Highlights
-
•
The molecular characteristics of MSI and MSS CRC tumors are quite different.
-
•
MSI CRC postoperative plasma had distinct mutational characteristics with MSS CRC.
-
•
The prognostic value of ctDNA in postoperative plasma of MSI CRC should be independently evaluated in MSI.
Keywords: Colorectal cancer, Microsatellite instability, Microsatellite stability, Surgery, Molecular residues
Abstract
The mutation in postoperative plasma (molecular residues) was an independently prognostic factor in colorectal cancer (CRC). The status of postoperative plasma mutation of microsatellite instability (MSI) CRC has not been systematically examined. In this study, we enrolled 30 MSI and 46 microsatellite stability (MSS) CRCs, and performed next generation sequencing on surgical tissues, postoperative plasma, and plasma during follow-up. Compared with MSS, MSI tumors had dissimilar genomic profiles, higher tumor mutation burden (TMB), and more frameshift mutations. In the postoperative plasma, more MSI CRCs were detected with tumor-derived mutations (77% in MSI vs 33% in MSS, p < 0.001). The numbers of postoperative mutations were proportional to MSI tissues (Spearman r = 0.47, p = 0.023), while not for MSS. More proportion of postoperative plasma samples of MSI CRCs harbored frameshift mutations than MSS (p = 0.007). For the follow-up plasma, 93% (14 out of 15) MSI CRCs harbored tumor-derived mutations; 33% (4/12) MSS were mutation-positive, lower than MSI (p = 0.003). Thus, considering that MSI CRC had extremely distinct mutational characteristics in tumor and postoperative plasma compared with MSS CRC, we propose that the prognostic value of molecular residue identification in postoperative plasma needs to be independently evaluated in MSI and MSS CRCs.
Introduction
Colorectal cancer (CRC), is a common malignancy and the fourth cause of cancer-related death worldwide [1,2]. CRC is genetically produced by microsatellite stability (MSS) and microsatellite instability (MSI) pathways. MSI type accounts for about 15%, characterized by frame-shift mutations and base-pair substitutions that are commonly found in short, tandemly repeated nucleotide sequences [3], [4], [5]. MSI is routinely caused by a deficiency of the DNA mismatch repair (MMR) system. In early-stage tumors, MSI is more frequent, accounting for around 20% of stage II, 12% of stage III [6], and 4% of stage IV CRCs [7].
Comprehensive sequencing studies have been recently performed on CRC. Through identifying genomic events, molecular subgroups of hypermutation, MSI-H/hypermutation, and MSS were defined for early-stage and resectable CRC [8], [9], [10]. Prior reports supported the favorable stage-adjusted prognosis of MSI-H compared to MSS CRC stage II/III patients [11,12]. MSI-H CRCs have been shown to contain a higher degree of immune infiltration [13], which carries prognostic significance [14]. Analysis of genomic data revealed that most MSI-H CRC harbored higher tumor mutation burden (TMB) [15] and increased numbers of neoantigens, correlating with immune infiltrates [16]. Therefore, disparate genomic characteristics and likely better outcomes of MSI CRC compared to their MSS counterparts were generally recognized.
Circulating tumor DNA (ctDNA), found in the peripheral blood [17], is a potential tool to characterize the genomic features of early-stage and advanced tumors [18]. In colon cancer, ctDNA harbor genomic alterations with comparable frequencies to tumor sequencing [19]. For patients treated with surgery, positive ctDNA status in postoperative plasma had the capacity of identifying those at high risk of recurrence. According to Tie et al., [20], postoperatively positive ctDNA detected using a 15-gene-panel in stage II colon cancer was an indicator of poor prognosis with the independence of other factors. ctDNA was postoperatively detected in only 1 out of 41 MSI CRCs in their study. Considering the complexity of molecular features, 15-gene-panel might not be sufficient to characterize the ctDNA status in the postoperative plasma of MSI patients.
In this study, a total of 76 patients with MSI and MSS CRC were analyzed. Next generation sequencing (NGS) using a 1021-gene-panel were performed on the surgical tissue, postoperative plasma, and follow-up plasma, in which tumor-derived mutations were tracked. We reported the significantly enriched altered genes in MSI compared with MSS tumor to further highlight the molecular distinction, and proposed that the higher ctDNA positive rate in MSI CRC might not be the biomarker for a worse outcome but probably provide the clues of higher immunogenicity retained by patients.
Materials and methods
Patients and samples
We recruited CRC patients initially diagnosed with primary CRC and treated with radical surgery in Sun Yat-sen University Cancer Center. Patients with a previous malignancy within the last 3 years were excluded. Surgical tissues were obtained. The plasma samples were collected one week after surgery and before adjuvant therapy (if necessary). Medical records were used to obtain the clinicopathologic characteristics, including sex, age, TNM stage, and treatment history.
Surgical tissues were frozen at −80 °C for the following DNA extraction. Peripheral blood samples were collected before radical surgery (preoperative), one week after surgery (postoperative), and 6 months after surgery (follow-up). At least 20 mL of peripheral blood was drawn into the EDTA Vacutainer tube (BD Diagnostics, Franklin Lakes, NJ, USA) and processed within 2 h. Plasma was separated by centrifugation at 1600 g for 10 min and transferred to new microcentrifuge tubes, then centrifuged at 16,000 g for 10 min to remove remaining cell debris. Peripheral blood lymphocytes (PBLs) from the first centrifugation were obtained for the extraction of germline DNA.
Identification of MMR status
MMR status was assessed by immunohistochemistry (IHC) for MLH1, MSH2, MSH6, and PMS2 proteins using standard protocols [21]. Primary monoclonal antibodies were MLH1 (clone G168–728, diluted 1:250; BD Biosciences Pharmingen, San Diego, CA), MSH2 (clone FE11, diluted 1:50; Oncogene Research Products, Cambridge, MA), MSH6 (clone GRBP.P1/2.D4, diluted 1:200; AbD Serotec, Raleigh, NC), and PMS2 (clone A16-4, diluted 1:200; BD Biosciences Pharmingen). Non-neoplastic colonic mucosa and colorectal tumors known to be deficient of MLH1, MSH2, MSH6, and PMS2 were used as external positive and negative controls, respectively. Tumors showing loss of expression in one or more of these proteins were considered dMMR, and those showing normal expression of the proteins were mismatch repair-proficient (pMMR).
DNA extraction and quality control
DNA was extracted from tissue samples using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). DNA concentration was measured using a Qubit fluorometer (Invitrogen, Carlsbad, CA USA) and the Qubit dsDNA HS (High Sensitivity) Assay Kit.
Cell-free circulating DNA (cfDNA) was isolated from 4.0–8.0 mL plasma using QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany), and PBL DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) (Qiagen, Hilden, Germany), as previously described [22]. DNA concentration was measured using the Qubit 3.0 fluorometer and the Qubit dsDNA HS (High Sensitivity) Assay Kit (Thermo Fisher Scientific Inc., Carlsbad, CA, USA). cfDNA sample was analyzed on the Agilent 2100 BioAnalyzer using the Agilent High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA).
Target capture and next-generation sequencing
The custom-designed biotinylated oligonucleotide probes (Roche NimbleGen, Madison, WI, USA) covering ∼1.0 Mbp coding region of genomic sequence of 1021 genes frequently mutated in CRC and other common solid tumors was designed. For library construction, 1.0 μg of PBL and tissue DNA were sheared to 300-bp fragments with a Covaris S2 ultrasonicator (Covaris, Woburn, MA, USA). For cfDNA, 20–80 ng sample was used for library construction. Libraries were constructed using the KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA). Captured libraries were measured using an Agilent 2100 Bioanalyzer and an Applied Biosystems 7500 real-time PCR system (Thermo Fisher Scientific Inc., Carlsbad, CA, USA). DNA sequencing was performed on the HiSeq3000 Sequencing System (Illumina, San Diego, CA, USA) with 2 × 100 bp paired-end reads.
Sequencing data analysis
Terminal adaptor sequences and low-quality reads were removed from raw sequencing data. The reads were aligned to the human genome build GRCh37 using BWA (a Burrows-Wheeler aligner) [23]. To mark PCR duplicates, Picard tools (http://broadinstitute.github.io/picard/) were used. Single nucleotide variation (SNV) and small insertion and deletion (Indel) were called using MuTect (version 1.1.4) [24] and GATK (version 3.4–46-gbc02625) [25], respectively. Preoperative PBL sequencing results were used to filter germline variations. All candidate somatic mutations identified by the bioinformatics pipeline were manually reviewed in the Integrative Genomics Viewer (IGV) [26] through assessing the quality of base calls, the mapping quality of the reads, and the overall read depth at each mutation site. Mutations were annotated to genes by ANNOVAR software [27] to identify the mutated protein-coding position and filtered intronic and silent changes. Variant allele fraction (VAF) = sequencing read count of altered alleles / (sequencing read count of reference alleles + sequencing read count of altered alleles) × 100%. For tissue, a mutation was identified according to these standards: VAF ≥ 1.0%, and at least 5 high-quality reads (Phred score ≥ 30, mapping quality ≥ 30, and without paired-end reads bias). For postoperative and follow-up plasma, tumor-derived ctDNA mutations were tracked and determined satisfying the condition of at least 2 high-quality reads (Phred score ≥ 30 and mapping quality ≥ 30). MSIsensor score analysis was performed for identification of MSI status for tumors [28].
Statistical analysis
Data were analyzed using R Package (Version 3.3.0) or Prism 5.0 (Graph Pad Software Inc., La Jolla, CA). The Fisher's exact test was used to compare proportions between two groups. Mann-Whitney test was used for TMB comparison in different groups. An unbiased analysis of enriched prevalence of altered genes between two groups was examined by plotting the log2 (odds ratio) and log2 (p value). Correlations of mutational prevalence were examined by the Spearman method. All reported p values were two-sided, and p < 0.05 was considered statistically significant.
Results
Clinicopathological characteristics and samples collection
We enrolled 76 CRC patients from February 2017 to December 2019 (Table 1 and Supplement TablesS1). Patients were initially diagnosed with stage I-III tumors and followed by radical surgery. A proportion of patients (n = 40) received standard adjuvant chemotherapy. Tumor tissues (n = 76) and post-operatively matched plasma samples (n = 76) were collected from each patient. Follow-up plasma samples of 27 patients after 6 months were collected.
Table 1.
Clinical characteristics of the CRC patients with MSI and MSS status.
| Patient Characteristics | All (n = 76) No. | MSI (n = 30) No. | MSS (n = 46) No. | p | |||
|---|---|---|---|---|---|---|---|
| Median age, years (range) | 58 (21–85) | 57 (21–76) | 62 (33–85) | 0.221 | |||
| Gender | 0.166 | ||||||
| Male | 43 | 57% | 20 | 67% | 23 | 50% | |
| Female | 33 | 43% | 10 | 33% | 23 | 40% | |
| Tumor site | 0.104 | ||||||
| Colon | 58 | 76% | 26 | 87% | 32 | 70% | |
| Rectum | 18 | 24% | 4 | 13% | 14 | 30% | |
| Stage | 0.09 | ||||||
| I | 10 | 13% | 2 | 7% | 8 | 17% | |
| II | 32 | 42% | 17 | 57% | 15 | 33% | |
| III | 34 | 45% | 11 | 37% | 23 | 50% | |
| Differentiation | 0.576 | ||||||
| Moderate | 59 | 78% | 22 | 67% | 37 | 80% | |
| Poor | 17 | 22% | 8 | 33% | 9 | 20% | |
| Adjuvant chemotherapy | 0.242 | ||||||
| Yes | 40 | 53% | 13 | 43% | 27 | 59% | |
| No | 36 | 47% | 17 | 57% | 19 | 41% | |
†CRC: colorectal cancer; MSI: microsatellite instability; MSS: microsatellite stability.
We purposely recruited 30 MSI patients in this cohort. The median diagnostic age of these MSI patients was 57 years old and the majority (67%) were male. Eighty-seven percent (n = 26) tumors occurred in the colon, and 93% (n = 28) were at stage II or III. More than half (n = 17) received radical operation without adjuvant chemotherapy. MSS patients had similar clinical features with MSI were recruited simultaneously (Table 1). Briefly, 70% were with colon tumor, half of the tumors were at stage III, and 59% were treated with surgery followed by adjuvant chemotherapy.
Comparison of somatic mutations in MSI and MSS tumors
We successfully performed high-throughput sequencing on 76 surgical tissues with a median depth of 985 × (range from 380 to 2326 ×) without duplications. In total, 2755 non-silent somatic mutations (SNV and small Indels) were detected. Each patient harbored at least one mutation, with a median of 73 (from 18 to 194) mutations in MSI and 6 (2 to 157) in MSS group. MSI tumors had remarkably higher TMB than MSS (p < 0.001) (Fig. 1A).
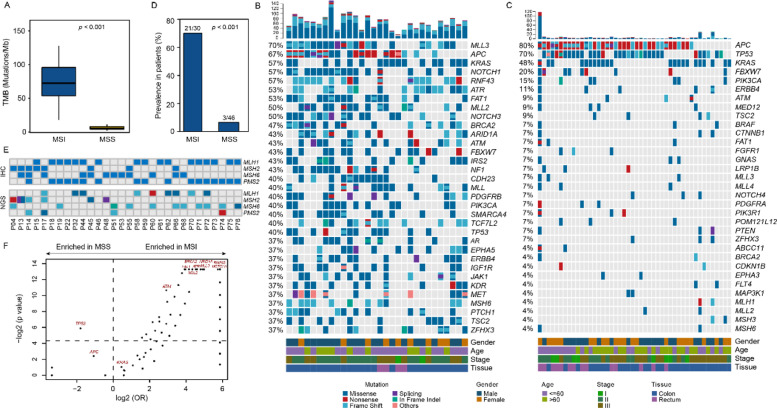
Fig. 1.
Genomic landscape of CRC tissue samples. (A) Tumor mutation burden (TMB) of MSI (microsatellite instability) and MSS (microsatellite stability) CRCs. TMB comparisons between two groups are performed by Mann-Whitney test. (B) Somatic mutation profile detected in tissues of MSI CRCs. The top graph shows numbers of alteration of each sample. Genes with somatic alterations occurring in more than eleven samples are shown in the middle heatmap. Different alteration types are indicated by different colors. Altered frequency in patients of each gene is shown on the left. Gender, age, stage, and lesion location are shown at the bottom. (C) Somatic mutations detected in tissues of MSS CRCs. (D) Frequency of altered classical MMR-related genes including MLH1, MSH2, MSH6, and PMS2. The comparion of prevalence between MSI and MSS groups is performed by Fisher's exact test. (E) The IHC (immunohistochemistry) and altered status of MMR-related genes in MSI CRCs. The top heatmap demonstrates the expression status of particular protein detected by IHC. The somatic alterations are showed in bottom heatmap, in which different colors indicate different mutational types. (F) Comparison of prevalence of all altered genes occurring in MSI or MSS patients. odd ratios (OR) and p values were calculated by Fisher's exact test. The spots above the horizontal dashed line in the middle of figure are mutated genes with significant differences. The longitudinal dashed line separates genes enriched in one group, as indicated by arrows at the top.
In the MSI tumors, MLL3 (affecting 70% of MSI patients), APC (67%), NOTCH1 (57%), RNF43 (57%), KRAS (57%), ATR (53%), and FAT1 (53%) were the most common mutated genes (Fig. 1B). In the MSS group, the recurrently mutated genes were APC (80%), TP53 (70%), KRAS (48%), FBXW7 (20%), PIK3CA (15%), ERBB4 (10%), and ATM (9%) which were commonly known as drivers in CRC (Fig. 1C). MLL3, the top mutated gene detected in MSI tumors, was enriched compared with MSS with statistical significance (p <0.001). Besides, somatic mutations in classical MMR-related genes including MLH1, MSH2, MSH6, and PMS2 were significantly enriched in the MSI group (p <0.001, Fig. 1D). Somatic mutations examined by NGS and loss of protein expression detected by IHC of classical MMR-related genes in MSI CRCs were showed in Fig. 1E. Mutations in both groups were comprehensively compared and numerous differentially mutated genes were identified, showing the dissimilarly mutational features between MSI and MSS patients (Fig. 1F).
Comparison of mutational types between MSI and MSS tumors
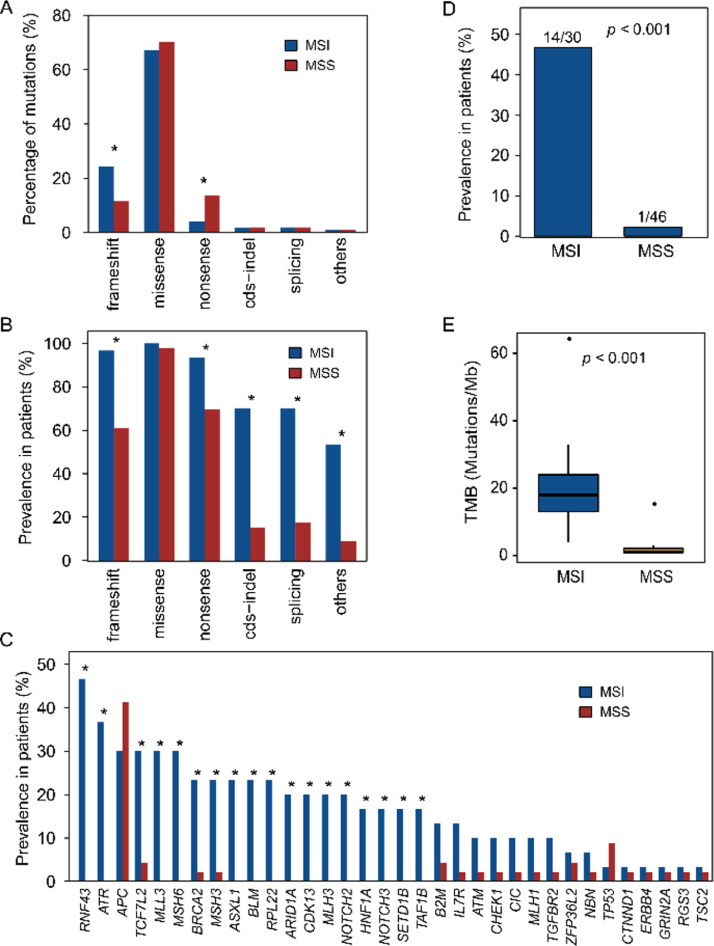
We next compared the mutational types between MSI and MSS tumors. Missense (67.1% of 2277), frameshift (24.3%), and nonsense (4.1%) were most popular in MSI tumors. MSI tumors had similar proportion of missense (67.1% vs 70.3%, p = 0.200), more frameshift (24.3% vs 11.5%, p < 0.001), and less nonsense mutations (4.1% vs 13.6%, p < 0.001) when compared with MSS tumors (Fig. 2A).
Fig. 2.
Mutational type between MSI and MSS tumor tissues. (A) Proportion of a variety of mutations detectable in MSI compared with MSS CRCs. (B) Differences in prevalence of mutations in patients between MSI and MSS CRCs. (C) Pervalence of genes with higher frameshift mutation frequency in MSI versus MSS. in A-C, asterisk above the bar indicates significant difference between two groups with p value less than 0.05 determined by Fisher's exact test. (D) Comparison of frequency in MMR-related genes harboring frameshift mutation between MSI and MSS patients. (E) TMB of frameshift mutations in MSI tumors compared with MSS tumors (Mann-Whitney test).
Ninety-seven percent of MSI patients harbored frameshift mutations (Fig. 2B), and RNF43 (affecting 47% of MSI patients), ATR (37%), APC (30%), MLL3 (30%), and MSH6 (30%) were the most frequent genes with frameshift mutations (Fig. 2C). Forty-seven percent of MSI tumors had somatic frameshift mutations in classical MMR-related genes (Fig. 2D). In MSS tumors, 61% had frameshift mutations (lower than MSI tumors, p < 0.001) (Fig. 2B), with most common genes of APC (affecting 41% of MSS patients) and TP53 (9%). Only one (2%) patient with MLH1 but no other MMR-related genes were found with frameshift mutations (p < 0.001) (Fig. 2D). In patients harboring frameshift mutations, compared with MSS, MSI tumors had significantly higher frameshift mutations burden (median 18 vs 1 mutation/Mb, p <0.001) (Fig. 2E).
Mutations in postoperative plasma of MSI and MSS patients
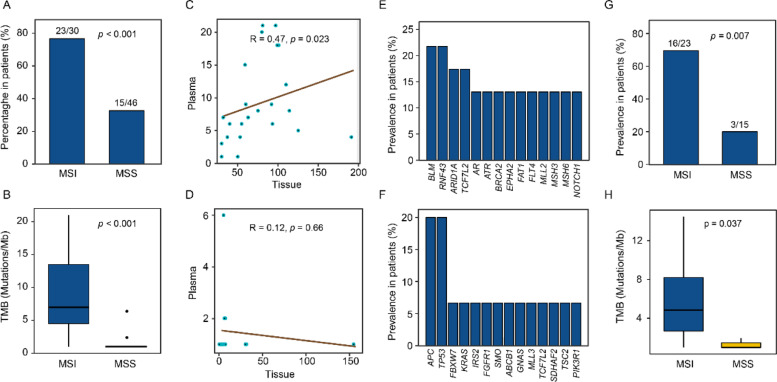
Somatic mutations derived from tumor tissues were tracked in postoperative plasma samples. A median sequencing depth of 2538 × (range from 956 to 5136 ×) without duplications was reached. For all patients, postoperative mutations were detected in 50% (38/76) of the plasma samples. Of these, 77% (23/30) of MSI and 32.6% (15/46) of MSS patients were found with tumor-derived mutations. The mutational detection rate of MSI patients was significantly higher than MSS (p < 0.001) (Fig. 3A). In mutation-positive samples, a median of 7 (1 to 21) mutations detected in MSI was higher than 1 (1 to 6) mutation in MSS (p <0.001) (Fig. 3B). We found that the number of mutations in plasma had a significant positive correlation with that in the paired tissues for the mutation-positive samples. Especially, the strong correlation was found in MSI (Spearman r = 0.47, p = 0.023), rather than in MSS group (Spearman r = 0.12, p = 0.66) (Fig. 3C and D). These results indicated that MSI patients had a higher proportion of mutation-positive and higher mutation burden in postoperative plasma.
Fig. 3.
Mutations detected in postopritive plasma. (A) The mutational detection rate in postoperative plasma of MSI and MSS patients. (B) Comparison of TMB in postopritive plasma between MSI and MSS CRCs (Mann-Whitney test). (C) Correlation of TMB between tissue and postoperative plasma in MSI patients. (D) Correlation of TMB between tissue and postoperative plasma in MSS patients. In C and D, Spearman r and p values are showed. (E) Genes with higher mutational frequency detected in MSI patients are presented. (F) Genes with higher mutational frequency detected in MSS patients are showed. (G) Percentage of postoperative samples harboring frameshift mutations in MSI patients compared with MSS patients (Fisher's exatct test). (H) Comparison of frameshift mutation burden between MSI and MSS postoperative plasmas (Mann-Whitney test).
In addition, recurrently mutated genes including BLM (5/23), RNF43 (5/23), ARID1A (4/23), and TCF7L2 (4/23) were identified in MSI patients (Fig. 3E), while APC (3/15), TP53 (3/15), FBXW7(1/15), and KRAS (1/15) genes in MSS were detected (Fig. 3F). Seventy percent (16/23) of mutation-positive plasma samples in the MSI group harbored frameshift mutations, with a higher mutation-positive proportion than MSS (3/15) (p = 0.007) (Fig. 3G). The frameshift mutation burden of MSI plasma was also higher than MSS (5 vs 1 mutation, p = 0.037) (Fig. 3H). These results suggested that there were differences in the mutational characteristics of postoperative plasma between the two groups.
Mutation tracking in plasma during follow-up
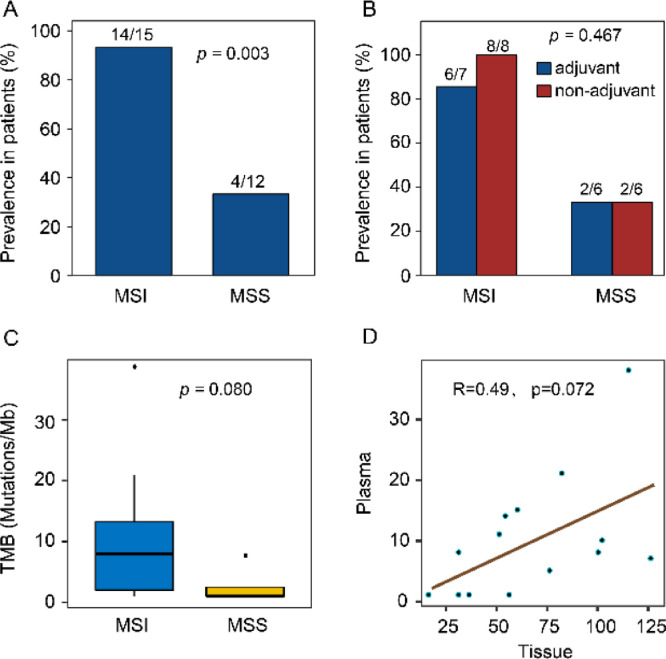
We collected plasma samples during follow-up from 15 MSI and 12 MSS CRCs for mutation tracking. A median depth of 2136 × (range from 872 to 4398 ×) without duplications was reached. Seven (47%) patients in MSI and 6 (50%) in the MSS group had received adjuvant chemotherapy with standard duration. In the MSI group, 14 (93%) samples were detected with tumor-derived mutations, with a median of 8 (range: 1–38) mutations (Fig. 4A-C). The number of mutations was marginally proportional between tumor and follow-up plasma (Spearman r = 0.49, p = 0.072) (Fig. 4D). Eighty-six percent (12/14) of MSI CRCs were detected with frameshift mutations. Among the 12 MSS samples, 4 (33%) with tumor-derived mutations were identified, with a lower positive proportion (Fisher's exact test, p = 0.003), and possibly fewer mutations (median: 1, Mann-Whitney test, p = 0.080) than MSI group (Fig. 4A and C).
Fig. 4.
Mutation tracking in plasma during follow-up. (A) Propotion of follow-up plasma samples with tumor-derived mutations in MSI and MSS patients. (B) The mutational detection rate in follow-up plasma of patients with and without postoperative adjuvant treatment. (C) TMB of follow-up plasma in MSI and MSS CRCs (Mann-Whitney test). (D) Correlation of TMB between tissue and follow-up plasma in MSI patients. Spearman r and p value are showed.
Four (1 MSI and 3 MSS) out of 76 patients were suffered progressed-disease (PD) within one year (median follow-up time was 28.6 months ranging from 2.5 to 35.9 months). Of the 3 MSS CRCs, 1 was found with recurrence in liver, 1 with lung, and 1 with multiple metastases. All of them were detected with ctDNA in postoperative plasma. Among the MSI patients (n = 23) harboring postoperatively tumor-derived mutations, only one (4%) experienced PD at the liver.
Discussion
Previous studies have proved the heterogeneity of CRC in terms of the molecular basis and clinical results. CRC was genetically grouped by MSI and MSS type according to the specific genomic characteristics [29]. MSI patients with operable colon cancers have a better prognosis than patients with MSS [30]. For advanced CRC, immune checkpoint inhibitors (ICIs) have been approved for use in MSI patients, while MSS CRCs showed inferior immunotherapy results [31]. We have further proved that the distinction of mutational characteristics of primary tumors in MSS and MSI CRCs. Although mutation profile analysis revealed that the prevalence of APC and KRAS was similar between the MSS and MSI groups, numerous genes, e.g., MLL3, NOTCH1, FBXW7, PIK3CA, and ERBB4, were enriched in MSI patients. Thus, under the principle of precision treatment, patients with MSI and MSS need to be treated differently.
The molecular features used for prognostic stratification in the MSI group have not yet been developed. In resected stage III patients receiving adjuvant FOLFOX, KRAS mutations were independently associated with shorter outcomes in MSS but not in MSI CRCs [32]. Those recurrently mutated and significantly enriched genes should be examined for the associations with resectable MSI CRC outcomes. For example, NOTCH pathway, including NOTCH1, NOTCH2, NOTCH3, NOTCH4, FBXW7, and CREBBP, which enriched in MSI CRCs, plays an extensive role in the immune system and operates at various levels by acting in conjunction with defined immunological signals such as cytokines, T cell antigen receptor and co-stimulatory receptor-mediated signaling [33]. In esophageal squamous cell carcinoma, NOTCH1 mutant patients had a better outcome than those individuals without mutations [34]. Somatic mutations in chromatin-regulating genes MLL, MLL2, MLL3, and ARID1A in 20% of pancreatic cancers had the associations with improved survival [35]. We found these genes were all enriched in the MSI group, especially for the most frequent MLL3, consistent with another study [36]. According to our observations, alterations in NOTCH pathway and chromatin-regulating genes were more common in the MSI CRCs, suggesting the interaction between these genes and MSI. Despite the lack of direct evidence, these clues might lead to the potential association between this interaction and a better prognosis in the MSI patients. In our study, however, one limitation is that we could not investigate the association between the mutational profile and outcomes of patients, due to the rare PD patients during the follow-up.
MSI CRCs had different mutational types in genes compared with MSS, with more frameshift and fewer nonsense mutations. Frameshift caused by small Indel mutations related to MSI status could develop a novel open reading frame and produce a large quantity of neoantigenic peptides. Analysis of tumor-specific neoantigens showed that enrichment of Indel mutations for high-affinity binders was three times that of non-synonymous SNV mutations [37]. Therefore, as the key substrate for the generation of anticancer immunity, more abundant frameshift mutations could be related to the stronger immunogenicity of MSI tumors.
The study of ctDNA in early-stage MSI CRC should not be ignored. Tie et al. analyzed 40 MSI tissues using a panel of 15 genes, and investigated one of the TP53, APC, or KRAS mutations in postoperative plasma to identify 1 patient with postoperative ctDNA [20]. Thus, a relatively low level of ctDNA positive rate (2.5%) was reported, statistically comparable to the positive rate of the overall population (7.9%, 14 out of 178). In our cohort, ctDNA positive (with at least one tumor-derived mutation) rate in postoperative plasma in MSI CRCs was significantly higher than that in MSS patients, and postoperative mutation burden was also higher than MSS plasma. Considering the mutational discrepancy between MSI and MSS CRCs, we propose that a comprehensive panel is more suitable for the examination of postoperative plasma ctDNA in MSI CRCs.
For early-stage CRC, tumor-derived mutations in postoperative plasma is an independent indicator of the worse outcome in patients undergoing surgery. The value of postoperative plasma ctDNA in the prediction of recurrence after surgical treatment of breast cancer, lung cancer, and colorectal cancer has been reported [20,[38], [39], [40]]. Among the 40 MSI patients reported by Tie et al. [20], the only one with ctDNA remained recurrence-free at 12 months follow-up, while the disease information of others could not be obtained. Therefore, the prognostic value of postoperative ctDNA in MSI CRCs was not elucidated. A large number of sequencing reads carrying tumor-specific mutations in postoperative plasma before adjuvant chemotherapy indicated the molecular residues of the primary MSI tumor in our study. We further found the high proportional frameshift mutations in postoperative plasma of MSI CRC. It is currently unclear how the MSI status significantly influences the prognosis of stage III CRCs, which represented a great proportion of the study population. Based on the substantial pieces of evidence of the better surgery prognosis of MSI CRCs, thus, we herein speculated that these molecular residues might not be an indicator of the worse outcome and even was probably the signal of residual immunogenicity, which might be associated with the better prognosis of MSI CRC. Due to the numbers of MSS and MSI patients who experienced recurrence were extremely small to derive any safe conclusions, this hypothesis requires further fundamental research of a large-scale cohort and long-term follow-up. Interestingly, during the 6 months of follow-up regardless of whether adjuvant chemotherapy was treated or not, the tumor-derived mutant ctDNA was detected in the majority of MSI rather than MSS patients. One feasible explanation for the high ctDNA level of chemo-treated patients is that the chemotherapy had less effect in reducing the number of mutated clones due to a higher baseline burden in the MSI.
In summary, we recruited MSI CRCs, collected their surgical tissue and postoperative plasma samples, and compared their clinical features and molecular characteristics in both tissues and plasmas with MSS CRCs, provided the preliminary insights into the postoperative ctDNA status of MSI CRCs. Our research reminds researchers that the postoperative ctDNA status of MSI may not be regarded as a poor prognostic factor as in MSS.
Conclusions
MSI CRC has distinct mutational characteristics in tumor and postoperative plasma compared with MSS CRC. The prognostic value of ctDNA in postoperative plasma of MSI CRC should be independently evaluated in MSI.
Author contributions
Liren Li conceived the study and designed the experiments. Wenhao Zhou prepared the research materials. Qian Li and Pansong Li did the bioinformatic analysis and performed initial exploratory analysis. Ling Yang and Xin Yi provided insight in methodological approaches and analysis. Liren Li and Xuefeng Xia supervised the study. All authors drafted the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81872267), and Natural Science Foundation of Guangdong Province (No. 2018A030313542)
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Research Ethics Committee of the Sun Yat-sen University Cancer Center (NO. B2019-068-01). We have received consents from all individual participants. The consent forms will be provided upon request.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100945.
Contributor Information
Liren Li, Email: lilr@sysucc.org.cn.
Wenhao Zhou, Email: zhouwh@sysucc.org.cn.
Qian Li, Email: liqian@geneplus.org.cn.
Pansong Li, Email: lips@geneplus.org.cn.
Ling Yang, Email: yangl@geneplus.org.cn.
Xuefeng Xia, Email: xiaxf@geneplus.org.cn.
Xin Yi, Email: yixin@geneplus.org.cn.
Desen Wan, Email: wands@sysucc.org.cn.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fedewa S.A., Ahnen D.J., Meester R.G.S., Barzi A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Ionov Y., Peinado M.A., Malkhosyan S., Shibata D., Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 4.Gryfe R., Kim H., Hsieh E.T.K., Aronson M.D., Holowaty E.J., Bull S.B. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342(2):69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 5.Pinol V., Castells A., Andreu M., Castellvi-Bel S., Alenda C., Llor X. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293(16):1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 6.Roth A.D., Tejpar S., Yan P., Fiocca R., Dietrich D., Delorenzi M. Stage-specific prognostic value of molecular markers in colon cancer: results of the translational study on the PETACC 3-EORTC 40993-SAKK 60-00 trial. J. Clin. Oncol. 2009;27(15_suppl):4002. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 7.Koopman M., Kortman G.A., Mekenkamp L., Ligtenberg M.J., Hoogerbrugge N., Antonini N.F. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br. J. Cancer. 2009;100(2):266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muzny D.M., Bainbridge M.N., Chang K., Dinh H., Drummond J., Fowler G. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donehower L.A., Creighton C.J., Schultz N., Shinbrot E., Chang K., Gunaratne P.H. MLH1 -silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J. Pathol. 2013;229(1):99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Sethi N.S., Hinoue T., Schneider B.G., Cherniack A.D., Sanchez-Vega F. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33(4):721–735. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gryfe R., Kim H., Hsieh E.T.K., Aronson M., Holowaty E.J., Bull S.B. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342(2):69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 12.Saridaki Z., Souglakos J., Georgoulias V. Prognostic and predictive significance of MSI in stages II/III colon cancer. World J. Gastroenterol. 2014;20(22):6809–6814. doi: 10.3748/wjg.v20.i22.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jass J.R., Do K.A., Simms L.A., Iino H., Wynter C.V.A., Pillay S.P. Morphology of sporadic colorectal cancer with DNA replication errors. Gut. 1998;42(5):673–679. doi: 10.1136/gut.42.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galon J., Costes A., Sanchezcabo F., Kirilovsky A., Mlecnik B., Lagorcepages C. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 15.Fabrizio D.A., George T.J., Jr, Dunne R.F., Frampton G., Sun J., Gowen K. Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J. Gastrointest. Oncol. 2018;9(4):610–617. doi: 10.21037/jgo.2018.05.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakis M., Mu X.J., Shukla S.A., Qian Z.R., Cohen O., Nishihara R. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15(4):857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leon S.A., Shapiro B., Sklaroff D.M., Yaros M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 18.Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 19.Strickler J.H., Loree J.M., Ahronian L.G., Parikh A.R., Niedzwiecki D., Pereira A.A.L. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2017;8(2):164–173. doi: 10.1158/2159-8290.CD-17-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016;8(346) doi: 10.1126/scitranslmed.aaf6219. 346ra92-ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J. Mol. Diagn.: JMD. 2008;10(4):293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X., Chu Y., Zhang R., Han Y., Zhang L., Fu Y. Technical validation of a next-generation sequencing assay for detecting clinically relevant levels of breast cancer–related single-nucleotide variants and copy number variants using simulated cell-free DNA. J. Mol. Diagn. 2017;19(4):525–536. doi: 10.1016/j.jmoldx.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis toolkit: a map reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G. Integrative genomics viewer. Nat. Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids. Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu B., Ye K., Zhang Q., Lu C., Xie M., McLellan M.D. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guinney J., Dienstmann R., Wang X., De Reynies A., Schlicker A., Soneson C. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parc Y. Prognostic significance of microsatellite instability determined by immunohistochemical staining of MSH2 and MLH1 in sporadic T3N0M0 colon cancer. Gut. 2004;53(3):371–375. doi: 10.1136/gut.2003.019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le D.T., Uram J.N., Wang H., Bartlett B., Kemberling H., Eyring A. Programmed death-1 blockade in mismatch repair deficient colorectal cancer. J. Clin. Oncol. 2016;34:103. [Google Scholar]
- 32.Taieb J., Malicot K.L., Shi Q., Penaultllorca F., Bouche O., Tabernero J. Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J. Natl. Cancer Inst. 2017;109(5):1–12. doi: 10.1093/jnci/djw272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leong K.G., Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107(6):2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 34.Qin H.D., Liao X.Y., Chen Y.B., Huang S.Y., Xue W.Q., Li F.F. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am. J. Hum. Genet. 2016;98(4):709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sausen M., Phallen J., Adleff V., Jones S., Leary R.J., Barrett M.T. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015;6:7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe Y., Castoro R.J., Kim H.S., North B., Oikawa R., Hiraishi T. Frequent alteration of MLL3 frameshift mutations in microsatellite deficient colorectal cancer. PLoS ONE. 2011;6(8):e23320. doi: 10.1371/journal.pone.0023320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turajlic S., Litchfield K., Xu H., Rosenthal R., McGranahan N., Reading J.L. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18(8):1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015;7(302) doi: 10.1126/scitranslmed.aab0021. 302ra133-302ra133. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhuri A.A., Chabon J.J., Lovejoy A.F., Newman A.M., Stehr H., Azad T.D. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7(12):1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinert T., Henriksen T.V., Christensen E., Sharma S., Salari R., Sethi H. Analysis of plasma cell-free DNA by ultra-deep sequencing in patients with stages I to III colorectal Cancer. JAMA Oncol. 2019;5(8):1124–1131. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.