Abstract
Septic cardiomyopathy is characterized by impaired contractive function with mitochondrial dysregulation. Songorine is a typical active C20-diterpene alkaloid from the lateral root of Aconitum carmichaelii, which has been used for the treatment of heart failure. This study investigated the protective role of songorine in septic heart injury from the aspect of mitochondrial biogenesis. Songorine (10, 50 mg/kg) protected cardiac contractive function against endotoxin insult in mice with Nrf2 induction. In cardiomyocytes, lipopolysaccharide (LPS) evoked mitochondrial reactive oxygen species (ROS) production and redistributed STIM1 to interact with Orai1 for the formation of calcium release-activated calcium (CRAC) channels, mediating calcium influx, which were prevented by songorine, likely due to ROS suppression. Songorine activated Nrf2 by promoting Keap1 degradation, having a contribution to enhancing antioxidant defenses. When LPS shifted metabolism away from mitochondrial oxidative phosphorylation (OXPHOS) in cardiomyocytes, songorine upregulated mitochondrial genes involved in fatty acid β-oxidation, tricarboxylic acid (TCA) cycle and electron transport chain in a manner dependent on Nrf2, resultantly protecting the capability of OXPHOS. Songorine increased luciferase report gene activities of nuclear respiratory factor-1 (Nrf1) and mitochondrial transcription factor A (Tfam) dependently on Nrf2, indicative of the regulation of Nrf2/ARE and NRF1 signaling cascades. Songorine promoted PGC-1α binding to Nrf2, and the cooperation was required for songorine to activate Nrf2/ARE and NRF1 for the control of mitochondrial quality and quantity. In support, the beneficial effects of songorine on cardioprotection and mitochondrial biogenesis were diminished by cardiac Nrf2 deficiency in mice subjected to LPS challenge. Taken together, these results showed that Nrf2 transcriptionally promoted mitochondrial biogenesis in cooperation with PGC-1α. Songorine activated Nrf2/ARE and NRF1 signaling cascades to rescue cardiomyocytes from endotoxin insult, suggesting that protection of mitochondrial biogenesis was a way for pharmacological intervention to prevent septic heart injury.
Keywords: Songorine, Septic cardiomyopathy, Mitochondrial biogenesis, Nrf2, PGC-1α
Abbreviations: ARE, antioxidant response element; CPT-1, carnitine palmitoyltransferase-1; CRAC, calcium release-activated calcium; ECAR, extracellular acidification rate; ETC, electron transport chain; GSK3β, glycogen synthase kinase 3β; HO-1, heme oxygenase-1; LPS, lipopolysaccharide; mtDNA, mitochondrial DNA; MCU, mitochondrial calcium uniporter; NAC, n-acetylcysteine; NAO, nonyl acridine orange; NRF1, nuclear respiratory factor-1; Nrf2, NF-E2-p45-related factor 2; NRVMs, neonatal rat ventricular myocytes; OCR, oxygen consumption rate; ROS, reactive oxygen species; OXPHOS, oxidative phosphorylation; Sod2, superoxide dismutase 2; TCA, tricarboxylic acid; TFAM, mitochondrial transcription factor A; 3-MA, 3-methyladenine
Graphical abstract
Highlights
-
•
Mitochondrial ROS increased calcium influx through CRAC channels.
-
•
Nrf2 interacted with PGC-1α to regulate mitochondrial biogenesis.
-
•
Songorine activated Nrf2 by promoting Keap1 degradation.
-
•
Songorine regulated Nrf2/ARE and PGC-1α cascades to protect cardiac function.
1. Introduction
Septic cardiomyopathy is an acute cardiac injury, characterized by impaired left ventricular systolic and diastolic function [1,2]. Sepsis is derived from dysregulated immune response, but most of the therapeutics targeting specific inflammatory cytokines have failed in clinical trials [3], indicating that some other mechanisms are needed to be addressed.
Accumulating evidence demonstrates the causative involvement of mitochondrial dysregulation [4]. Mitochondrion is a semi-self-replicating organelle containing its own genome. The mitochondrial DNA (mtDNA) encodes 13 essential components of the OXPHOS machinery, while most other proteins in mitochondria are encoded by the nuclear genomes. The heart is an organ with high energy requirements, largely depending on the capacity of mitochondrial biogenesis. Mitochondrial biogenesis regulates the mass, distribution and activity of mitochondria and maintains the stability of intracellular environment. Recently, dysregulation of mitochondria emerges as a common pathology for ischemic injury, metabolic disorders, neurodegeneration and heart failure [5].
Mitochondrial biogenesis is regulated by a range of transcription factors in association with transcriptional co-activators. Nuclear respiratory factor-1 or 2 (NRF1 or NRF2) transcriptionally regulates the mitochondrial electron transport chain (ETC) subunits encoded by the nuclear genome [6]. Specifically, NRF1 activates gene-encoded factors that mediate replication and transcription of the mitochondrial genome [7]. The antioxidant response element (ARE), as a DNA regulatory element located in various genes including NRF1, Hmox1 and Sod2, primarily binds to nuclear factor erythroid 2-related factor 2 (Nrf2) to activate these genes. In response to oxidative stress, Nrf2 enters the nucleus and activates ARE to enhance antioxidant defenses. The involvement of Nrf2 in mitochondrial biogenesis is revealed by the action of heme oxygenase (HO)-1, which is the targeting gene of Nrf2. In cardiomyocytes, HO-1 enhanced endogenous carbon monoxide (CO) generation to permit Nrf2 binding to ARE in the NRF1 promoter, leading to gene induction encoding mitochondrial biogenesis [8]. Similar regulation was also observed in mouse liver subjected to sepsis [9]. These events suggest the potential role of Nrf2/ARE and NRF1 signaling cascades in mitochondrial biogenesis.
PGC-1α is a member of a family of transcriptional co-regulators, and has been characterized as a broad regulator of cellular metabolism. In combination with other transcriptional factors, PGC-1α promotes mitochondrial fatty acid oxidation and biogenesis in the heart [10]. Nrf2 knockdown inhibits mitochondrial biogenesis with downregulation of PGC-1α [11], while PGC-1α potentiates Nrf2-mediated antioxidant response [12], indicative of the presence of Nrf2/PGC-1α feedback loop. Mitochondrial reactive oxygen species (ROS) is a driving force for mitochondrial dysfunction in septic cardiomyopathy [4], while PGC-1α is responsive to the alternations in cellular energetic demands and redox status [13]. Therefore, finding the role of Nrf2 in the cooperation with PGC-1α would provide important insight into the action of antioxidant defenses from the aspect of mitochondrial biogenesis.
Radix Aconiti Lateralis Preparata (Fuzi in Chinese) is the processed lateral roots of Aconitum carmichaelii Debx. (Ranunculaceae), and has been used for the treatment of chronic heart failure, hypotension and acute myocardial infarction owing to the potential cardiotonic action [14]. Songorine is a napelline-type C20-diterpene alkaloid in Aconitum carmichaelii Debx. (Supplement Fig. 1), exhibiting anti-arrhythmic and anti-inflammatory effects [15,16] with low toxicity [17], indicative of the therapeutic potential in cardioprotection. In the present study, we investigated the cardioprotective role of songorine during sepsis, and found that Nrf2 induction promoted mitochondrial biogenesis in a manner that was in cooperation with PGC-1α.
2. Materials and methods
2.1. Reagents
Songorine (purity ≥ 98%) was obtained from Chengdu Biopurify Phytochemicals Ltd. (Chengdu, China). Lipopolysaccharide (LPS, L2880), N-acetyl-l-cysteine (NAC, A9165), β-nicotinamide mononucleotide (NMN, N3501) and 5 bromo-2′-deoxyuridine (5-BrdU, B5002) were purchased from Sigma (St. Louis, MO, USA). 3-methyladenine (3-MA, HY-19312), cycloheximide (HY-12320), ML385 (HY-100523), MG-132 (HY-13259), 2-deoxy-d-glucose (2-DG, HY-13966), FCCP (HY-100410) and SR18292 (HY-101491) were provided by Med Chem Express (Brea, CA, USA).
2.2. Animals and treatments
All animal experiments were carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals, following protocols approved by Pharmaceutical Animal Experimental of China Pharmaceutical University. Male C57BL/6 mice (6 to 8-week-old) were purchased from the Laboratory Animal Center of Yangzhou University (Yangzhou, China). The mice were housed in a constant temperature (24 ± 2 °C) and a 12-h light/dark cycle, with free access to standard food and water.
To measure mortality rate, mice were intraperitoneally injected with LPS (30 mg/kg), and songorine (10 or 50 mg/kg) was intraperitoneally administrated 1 h before LPS challenge and then administered for 3 consecutive days. 72-hour mortality was recorded. For heart injury, mice were injected with LPS (10 mg/kg), and songorine (10 or 50 mg/kg) was administrated 1 h before and 12 h after LPS treatment. 24 h later, mice were euthanized for the collection of the hearts. Furthermore, the mice were treated with songorine (50 mg/kg) 1 h before and 24 h after LPS treatment, as the Pre-songorine group, while the group of Post-songorine was administrated 2 h and 24 h after LPS treatment.
For the cardiac-specific Nrf2 knockdown in mice, AAV9-shNfe2l2 and AAV9-shNC (negative control) viruses were designed by Hanbio biotechnology (Shanghai, China). Male C57BL/6 mice were injected with 200 μL of AAV9-shNfe2l2 at a concentration of 8.5 × 1011 viral genomes (vg)/mL or AAV9-shNC at a concentration of 6.5 × 1011 vg/mL through the caudal vein. Three weeks later, cardiac Nrf2 protein expression was examined to confirm the efficiency of knockdown. The target sequence of AAV9-shNfe2l2 is 5′-CGAGAAGUGUUUGACUUUA-3′, and the sequence of AAV9-shNC is 5′-UUCUCCGAACGUGUCACGUAA-3'.
2.3. Echocardiography
After 24 h of LPS injection, echocardiography was performed using the Vevo 3100LT micro-ultrasound system. The mice were anesthetized with 1.5%–2% isoflurane, and the position and direction of the ultrasound beam was slowly adjusted to obtain an echocardiography of the left ventricle. M-mode images were acquired for the evaluation of the left ventricular function parameters.
2.4. Transmission electron microscopy
To observe the morphology of mitochondria in myocardial cells, the fresh heart was rapidly fixed with electron microscope fixation solution at 4 °C for 2–4 h. After washing with 0.1 M PBS, the heart was fixed with 1% osmic acid at room temperature for 2 h. After gradient dehydration, and the tissue was embedded and sliced, followed by staining with 3% uranyl acetate and lead citrate for 15 min. Transmission electron microscopy (HITACHI, HT7700) was used for images acquisition and analysis. Mitochondrial number and volume density were measured using Image J software.
2.5. Primary neonatal rat ventricular myocytes (NRVMs) isolation and culture
NRVMs were isolated and cultured as described previously [18]. In short, NRVMs were isolated from 1-2-day-old sprague-dawley rats (B&K Universal Group Ltd., Shanghai, China). The heart tissue was cut into pieces and washed with precooled phosphate buffer saline (PBS), and then digested continuously with 0.1% type II collagenase (Biofroxx, 2275GR001) in 37 °C water bath for 5–7 times. After removing erythrocytes using red blood cell lysate, the harvested cells were pre-incubated with Dulbecco's Minimum Essential Medium (DMEM, KeyGEN BioTECH, China) containing 10% (vol/vol) fetal bovine serum (FBS, Gibco, USA) for 2 h. Cardiomyocytes were isolated and purified by differential adhesion method, and then grown in DMEM supplemented with 100 μM 5-BrdU to inhibit fibroblast proliferation. The cells were used for experiments after 3–4 days culture.
H9C2 cells were obtained from Stem Cell Bank, Chinese Academy of Sciences. NRVMs and H9C2 cells cultured in the DMEM supplemented with 10% (vol/vol) FBS at 37 °C with 5% CO2 in air atmosphere.
2.6. Cytotoxicity assay
NRVMs were seeded in a 96-well plate and incubated with songorine at given concentrations for 24 h. Cell survival was evaluated by commercial Cell Counting Kit-8 (CCK8) (Dojindo, Japan).
2.7. Transfection of NRVMs
To specifically suppress Nfe2l2, Nrf1 or Ppargc1α expression, NRVMs were transfected with Nfe2l2, Nrf1 or Ppargc1α short hairpin RNA (shRNA) or negative control (NC) shRNA with Lipofectamine® 3000 transfection reagent (InvitrogenTM,L3000008, USA) at 70–80% confluence. The shRNA sequences are as follows: shNfe2l2: 5′-GCAAGAAGCCAGATACAAAGA-3′, shNrf1: 5′- GGCTGCTGGGAAGTACAAACA-3′, shPpargc1α: 5′-GCAACATGCTCAAGCCAAACC-3', shNC: 5'-TTCTCCGAACGTGTCACGT-3' (Shanghai GenePharma Co., Ltd). For Nfe2l2 overexpression, NRVMs were transfected with pEX3-Nfe2l2 or pEX3-NC plasmids using Lipofectamine® 3000 transfection reagent. The cells were cultured for 48 h and then treated with indicated reagents in the presence of LPS (1 μg/mL) for 24 h.
2.8. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR)
NRVMs were seeded in a seahorse XF96 cell culture microplates (Agilent Technologies, USA) at a density of 5 × 104 cells/well and then treated with indicated agents at the given concentrations in the presence of LPS (1 μg/mL). Before the measurement of OCR, cells were incubated with glucose (10 mM), pyruvate (1 mM) and l-glutamine (1 mM)-supplemented XF base medium minimal DMEM at 37 °C in a CO2-free incubator for 1 h. OCR was detected with seahorse XFe96 extracellular flux assay kit under XF calibrant and then supplement with 2.5 μM oligomycin, 1 μM FCCP and 0.5 μM rotenone/antimycin A to the appropriate port of the injector plate. For the detection of ECAR, NRVMs were incubated with l-glutamine (1 mM)-supplemented XF base medium minimal DMEM and the injector plate was added with 10 mM glucose, 1 μM oligomycin and 50 mM 2-DG. The measurement of OCR and ECAR were run on the seahorse XFe96 analyzer (Agilent Technologies, USA) and the results were analyzed using Wave 2.6.1 software.
2.9. NAD+/NADH quantification and ATP content detection
NRVMs were pretreated with songorine (20 μM) or NAC (2 mM), followed by LPS (1 μg/mL) for 24 h. After washing with precooled PBS, intracellular NAD+/NADH ratio was measured according to the manufacturer's protocol of NAD+/NADH quantification kit (Sigma, MAK037), and intracellular ATP contents were assay with a commercial kit (Beyotime, S0027).
2.10. The assay of ROS, mitochondrial membrane potential (Δψm), mitochondrial number and calcium contents
The treated NRVMs were incubated with 10 μM DCFH-DA (Beyotime, S0033S) at 37 °C for 20 min in darkness. After washing, intracellular ROS production was measured by a microplate reader (BERTHOLD Technologies, Germany). For the observation of mitochondrial ROS, NRVMs were treated with 5 μM MitoSOX™ red mitochondrial superoxide indicator (Invitrogen™, M36008) reagent working solution at 37 °C for 10 min and 50 nM Mito-tracker green (Beyotime, C1048) working reagent at 37 °C for 30 min in darkness. Mitochondrial ROS production was also measured using MitoSOX™ red mitochondrial superoxide indicator by a microplate reader (BERTHOLD Technologies, Germany) in H9C2 cells at the time indicated. To visualize ROS production in the heart tissue, the tissue slice was stained with 5 μM dihydroethidium (DHE, Beyotime, S0063) and DAPI (Bioworld Technology, BD5010). Images were viewed with confocal scanning microscopy (Zeiss, LSM 800).
For the analysis of mitochondrial membrane potential (Δψm), NRVMs were loaded with 10 μM TMRM(Invitrogen™, T668) dilution at 37 °C for 30 min in darkness. For the detection of intracellular calcium content, NRVMs were incubated with 5 μM Fluo8-AM (Abcam, ab142773) at 37 °C for 1 h in darkness. After washing with HBSS, cells were observed by confocal scanning microscopy (Zeiss, LSM 800). Intracellular calcium content was also measured using Fluo8-AM by a microplate reader in H9C2 cells at the time indicated. To quantitate mitochondrial number, the treated NRVMs were cultured with 0.1 μM nonyl acridine orange (NAO, Invitrogen™, A1372) at 37 °C for 30 min in darkness. According to the manufacturer's instructions, cellular fluorescence was imaged using confocal scanning microscopy and flow cytometry analysis (Beckman Coulter, Inc., USA), respectively. Mitochondrial number was also measured using NAO by a microplate reader in H9C2 cells exposed to LPS insult.
2.11. Immunofluorescence staining
For detection the protein expression levels of Nrf2, SIRT1 and F4/80 in heart of LPS-treated mice, the heart tissue was embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek), cut into 5 μm slice and then fixed with 4% paraformaldehyde. After fixation, the slide was permeabilized with 0.3% Triton X-100 and blocked with goat serum before being incubated overnight with the primary antibodies (anti-Nrf2, 1:400, Abcam, ab137550; anti-SIRT1, 1:400, Cell Signaling Technology, 9475S and anti-F4/80, 1:100, Abcam, ab100790) overnight at 4 °C in a humidified chamber. After washing, the donkey anti-rabbit IgG H&L (Alexa Fluor® 594) (1:500, Abcam, ab150076) or goat anti-rabbit IgG H&L (Alexa Fluor® 488) (1:500, Abcam, ab150077) was incubated at room temperature for 2 h. And then the slide was staining with DAPI (Bioworld Technology, BD5010) for 10 min and added anti-fluorescence quenching agent (Beyotime, P0126). Protein expression was observed by a confocal scanning microscope.
For the colocalization of Orai1 and STIM1, NRVMs were pre-treated with 1 μM ER-tracker red (Beyotime, C1041) for 30 min at 37 °C, and then fixed with 4% paraformaldehyde for 15 min and permeated with 0.1% Triton X-100 for 10 min. After blocking with 5% bovine serum albumin (BSA), the cells were incubated with anti-Orai1 (1:300, Sigma, SAB3500126), anti-STIM1 (1:800, Cell Signaling Technology, 5668S), anti-Nrf2 (1:300, Abcam, ab89443) or anti-NRF1 (1:300, Cell Signaling Technology, 69432s) overnight at 4 °C, followed by incubation with goat anti-rabbit IgG H&L (Alexa Fluor® 488) (1:500, Abcam, ab150077) and goat anti-mouse IgG H&L (Alexa Fluor® 647) (1:500, Abcam, ab150115) for 2 h at room temperature and DAPI (Bioworld Technology, BD5010) for 10 min at 37 °C. The cells were visualized under a confocal scanning microscope.
2.12. Western blot analysis and immunoprecipitation
Primary NRVMs and heart tissues were lysed with RIPA buffer supplemented with 1 mM PMSF, and the supernatants were collected by centrifugation at 12000 g for 15 min at 4 °C. After that, the protein concentration was determined by the BCA protein assay kit (Thermo Scientific, 23225). The same amounts of protein were separated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes. After blocking with 5% non‐fat milk at room temperature for 2 h, the membranes were immunoblotted with primary antibody (anti-Keap1, 1:1000, Cell Signaling Technology, 4678S; anti-STIM1, 1:1000, Cell Signaling Technology, 5668S; anti-MTCO1, 1:2000, Abcam, ab14705; anti-GAPDH, Abcam, ab181602; anti-Nrf2, 1:1000, Abcam, ab137550, 110KD or ab89443, 68KD; anti-Orai1, 1:2000, Abcam, ab244352; anti-PGC-1α, 1:1000, Abcam, ab54481; anti-SIRT1, 1:1000, Abcam, ab189494; anti-VDAC1, 1:1000, Abcam, ab15895) overnight at 4 °C, and then followed by incubation with the HRP-conjugated secondary antibody (goat anti-rabbit IgG (H + L) HRP, 1:10000, Bioworld Technology, BS13278; goat anti-mousse IgG (H + L) HRP, 1:10000, Bioworld Technology, BS12478) at room temperature for 1 h. Protein bands were visualized using an ECL kit and Image-Pro Plus 6.0 software was applied to quantify the band intensity values.
For immunoprecipitation, the treated NRVMs and heart tissues were lysed on ice for 30 min, and then centrifuged at 12000 g for 15 min to collect the supernatant. Next, the lysate was combined with anti-Orai1 (1:100, Abcam, ab244352) or anti-Nrf2 (1:200, Abcam, ab137550) at 4 °C overnight, and then co-precipitated with protein A + G agarose beads (Med Chem Express, HY-K0202) for 4 h. After washing with PBS for 5 times, the beads were boiled in 1% SDS loading buffer and western blot was performed with indicated antibodies.
2.13. Quantitative real-time PCR
Total RNA was obtained from primary NRVMs or heart tissues using RNA isolater (Vazyme, Nanjing, China) according to the manufacturer's protocol. Monitor the absorbance at 260/280 nm to determine the purity and concentration of extracted RNA. cDNA was synthesized from the total RNA (1 μg) by using the HiScript® Q RT SuperMix for qPCR (Vazyme) and then quantitative real-time PCR (qRT-PCR) was performed on LightCycler 480 II (Roche) using ChamQ SYBR Color qPCR Master Mix (Vazyme). Actb (mouse) or Rn18s (rat) was used to normalize relative mRNA levels, and data was analyzed using the 2−ΔΔCt method. Primer sequences were shown in Supplement Table 1.
2.14. Luciferase reporter assay
The Nrf1 and mitochondrial transcription factor A (Tfam) promoter (-2,000 bps relative to the TSS) were synthesized and inserted in the pGL3-basic vector between Kpnl and Xhol sites (Genebay Biotech Co., Ltd., Nanjing, China). Combined with these plasmids, additional pRL-SV40-Renilla vectors (as a normalized control) were co-transfected into H9C2 cells in the presence or absence of pEX3-Nfe2l2, shNfe2l2, shNrf1 or shPpargc1α using Lipofectamine® 3000 transfection reagent (InvitrogenTM, L3000008, USA) at 70–80 % confluence. 48 h after transfection, the cells were treated with indicated reagents for 24 h and lysed with passive lysis buffer (Promega, E1941). The activities of firefly luciferase and renilla luciferase were detected by the dual-luciferase substrate system (Promega, E2920) according to the manufacturer's instruction.
2.15. Statistical analysis
All of the experimental results are expressed as the means ± SEM (n≥5). After tested for normal distribution using the Shapiro-Wilk test, data were analyzed by unpaired Student's t-test (2 groups). And statistical significance was also determined by one-way ANOVA followed by Tukey's test (>2 two groups). All experiments were randomized and blinded, and p < 0.05 was considered statistically significant. Statistical analysis was performed using GraphPad Prism 8.0 software.
3. Results
3.1. Songorine protected cardiac function with Nrf2 induction during sepsis
As female C57BL/6 mice were reported to be resistant to cardiac injury in sepsis [19], we prepared sepsis model in male C57BL/6 mice. Because low doses of LPS (10 mg/kg) failed to induce mice death within 72 h, we challenged mice with high dose of LPS (30 mg/kg, intraperitoneally), referring to the reported study [20], and songorine was administrated 1 h before LPS challenge and then administered for 3 consecutive days. 72-hour mortality in the sepsis-induced mice was 50%, whereas songorine administration prolonged the survival, as the mortality dropped to 37.5% (10 mg/kg), and no death was observed when treated with high dose (50 mg/kg) (Fig. 1A). Since the characteristic of myocardial dysfunction in sepsis is the loss of contractive force, we examined cardiac function by echocardiography in mice at 24 h after LPS injection (10 mg/kg). Songorine restored the loss of ejection fractions (EF) and shortening fraction (FS) (Fig. 1B) with a decrease in left ventricular end-systolic and end-diastolic volumes (LVVs, LVVd) (Fig. 1C), while the LV mass was not influenced by LPS or songorine (Fig. 1D), demonstrating protective effects of songorine on cardiac contractive function. Post-treatment may be a more clinically relevant means of intervention. We examined heart function at 48 h after LPS insult under the conditions of pre- or post-treatment of songorine, and further confirmed the protective effects of songorine when the impaired contractive function appeared to be improved, in spite of no significant difference in LVVd between the control and LPS treatments (Supplement Fig. 2A–C). The view of transmission electronic microscopy showed notable cytoarchitectural aberrations, especially the disrupted mitochondria, indicated by decline of mitochondrial number and volume density, formation of internal vesicles and loss of cristae, which were alleviated in songorine-treated mice (Fig. 1E). The view of DHE probe and F4/80 staining showed that songorine administration suppressed ROS production and reduced macrophage infiltration into the heart tissue (Fig. 1F and G). Immunofluorescence staining showed that songorine increased Nrf2 protein induction in the heart (Fig. 1H). Hmox1 and Nqo1 are Nrf2 targeting genes. LPS mildly increased the gene expression of Hmox1 and Nqo1, properly an adaptive response to oxidative stress [8], which was potentiated by songorine (Fig. 1I). Superoxide dismutase 2 (Sod2) and catalase are anti-oxidant enzymes transcriptionally activated by Nrf2. The impaired gene expression of Sod2 and Cat was also restored by songorine (Fig. 1J). These results raised the possibility that songorine activated Nrf2 and reinforced antioxidant enzyme defenses to protect the heart.
Fig. 1.
Songorine protected heart function with Nrf2 activation in septic heart. (A) songorine was intraperitoneally administrated 1 h before LPS injection (30 mg/kg) and then administered for 3 consecutive days. The mortality in mice within 72 h was recorded (n = 8). B–I: mice were administrated with songorine (i.p.) 1 h before and 12 h after LPS challenge (10 mg/kg), and the cardiac function was examined after 24 h: (B) ejection fractions (EF), shortening fraction (FS), (C) left ventricular end-systolic volumes (LVVs), left ventricular end-diastolic volumes (LVVd) and (D) left ventricular mass (LV Mass) (n = 6); (E) the morphology of mitochondria in the heart was viewed using transmission electronic microscopy (n = 5, scale bar: 5 μm); (F–H) ROS production, F4/80 staining and Nrf2 protein staining in the heart were observed by immunofluorescence (n = 5, scale bar: 50 μm or 100 μm). (I-J) genes expression of Hmox1, Nqo1, superoxide dismutase 2 (Sod2) and Cat were detected by Q-PCR. Data are presented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS treatment; #p < 0.05, ###p < 0.001 vs. indicated treatment; ns: no significant difference. p values are determined by one-way ANOVA followed by Tukey's test.
3.2. Songorine combated oxidative stress and prevented calcium overload in cardiomyocytes
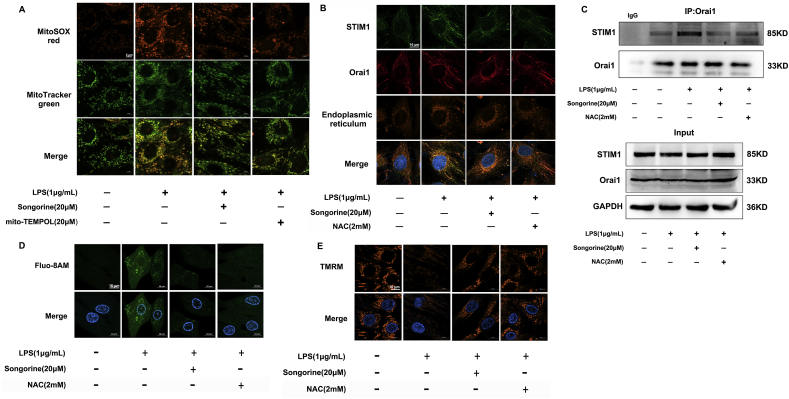
Next, we examined the potency of songorine in suppression of oxidative stress. In primary neonatal rat ventricular myocytes (NRVMs), songorine concentration-dependently reduced LPS-evoked ROS production with an IC50 value of about 20 μM (Supplement Fig. 3A). No impact on cell survival was observed when incubated with songorine at concentrations ranging from 1 μM to 80 μM (Supplement Fig. 3B). Therefore, we chose 20 μM as the working concentration in following experiments to prevent the potential cytotoxicity. As LPS is capable of boosting mitochondrial ROS production in macrophages [21], we viewed mitochondrial ROS production in NRVMs using MitoSOXTM fluorescent probe, and showed that songorine, as well as mito-TEMPOL (a mitochondrial ROS scavenger), decreased mitochondrial ROS generation in response to LPS (Fig. 2A). Calcium release-activated calcium (CRAC) channels mediate calcium influx in response to mitochondrial ROS [22]. STIM proteins are endoplasmic reticulum (ER)-resident Ca2+-sensing molecules. Upon the depletion of intracellular calcium stores, they translocate to the junction of ER to interact with Orai1/2 for the formation of CRAC channels [23]. Although there are different isoforms of STIM and Orai proteins, STIM1 and Orai1 are capable of forming CRAC in neonatal cardiomyocytes [24]. The confocal microscopic image showed that LPS redistributed STIM1 to interact with Orai1 in cardiomyocytes (Fig. 2B), and the interaction was further confirmed by the examination of immunoprecipitation (Fig. 2C). Songorine prevented the formation of CRAC channels (Fig. 2B and C), and resultantly reduced calcium influx in LPS-stimulated cardiomyocytes (Fig. 2D). ROS scavenger n-acetylcysteine (NAC) exhibited effects similar to songorine, suggestive of the involvement of ROS in CRAC channel activation and calcium overload (Fig. 2B–D). We further confirmed the inhibitory effects of songorine on mitochondrial ROS production and calcium load in H9C2 cells at the time ranging from 0.5 to 12 h (Supplement Fig. 3C and D), indicative of the lasting effects. Consistently, post-treatment with songorine also decreased mitochondrial ROS production and intracellular calcium overload in the presence of LPS (Supplement Fig. 3E and F). ROS suppression and calcium handling should confer protection against mitochondrial dysfunction. As expected, songorine prevented the collapse of mitochondrial membrane potential (Δψm) in NRVMs exposed to LPS (Fig. 2E).
Fig. 2.
Songorine prevented ROS-associated calcium overload. Primary neonatal rat ventricular myocytes (NRVMs) were stimulated with LPS (1 μg/mL) for 24 h. (A) mitochondrial ROS production in NRVMs (scale bar: 5 μm); (B) the view of co-localization of STIM1 and Orai1 in NRVMs (scale bar: 10 μm, green: STIM1, red: Orai1, yellow: endoplasmic reticulum); (C) the binding of STIM1 to Orai1 determined by immunoprecipitation in NRVMs; (D) calcium influx and (E) mitochondrial membrane potential (Δψm) in NRVMs (scale bar: 10 μm). NAC, n-acetylcysteine. All data are repeated in five independent experiments. p values are determined by one-way ANOVA followed by Tukey's test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Songorine protected Nrf2 protein by promoting Keap1 degradation
As songorine mediated Nrf2 induction in mouse heart during sepsis, we tested whether songorine activated Nrf2 to protect heart function. In the absence of activators, synthesized Nrf2 protein is continuously degraded by Keap1-dependent ubiquitination in the cytosol [25], and thus we examined the effect of songorine on Keap1 protein expression in NRVMs. When protein synthesis was inhibited by cycloheximide, Keap1 protein expression remained relatively stable, but songorine treatment induced a continuous degradation from 4 to 12 h (Fig. 3A). In contrast, the continuous degradation of exogenous Nrf2 protein was prevented by songorine (Fig. 3B). Keap1 degradation by songorine was abrogated by proteasome inhibitor MG-132 (Fig. 3C), suggesting that songorine impaired Keap1 stability via proteasomal degradation. Concordantly, songorine significantly increased Nrf2 protein expression in normal and LPS-stimulated cardiomyocytes (Fig. 3D). These results indicated that songorine promoted Keap1 degradation to protect Nrf2 stability. Similar to the regulation in the heart of septic mice (Fig. 1J), songorine restored gene expression of Sod2 and Cat in cardiomyocytes exposed to LPS, but these actions were blocked by Nfe2l2 silencing (Fig. 3E, Supplement Fig. 4A), suggesting that songorine combated oxidative stress in a manner dependent on Nrf2.
Fig. 3.
Songorine preserved Nrf2 protein abundance in cardiomyocytes. (A) Keap1 protein degradation when protein synthesis was inhibited by cycloheximide in primary neonatal rat ventricular myocytes (NRVMs); (B) exogenous Nrf2 protein stability when protein synthesis was inhibited by cycloheximide in NRVMS; (C, D) Keap1 and Nrf2 protein expression in NRVMs stimulated with LPS for 24 h; (E) gene expression of superoxide dismutase 2 (Sod2) and Cat in NRVMs exposed to LPS for 24 h. All data are presented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS or LPS + shNC treatment; ###p < 0.001 vs. indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test.
3.4. Songorine rescued mitochondrial respiration dependently on Nrf2
The energy metabolism of cardiomyocytes mainly relies on mitochondrial OXPHOS. Although the basal oxygen consumption rate (OCR) was little affected, LPS reduced maximum OCR, indicative of the impairment in the content or electron transport activity of mitochondria (Fig. 4A). Meanwhile, extracellular acidification rate (ECAR) was elevated (Fig. 4B). These results indicated that metabolism was shifted away from mitochondrial OXPHOS. However, the altered metabolism was reversed by songorine and ROS scavenger NAC (Fig. 4A and B). Increased ATP production by songorine was reduced by OXPHOS uncoupler FCCP but not hexokinase inhibitor 2-DG (Fig. 4C), suggesting that songorine protected ATP generation from mitochondrial source. As fatty acid β-oxidation and the TCA cycle provide reducing equivalent to the respiratory chain, we investigated the regulation of genes involved in fatty acid β-oxidation, TCA cycle and OXPHOS in NRVMs. Carnitine palmitoyltransferase-1 (CPT-1) facilitates the uptake of fatty acetyl coenzyme A into mitochondria, and Ehhadh is α-subunit of mitochondrial trifunctional protein in fatty acid β-oxidation. qRT-PCR revealed that downregulation of Cpt1α and Ehhadh by LPS were normalized by songorine (Fig. 4D). Similarly, songorine increased gene induction involved in the TCA cycle (Fh, Idh3a and Sdha) and OXPHOS (Cycs, Ndufv1, Ndufs1) (Fig. 4E and F). Nrf2 inhibitor ML385 diminished the effects of songorine, suggestive of the role of Nrf2 in the regulation of mitochondrial genes. ND1 and COX1 are encoded by mitochondrial DNA, and the induction by songorine was Nrf2-dependent, indicative of the ability to regulate mitochondrial biogenesis (Fig. 4G). These results showed that songorine preserved mitochondrial genes induction encoded by both nuclear and mitochondrial genomes. Mitochondria are a dynamic network within the cell, and the quality and quantity control are essential for OXPHOS function. To quantify mitochondrial number, we utilized the fluorescent dye of nonyl acridine orange (NAO) that yields a fluorescence signal proportional to mitochondrial number [26]. LPS stimulation impaired mitochondrial mass with morphological derangements, which were normalized by songorine in a manner that was dependent on Nrf2 (Fig. 4H). Autophagy is a rescue way when intracellular components are impaired. Autophagy inhibitor 3-methyladenine (3-MA) attenuated the protective effects of songorine on mitochondrial mass (Supplement Fig. 5), suggestive of the potential effect of autophagy in mitochondrial repairment. Given the involvement of Nrf2/ARE in mitochondrial biogenesis [11], these data indicated that in addition to combating oxidative stress, regulation of mitochondrial biogenesis via Nrf2 induction was a way for songorine to protect mitochondrial integrity and function.
Fig. 4.
Songorine protected mitochondrial respiration. Primary neonatal rat ventricular myocytes (NRVMs) were treated with LPS (1 μg/mL) for 24 h: (A, B) oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured in NRVMs; (C) ATP production in NRVMs; (D) gene expression of Cpt1α and Ehhadh were detected in NRVMs; (E) gene expression of Fh, Idh3a and Sdha were detected in NRVMs; (F) gene expression of Cycs, Ndufv1 and Ndufs1 were detected in NRVMs; (G) gene expression of ND1 and COX1 were detected in NRVMs; (H) mitochondrial mass staining with nonyl acridine orange (NAO) in NRVMs (scale bar: 20 μm). NAC, n-acetylcysteine. All data are presented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS or LPS + shNC treatment; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. indicated treatment; ns: no significant difference. p values are determined by one-way ANOVA followed by Tukey's test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Songorine induction of Nrf2 regulated NRF1 and mitochondrial biogenesis
In addition to the regulation of electron transport chain (ETC) subunits encoded by the nuclear genome, NRF1 also binds to the promoter of TFAM that is directly responsible for mtDNA replication [27]. Although gene induction of Nrf1 and Tfam were not significantly influenced by LPS, songorine potentiated both gene expression in NRVMs. Nrf2 inhibitor ML385 blocked the effects of songorine (Fig. 5A), indicative of the involvement of Nrf2/ARE in NRF1 signaling cascades. The view of confocal microscopy showed that songorine promoted Nrf2 transport into the nucleus with co-location of NRF1 in cardiomyocytes, but the interaction was blocked by ML385 (Fig. 5B). In line with this regulation, songorine increased luciferase report activity of Nrf1 in LPS-treated H9C2 cells, but the action was blocked by Nfe2l2 knockdown (Fig. 5C, Supplement Fig. 4A). Meanwhile, songorine induction of Tfam promoter activity was abrogated by Nrf1 knockdown (Fig. 5D, Supplement Fig. 4B). These results provided evidence that songorine transcriptionally activated TFAM through Nrf2/ARE and NRF1 signaling cascades, implicating in mitochondrial biogenesis.
Fig. 5.
Songorine regulated NRF1 and TFAM activity. (A) Nrf1 and Tfam gene expression in primary neonatal rat ventricular myocytes (NRVMs) exposed to LPS for 24 h; (B) the view of Nrf2 transport into the nucleus with co-location of NRF1 in NRVMs stimulated with LPS for 24 h (scale bar: 5 μm, green: NRF1, red: Nrf2) (n = 5); (C, D) luciferase report activity of Nrf1 and Tfam in H9C2 cells stimulated with LPS for 24 h. All data are presented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS or LPS + shNC treatment; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. PGC-1α was required for songorine induction of mitochondrial biogenesis
PGC-1a is a metabolic coactivator and characterized by interacting with transcription factors to regulate mitochondrial biogenesis and respiration. In NRVMs, LPS impaired gene and protein expression of PGC-1α, whereas these were rescued by songorine in a manner dependent on Nrf2 (Fig. 6A). Increased NAD+/NADH ratio and SIRT1 induction might be a reason for PGC-1α protein induction by songorine (Supplement Fig. 6A and B), because NAD+ is required for SIRT1 to deacetylate of PGC-1α for activation [13]. In support of this, NAD+ precursor β-nicotinamide mononucleotide (NMN) preserved PGC-1α protein abundance by improving its stability (Supplement Fig. 6C). These results suggested a link between redox status and mitochondrial biogenesis. To know if Nrf2 co-activated PGC-1α via direct interaction, we examined the affinity of PGC-1α to Nrf2. The immunoprecipitation assay showed when LPS impaired the interaction in cardiomyocytes, songorine increased PGC-1α binding to Nrf2, which was blocked by Nrf2 inhibitor ML385 (Fig. 6B). Nrf2 overexpression increased luciferase report activity of Nrf1, but this action was blocked by Ppargc1α silencing (Fig. 6C, Supplement Fig. 4C). Similar regulation was also observed in the regulation of the activity of Tfam promoter (Fig. 6D). In LPS-treated cardiomyocytes, Nrf2 overexpression increased gene expression of Nrf1 and Tfam, whereas the enhancing effect was lost in the presence of PGC-1α inhibitor SR18292, which inactivates PGC-1α by preserving acetylation [28] (Fig. 6E). To quantify mitochondrial number using flow cytometry, we treated NRVMs with NAO and chose the mean fluorescence intensity of 8 × 105 as an arbitrary reference point. In the untreated cardiomyocytes, only 25.28 % of cells had a signal below this threshold; however, the ratio raised to 79.91% after LPS treatment (Fig. 6F). Forced Nrf2 expression significantly increased the fluorescence signal, but the effect was diminished by co-transfecting with shPpargc1α (Fig. 6F). These results indicated that co-activation of PGC-1α was required for Nrf2 to promote mitochondrial biogenesis. Mitochondria biogenesis is mediated by both mitochondrial and nuclear genome. MTCO1 and VDAC1 are encoded by mitochondrial and nuclear DNA, respectively. LPS triggered a decrease in the abundance of the two proteins. Nrf2 overexpression restored protein expression of MTCO1 and VDAC1, but the effects were attenuated in the presence of PGC-1α inhibitor SR18292 (Fig. 6G). The protein induction of MTCO1 and VDAC1 by songorine were also impaired by Nrf2 inhibitor ML385 and PGC-1α inhibitor SR18292, respectively (Fig. 6H). Together, these results indicated that co-activation of Nrf2 and PGC-1α was required for songorine to protect mitochondrial biogenesis.
Fig. 6.
PGC-1α was required for songorine induction of mitochondrial biogenesis. (A) PGC-1α gene and protein expression in primary neonatal rat ventricular myocytes (NRVMs) stimulated with LPS for 24 h; (B) the immunoprecipitation assay of the binding of PGC-1α to Nrf2 in NRVMs stimulated with LPS for 24 h; (C, D) luciferase report activities of Nrf1 and Tfam in H9C2 cells stimulated with LPS for 24 h; (E) gene expression of Nrf1 and Tfam in NRVMs exposed to LPS for 24 h; (F) mitochondrial number labeled with nonyl acridine orange (NAO) was viewed using flow cytometry in NRVMs stimulated with LPS for 24 h; (G, H) MTCO1 and VDAC1 protein expression in NRVMs treated with LPS for 24 h. All data are presented as mean ± SEM (n = 5); *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS or LPS + pEX3-NC treatment; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.7. Cardiac Nrf2 deficiency attenuated protective effects of songorine during sepsis
To further confirm Nrf2-dependent role in songorine action in vivo, cardiac Nrf2 was knocked down by tail vein injection of AAV9-shNfe2l2 in mice. The mRNA level of Nfe2l2 was markedly reduced and the effect of songorine on Nrf2 protein abundance was blocked by cardiac knockdown of Nfe2l2, confirming the efficiency of Nrf2 depletion (Supplement Fig. 7A and B). The data of echocardiography showed that the protective effects of songorine on heart function was lost in Nrf2 deficient heart (Fig. 7A). The test of immunoprecipitation showed that songorine increased SIRT1 expression in the septic heart dependent on Nrf2 (Fig. 7B). The binding of Nrf2 to PGC-1α was impaired by LPS challenge, whereas songorine increased the interaction in the septic heart, largely owing to the increased Nrf2 protein abundance (Fig. 7C). Different from what was observed from LPS-stimulated cardiomyocytes (Fig. 7C and D), LPS challenge in mice impaired cardiac gene induction of Nrf1 and Tfam, which were normalized by songorine in a manner that was Nrf2 dependent (Fig. 7D). More pathogenic factors converged in septic heart injury might be a possible reason for the discrepancy. Compared to the AAV9-shNC, OXPHOS mRNAs encoded by the mitochondrial (ND1 and COX1) and the nuclear genome (Cycs, Ndufv1, Ndufs1) were all significantly lower in the septic heart, and were rescued by songorine in a manner dependent on Nrf2 (Fig. 7E and F). Concordantly, the promotion effect of songorine on the protein expression of MTCO1 and VDAC1 were also attenuated in Nrf2 deficient heart (Fig. 7F). Collectively, these results reproduced the findings in vitro, and confirmed that songorine regulated Nrf2/ARE and PGC-1α cascades to promote mitochondrial biogenesis.
Fig. 7.
Cardiac Nrf2 deficiency attenuated protective effects of songorine. Male C57BL/6 mice were injected with AAV9-shNfe2l2 or AAV9-shNC through the caudal vein. Mice were administrated with songorine (i.p.) 1 h before and 12 h after LPS challenge (10 mg/kg), and the heart was examined 24 h later: (A) ejection fraction (EF), shortening fractional (FS), left ventricular end-systolic volumes (LVVs), left ventricular end-diastolic volumes (LVVd) and left ventricular mass (LV Mass) (n = 7); (B) immunofluorescence staining of cardiac SIRT1 protein (scale bar: 100 μm), one presentation of five mice; (C) immunoprecipitation examination of the binding of PGC-1α to Nrf2 (n = 5); (D-F) cardiac genes expression of Nrf1 and Tfam,ND1 and COX1, Cycs, Ndufv1 and Ndufs1were detected; (G) MTCO1 and VDAC1 protein expression in heart tissue. All data are presented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS + AAV9-shNC treatment; #p < 0.05,##p < 0.01, ###p < 0.001 vs. indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test.
4. Discussion
Although LPS induced global mitochondrial dysfunction in the heart, we showed that this impairment was reversible by pharmacological intervention from the aspect of mitochondrial biogenesis. More than maintenance of redox homeostasis [29], Nrf2 activated AREs/PGC-1α cascades to protect mitochondrial quantity and metabolism in cardiomyocytes during sepsis. This finding suggests that regulation of mitochondrial biogenesis is a way for Nrf2/ARE signaling to attenuate septic heart injury.
Although more important organs are affected by sepsis, myocardial injury is the leading cause of mortality and morbidity during septic shock [30,31]. Cardiac contractive function is supported by high energy consumption, largely depending on mitochondrial OXPHOS. However, damaged mitochondria impair respiration and electron transfer, and thus fail to support heart function in septic cardiomyopathy [28]. Songorine treatment increased mitochondrial density with improved morphologic integrity, providing a structural integrity to protect mice survival during sepsis. It is well known that LPS increases ROS production mainly form NADPH oxidase in activated macrophages [32], however, recently it is found that LPS promotes ROS production from mitochondria owing to reverse electron transport as mitochondrial ATP synthase is inhibited [21]. This finding elucidates another important source for ROS generation in the context of altered metabolism. In line with this, mitochondrial ROS production also increases substantially in ischemic heart in a similar way [33]. Protection of mitochondrial function should be a way for songorine to reduce mitochondrial ROS production.
ROS-induced mitochondrial dysfunction and abnormal calcium handling are important mediators. Voltage dependent L-type Ca2⁺ channel-mediated Ca2⁺ influx in heart diseases is well established, while store-operated calcium entry emerges as another regulatory mechanism for Ca2+ cycling and the alternation is implicated in heart failure and hypertrophy [34]. Calcium release-activated calcium (CRAC) channels are formed in response to the depletion of Ca2⁺ stores within the endoplasmic or sarcoplasmic reticulum, facilitating calcium influx. In LPS-stimulated cardiomyocytes, we observed the formation of CRAC channels and subsequent calcium overload. Suppression of mitochondrial ROS production by songorine had a contribution to preventing CRAC channels formation and calcium influx. In support of this, a recent study reported that mitochondrial ROS activates CRAC channels to facilitate calcium influx in activated macrophage [22]. Endotoxin is proposed to induce calcium overload via the leakage from sarcoplasmic reticulum [35]. Sarcoplasmic reticulum calcium leak via type 2 ryanodine receptor channels is also responsible for mitochondrial calcium overload in heart failure [36]. Because calcium content within sarcoplasmic reticulum is continuously refilled by CRAC channels, we speculate that CRAC channels-mediated calcium influx provides a way to refill calcium content when stored-calcium is depleted. Elevated level of cytosolic calcium promotes calcium uptake by mitochondria through a specific channel in the mitochondrial inner membrane, termed mitochondrial calcium uniporter (MCU) [37], which is a multiprotein complex that regulates mitochondrial calcium [38]. Mitochondrial calcium overload is an important cause of ROS production and mitochondrial dysfunction [37,39], and CRAC channels act as a regulator of calcium signaling, implicated in brain injury and neuronal death [40,41]. Nrf2 upregulates genes encoding antioxidant and detoxifying enzymes or proteins, including Sod2 and catalase. Mitochondrial ROS is generated in the form of superoxide, which are consequently moved by Sod2 and catalase. Therefore, we reasoned that ROS suppression and calcium handing by songorine induction of Nrf2 conferred protection to prevent mitochondrial dysfunction.
LPS-activated macrophages are characteristic of highly aerobic glycolysis with a broken TCA cycle [42]. In cardiomyocytes, we also observed that LPS challenge shifted bioenergy away from mitochondria, similar to what happened in classically activated macrophages. LPS impaired mitochondrial genes encoding OXPHOS, and the reduced mitochondrial capacity of bioenergy is consistent with the finding in the heart of patients who died from sepsis [43]. Although songorine induction of mitochondrial genes were dependent on Nrf2 activation, the actions cannot be simply explained by its antioxidative effects, because mitochondrial biogenesis in the case of low ATP production is a response to replenish damaged mitochondria. Songorine increased mitochondrial mass dependently on Nrf2, indicative of the potential of Nrf2/ARE in the regulation of mitochondrial biogenesis.
ARE is a promoter element implicating in antioxidant and detoxify genes, and Nrf2 is an activator of ARE by binding to AREs in the target gene promoters. Endogenous CO generated by HO-1 permits Nrf2 occupancy of AREs in the Nrf1 promoters to protect cardiomyocytes from apoptosis, which was proposed to be regulated by Akt [8]. However, we demonstrated that songorine promoted Nrf2 nuclear translocation for binding to Nrf1 promoter. This action is unlikely to be a result derived from HO-1 activation, because HO-1 is the downstream target gene of Nrf2. Although LPS evoked massive ROS production in cardiomyocytes, it failed to impact the activity of Nrf1 promoter, suggesting that Nrf2 transcriptionally regulated NRF1 induction in a manner that was independent from its role in ROS suppression. NRF1, as well as NRF2, regulates gene expression of mitochondrial complexes in the ETC, encoded by the nuclear genome. Specifically, NRF1 indirectly regulates mitochondrially encoded genes in the ETC via activating TFAM, which is a key enhancer protein ensuring mtDNA replication by mtDNA polymerase γ [11]. In fact, mitochondrial biogenesis is a compensatory response secondary to the damaged respiratory apparatus and low ATP production, aiming to replenish mitochondrial components. Meanwhile, mitophagy is also activated to rescue cardiomyocytes [44]. Songorine potentiated NRF1 and TFAM induction dependently of Nrf2, providing a rational mechanism through which songorine upregulated both nucleus and mitochondria-encoded genes involved in mitochondrial function.
Interestingly, we found that songorine increased PGC-1α and SIRT1 protein abundance, largely due to metabolic regulation. SIRT1 is an NAD+-responsive deacetylase that serves as a redox rheostat, and therefore, declining NAD+ was the main reason of SIRT1 inhibition in LPS-stimulated cardiomyocytes. Although SIRT1 is predominantly located in the nucleus, it shuttles in and out of the nucleus to sense the change of NAD+ levels and activates nuclear PGC-1α by deacetylation. In aging mice, declining nuclear NAD+ is shown to disrupt mitochondrial biogenesis in an SIRT1-dependent manner [45]. PGC-1α protein can be regulated at the post-transcriptional level. Glycogen synthase kinase 3β (GSK3β) inhibits PGC-1α via intranuclear proteasomal degradation [46]. In contrast, we showed that NAD+ supplement preserved PGC-1α protein abundance by improving its stability in the setting of LPS challenge, a regulation presumably due to SIRT1-mediated PGC-1α deacetylation. In this context, increased NAD+/NADH ratio and SIRT1 protein expression by songorine had a contribution to preserving PGC-1α induction. There are data that confirm overlapping functions between PGC-1α and Nrf2 [11]. PGC-1α interacts with specific transcriptional factors to exert its biological functions, including mitochondrial biogenesis [47]. Therefore, we speculated that Nrf2 transcriptionally regulated mitochondrial genes in cooperation with PGC-1α. Supporting this notion, we observed that songorine activated Nrf2 with binding to PGC-1α. Forced Nrf2 expression enhanced gene induction of Nrf1 and Tfam with increased mitochondrial number in a manner dependent on PGC-1α. These events provided evidence to support the existence of Nrf2/ARE and PGC-1α signaling cascades in the process of mitochondrial biogenesis. In view of metabolic regulation of PGC-1α by NAD+-dependent SIRT1 activation, it is rational to believe that preventing metabolic shift by songorine had a contribution to improving mitochondrial biogenesis from the aspect of metabolic and redox homeostasis.
In septic heart, we also observed that songorine protected heart function and rescued gene encoding mitochondrial OXPHOS dependently on Nrf2, and the induction of PGC-1α, NRF1 and TFAM suggested the involvement of mitochondrial biogenesis. These results from septic heart provided evidence in vivo to support the findings observed in vitro. We should note that although songorine induction of Nrf2 promoted mitochondrial biogenesis, this was not the only reason for its protection in vivo. Together with mitochondrial biogenesis, anti-arrhythmic and anti-inflammatory effects of songorine should have a contribution to cardioprotection [15,16]. In fact, sepsis is a systemic inflammatory response that affects vessel endothelium and multiple organs. Macrophage activation is supported by metabolic remodeling [42]. Logically, protection of mitochondrial biogenesis by songorine should have a contribution to restraining inflammatory response via metabolic inactivation of macrophages. Intervention to inactivate TLR4 and downstream NF-κB signaling also confer protection in cardiomyocytes [48,49]. Therefore, a comprehensive study considering systemic regulation from different aspects is needed for the full understanding of the cardioprotective role of songorine during sepsis.
5. Conclusions
Overall, we concluded that impaired mitochondrial respiration is the main course of heart failure during septic cardiomyopathy and promotion of mitochondrial biogenesis is a way to improve heart function. Songorine activated Nrf2/ARE and NRF1 signaling cascades to rescue cardiomyocyte from endotoxin insult by promoting mitochondrial biogenesis in a manner that was incorporation with PGC-1α (the diagrammatic summary is shown in Fig. 8). This finding suggests the possible transformation of songorine to clinical application for the treatment of septic heart injury.
Fig. 8.
Schematic regulatory mechanism of songorine action. Songorine activated Nrf2 that transcriptionally regulated NRF1 induction in cooperation with PGC-1α. NRF1 directly and indirectly (in TFAM induction) upregulated nuclearly and mitochondrially encoded OXPHOS genes to promote mitochondrial biogenesis. In addition, songorine reduced CRAC channels-mediated calcium influx to protect mitochondrial function by suppressing mitochondrial ROS production.
Declaration of competing interest
None.
Acknowledgements
All of the authors thank Li Guo, Wei Jiang and Ping Zhou for technical support from State Key Laboratory of Natural Medicines of China Pharmaceutical University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101771.
Formatting of funding sources
This study is supported by the National Natural Science Foundation of China (Nos. 81673592, 81722048), “Double First-Class University project (CPU2018GY09) and the China Postdoctoral Science Foundation (2018M642379).
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplement Fig. 1.

Chemical structure of songorine.
Supplement Fig. 2.
Songorine protected heart function in septic heart. Mice were treated with songorine (50 mg/kg) 1 h before and 24 h after LPS treatment, as the Pre-songorine group, while the group of Post-songorine was administrated with songorine (50 mg/kg) 2 h and 24 h after LPS treatment (n = 9). The cardiac function was examined 48 h after LPS treatment: (A) ejection fractions (EF), shortening fraction (FS), (B) left ventricular end-systolic volumes (LVVs), left ventricular end-diastolic volumes (LVVd) and (C) left ventricular mass (LV Mass). *p < 0.05, ***p < 0.001vs. LPS treatment; #p < 0.05, ###p < 0.001vs. indicated treatment; ns: no significant difference. p values are determined by one-way ANOVA followed by Tukey's test.
Supplement Fig. 3.
Songorine inhibited ROS production and calcium overload in cardiomyocytes. (A) ROS production in primary neonatal rat ventricular myocytes (NRVMs) exposed to LPS for 24 h; (B) cell survival in songorine-treated NRVMs was measures by a microplate reader. The data are presented as mean ± SEM (n = 5), *p < 0.05, ***p < 0.001vs. Control treatment. (C–F) H9C2 cells were treated with songorine (20 μM) and exposed to LPS at the indicated time points. (C) mitochondrial ROS production and (D) calcium content were measured by a microplate reader. H9C2 cells were post-treated with songorine (20 μM) 2 h after LPS stimulation, (E) mitochondrial ROS production and (F) calcium content were measured by a microplate reader 24 h later. All data are presented as mean ± SEM (n = 5 or 6). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS treatment; ###p < 0.001vs. indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test.
Supplement Fig. 4.
Short hairpin RNA (shRNA) plasmid screening. Primary neonatal rat ventricular myocytes (NRVMs) were transfected with shNC, shNfe2l2, shNrf1 or shPpargc1α plasmids using Lipofectamine 3000 transfection reagent for 48 h. (A-C) The gene expression of Nfe2l2,Nrf1 and Ppargc1α were tested by qRT-PCR. All data are presented as mean ± SEM (n = 5). ***p < 0.001vs. shNC treatment. p values are determined by one-way ANOVA followed by Tukey's test.
Supplement Fig. 5.

Songorine protected mitochondrial mass in the presence of autophagy inhibitor. Mitochondrial number was detected with nonyl acridine orange (NAO) and measured using a microplate reader in primary neonatal rat ventricular myocytes (NRVMs) stimulated with LPS for 24 h (3-MA, 3-methyladenine). All data are presented as mean ± SEM (n = 5). ***p < 0.001 vs. LPS treatment; ##p < 0.01, ###p < 0.001 vs. indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test.
Supplement Fig. 6.
Songorine improved PGC-1α protein stability. (A) NAD+/NADH ratio in primary neonatal rat ventricular myocytes (NRVMs) after LPS stimulation; (B) SIRT1 protein expression in NRVMs; (C) PGC-1α protein expression in the indicated time. All data are presented as mean ± SEM (n = 5). *p < 0.05, **p < 0.01, ***p < 0.001 vs. LPS treatment; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. Control or indicated treatment. p values are determined by one-way ANOVA followed by Tukey's test.
Supplement Fig. 7.
Gene and protein level of Nrf2 in Nrf2 knockdown heart. Male C57BL/6 mice were injected with AAV9-shNfe2l2 or AAV9-shNC through the caudal vein: (A) the cardiac mRNA level of Nfe2l2. The data are presented as mean ± SEM (n = 5). ###p < 0.001 vs. indicated treatment. After tested for normal distribution using the Shapiro-Wilk test, data were analyzed by unpaired Student's t-test (2 groups). (B) immunofluorescence staining of cardiac Nrf2 protein (scale bar: 100 μm), one presentation of five mice.
References
- 1.Pulido J.N., Afessa B., Masaki M. Clinical spectrum, frequency, and significance of myocardial dysfunction in severe sepsis and septic shock [J] Mayo Clin. Proc. 2012;87(7):620–628. doi: 10.1016/j.mayocp.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin L., Derwall M., Al Zoubi S. The septic heart: current understanding of molecular mechanisms and clinical implications [J] Chest. 2019;155(2):427–437. doi: 10.1016/j.chest.2018.08.1037. [DOI] [PubMed] [Google Scholar]
- 3.Kumar V. Sepsis roadmap: what we know, what we learned, and where we are going [J] Clin. Immunol. 2020;210:108264. doi: 10.1016/j.clim.2019.108264. [DOI] [PubMed] [Google Scholar]
- 4.Durand A., Duburcq T., Dekeyser T. Involvement of mitochondrial disorders in septic cardiomyopathy [J] Oxid. Med. Cell Longev. 2017;2017:4076348. doi: 10.1155/2017/4076348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy M.P., Hartley R.C. Mitochondria as a therapeutic target for common pathologies [J] Nat. Rev. Drug Discov. 2018;17(12):865–886. doi: 10.1038/nrd.2018.174. [DOI] [PubMed] [Google Scholar]
- 6.Scarpulla R.C., Vega R.B., Kelly D.P. Transcriptional integration of mitochondrial biogenesis [J] Trends Endocrinol. Metabol. 2012;23(9):459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleyzer N., Vercauteren K., Scarpulla R.C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators [J] Mol. Cell Biol. 2005;25(4):1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piantadosi C.A., Carraway M.S., Babiker A. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1 [J] Circ. Res. 2008;103(11):1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacGarvey N.C., Suliman H.B., Bartz R.R. Activation of mitochondrial biogenesis by heme oxygenase-1-mediated NF-E2-related factor-2 induction rescues mice from lethal Staphylococcus aureus sepsis [J] Am. J. Respir. Crit. Care Med. 2012;185(8):851–861. doi: 10.1164/rccm.201106-1152OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn G.W., Vega R.B., Kelly D.P. Mitochondrial biogenesis and dynamics in the developing and diseased heart [J] Genes Dev. 2015;29(19):1981–1991. doi: 10.1101/gad.269894.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1α signaling pathways [J] Front. Genet. 2019;10:435. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aquilano K., Baldelli S., Pagliei B. p53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance [J] Antioxidants Redox Signal. 2013;18(4):386–399. doi: 10.1089/ars.2012.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantó C., Gerhart-Hines Z., Feige J.N. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity [J] Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou G.H., Tang L.Y., Zhou X.D. A review on phytochemistry and pharmacological activities of the processed lateral root of Aconitum carmichaelii Debeaux [J] J. Ethnopharmacol. 2015;160:173–193. doi: 10.1016/j.jep.2014.11.043. [DOI] [PubMed] [Google Scholar]
- 15.Shakhidoyatova N.K., Dzhakhangirov F.N., Sultankhodzhaev M.N. Antiarrhythmic activity of diterpenoid alkaloids of the napelline type and their acylated derivatives [J] Pharm. Chem. J. 2001;35(5):266–267. doi: 10.1023/A:1011917406301. [DOI] [Google Scholar]
- 16.Nesterova Y.V., Povetieva T.N., Suslov N.I. Anti-inflammatory activity of diterpene alkaloids from Aconitum baikalense [J] Bull. Exp. Biol. Med. 2014;156(5):665–668. doi: 10.1007/s10517-014-2421-4. [DOI] [PubMed] [Google Scholar]
- 17.Khan H., Nabavi S.M., Sureda A. Therapeutic potential of songorine, a diterpenoid alkaloid of the genus Aconitum [J] Eur. J. Med. Chem. 2018;153:29–33. doi: 10.1016/j.ejmech.2017.10.065. [DOI] [PubMed] [Google Scholar]
- 18.Akao M., Ohler A.O., Rourke B. Mitochondrial ATP-sensitive potassium channels inhibit apoptosis induced by oxidative stress in cardiac cells [J] Circ. Res. 2001;88(12):1267–1275. doi: 10.1161/hh1201.092094. [DOI] [PubMed] [Google Scholar]
- 19.Kokkinaki D., Hoffman M., Kalliora C. Chemically synthesized Secoisolariciresinol diglucoside (LGM2605) improves mitochondrial function in cardiac myocytes and alleviates septic cardiomyopathy [J] J. Mol. Cell. Cardiol. 2019;127:232–245. doi: 10.1016/j.yjmcc.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recknagel P., Gonnert F.A., Halilbasic E. Mechanisms and functional consequences of liver failure substantially differ between endotoxaemia and faecal peritonitis in rats [J] Liver Int. 2013;33(2):283–293. doi: 10.1111/liv.12012. [DOI] [PubMed] [Google Scholar]
- 21.Mills E.L., Kelly B., Logan A. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages [J] Cell. 2016;167(2):457–470. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soberanes S., Misharin A.V., Jairaman A. Metformin targets mitochondrial electron transport to reduce air-pollution-induced thrombosis [J] Cell Metabol. 2019;29(2):335–347. doi: 10.1016/j.cmet.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prakriya M., Lewis R.S. Store-operated calcium channels [J] Physiol. Rev. 2015;95(4):1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javier A.M., Isabel M.G., Alejandro D.R. The complex role of store operated calcium entry pathways and related proteins in the function of cardiac, skeletal and vascular smooth muscle cells [J] Front. Physiol. 2018;9:257. doi: 10.3389/fphys.2018.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi A., Kang M.I., Okawa H. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2 [J] Mol. Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson J., Duchen M.R., Heales S.J.R. Intracellular distribution of the fluorescent dye nonylacridine orange responds to the mitochondrial membrane potential: implications for assays of cardiolipin and mitochondrial mass [J] J. Neurochem. 2002;82(2):224–233. doi: 10.1046/j.1471-4159.2002.00945.x. [DOI] [PubMed] [Google Scholar]
- 27.Kang D., Kim S.H., Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions [J] Mitochondrion. 2007;7(1–2):39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Vanasco V., Saez T., Magnani N.D. Cardiac mitochondrial biogenesis in endotoxemia is not accompanied by mitochondrial function recovery [J] Free Radic. Biol. Med. 2014;77:1–9. doi: 10.1016/j.freeradbiomed.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Shanmugam G., Challa A.K., Litovsky S.H. Enhanced Keap1-Nrf2 signaling protects the myocardium from isoproterenol-induced pathological remodeling in mice [J] Redox Biol. 2019;27:101212. doi: 10.1016/j.redox.2019.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudiger A., Singer M. Mechanisms of sepsis-induced cardiac dysfunction [J] Crit. Care Med. 2007;35(6):1599–1608. doi: 10.1097/01.CCM.0000266683.64081.02. [DOI] [PubMed] [Google Scholar]
- 31.Merx M.W., Weber C. Sepsis and the heart [J] Circulation. 2007;116(7):793–802. doi: 10.1161/CIRCULATIONAHA.106.678359. [DOI] [PubMed] [Google Scholar]
- 32.Jha A.K., Huang S.C.C., Sergushichev A. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization [J] Immunity. 2015;42(3):419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Chouchani E.T., Pell V.R., Gaude E. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS [J] Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Čendula R., Dragún M., Gažová A. Changes in STIM isoforms expression and gender-specific alterations in Orai expression in human heart failure [J] Physiol. Res. 2019;68(Suppl 2):S165–S172. doi: 10.33549/physiolres.934300. [DOI] [PubMed] [Google Scholar]
- 35.Hassoun S.M., Marechal X., Montaigne D. Prevention of endotoxin-induced sarcoplasmic reticulum calcium leak improves mitochondrial and myocardial dysfunction [J] Crit. Care Med. 2008;36(9):2590–2596. doi: 10.1097/CCM.0b013e3181844276. [DOI] [PubMed] [Google Scholar]
- 36.Santulli G., Xie W.J., Reiken S.R. Mitochondrial calcium overload is a key determinant in heart failure [J] Proc. Natl. Acad. Sci. U.S.A. 2015;112(36):11389–11394. doi: 10.1073/pnas.1513047112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhuri D., Sancak Y., Mootha V.K. MCU encodes the pore conducting mitochondrial calcium currents. [J]. Elife. 2013;2 doi: 10.7554/eLife.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J.C., Liu J., Holmström K.M. MICU1 serves as a molecular gatekeeper to prevent in vivo mitochondrial calcium overload [J] Cell Rep. 2016;16(6):1561–1573. doi: 10.1016/j.celrep.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joiner M.L.A., Koval O.M., Li J.D. CaMKII determines mitochondrial stress responses in heart [J] Nature. 2012;491(7423):269–273. doi: 10.1038/nature11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuma A., Kim J.Y., Kacimi R. Microglial calcium release-activated calcium channel inhibition improves outcome from experimental traumatic brain injury and microglia-induced neuronal death [J] J. Neurotrauma. 2019;36(7):996–1007. doi: 10.1089/neu.2018.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth A.B., Hori K., Novakovic M.M. CRAC channels regulate astrocyte Ca2+ signaling and gliotransmitter release to modulate hippocampal GABAergic transmission [J] Sci. Signal. 2019;12(582) doi: 10.1126/scisignal.aaw5450. eaaw5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P.S., Wang H.P., Li X.Y. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming [J] Nat. Immunol. 2017;18(9):985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 43.Matkovich S.J., Al Khiami B., Efimov I.R. Widespread down-regulation of cardiac mitochondrial and sarcomeric genes in patients with sepsis [J] Crit. Care Med. 2017;45(3):407–414. doi: 10.1097/CCM.0000000000002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y.X., Yao X., Zhang Q.J. Beclin-1-dependent autophagy protects the heart during sepsis [J] Circulation. 2018;138(20):2247–2262. doi: 10.1161/CIRCULATIONAHA.117.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomes A.P., Price N.L., Ling A.J.Y. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. J. Cell. 2013;155(7):1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson R.M., Barger J.L., Edwards M.G. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response [J] Aging Cell. 2008;7(1):101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J.D. Minireview: the PGC-1 coactivator networks: chromatin-remodeling and mitochondrial energy metabolism [J] Mol. Endocrinol. 2009;23(1):2–10. doi: 10.1210/me.2008-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tavener S.A., Kubes P. Is there a role for cardiomyocyte toll-like receptor 4 in endotoxemia? [J] Trends Cardiovasc. Med. 2005;15(5):153–157. doi: 10.1016/j.tcm.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Avlas O., Fallach R., Shainberg A. Toll-like receptor 4 stimulation initiates an inflammatory response that decreases cardiomyocyte contractility [J] Antioxidants Redox Signal. 2011;15(7):1895–1909. doi: 10.1089/ars.2010.3728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.