Abstract
Background
Behr syndrome is a clinically distinct, but genetically heterogeneous disorder characterized by optic atrophy, progressive spastic paraparesis, and motor neuropathy often associated with ataxia. The molecular diagnosis is based on gene panel testing or whole-exome/genome sequencing.
Methods
Here, we report the clinical presentation of two siblings with a novel genetic form of Behr syndrome. We performed whole-exome sequencing in the two patients and their mother.
Results
Both patients had a childhood-onset, slowly progressive disease resembling Behr syndrome, starting with visual impairment, followed by progressive spasticity, weakness, and atrophy of the lower legs and ataxia. They also developed scoliosis, leading to respiratory problems. In their late 30’s, both siblings developed a hypertrophic cardiomyopathy and died of sudden cardiac death at age 43 and 40, respectively. Whole-exome sequencing identified the novel homozygous c.627_629del; p.(Gly210del) deletion in UCHL1.
Conclusions
The presentation of our patients raises the possibility that hypertrophic cardiomyopathy may be an additional feature of the clinical syndrome associated with UCHL1 mutations, and highlights the importance of cardiac follow-up and treatment in neurodegenerative disease associated with UCHL1 mutations.
Keywords: Neurogenetics, Behr syndrome, Hereditary spastic paraplegia, Ataxia, Whole exome sequencing
Introduction
Hereditary spastic paraplegias (HSP) are a clinically and genetically diverse group of disorders characterized by progressive lower limb spasticity. Inheritance can be dominant, recessive or X-linked, or rarely mitochondrial [1]. Additional neurological manifestations include optic atrophy, cerebellar ataxia, and peripheral neuropathy [2]. SPG79 is caused by homozygous or compound heterozygous mutations in UCHL1, which encodes Ubiquitin C‐terminal hydrolase‐L1. In addition to spasticity, ataxia, and peripheral neuropathy, SPG79 is characterized by severe optic atrophy in the first decade of life. To date, mutations in UCHL1 have been described in only three families [3–5].
Using whole exome sequencing, we have identified a further family with SPG79, carrying a novel homozygous deletion in UCHL1. Both siblings presented in childhood with motor developmental delay, optic atrophy leading to progressive visual loss, cerebellar ataxia, spastic paraparesis, and motor neuropathy. Intriguingly, both siblings also developed hypertrophic cardiomyopathy, a potentially modifiable clinical feature which has not been described in association with SPG79 before.
Patients and methods
Patient 1 (P1) and Patient 2 (P2) were born to British parents from the North of England. There is no information on consanguinity. P1 was the product of a normal pregnancy and was born at full-term, and had normal early development. She first presented aged 8 years with visual problems and optic atrophy was detected. Her motor function gradually deteriorated over the next years and she developed progressive lower limb spasticity, foot deformities, and severe scoliosis requiring spinal surgery aged 16 years. Cognitive function was normal. She became non-ambulant following a femoral fracture aged 38 years. From the age of 39, she developed nocturnal hypoventilation and was commenced on overnight non-invasive ventilation.
On examination aged 41 years (Fig. 1a), she had facial dysmorphism including low nasal bridge, micrognathia, and large, low-set ears, bilateral optic atrophy and severely reduced visual acuity, but no ophthalmoparesis or nystagmus. Deep tendon reflexes were absent in the upper and lower limbs, and there was length-dependent atrophy and sensory loss in-keeping with a peripheral neuropathy. She had marked lower limb spasticity, extensor plantar responses and ankle clonus, and a severe scoliosis.
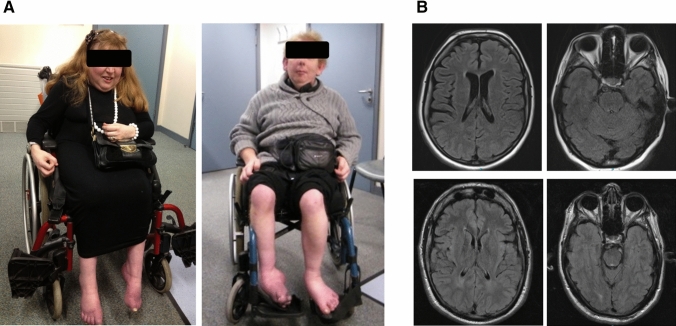
Fig. 1.
a P1 aged 40 years (left) and P2 aged 38 years. Both patients have characteristic facial features low-set ears and, and presented with distal upper and lower limb wasting and equinovarus deformity due to peripheral neuropathy and spastic paraparesis. b Axial T2 FLAIR MR images of P1 (top panels) and P2 (bottom panels). MRI brain in P1 aged 41 years showed mild global atrophy. MRI brain in P2 aged 34 demonstrated no significant abnormality apart from mild increased T2 signal in the peritrigonal white matter bilaterally
P2, her younger brother, also presented in early childhood with reduced visual acuity. He developed lower limb spasticity, contractures, and foot deformity. He became non-ambulant aged 8 years following surgical correction of his foot deformity. His visual acuity continued to deteriorate due to optic atrophy, so that he was functionally blind. Cognitive function was also unaffected in P2.
Examination of P2 aged 33 (Fig. 1a) revealed bilateral optic atrophy, both pendular and gaze induced nystagmus, ophthalmoparesis, facial dysmorphism including low nasal bridge, micrognathia and large, low-set ears, and facial hypotonia. Deep tendon reflexes were pathologically reduced in upper and lower limbs, and plantar responses were extensor. Sensation was intact. He had severe finger–nose ataxia, dysdiadochokinesia, and truncal ataxia.
Detailed laboratory work-up (including acylcarnitines, urinary organic acids, very long chain fatty acids, pristanic and phytanic acids, lysosomal enzymes, copper, caeruloplasmin, and acanthocytes) was normal. CSF constituents were normal in both cases. MRI brain in P1 aged 41 years showed mild global atrophy and a small focus of non-specific periventricular T2 signal change at the right frontal horn (Fig. 1b). MRI brain in P2 aged 34 (Fig. 1b) demonstrated no significant abnormality apart from mild increased T2 signal in the peritrigonal white matter bilaterally. Unfortunately, neurophysiological testing was not technically possible due to the degree of foot deformity and swelling. Direct sequencing of FXN revealed no candidate pathogenic variants. Fifty-six gene inherited peripheral neuropathy panel (Bristol genetics laboratory) which also failed to identify any pathogenic variants and a comparative genomic hybridization (CGH) array was normal.
Given the symptomatic features of a possible mitochondrial disorder, in particular optic atrophy, a cardiac evaluation was carried out in both patients. Echocardiogram in P2 aged 33 years revealed a small, globally hypertrophied left-ventricular cavity, left-ventricular systolic ejection gradient (36 mmHg), and impaired diastolic left-ventricular function. Echocardiogram in P1 aged 41 years was technically difficult due to severe scoliosis, but also demonstrated cardiomyopathy, with moderately impaired left-ventricular function. The left-ventricular posterior wall was hypokinetic and the interventricular septum was dyskinetic. There was no significant family history of sudden cardiac death, arrhythmias, heart failure, or young stroke.
Both siblings were commenced on ACE inhibitors and beta-blockers and their cardiac function stabilized. Despite this, both siblings developed progressive shortness of breath and apnoeic episodes. P1 died aged 43 years, P2 aged 40 years, both due to sudden cardiac death.
Whole exome sequencing
WES was performed on genomic DNA in P1 and P2 and on their mother. Their father was deceased at the time of study. The genomic DNA was exome-enriched using Illumina TruSeq 62 Mb exome capture and sequenced on an Illumina HiSeq 2000 with 100 bp paired-end reads. Standard filtering criteria were applied, including minor allele frequency of less than 1% and high-to-moderate variant effect predictor (i.e., nonsense, splice site, frame-shift, in-frame, and non-synonymous variants). Rare homozygous and compound heterozygous variants were defined, and protein altering and/or putative ‘disease causing’ mutations, along with their functional annotation, were identified using ANNOVAR. Candidate genes were prioritized if previously associated with a disease phenotype. Genomic and phenotypic data have been submitted to RD-Connect Genome-Phenome Platform (GPAP, https://platform.rd-connect.eu), where they can be accessed under a controlled access agreement.
Detection of homozygous regions
As part of the RD-Connect GPAP standard analysis pipeline, runs of homozygosity (ROH) were identified in each individual using PLINK v.1.90 [6], applying optimized parameters as defined by Kancheva et al. [7]. Only ROH with a minimum length of 1 Mbp were identified, to exclude common shorter ROH. Consanguinity assessment was performed through application of ROH thresholds for WES as described in Matalonga et al. [8].
Results
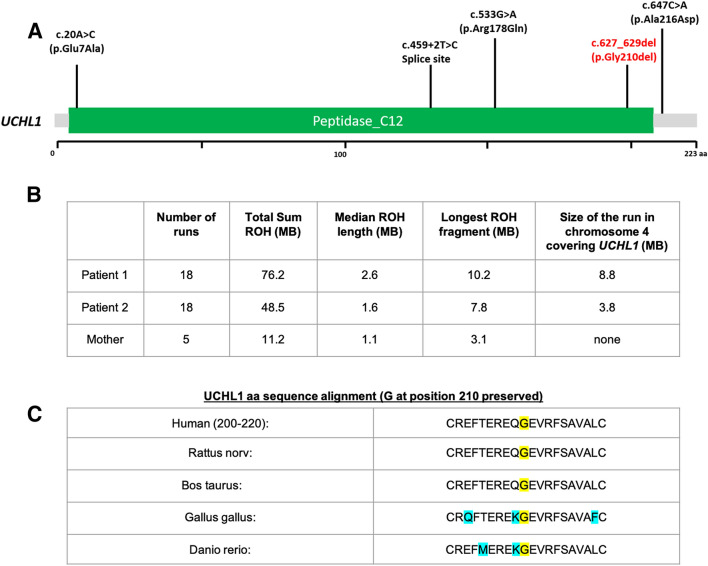
WES analysis revealed the presence of a homozygous c.627_629del; p.(Gly210del) UCHL1 variant (NM_004181.5) in P1 and P2 (Fig. 2a). This variant is reported only once in heterozygous form in gnomAD (allele frequency 0.000004), but never in homozygous form. Their unaffected mother was heterozygous for the variant. This variant could disrupt substrate binding at the distal UCHL1 site [9]. Additionally, we assessed IBD regions by the detection of > 1 Mbp runs of homozygosity (ROH). Results have shown that both patients carry an increased number of large ROH segments, 18 (P1 and P2) versus 5 from their mother, with a total ROH size of 76 MB (P1) and 48 MB (P2) respectively, underlying a possible recent consanguinity (Fig. 2b) [8]. Exome data were also analyzed for variants in genes associated with hypertrophic cardiomyopathy, but no potentially causative variants were detected.
Fig. 2.
a Pathogenic mutations identified in UCLH1. b Consanguinity analysis through run of homozygosity (ROH) > 1 Mbp detection in autosomal chromosomes. c Conservation of the amino acid affected by the c.627_629del; p.(Gly210del) UCHL1 mutation
Discussion
Here, we describe two siblings with clinical presentation comprising progressive spasticity, motor neuropathy, ataxia, and optic atrophy, resembling Behr syndrome, and with the additional clinical finding of hypertrophic cardiomyopathy. Whole exome sequencing identified a novel homozygous deletion of three nucleotides in UCHL1. Paternal data were not available to perform complete segregation studies and confirm his carrier status. However, normal CGH array results excluded a large chromosomal rearrangement in the paternal allele, and although a deletion in UCHL1 cannot be discarded, the possible recent consanguinity identified by ROH analyses support the homozygous status assumption. Ubiquitin C‐terminal hydrolase‐L1 (UCHL1) is critically important in maintaining free ubiquitin levels through the addition or removal of ubiquitin to poly-ubiquitin chains, and thus has a central role in cytoplasmic protein degradation [10]. It is one of the most abundant proteins in the nervous system, being highly expressed in both the central and peripheral nervous system [11–13]. Through its close interaction with the neuronal cytoskeleton, UCHL1 is thought to have important roles in axonal repair after injury, axonal transport, and synaptic function [14–16].
Mutations in UCHL1 have been described in three families with autosomal recessive spastic paraplegia (SPG79) [3–5]. The phenotype of the patients in the present study had many similarities to the previously reported cases (Table 1). In all cases described to date, the first symptom was of childhood-onset visual loss due to optic atrophy. The subsequent disease course was characterized by varying degrees of spasticity and cerebellar ataxia. Additional, less consistently described features include epilepsy, cognitive impairment, facial myokymia, fasciculations, and reduced peripheral sensation, reflecting the widespread expression of UCHL1 in most neuronal cells. The clinical presentation of our patients and all previously reported cases were compatible with Behr syndrome (Table 1).
Table 1.
Clinical and molecular findings in patients with SPG79 in the patients described in this study and in the patients described by Bliguvar et al. [4], Rydning et al. [5], and Bhowmik et al. [3]
| Paper | Bilguvar et al. [4] | Rydning et al. [5] | Bhowmik et al. [3] | Present family | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | NG 1024–1 | NG 1024–2 | NG 1024–3 | Patient III-4 | Patient III-5 | Patient III-6 | P1 | P2 | P1 | P2 |
| Age at examination | 28 y | 33 y | 34 y | 65 y | 62 y | 62 y | 10 y | 7 y |
41 y (died at 43) |
33 y (died at 40) |
| Age at onset | 5 y | 7 y | 5 y | 10 y | 10 y | 10 y | 2 y | 2 y | 8 y | 8 y |
| First symptom | Visual loss | Visual loss | Gait imbalance | Visual loss | Visual loss | Visual loss | Delayed development and seizures | Delayed development and abnormal gait | Visual loss | Visual loss |
| Optic atrophy | + | + | + | + | + | + | + | + | + | + |
| Nystagmus | + | + | + | + | + | + | + | + | − | + |
| Ophthalmoparesis | NA | NA | NA | − | + | − | − | − | − | + |
| Pyramidal signs | + | + | + | + | + | + | + | + | + | + |
| Spasticity | + | + | + | − | + | + | + | + | + | + |
| Peripheral motor neuropathy | + | + | + | + | + | + | + | + | + | + |
| Pes cavus | NA | NA | NA | + | + | + | − | − | + | + |
| Sensory loss | + | + | + | + | + | + | − | − | − | − |
| Cerebellar signs | + | + | + | + | + | + | + | + | + | + |
| Lost ambulation | + | + | + | + | + | + | + | + | + | + |
| Seizures | − | − | + | − | − | − | + | − | − | − |
| Cognitive impairment | NA | + | + | − | − | − | + | + | − | − |
| Facial dysmorphism | NA | NA | NA | NA | NA | NA | + | + | + | + |
| Chest deformity | NA | NA | NA | − | + | + | + | + | + (Scoliosis) | + (Scoliosis) |
| Mutations reported | c.20A > C (p.Glu7Ala) homozygous | c.533G > A (p.Arg178Gln); c.647C > A (p.Ala216Asp) compound heterozygous | c.459 + 2 T > C (homozygous) | c.627_629del (homozygous) | ||||||
y years, NA not available, + present,− absent
First described by Carl Behr in 1909, the classical Behr syndrome (MIM 210000) comprises childhood-onset optic atrophy combined with various neurological symptoms including spastic paraparesis, ataxia, mild ophthalmoparesis, peripheral neuropathy, and a variable degree of learning difficulties [17]. While the optic atrophy in these patients remains stable, other neurological signs progress during childhood becoming more prominent in the second or third decades [17].
Autosomal recessive OPA3 mutations have been identified in Iraqi Jewish patients presenting with Behr syndrome and 3-methylglutaconic aciduria (Costeff syndrome) [18] and more recently autosomal dominant [19] and recessive [20] OPA1 mutations were shown in Behr syndrome without metabolic abnormalities. We reported six patients with Behr syndrome and autosomal recessive mutation in C12orf65 [21], and a homozygous C12orf19 mutation has also been reported to cause this phenotype [22], illustrating further genetic heterogeneity of this phenotype. All previously reported genetic forms of Behr syndrome affect mitochondrial proteins involved in different mitochondrial functions. The detection of UCHL1 mutations in this phenotype adds another molecular mechanism, protein de-ubiquitination contributing to Behr syndrome and raising the possibility that this gene is also contributing to mitochondrial protein quality control.
In contrast to previously described cases, however, the siblings in this study both presented with the additional clinical feature of hypertrophic cardiomyopathy. Specifically, both patients demonstrated globally hypertrophied left ventricles in the absence of acquired causes of ventricular dysfunction (e.g., hypertension, myocardial ischaemia, valve dysfunction, prior exposure to toxins, and environmental pathogens). In addition, detailed family history failed to identify other family members known or suspected to be affected by a myocardial disease. The tissue distribution of UCHL1 is predominantly within the nervous system, although it is also expressed at low levels in gonadal cells [23]. Interestingly, UCHL1 is also upregulated in other cells under specialized conditions, for example in fibroblasts during wound healing and in pancreatic, colorectal, medullary thyroid, and breast cancer cells [24–27]. In these conditions, it is suggested that deregulation of the ubiquitin–proteasome system (UPS) may lead to uncontrolled cell growth. Furthermore, the UPS plays a key role in maintaining protein homeostasis and cardiac function, and UCHL1 has been shown to play a key role in this regulation. Knockdown of UCHL1 in cardiomyocytes and mouse hearts rescued cardiac hypertrophy induced by agonist or pressure overload, and overexpression resulted in the opposite effect [28]. We have not carried out functional studies of mutated UCHL1 protein (p.Gly210del). The Glycine residue at position 210 is close to the C-terminus, which regulates protein aggregation and stability [29]. The removal of the final four amino acids from the C-terminus renders the protein insoluble and abolishes binding to ubiquitin substrates [30]. Glycine at position 210 is located just upstream of the final beta-sheet and may have an impact on protein folding and stability. Further studies will be required to ascertain how the c.627_629del; p.(Gly210del) UCHL1 mutation might lead to clinical hypertrophic cardiomyopathy. We recognize that it is not possible to completely exclude an alternative genetic or acquired cause for hypertrophic cardiomyopathy in these siblings, or an unrecognized cardiac disease in previous family members. Nonetheless, the presentation in these cases raises the possibility that hypertrophic cardiomyopathy may be an additional feature of the clinical syndrome associated with UCHL1 mutations. The sudden cardiac death in both patients highlights the importance of cardiac follow-up and treatment in neurodegenerative disease associated with UCHL1 mutations.
In summary, we have reported on a novel UCHL1 deletion, resulting in a progressive neurodegenerative syndrome. The additional feature of hypertrophic cardiomyopathy has not been previously described. Cardiac evaluation is, therefore, essential in cases with this rare neurological phenotype.
Author contributions
GM has collected clinical data and drafted the manuscript. HL and AP performed bioinformatics analysis of exome sequencing data. BB and PFC performed clinical follow-up examination of the patients. AL collected clinical information and drafted Table 1; SL, SB, and LM performed the homozygosity analysis and participated in drafting the manuscript; RH performed clinical follow-up examinations of the patients, initiated genetic testing, and drafted the manuscript.
Funding
The study was supported by the Newton Fund UK/Turkey (MR/N027302/1 to HL and RH), the Medical Research Council (UK) (MR/N025431/1 to RH), the Wellcome Investigator fund (109915/Z/15/Z to RH), the Lily Foundation UK (to RH), the European Research Council (309548 to RH), and the Wellcome Trust Pathfinder Scheme (201064/Z/16/Z to HL and RH). HL receives support from the Canadian Institutes of Health Research (Foundation Grant FDN-167281), the Canadian Institutes of Health Research and Muscular Dystrophy Canada (Network Catalyst Grant for NMD4C), the Canada Foundation for Innovation (CFI-JELF 38412), and the Canada Research Chairs program (Canada Research Chair in Neuromuscular Genomics and Health, 950-232279). The study was further supported by the Horizon 2020 research and innovation program via grant 779257 “Solve-RD”. Data were analyzed using the RD-Connect Genome-Phenome Analysis platform developed under FP7/2007-2013 funded project (grant agreement no. 305444).
Data availability
Whole exome sequencing data of the patients and their mother have been uploaded to the RD-CONNECT platform for data sharing and storage.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest.
Ethics approval
Informed consent was obtained from each participant approved by the Yorkshire and The Humber–Leeds Bradford Research Ethics Committee (13/YH/0310).
Consent to participate
We obtained informed consent from both patients and their mother for this study.
Consent for publication
The patients died, therefore we could not obtain consent for this publication.
References
- 1.Finsterer J, Löscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318:1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Parodi L, Fenu S, Stevanin G, Durr A. Hereditary spastic paraplegia: more than an upper motor neuron disease. Rev Neurol (Paris) 2017;173:352–360. doi: 10.1016/j.neurol.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Bhowmik AD, Patil SJ, Deshpande DV, Bhat V, Dalal A. Novel splice-site variant of UCHL1 in an Indian family with autosomal recessive spastic paraplegia-79. J Hum Genet. 2018;63:927. doi: 10.1038/s10038-018-0463-6. [DOI] [PubMed] [Google Scholar]
- 4.Bilguvar K, Tyagi NK, Ozkara C, Tuysuz B, Bakircioglu M, Choi M, Delil S, Caglayan AO, Baranoski JF, Erturk O, et al. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:3489–3494. doi: 10.1073/pnas.1222732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rydning SL, Backe PH, Sousa MML, Iqbal Z, Øye A-M, Sheng Y, Yang M, Lin X, Slupphaug G, Nordenmark TH, et al. Novel UCHL1 mutations reveal new insights into ubiquitin processing. Hum Mol Genet. 2017;26:1031–1040. doi: 10.1093/hmg/ddx072. [DOI] [PubMed] [Google Scholar]
- 6.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kancheva D, Atkinson D, De Rijk P, Zimon M, Chamova T, Mitev V, Yaramis A, Maria Fabrizi G, Topaloglu H, Tournev I, Parman Y, Parma Y, Battaloglu E, Estrada-Cuzcano A, Jordanova A. Novel mutations in genes causing hereditary spastic paraplegia and Charcot-Marie-Tooth neuropathy identified by an optimized protocol for homozygosity mapping based on whole-exome sequencing. Genet Med. 2016;18(6):600–607. doi: 10.1038/gim.2015.139. [DOI] [PubMed] [Google Scholar]
- 8.Matalonga L, Laurie S, Papakonstantinou A, Piscia D, Mereu E, Bullich G, Thompson R, Horvath R, Pérez-Jurado L, Riess O, et al. Improved diagnosis of rare disease patients through systematic detection of runs of homozygosity. J Mol Diagn. 2020 doi: 10.1016/j.jmoldx.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudreaux DA, Maiti TK, Davies CW, Das C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc Natl Acad Sci U S A. 2010;107:9117–9122. doi: 10.1073/pnas.0910870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT. The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/S0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 11.Ann ES, Mizoguchi A, Okajima S, Ide C. Motor axon terminal regeneration as studied by protein gene product 9.5 immunohistochemistry in the rat. Arch Histol Cytol. 1994;57:317–330. doi: 10.1679/aohc.57.317. [DOI] [PubMed] [Google Scholar]
- 12.Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40:1542–1547. doi: 10.1111/j.1471-4159.1983.tb08124.x. [DOI] [PubMed] [Google Scholar]
- 13.Lin WM, Hsieh ST, Huang IT, Griffin JW, Chen WP. Ultrastructural localization and regulation of protein gene product 9.5. NeuroReport. 1997;8:2999–3004. doi: 10.1097/00001756-199709290-00002. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci U S A. 2010;107:1636–1641. doi: 10.1073/pnas.0911516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genç B, Jara JH, Schultz MC, Manuel M, Stanford MJ, Gautam M, Klessner JL, Sekerkova G, Heller DB, Cox GA, et al. Absence of UCHL 1 function leads to selective motor neuropathy. Ann Clin Transl Neurol. 2016;3:331–345. doi: 10.1002/acn3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 17.Behr C. Die komplizierte, hereditär-familiäre optikusatrophie des kindesalters: ein bisher nicht beschriebener symptomkompleks. Klinische Monatsblätter für Augenheilkunde. 1909;47:138–160. [Google Scholar]
- 18.Anikster Y, Kleta R, Shaag A, Gahl WA, Elpeleg O. Type III 3-methylglutaconic aciduria (optic atrophy plus syndrome, or Costeff optic atrophy syndrome): identification of the OPA3 gene and its founder mutation in Iraqi Jews. Am J Hum Genet. 2001;69:1218–1224. doi: 10.1086/324651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marelli C, Amati-Bonneau P, Reynier P, Layet V, Layet A, Stevanin G, Brissaud E, Bonneau D, Durr A, Brice A. Heterozygous OPA1 mutations in Behr syndrome. Brain. 2011;134:e169. doi: 10.1093/brain/awq306. [DOI] [PubMed] [Google Scholar]
- 20.Bonneau D, Colin E, Oca F, Ferré M, Chevrollier A, Guéguen N, Desquiret-Dumas V, N'Guyen S, Barth M, Zanlonghi X, et al. Early-onset Behr syndrome due to compound heterozygous mutations in OPA1. Brain. 2014;137:e301. doi: 10.1093/brain/awu184. [DOI] [PubMed] [Google Scholar]
- 21.Pyle A, Ramesh V, Bartsakoulia M, Boczonadi V, Gomez-Duran A, Herczegfalvi A, Blakely EL, Smertenko T, Duff J, Eglon G, Moore D, Yu-Wai-Man P, Douroudis K, Santibanez-Koref M, Griffin H, Lochmüller H, Karcagi V, Taylor RW, Chinnery PF, Horvath R. Behr’s Syndrome is typically associated with disturbed mitochondrial translation and mutations in the C12orf65 gene. J Neuromuscul Dis. 2014;1(1):55–63. doi: 10.3233/JND-140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleffner I, Wessling C, Gess B, Korsukewitz C, Allkemper T, Schirmacher A, Young P, Senderek J, Husstedt IW. Behr syndrome with homozygous C19ORF12 mutation. J Neurol Sci. 2015;357(1–2):115–118. doi: 10.1016/j.jns.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson KD, Deshpande S, Larsen CN. Comparisons of neuronal (PGP 9.5) and non-neuronal ubiquitin C-terminal hydrolases. Biochem Soc Trans. 1992;20:631–637. doi: 10.1042/bst0200631. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi Y, Nakayama S, Torikoshi Y, Tanaka S, Ishihara H, Taguchi T, Tamaki Y, Noguchi S. High expression of ubiquitin carboxy-terminal hydrolase-L1 and -L3 mRNA predicts early recurrence in patients with invasive breast cancer. Cancer Sci. 2006;97:523–529. doi: 10.1111/j.1349-7006.2006.00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano T, Miyauchi A, Matsuzuka F, Yoshida H, Nakata Y, Kuma K, Amino N. PGP9.5 mRNA could contribute to the molecular-based diagnosis of medullary thyroid carcinoma. Eur J Cancer Oxf Engl. 2004;40:614–618. doi: 10.1016/j.ejca.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Tezel E, Hibi K, Nagasaka T, Nakao A. PGP9.5 as a prognostic factor in pancreatic cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2000;6:4764–4767. [PubMed] [Google Scholar]
- 27.Yamazaki T, Hibi K, Takase T, Tezel E, Nakayama H, Kasai Y, Ito K, Akiyama S, Nagasaka T, Nakao A. PGP9.5 as a marker for invasive colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2002;8:192–195. [PubMed] [Google Scholar]
- 28.Bi HL, Zhang XL, Zhang YL, Xie X, Xia YL, Du J, Li HH. The deubiquitinase UCHL1 regulates cardiac hypertrophy by stabilizing epidermal growth factor receptor. Sci Adv. 2020;6(16):eaax4826. doi: 10.1126/sciadv.aax4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop P, Rocca D, Henley JM. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem J. 2016;473(16):2453–2462. doi: 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishop P, Rubin P, Thomson AR, Rocca D, Henley JM. The ubiquitin C-terminal hydrolase L1 (UCH-L1) C-terminus plays a key role in protein stability, but its farnesylation is not required for membrane association in primary neurons. J Biol Chem. 2014;289(52):36140–36149. doi: 10.1074/jbc.M114.557124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole exome sequencing data of the patients and their mother have been uploaded to the RD-CONNECT platform for data sharing and storage.