Highlights
-
•
Established a new in vitro tumor model called novel conditionally reprogrammed (termed i-CR).
-
•
Accomplished personalized drug tests within 2–3 weeks.
-
•
Achieved 100% sensitivity, 85.7% specificity, 91.7% positive predictive value, and 100% negative predictive value.
-
•
i-CR guided a inoperable patient with metastases converted to radical surgery.
Keywords: Conditional reprogramming, Colorectal cancer, Chemotherapy, Pre-clinical, In vitro drug test
Abbreviations: PDO, patient-derived organoids; CR, conditional reprogramming; I-CR, individualized conditional reprogramming; CRC, colorectal cancer; NGS, next-generation sequencing; WES, whole-genome sequencing; CNV, copy number variation; EPCAM, epithelial cell adhesion molecule; EDU, 5-ethynyl-2´-deoxyuridine; MI, maximum inhibition; DSI, drug sensitivity index; TGI, tumor growth inhibition; MDT, multidisciplinary team
Abstract
Background
In vitro patient tumor models such as patient-derived organoids (PDO) and conditionally reprogrammed (CR) cell culture are important for translational research and pre-clinical drug testing. In this study we present a personalized drug sensitivity test for late stage, potentially operable colorectal cancer (CRC) using patient-derived primary tumor cells isolated with i-CR technology, an optimized CR method. We explored the clinical feasibility of using i-CR platform to guide CRC chemotherapy, and established the correlation between in vitro drug sensitivity and patient clinical response.
Methods
Primary CRC tumor cells were isolated and cultured with the i-CR technology. NGS was performed and the WES and CNV results of i-CR cells were compared with that of the original patient tumor samples. In vitro drug screenings were done with guideline chemotherapy drugs for CRC. In vivo drug response was examined with paired PDX mouse models. A double-blind co-clinical cohort study was carried out and the clinical outcomes of the enrolled patients were compared with the i-CR results.
Results
i-CR platform could be used to rapidly propagate primary colorectal tumor cells that represent individual patient tumors effectively by keeping the clonal heterogeneity and the genetic characteristics. Chemotherapy drug screenings with i-CR cells were comparable with that of PDX models. More importantly, i-CR results showed high accordance with the clinical outcomes of the enrolled CRC patients.
Conclusion
i-CR platform was capable to test and optimize therapeutic regimens pre-clinically, study cancer cell biology, and model tumor re-emergence to identify new targeted therapeutics from an effective personalized medicine standpoint.
Background
Colorectal cancer (CRC) is one of the most common cancers worldwide and CRC ranks among the highest in terms of the cancer-related death [1], [2], [3], [4]. As the most lethal gastrointestinal cancer malignancy, the projected 5-year mortality rate for late stage CRC is >70%. The treatments for CRC patients include surgery, radiation therapy, chemotherapy, targeted therapy and immunotherapy [5], [6], [7], [8]. Radical surgery and adjuvant chemotherapy and targeted therapy can yield a clinical response even in stage III-IV disease. Unfortunately, the response rate to guideline therapies remains below expectation, and relapse and chemo-resistance are observed in a majority of the patients [7,9]. In the era of precision medicine, it is essential to develop new technologies for finding tailored therapies for CRC patients.
In the past decade, the progress made in gene sequencing technology, especially the emergence of NGS, and the development of targeted therapeutics have been pushing the boundaries of precision medicine. However, recent studies showed that there are still over 70% of the patients cannot benefit from gene sequencing results in terms of personalized therapy [10]. Even for the small fraction of cancer-related mutations that were validated clinically, the matched targeted therapies are still mostly transient and partial. The complexity and diversity of cancer systems prevented the wider success of gene sequencing. It is therefore important to develop in vitro platforms that can reliably test and predict patient responses to novel therapeutics [11].
For a long time, scientists have undertaken the challenge to develop methods for propagating and studying primary tumors outside of the human body. Traditional established cell lines of patient tumor tissues have been widely used in this regard. Conversely, functional drug testing with traditional tumor cell lines or primary cell cultures is associated with cloning bias and the loss of tumor cell heterogeneity remains a major obstacle for studying drug efficacy and drug resistance [12]. In addition, the establishment of tumor cell lines is hindered by the low rate of success (1–10%, depending on the tissue of origin and state of disease progression). Patient-derived xenograft (PDX) [13], [14], [15] and tumor organoids [16], [17], [18] have been considered more clinically relevant models for original patient tumors. Yet the use of such systems still facing hurdles such as high cost, long duration and technical difficulties, etc. [19,20].
Conditional reprogramming (CR) is an emerging primary cell culture technology first reported by Liu et al. [21,22]. CR system allows expansion of epithelial cells in vitro with high efficiency and can be used to propagate normal or tumor cells from various different tissues. Genetic analysis by whole exome sequencing (WES) and copy number variations (CNVs) suggests CR cells are capable of keeping the tumor heterogeneity [23], [24], [25]. CR tumor cell cultures therefore can be excellent cancer models for their ability to maintain the all-around characteristics of the human cancer. Recently an optimized CR-based primary tumor cell culture system termed i-CR was developed [26]. The i-CR technology showed promise in translational medicine and personalized cancer therapy. In the present study, we thoroughly evaluated i-CR technology for its value as a bona fide pre-clinical tumor model. Moreover, we explored the capability of i-CR technology in vitro drug sensitivity test for late-stage colorectal cancer patients in a co-clinical cohort study, in which clinical patient responses to chemotherapies were matched to personalized treatment in parallel laboratory i-CR studies. It was demonstrated i-CR system could be an excellent pre-clinical test platform for personalized therapy.
Methods
Generation of patient-derived primary cultures
This study was approved by the Medical Ethics Committee of Peking University Cancer Hospital. All specimens were collected from patients with written consent.
Swiss 3T3 fibroblast cells were purchased from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai, China. The isolation and cultivation of i-CR primary tumor cells and normal primary epithelial cells were similar to the previously reported method [20] with modifications [26]. In brief, human tumor samples were obtained and immediately transferred into Tissue Preservation Solution (Percans Oncology, Beijing, China) at 4 °C. The tissue was rinsed twice with cold PBS and minced with surgical scissors in a sterile Petri dish. It was then subjected to enzymatic dissociation with a combination of collagenase I, DNase and dispase. Final cell suspensions were filtered through 100-μm cell strainers, followed by pelleting and resuspension in the complete i-CR medium or the selective medium. The complete medium consisted of DMEM/F-12 basal medium, 2% FBS, 10 ng/mL human EGF (ThermoFisher), 10 μM Y-27,632 (Selleckchem), 10 ng/mL bFGF (ThermoFisher), 10 mM Nicotinamide (Sigma), 1 fold of Insulin-Transferrin-Selenium (ThermoFisher), 1 fold of Non-essential amino acid (ThermoFisher), 25 ng/mL mouse Wnt3a (Peprotech), 500 ng/mL human R-spondin-1 (Peprotech), 100 ng/mL Noggin (Peprotech), 100 μg/mL Primocin (Vivogen). The selective medium was the complete medium minus Wnt3a, R-spondin-1 and Noggin. Isolated cells were seeded onto a layer of lethally irradiated (40 Gy) Swiss 3T3 fibroblasts feeder cells and incubated for at 37 °C in 5% CO2.
Establishment of PDX models
All animal experiments were performed under sterile conditions at Percans Oncology Inc. specific-pathogen free facility and carried out in accordance with the National Institutes of Health guide for the care and use of laboratory animals. The procedure of PDX model establishment was referred to previous reports [14,26]. Briefly, 6–8 week-old female NOD/SCID mice (Beijing HFK Bio-Technology Co., LTD, Beijing, China) were used for the studies. Tumor samples obtained from patients were immediately transferred into tissue preservation solution (Percans Oncology, Beijing, China) and sliced into small fragments. The mice were inoculated with the fragments subcutaneously at one flank to produce xenografts called passage 1 (P1). The serial xenografts of different passages were generated using the same procedure.
STR analysis
Genomic DNA was prepared with Axygen genomic DNA preparation kit and used for PCR assay. Short tandem repeat (STR) analysis was performed with 20 STR loci (including the 8 loci recommended by ATCC) and Amelogenin locus. Amplified fragments were detected with the ABI3730XL genetic analyzer (Applied Biosystems). Data analysis was done with DSMZ tools (Leibniz-Institut DSMZ, Germany).
Whole-exome sequencing (WES) and copy-number analysis
Whole-exome enrichment was performed using the TruSeq Exome Enrichment Kit (Illumina). Captured DNA libraries were sequenced with the Illumina HiSeq 2500 Genome Analyzer, yielding 200 (2 × 100) base pairs from the final library fragments. All sequencing reads were trimmed and filtered using Trimmomatic 36, followed with alignments of resulting reads to hg19 reference genome with Burrows-Wheeler Aligner (BWA, http://bio-bwa.Sourceforge.net/). Then Genome Analysis Tool Kit (GATK) was used for base quality score recalibration, indel realignment, and duplicate removal, reads with quality below 20 were discarded.
Four popular somatic SNV callers, i.e. Varscan, SomaticSniper, Strelka and MuTect2 were run on above pre-processed sequencing data and with default parameters recommended by the developers. We set the somatic quality threshold of SomaticSniper to 30. Raw call sets generated by Varscan and SomaticSniper were filtered by pipelines proposed by the developers, and those generated by Strelka and MuTect2 were processed with built-in post-calling filters for either tool. Various genome databases (Human Genome Mutation Database (HGMD), HapMap data, Single Nucleotide Polymorphism database (dbSNP), 1000 genomes, and COSMIC v70) were used to search for previously described mutations and/or polymorphisms, and co-segregation studies were performed for candidate gene mutations.
In vitro drug screening with i-CR models
Approximately 1500–5000 per well isolated tumor cells were seeded into a 96-well black-walled clear-bottom microplate (Corning, USA), which was layered with feeder cells 24 h before. The cells were cultured until small colonies became readily visible (2–5 days).
All drugs and drug combinations were first dissolved in DMSO as 1000x stock, and then added to each well according to specific study design. Typically, the drugs were tested in vitro at a maximum concentration C0, which is assigned according to the reported steady-state drug concentration in human serum [27] and further adjusted based on empirical evidence. Serial dilutions were performed when needed. The cells were continuously cultured for 7 days in the presence of the drugs or DMSO (as control). One micro molar of EdU was added for the last 24 h. EdU staining uses a “click” reaction to directly label active DNA synthesis and measure cells in S phase. [28].Test plates were fixed and stained with Cell Quantitative Detection Kit (Percans Oncology, Beijing, China) according to the manufacturer's instruction. After staining, test plates were scanned with Arrayscan XTI 800, and images were acquired and analyzed with the built-in Bioapplication software package.
The effectiveness of each therapeutic regimen was evaluated and quantified using the formula: Maximum Inhibition (MI) = N0/Nd, where N0 and Nd denotes the number of EpCAM+EdU+ epithelial cells in the wells of control or with the drug at concentration C0, respectively.
The Drug Sensitivity Index (DSI) was a novel concept based on the well-established “log-kill” model [29]. It was calculated using the formula: , where MIC0, MI1/2C0, and MI1/4C0 are MI values observed when cells were treated at drug concentrations C0, 1/2C0 and 1/4C0, respectively.
In vivo drug screening with the PDX model
When the average tumor size reached approximately 250–300 mm3 in the mice, the animals were randomly allocated into different groups, with 5 mice per group. The day of randomization was defined as study day 0. Tumor volume is expressed in mm3 using the following formula: V (volume) = (a × b2)/2 where a and b are the long and short diameters of the tumor, respectively. The list of drugs and their dosing regimens were presented in Supplementary Table S1. Tumor suppression was expressed as Tumor Growth Inhibition (TGI), which is calculated according to the formula: TGI = (1-(Ti-T0)/(Vi-V0))*100, where Ti as the mean tumor volume of the treatment group on the measurement day; T0 as the mean tumor volume of the treatment group at D0; Vi as the mean tumor volume of control group at the measurement day; V0 as the tumor volume of the control group at D0.
Clinical validation of chemosensitivity assays
This study was approved by the Medical Ethics Committee of Peking University Cancer Hospital. All specimens were collected from patients with written consent.
Patients were pathology confirmed, stage IV colorectal cancer patients who underwent emergency surgery or palliative resection to remove the tumor, and can tolerate chemotherapy with measurable tumor lesion thereafter. Adult patients between 18 and 70 years old, male or female, informed patient consent, treatment-naïve, stage IV CRC patients with pathology confirmation with resectable tumor, estimated survival time no less than 6 months, and the patient has at least one measurable disease lesion (according to RECIST1.1). Patients met the following criteria should be excluded from the study: history of any prior anti-cancer treatment; participant of any other clinical study within 6 months, women currently breast feeding or pregnant, severe liver or kidney function impairment (liver function: TBIL ≤1.5 × ULN, ALT & AST≤2.5 × ULN), kidney function Cr ≤1.5 × ULN and creatinine clearance rate≥ 50 mL/min (according to the Cockcroft-Gault formula), patients with cognitive impairment, psychological disease, or poor compliance, allergic to known chemotherapy ingredients, or other conditions researchers deemed not suitable for study participation.
All drugs used are approved by the CFDA (China Food and Drug Administration) for the treatment of colorectal cancer, generally according to current guidelines or recommendation from MDT (multidisciplinary team) panels. Evaluate the correlation between MI as well as DSI as measured by CR-based ex vivo drug sensitivity test with patient clinical response as measured by imaging data. Any adverse or severe event associated with image analysis, laboratory analysis, as well as during clinical follow up period. CEA serum levels and computed tomography (CT) scans of the chest, abdomen and pelvis were performed at baseline and then repeated at least every 8 weeks during treatment (or earlier for patients with suspected disease progression). CT scans were centrally reviewed by a single radiologist to document response to treatment according to the RECIST criteria (version 1.1). This clinical and radiological evaluation was conducted blindly from CR results.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 software (SPSS) or Graphpad Prism version 6.0 (Graphpad software). One-way ANOVA was used to compare differences in MI between different groups. A two-sided p < 0.05 was considered statistically significant.
Results
Cultivation of primary tumor cells with i-CR system
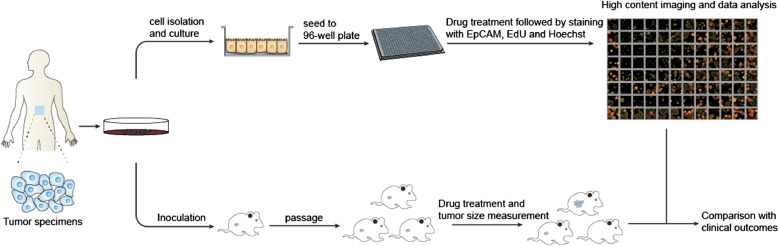
Tumor specimens were collected from 47 stage III-IV colorectal cancer patients with age ranging from 27 to 87. Metastasis was reported in most of these patients (Supplementary Table S1). The samples were subjected to treatments for the establishment of i-CR cell cultures and PDX models as shown in Fig. 1.
Fig. 1.
Flowchart of i-CR-based drug sensitivity test and data analysis. Patient tumor samples were collected and subjected to i-CR primary tumor cell culture and PDX model formation according to Material and Methods. High-content drug tests and data analysis were done with i-CR cells, the results were validated with PDX models and further compared with clinical outcomes.
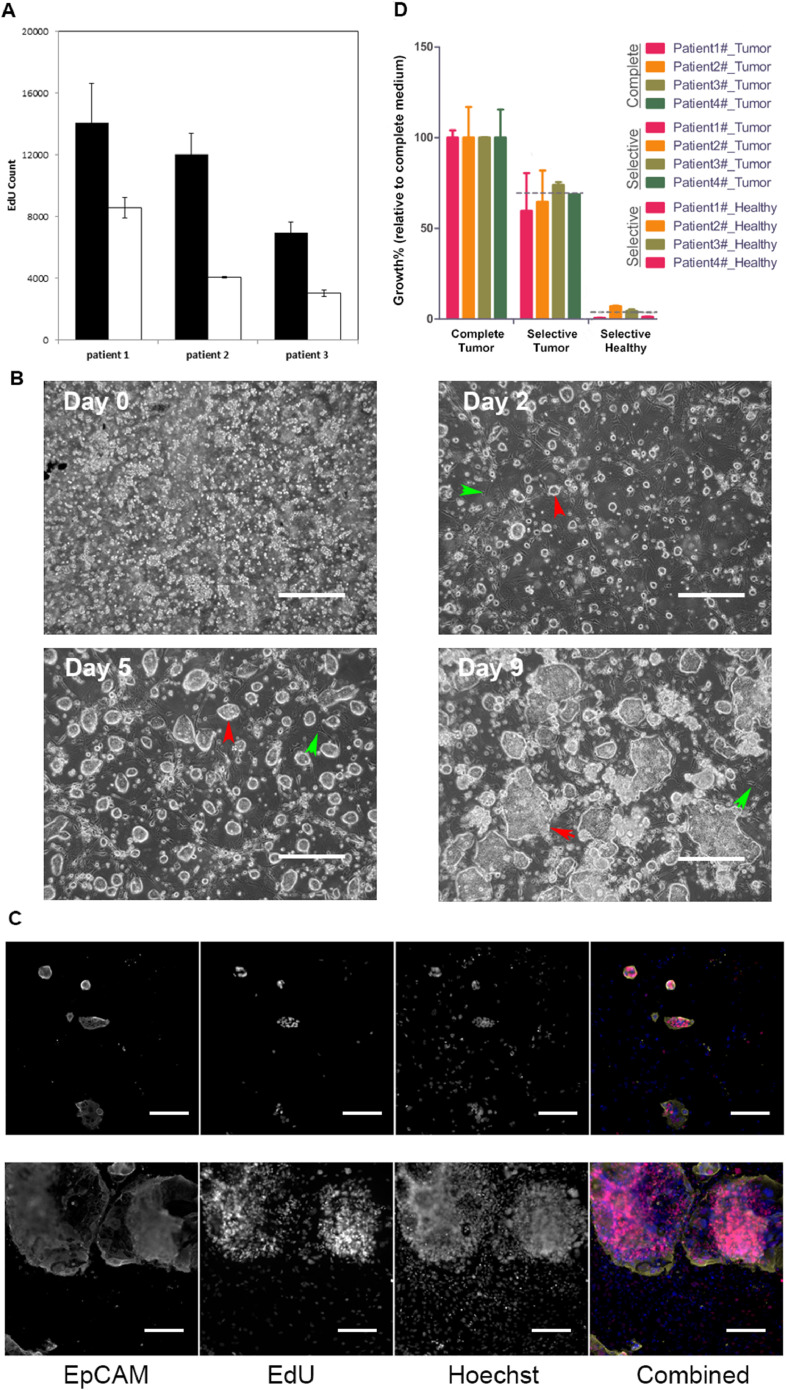
The newly developed system, i-CR, showed significant advantage in culturing CRC tumor cells versus the original CR (Fig. 2A). The morphology of the cultured i-CR cells was comparable to the patient tissue samples. As shown in Fig. 2B, small clusters of cells were isolated from tumor tissues and seeded onto tissue culture plates (96-well). The cells were continually cultured in i-CR medium until near confluent (normally 8–10 days). When performing drug testing, the cells were allowed to recover for 1–2 days after plating before the treatment of drugs. The cells were then incubated for 7 days, and EdU reagent was added for the last 24 h. After fluorescent labeling, the plates were assayed with a high-content imaging system. A typical analysis of cell growth result was shown in Fig. 2C.
Fig. 2.
Development of i-CR system. A) Growth comparison of tumor cells from CRC patients using i-CR system and conventional CR system. B) Reverse-phase microscopic images of the cultured i-CR cells at different time points after isolation. Red arrow: tumor cell colonies; green arrow: feeder cells. The scale bar equals to 200 μm. C) Fluorescent microscopic images the cultured i-CR cells at day 2 (upper panel) and day 7 (lower panel) after isolation. In the combined images, the yellow color represents EpCAM staining, the red color represents EdU labeling and the blue color represents Hoechst staining. The scale bar equals to 200 μm. D) Comparison of tumor and normal cell growth in complete medium and selective medium. The samples were from four patients and all experiments were repeated three times. p < 0.05.
As shown in Fig. 2D, equal numbers of isolated CRC tumor cells and normal colon epithelial cells from four patients were cultured under i-CR condition in complete medium or in selective medium, respectively. The tumor cells expand rapidly in both complete medium and selective medium. The total cell number of the tumor cells cultured in selective medium was 70% of that in complete medium. Normal epithelial cell growth, however, was hindered in selective medium, with cell number reached only 4% of that in complete medium.
The ability of i-CR system to quickly expand CRC tumor cells is crucial for rapid pre-clinical drug test. In addition, i-CR cells potentially can be cultured indefinitely, providing enough cells for more in vitro drug screening such as targeted drugs and combination therapy. To ensure that the i-CR culture remained uncontaminated after prolonged passaging, we performed STR analysis at 20 genetic loci and the Amelogenin locus. The STR results shown in Figure S1 indicated the cultures in passages 0, 4 and 10 were identical.
Genetic analysis of i-CR cultured cells and patient tumor tissues
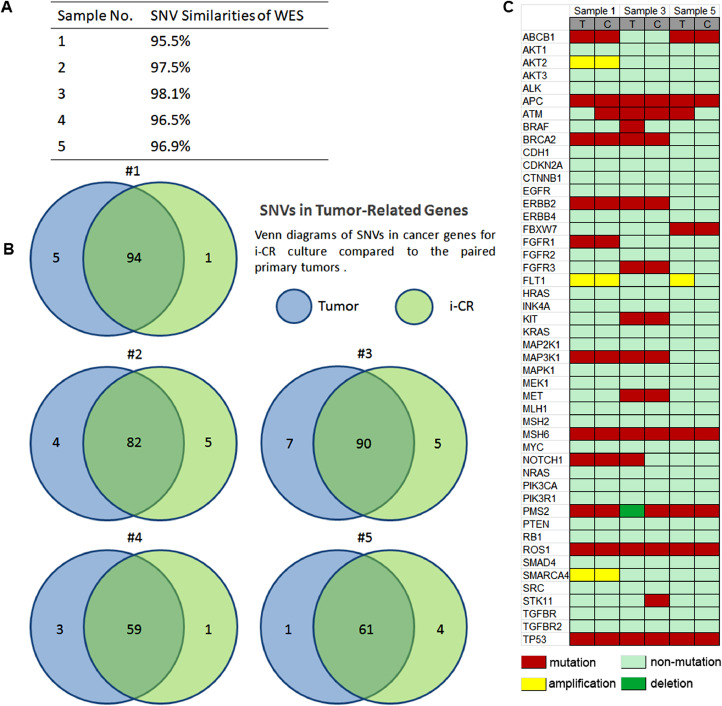
DNA isolated from resected CRC tumor tissues and the derived i-CR cultured cells was used for WES and copy number analysis. Five pairs of samples, which include three pairs of patient tissue-i-CR cells and two pairs of PDX tissue-i-CR cells, were tested. To investigate whether i-CR cells maintained the genetic heterogeneity of their original tissues, we examined the single nucleotide variations (SNVs) of each sample against the reference genome and the results are summarized in Fig. 3A. In all, i-CR cells shared 96.9% of their SNVs with primary tumors. The high concordance of SNVs indicated the genomic heterogeneity of the primary tumors was mostly maintained in the i-CR cultures. This observation was supported by comparing the SNVs of tumor-related genes of all the samples. The results were presented as Venn diagram in Fig. 3B. High similarities were shown between i-CR cells and the paired tumor tissues. Genes related to human colorectal cancer were selected and their expression profiles were analyzed [30,31]. The results were shown in Fig. 3C. Next, we analyzed the copy number variations (CNVs) of the i-CR samples and the corresponding tumor tissues. CNVs were obtained by comparing copy number profiles of our samples against the reference profiles and the representative result was shown in Supplementary Table S2. The CNV profiles of i-CR cells and primary tumors appeared to be highly conserved, consistent with the supposition that i-CR cells largely maintained the genomic heterogeneity of the primary tumors.
Fig. 3.
Genetic analysis of i-CR primary tumor cells. A) SNV similarities between i-CR cells and patient tumor tissues. B) Venn diagrams of SNVs in cancer-related genes for i-CR cultured cells compared to that of patient tumor tissues. C) Heatmap of genetic profiles of cancer genes of CRC. T: tumor, C: cultured cells.
Maximum inhibition (MI) and drug sensitivity index (DSI) as measurements for in vitro drug response
The drug response of i-CR tumor cells was calculated as MI and DSI values as described in Material and methods. The MI values of the therapeutic regimens for each patient were shown in Fig. S2A and Table S3. MI is a more intuitive indication of the inhibition effect of the drug treatments. Higher MI value represents more effective inhibition. DSI is a novel in vitro drug sensitivity criteria used in this study. The calculation of DSI takes into account of the populational difference of tumor cells in terms of drug sensitivity [29]. Using the derived mathematical formula, we calculated the DSI values of the drugs. The results are listed in Fig. S2B. And Table S4.
Validation of in vitro drug screening results of i-CR cells with paired PDX models
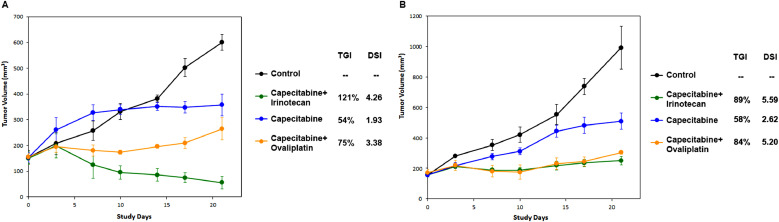
Next, we sought to validate in vitro screening results with paired PDX models in vivo. Fig. 4 showed the PDX tumor growth of two different CRC patients. The mouse models were treated with capecitabine, oxaliplatin, irinotecan or in combinations of capecitabine + oxaliplatin (XELOX), capecitabine + irinotecan (XELIRI). In the PDX model of patient A, the tumor growth inhibition (TGI) values of different regimens were 54% (capecitabine), 75% (XELOX), and 121% (XELIRI), respectively. The corresponding DSI values in paired i-CR culture were 1.93, 3.38, and 4.26, respectively (Fig. 4A). In the PDX model of patient B, the tumor growth inhibition (TGI) values of different regimens were 58% (capecitabine), 84% (XELOX), and 89% (XELIRI), respectively. The corresponding DSI values in paired i-CR culture were 2.62, 5.2, and 5.59, respectively (Fig. 4B). Results from PDX models are consistent with that of i-CR. Similar results were repeated with other patient samples and the results were presented in Table 1.
Fig. 4.
In vivo drug sensitivity test in PDX models and the correlation of tumor growth inhibition in PDXs with DSI measurements in the paired i-CR systems. A and B are two representatives of the patient samples (Patient NYZ102 and NYZ092, respectively).
Table 1.
In vitro and in vivo drug tests.
| Patient ID | Drug test index | Capecitabine/5-FU | XELOX | FOLFIRI |
|---|---|---|---|---|
| NYZ081 | MI7 | 59.91 | 283.5 | 203.4 |
| DSI | 5.11 | 7.17 | 7.77 | |
| TGI | 82% | 91% | 90% | |
| NYZ092 | MI7 | 29.68 | 297.9 | 270.4 |
| DSI | 2.62 | 5.2 | 5.59 | |
| TGI | 58% | 84% | 89% | |
| NYZ093 | MI7 | 2.52 | 4.78 | 0.75 |
| DSI | 0.06 | 0.61 | 0.23 | |
| TGI | 62% | 72% | 72% | |
| NYZ094 | MI7 | 2.72 | 29.4 | 6.09 |
| DSI | 1.34 | 2.1 | 1.13 | |
| TGI | 78% | 88% | 80% | |
| NYZ109 | MI7 | 18.37 | 26.22 | 7.23 |
| DSI | 2.49 | 3.44 | 3.41 | |
| TGI | 43% | 61% | 90% | |
| NYZ119 | MI7 | 113.6 | 242.7 | 615.0 |
| DSI | 3.11 | 4.16 | 4.07 | |
| TGI | 54% | 66% | 101% | |
| NYZ113 | MI7 | 661.9 | 800.5 | 2801 |
| DSI | 6.04 | 7.06 | 7.21 | |
| TGI | 40% | 83% | 73% | |
| NYZ102 | MI7 | 12.56 | 41.79 | 28.02 |
| DSI | 1.93 | 3.38 | 4.26 | |
| TGI | 54% | 75% | 121% | |
| NYZ053 | MI7 | 164.8 | 532.6 | 391.5 |
| DSI | 4.51 | 5.63 | 6.46 | |
| TGI | 107% | 83% | 103% |
Clinical parameters of patient tumor samples affecting the establishment of i-CR culture
Out of the total 47 CRC patient tumor samples collected in this study, 20 were successfully cultured in i-CR system. Significant number of contamination-free tumor cells were isolated. And the cells proliferated at a satisfactory rate for high-content drug screening. The overall success rate was 42.6%. Twenty-two samples were also subjected to creating PDX models, with 16 being successful. The success rate was 72.7%. PDX and i-CR overlapped in 10 models (Table S1). PDX models are known to be easier to establish from aggressive, high-grade and metastatic tumors [32]. We investigated the factors that affect the success rate of i-CR culture. Comparisons were made on the origin, the pathological profile and the characteristics of the tumor samples, and the results were summarized in Table 2. Three factors appeared to significantly affect the success rate of i-CR culture. Biopsy samples from tumor deposits (TDs, focal aggregates of tumor cells located in the pericolic or perirectal fat and not associated with lymph nodes) had extremely low success rate (one out of ten succeeded). Also, all five mucinous adenocarcinoma samples failed. Furthermore, out of 25 tumor samples that were previously treated with chemo or radio-therapy, only 7 succeeded. Other parameters, such as tumor stage, primary tumor site, BRAF mutation profile, etc., did not seem to affect i-CR culture.
Table 2.
Summary of parameters affecting i-CR model establishment.
| Parameters | Success(N = 20) Number(%) | No Success(N = 27) Number(%) | P-value |
|---|---|---|---|
| Sex | 0.358 | ||
| male | 14(70.0%) | 22(81.5%) | |
| female | 6(30.0%) | 5(18.5%) | |
| Age | 0.582 | ||
| <65 | 11 (55.0%) | 17 (63.0%) | |
| ≥65 | 9 (45.0%) | 10 (37.0%) | |
| Biopsy position | 0.025 | ||
| primary | 17 (85.0%) | 13 (48.1%) | |
| metastasis | 2 (10.0%) | 5 (18.5%) | |
| deposit | 1 (5.0%) | 9 (33.3%) | |
| Type of tumor | 0.042 | ||
| adenocarcinoma | 20 (100.0%) | 22 (81.5%) | |
| mucinous adenocarcinoma | 0 (0.0%) | 5 (18.5%) | |
| Tumor stage | 0.355 | ||
| II | 0 (0.0%) | 1 (3.8%) | |
| III | 1 (5.0%) | 0 (0.0%) | |
| IV | 19 (95.0%) | 26 (96.2%) | |
| Primary location | 0.999 | ||
| Rectum | 8 (40.0%) | 11 (40.7%) | |
| Left Colon | 6 (30.0%) | 8 (29.6%) | |
| Right Colon | 6 (30.0%) | 8 (29.6%) | |
| Pre treatment | 0.031 | ||
| No | 13 (65.0%) | 9 (33.3%) | |
| Chemotherapy/radiotherapy | 7 (35.0%) | 18 (66.7%) | |
| Kras/Nras | 0.967 | ||
| unknown | 1 (5.0%) | 0 (0.0%) | |
| wild type | 14 (70.0%) | 19 (70.0%) | |
| mutant type | 5 (25.0%) | 8 (30.0%) | |
| BRAF | 0.497 | ||
| unknown | 1 (5.0%) | 0 (0.0%) | |
| wild type | 13 (65.0%) | 19 (70.4%) | |
| mutant type | 6 (30.0%) | 8 (29.6%) | |
| Outcomes | 0.275 | ||
| unknown | 0 (0.0%) | 8 (29.6%) | |
| alive | 16 (80.0%) | 16(59.3%) | |
| dead | 4(20.0%) | 3(11.1%) |
The clinical outcomes of i-CR success and i-CR failure patients were monitored and compared. As shown in Figure S3, tumorigenicity appeared to be an independent predictor of poor FPS (Breslow (Generalized Wilcoxon), P < 0.0001) for stage IV colorectal cancer patients. The patient group with success i-CR culture had a statistically significant (p = 0.02) shorter PFS (progression free survival) when compared with the i-CR failure group. Therefore, there is a reciprocal relationship between the success rate of i-CR model generation and the disease outcome. The higher the rate is, the worse the prognosis.
Comparison of i-CR drug sensitivity tests with clinical outcomes of CRC patients
Genetic analysis and functional characterization proved that i-CR culture can be an excellent tumor model in vitro drug testing. Next, we examined its clinical predictive value for CRC patients’ responses to chemotherapeutic agents. Of the 20 patients with i-CR model established, 2 did not receive further chemotherapy treatment, 1 received additional targeted therapy. For the rest 17 eligible patients, the responses to the treatment regimens they received were compared with i-CR data, as long as PDX results. As shown in Table S5, the accordance rate between i-CR tests and clinical responses is 94.1% (16/17). The accordance rate for PDXs and clinical results is 66.7% (6/9).
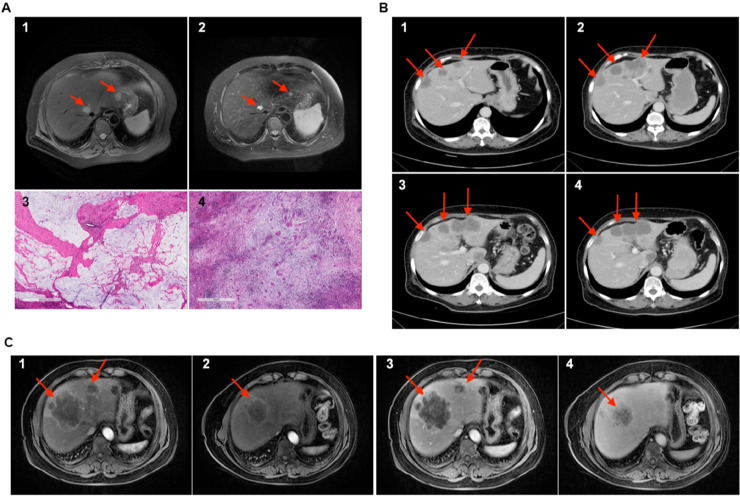
Patient NYZ113 has ulcerated moderately-differentiated adenocarcinoma of the rectum and further found to be accompanied by liver metastasis (pathological stage: pT3N1bM1a), and received rectal cancer resection. The corresponding i-CR model analysis predicted the patient to be hypersensitive to 5-FU+ Oxaliplatin treatment (Table 1). Clinically, after receiving 3 cycles of postoperative CAPOX regimen, the patient showed very good prognosis, as evidenced by the dramatic decrease of two tumor markers (CEA and CA19–9) and liver CT scan (Fig. 5A). Liver pathology indicated that one lesion was more than 90% tumor necrosis (Fig. 5A, 3) and another was pathological complete response (Fig. 5A, 4). No recurrence has been found in this patient during the follow-ups. In another example, patient NYZ132 was diagnosed with hepatic metastasis of sigmoid carcinoma (clinical stage: cT4aN2Ma). After sigmoid colon resection, the patient received 4 cycles of post-operative XELOX regimen. With the abdominal CT scan showed progression of liver metastasis lesions (Fig. 5B, 1 and 2), the chemotherapy regimen was changed to FOLFIRI. The patient showed favorable prognosis with the new regimen (Fig. 5B, 3 and 4), which was highly consistent with drug sensitivity results done with i-CR cultures (Table S3 and S4). In the case of patient NYZ080 (rectal cancer with hepatic metastasis), tests with i-CR culture indicated that the tumor was sensitive to FOLFIRI (Table S3 and S4). This result was verified by the clinical outcome. After three cycles of FOLFIRI regimen, nuclear magnetic resonance (NMR) gave an overall evaluation of PR (partial response). And the liver metastatic tumor was transformed from an unresectable lesion (Fig 5C, 1 and 3) to a borderline resectable lesion (Fig. 5C, 2 and 4).
Fig. 5.
Clinical results for CRC patients. A) the upper panel is the CT scan of patient NYZ113, the lower panel is the pathology staining (scale bar = 600 μm). B) CT scan of patient NYZ132. C) CT scan of patient NYZ080.
Furthermore, the results of i-CR drug sensitivity tests of six patients (NYZ53 (Ras wt;BRAF V600E mt; MSS), NYZ93 (Ras mt;BRAF V600E wt; MSS), NYZ94 (Ras wt;BRAF V600E wt; MSS), NYZ102 (Ras mt;BRAF V600E wt; MSS), NYZ109 (Ras mt;BRAF V600E wt; MSS), NYZ124 (Ras wt;BRAF V600E wt; MSS)) indicated that they were resistant to all the first-line therapeutic agents. Clinical responses proved this is the case. All six patients progressed quickly during the period of scheduled treatments. Figure S4 are two representatives of these patients (NYZ124 and NYZ109), showing rapid progression of the disease. Overall, with the exception of one patient (NYZ092), the clinical outcomes of eligible patients showed excellent consistency with the respective i-CR data. Among the i-CR models that we analyzed, as well as the parallel clinical treatment outcomes, we report that the i-CR platform holds 100% sensitivity, 85.7% specificity, 91.7% positive predictive value, and 100% negative predictive value in forecasting response to chemotherapy in patients (Student's t-test, p < 0.0001). The entire evaluation process can be seamlessly integrated into conventional and neoadjuvant chemotherapy programs.
Discussion
Human cancers are histologically complex and genetically diverse [33]. A good cancer model should be able to largely maintain the all-around characteristics of the human cancer [10,34]. Cancer cell lines and animal models are traditional key components of oncology research. However, the generation of cancer lines involves long-term cloning and adaptation of cloned cells to the culturing condition. They are very often unable to recapitulate the patient's tumor. PDX models are good at mimicking in vivo characteristics of the human tumor, but their application is limited by high cost and long duration [19]. Researchers have long been trying to develop ways to culture primary human tumor tissues. Progress has been made in technologies such as conditional reprogramming and organoid generation. Studies on CR system, especially the application with CRC and prostate cancer cells showed great potential of this technology in fighting cancer [23,26]. However, rapid expanding normal (or healthy) cells sometimes overshadow tumor cells’ drug response. The selective medium in i-CR system was developed based on the idea that genetically altered tumor cells have different growth requirement than normal epithelial cells. By changing compositions of the growth medium, we established a culture system that only allows the rapid growth of tumor cells. In the presented study we examined i-CR technology and evaluated its potential application in pre-clinical chemotherapy drug sensitivity test for colorectal cancer patients. The i-CR system was modified from the original CR method and the culturing conditions were optimized for primary tumor cells especially colorectal cancer. We noticed the success of generating an i-CR model correlates with the tumorigenicity of the disease specimen (Fig. S3). We categorized the main factors that affect the successful establishment of an i-CR model. Taking into consideration of the issues listed in Table 2, we showed that the success rate of i-CR to generate primary tumor models from the patient tumor tissues was on-par with other methods such as organoid preparation and PDX. Furthermore, we developed a selective medium, with which the tumor cells proliferated rapidly but the normal cells failed to do so. This allowed us to monitor unmasked drug response without the interference of the typically overgrown normal cells. Moreover, extended sequencing tests on the cultured i-CR cells and the paired tumor tissues indicated the genetic heterogeneity of the tumors was well maintained.
A persistent problem faced by researchers in vitro drug tests is how to score the drug response more precisely and accurately. Since the tests are done with cultured cells, how to relate the results with clinical outcomes is critical. To address this problem, we developed an algorism (drug sensitivity index, DSI) that overcomes the populational differences in their sensitivities to a drug by the cultured cells. Tumors are heterogeneic and each tumor harbors cells with different sensitivities to a drug treatment. It is clinically valuable to take into consideration of sensitivities of cultured tumor cells to different dosages of a drug. Therefore, we introduced DSI (drug sensitivity index) as the criteria when assessing the efficacy of a drug or drug combination in our in vitro drug screening. The mathematical calculation for DSI was based on the well-established “log-kill” model in vitro studies [29]. One further assumption was made that the distribution of cell population as a function of drug sensitivity is exponential, the greater part of the cells are highly sensitive to lower dosages in the sensitivity spectrum, the cells only sensitive to high dosages account to minor fractions of the whole population [29,35]. We then simplified the equation to make it more feasible for high-content drug screens without sacrificing the capacity of the concept.
Another key point in the in vitro sensitivity test is the effective drug concentrations in cell cultures could be far off from clinical regimens. To make the tests results more clinically relevant, we performed our tests at the human steady-state serum concentration of the drug as a starting point [27]. Steady-state serum concentration is reached when the concentration of the drug in the body stays consistent, usually during which the drug is given continuously or repeatedly. By using steady sate concentration in tissue culture, we can mimic the amount of exposure of the tumor cells to the drug. This approach worked particularly well in avoiding false negative results. Further modification and optimization of the drug concentration were done based on empirical evidence. The protocol for each drug regimen was then verified with PDX models and ultimately compared with clinical outcomes.
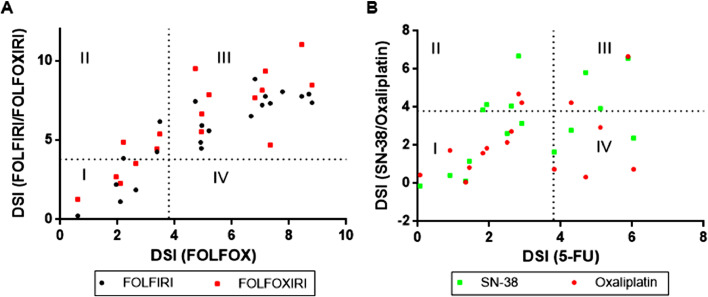
The choice of agents used in the traditional chemotherapy for CRC includes fluoropyrimidine or its analogs 5-Fluorouracil (5-FU) and capecitabine, oxaliplatin and irinotecan [5,6]. Over the past 20 years, combination chemotherapy or systemic chemotherapy, which uses two or more agents in various combinations and schedules, has shown to be superior to the single agents and are widely accepted in the clinical guidelines [6]. However, despite the improved efficacy, difficulties still exist in decision-making in clinical applications, mainly due to the complexity of the disease and the individualities of the patients. Especially for late stage patients, when the aim of the treatments shifts from cure to palliation, it becomes more important to avoid over-treatment [36,37]. Besides disease progression and adverse side effects, high medical cost is a significant problem for majority of the patients. Therefore, it is critical to select the best chemotherapy regimens for individual patients. To achieve this, an appropriate pre-clinical drug testing method is essential. In the presented study, we successfully cultured primary tumor cells from 20 late-stage CRC patients, performed in vitro chemotherapy drug tests and demonstrated the concordance between i-CR system and clinical outcomes. As shown in Fig. 6, drug responses for chemotherapy agents (Fig. 6A for combined therapies and Fig. 6B for single agents) were compared. The dash lines were theoretical drug sensitivity thresholds (DSI= 3.8, based on statistical analysis of previous studies, Figure S5). FOLFOX and FOLFIRI (Fig. 6A, black circles) showed similar sensitivity profiles, as quadrant I being the non-responders to both regimens and quadrant III being the responders. Quadrant II and IV are the ones sensitive to one of the two regimens [38], [39], [40]. Single agents, on the other hand, had larger discrepancies (Fig. 6B). Interestingly, FOLFOXIRI (Fig. 6A, red squares) did not show significant advantage over two-agent regimens in terms of drug response rate among the patients. This observation is contrary to published clinical data [41]. One conceivable reason is the small sample size. Another possibility is when we removed the original tumor samples that failed to establish i-CR models, a bias in drug responses toward different treatments may be introduced. In any event, further studies are needed to explore the full potential of the technology. Nevertheless, Fig. 6 clearly demonstrated the enormous clinical value of i-CR system. For patients in quadrant I, when none of the chemotherapy regimens were predicted as effective, alternative approaches should be considered as soon as possible. And for patients in quadrant II and IV, i-CR tests could be the guidance for selecting the best therapies. The ability for tumor cells to expand rapidly in i-CR culture permits timely clinical decisions. To the best of our knowledge, the i-CR system described here is the first for forecasting patients’ clinical responses to chemotherapies or targeted agents within 10–14 days. Although i-CR still cannot be applied directly to screening treatments related to tumor environment such as anti-angiogenic drugs and immunotherapy, the predictive value of the technology to identify chemo-resistant patients can be used to demonstrate feasibility of alternative therapies [26].
Fig. 6.
Quadrant maps of i-CR responses for CRC chemotherapy agents. The dashed lines are the putative sensitivity threshold set at DSI = 3.8.
Conclusion
i-CR platform was capable to test and optimize therapeutic regimens pre-clinically, study cancer cell biology, and model tumor re-emergence to identify new targeted therapeutics from an effective personalized medicine standpoint.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Peking University cancer hospital (approval no. 2017YJZ16). All specimens were collected from patients with written consent. All animal experiments were performed under sterile conditions at Percans Oncology Inc. specific-pathogen free facility and carried out in accordance with the National Institutes of Health guide for the care and use of laboratory animals.
Consent for publication
All authors have agreed to the publication of this manuscript.
Availability of data and materials
The datasets used in the current study are available from the corresponding author on reasonable request.
Funding
This work was supported the National Natural Science Foundation of China (81773214).
Author contributions
Yingjie Li:Writing - Original Draft, Writing- Reviewing and Editing, Project administration, Data Curation, Resources, Formal analysis, Investigation. Dagang Guo:Writing - Original Draft, Data Curation, Resources, Methodology, Investigation. Yingjie Li and Dagang Guo contributed equally to this manuscript. Yihong Zhang: Resources, Methodology. Lin Wang: Resources. Tingting Sun:Software, Formal analysis. Zhongwu Li: Validation, Visualization. Xiaoyan Zhang: Validation, Visualization. Shuai Wang:Validation, Visualization. Yiyou Chen:Funding acquisition, Project administration. Aiwen Wu:Conceptualization, Project administration, Supervision, Project administration, Funding acquisition.
Acknowledgments
We thank Center for Molecular Diagnostics, Peking University Cancer Hospital & Institute for the assistance in data collection and analysis. Part of the analysis was performed on the Computing Platform of School of Public Health, Peking University.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100935.
Appendix. Supplementary materials
References
- 1.Ferlay J., Soerjomataram I., Ervik M. International Agency for Research on Cancer; Lyon, France: 2013. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 11. [Google Scholar]
- 2.Rawla P., Sunkara T., Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 4.GBD 2017 Colorectal Cancer Collaborators The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol. Hepatol. 2019;4(12):913–933. doi: 10.1016/S2468-1253(19)30345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cremolini C., Schirripa M., Antoniotti C. First-line chemotherapy for mCRC—a review and evidence-based algorithm. Nat. Rev. Clin. Oncol. 2015;12(10):607–619. doi: 10.1038/nrclinonc.2015.129. [DOI] [PubMed] [Google Scholar]
- 6.Marques R.P., Duarte G.S., Sterrantino C. Triplet (FOLFOXIRI) versus doublet (FOLFOX or FOLFIRI) backbone chemotherapy as first-line treatment of metastatic colorectal cancer: a systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2017 doi: 10.1016/j.critrevonc.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Sveen A., Kopetz S., Lothe R.A. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat. Rev. Clin. Oncol. 2020;17(1):11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koi M., Carethers J.M. The colorectal cancer immune microenvironment and approach to immunotherapies. Fut. Oncol. 2017;13(18):1633–1647. doi: 10.2217/fon-2017-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dörr N.M., Bartels M., Morgul M.H. Current treatment of colorectal liver metastasis as a chronic disease. Anticancer Res. 2020;40(1):1–7. doi: 10.21873/anticanres.13921. [DOI] [PubMed] [Google Scholar]
- 10.Friedman A.A., Letai A., Fisher D.E., Flaherty K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer. 2015;15(12):747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandori C., Kemp C.J. Personalized cancer models for target discovery and precision medicine. Trends Cancer. 2018;4(9):634–642. doi: 10.1016/j.trecan.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt M.W. The influence of subclonal resistance mutations on targeted cancer therapy. Nat. Rev. Clin. Oncol. 2016;13:335–347. doi: 10.1038/nrclinonc.2015.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bousquet G. Patient-derived xenograft: an adjuvant technology for the treatment of metastatic disease. Pathobiology. 2016;83:170–176. doi: 10.1159/000444533. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Wang Y., Cao Z., Gao Y. Changes in the urinary proteome in a patient-derived xenograft (PDX) nude mouse model of colorectal tumor. Sci. Rep. 2019;9(1):4975. doi: 10.1038/s41598-019-41361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao H. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 16.Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18(7):407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 17.Aberle M.R., Burkhart R.A., Tiriac H. Patient-derived organoid models help define personalized management of gastrointestinal cancer. Br. J. Surg. 2018;105(2):e48–e60. doi: 10.1002/bjs.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020;13(1):4. doi: 10.1186/s13045-019-0829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrne A.T. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer. 2017;17:254–268. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 20.Bleijs M., van de Wetering M., Clevers H., Drost J. Xenograft and organoid model systems in cancer research. EMBO J. 2019;38(15) doi: 10.15252/embj.2019101654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X., Ory V., Chapman S. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012;180(2):599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Krawczyk E., Suprynowicz F.A. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 2017;12(2):439–451. doi: 10.1038/nprot.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timofeeva O.A., Palechor-Ceron N., Li G. Conditionally reprogrammed normal and primary tumor prostate epithelial cells: a novel patient-derived cell model for studies of human prostate cancer. Oncotarget. 2017;8(14):22741–22758. doi: 10.18632/oncotarget.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palechor-Ceron N., Krawczyk E., Dakic A. Conditional reprogramming for patient-derived cancer models and next-generation living biobanks. Cells. 2019;8(11) doi: 10.3390/cells8111327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Correa B.R.S., Hu J., Penalva L.O.F. Patient-derived conditionally reprogrammed cells maintain intra-tumor genetic heterogeneity. Sci. Rep. 2018;8(1):4097. doi: 10.1038/s41598-018-22427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Liao H., Zheng T. Conditionally reprogrammed colorectal cancer cells combined with mouse avatars identify synergy between EGFR and MEK or CDK4/6 inhibitors. Am. J. Cancer Res. 2020;10(1):249–262. [PMC free article] [PubMed] [Google Scholar]
- 27.Brunton, L.L., Hilal-Dandan, R. and Knollmann, B.C. Goodman & Gilman's the pharmacological basis of therapeutics (13th edition) McGraw-Hill Education ( 2018 )
- 28.Salic A., Mitchison T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakobsen A., Mortensen L.S. On the importance of sensitivity to the dose-effect relationship in chemotherapy. Acta Oncol. (Madr) 1997;36(4):375–381. doi: 10.3109/02841869709001283. [DOI] [PubMed] [Google Scholar]
- 30.Han S.-.W. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS ONE. 2013;8(5):e64271. doi: 10.1371/journal.pone.0064271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlachogiannis G., Hedayat S., Vatsiou A. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bousquet G., Janin A. Patient-derived xenograft: an adjuvant technology for the treatment of metastatic disease. Pathobiology. 2016;83(4):170–176. doi: 10.1159/000444533. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Kamb A. What's wrong with our cancer models? Nat. Rev. Drug Discov. 2005;4(2):161–165. doi: 10.1038/nrd1635. [DOI] [PubMed] [Google Scholar]
- 35.Yadav B., Pemovska T., Szwajda A. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. Jun 2014;5(4):51–93. doi: 10.1038/srep05193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marley A.R., Nan H. Epidemiology of colorectal cancer. Int. J. Mol. Epidemiol. Genet. 2016;7(3):105–114. [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J.H. Chemotherapy for colorectal cancer in the elderly. World J. Gastroenterol. 2015;21(17):5158–5166. doi: 10.3748/wjg.v21.i17.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Gramont A. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J. Clin. Oncol. 2000;18:2938–2947. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 39.Goldberg R.M. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J. Clin. Oncol. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 40.Douillard J.Y., Cunningham D., Roth A.D., Navarro M., James R.D., Karasek P., Jandik P., Iveson T., Carmichael J., Alakl M. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicenter randomized trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 41.Cremolini C. Upfront FOLFOXIRI plus bevacizumab and reintroduction after progression versus mFOLFOX6 plus bevacizumab followed by FOLFIRI plus bevacizumab in the treatment of patients with metastatic colorectal cancer (TRIBE2): a multicenter, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2020;21(4):497–507. doi: 10.1016/S1470-2045(19)30862-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the current study are available from the corresponding author on reasonable request.