Abstract
JAK/STAT signaling pathway is involved in many diseases, including autoimmune diseases, which are characterized by a close interconnection between immune and bone system. JAK/STAT pathway is involved in bone homeostasis and plays an important role in proliferation and differentiation of some cell types, including osteoblasts and osteoclasts. Different molecules, such as cytokines, hormones, and growth factors are responsible for the activation of the JAK/STAT pathway, which leads, at the nuclear level, to start DNA transcription of target genes. Bone cells and remodeling process are often influenced by many cytokines, which act as strong stimulators of bone formation and resorption. Our aim, through careful research in literature, has been to provide an overview of the role of the JAK/STAT pathway in bone remodeling and on bone cells, with a focus on cytokines involved in bone turnover through this signal cascade. The JAK/STAT pathway, through the signal cascade activation mediated by the interaction with many cytokines, acts on bone cells and appears to be involved in bone remodeling process. However, many other studies are needed to completely understand the molecular mechanism underlying these bone process.
Keywords: JAK/STAT pathway, Bone, Osteoblast, Osteoclast, Cytokine
Introduction
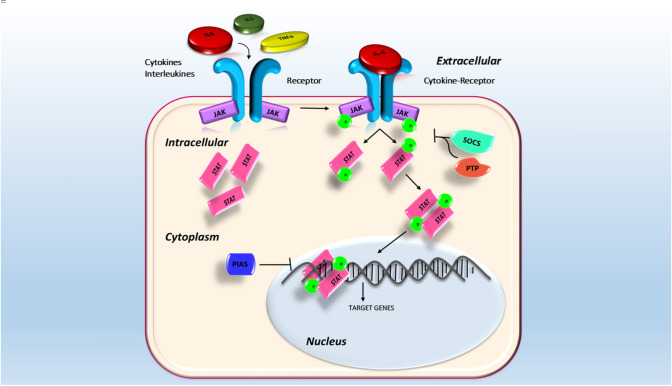
The Janus kinases (JAKs) are a family of protein tyrosine kinases (PTKs), named JAK1, JAK2, JAK3, and TYK2, that act on signal transducer and activator of transcription (STAT). The expression of JAK3 appears to be mainly in the hematopoietic cells. In contrast, the expression of the other members, JAK1, JAK2, and TYK2, is ubiquitous. After their activation, JAKs induce the phosphorylation of some STAT elements in the cytoplasm, which, subsequently their dimerization, are translocated in the nucleus. In the nucleus, STAT dimers bind to specific areas of DNA leading to the regulation of target genes responsible for the regulation of migration, proliferation, and apoptosis [1–3]. JAK/STAT signaling pathway (Fig. 1) plays a key role in several cytokines, immune system regulators, hormones, and hematopoiesis factors [4]. A specific receptor is located on the surface of target cells that bind specific cytokines. These receptors, which can be composed of multiple subunits, are substantially associated with JAK monomer [5–9]. Initially, the JAK monomers appear to be inactive; consequently, the binding between the ligand and its receptor induces a JAK transphosphorylation receptor-associated and therefore its activation. Each member of the JAK family consists of four domains, including the SH2 domain, which is responsible for the link between the receptor and STAT member. In the cytoplasmic compartment, some receptor tyrosine residues are subject to an activated JAK-induced phosphorylation process. This process involves the formation of docking sites for the subsequent binding of the STAT components. The STAT family consists of seven members, named STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6, and each of these participates in the signal cascade depending on which cytokine binds to its receptor on the cell surface [10, 11]. Each STAT member is characterized by some domains, which perform specific functions in the activation and transcription process. N-terminal and SH2 domains are responsible for the binding and interactions between dimers and proteins. Besides N-terminal domain is involved in STAT phosphorylation. STATs link DNA of target genes through the DNA binding domain, forming a protein-DNA complex. Lastly, the C-terminal transcription domain contains highly conserved phosphorylated tyrosine (Y) and serine (S) residues required for STATs activation [12–15]. Through careful research in the literature, our purpose is to review how the JAK/STAT signaling pathway is involved in bone remodeling, how it acts at the cellular level, especially in osteoblasts and osteoclasts, and finally in which cytokines involved in the pathway may affect bone homeostasis.
Fig. 1.
JAK/STAT pathway signaling. The binding of the cytokine to its receptor induces the activation of the JAKs, which phosphorylate the STATs elements. After the formation of STAT dimers, they migrate to the nucleus and, by binding to the target genes, they modify the transcription of DNA. Some elements can regulate the signaling cascade
JAK/STAT negative factors
The activation of the JAK/STAT pathway is negatively regulated by a class of proteins called suppressor of cytokine signaling (SOCs), protein tyrosine phosphatase (PTP), and protein inhibitors of activated STAT (PIAS). SOCs protein class includes eight members named as follows: SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6, SOCS7 and CIS (cytokine-inducible SH2-containing protein) [16]. Each member of the family is made up of three common domains: a central SH domain, a C-terminal SOCS box, and a variable N-terminal domain. The conserved central domain SH2 is responsible for binding SOCs proteins with cytokine receptors. In this way, the activity of the JAK kinases is blocked and the whole signaling complex undergoes proteasome-mediated degradation. In addition, two members of the SOCS class, SOCS1, and SOCS3 contain a kinase inhibitory region (KIR), so, when they hook to the receptors, the catalytic activity of JAKs is inhibited [17–19]. The N-terminal domain is variable in the different members of SOCS, with different lengths in each of them. In fact, it has been seen that the N-terminal region of SOCS4-7 is longer than the other member of the family [20]. In the end, a highly conserved C-terminal SOCS box domain can enroll factors to the formation of ligase complex that leads to proteasomal degradation of ubiquitinylated proteins [21, 22].
PTP proteins belong to a large family of protein tyrosine phosphatase, whose function is mainly to cut out the phosphate group from tyrosine residues of phosphorylated proteins. PTPs have a catalytic domain within which a site-active sequence is positioned. Based on the differences in the aminoacidic sequence in the catalytic domain, they are divided into 4 classes (I, II, III, and IV) [23–26]. Protein inhibitors of activated STAT (PIAS), as the name says, act on STATs proteins by inhibiting their transcriptional activity. The members of the PIAS family are five: PIAS1, PIASxα, PIASxβ, PIAS3, and PIAS4 (PIASγ) [27, 28]. Some studies show that every protein can interact with a particular STAT member. For example, PIAS1 and PIAS4 are capable of interacting with STAT1, while PIAS3 with STAT3 and STAT5. When PIAS proteins connect to STAT members, they induce the blockade of the link between STATs and DNA. Thus, the transcription co-factors of the STAT target gene are not recruited. In this way, PIAS proteins negatively regulate the STATs transcriptional activity [29–32].
JAK/STAT and bone cells
Many studies show that the JAK/STAT signaling pathway is involved in many diseases, including autoimmune diseases, and that its inhibition may be a therapeutic strategy. In autoimmune diseases, such as rheumatoid arthritis, the immune system, and the bone system are closely interconnected [33, 34]. The JAK/STAT pathway not only appears to be involved in bone homeostasis and response but also in the differentiation processes of some cells type. In particular, even the cytokines involved in the signaling pathway activation can act on this variety of cells, for example on osteoblasts, osteoclasts and osteocytes [35, 36]. The bone and the cells that regulate its remodeling are often subject to and/or influenced by the presence of cytokines belonging to the immune system. Many of them are strong modulators of bone metabolism, to influence the processes of bone formation and resorption. The bone physiological remodeling mechanism is due to a perfect relationship between osteoblasts and osteoclasts. These cells are responsible for bone formation and resorption mechanisms, respectively, while the role of mechanosensing is attributed to osteocytes [36]. Although both cells type cooperates in this process, they have different origins; while osteoblasts and osteocytes are of mesenchymal origin, osteoclasts, on the other hand, derive from hematopoietic stem cells [37, 38].
Osteoblasts are responsible for bone formation process, protein production, and matrix mineralization [39, 40]. During the differentiation process of osteoblasts, two transcriptional factors are very expressed. Shreds of evidence that these two factors, Runt-related transcription factor 2 (Runx2) and osterix (Osx), are necessary for the regulation of differentiation and are involved in maintaining of osteoblasts functionality [41, 42]. However, the role of osteoblasts also concerns the regulation of osteoclast differentiation. Indeed, when these two types of cells interact, osteoblasts produce several soluble factors, among which macrophage colony-stimulating factors (M-CSF), receptor activator of nuclear factor-κB ligand (RANKL), and osteoprotegerin (OPG) [43]. During the osteoclastogenesis process, RANKL is an essential factor, as it binds to the RANK receptor present on the surface of the precursors of the osteoclasts. This interaction also induces an increase in bone resorption capacity. In contrast, the RANK/RANKL interaction is negatively regulated by the presence of OPG. This molecule has a binding affinity with RANKL, thus, it prevents further binding (leads to a lower binding interaction) between the ligand and its receptor on osteoclasts [44, 45].
Recently, several studies have shown that different factors are involved during the osteoclastogenesis process [46]. This process leads to maturation and differentiation of osteoclasts from the monocyte/macrophage lineage. These cells are induced to differentiate starting from two factors, nuclear factor of activated T cells, cytoplasmic 1 (NFATc1) and c-Fos, whose expression is promoted by macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor-κB ligand (RANKL). The main osteoclasts signaling pathway provides a signal cascade that includes several factors including TNF receptor-associated factors (TRAFs), mitogen-activated protein kinases (MAPKs), and consequently the activation of c-Fos factor that induces the activity of NFATc1 [47]. The role of NFATc1 is considered significant for the promotion of differentiation and activity of osteoclasts [48–51]. Indeed, some studies have reported that downregulation of NFATc1 due to PIAS3 compromises the osteoclastogenesis process, confirming that this factor is necessary for the differentiation of active osteoclasts [52–54].
Implication of JAK/STAT in bone metabolism
JAKs and STATs proteins, according to many studies, play an important role in the proliferation and differentiation of osteoblasts and osteoclasts. A murine study conducted on osteoblasts [55] has demonstrated that phosphorylation of JAKs proteins occurs in the presence of oncostatin-M. This indicates the involvement of the three members, JAK1, JAK2, and TYK2, in the bone formation process [1, 56–59]. Nevertheless, despite the expression of JAK3 is mainly limited to the leukocytes, its probable role in the bone remodeling process has recently been highlighted [37]. Other animal studies, conducted on JAK1-deficient mice, suggest that the absence of the JAK1 factor induces lower bone growth and significantly reduced body mass [60]. Experimental evidence shows that the role of the JAK2 member is determined by coupling with the STAT5B factor [61, 62]. A JAK2/STAT5B signaling pathway is very important in the mechanism of growth hormone (GH) signaling which, in turn, regulates osteoblasts differentiation. Furthermore, STAT5B is considered a transcriptional promoter of insulin-like growth factor 1 (IGF-1), which is also produced in osteoblasts and is involved as a mediator during the bone growth process [63–65]. Also, other studies have highlighted the role of JAK2/STAT5B in osteoblastogenesis for the implication of some transcriptional factors and proteins, such as Runx-2, BMP-7, Tbx-3 [66–69]. Nevertheless, due to the strong interaction between JAK2/STAT5B, it is not easy to associate a respective function to each molecule. As for the three members, STAT2, STAT4, and STAT6, they do not seem to be directly involved in bone remodeling. Although, studies show that STAT2 could be implicated in bone homeostasis through STAT1; while STAT4 and STAT6 are involved in inflammatory arthritis [70]. It has been shown that STAT 3 inhibition induced a reduction of RANKL levels and decrease bone resorption through a decline of osteoclast activity mediated by RANKL in several experimental animal models of inflammatory arthritis [71, 72]; accordingly, in vitro studies showed that the STAT 3 inhibition induced a reduction of RANKL-mediated osteoclast differentiation from monocytes in mice and human [73, 74]. Among all the STATs proteins, STAT1 and STAT3 play a more important role in bone maintenance. Some studies show the critical role of STAT1 in the inhibition of the osteoclastogenesis process [75, 76]. On the contrary, other studies carried out on STAT1-deficient animal models highlight the suppressor function that STAT1 has in bone formation, through its relationship with Runx2 [77].
To confirm this last hypothesis, some researchers found that the activity of osteoblasts, in the bone formation, is accelerated in the absence of STAT1 during fracture healing [78]. Further, in ovariectomized rat, STAT1/3 inhibition is associated to an increased osteoblast activity, expressed as enhanced osteocalcin al alkaline phosphatase production [79]. The key role as a transcription factor for bone cells is certainly assigned to STAT3 [33–35]. The involvement of STAT3, in addition to survival and cellular functionality, also concerns the pathways of many cytokines and growth factors [80]. A team of researchers observed swift activation and induction of STAT3 signaling in mesenchymal stem cells (MSC), important regulators of osteoblast differentiation [81]. It has been widely discussed that STAT3 partakes in bone maintenance through its expression in osteoblasts and its inactivation effectively decreases bone formation in vivo [82–84]. To confirm this, animal studies were conducted on knockout mice engineered on the CRE-loxP system, highlighting that the inactivation of STAT3 in osteoblasts leads to a lower BMD. Moreover, in a very recent study conducted by Davidson et al., it is highlighted that the participation of STAT3, during the regulation of the osteoclastogenic process, occurs in a different way in female than male osteoclastic cells. So, these data suggest that the STAT3 signal may have a key role in bone turnover [46, 84, 85]. Initially, an inactivating STAT3 mutation was found, which leads to a disease called Job syndrome. Among the characteristics found in patients with this syndrome, a reduced BMD and presence of fractures are not to be underestimated. Besides, the mutation results in increased bone resorption and a greater number of osteoclasts [86–89]. Some studies show that inhibition of STAT3 induces a decrease in the osteoclastogenesis process [52, 53]. Young et al., in their animal experiment, show that STAT3 is involved in osteoclastogenesis in vivo and it can regulate the NFATc1 factor [90].
JAK/STAT, cytokines and bone cells
Many molecules, including cytokines, are involved in the cascade of the JAK/STAT signaling pathway. The recognition of cytokines with their receptors on the cell surface induces the activation of JAKs proteins. Consequently, the cascade activation mechanism of other components begins, such as the STATs, which, by binding to DNA, regulate the transcription processes of many genes [91]. The cytokines that are part of this pathway are more than 30, including numerous interleukins, tumor necrosis factor-α (TNF-α), interferon-α/γ (IFNα/γ), and oncostatin M (OSM), each of which is implicated in many pathologies [92]. Some cytokines (Table 1) are potentially involved in normal bone remodeling [93].
Table 1.
Positive/Negative effect of citokines on bone cells, processes and bone remodeling
| Citokine | Effect on Osteoblast and bone formation | Effect on osteoclast and bone resorption | Bibliographic references |
|---|---|---|---|
| IL-1 | – |
Positive effect (↑ osteoclastogenesis) (↑ bone resorption) |
[37, 99, 100] |
| IL-3 |
Positive effect (↑ osteoblastogenesis) (↑ bone formation) |
Negative effect (↓ osteoclastogenesis) (↓ bone resorption) |
[101–105] |
| IL-4 | – |
Negative effect (↓ osteoclastogenesis) (↓ bone resorption) (↓ osteoclast activity) |
[47, 106, 107] |
| IL-6 ● |
Positive effect (↑ osteoblastogenesis) (↑ bone formation) |
Negative/Positive effect ● (↓ osteoclastogenesis) (↑bone resorption) (↑ osteoclasts formation) |
[94–98] |
| IL-7 ● | – |
Negative/Positive effect ● (↓ osteoclastogenesis) (↓ bone resorption) (↓ osteoclast activity) (↑ osteoclasts formation) |
[47, 106–108] |
| IL-12 | – |
Negative effect (↓ osteoclastogenesis) |
[109–111] |
| IL-15 | – |
Positive effect (↑ osteoclastogenesis) |
[90, 112] |
| IL-17 |
Positive effect (↑ osteoblastogenesis) (↑ bone formation) |
– | [113–115] |
| IL-23 | – |
Negative effect (↓ osteoclastogenesis) |
[114, 116] |
| IL-27 |
Involvement in osteoblasts (to be studied) |
Negative effect (↓ osteoclastogenesis) |
[114, 116] |
↑, Stimulation, increase, promotion; ↓, Inhibition, decrease, induction; ●, uncertain/contrasting effect
Interleukin-6
Interleukin-6 plays a key role in bone turnover. Recently, experimental evidence has shown that IL-6 presents two forms, cis and trans, and that IL-6 trans form promotes bone formation [94]. Other studies suggest that, with the activation of STAT3 and the involvement of receptor subunit gp130 to transduce signals, IL-6 can mightily inhibit RANKL-induced osteoclasts differentiation [95]. In contrast, IL-6 stimulates the production of RANKL in osteoblasts and it could also induce an increase in osteoclasts and bone resorption [96–98].
Interleukin-1
Similarly, IL-1 also stimulates the process of osteoclastogenesis and bone resorption [99, 100]. A study has shown that the formation of TRAP-positive multinucleated cells (MNCs) is inhibited by using a molecule that blocks the action of IL-1 and IL-6 [37].
Interleukin-3
IL-3 could inhibit the process of osteoclast differentiation and bone resorption. On the other hand, it stimulates the process of osteoblastogenesis and consequently an increased bone formation. Moreover, IL-3 probably modulates, through the activation of JAK2/STAT5, a greater expression of both the soluble and membrane form of RANKL in calvaria osteoblasts, despite this does not involve the maturation of multinucleated osteoclasts [101–105].
Interleukin-4 and Interleukin-7
Some studies suggest that IL-4 and IL-7 negatively regulate the osteoclastogenesis process [106, 47–107]. Therefore IL-4, through the phosphorylation of STAT6, inhibits the osteoclasts activity and bone resorption. However, a team of researchers has found that, through STAT5 signaling, IL-7 induces the formation of osteoclasts [108].
Interleukin-12
Several studies indicate that IL-12 also has inhibitory effects on the osteoclastogenesis process; however, the mechanism by which this inhibitory effect occurs is still being investigated. Nevertheless, it has been shown that this interleukin induces STAT phosphorylation through two of the JAKs proteins, JAK2 and TYK2, but its negative effect appears to involve both interferon g and T cells [109–111].
Interleukin-15 and Interleuikin-17
In an animal study, in which the mice were defective of the IL-15 receptor, increased bone mass was shown, suggesting the role of IL-15 in the formation of osteoclasts [90–112]. IL-17 is of great interest in the involvement of bone and inflammatory diseases. Recent studies have indicated that Il-17 may be directly involved in the process of osteoblastogenesis and its implication in the rapid differentiation and maturation of osteoblasts results. The role of IL-17 has been investigated in bone cells of spondyloarthritis patients. Therefore, the researchers found that IL-17A induces the activity of osteoblasts through JAK2/STAT3 signaling. Furthermore, the activation of osteoblasts is drastically reduced by blocking JAK2 and IL-17A through an inhibitor [112–115].
Interleukin-23 and Interleukin-27
Finally, IL-23 and IL-27 are also part of the gp130 family of cytokines. However, their functionality at the level of the bone compartment is still being studied. Despite this, IL-27 has been seen to induce the transcription process in osteoblasts through STAT3. Moreover, both seem to participate in the process of osteoclastogenesis by exhibiting their inhibitory effect and reducing the number of multinucleated osteoclasts [114–116].
Conclusions
Through careful research in the literature, we wanted to focus our attention on the molecular mechanisms that affect the involvement of the JAK/STAT pathway at the cellular level, specifically at the level of bone cells, osteoblasts, and osteoclasts, and how they behave in the remodeling process bone. At the same time, the knowledge of the interaction between the immune system and the bone system has allowed us to evaluate also how cytokines, through the JAK/STAT signaling pathway, influence and participate in the mechanisms that govern bone turnover. It should be underlined that JAK/STAT inhibition showed conflicting and often opposite effects both in experimental and clinical models, in a large variety of physio-pathological relevant processes and in different tissues, including bone, probably because they can act by nonspecific mechanisms. Further, the different effects depending on cellular and tissue context and the complex interaction with other regulatory signaling pathways. The development of more specific JAK inhibitors, which target individual members of the STAT family, could contribute better to understand the role of JAK/STAT signaling in physiological and pathological conditions, including skeletal disorders. On the other hand, many studies will still be needed that allows scientists to investigate the functional mechanisms of the JAK/STAT pathway and its involvement in the bone system. This will allow us to develop new molecules and improve existing ones by inhibiting and/or blocking the JAK/STAT cascade with a view to new therapeutic strategies.
Acknowledgements
This study has been supported by the Department of Medical and Surgical Sciences, University of Foggia Medical School.
Funding
Open access funding provided by Università di Foggia within the CRUI-CARE Agreement.. This work was not funded.
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This work does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Eliana Rita Sanpaolo, Cinzia Rotondo have contributed equally to the achievement of the manuscript.
Contributor Information
Eliana Rita Sanpaolo, Email: eliana.sanpaolo@unifg.it.
Cinzia Rotondo, Email: cinzia.rotondo@gmail.com.
Daniela Cici, Email: daniela.cici@gmail.com.
Ada Corrado, Email: ada.corrado@unifg.it.
Francesco Paolo Cantatore, Email: francescopaolo.cantatore@unifg.it.
References
- 1.Kawamura M, Mcvicar DW, Johnston JA, Blake TB, Chen YQ, Lal BK, Lloyd AR, Kelvin DJ, Staples JE, Ortaldo JR. Molecular cloning of L-JAK, a Janus family protein-tyrosine kinase expressed in natural killer cells and activated leukocytes. Proc Natl Acad Sci USA. 1994;91:6374–6378. doi: 10.1073/pnas.91.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Shea JJ, Pesu M, Borie DC, Changelian PS. A new modality for immunosuppression: targeting the JAK/STAT pathway. Nat Rev Drug Discov. 2004;3:555–564. doi: 10.1038/nrd1441. [DOI] [PubMed] [Google Scholar]
- 3.Jaime-figueroa S, De Vicente J, Hermann J, Jahangir A, Jin S, Kuglstatter A, Lynch SM, Menke J, Niu L, Patel V, Shao A, Soth M, Vu MD, Yee C. Discovery of a series of novel 5Hpyrrolo[2,3-b]pyrazine-2-phenyl ethers, as potent JAK3 kinase inhibitors. Bioorg Med Chem Lett. 2013;23:2522–2526. doi: 10.1016/j.bmcl.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 4.O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, Mclnnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon-alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 6.Wilks AF. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:1603–1607. doi: 10.1073/pnas.86.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zurcher G, Ziemiecki A. Two novel proteintyrosine kinases, each with a second phosphotransferase- related catalytic domain, define a new class of protein kinase. Mol Cell Biol. 1991;11:2057–2065. doi: 10.1128/mcb.11.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski JJ. tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene. 1990;5:1329–1336. [PubMed] [Google Scholar]
- 9.David M, Petricoin EF, Igarashi KI, Feldman GM, Finbloom D, Larner AC. Prolactin activates the Interferon-Regulated p91 transcription factor and the Jak2 Kinase by tyrosine phosphorylation. Proc Natl Acad Sci USA. 1994;91:7174–7178. doi: 10.1073/pnas.91.15.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Pardoll DM, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boengler K, Hilfikerkleiner D, Drexler H, Heusch G, Schulz R. The myocardial JAK/STAT pathway: from protection to failure. Pharmacol Ther. 2008;120:172–185. doi: 10.1016/j.pharmthera.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Saeid A, Najmaldin S, Mohammad A, Asghari F, Salari F, Rahim F. STATs: an old story, yet mesmerizing. Cell J. 2015;17:395–411. doi: 10.22074/cellj.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Liang X, Shaikh AS, Zang J, Xu W, Zhang Y. JAK/STAT signal transduction: promising attractive targets for immune, inflammatory and hematopoietic diseases. Curr Drug Targ. 2018;19:487–500. doi: 10.2174/1389450117666161207163054. [DOI] [PubMed] [Google Scholar]
- 14.Darnel JR. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 15.Kim SK, Park KY, Yoon WC, Park SH, Park KK, Yoo DH, Choe JY. Melittin enhances apoptosis through suppression of IL-6/sIL-6R complex-induced NF-κB and STAT3 activation and Bcl-2 expression for human fibroblast-like synoviocytes in rheumatoid arthritis. Joint Bone Spine. 2011;78:471–477. doi: 10.1016/j.jbspin.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Sign. 2017;15:1–13. doi: 10.1186/s12964-017-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS box encodes a hierarchy of affinities for Cullin5: implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker BJ, Akhtar LN, Benveniste EN. SOCS1 and SOCS3 in the control of CNS immunity. Trends Immunol. 2009;30:392–400. doi: 10.1016/j.it.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 20.Bullock AN, Rodriguez M, Debreczeni JE, Songyang Z, Knapp S. Structure of the Socs4-elonginb/C complex reveals a distinct socs box interface and the molecular basis for socs-dependent EGFR degradation. Structure. 2007;15:1493–1504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vuong BQ, Arenzana TL, Showalter MB, Losman J, Chen XP, Mostecki J, Banks AS, Limnander A, Fernandez N, Rothman PB. SOCS-1 localizes to the microtubule organizing complex-associated 20S proteasome. Mol CelL Biol. 2004;24:9092–9101. doi: 10.1128/MCB.24.20.9092-9101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu ZH, Zhang ZY. Regulatory mechanisms and novel therapeutic targeting strategies for protein tyrosine phosphatases. Chem Rev. 2017;118:1069–1091. doi: 10.1021/acs.chemrev.7b00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Zhang Y, Ge L, Lin Y, Kwok HF. The roles of protein tyrosine phosphatases in hepatocellular carcinoma. Cancers. 2018;10:82–102. doi: 10.3390/cancers10030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang ZY. Chemical and mechanistic approaches to the study of protein tyrosine phosphatases. Acc Chem Res. 2003;36:385–392. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- 27.Heppler LN, Frank DA. Targeting oncogenic transcription factors: therapeutic implications of endogenous STAT inhibitors. Trends Cancer. 2017;3:816–827. doi: 10.1016/j.trecan.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 29.Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- 30.Dagvadorj A, Tan SH, Liao Z, Xie J, Nurmi M, Alanen K, Rui H, Mirtti T, Nevalainen MT. N-terminal truncation of Stat5a/b circumvents PIAS3-mediated transcriptional inhibition of Stat5 in prostate cancer cells. Int J Biochem Cell Biol. 2010;42:2037–2046. doi: 10.1016/j.biocel.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kipp M, Göhring F, Ostendorp T, van Drunen CM, van Driel R, Przybylski M, Fackelmayer FO. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J, Zhang F, Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep. 2015;5:17663–17672. doi: 10.1038/srep17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orsolini G, Bertoldi I, Rossini M. Osteoimmunology in rheumatoid and psoriatic arthritis: potential effects of tofacitinib on bone involvement. Clin Rheumatol. 2020;39:727–736. doi: 10.1007/s10067-020-04930-x. [DOI] [PubMed] [Google Scholar]
- 34.Maruotti N, Corrado A, Rotondo C, Cantatore FP. Janus kinase inhibitors role in bone remodeling. J Cell Physiol J Cell Physiol. 2020;235:1915–1920. doi: 10.1002/jcp.29149. [DOI] [PubMed] [Google Scholar]
- 35.Li J. JAK-STAT and bone metabolism. JAK-STAT. 2013;2(3):e23930. doi: 10.4161/jkst.23930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corry KA, Zhou H, Brustovetsky T, Himes ER, Bivi N, Hornd MR, Kitase Y, Wallace JM, Bellido T, Brustovetsky N, Li J. Stat3 in osteocytes mediates osteogenic response to loading. Bone Rep. 2019;11:100218. doi: 10.1016/j.bonr.2019.100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheon YH, Kim JY, Baek JM, Ahn SJ, Jun HY, Erkhembaatar M, Kim MS, Lee MS, Oh J. WHI-131 promotes osteoblast differentiation and prevents osteoclast formation and resorption in mice. J Bone Miner Res. 2016;31:403–415. doi: 10.1002/jbmr.2612. [DOI] [PubMed] [Google Scholar]
- 38.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 39.Corrado A, Sanpaolo ER, Di Bello S, Cantatore FP. Osteoblast as a target of anti-osteoporotic treatment. Postgrad Med. 2017;129:858–865. doi: 10.1080/00325481.2017.1362312. [DOI] [PubMed] [Google Scholar]
- 40.Maruotti N, Corrado A, Cantatore FP. Osteoblast role in osteoarthritis pathogenesis. J Cell Physiol. 2017;232:2957–2963. doi: 10.1002/jcp.25969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KG, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 42.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc fingercontaining transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 43.Corrado A, Neve A, Macchiarola A, Gaudio A, Marucci A, Cantatore FP. RANKL/OPG ratio and DKK-1 expression in primary osteoblastic cultures from osteoarthritic and osteoporotic subjects. J Rheumatol. 2013;40:684–694. doi: 10.3899/jrheum.120845. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Wang Z, Duan N, Zhu G, Schwarz EM, Xie C. Osteoblast–osteoclast interactions. Connect Tissue Res. 2018;59:99–107. doi: 10.1080/03008207.2017.1290085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corrado A, Neve A, Cantatore FP. Expression of vascular endothelial growth factor in normal, osteoarthritic and osteoporotic osteoblasts. Clin Exp Med. 2013;13:81–84. doi: 10.1007/s10238-011-0170-5. [DOI] [PubMed] [Google Scholar]
- 46.Davidson KD, Himes RE, Takigawa S, Chen A, Horn MR, Meijome T, Wallace MT, Kacena MA, Yokota H, Nguyen AV, Li J. The loss of STAT3 in mature osteoclasts has detrimental effects on bone structure. PLoS ONE. 2020;15(7):e0236891. doi: 10.1371/journal.pone.0236891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee ZH, Kim HH. Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochem Biophys Res Commun. 2003;305:211–214. doi: 10.1016/s0006-291x(03)00695-8. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi N, Yamana H, Yoshiki S, Roodman GD, Mundy GR, Jones SJ, Boyde A, Suda T. Osteoclast-like cell formation and its regulation by osteotropic hormones in mouse bone marrow cultures. Endocrinology. 1988;122:1373–1382. doi: 10.1210/endo-122-4-1373. [DOI] [PubMed] [Google Scholar]
- 49.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, Tsuda E, Morinaga T, Higashio K, Udagawa N, Takahashi N, Suda T. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 51.Byung-Chul J, Jung Ha K, Kabsun K, Inyoung K, Semun S, Nacksung K. ATF3 modulates calcium signaling in osteoclast differentiation and activity by associating with c-Fos and NFATc1 proteins. Bone. 2017;95:33–40. doi: 10.1016/j.bone.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Hikata T, Takaishi H, Takito J, Hakozaki A, Furukawa M, Uchikawa S, Kimura T, Okada Y, Matsumoto M, Yoshimura A, Nishimura R, Reddy SV, Asahara H, Toyama Y. PIAS3 negatively regulates RANKL-mediated osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblasts. Blood. 2009;113:2202–2212. doi: 10.1182/blood-2008-06-162594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim K, Lee J, Kim JH, Jin HM, Zhou B, Lee SY, Kim N. Protein inhibitor of activated STAT 3 modulates osteoclastogenesis by down-regulation of NFATc1 and osteoclast-associated receptor. J Immunol. 2007;178:5588–5594. doi: 10.4049/jimmunol.178.9.5588. [DOI] [PubMed] [Google Scholar]
- 54.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 55.Levy JB, Schindler C, Raz R, Levy DE, Baron R, Horowitz MC. Activation of the JAK-STAT signal transduction pathway by oncostatin-M cultured human and mouse osteoblastic cells. Endocrinology. 1996;137:1159–1165. doi: 10.1210/endo.137.4.8625884. [DOI] [PubMed] [Google Scholar]
- 56.Verbsky JW, Bach EA, Fang YF, Yang L, Randolph DA, Fields LE. Expression of Janus kinase 3 in human endothelial and other non-lymphoid and non-myeloid cells. J Biol Chem. 1996;271:13976–13980. doi: 10.1074/jbc.271.24.13976. [DOI] [PubMed] [Google Scholar]
- 57.Tortolani PJ, Lal BK, Riva A, Johnston JA, Chen YQ, Reaman GH, Beckwith M, Longo D, Ortaldo JR, Bhatia K, McGrath I, Kehrl J, Tuscano J, McVicar DW, O’Shea JJ. Regulation of JAK3 expression and activation in human B cells and B cell malignancies. J Immunol. 1995;155:5220–5226. [PubMed] [Google Scholar]
- 58.Sharfe N, Dadi HK, O’Shea JJ, Roifman CM. Jak3 activation in human lymphocyte precursor cells. Clin Exp Immunol. 1997;108:552–556. doi: 10.1046/j.1365-2249.1997.4001304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Musso T, Johnston JA, Linnekin D, Varesio L, Rowe TK, O’Shea JJ, McVicar DW. Regulation of JAK3 expression in human monocytes: phosphorylation in response to interleukins 2, 4, and 7. J Exp Med. 1995;181:1425–1431. doi: 10.1084/jem.181.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 61.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 62.Neubauer H, Cumano A, Müller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 63.Joung YH, Lim EJ, Pramod D, Chung SC, Jang JW, Do Park J, Lee HK, Kim HS, Cho BW, Park T, Chung S, Park JH, Yang YM. HSE enhances BMP7 and GH signaling through the activation of Jak2/STAT5B in osteoblastic MC3T3-E1 cells. Mol Med Rep. 2013;8:891–896. doi: 10.3892/mmr.2013.1593. [DOI] [PubMed] [Google Scholar]
- 64.Laviola L, Natalicchio A, Giorgino F. The IGF-I signaling pathway. Curr Pharm Des. 2007;13:663–669. doi: 10.2174/138161207780249146. [DOI] [PubMed] [Google Scholar]
- 65.Rosen CJ. Insulin-like growth factor I and bone mineral density: experience from animal models and human observational studies. Best Pract Res Clin Endocrinol Metab. 2004;18:423–435. doi: 10.1016/j.beem.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Miyazono K, Maeda S, Imamura T. Coordinate regulation of cell growth and differentiation by TGFbeta superfamily and Runx proteins. Oncogene. 2004;23:4232–4237. doi: 10.1038/sj.onc.1207131. [DOI] [PubMed] [Google Scholar]
- 67.Ziros PG, Georgakopoulos T, Habeos I, Basdra EK, Papavassiliou AG. Growth hormone attenuates the transcriptional activity of Runx2 by facilitating its physical association with Stat3β. J Bone Miner Res. 2004;19:1892–1904. doi: 10.1359/JBMR.040701. [DOI] [PubMed] [Google Scholar]
- 68.Govoni KE, Lee SK, Chadwick RB, Yu H, Kasukawa Y, Baylink DJ, Mohan S. Whole genome microarray analysis of growth hormone-induced gene expression in bone: T-box3, a novel transcription factor, regulates osteoblast proliferation. Am J Physiol Endocrinol Metab. 2006;291:E128–E136. doi: 10.1152/ajpendo.00592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Govoni KE, Linares GR, Chen ST, Pourteymoor S, Mohan S. T-box 3 negatively regulates osteoblast differentiation by inhibiting expression of osterix and runx2. J Cell Biochem. 2009;106:482–490. doi: 10.1002/jcb.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker JG, Ahern MJ, Coleman M, Weedon H, Papangelis V, Beroukas D, Roberts-Thomson PJ, Smith MD. Expression of Jak3, STAT1, STAT4, and STAT6 in inflammatory arthritis: unique Jak3 and STAT4 expression in dendritic cells in seropositive rheumatoid arthritis. Ann Rheum Dis. 2006;65:149–156. doi: 10.1136/ard.2005.037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.LaBranche TP, Jesson MI, Radi ZA, Storer CE, Guzova JA, Bonar SL, Thompson JM, Happa FA, Stewart ZS, Zhan Y, Bollinger CS, Bansal PN, Wellen JW, Wilkie DP, Bailey SA, Symanowicz PT, Hegen M, Head RD, Kishore N, Mbalaviele G, Meyer DM. JAK inhibition with Tofacitinib suppresses arthritic joint structural damage through decreased RANKL production. Arthritis Rheum. 2012;64(11):3531–3542. doi: 10.1002/art.34649. [DOI] [PubMed] [Google Scholar]
- 72.Vidal B, Cascão R, Finnilä MAJ, Lopes IP, da Glória VG, Saarakkala S, Zioupos P, Canhão H, Fonseca JE. Effects of tofacitinib in early arthritis-induced bone loss in an adjuvant-induced arthritis rat model. Rheumatology (Oxford) 2019;58(2):371. doi: 10.1093/rheumatology/key377. [DOI] [PubMed] [Google Scholar]
- 73.Li CH, Xu LL, Jian LL, Yu RH, Zhao JX, Sun L, Du GH, Liu XY. Stattic inhibits RANKL-mediated osteoclastogenesis by suppressing activation of STAT3 and NF-κB pathways. Int Immunopharmacol. 2018;58:136–144. doi: 10.1016/j.intimp.2018.03.021. [DOI] [PubMed] [Google Scholar]
- 74.Park JS, Kwok SK, Lim MA, Kim EK, Ryu JG, Kim SM, Oh HJ, Ju JH, Park SH, Kim HY, Cho ML. STA-21, a promising STAT-3 inhibitor that reciprocally regulates Th17 and Treg cells, inhibits osteoclastogenesis in mice and humans and alleviates autoimmune inflammation in an experimental model of rheumatoid arthritis. Arthritis Rheumatol. 2014;66(4):918–929. doi: 10.1002/art.38305. [DOI] [PubMed] [Google Scholar]
- 75.Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T. RANKL maintains bone homeostasis through c-Fos-dependent induc-tion of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 76.Takayanagi H, Ogasawara K, Hida S, Chiba T, Murata S, Sato K, Takaoka A, Yokochi T, Oda H, Tanaka K, Nakamura K, Taniguchi T. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 77.Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H. Stat1 functions as acytoplasmic attenuator of Runx2 in the transcriptionalprogram of osteoblast differentiation. Genes Dev. 2003;17:1979–1991. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tajima K, Takaishi H, Takito J, Tohmonda T, Yoda M, Ota N, Kosaki N, Matsumoto M, Ikegami H, Nakamura T, Kimura T, Okada Y, Horiuchi K, Chiba K, Toyama Y. Inhibition of STAT1 Accelerates Bone Fracture Healing. Wiley Periodicals Inc J OrthopRes. 2010;28:937–941. doi: 10.1002/jor.21086. [DOI] [PubMed] [Google Scholar]
- 79.Xu L, Zhang L, Zhang H, Yang Z, Qi L, Wang Y, Ren S. The participation of fibroblast growth factor 23 (FGF23) in the progression of osteoporosis via JAK/STAT pathway. J Cell Biochem. 2018;119(5):3819–3828. doi: 10.1002/jcb.26332. [DOI] [PubMed] [Google Scholar]
- 80.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Stat3 activation of NF-kappaB p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci USA. 2006;103:7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nicolaidou V, Wong MM, Redpath AN, Ersek A, Baban DF, Williams LM, Cope AP, Horwood NJ. Monocytes induce STAT3 activation in human mesenchymal stem cells to promote osteoblast formation. PLoS ONE. 2012;7(7):e39871. doi: 10.1371/journal.pone.0039871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pan J, Fukuda K, Saito M, Matsuzaki J, Kodama H, Sano M, Takahashi T, Kato T, Ogawa S. Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ Res. 1999;84:1127–1136. doi: 10.1161/01.res.84.10.1127. [DOI] [PubMed] [Google Scholar]
- 83.Zhou H, Newnum AB, Martin JR, Li P, Nelson MT, Moh A, Fu Y, Yokota H, Li J. Osteoblast/osteocyte-specific inactivation of Stat3 decreases load-driven bone formation and accumulates reactive oxygen species. Bone. 2011;49:404–411. doi: 10.1016/j.bone.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 84.Itoh S, Udagawa N, Takahashi N, Yoshitake F, Narita H, Ebisu S, Ishihara K. A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone. 2006;39:505–512. doi: 10.1016/j.bone.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 85.Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z, Kano A, Iwamoto Y, Li E, Craft JE, Bothwell ALM, Fikrig E, Koni P, Flavell RA, Fu XY. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci USA. 2003;100:1879–1884. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grimbacher B, Holland SM, Gallin JI, Greenberg F, Hill SC, Malech HL, Miller JA, O’Connell AC, Puck JM. Hyper-IgE syndrome with recurrent infections–an autosomal dominant multisystem disorder. N Engl J Med. 1999;340:692–702. doi: 10.1056/NEJM199903043400904. [DOI] [PubMed] [Google Scholar]
- 87.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, Anderson VL, Darnell DN, Welch PA, Kuhns D, Frucht DM, Malech HL, Gallin JI, Kobayashi SD, Whitney AR, Voyich JM, Musser JM, Woellner C, Schäffer AS, Puck JM, Grimbacher B. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 88.Grimbacher B, Puck JM, Holland SM. Hyper-IgE recurrent infections syndrome. In: Ochs HD, Smith CIE, Puck JM, editors. Primary immunodeficiency diseases: a molecular & cellular approach. New York City: Oxford University Press Inc; 2007. pp. 496–504. [Google Scholar]
- 89.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, Metin A, Karasuyama H. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 90.Yang Y, Chung MR, Zhou S, Gong X, Xu H, Hong Y, Jin A, Huang X, Zou W, Dai Q, Jiang J. STAT3 controls osteoclast differentiation and bone homeostasis by regulating NFATc1 transcription. J Biol Chem. 2019;294:15395–15407. doi: 10.1074/jbc.RA119.010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gadina M, Johnson C, Schwartz D, Bonelli M, Hasni S, Kanno Y, O’Shea JJ. Translational and clinical advances in JAK-STAT biology: the present and future of jakinibs. J Leukoc Biol. 2018;104:499–514. doi: 10.1002/JLB.5RI0218-084R. [DOI] [PubMed] [Google Scholar]
- 92.Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol. 2015;194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Manolagas SC, Jilka RL. Bone marrow, cytokines, and bone remodeling. Emerging insights into the pathophysiology of osteoporosis. N Engl J Med. 1995;332:305–311. doi: 10.1056/NEJM199502023320506. [DOI] [PubMed] [Google Scholar]
- 94.McGregor NE, Murat M, Elango J, Poulton IJ, Walker EC, Crimeen-Irwin B, Ho PWM, Gooi JH, Martin TJ, Sims NA. IL-6 exhibits both cis and trans signaling in osteocytes and osteoblasts, but only trans signaling promotes bone formation and osteoclastogenesis. J Biol Chem. 2019;294:7850–7863. doi: 10.1074/jbc.RA119.008074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bellido T, Stahl N, Farruggella TJ, Borba V, Yancopoulos GD, Manolagas SC. Detection of receptors for interleukin-6, interleukin-11, leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor in bone marrow stromal/osteoblastic cells. J Clin Invest. 1996;97:431–437. doi: 10.1172/JCI118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kishimoto T. The biology of interleukin-6. Blood. 1989;74:1–10. [PubMed] [Google Scholar]
- 97.Wu Q, Zhou X, Huang D, Ji Y, Kang F. IL-6 enhances osteocyte-mediated osteoclastogenesis by promoting JAK2 and RANKL activity in vitro. Cell Physiol Biochem. 2017;41(1360):1369. doi: 10.1159/000465455. [DOI] [PubMed] [Google Scholar]
- 98.Romas E, Udagawa N, Zhou H, Tamura T, Saito M, Taga T, Hilton DJ, Suda T, Ng KW, Martin TJ. The role of gp130-mediated signals in osteoclast development: regulation of interleukin 11 production by osteoblasts and distribution of its receptor in bone marrow cultures. J Exp Med. 1996;183:2581–2591. doi: 10.1084/jem.183.6.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ishimi Y, Miyaura C, Jin CH, Akatsu T, Abe E, Nakamura Y, Yamaguchi A, Yoshiki S, Matsuda T, Hirano T. IL-6 is produced by osteoblasts and induces bone resorption. J Immunol. 1990;145:3297–3303. [PubMed] [Google Scholar]
- 100.Kim JH, Jin HM, Kim K, Song I, Youn BU, Matsuo K, Kim K. The mechanism of osteoclast differentiation induced by IL-1. J Immunol. 2009;183:1862–1870. doi: 10.4049/jimmunol.0803007. [DOI] [PubMed] [Google Scholar]
- 101.Khapli SM, Mangashetti LS, Yogesha SD, Wani MR. IL-3 acts directly on osteoclast precursors and irreversibly inhibits receptor activator of NF-kB ligand-induced osteoclast differentiation by diverting the cells to macrophage lineage. J Immunol. 2003;171:142–151. doi: 10.4049/jimmunol.171.1.142. [DOI] [PubMed] [Google Scholar]
- 102.Yogesha SD, Khapli SM, Srivastava RK, Mangashetti LS, Pote ST, Mishra GC, Wani MR. IL-3 inhibits TNF-a-induced bone resorption and prevents inflammatory arthritis. J Immunol. 2009;182:361–370. doi: 10.4049/jimmunol.182.1.361. [DOI] [PubMed] [Google Scholar]
- 103.Singh K, Piprode V, Mhaske ST, Barhanpurkar-Naik A, Wani MR. IL-3 differentially regulates membrane and soluble RANKL in osteoblasts through metalloproteases and the JAK2/STAT5 pathway and improves the RANKL/OPG ratio in adult mice. J Immunol. 2018;200:595–606. doi: 10.4049/jimmunol.1601528. [DOI] [PubMed] [Google Scholar]
- 104.Gupta N, Barhanpurkar AP, Tomar GB, Srivastava RK, Kour S, Pote ST, Mishra GC, Wani MR. IL-3 inhibits human osteoclastogenesis and bone resorption through downregulation of c-Fms and diverts the cells to dendritic cell lineage. J Immunol. 2010;185:2261–2272. doi: 10.4049/jimmunol.1000015. [DOI] [PubMed] [Google Scholar]
- 105.Barhanpurkar AP, Gupta N, Srivastava RK, Tomar GB, Naik SN, Joshi SR, Pote ST, Mishra GC, Wani MR. IL-3 promotes osteoblast differentiation and bone formation in human mesenchymal stem cells. Biochem Biophys Res Commun. 2012;418:669–675. doi: 10.1016/j.bbrc.2012.01.074. [DOI] [PubMed] [Google Scholar]
- 106.Abu-Amer Y. IL-4 abrogates osteoclastogenesis through STAT6-dependent inhibition of NF-kappaB. J Clin Invest. 2001;107:1375–1385. doi: 10.1172/JCI10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yamada A, Takami M, Kawawa T, Yasuhara R, Zhao B, Mochizuki A, Miyamoto Y, Eto T, Yasuda H, NakamichI Y, Kim N, Katagiri T, Suda T, Kamijo R. Interleukin-4 inhibition of osteoclast differentiation is stronger than that of interleukin-13 and they are equivalent for induction of osteoprotegerin production from osteoblasts. Immunology. 2007;120:573–579. doi: 10.1111/j.1365-2567.2006.02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim J-H, Sim JH, Lee S, Seol MA, Ye S-K, Shin HM, Lee EB, Lee YJ, Choi YJ, Yoo W-H, Kim JH, Kim W-U, Lee D-S, Kim J-H, Kang I, Kang SW, Kim H-R. Interleukin-7 induces osteoclast formation via STAT5, independent of receptor activator of NF-kappaB ligand. Front Immunol. 2017;8:1376. doi: 10.3389/fimmu.2017.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagata N, Kitaura H, Yoshida N, Nakayama K. Inhibition of RANKL-induced osteoclast formation in mouse bone marrow cells by IL-12: involvement of IFN-gamma possibly induced from non-T cell population. Bone. 2003;33:721–732. doi: 10.1016/s8756-3282(03)00213-8. [DOI] [PubMed] [Google Scholar]
- 110.Horwood NJ, Elliott J, Martin TJ, Gillespie MT. IL- 12 alone and in synergy with IL-18 inhibits osteoclast formation in vitro. J Immunol. 2001;166:4915–4921. doi: 10.4049/jimmunol.166.8.4915. [DOI] [PubMed] [Google Scholar]
- 111.Zou J, Presky DH, Wu CY, Gubler U. Differential associations between the cytoplasmic regions of the interleukin-12 receptor subunits beta1 and beta2 and JAK kinases. J Biol Chem. 1997;272:6073–6077. doi: 10.1074/jbc.272.9.6073. [DOI] [PubMed] [Google Scholar]
- 112.Djaafar S, Pierroz DD, Chicheportiche R, Zheng XX, Ferrari SL, Ferrari-Lacraz S. Inhibition of T cell-dependent and RANKL-dependent osteoclastogenic processes associated with high levels of bone mass in interleukin-15 receptor-deficient mice. Arthritis Rheum. 2010;62:3300–3310. doi: 10.1002/art.27645. [DOI] [PubMed] [Google Scholar]
- 113.Raychaudhuri SP, Raychaudhuri SK. Mechanistic rationales for targeting interleukin-17A in spondyloarthritis. Arthritis Res Ther. 2017;19:51. doi: 10.1186/s13075-017-1249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lubberts E. The IL-23-IL-17 axis in inflammatory arthritis. Nat Rev Rheumatol. 2015;11:415–429. doi: 10.1038/nrrheum.2015.53. [DOI] [PubMed] [Google Scholar]
- 115.Jo S, Wang SE, Lee YL, Kang S, Lee B, Han J, Sung IH, Park YS, Bae SC, Kim TH. IL-17A induces osteoblast differentiation by activating JAK2/STAT3 in ankylosing spondylitis. Arthritis Res Ther. 2018;20:115. doi: 10.1186/s13075-018-1582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kamiya S, Nakamura C, Ono TFK, Ohwaki T, Wada TYS. Effects of IL-23 and IL-27 on osteoblasts and osteoclasts: inhibitory effects on osteoclast differentiation. J Bone Miner Metab. 2007;25:277–285. doi: 10.1007/s00774-007-0766-8. [DOI] [PubMed] [Google Scholar]