18.36 | GSM4451325 | ||
| 20 | 17.51 | GSM4451326 | ||
| 21 | 14.14 | GSM4451327 | ||
| 22 | 4.31 | GSM4451343 | ||
| 23 | 5.88 | GSM4451344 | ||
| 24 | 5.19 | GSM4451345 | ||
| 25 | 4.31 | GSM4451346 | GSM4451349 | GSM4451352 |
| 26 | 5.88 | GSM4451347 | GSM4451350 | GSM4451353 |
| 27 | 5.19 | GSM4451348 | GSM4451351 | GSM4451354 |
| 28 | 4.14 | GSM4451355 | GSM4451328 | |
| 29 | 5.5 | GSM4451356 | GSM4451329 | |
| 30 | 5.28 | GSM4451357 | GSM4451330 | |
Fig. 2.
Distribution of signal intensity. The log10 transformed signal intensity of each sample is shown as boxplot. The upper panel is raw intensity (before pre-processing) and the lower panel is after pre-processing. The abbreviation “dpp” corresponds days post-partum (days-old) and “cddp” corresponds corresponding days post-partum, which is age-matched sample to the in vivo derived testes. Rep means biological replicate index of an experiment.
2. Experimental Design, Materials and Methods
2.1. Mice
Acr-Gfp transgenic mice (C57BL/6 strain) [2] were obtained from RIKEN BRC through the National Bio-Resource Project of MEXT, Japan. Male homozygous Acr-Gfp transgenic mice were bred with female ICR, C57BL/6 (CLEA Japan, Tokyo, Japan), or ICRxC57BL/6 F1. Thus, male mice of Acr-Gfp (+/+) or (+/-) background were used for analysis. Mice were housed in the TSRI Specific Pathogen Free (SPF) facility with a 14-hour light cycle at 24 ± 1°C and 55 ± 5% air conditions and were given hard pellet food (Oriental Yeast, Tokyo, Japan) and acidified water (pH 2.8–3.0) ad libitum.
2.2. Organ culture
Testes were extracted from 7 dpp male Acr-Gfp transgenic mice. The extracted testes were decapsulated and cut into 4-8 fragments by forceps under a microscope in the culture medium (α-minimum essential medium (Thermo Fisher Scientific Inc., Wilmington, NC, USA)), supplemented with 40 mg/mL AlbuMAX (Thermo Fisher Scientific Inc.). The testis fragments were then patterned onto a 1.5% (w/v) agarose gel block (about 10 mmW × 10 mmD × 5 mmH) that was pre-soaked for at least two days in advance and then half-submerged in the culture medium in a well of a 12 well tissue-culture plate (Greiner Bio-One, Kremsmünster, Austria). The testes fragments were covered with the Polydimethylsiloxane (PDMS)-ceiling chip [3] and cultured in 5% CO2 at 34°C with once a week medium replacement.
2.3. RNA extraction
Fresh in vivo-derived testes were maintained in RNA later (Thermo Fisher Scientific Inc.) on ice for at least 3 h until RNA extraction. The in vivo-derived testes and in vitro cultured testes were homogenized in TRIzol Reagent (Thermo Fisher Scientific Inc.) with the PT1300D polytron homogenizer (KINEMATICA AG, Luzern, Switzerland), and the aqueous phase was collected according to the manufacturer's instructions. RNA was extracted from the corrected aqueous phase using the NucleoSpin RNA (MACHEREY-NAGEL, Düren, Germany), according to the manufacturer's instructions. The quality of the RNA was confirmed by NanoDrop (ND-1000, Thermo Fisher Scientific Inc.) and RNA 6000 nano kit of Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA samples with high RNA integrity values (> 9) were used in the microarray analyses.
2.4. Microarray
Total RNA was amplified by in vitro transcription method and the resulting cRNAs were biotinylated using the Illumina TotalPrep RNA Amplification Kit (Illumina, Inc., San Diego, CA, USA). The biotinylated cRNA was hybridized to MouseWG-6 v2.0 Expression beadchip (Illumina, Inc.). Signal intensity was measured using the Illumina BeadArray reader (Illumina, Inc.).
2.5. Data pre-processing
Raw intensity binary data (idat files) were converted to text data using BeadStudio (I llumina, Inc.) without background correction and normalization. Raw intensity text data were subjected to background correction, variance-stabilizing transformation (VST), and quantile normalization using lumiExpresso function implemented in the lumi package of R.
2.6. Differential expression analysis
The probes whose detection p-value was greater than 0.01 in any of the samples were removed. A linear model was fitted to the expression data for each probe using the lmFit function implemented in the limma package of R. Then, p-values of empirical Bayes moderated t-statistics test between in vitro cultured- and in vivo-derived-testes at each time point were computed by contrasts.fit and eBayes functions implemented in the limma package of R, followed by p-values adjustment by Benjamin-Hochberg method. The probes were considered to be differentially expressed genes if their adjusted p-value was less than 0.05.
2.7. Scripts
Scripts and data for R analysis used in this paper are available on GitHub (https://github.com/RIKEN-CFCT/ivs_data_in_brief).
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgments
Ethics Statement
All animal experiments conformed to the ARRIVE guidelines and the National Institutes of Health Guide for the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation (Animal Research Center of Yokohama City University, Yokohama, Japan)(Approval number: 17-A-17-072).
Acknowledgments
We thank K. Katagiri for technical assistance and J. H. P. Lim for English proofreading. We appreciate H. Kimura for development of the PDMS-ceiling chip device. This study was funded by a research grant from the Ministry of Education, Culture, Sport, Science, and Technology of Japan for the RIKEN Center for Integrative Medical Sciences, a Grant-in-Aid for Scientific Research on Innovative Areas, ‘Mechanisms regulating gamete formation in animals’ 25114007 (to T.O.) and ‘Ensuring integrity in gametogenesis’ 18H05546 (to T.O.) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2020.106482.
Contributor Information
Takehiko Ogawa, Email: ogawa@yokohama-cu.ac.jp.
Takahiro Suzuki, Email: takahiro.suzuki.aa@riken.jp.
Appendix. Supplementary materials
References
- 1.Abe T., Nishimura H., Sato T., Suzuki H., Ogawa T., Suzuki T. Time-course microarray transcriptome data of in vitro cultured testes and corresponding in vivo testes. Biochem. Biophys. Res. Commun. 2020;530:732–738. [Google Scholar]
- 2.Nakanishi T., Ikawa M., Yamada S., Parvinen M., Baba T., Nishimune Y., Okabe M. Real-time observation of acrosomal dispersal from mouse sperm using GFP as a marker protein. FEBS Lett. 1999;449:277–283. doi: 10.1016/s0014-5793(99)00433-0. [DOI] [PubMed] [Google Scholar]
- 3.Kojima K., Nakamura H., Komeya M., Yamanaka H., Makino Y., Okada Y., Akiyama H., Torikai N., Sato T., Fujii T., Kimura H., Ogawa T. Neonatal testis growth recreated in vitro by two‐dimensional organ spreading. Biotechnol. Bioeng. 2018;115 doi: 10.1002/bit.26822. 3030-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.