Highlights
-
•
NGS-based clinical studies have reported detection of clinically relevant alterations in ∼30% of patients.
-
•
To increase the detection of potential targets, we integrated whole-exome sequencing with a multiplex PCR-based NGS assay for fusion detection.
-
•
The targeted transcriptome sequencing was performed using a very low RNA input from archival cancer tissues.
-
•
With this integrated approach, we demonstrate a significant increase in detection of targetable genomic alterations in cancer patients.
Keywords: Anchored multiplex PCR-based next-generation sequencing, Whole-exome sequencing, RNA Sequencing, Novel fusion, Oncogenic
Abstract
Background
Frequency of clinically relevant mutations in solid tumors by targeted and whole-exome sequencing is ∼30%. Transcriptome analysis complements detection of actionable gene fusions in advanced cancer patients. Goal of this study was to determine the added value of anchored multiplex PCR (AMP)-based next-generation sequencing (NGS) assay to identify further potential drug targets, when coupled with whole-exome sequencing (WES).
Methods
Selected series of fifty-six samples from 55 patients enrolled in our precision medicine study were interrogated by WES and AMP-based NGS. RNA-seq was performed in 19 cases. Clinically relevant and actionable alterations detected by three methods were integrated and analyzed.
Results
AMP-based NGS detected 48 fusions in 31 samples (55.4%); 31.25% (15/48) were classified as targetable based on published literature. WES revealed 29 samples (51.8%) harbored targetable alterations. TMB-high and MSI-high status were observed in 12.7% and 1.8% of cases. RNA-seq from 19 samples identified 8 targetable fusions (42.1%), also captured by AMP-based NGS. When number of actionable fusions detected by AMP-based NGS were added to WES targetable alterations, 66.1% of samples had potential drug targets. When both WES and RNA-seq were analyzed, 57.8% of samples had targetable alterations.
Conclusions
This study highlights importance of an integrative genomic approach for precision oncology, including use of different NGS platforms with complementary features. Integrating RNA data (whole transcriptome or AMP-based NGS) significantly enhances detection of potential targets in cancer patients. In absence of fresh frozen tissue, AMP-based NGS is a robust method to detect actionable fusions using low-input RNA from archival tissue.
Introduction
Next generation sequencing (NGS) has revolutionized the field of molecular cancer diagnostics, playing a pivotal role in detecting known and novel targetable genomic alterations in patients with cancer [1], [2], [3]. In the era of precision oncology, ongoing efforts are focused on detecting therapeutic targets through various assays analyzing the genome and transcriptome. NGS-based clinical trials have reported clinically relevant alterations in ∼30% of patients with advanced cancer, but only a subset who receive targeted treatment show an objective benefit in overall survival [4,5].
In order to expand the landscape of therapeutic targets in difficult tumors, transcriptomic and epigenomic analysis becomes necessary. Yet, it is technically challenging if fresh frozen tissue is unavailable [6,7]. There are recent efforts to analyze whole transcriptomes from formalin-fixed paraffin-embedded (FFPE) tissue with encouraging results, but it may take some time for these assays to become clinically available [7,8]. Targeted NGS-based platforms that utilize FFPE can detect targetable fusions but knowledge of both fusion partners is required, and they do not detect novel fusion partners [9,10].
Gene fusions have been recognized as oncogenic drivers, in addition to their role as diagnostic markers in certain tumor types [11]. These fusions result in altered function of involved genes either by amplifying the oncogenic partner or inactivating the tumor suppressor [12]. Identification of potential actionable fusions complements the targets detected by whole exome analysis, as fusions and oncogenic driver mutations are typically mutually exclusive [12]. For instance, in non-small cell lung cancers harboring EML4-ALK fusion, EGFR mutation will rarely be detected, and both of these molecular alterations have FDA-approved drugs [13], [14], [15]. In a recent study, it was determined by RNA-seq from fresh frozen tumor samples that 37% of 500 advanced metastatic pan-cancer patients harbored a putative driver fusion, or an inactivating fusion in a tumor suppressor gene [16].
Our clinical whole-exome sequencing (WES) assay (EXaCT-1), a test approved by the Department of Health at New York State (NYS-DOH ID#43032), detects somatic mutations, indels and copy number alterations (CNA), as well as tumor mutational burden (TMB) and microsatellite instability (MSI) but not gene fusions [3]. In our prior pan-cancer study, fresh frozen tissue was available in less than a third of cohort for transcriptomic analysis [2]. Often, the tissue received for pathological diagnosis of metastatic biopsies is small, which makes tissue triaging for ancillary/molecular studies very challenging at times.
With the goal of increasing the detection rate of actionable alterations, we expanded the genomic characterization of tumors from patients enrolled in our WES-based precision medicine study [2,17], using the anchored multiplex PCR (AMP)-based NGS assay (ArcherⓇ FusionPlexⓇ Solid Tumor Kit) for fusion detection. This assay requires very low input RNA – as low as 20 ng, from FFPE (archival) material. In a subset of cases, RNA-seq data from fresh frozen tissue was available. We then compared the results to assess the projected benefit of detecting targetable alterations using these assays alone and in combination.
Materials and methods
Study samples and patient characteristics
Study samples were obtained from cancer patients who were prospectively enrolled in our Institutional Review Board (IRB)-approved Precision Medicine Study (IRB# 1305013903). Tumor DNA for WES was obtained from fresh frozen or formalin-fixed paraffin embedded (FFPE) tissue. Germline DNA was obtained from blood samples (buffy coat mononuclear cells), buccal swabs, or benign tissue as described previously [2]. Hematoxylin and eosin (H&E) stained slides from each case were reviewed by board certified pathologists (JMM, BR, RR, DP, FK). All the samples submitted for either frozen or FFPE were macro-dissected after H&E stained slides were reviewed by the pathologists and annotated for tumor rich areas. Fifty-six (56) tumor samples with sufficient FFPE material available for RNA extraction were selected. Selection was based on WES results, i.e. cases with and without clinically actionable (Tier1) variants were chosen (see below). This was done to have a sufficient number of samples to compare with the AMP-based NGS assay.
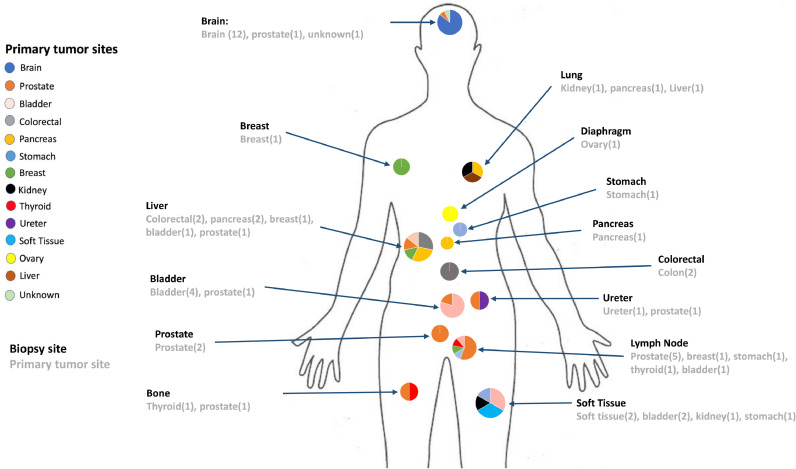
Briefly, the study cohort included 56 tumor samples from 55 cancer patients and diagnoses from 14 different tumor types. The most common primary tumor sites from these samples were prostate (12), brain (12), bladder (8), colorectal (4), pancreas (4), stomach (3), breast (3), and the remaining 10 samples from other primary sites. A single patient with bladder urothelial carcinoma had 2 samples from different metastatic sites (Fig. 1). Age at diagnosis ranged from 4 weeks to 89 years with a median of 63 years. Thirty-two patients were male and 23 were female, with a M:F sex ratio of 1.39. The clinical characteristics of the patients are detailed in Supplementary Table 1.
Fig. 1.
Tumor types and corresponding biopsy sites of study cohort. 56 tumor samples from 55 patients and 14 different tumor origins underwent gene fusion assay in addition to whole-exome sequencing. Tumor types are listed on the left and the diagram illustrates anatomic sites (bold) and tumor origin (light gray).
RNA extraction from FFPE tissue
To perform AMP-based targeted NGS, RNA was extracted from FFPE using Promega MaxwellⓇ RSC RNA FFPE Kit (Promega Corporation, Madison, WI, USA). RNA integrity was verified using the Agilent Bioanalyzer 2100 (Agilent Technologies). All samples passed quality control (QC) and were above the 20% cut-off for DV200 (% of RNA fragments >200 nucleotides), which ranged from 32% to 91%, with a mean value of 63%. The RNA integrity number (RIN) of FFPE samples ranged from 1.0 to 6.9 with a mean value of 2.3 (Supplementary Figure 1).
RNA extraction from fresh frozen tissue
RNA was extracted from fresh frozen material to perform whole transcriptome analysis (RNA-seq) using Promega Maxwell 16 MDx instrument, (Maxwell 16 LEV simplyRNA Tissue Kit). RNA-seq requires 100 to 1000 ng of total RNA input. All fresh frozen samples passed QC with DV200 ranging from 32% to 90%, with a mean value of 59%. The RNA integrity number (RIN) of fresh frozen samples ranged from 1.0 to 7.8 with a mean value of 3.0 (Supplementary Table 1).
AMP-based targeted NGS (FFPE tissue)
The AMP-based NGS panel (ArcherⓇ FusionPlexⓇ Solid Tumor Kit) targets 54 cancer-related genes (Supplementary Table 2). It is an Anchored Multiplex PCR (AMP™)-based technique, which detects fusions associated with the genes in the panel, without prior knowledge of fusion partners or breakpoints [18]. Input of 20–250 ng total RNA was reverse transcribed to cDNA using random primers. Double-stranded cDNA was processed with end repair and dA tailing, followed by ligation with a half-functional Archer MBC (molecular barcode) adapter to both ends of the cDNA molecules. Ligated fragments were amplified with 15 cycles of the first round of PCR (PCR1) using FusionPlex gene-specific panel primers (GSP1 pool) that contain a primer complementary to a portion of the universal ligated adapter. PCR1 amplicons were amplified with a second round of 20 cycles of PCR2 using a combination of GSP2 pool of nested gene-specific panel primers (3′ downstream of GSP1), which are tagged with the second adapter sequence specific for Illumina and a second nested primer against the ligated universal adapter. After a final cleanup using AMPure beads (Agencourt Bioscience), the final libraries were quantified using KAPA Universal Library Quantification Kit (Roche, CA), and sequenced on the Illumina NextSeq Mid Output Kit to obtain approximately 4 million reads per sample (2 × 151 cycles) with 20% PhiX control spike-in. Sequencing data generated was processed through the ArcherDx Analysis software package (v5.1.8, ArcherDx) for fusion detection. Workflow for ArcherⓇ FusionPlexⓇ Solid Tumor fusion assay is detailed in Supplementary Figure 2. Fusions were reported if they were within ±15 base pairs of the exon boundaries of two genes, had a minimum of 5 supporting reads, and had breakpoints further than 100 kb apart with the exception of clinically significant genes with available specific inhibitors and in-frame fusion [18].
RNA-Seq (Frozen tissue)
Libraries were prepared for RNA sequencing using TruSeq RNA Library Preparation Kit v2 or riboZero as previously described [19]. The normalized cDNA libraries were pooled and sequenced on Illumina HiSeq2500 sequencer with pair-end 75 cycles. Paired-end sequencing was then performed on the Illumina HiSeq 2500 system. Samples run on the Illumina platform generated an average of 45 million reads per sample. All reads were independently aligned with STAR_2.4.0f for sequence alignment against the human genome build hg19, downloaded via the UCSC genome browser [http://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/]x, and SAMTOOLS v0.1.19 for sorting and indexing reads [20,21]. For fusion analysis, we used STAR-fusion (STAR-Fusion_v0.5.1) and FusionSeq [22,23]. Fusions with significant support of junction reads and spanning pairs were selected and manually reviewed. Oncofuse [24] was also used to estimate their oncogenic potential by looking at the protein domains lost or retained after fusion event. AGFusion was used to annotate and visualize the protein domain architecture [25]. We used Clinker software to visualize selected fusions and its supporting reads, and circos plot for showing the detected fusions [26,27].
Whole exome sequencing and data analysis
Whole Exome Sequencing (WES) (EXaCT-1 test) was performed on each patient's tumor/matched germline DNA pair using previously described protocols [2,3,17]. Out of 56 samples, WES was performed on available fresh frozen tumor tissue in 19 samples and for the remaining 37 samples we used clinical FFPE (archival) tissue. In these 37 samples, RNA was extracted from the same FFPE block and used for AMP-based NGS. In the 19 samples where frozen tissue was used for WES, the corresponding clinical FFPE blocks were used for RNA extraction. The EXaCT-1 test allows for assessment of >21,000 genes through the development and implementation of novel computational approaches for simultaneous detection of somatic point/indel mutations and copy-number alterations (CNAs). This test uses the Agilent HaloPlex capture platform (Agilent Technologies, Santa Clara, CA, USA) on the Illumina HiSeq 2500 system for sequencing. Tumor mutational burden (TMB) [28] and microsatellite-instability (MSI) [29] were calculated based on an established in-house bioinformatics pipeline, described in detail previously [28, 30]. Cutoff values for TMB-high (>10 mut/MB), and MSI (MSISensor score >6) were applied.
WES alterations were categorized based on their actionability and clinical or biologic relevance into a 3-tier system as described in one of our previous WES-based studies [2]. Briefly, Tier1 includes variants with strong evidence of clinical utility (i.e. FDA-approved therapy or well-powered studies with consensus from experts), Tier 2 includes variants with potential clinical relevance, and Tier 3 include variants of undetermined clinical significance. (Supplementary Table 3, 4).
We defined copy number gain when genomic alteration leads to increased copies in tumor relative to the control sample (log2 copy number ratio between 0.5 and 1.0), copy number loss when genomic alteration leads to decreased copies in tumor relative to the control sample (log2 copy number ratio between −0.5 and −1.0), amplification as high copy number gain (log2 copy number ratio >= 1.0) and deletion when extensive copy number loss, which corresponds to homozygous deletions (log2 copy number ratio <= −1.0).
Fluorescence in situ hybridization (FISH)
Break-apart assays (performed at the FISH core facility at the Englander Institute for Precision Medicine of Weill Cornell Medicine): Dual color FISH probes were designed against loci of interest using BAC clones (https://bacpacresources.org) and labeled with fluorophores (5-Fluorescein dUTP for green and 5-TAMRA dUTP for orange) by Enzo Life Sciences. FGFR3 break apart was designed using BAC clone RP11–262P20 (orange) and RP11-20I20 (green). NTKR1 break apart was designed using RP11-66D17 (green) and RP11-110J1 (orange). A break apart was determined as one individual green signal, one individual red signal per nucleus. The unrearranged allele was determined by one individual yellow signal (green and red overlapping signals). Prior to use, all clones were validated on metaphase spreads. A minimum of 100 nuclei were observed per case using a fluorescence microscope (Olympus BX51; Olympus Optical, Tokyo, Japan). Cytovision and Fiji software were used for imaging.
Fusion assays (performed at the Molecular Cytogenetics core facility at Memorial Sloan Kettering Cancer Center): FGFR3 fusions were designed using either RP5–1091E12 or RP11-339F13 (red). FGFR3 fusion partners included GNAS (green RP5-907D15 or RP4-543J19) and ANKRD30BL (green RP11-1L22). A three-color FISH probe was used for the NTRK1-TPM3 fusion. This included NTKR1 red (RP11-1059C21 or RP11-1038N13), NTRK1 orange (RP11-66D17 or RP11-284F21), and TPM3 green (RP11-1115K24 or RP11-1043P24). The fusion was measured as one orange spectrum signal (overlapping red and green signals), and individual unfused separate red and green signals per nucleus.
Results
Alterations detected by whole exome sequencing (WES)
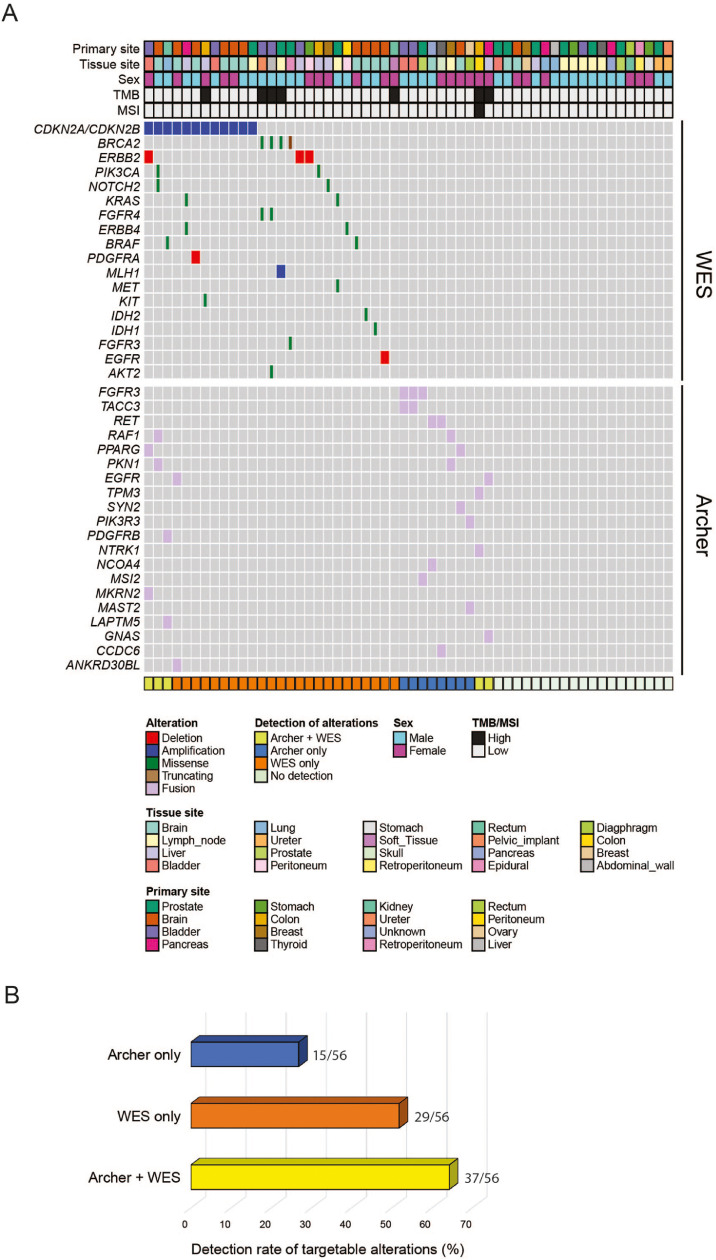
All samples underwent matched tumor/normal WES. Mean coverage was 95x. Mean tumor purity estimated by CLONET [31] for all patient samples was 72% (range: 20–100%). In cases where CLONET data was unavailable, the pathologist assessment of tumor content was incorporated. By WES, 29/56 tumor samples (51.8%) had alterations, which were classified as targetable (Tier1, see Methods). Of these, 13/29 patient samples harbored somatic mutations in either BRCA2, PIK3CA, NOTCH2, FGFR4, IDH1, ERBB4 and MET genes, or copy number alterations in EGFR or ERBB2 genes. Twelve had copy number loss in CDKN2A and CDKN2B genes, 7 tumors had high tumor mutational burden and 1 out of 7 samples with TMB-high showed MSI (Supplementary Table 1). Among them, 5 tumors (8.9%, 5/56) had no targetable fusions on AMP-based NGS. The remaining 2 patients had fusions in NTRK3, NTRK1 and EGFR genes. When WES data was combined with AMP-based NGS results, the proportion of patient samples with targetable alterations increased from 51.8% (29/56) to 66.1% (37/56) (Fig. 2). Interestingly, 4 of these 29 patient samples (13.8%) whose tumors had actionable alterations by WES, also demonstrated targetable fusions on AMP-based NGS, e.g. WCM1230, a case of colon cancer with TPM3-NTRK1 fusion, MSI-High (MSISensor Score 9.14), and TMB-High (Supplementary Table 1). Among tumors with no targets detected by WES, 7/29 (24.1%) harbored targetable fusions, detected by AMP-based NGS.
Fig. 2.
Integrative genomic landscape of whole-exome sequencing and actionable gene fusion detection.(A) CoMut plot representation of individual Tier1 mutations, copy number alterations by WES and fusions by AMP-based NGS in 56 tumor samples in the study cohort of 55 patients. Top: primary site, metastatic sites, gender and TMB/MSI profile. Left: individual gene alterations detected by WES and AMP-based assay. Bottom: Detection of targetable alteration by individual assays alone and in combination. (B) Percentage of molecular targets identified by individual NGS assays, alone and in combination. A significant increase in actionable genomic alterations is seen when results from both WES and AMP-based NGS are integrated.
Gene fusions and novel gene partners detected by AMP-based targeted NGS
The AMP-based NGS assay detected 48 fusions in 31/56 tumors (55.4%): 18 samples showed a single fusion event, 10 showed 2 distinct fusions, while the remaining 3 tumors showed 3 separate fusions. 18 out of the 48 fusions (37.5%) contained intronic regions. The total number of supporting reads for the detected fusions ranged from 5 to 2254 (mean=98 reads; median=10 reads). A TPM3-NTRK1 fusion detected in a case of colon cancer showed the highest read count of 2254 and skewed the aforementioned results.
The detected fusions comprised 36 distinct types, of which 83.3% (30/36) were present in single cases, and the other 16.7% (6/36) were detected in multiple samples (Table 1 and Fig. 2).
Table 1.
Fusions detected by AMP-based NGS assay the in study cohort.
| Fusion partners | Novelty | n = 48 | Primary tumor sites | |||
|---|---|---|---|---|---|---|
| 1 | TMPRSS2-ERG | Partners Reported | 6 | Prostate | ||
| 2 | ETV4-ETV1 | Novel gene partner | 3 | Prostate | Brain | Ovary |
| 3 | PKN1-RAF1 | Novel gene partner | 3 | Colorectal | Brain | Breast |
| 4 | FGFR3-TACC3 | Partners Reported | 2 | Bladder | ||
| 5 | KIAA2022-MAST2 | Novel gene partner | 2 | Prostate | Bladder | |
| 6 | TFE3-SYNE1 | Novel gene partner | 2 | Stomach | Breast | |
| 7 | ABCA13-NONE-FGFR2 | Novel gene partner | 1 | Bladder | ||
| 8 | BRD4-ARHGDIB | Novel gene partner | 1 | Pancreas | ||
| 9 | CCDC6-RET | Partners Reported | 1 | Thyroid | ||
| 10 | CCDC82-MAML2 | Partners Reported | 1 | Bladder | ||
| 11 | CTNNBL1-MAST2 | Novel gene partner | 1 | Soft Tissue | ||
| 12 | EGFR-ANKRD30BL-EGFR | Novel gene partner | 1 | Brain | ||
| 13 | EIF4H-ETV1 | Novel gene partner | 1 | Prostate | ||
| 14 | ELF5-NOTCH2 | Novel gene partner | 1 | Brain | ||
| 15 | ETV6-C4orf21 | Novel gene partner | 1 | Bladder | ||
| 16 | EWSR1-PCGF3 | Novel gene partner | 1 | Soft Tissue | ||
| 17 | EWSR1-PRKCSH | Novel gene partner | 1 | Pancreas | ||
| 18 | FGFR3-MSI2 | Novel gene partner | 1 | Prostate | ||
| 19 | GNAS-EGFR | Novel gene partner | 1 | Pancreas | ||
| 20 | INSR-PDE3A | Novel gene partner | 1 | Bladder | ||
| 21 | LAPTM5-PDGFRB | Novel gene partner | 1 | Kidney | ||
| 22 | LOC101929724-NTRK3 | Novel gene partner | 1 | Brain | ||
| 23 | MAST2-ETV4 | Novel gene partner | 1 | Brain | ||
| 24 | MKRN2-PPARG | Novel gene partner | 1 | Bladder | ||
| 25 | NCOA4-RET | Partners Reported | 1 | Unknown | ||
| 26 | PIK3R3-MAST2 | Novel gene partner | 1 | Ovary | ||
| 27 | PKN2-NOTCH2 | Novel gene partner | 1 | Breast | ||
| 28 | RNF157-AS1-TFEB | Novel gene partner | 1 | Pancreas | ||
| 29 | SLC45A3-ERG | Partners Reported | 1 | Prostate | ||
| 30 | SLC45A3-ETV1 | Partners Reported | 1 | Prostate | ||
| 31 | SYN2-PPARG | Novel gene partner | 1 | Brain | ||
| 32 | TFE3-DMD | Novel gene partner | 1 | Breast | ||
| 33 | TFE3-DNAH14 | Novel gene partner | 1 | Brain | ||
| 34 | TPM3-NTRK1 | Partners Reported | 1 | Colorectal | ||
| 35 | TYW1-TFE3 | Novel gene partner | 1 | Pancreas | ||
| 36 | ZNF254-RELA | Novel gene partner | 1 | Peritoneum | ||
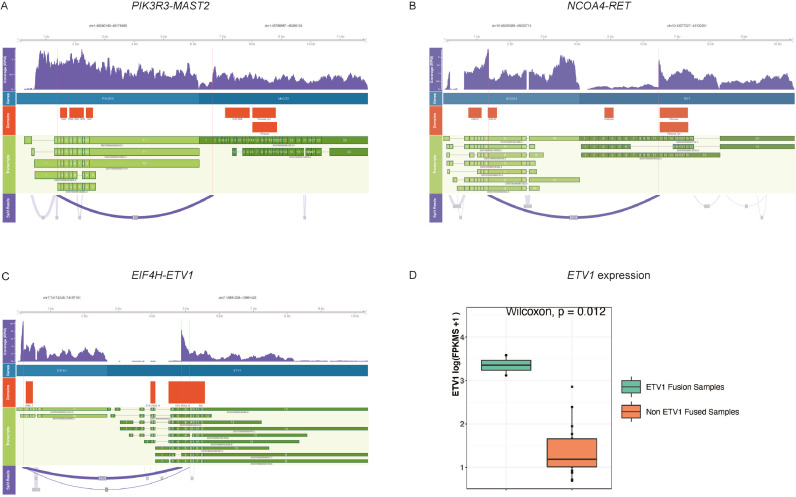
Twenty eight out of 36 (77.8%) gene fusion partners were novel at the time of this publication - 3 online fusion databases were interrogated (Quiver-Archer, TCGA and Cosmic). Examples of known gene fusions detected include TMPRSS2-ERG (n = 6; all prostate) [32], NCOA4-RET (n = 1; unknown primary) [33], TPM3-NTRK1 (n = 1; colorectal) [34], and FGFR3-TACC3 (n = 2; bladder) [35] (Fig. 3), and examples of novel gene partners include PKN1-RAF1 (n = 3; colon, brain, breast); KIAA2022-MAST2 (n = 2; bladder, prostate); TFE3-SYNE1 (n = 2; stomach, breast), FGFR3-MSI2 (n = 1; prostate), GNAS-EGFR (n = 1; pancreas) and ETV4-ETV1 (n = 3; prostate, brain, ovary). Two of the three ETV4-ETV1 fusion positive tumors had RNA-seq data. Expression of ETV1 gene in 2 ETV1 fusion-positive tumors was significantly higher when compared with 17 ETV1-fusion negative tumors (Fig. 4 and Supplementary Table 5). Of the detected three ETV4-ETV1 fusions, two of them in WCM331 and WCM1004 were out-of-frame, and one in WCM1401 was in-frame (Supplementary Table 7).
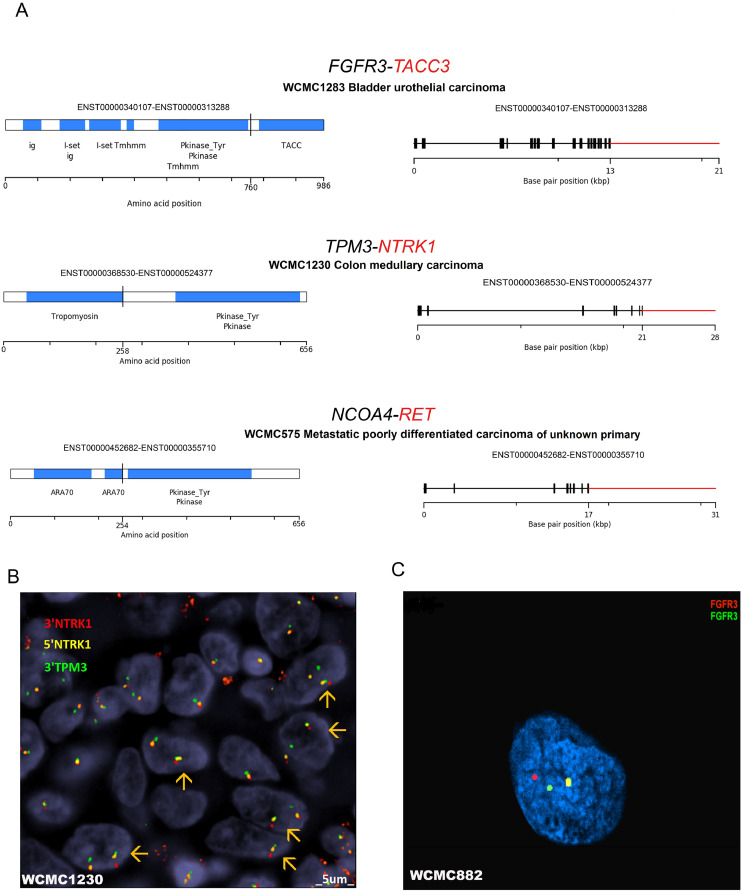
Fig. 3.
Selected known fusions detected in cancer patients enrolled in the Englander Institute for Precision Medicine.(A) Selected examples of known fusions detected by using AMP-based NGS are illustrated. Genes in red color are covered by the ArcherⓇ FusionPlexⓇ Solid Tumor panel, and genes in black color were partners detected by the assay. The column on the left shows the protein domain structure of putative fusions and the vertical black line is the detected fusion breakpoint. The column on the right shows the exon structure of the same fusions. (B) Fluorescence in situ hybridization (FISH) fusion assay for TPM3-NTRK1 is positive in a case of colon cancer. In each nucleus the FISH assay demonstrates a yellow signal (superimposed red-green fused allele) and separate red and green signals (unfused allele) in represented nuclei. (C) FISH break-apart assay for FGFR3 is positive in a case of prostate adenocarcinoma. An FGFR3-MSI2 gene fusion was detected by ArcherⓇ FusionPlexⓇ Solid Tumor. This FISH assay demonstrates a yellow signal (nontranslocated allele) and separate red (5′) and green (3′) signals in the translocated allele of FGFR3 in the representative nucleus (see alsoTable 1and Supplementary Tables).
Fig. 4.
Schematic representation of one known fusion and two novel fusion partners. Coverage tracks with annotated breakpoints, genes partners, protein domains, gene transcripts and split reads involving (A)PIK3R3-MAST2,(B)NCOA4-RET and (C)EIF4H-ETV1. To further support the latter, (D) RNA-seq expression was compared between 2 ETV1 fusion-positive and 17 non-ETV1 fused samples (p = 0.012) (see also Supplementary Table 5).
Visualization of selected known fusions and novel gene partners that were generated from RNA-seq are illustrated in Fig. 4, which shows read coverage tracks, gene boundary tracks, protein domain tracks, transcript tracks and a sashimi plot that indicates the fusion breakpoints. The breakpoints are also highlighted by the vertical lines.
Fifteen fusions detected in 15/56 samples (26.8%) from 12 patients were deemed targetable either by FDA-approved specific inhibitors or by their use in clinical trials, or by drugs that have proven therapeutic benefit in preclinical studies (Table 2). Nine out of these 15 fusions (60.0%) demonstrated novel gene partners while the other 6 (40.0%) have been previously reported. Of the 15 cases with targetable fusions, 4 cases harbored FGFR fusions (3 bladder and 1 prostate) and 2 had RET fusions (thyroid carcinoma, and carcinoma of unknown primary).
Table 2.
Targetable fusions detected by AMP-based NGS assay in the study cohort.
| PATIENT | CASE ID | FUSION | Novel Partners/Reported* | READS | TUMOR TYPE | PRIMARY SITE | TUMOR SITE |
|---|---|---|---|---|---|---|---|
| 1 | WCM1230 | TPM3-NTRK1 | Reported before | 2254 | Medullary carcinoma | Colon | Colon |
| WCM1230 | PKN1-RAF1 | Novel gene partner | 7 | Medullary carcinoma | Colon | Colon | |
| 2 | WCM1283 | INSR-PDE3A | Novel gene partner | 6 | Urothelial carcinoma | Bladder | Bladder |
| WCM1283 | FGFR3-TACC3 | Reported before | 8 | Urothelial carcinoma | Bladder | Bladder | |
| 3 | WCM1304 | FGFR3-TACC3 | Reported before | 27 | Urothelial carcinoma | Bladder | Bladder |
| 4 | WCM1408 | SYN2-PPARG | Novel gene partner | 16 | Teratoma | Brain | Brain |
| 5 | WCM1437 | PKN1-RAF1 | Novel gene partner | 9 | Glioblastoma | Brain | Brain |
| 6 | WCM1442 | LAPTM5-PDGFRB | Novel gene partner | 6 | Renal cell carcinoma | Kidney | Lung |
| 7 | WCM1532 | PKN1-RAF1 | Novel gene partner | 6 | Invasive ductal carcinoma | Breast | Lymph node |
| 8 | WCM271 | CCDC6-RET | Reported before | 56 | Papillary carcinoma | Thyroid | Skull |
| 9 | WCM331 | PIK3R3-MAST2 | Novel gene partner | 57 | Serous carcinoma | Ovary | Diaphragm |
| 10 | WCM511 | MKRN2-PPARG | Reported before | 29 | Urothelial carcinoma | Bladder | Bladder |
| WCM511 | ABCA13-FGFR2 | Novel gene partner | 10 | Urothelial carcinoma | Bladder | Bladder | |
| 11 | WCM575 | NCOA4-RET | Reported before | 169 | Metastatic poorly differentiated carcinoma | Unknown | Brain |
| 12 | WCM882 | FGFR3-MSI2 | Novel gene partner | 17 | Adenocarcinoma | Prostate | Prostate |
Checked with TCGA, Quiver-Archer and Cosmic databases, date 1.2.2019.
Next, we performed FISH to validate a subset of fusions detected by AMP-based NGS. In WCM1230, a colon adenocarcinoma harboring TPM3-NTRK1, we observed fused signals of 5′ NTRK1 and 3′ TPM3 (Fig. 3B), and also NTRK1 separate red and green signals by FISH break-apart assay (not shown). A break-apart FISH assay for FGFR3 was also used to confirm gene rearrangement in two samples with FGFR3 fusions detected by AMP-based NGS: WCM882 (prostate adenocarcinoma) that harbors a novel FGFR3-MSI2 fusion partner (Fig. 3C) and WCM1283 (bladder urothelial carcinoma) that harbors known FGFR3-TACC3 fusion (Supplementary Figure 3A). In both cases, break-apart signals of FGFR3 allele were observed. Although we tried to validate a novel GNAS-EGFR fusion detected in WCM1364 (neuroendocrine tumor of pancreas), a fused signal of probes recognizing GNAS and EGFR was not observed (Supplementary Figure 3B).
Gene fusions and novel gene partners detected by RNA-seq
Whole-transcriptome data from frozen tissue was available in 19 of 56 study samples. Gene fusions were detected in 16/19 (84.2%) samples, 13 of which harbored fusion events that were also detected by AMP-based NGS, and 8 are considered as potential targets. Details of those matched gene fusions and gene breakpoints are provided in Supplementary Table 6. Overall, RNA-seq had lower supporting reads (mean 12, median 4) when compared to AMP-based NGS (mean 97, median 10). When RNA-seq results were combined with AMP-based NGS, 52.6% (10/19) of patient samples showed druggable fusions. When all 3 assays (WES, AMP-based NGS and RNA-seq) were combined, 68.4% (13/19) of patient samples had targetable molecular alterations. No reciprocal gene fusions were found in any of the samples, neither in AMP-based NGS nor in RNA-seq data.
Discussion
The advent of next-generation sequencing (NGS) has revolutionized the field of precision oncology and novel molecular alterations driving oncogenic transformation continue to be discovered. These oncogenic drivers can be single nucleotide variations, copy number changes or gene fusions, and may potentially be targeted by selective inhibitors, forming the basis of recent precision medicine-based clinical trials [36], [37], [38], [39], [40]. The aim of our study was to determine the increase of molecular targets and added potential benefit when whole-exome sequencing (WES) of solid tumors [2,3] is combined with gene fusion detection through an Anchored Multiplex PCR assay (ArcherⓇ FusionPlexⓇ Solid Tumor Kit). Both assays use archival FFPE tissue in contrast to whole transcriptome analysis (RNA-seq), which requires fresh frozen tissue for confident gene fusion detection. To put this in perspective, in our previous pan-cancer WES-based precision medicine study [2], 31% of cases had available fresh frozen tissue for transcriptomic studies. It becomes challenging to perform gene fusion assays when the latter is unavailable or when tissue is limited, e.g. small biopsy tumor tissue that is used for standard-of-care histopathology diagnosis and analysis. Hence, a robust NGS assay that uses small RNA input from FFPE enhances the ability to detect potentially targetable fusions. Further, the RNA input requirement for AMP-based NGS assays is as low as 20 ng from FFPE material, which is significantly lower than 100–1000 ng RNA from fresh frozen tissue required for RNA-seq. Previous studies have demonstrated that AMP-based NGS assays can detect novel fusions or novel fusion partners [41], [42], [43], [44], [45], [46].
Past NGS-based clinical trials have reported clinically significant molecular events in approximately a third of patients [4,5]. For the purpose of our study, the group with targetable alterations by WES was enriched, and Tier1 alterations [2] were present in 51.8% of patients (see Materials and Methods). We observed a significant increase in detection of potential targets, from 51.8% to 66.1% when WES results were integrated with the AMP-based NGS fusion assay. This significant boost of possible druggable targets may have a significant clinical impact in the treatment of brain cancer and advanced solid tumors. Gene fusions detected by ArcherⓇ FusionPlexⓇ Solid Tumor Kit were also identified by RNA-seq in 75% (12/16) of cases. Whole transcriptome analysis from fresh frozen tissue was available in 19 patients and, when coupled with WES, it also showed a significant increase in likely targetable alterations from 36.8% (7/19) to 63.1% (12/19), comparable to the increment of molecular targets above. The low RNA input and possibility of utilization of FFPE tissue establishes an AMP-based NGS test as an excellent alternative for identifying clinically relevant gene fusions in solid tumor specimens [47,48]. Moreover, detection of novel gene fusions or novel fusion partners that may translate into drug targets is an advantage of this technology and targeted AMP-based NGS has already been implemented in clinical molecular laboratories [49]. Whole-transcriptome analysis (RNA-seq) on FFPE material is an area of active research that has reported encouraging results [50], however further studies and validation are needed to be incorporated clinically.
A study of gene fusion analysis on 81 bone and soft tissue tumors, using a similar AMP-based NGS assay showed a concordance rate of 90% in fusion detection when results were compared with conventional methods such as fluorescence in situ hybridization (FISH) and RT-PCR [51]. In our study, the high sensitivity of AMP-based NGS to detect gene fusions (with very low number of reads) explains discrepant results when compared with RNA-seq. The AMP-based assay is a targeted panel that incorporates a pre-amplification step to enhance the initial signal, thus generating a higher number of fusion reads (compared to RNA-seq), making it feasible to work with minimal RNA input [41,52].
ArcherⓇ FusionPlex has been recently validated for accuracy, precision, specificity and sensitivity against four orthogonal assays using two different methodologies on 109 clinical samples, in Sussman et al. [53]. In our study, among the 19 cases with both RNA-seq and AMP-based NGS data, only a single fusion PLEC-NOTCH1 (case WCM1073; oligodendroglioma) found by RNA-seq with 3 reads was not detected by AMP-based NGS. In comparison, RNA-seq did not reveal several fusions detected by AMP-based NGS (Supplementary Table 5).
Developments in massive parallel sequencing technology have led to a significant increase in the detection of gene fusions, which has raised concerns about their biological meaningfulness. Recently, it has been shown that the majority of fusions detected through NGS are passenger fusions and purely random events [54]. In an attempt to overcome this issue we used Oncofuse [24] and AGFusion [25], both bioinformatics pipelines to decipher the oncogenic potential of fusions and to visualize the protein domains. We concluded that new gene partners (detected by ArcherⓇ FusionPlexⓇ) that fuse with targetable genes result in a chimeric targetable protein.
After integration of WES targetable alterations with actionable fusions detected by AMP-based NGS, the frequency of drug targets in our study increased to 66.1%. However, we enriched our study set with cases harboring Tier1 potentially targetable alterations by WES, including detection of MSI-H and tumors with high TMB. Molecular data from 948 solid cancer patients from the MOSCATO 01 trial identified actionable alterations in 49% of patient samples when multiple assays were integrated, including targeted sequencing, array comparative genomic hybridization (aCGH), WES, RNA-seq, immunohistochemistry and FISH [5]. Thirty-three percent of patients who received matched targeted treatment obtained a progression-free survival rate above the predefined threshold. Another clinical trial on metastatic breast cancer found that 46% of patients had at least one actionable alteration when targeted DNA sequencing was integrated with aCGH [4]. Nine percent of matched target-treated patients in their cohort had an objective response, and 21% had stable disease. In their study, the authors advocate for the use of combined therapies targeting different drivers because this approach potentially delays drug resistance. On the other hand, there have been conflicting studies where researchers have found no significant difference in terms of survival in patients treated with targeted agents and have discouraged the off-label use of molecularly target agents [55]. Compared to the aforementioned studies, the percentage of molecular targets (49.1%) detected in our cohort selection exclusively on WES is higher. Explanations would include that most of the above-mentioned studies used targeted DNA sequencing panels for mutation and copy number detection instead of WES, lacking TMB/MSI data as potential targetable biomarkers in their cohorts, and that in our study we selected more Tier1 cases. In our cohort, seven patient samples (12.7%, 7/56) had high TMB/MSI-H by WES alone, which are potential targets for immunotherapy. Of these seven patient samples, five had no targetable alterations by AMP-based NGS.
Of particular clinical relevance, we found 30 novel gene partners, 5 of which assemble with known drug targets including RAF1, FGFR2, FGFR3, EGFR, NTRK3, and RET (Fig. 3 and Table 1).
Three ETV4-ETV1 fusions were detected (prostate, brain, ovary); two were out-of-frame and one was in-frame. Archer demonstrated that ETV4 fused with ETV1 as the 5′ partner in all ETV4-ETV1 fusions detected in our cohort, while ETV4 is reportedly the 3′ partner when that fuses with TMPRSS2 in prostate cancers that harbor such fusion event. While TMPRSS2 fusions with ETS transcriptional factors, including ERG, ETV1 and ETV4 gene, are well-studied oncogenic alterations defining a distinct subsets of prostate cancers [56], biological consequences of ETV4-ETV1 fusion events are unclear. Further functional studies would be warranted in order to clarify the potential oncogenic role of ETV4-ETV1 fusions.
In the FGFR3-TACC3 in-frame fusion detected in 2 bladder cancer samples, the coiled-coil domain of TACC3 causes constitutive phosphorylation of key activating FGFR3 tyrosine residues without the need for ligand binding, leading to tyrosine kinase domain dimerization, auto-phosphorylation and increased and altered levels of FGFR3 activation, fusion protein phosphorylation, downstream signaling, cellular transformation, proliferation, and viability [57]. On the other hand, ABCA13-FGFR2 and FGFR3-MSI2 fusions were predicted to be out-of-frame, and their functional consequences are unknown. Although these FGFR2 and FGFR3 fusions with novel gene partners could still be explored as potential biomarkers, further investigation would be required.
One limitation of our retrospective study is the lack of data regarding targeted treatment and patient response. At the time of tumor tissue collection, most of the patients received standard-of-care treatment (e.g. surgery, chemotherapy). Alternative options (i.e. NGS-based) were only considered in a few patients. One patient with papillary thyroid carcinoma and multiple metastases (brain, lung, bone, spleen, lymph nodes) (case WCM271) did receive the tyrosine kinase inhibitor cabozantinib based on the detection of CCDC6-RET. After 15 months of treatment the patient died of disease. Another patient with metastatic urothelial carcinoma of the ureter (WCM1304) with an FGFR3-TACC3 fusion received the FGFR3 inhibitor Ertafitinib (JNJ42756493), which was well tolerated. Fusion of FGFR genes cause receptor dimerization and subsequent ligand-independent constitutive kinase activity which are highly sensitive to FGFR selective inhibitors [58, 59]. However, patient had disease progression and the therapy was changed after 3 months of treatment to which he had a complete response. Another patient with localized medullary carcinoma of the colon (pathologic stage pT2 pN1b) that harbors TPM3-NTRK1 fusion (case WCM1230) underwent surgery only and has not required additional/targeted therapy. No local recurrence or distant metastases have yet been detected after surgical removal. Available treatment data to date is provided in Supplementary Table 1. As part of the IRB-approved precision medicine clinical study, new molecular targets detected by the ArcherⓇ FusionPlexⓇ Solid Tumor assay or RNA-seq are communicated to clinicians and treatment decisions are discussed at regular precision medicine tumor boards.
Another factor to consider is the molecular complexity of certain tumors, which may affect downstream fusion detection and interpretation. One such example is tumors that show bi-allelic inactivation of genes, specifically CDK12 mutant tumors have been strongly linked to structural variants. Such tumors show high fusion burden or large number of fusions which are generated by tandem duplications and have the potential to be therapeutically targeted [60,61]. One additional consideration is that we have used selected genes included in ArcherⓇ FusionPlexⓇ Solid Tumor assay. Although it covers most common targetable and oncogenic fusions described in solid cancers, the possibility exists that tumor samples in our study cohort may harbor rare unexpected fusion events that are not covered by the gene panel.
In summary, we demonstrate in this pilot study that integration of WES with AMP-based NGS fusion assays increases the detection of targetable genomic alterations in cancer patients, which offers additional therapeutic options to treating oncologists. In the absence of available fresh frozen tissue for whole transcriptome tumor analysis, FFPE archival material can be utilized effectively since the AMP-based NGS technology is highly sensitive, requires minimal RNA input, and is able to detect novel gene fusions and gene partners of potential clinical relevance.
Authors' contributions
Shaham Beg: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis, Manuscript preparation, Manuscript editing, Manuscript review
Rohan Bareja: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis, Manuscript preparation
Kentaro Ohara: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
Kenneth Wha Eng: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
David J. Pisapia: Data acquisition
David C. Wilkes: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
Wael Al Zoughbi: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
Sarah Kudman: Data analysis and interpretation, Statistical analysis
Rema Rao: Manuscript preparation
Jyothi Manohar: Data acquisition
Troy Kane: Data analysis and interpretation
Michael Sigouros: Data analysis and interpretation
Wei Zhang: Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
Jenny Zhaoying Xiang: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
Francesca Khani: Manuscript preparation, Manuscript editing, Manuscript review
Brian D. Robinson: Data acquisition
Bishoy M. Faltas: Data acquisition, Manuscript preparation, Manuscript editing, Manuscript review
Cora N. Sternberg: Manuscript preparation, Manuscript editing, Manuscript review
Andrea Sboner: Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis
Himisha Beltran: Data acquisition, Manuscript preparation, Manuscript editing, Manuscript review
Olivier Elemento: Study concepts, Study design, Manuscript preparation, Manuscript editing, Manuscript review
Juan Miguel Mosquera: Study concepts, Study design, Data acquisition, Quality control of data and algorithms, Data analysis and interpretation, Statistical analysis, Manuscript preparation, Manuscript editing, Manuscript review
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
Acknowledgments
The Clinic team at Englander Institute for Precision Medicine (Noah Greco) coordinated clinical activities. Logistic support was provided by the Institutional Biobank at Weill Cornell Medicine (Maria T Salpietro, Christopher Louie, Alyssa F. Polak, Katelyn Hadley). This work was also supported in part by the Translational Research Program of the Department of Pathology and Laboratory Medicine (Mai Ho, Leticia Dizon, Bing He). FISH fusion assays were performed at the Molecular Cytogenetics core facility of the Memorial Sloan Kettering Cancer Center.
Ethics approval and consent to participate
This study obtained approval from the ethics committee in United States from Weill Cornell Medicine through Institutional Review Board (IRB)–approved Precision Medicine Trial (IRB No. 1305013903). All participants provided written informed consent to participate. The study was performed in accordance with the Declaration of Helsinki.
Funding
This work was supported by the Caryl and Israel Englander Institute for Precision Medicine (EIPM) of Weill Cornell Medicine and NewYork-Presbyterian.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100944.
Appendix. Supplementary materials
References
- 1.LeBlanc V.G., Marra M.A. Next-generation sequencing approaches in cancer: where have they brought us and where will they take us? Cancers. 2015;7(3):1925–1958. doi: 10.3390/cancers7030869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran H. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1(4):466–474. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rennert H. Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care. NPJ Genom. Med. 2016;1:16019. doi: 10.1038/npjgenmed.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andre F. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 5.Massard C. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 6.Webster A.F. Mining the archives: a cross-platform analysis of gene expression profiles in archival formalin-fixed paraffin-embedded tissues. Toxicol. Sci. 2015;148(2):460–472. doi: 10.1093/toxsci/kfv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteve-Codina A. A comparison of RNA-Seq results from paired formalin-fixed paraffin-embedded and fresh-frozen glioblastoma tissue samples. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0170632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossel Ben-Moshe N. mRNA-seq whole transcriptome profiling of fresh frozen versus archived fixed tissues. BMC Genom. 2018;19(1):419. doi: 10.1186/s12864-018-4761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams H.L. Validation of the Oncomine focus panel for next-generation sequencing of clinical tumour samples. Virchows. Arch. 2018;473(4):489–503. doi: 10.1007/s00428-018-2411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solomon J.P., Hechtman J.F. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. 2019;79(13):3163–3168. doi: 10.1158/0008-5472.CAN-19-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mertens F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015;15(6):371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23(1):227–238. doi: 10.1016/j.celrep.2018.03.050. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J.J. Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin. Cancer Res. 2014;20(5):1383–1392. doi: 10.1158/1078-0432.CCR-13-0699. [DOI] [PubMed] [Google Scholar]
- 14.Lo Russo G. Concomitant EML4-ALK rearrangement and EGFR mutation in non-small cell lung cancer patients: a literature review of 100 cases. Oncotarget. 2017;8(35):59889–59900. doi: 10.18632/oncotarget.17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soda M. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 16.Robinson D.R. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sailer V. Integrative molecular analysis of patients with advanced and metastatic cancer. JCO Precis. Oncol. 2019;3 doi: 10.1200/PO.19.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winters J.L. Development and verification of an RNA Sequencing (RNA-Seq) Assay for the detection of gene fusions in tumors. J. Mol. Diagn. 2018;20(4):495–511. doi: 10.1016/j.jmoldx.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Beltran H. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin A. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stransky N. The landscape of kinase fusions in cancer. Nat. Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sboner A. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11(10):R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shugay M. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29(20):2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 25.Murphy C., Elemento O. AGFusion: annotate and visualize gene fusions. bioRxiv. 2016 [Google Scholar]
- 26.Schmidt B.M. Clinker: visualizing fusion genes detected in RNA-seq data. Gigascience. 2018;7(7) doi: 10.1093/gigascience/giy079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y. OmicCircos: a Simple-to-Use R package for the circular visualization of multidimensional Omics data. Cancer Inform. 2014;13:13–20. doi: 10.4137/CIN.S13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez E.M. Cancer-specific thresholds adjust for whole exome sequencing-based tumor mutational burden distribution. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.18.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu B. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson B.D. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat. Commun. 2019;10(1):2977. doi: 10.1038/s41467-019-10873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prandi D. Unraveling the clonal hierarchy of somatic genomic aberrations. Genome Biol. 2014;15(8):439. doi: 10.1186/s13059-014-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Perdomo H.A. Association between TMPRSS2:ERG fusion gene and the prostate cancer: systematic review and meta-analysis. Cent. Eur. J. Urol. 2018;71(4):410–419. doi: 10.5173/ceju.2018.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li A.Y. RET fusions in solid tumors. Cancer Treat. Rev. 2019;81 doi: 10.1016/j.ctrv.2019.101911. [DOI] [PubMed] [Google Scholar]
- 34.Gatalica Z. Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol. 2019;32(1):147–153. doi: 10.1038/s41379-018-0118-3. [DOI] [PubMed] [Google Scholar]
- 35.Costa R. FGFR3-TACC3 fusion in solid tumors: mini review. Oncotarget. 2016;7(34):55924–55938. doi: 10.18632/oncotarget.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyman D.M. AKT inhibition in solid tumors with AKT1 mutations. J. Clin. Oncol. 2017;35(20):2251–2259. doi: 10.1200/JCO.2017.73.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyman D.M. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barroilhet L., Matulonis U. The NCI-MATCH trial and precision medicine in gynecologic cancers. Gynecol. Oncol. 2018;148(3):585–590. doi: 10.1016/j.ygyno.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y., Chi P. Basket trial of TRK inhibitors demonstrates efficacy in TRK fusion-positive cancers. J. Hematol. Oncol. 2018;11(1):78. doi: 10.1186/s13045-018-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drilon A. Efficacy of Larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Z. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 2014;20(12):1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 42.Loke B.N. Novel exon-exon breakpoint in CIC-DUX4 fusion sarcoma identified by anchored multiplex PCR (Archer FusionPlex Sarcoma Panel) J. Clin. Pathol. 2017;70(8):697–701. doi: 10.1136/jclinpath-2016-204247. [DOI] [PubMed] [Google Scholar]
- 43.Guseva N.V. Anchored multiplex PCR for targeted next-generation sequencing reveals recurrent and novel USP6 fusions and upregulation of USP6 expression in aneurysmal bone cyst. Genes Chromosomes Cancer. 2017;56(4):266–277. doi: 10.1002/gcc.22432. [DOI] [PubMed] [Google Scholar]
- 44.Lam S.W. Molecular analysis of gene fusions in bone and soft tissue tumors by anchored multiplex PCR-based targeted next-generation sequencing. J. Mol. Diagn. 2018;20(5):653–663. doi: 10.1016/j.jmoldx.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Seager M., Aisner D.L., Davies K.D. Oncogenic gene fusion detection using anchored multiplex polymerase chain reaction followed by next generation sequencing. J. Vis. Exp. 2019;(149) doi: 10.3791/59895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benini S. Identification of a novel fusion transcript EWSR1-VEZF1 by anchored multiplex PCR in malignant peripheral nerve sheath tumor. Pathol. Res. Pract. 2020;216(1) doi: 10.1016/j.prp.2019.152760. [DOI] [PubMed] [Google Scholar]
- 47.Han Y. Advanced applications of RNA sequencing and challenges. Bioinform Biol Insights. 2015;9(Suppl 1):29–46. doi: 10.4137/BBI.S28991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nazarov P.V. RNA sequencing and transcriptome arrays analyses show opposing results for alternative splicing in patient derived samples. BMC Genomics. 2017;18(1):443. doi: 10.1186/s12864-017-3819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hechtman J.F. Pan-Trk Immunohistochemistry Is an efficient and reliable screen for the detection of NTRK fusions. Am. J. Surg. Pathol. 2017;41(11):1547–1551. doi: 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cieslik M. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015;25(9):1372–1381. doi: 10.1101/gr.189621.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam S.W. Molecular analysis of gene fusions in bone and soft tissue tumors by anchored multiplex PCR-based targeted next-generation sequencing. J. Mol. Diagn. 2018;20(5):653–663. doi: 10.1016/j.jmoldx.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Fletcher, C.D.M. and ebrary Inc., WHO classification of tumours of soft tissue and bone, in World Health Organization Classification of Tumours. p. 1 online resource (471 pages).
- 53.Sussman R.T. Validation of a next-generation sequencing assay targeting RNA for the multiplexed detection of fusion transcripts and oncogenic isoforms. Arch. Pathol. Lab. Med. 2020;144(1):90–98. doi: 10.5858/arpa.2018-0441-OA. [DOI] [PubMed] [Google Scholar]
- 54.Johansson B. Most gene fusions in cancer are stochastic events. Genes Chromosomes Cancer. 2019;58(9):607–611. doi: 10.1002/gcc.22745. [DOI] [PubMed] [Google Scholar]
- 55.Le Tourneau C. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 56.Tomlins S.A. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10(2):177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson K.N. Oncogenic driver FGFR3-TACC3 is dependent on membrane trafficking and ERK signaling. Oncotarget. 2018;9(76):34306–34319. doi: 10.18632/oncotarget.26142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrell B., Breeze A.L. Structure, activation and dysregulation of fibroblast growth factor receptor kinases: perspectives for clinical targeting. Biochem. Soc. Trans. 2018;46(6):1753–1770. doi: 10.1042/BST20180004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babina I.S., Turner N.C. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer. 2017;17(5):318–332. doi: 10.1038/nrc.2017.8. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y.M. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell. 2018;173(7):1770–1782.e14. doi: 10.1016/j.cell.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quigley D.A. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174(3):758–769.e9. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.