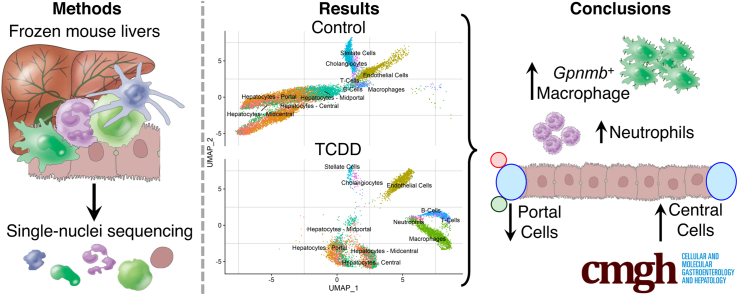
Abstract
Background and Aims
Characterization of cell specific transcriptional responses to hepatotoxicants is lost in the averages of bulk RNA-sequencing (RNA-seq). Single-cell/nuclei RNA-seq technologies enable the transcriptomes of individual cell (sub)types to be assessed within the context of in vivo models.
Methods
Single-nuclei RNA-sequencing (snSeq) of frozen liver samples from male C57BL/6 mice gavaged with sesame oil vehicle or 30 μg/kg 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) every 4 days for 28 days was used to demonstrate the application of snSeq for the evaluation of xenobiotics.
Results
A total of 19,907 genes were detected across 16,015 nuclei from control and TCDD-treated livers. Eleven cell (sub)types reflected the expected cell diversity of the liver including distinct pericentral, midzonal, and periportal hepatocyte subpopulations. TCDD altered relative proportions of cell types and elicited cell-specific gene expression profiles. For example, macrophages increased from 0.5% to 24.7%, while neutrophils were only present in treated samples, consistent with histological evaluation. The number of differentially expressed genes (DEGs) in each cell type ranged from 122 (cholangiocytes) to 7625 (midcentral hepatocytes), and loosely correlated with the basal expression level of Ahr, the canonical mediator of TCDD and related compounds. In addition to the expected functions within each cell (sub)types, RAS signaling and related pathways were specifically enriched in nonparenchymal cells while metabolic process enrichment occurred primarily in hepatocytes. snSeq also identified the expansion of a Kupffer cell subtype highly expressing Gpnmb, as reported in a dietary NASH model.
Conclusions
We show that snSeq of frozen liver samples can be used to assess cell-specific transcriptional changes and population shifts in models of hepatotoxicity when examining freshly isolated cells is not feasible.
Keywords: Transcriptomics, TCDD, Hepatotoxicant, Cellular Heterogeneity
Abbreviations used in this paper: AhR, aryl hydrocarbon receptor; DEG, differentially expressed gene; GEO, Gene Expression Omnibus; GSKB, Gene Set Knowledgebase; KC, Kupffer cell; lncRNA, long noncoding RNA; mRNA, messenger RNA; NAM, nonalcoholic steatohepatitis–associated macrophage; NASH, nonalcoholic steatohepatitis; NPC, nonparenchymal cell; PND, postnatal day; RNA-seq, RNA sequencing; scSeq, single-cell RNA sequencing; snSeq, single-nuclei RNA sequencing; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin
Graphical abstract
Summary.
The application of single-nuclei RNA sequencing is demonstrated for the evaluation of a hepatotoxicant highlighting key considerations in study design. The potent aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin is shown to elicit cell-specific responses and alter relative population sizes.
The liver is particularly susceptible to toxicity due to its close association with the gastrointestinal tract and xenobiotic metabolism capacity.1 Liver toxicity is the primary driver of drug candidate attrition and a common cause of market withdrawal.2 Similarly, environmental contaminant exposure is implicated in liver damage in humans, evidenced by increased levels of the liver damage biomarker alanine aminotransferase in epidemiological studies.3, 4, 5 Moreover, environmental contaminants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) are potential contributing factors in the etiology of complex metabolic diseases such as obesity, type II diabetes, and nonalcoholic fatty liver disease.3, 4, 5 For example, aryl hydrocarbon receptor (AhR) agonists such as TCDD and related compounds promote hepatic lipid accumulation (steatosis) and its progression to steatohepatitis (NASH) with fibrosis in mice,6, 7, 8 while epidemiological studies suggest an association with metabolic disease in humans.9,10
Although bulk “omic” strategies have uncovered important knowledge on the mechanisms of hepatotoxicants, key molecular events, as well as the significance of temporal, spatial, and cellular heterogeneity, remain poorly understood within the in vivo context of a tissue.11 Single-cell transcriptomic technologies provide the opportunity to investigate the transcriptomic responses to exogenous agents while also considering cellular heterogeneity and putative cell-cell interactions.12 Single-cell RNA sequencing (scSeq) can query the transcriptome at unprecedented resolution, characterizing rare cell types and developmental processes,13,14 and further elucidate the significance of tissue spatial organization.15,16 scSeq analysis of a diet-induced NASH model revealed the expansion of a novel Kupffer cell (KC) subtype termed NASH-associated macrophages (NAMs), as well as altered vascular signaling.17 It is also established that drugs and toxicants elicit spatial (zonal) toxicities as in the case of acetaminophen, which primarily affects the centrilobular region due to higher expression levels of xenobiotic metabolizing enzymes.18 Conversely, TCDD elicits periportal hepatotoxicity despite preferential accumulation in the centrilobular region due to sequestration by induced CYP1A2 levels.19,20 It remains unclear how transcriptional networks are implicated in these zonal toxicities. Single-cell analysis is expected to further elucidate the role of specific cell (sub)populations in toxicity and models of liver disease progression.
Preclinical drug or chemical toxicity assessments typically involve dose-response designs that present numerous challenges for implementing scSeq, primarily the required use of freshly isolated cells. Ongoing efforts to minimize experimental animal use also demand that studies maximize the extraction of relevant data including gross pathology, clinical chemistry, and histopathology. The impact of diurnal rhythm in large studies places additional logistic constraints regarding sample collection within a specific time window.21 Most importantly, it is difficult to predict how treatment and/or disease pathologies impact cell populations, structure and viability, digestion efficacy, and cell type selection for single-cell analysis without extensive a priori validation. Nuclei-based approaches address many of these challenges and produce results comparable to single-cell approaches.14,22, 23, 24 Notably, single-nuclei RNA sequencing (snSeq) can be performed on frozen samples, providing the best solution for traditional toxicology assessments. In the presented study, snSeq was used to evaluate the hepatic effects of TCDD. snSeq analysis using frozen liver samples showed TCDD-elicited cell population shifts and cell-specific differential gene expression consistent with the progression of hepatic steatosis to NASH with fibrosis.
Results
snSeq Identifies Major Liver Cell (Sub)types in Vehicle and TCDD-Treated Mice
We adapted a scSeq protocol for frozen cancer tissue biopsy samples that was compatible with the 10x Genomics (Pleasanton, CA) technology to characterize gene expression in nuclei isolated from mouse liver samples treated with either sesame oil vehicle or 30 μg/kg TCDD. A total of 16,015 individual nuclei transcriptomes (9981 and 6034 in vehicle and TCDD-treated, respectively) were characterized across 2 biological replicates per treatment group after quality control and doublet removal. The average number of expressed genes detected in each sample was 17,920, consistent with our published bulk RNA sequencing (RNA-seq) assessments collected using a similar study design.6,25 The median number of unique detected genes in individual nuclei was 1694, with a median unique molecular identifier (transcript) count of 3385 per nuclei. As expected with a nuclear preparation, there was negligible expression of mitochondrial genes, and long noncoding RNAs (lncRNAs) such as Malat1 and Gm42416 were the most abundantly expressed genes.23,24 The number of unique expressed genes and median unique molecular identifier count (1500 and 3805, respectively), as well as total expressed genes (19,907), was similar to a published 10x Genomics liver single-cell dataset (19,349),16 lending further confidence in our snSeq approach. Cell Ranger (10x Genomics) detected some ambient RNA contamination in some samples, likely owing to the lysis of cells during nuclei isolation.26 However, neither of the common indicators of ambient RNA contamination (mitochondrial or hemoglobin gene expression—Mt or Hba/Hbb, respectively) were elevated in our samples (https://doi.org/10.6084/m9.figshare.12728411). Consequently, ambient RNA decontamination tools were not used.
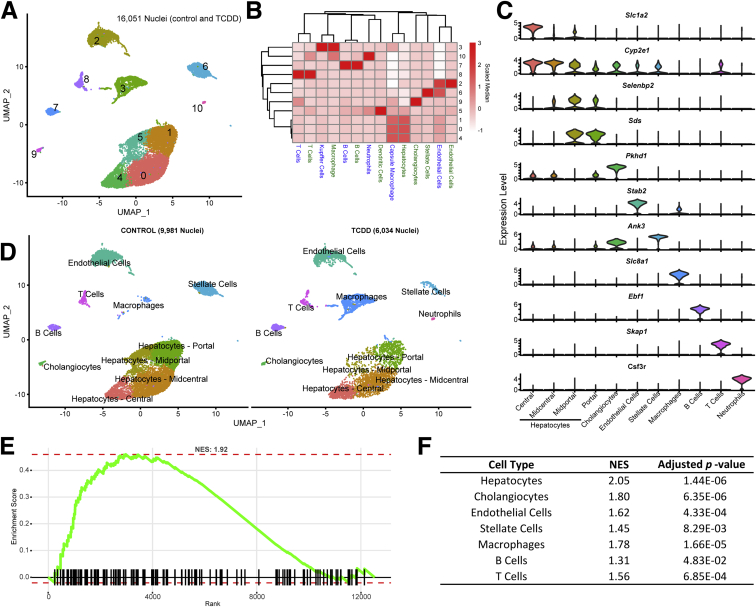
Integration and clustering of nuclei transcriptomic profiles identified 11 clusters, of which only the neutrophil cluster was unique to TCDD treatment (Figure 1A and D). Nuclei clusters were initially annotated by integrating our dataset with 2 published scSeq datasets and comparing annotation assignments (Figure 1B).15,17 Initial annotations were subsequently corroborated using the expression of known markers for specific cell types based on published reports or panglaoDB.27 For example, nuclei with higher levels of Stabilin 2 (Stab2) (Figure 1B and C), an expression marker for hepatic sinusoidal endothelial cells,28 were identified as “Endothelial Cell” nuclei in our dataset, as reported by Xiong et al17 in a diet-induced NASH model. Similarly, cholangiocyte expression profiles aligned with a distinct nuclei cluster (Figure 1B).16 Although the classical cholangiocyte marker, SRY-Box 9 (Sox9), was not expressed in our nuclei, the similarity between the expression profiles in our dataset and cells expressing Sox9 reported by Xiong et al17 suggested that these nuclei are indeed cholangiocytes. Examination of distinguishing markers for each nuclei cluster (Figure 1C) indentified Pkhd1 (also known as fibrocystin) as one of the distinguishing markers for cholangiocytes. Pkhd1 has been shown to be expressed in rat cholangiocytes.29 Other distunguishing markers such as Ebf1, important in B cell development,30 and the T cell adaptor Skap1,31 were also in agreement with cluster annotations.
Figure 1.
snSeq analysis of nuclei isolated from frozen liver samples of mice gavaged every 4 days for 28 days with sesame oil vehicle or 30 μg/kg TCDD. (A) UMAP visualization of nuclei isolated from vehicle and TCDD-treated liver samples clustered based on gene expresion profile similarity. (B) Label transfer prediction from published liver scSeq data—Bahar Halpern et al15 in blue, Xiong et al16 in green—was used to identify similar clusters for annotation. (C) Expression distribution for distinguishing (largest average fold-change, adjusted P ≤ .05) marker genes within each specific cell type. Color of clusters in panel C correspond to cell type clusters in panel A. (D) UMAP visualization of annotated nuclei in control and TCDD-treated samples. Gene set enrichment analysis of nuclear biased genes (first quartile)31 using genes ranked from most to least abundantly expressed in our nuclei dataset compared with Xiong et al’s16 whole-cell data was performed for (E) all cell types and (F) paired cell types identified in both datasets. NES, normalized enrichment score.
Our dataset suggests that Sox9 may be less abundant in nuclei sample preparations, consistent with previous reports indicating some genes exhibit nuclear or cytosolic biases.23,24,32 Comparison of our dataset with Xiong et al17 using a similar model and the same technology shows excellent concordance in identifying genes expressed in liver cells (16,789; 84%–87%) despite Xiong et al’s enrichment for nonparenchymal cells. However, it is evident that relative levels of certain genes differ, particularly lncRNA’s. Comparisons of nuclear and cytoplasmic gene expression in liver cells (mixed cell types) identified Mlxipl (ChREBP) and Nrlp6 as retained nuclear genes, as well as the lncRNA Neat1.32 In agreement, our dataset exhibited greater expression of these genes compared with whole-cell liver expression data Xiong et al17 (https://doi.org/10.6084/m9.figshare.12728441). We performed gene set enrichment analysis on ranked genes from nuclear-biased (elevated fold change in our vehicle control dataset compared with control whole cell data) to cytosolic/whole cell–biased (elevated in control single-cell data compared with our vehicle control dataset) using nuclear biased genes (top first percentile) identified by Bahar Halpern et al32 as the gene set. Figure 1E and F shows our single-nuclei gene expression dataset is indeed enriched in nuclear biased genes in all cell types.
TCDD Shifted Cell Type Proportions
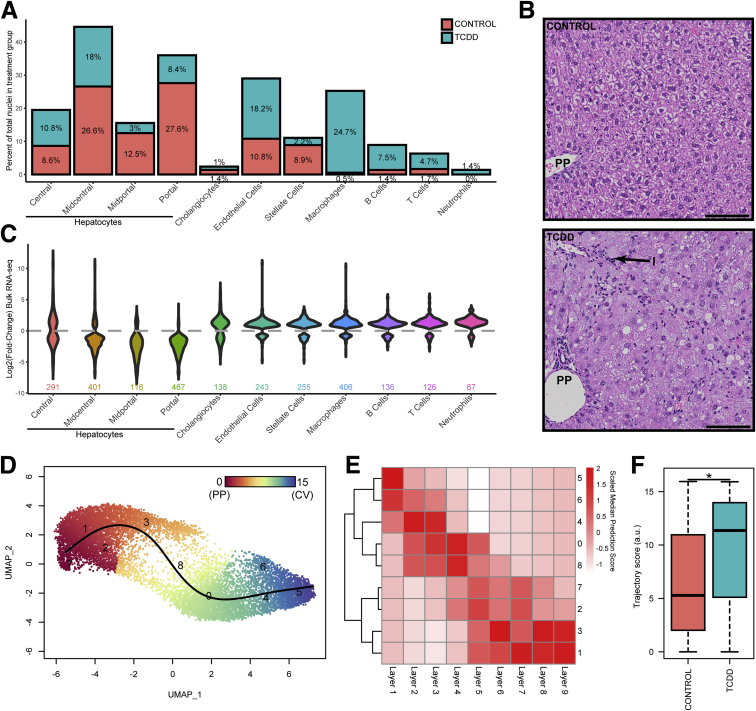
TCDD elicited shifts in the relative proportions of nuclei clusters (Figure 2A). Macrophages increased from 0.5% in control samples to 24.7% in TCDD-treated samples, accompanied by other immune cells such as B cells (1.4% to 7.5%), T cells (1.7% to 4.7%), and neutrophils (0.0% to 1.4%). This is consistent with TCDD-induced inflammation (Figure 2B) and elevated levels of proinflammatory cytokines, including tumor necrosis factor-α and interleukin-6.7 Overall, the relative proportion of hepatocytes in TCDD-treated samples was reduced by 46.6% (75.3% in control samples compared with 40.2% in treated samples). All zones were reduced, except for central hepatocytes, which increased from 8.6% to 10.8% (Figure 2A). Note that distinguishing between decreases in relative proportion of cell types reflecting cell death, loss of cell types during processing, or a true shift in relative levels is challenging. Previous studies using the same study design did not report cell death from histological evaluation.33 These challenges are further explored in the subsequent discussion.
Figure 2.
Hepatic cell population shifts in response to TCDD. (A) Percentage of nuclei represented in cell type–specific clusters in control and treated samples. (B) Representative liver photomicrographs from control and TCDD-treated samples using the same study design33 showing periportal (PP) immune cell infiltration (I). Scale bar = 100 μm. (C) Violin plots of fold-change expression distribution for genes defining cell type clusters (adjusted P ≤ .05) in TCDD-elicited bulk RNA-seq dataset. (D) Reclustering and trajectory analysis of only hepatocyte nuclei from vehicle control and TCDD treatment groups using a resolution of 1.2 to identify 9 clusters in order to (E) compare snSeq gene expression with the 9 distinct spatially resolved hepatocyte layers defined Halpern et al.16 (F) Nuclei distribution along the trajectory determined in control and TCDD-treated samples. The asterisk indicates a significant difference in distribution with TCDD causing a shift in the hepatocyte nuclei population toward the central cluster/layer as determined by the Kolmogorov-Smirnov test (P ≤ .05).
To further investigate cell population shifts, we re-examined our published hepatic bulk RNA-seq (same species, strain, sex, age, treatment regimen, dose, and duration of exposure)6 for TCDD-elicited fold-change distribution of genes identified as cell type markers (adjusted P ≤ .05) (Figure 2C). Overall, marker gene expression for midcentral, midportal, and portal hepatocytes was repressed in our bulk RNA-seq dataset, as evidenced by the bulk RNA-seq log2(fold-change) distribution ≤0 (wider below the gray line). Conversely, marker genes for cholangiocytes, endothelial cells, stellate cells, and immune cells were induced. Comparing the changes in relative proportions (Figure 2A) to differential gene expression in bulk RNA-seq (Figure 2C) confirms the impact of TCDD on cell population shifts in bulk RNA-seq analysis. Specifically, central hepatocyte marker genes were repressed despite a modest increase in relative cell numbers suggesting TCDD effects on gene expression. In contrast, macrophage marker induction concommitant with increased macrophage number makes it difficult to distinguish a direct effect on gene expression from an increase in the number of infiltrating cells with basal gene expression. The limited ability to delineate these effects using bulk RNA-seq demonstrates another advantage of a single-cell/nuclei analysis.
Characterization of Spatially Resolved Hepatocytes
To investigate zonal gene expression using snSeq data, hepatocyte nuclei from both treatment groups were selected, reintegrated, and reclustered to obtain 9 clusters, guided by the 9 hepatocyte zones (ie, layers) defined by Bahar Halpern et al (Figure 2D).16 Our hepatocyte nuclei clusters demonstrate high similarity to the transcriptomes of the different cell layers defined by Bahar Halpern et al (Figure 2E).16 Specifically, the nuclei cluster labeled 5 was most similar to layer 1 cells, while clusters 1 and 3 were most like layers 8 and 9. Zonal trajectory assessed using Slingshot34 shows a transition in gene expression profiles from cluster 5 to 1, reflecting a gradient from central (layer 1) to portal (layer 9) (Figure 2D and E). Cluster 7 was less defined, though closer examination identified the expression of portal markers, along with elevated expression of metallothioneins (Mt1 and Mt2) equally expressed in control and treated samples (https://doi.org/10.6084/m9.figshare.12728507). Comparing zonal distribution between control and treated nuclei revealed a shift toward central hepatocyte expression, suggesting that TCDD caused a loss of portal hepatocyte gene expression, or possibly a loss of portal hepatocytes during sample preparation (Figure 2F). TCDD elicits periportal hepatotoxicity, and therefore the loss of identity from de- or transdifferentation cannot be distinguished from cell-specific cytotoxicity.
Differential Gene Expression Elicited by TCDD Is Largely Cell Type Specific
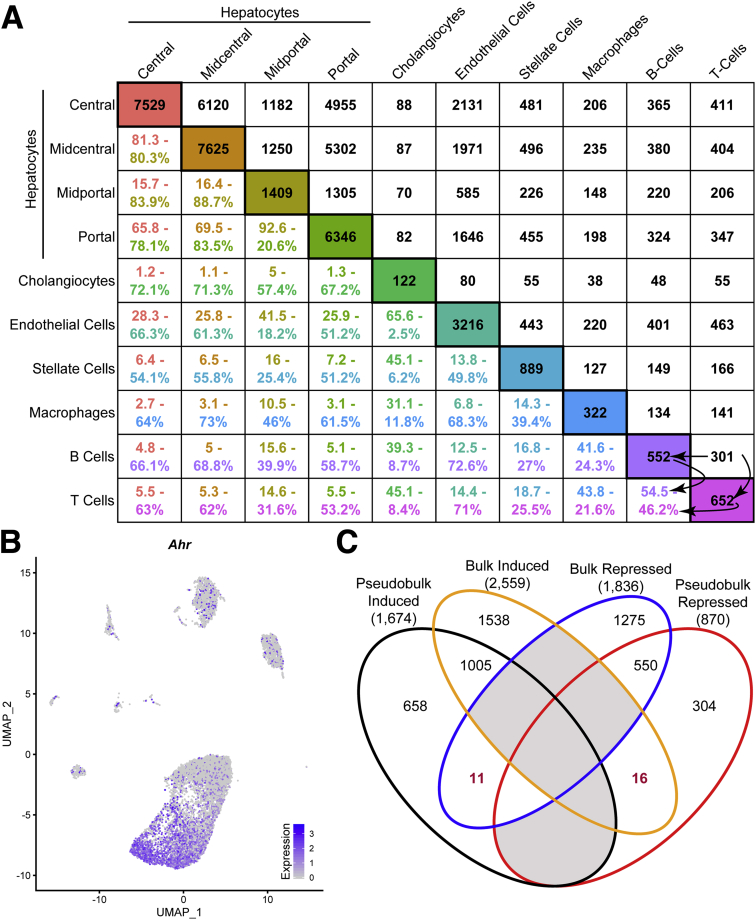
Of the 19,907 genes detected across all liver cell types, 10,951 were identified as differentially expressed, comparable to the 9313 (of 19,935) DEGs reported in our comparable bulk RNA-seq study.6 Hepatocyte subtypes exhibited the greatest similarity in differential gene expression with 92.6% (n = 1305 of 1409) of midportal DEGs overlapping with portal DEGs (Figure 3A). However, this only represents 20.6% (n = 1306 of 6346) of portal DEGs, suggesting that midportal cells may be a subset of portal hepatocytes. Cholangiocytes compared with macrophages shared the fewest overlapping DEGs, with only 38 in common (31.1% and 11.8% of DEGs, respectively). Only 28 genes were differentially expressed in all cell types, with 26 repressed in every cell type. Only cadherin 18 (Cdh18) was induced in all cells, while dihydropyrimidine dehydrogenase (Dpyd) was induced in hepatocytes but repressed in all other cell types (https://doi.org/10.6084/m9.figshare.12728522).
Figure 3.
TCDD-elicited differential gene expression in distinct hepatic cell subtypes. (A) Cross-comparisons of DEGs (adjusted P ≤ .05) between cell (sub)types. The number of DEGs (adjusted P ≤ .05) determined for each nuclei cluster is provided in the diagonal boxes color-coded according to cell subtypes identified in Figure 1A. The number of common DEGs determined between each pair of cell types is provided in upper right portion of table in black font. The percentage of common DEGs between cell types is provided in the bottom left portion of table using 2 different colored fonts. The upper colored font represents the percent overlap of DEGs relative to the cell subtype for the column, while the lower colored font represents the percent overlap of DEGs relative to the cell subtype for the row. An example of percent calculations is shown between B cells and T cells using arrows (eg, 301/552 = 0.545). (B) UMAP visualization of Ahr expression in all nuclei from vehicle control samples. (C) Comparison of pseudobulk and bulk RNA-seq analyses for up- and downregulated genes detected in both datasets.
The number of DEGs in hepatocytes correlated with basal Ahr expression levels in control samples. Central and midcentral hepatocytes exhibited the highest basal Ahr expression levels (detected in 79% and 65% of nuclei, respectively) (Figure 3B) and exhibited the most DEGs. Portal hepatocytes also exhibited a large number of DEGs (6346) despite a lower AhR expression level (detected in only 17% of portal nuclei) suggesting secondary or tertiary factors (eg, chromatin accessibility, metabolite gradient, cell-cell interactions) contributed to TCDD responsiveness. Induction of classic AhR target genes was not detected in all cell types (https://doi.org/10.6084/m9.figshare.12728522) and exhibited fold-change differences between cell types. For instance, Cyp1a1 was induced 2736-fold in portal hepatocytes, consistent with reports in bulk RNA-seq analyses. The 1177-fold repression of Ces3b reported in bulk RNA-seq was comparable to 3089-fold repression observed in central hepatocytes. Many genes went from undetected to detected, confounding fold-change estimates (ie, division by zero), though these genes were typically significantly induced in bulk RNA-seq. For example, Gpnmb was induced 1206-fold in our bulk RNA-seq dataset but only detected in macrophage following TCDD treatment in the snSeq dataset. A large number of zeroes is expected in scSeq/snSeq resulting from either absent expression or dropouts.
To compare single-nuclei with bulk RNA-seq data, snSeq data were first converted to “pseudo-bulk” data by summing read counts across all nuclei to approximate bulk expression. Differential gene expression analysis for our pseudo-bulk data identified 2812 DEGs (adjusted P ≤ .05 and |fold change| ≥ 2), 2544 of which were also detected in bulk RNA-seq, with 1005 induced and 550 repressed in both analyses (Figure 3C). Only 27 genes showed divergent expression (eg, induced in one dataset and repressed in the other). Owing to the substantial technological differences in determining transcript counts, some differences are expected. In addition, some genes may differ due to diurnal rhythm (eg, Alas1) or cell type biases (eg, macrophage genes Adgre4 and Clec4f).
TCDD Elicits Unique Functional Changes in Cell Populations
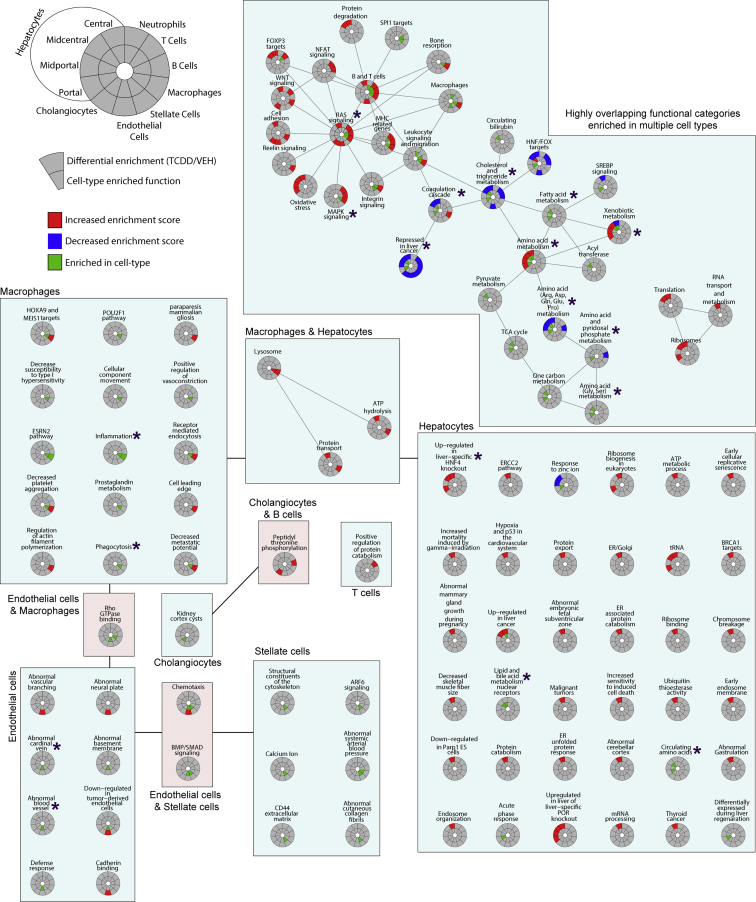
To identify enriched cellular pathways, processes, and functions associated within specific cell (sub)types, all constitutively expressed genes and DEGs were analyzed in control and treated samples. Enrichment scores were calculated for 6513 gene sets obtained from the Gene Set Knowledgebase (GSKB) for each nucleus independent of cell type classification. UMAP visualization of functional enrichment highlights TCDD-elicited shifts in enriched functions within specific cell types, most notably hepatocytes (https://doi.org/10.6084/m9.figshare.12728528). Pathways, processes, and functions enriched within specific cell (sub)types were consistent with their known physiological roles (Figure 4, inner ring). For instance, macrophages were enriched in phagocytosis and inflammation gene sets while endothelial cells were enriched in vasculature related genes. Hepatocytes were largely enriched in genes associated with the metabolism of carbohydrates, lipids, amino acids, and bile acids, as well as 1 carbon metabolism, and were consistent with their expected zonal distribution. Specifically, amino acid metabolism and the coagulation cascade were enriched in the portal region, while lipid, bile acid, and phase I xenobiotic metabolism were centrally enriched (Figure 4), as reported in a recent zonal proteomics study.35
Figure 4.
Gene set enrichment analysis of single-nuclei transcriptomes. Circos plots of functional enrichment analysis for each cell type indicates marker functions (adjusted P ≤ .05 compared with other cell types; inner ring; green) and functions showing increased (red) or decreased (blue) enrichment scores following TCDD treatment (outer ring). Enrichment scores for 6513 gene sets were determined using Gene Set Variation Analysis, grouped based on gene membership similarity using Enrichment Map, and summarized using AutoAnnotate, followed by manual curation, as described in the Materials and Methods. Asterisks indicate functions discussed in the manuscript.
We next examined nuclei for functional enrichment following TCDD treatment using enrichment score differences (fold-change) between control and treated nuclei (Figure 4, outer ring). RAS signaling had the most highly connected node, suggesting high similarity in gene membership with other nodes, particularly those associated with nonparenchymal cells. Metabolism pathways were largely associated with hepatocytes, including repression of cholesterol and triglyceride metabolism, and metabolism of selected amino acids, which were either induced or repressed (eg, Arg, Asp, Gln, Glu, Pro). Not surprisingly, xenobiotic metabolism was induced in all hepatocytes except central hepatocytes. This is due to the repression of several cytochrome P450s and high constitutive expression of phase I metabolism genes in central hepatocytes. Other notable gene sets with increased enrichment following treatment in hepatocytes included (1) “up-regulated in liver-specific HNF4α knockout,” suggesting TCDD repressed HNF4 signaling; and (2) “repressed in liver cancer,” suggesting transcriptomic inhibition consistent with the hepatocarcinogenicity of TCDD.
Emergence of New Macrophage (Sub)types Following Treatment
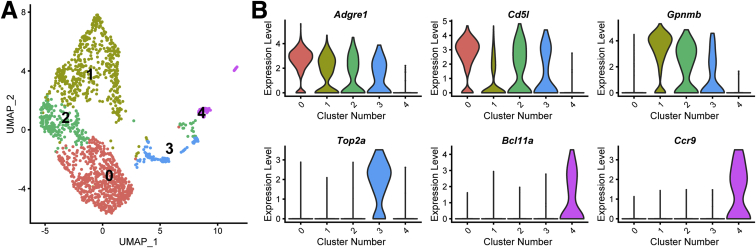
A hallmark of TCDD exposure is the infiltration of immune cells into the liver.33 Figures 1D and 2B show the increased presence of macrophages and neutrophils following treatment. Resident macrophages in a normal liver largely consist of KCs marked by high Adgre1 (F4/80) expression. Upon injury, motile monocyte-derived macrophages with high levels of Itgam (Cd11b) and Ccr2 expression, are recruited.17 Further analysis of macrophage nuclei identified 5 distinct clusters, of which 4 expressed high levels of Adgre1 and Cd5l (Figure 5), while none exhibited high levels of Itgam (not detected) or Ccr2 (expressed in ≤4% of control macrophages). Macrophage clusters 1, 2, and 3 not only expressed high levels of Adgre1 and Cd5l, but also expressed high levels of Gpnmb, which was not present in KCs from control samples. Interestingly, comparable macrophage clusters were reported in a diet-induced NASH mouse model, described as NAMs, that also exhibited high Trem2 expression.17 Although highly induced in our bulk RNA-seq dataset, we did not identify a single macrophage population among the 4 identified here expressing high Trem2 levels, nor Cd9, as reported in NAMs by Xiong et al.16 This is likely another example of a gene biased to the cytoplasm, as both genes are more abundant in the cytosol than nuclei in liver cells.32 However, segregating nuclei expressing Gpnmb does reveal significant enrichment in nuclei expressing Trem2 and Cd9 (https://doi.org/10.6084/m9.figshare.12728588). KC cluster 3 was also determined to express high levels of the cell cycle gene Top2a, possibly identifying a proliferating NAM subpopulation as these nuclei also abundantly expressed the NAM marker Gpnmb. The fourth and smallest cluster expressed high levels of Bcl11a and Ccr9 (Figure 5B), and likely represent plasmacytoid dendritic cells as previously reported.15
Figure 5.
Clustering of macrophage nuclei transcriptomes. (A) UMAP plot of macrophage nuclei determined at 0.2 resolution using Seurat. (B) Violin plots of the expression levels of unique genes that distinguish macrophage subtypes.
Discussion
Single-cell/nuclei transcriptomics not only has provided novel insight into molecular mechanisms involved in cellular development and normal cell function, but has also has identified and characterized rare cell (sub)types.13, 14, 15, 16, 17,36 It has the potential to further elucidate the mechanisms of toxicity elicited by exogenous agents that disrupt normal cellular functions. Most importantly, it enables the assessment of cell type–specific responses, thus integrating systemic (multiorgan) and local events within the context of a tissue or organ. In this study, we used a nuclei-based strategy to minimize potential biases in cell (sub)types due to toxicity that was also logistically feasible within a large dose-response and time-course study. Similar strategies have been used for neurons which vary wildly in shape, density, and other cellular characteristics, as well as postmortem human tissues with good success.37,38
Single-cell analyses have been performed on mouse and human liver samples, though most involved enrichment for either hepatocytes or nonparenchymal cells (NPCs).15, 16, 17,39 Using a single-nuclei approach, we captured the expected hepatic cell types identified in previous studies with hepatocyte nuclei highly represented, given that they compose ≥80% of the liver cell population. Consequently, immune cells such as the plasmacytoid dendritic cells and capsule macrophages identified in NPC-enriched samples, were difficult to identify, particularly in control samples.15 A potential solution to counter this unequal hepatocyte sampling bias could be fluorescence-activated cell sorting separation of nuclei based on DNA, as NPCs are enriched in the diploid (2N) population while hepatocyte nuclei are often polyploid.40 However, TCDD altered liver cell ploidy (https://doi.org/10.6084/m9.figshare.12728540),41 which may bias the relative proportions of distinct cell types, and therefore we did not separate nuclei by ploidy in this study. While our snSeq dataset could not estimate the total number of each cell type, the emergence of new cell populations and the transcriptomic changes of resident cells could be distinguished. Distinguishing the loss of cell (sub)types due to toxicity from de/transdifferentiation represents a unique challenge.
Our single-nuclei strategy highlighted several advantages over single-cell analysis to assess hepatotoxicants. First, nuclei isolated from frozen samples could clearly be classified as distinct known hepatic cell types. Second, despite higher representation of nuclear-biased genes, TCDD-elicited differential expression determined by bulk RNA-seq was largely recapitulated in a pseudo-bulk version of our snSeq dataset, with only 38 genes showing contrary results, which could be due to other confounding factors (eg, different technologies, diurnal rhythm). However, our dataset was enriched with nuclear-biased genes while other cell type markers, such as the cholangiocyte marker Sox9, were not detected. Single-nuclei droplets may also be more susceptible to ambient RNA contamination, though our mitochondrial gene expression content was low. Despite differences in messenger RNA (mRNA) capture, sequencing, counting, and analysis, the strong correspondence indicates that snSeq is a viable alternative, with distinct advantages over scSeq analysis.
Hepatotoxicants can elicit zonal toxicity based on their mode of action and the functional gradient of the lobule chords (central vein to portal vein). Toxicants not requiring bioactivation commonly damage periportal regions when first encountered via the portal vein. Conversely, toxicants requiring bioactivation are more often toxic to pericentral regions due to higher basal expression of phase I metabolism enzymes (eg, cytochrome P450s). TCDD is a periportal hepatotoxicant, though damage spans the periportal to pericentral regions (panacinar) with increasing dose. Interestingly, pericentral hepatocytes were most responsive to TCDD, followed by periportal hepatocytes. TCDD primarily accumulates in central hepatocytes due to the induction of CYP1A2, a phase I metabolism enzyme that sequesters TCDD.20 The high dose of TCDD that caused panacinar damage together with the higher basal expression of Ahr in central hepatocytes likely coalesced to elicit the large number of DEGs in this region. TCDD also reduced the portal hepatocyte representation. Emerging spatial transcriptomics technologies are expected to provide further insights into the apparent decrease in periportal cells, as well as other cell-specific changes.
Ultimately, snSeq provided novel insight into cell-specific gene expression and the roles of distinct cell populations. RAS/MAPK signaling was activated in hepatic NPCs in response to TCDD. Previous studies have linked RAS/MAPK signaling to inflammation in human NAFLD, likely through the production of growth factors, cytokines, chemokines, and adhesion molecules.42 Moreover, TCDD repressed hepatocyte proliferation,43 and RAS induction by TCDD in HepG2 cells does not activate the downstream effector, MAPK.44 The lack of hepatocyte enrichment is also consistent with the inhibition of hepatocyte proliferation and infiltration of immune cells. Further investigation of macrophage subpopulations identified a high Gpnmb-expressing KC population following TCDD treatment, which was also reported in a diet-induced NASH model17 and in humans with steatosis or NASH,45 suggesting conserved molecular events across disparate etiologies and species. In agreement with the loss of liver-specific gene expression,46 we find enriched genes that were also upregulated in a liver-specific HNF4 knockout model, a transcription factor essential for liver-specific gene expression.47 Similarly, there was decreased expression of genes repressed in liver cancer, consistent with TCDD being a known mouse liver carcinogen. Decreased enrichment was observed in most cell types, as the gene set was produced from whole-tissue expression profiling, highlighting some caveats when examining functional changes using single-cell/nuclei data.
In conclusion, snSeq represents a valuable strategy to characterize cell-specific responses of hepatotoxicants. Despite known biases in nuclear and cytosolic transcript levels, single-nuclei transcriptomic data accurately characterize the differential expression elicited by TCDD as reflected in (1) the consistency with bulk gene expression data, (2) the identification of NASH associated macrophages, and (3) changes in functional enrichment of pathways associated with TCDD-elicited hepatotoxicity. Furthermore, the analysis of frozen samples enables comprehensive dose- and time-dependent investigation of cell-specific adverse effects.
Materials and Methods
Animals and Treatment
Male C57BL/6 mice aged postnatal day (PND) 25 from Charles River Laboratories (Portage, MI) were housed in Innocages (Innovive, San Diego, CA) with ALPHA-dri bedding (Shepherd Specialty Papers, Chicago, IL) at 30%–40% humidity and a 12-hour light/dark cycle. Animals were fed ad libitum Harlan Teklad 22/5 Rodent Diet 8940 (Harlan Teklad, Madison, WI), with free access to Aquavive water (Innovive, San Diego, CA). PND 28 mice were orally gavaged with sesame oil vehicle (Sigma-Aldrich, St Louis, MO) or 30 μg/kg TCDD (AccuStandard, New Haven, CT) every 4 days for 28 days (7 treatments total). On day 28 (PND 52), animals were euthanized by CO2 asphixiation, and livers were immediately collected, frozen in liquid nitrogen, and stored at –80°C. All procedures were approved by the Michigan State University Institutional Animal Care and Use Committee.
Nuclei Isolation
Nuclei were isolated from frozen liver samples (∼200 mg) as previously described (https://doi.org/10.17504/protocols.io.3fkgjkw). Briefly, livers were diced in EZ Lysis Buffer (Sigma-Aldrich), homogenized using a disposable dounce homogenizer, and incubated on ice for 5 minutes. The homogenate was filtered using a 70-μm cell strainer, transferred to microcentrifuge tube, and centrifuged at 500 g and 4°C for 5 minutes. The supernatant was removed, and fresh EZ lysis buffer was added for an additional 5 minutes on ice following by centrifugation at 500 g and 4°C for 5 minutes. The nuclei pellet was washed twice in nuclei wash and resuspend buffer (1× phosphate-buffered saline, 1% bovine serum albumin, 0.2-U/μL RNAse inhibitor) with 5-minute incubations on ice. Following the washes, the nuclei pellet was resuspended in nuclei wash and resuspend buffer containing DAPI (10 μg/mL). The resuspended nuclei were filtered with 40-μm strainer and immediately underwent fluorescence-activated cell sorting using a BD FACSAria IIu (BD Biosciences, San Jose, CA) with 70-μm nozzle at the MSU Pharmacology and Toxicology Flow Cytometry Core (drugdiscovery.msu.edu/facilities/flow-cytometry-core; https://doi.org/10.6084/m9.figshare.12728540). Sorted nuclei were immediately processed for snSeq.
snSeq and Data Analysis
Libraries were prepared using the 10x Genomics Chromium Single Cell 3′ v3 kit and submitted for 150-bp paired-end sequencing at a depth ≥50,000 reads/cell using the HiSeq 4000 at Novogene (Beijing, China). Raw sequencing data were deposited in the Gene Expression Omnibus (GEO) (GSE148339). Following sequencing quality control, CellRanger v3.0.2 (10x Genomics) was used to align reads to a custom reference genome (mouse mm10 release 93 genome build) which included introns and exons to consider pre-mRNA and mature mRNA present in the nuclei.
Raw counts were further analysed using Seurat v3.1.1.48 Each sample was filtered for (1) genes expressed in at least 3 nuclei, (2) nuclei that express at least 100 genes, and (3) ≤1% mitochondrial genes. Additional quality control was performed using the scater package (v1.10.1). The DoubletFinder v2.0.2 package excluded putative doublets from subsequent analyses. Clustering of nuclei was performed using Seurat integration tools at a resolution of 0.2 and annotated using a semiautomated strategy by comparison to 3 published datasets (isolated hepatocytes sequenced by massively parallel scSeq [MARS-seq; GSE84498]; isolated hepatic NPCs by MARS-Seq [GSE108561]; isolated NPCs using the 10x Genomics platform [GSE129516]). Marker genes for individual nuclei clusters were also manually examined to verify annotation. Trajectory inference of hepatocytes was performed using Slingshot.34
Gene Set Enrichment Analysis
Gene set enrichment analysis was performed on transcriptomes of individual nuclei using the Gene Set Variation Analysis (GSVA v1.30.0) package49 with a minimum of 10 genes and maximum of 300 gene per gene set. Mouse gene sets were obtained from the GSKB (ge-lab.org/gskb/) filtered to only include MSIGDB, GENESIGDB, SMPDB, GO, KEGG, REACTOME, EHMN, MIRNA, MICROCOSM, MIRTARBASE, MPO, PID, PANTHER, BIOCARTA, INOH, NETPATH, WIKIPATHWAYS, MOUSECYC, TF, and TFACTS gene sets (6513 total). Human-based (eg, HPO), exposure-based (eg, DRUGBANK), or gene sets with limited annotation (eg, LIT) were excluded. Uniform Manifold Approximation and Projection (UMAP) reduction using the first 30 principal components analysis dimensions was used for visualization using original cell type annotations. For the visualization of marker and differentially enriched pathways, a network of gene sets was generated using the Enrichment Map plugin for Cytoscape.50 Nodes were summarized using AutoAnnotate51 and subsequently manually curated. Nodes were redrawn using the Enhanced Graphics package.
Data and Code Availability
Raw and processed single-nuclei sequencing data is deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession ID GSE148339. The final Seurat object including clustering, prediction scores, and annotation can be downloaded from the GEO repository or a compressed version from Dataverse (https://doi.org/10.7910/DVN/WGFSNO). Publically available datasets used to compare annotation were obtained from GEO under accession IDs GSE84498 GSE108561, and GSE129516. Bulk RNA-seq data for comparison with pseudobulk analysis was also obtained from GEO under accession ID GSE87543. Gene sets for gene set enrichment analysis were downloaded from the GSKB (ge-lab.org/gskb/). Analysis code for the processing and analysis of snSeq data can be obtained at https://zacharewskilab.github.io/snseq-2doses-analysis/. All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors would like to thank Dr Luciano Martelotto (University of Melbourne) for his help in optimizing the nuclei isolation protocol for single-nuclei RNA sequencing, as well as the labs of Dr Jiande Lin (University of Michigan) and Dr Shalev Itzkovitz (Weizmann Institute of Science) for sharing processed datasets used to semi-automate cluster annotation.
CRediT Authorship Contributions
Rance Nault, (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Methodology: Lead; Visualization: Lead; Writing – original draft: Lead)
Kelly A. Fader (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Sudin Bhattacharya (Funding acquisition: Supporting; Project administration: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Tim R. Zacharewski, (Conceptualization: Equal; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the National Institute of Environmental Health Sciences Superfund Research Program (P42ES004911) (to Tim R. Zacharewski) and the National Human Genome Research Institute (R21HG010789) to Tim R. Zacharewski and Sudin Bhattacharya. Tim R. Zacharewski and Sudin Bhattacharya are partially supported by AgBioResearch at Michigan State University.
References
- 1.Gu X., Manautou J.E. Molecular mechanisms underlying chemical liver injury. Expert Rev Mol Med. 2012;14:e4. doi: 10.1017/S1462399411002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hornberg J.J., Laursen M., Brenden N., Persson M., Thougaard A.V., Toft D.B., Mow T. Exploratory toxicology as an integrated part of drug discovery. Part I: why and how. Drug Discov today. 2014;19:1131–1136. doi: 10.1016/j.drudis.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Yorita Christensen K.L., Carrico C.K., Sanyal A.J., Gennings C. Multiple classes of environmental chemicals are associated with liver disease: NHANES 2003-2004. Int J Hyg Environ Health. 2013;216:703–709. doi: 10.1016/j.ijheh.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cave M., Appana S., Patel M., Falkner K.C., McClain C.J., Brock G. Polychlorinated biphenyls, lead, and mercury are associated with liver disease in American adults: NHANES 2003-2004. Environ Health Perspect. 2010;118:1735–1742. doi: 10.1289/ehp.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heindel J.J., Blumberg B., Cave M., Machtinger R., Mantovani A., Mendez M.A., Nadal A., Palanza P., Panzica G., Sargis R., Vandenberg L.N., Vom Saal F. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol. 2017;68:3–33. doi: 10.1016/j.reprotox.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fader K.A., Nault R., Kirby M.P., Markous G., Matthews J., Zacharewski T.R. Convergence of hepcidin deficiency, systemic iron overloading, heme accumulation, and REV-ERBalpha/beta activation in aryl hydrocarbon receptor-elicited hepatotoxicity. Toxicol Appl Pharmacol. 2017;321:1–17. doi: 10.1016/j.taap.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nault R., Fader K.A., Ammendolia D.A., Dornbos P., Potter D., Sharratt B., Kumagai K., Harkema J.R., Lunt S.Y., Matthews J., Zacharewski T. Dose-dependent metabolic reprogramming and differential gene expression in TCDD-elicited hepatic fibrosis. Toxicol Sci. 2016;154:253–266. doi: 10.1093/toxsci/kfw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierre S., Chevallier A., Teixeira-Clerc F., Ambolet-Camoit A., Bui L.C., Bats A.S., Fournet J.C., Fernandez-Salguero P., Aggerbeck M., Lotersztajn S., Barouki R., Coumoul X. Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol Sci. 2014;137:114–124. doi: 10.1093/toxsci/kft236. [DOI] [PubMed] [Google Scholar]
- 9.Taylor K.W., Novak R.F., Anderson H.A., Birnbaum L.S., Blystone C., Devito M., Jacobs D., Kohrle J., Lee D.H., Rylander L., Rignell-Hydbom A., Tornero-Velez R., Turyk M.E., Boyles A.L., Thayer K.A., Lind L. Evaluation of the association between persistent organic pollutants (POPs) and diabetes in epidemiological studies: a national toxicology program workshop review. Environ Health Perspect. 2013;121:774–783. doi: 10.1289/ehp.1205502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksen G.L., Ketchum N.S., Michalek J.E., Swaby J.A. Serum dioxin and diabetes mellitus in veterans of Operation Ranch Hand. Epidemiology. 1997;8:252–258. doi: 10.1097/00001648-199705000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Hart S.N., Li Y., Nakamoto K., Subileau E.A., Steen D., Zhong X.B. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos. 2010;38:988–994. doi: 10.1124/dmd.109.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 13.Su X., Shi Y., Zou X., Lu Z.N., Xie G., Yang J.Y.H., Wu C.C., Cui X.F., He K.Y., Luo Q., Qu Y.L., Wang N., Wang L., Han Z.G. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genomics. 2017;18:946. doi: 10.1186/s12864-017-4342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habib N., Li Y., Heidenreich M., Swiech L., Avraham-Davidi I., Trombetta J.J., Hession C., Zhang F., Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahar Halpern K., Shenhav R., Massalha H., Toth B., Egozi A., Massasa E.E., Medgalia C., David E., Giladi A., Moor A.E., Porat Z., Amit I., Itzkovitz S. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 2018;36:962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahar Halpern K., Shenhav R., Matcovitch-Natan O., Toth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E., Brandis A., Giladi A., Avihail A.S., David E., Amit I., Itzkovitz S. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong X., Kuang H., Ansari S., Liu T., Gong J., Wang S., Zhao X.Y., Ji Y., Li C., Guo L., Zhou L., Chen Z., Leon-Mimila P., Chung M.T., Kurabayashi K., Opp J., Campos-Perez F., Villamil-Ramirez H., Canizales-Quinteros S., Lyons R., Lumeng C.N., Zhou B., Qi L., Huertas-Vazquez A., Lusis A.J., Xu X.Z.S., Li S., Yu Y., Li J.Z., Lin J.D. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol Cell. 2019;75:644–660.e5. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anundi I., Lahteenmaki T., Rundgren M., Moldeus P., Lindros K.O. Zonation of acetaminophen metabolism and cytochrome P450 2E1-mediated toxicity studied in isolated periportal and perivenous hepatocytes. Biochem Pharmacol. 1993;45:1251–1259. doi: 10.1016/0006-2952(93)90277-4. [DOI] [PubMed] [Google Scholar]
- 19.Boverhof D.R., Burgoon L.D., Tashiro C., Sharratt B., Chittim B., Harkema J.R., Mendrick D.L., Zacharewski T.R. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- 20.Santostefano M.J., Richardson V.M., Walker N.J., Blanton J., Lindros K.O., Lucier G.W., Alcasey S.K., Birnbaum L.S. Dose-dependent localization of TCDD in isolated centrilobular and periportal hepatocytes. Toxicol Sci. 1999;52:9–19. doi: 10.1093/toxsci/52.1.9. [DOI] [PubMed] [Google Scholar]
- 21.Fader K.A., Nault R., Doskey C.M., Fling R.R., Zacharewski T.R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin abolishes circadian regulation of hepatic metabolic activity in mice. Sci Rep. 2019;9:6514. doi: 10.1038/s41598-019-42760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C., Duong T.E., Gao D., Chun J., Kharchenko P.V., Zhang K. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36:70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng W., Jiang S., Kong X., El-Ali N., Ball A.R., Jr., Ma C.I., Hashimoto N., Yokomori K., Mortazavi A. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 2016;44:e158. doi: 10.1093/nar/gkw739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K., Weitz D.A., Rozenblatt-Rosen O., Zhang F., Regev A. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nault R., Fader K.A., Zacharewski T. RNA-Seq versus oligonucleotide array assessment of dose-dependent TCDD-elicited hepatic gene expression in mice. BMC Genomics. 2015;16:373. doi: 10.1186/s12864-015-1527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez M., Rahmani E., Jew B., Garske K.M., Miao Z., Benhammou J.N., Ye C.J., Pisegna J.R., Pietiläinen K.H., Halperin E., Pajukanta P. Enhancing droplet-based single-nucleus RNA-seq resolution using the semi-supervised machine learning classifier DIEM. Sci Rep. 2020;10:11019. doi: 10.1038/s41598-020-67513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franzén O., Gan L.-M., Björkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019;2019:baz046. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nonaka H., Tanaka M., Suzuki K., Miyajima A. Development of murine hepatic sinusoidal endothelial cells characterized by the expression of hyaluronan receptors. Dev Dyn. 2007;236:2258–2267. doi: 10.1002/dvdy.21227. [DOI] [PubMed] [Google Scholar]
- 29.Masyuk T.V., Huang B.Q., Ward C.J., Masyuk A.I., Yuan D., Splinter P.L., Punyashthiti R., Ritman E.L., Torres V.E., Harris P.C., LaRusso N.F. Defects in cholangiocyte fibrocystin expression and ciliary structure in the PCK rat. Gastroenterology. 2003;125:1303–1310. doi: 10.1016/j.gastro.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z., Cotta C.V., Stephan R.P., deGuzman C.G., Klug C.A. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. EMBO J. 2003;22:4759–4769. doi: 10.1093/emboj/cdg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith X., Taylor A., Rudd C.E. T-cell immune adaptor SKAP1 regulates the induction of collagen-induced arthritis in mice. Immunol Lett. 2016;176:122–127. doi: 10.1016/j.imlet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahar Halpern K., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., Itzkovitz S. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 2015;13:2653–2662. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fader K.A., Nault R., Zhang C., Kumagai K., Harkema J.R., Zacharewski T.R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-elicited effects on bile acid homeostasis: Alterations in biosynthesis, enterohepatic circulation, and microbial metabolism. Sci Rep. 2017;7:5921. doi: 10.1038/s41598-017-05656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Moshe S., Shapira Y., Moor A.E., Manco R., Veg T., Bahar Halpern K., Itzkovitz S. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab. 2019;1:899–911. doi: 10.1038/s42255-019-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacono G., Massoni-Badosa R., Heyn H. Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biol. 2019;20:110. doi: 10.1186/s13059-019-1713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakken T.E., Hodge R.D., Miller J.A., Yao Z., Nguyen T.N., Aevermann B., Barkan E., Bertagnolli D., Casper T., Dee N., Garren E., Goldy J., Graybuck L.T., Kroll M., Lasken R.S., Lathia K., Parry S., Rimorin C., Scheuermann R.H., Schork N.J., Shehata S.I., Tieu M., Phillips J.W., Bernard A., Smith K.A., Zeng H., Lein E.S., Tasic B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018;13 doi: 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnaswami S.R., Grindberg R.V., Novotny M., Venepally P., Lacar B., Bhutani K., Linker S.B., Pham S., Erwin J.A., Miller J.A., Hodge R., McCarthy J.K., Kelder M., McCorrison J., Aevermann B.D., Fuertes F.D., Scheuermann R.H., Lee J., Lein E.S., Schork N., McConnell M.J., Gage F.H., Lasken R.S. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacParland S.A., Liu J.C., Ma X.Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., Gupta R., Cheng M.L., Liu L.Y., Camat D., Chung S.W., Seliga R.K., Shao Z., Lee E., Ogawa S., Ogawa M., Wilson M.D., Fish J.E., Selzner M., Ghanekar A., Grant D., Greig P., Sapisochin G., Selzner N., Winegarden N., Adeyi O., Keller G., Bader G.D., McGilvray I.D. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Digernes V., Bolund L. The ploidy classes of adult mouse liver cells. A methodological study with flow cytometry and cell sorting. Virchows Arch B Cell Pathol Incl Mol Pathol. 1979;32:1–10. [PubMed] [Google Scholar]
- 41.Moreno-Marin N., Merino J.M., Alvarez-Barrientos A., Patel D.P., Takahashi S., Gonzalez-Sancho J.M., Gandolfo P., Rios R.M., Munoz A., Gonzalez F.J., Fernandez-Salguero P.M. Aryl hydrocarbon receptor promotes liver polyploidization and inhibits PI3K, ERK, and Wnt/beta-catenin signaling. iScience. 2018;4:44–63. doi: 10.1016/j.isci.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jimenez-Castro M.B., Cornide-Petronio M.E., Gracia-Sancho J., Casillas-Ramirez A., Peralta C. Mitogen activated protein kinases in steatotic and non-steatotic livers submitted to ischemia-reperfusion. Int J Mol Sci. 2019;20:1785. doi: 10.3390/ijms20071785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson D.P., Li H., Mitchell K.A., Joshi A.D., Elferink C.J. Ah receptor-mediated suppression of liver regeneration through NC-XRE-driven p21Cip1 expression. Mol Pharmacol. 2014;85:533–541. doi: 10.1124/mol.113.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi M., Hankinson O. 2,3,7,8Tetrachlorodibenzopdioxin suppresses the growth of human liver cancer HepG2 cells in vitro: Involvement of cell signaling factors. Int J Oncol. 2018;53:1657–1666. doi: 10.3892/ijo.2018.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katayama A., Nakatsuka A., Eguchi J., Murakami K., Teshigawara S., Kanzaki M., Nunoue T., Hida K., Wada N., Yasunaka T., Ikeda F., Takaki A., Yamamoto K., Kiyonari H., Makino H., Wada J. Beneficial impact of Gpnmb and its significance as a biomarker in nonalcoholic steatohepatitis. Sci Rep. 2015;5:16920. doi: 10.1038/srep16920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nault R., Fader K.A., Harkema J.R., Zacharewski T. Loss of liver-specific and sexually dimorphic gene expression by aryl hydrocarbon receptor activation in C57BL/6 mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohguchi H., Tanaka T., Uchida A., Magoori K., Kudo H., Kim I., Daigo K., Sakakibara I., Okamura M., Harigae H., Sasaki T., Osborne T.F., Gonzalez F.J., Hamakubo T., Kodama T., Sakai J. Hepatocyte nuclear factor 4alpha contributes to thyroid hormone homeostasis by cooperatively regulating the type 1 iodothyronine deiodinase gene with GATA4 and Kruppel-like transcription factor 9. Mol Cell Biol. 2008;28:3917–3931. doi: 10.1128/MCB.02154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kucera M., Isserlin R., Arkhangorodsky A., Bader G.D. AutoAnnotate: a Cytoscape app for summarizing networks with semantic annotations. F1000Res. 2016;5:1717. doi: 10.12688/f1000research.9090.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw and processed single-nuclei sequencing data is deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession ID GSE148339. The final Seurat object including clustering, prediction scores, and annotation can be downloaded from the GEO repository or a compressed version from Dataverse (https://doi.org/10.7910/DVN/WGFSNO). Publically available datasets used to compare annotation were obtained from GEO under accession IDs GSE84498 GSE108561, and GSE129516. Bulk RNA-seq data for comparison with pseudobulk analysis was also obtained from GEO under accession ID GSE87543. Gene sets for gene set enrichment analysis were downloaded from the GSKB (ge-lab.org/gskb/). Analysis code for the processing and analysis of snSeq data can be obtained at https://zacharewskilab.github.io/snseq-2doses-analysis/. All authors had access to the study data and reviewed and approved the final manuscript.