Abstract
Survival in high-risk neuroblastoma (HR-NB) patients remains poor despite multimodal treatment. We aimed to identify HR-NB patients with worse outcomes by analyzing the genomic instability derived from segmental chromosomal aberrations. We calculated 3 genomic instability indexes for primary tumor SNP array profiles from 127 HR-NB patients: (1) Copy number aberration burden (%gainslength+%losseslength), (2) copy number load (CNL) (%gainslength-%losseslength) and (3) net genomic load (NGL) (%gainsamount-%lossesamount). Tumors were classified according to positive or negative CNL and NGL genomic subtypes. The impact of the genomic instability indexes on overall survival (OS) was assessed with Cox regression. We identified 38% of HR-NB patients with poor 5-year OS. A negative CNL genomic background was related to poor prognosis in patients ≥18 months showing tumors with homogeneous MYCN amplification (9.5% survival probability, P < 0.05) and patients with non-MYCN amplified NB (18.8% survival probability related to >2.4% CNL, P < 0.01). A positive CNL genomic background was associated with worse outcome in patients with heterogeneous MYCN amplification (22.5% survival probability, P < 0.05). We conclude that characterizing a tumor genomic background according to predominance of genome gained or lost contributes toward improved outcome prediction and brings greater insight into the tumor biology of HR-NB patients.
Keywords: Copy number load, Copy number aberration burden, Net genomic load, Genomic imbalance, Segmental chromosomal aberrations
Abbreviations: CNA, copy number aberration; CNL, copy number load; dNGL, decreased net genomic load; EFS, event-free survival; GI, genomic instability; hetMNA, heterogeneous MYCN-amplification; HR, high-risk; HR-NB, high-risk neuroblastoma; homMNA, homogeneous MYCN-amplification; iNGL, increased net genomic load; MNA, MYCN-amplification; nCNL, negative copy number load; NGL, net genomic load; NB, neuroblastoma; OS, overall survival; pCNL, positive copy number load; SCA, segmental chromosomal aberration; SNPa, single nucleotide polymorphism array; UHR, ultra-high-risk
Introduction
In neuroblastoma (NB), the most common extracranial solid tumor in children [1], stratification of patients into risk groups for relapse and death is pivotal for treatment decisions [2]. The International Neuroblastoma Risk Group uses a classification system based on age, stage, tumor histology, MYCN amplification (MNA), 11q deletion and ploidy to define very low-, low-, intermediate-, and high-risk (HR) groups according to 5-year event-free survival (EFS). Patients with HR disease deserve special attention, as survival prognosis remains poor despite multimodal treatment [3], including ALK inhibitors against common ALK variants in NB [4]. Furthermore, in recent years there has been much interest in characterizing a subgroup of HR-NB patients with particularly poor outcome, known as ultra-high-risk (UHR). Although there is no consensus about criteria to define UHR patients, current definitions are 10% to 15% 5-year EFS or death within 18 months of diagnosis (reviewed in [5]). Identifying these patients at diagnosis could be useful to offer them therapeutic alternatives.
Tumor genetics holds great promise for discovering potential risk substratification factors. In contrast to its low mutational burden [6], NB is characterized by abundant copy number aberrations (CNA), which are strongly associated with outcome. Specifically, segmental chromosomal aberrations (SCAs) are linked to worse prognosis than tumors harboring exclusively whole chromosomal gains or losses [7]. In fact, in an ongoing trial, treatment intensity is being adjusted for low and intermediate risk patients according to the global copy number profile (Clinicaltrials.gov Identifier NCT01728155); however, this strategy is not applicable to HR-NB patients as most HR-NB tumors present a SCA profile. MNA is strongly associated with poor outcome in the global NB population, but not necessarily within HR-NB [5, 8]. The best-characterized subsets of HR-NB patients with worse outcome are defined by telomere maintenance status mediated by single aberrations in TERT and ATRX intragenic deletions [6, [9], [10], [11]]. An extensive study searching for specific SCAs or combinations to discriminate patients with worse outcome revealed distal 6q losses as predictive of survival [8]. Tumor mutational burden may be useful in combination with other factors to optimize risk stratification [12]. However, approaches that consider whole SCAs present at the tumor cells for stratification are scarce and their measure of genomic instability is confined to quantifying the number of SCAs [13]. For other types of cancer, genomic instability measured by the length of the altered genome encompassed by CNAs, or the CNA burden, is related to biological features and predictive of survival independent of the length or position of breakpoints [14], [15], [16].
Aberration patterns in HR-NB tumors are complex, with an abundance of SCA gain and loss. Gains and losses have distinct effects on allele frequency and DNA amount [16] which may affect patient outcome through gene-dosage mechanisms [17]. Changes in the copy number of large regions of the genome result in an unbalanced genome with different copy numbers for several genes, and although these copy-number changes have minimal phenotypic consequences individually, they would impact on cell fitness as the result of a cumulative effect [18,19]. Therefore, we considered the imbalance between gains and losses could reflect an imbalance at gene function level. First, we distinguished 2 genomic subtypes based on whether copy number gain or loss was predominant in the primary tumor, which we termed copy number load (CNL). Second, we considered that the somy of the SCAs contributes to an increased or decreased amount of DNA in tumor cells, which we refer to as net genomic load (NGL). Our final aim was to investigate whether these indexes could add valuable information to identify HR-NB patients with worse prognosis. To address these questions, genomic profiles obtained with single nucleotide polymorphism arrays (SNPa) were reanalyzed to calculate the indexes proposed in primary tumors from HR-NB patients with SCAs tumor profiles.
Material and methods
Patients and tumor samples
A total of 127 treatment-naïve tumor profiles analyzed by SNPa from HR-NB patients were reviewed. Samples had been referred to the Molecular Pathology Laboratory (Medical School, University of Valencia/Incliva Institute) for biological analysis. The present study was approved by INCLIVA's Clinical Research Ethics Committee (reference B.0000339). Participants or their family members/legal guardians provided written informed consent for histological and genetic studies performed in our laboratory. Briefly, the HR-NB group comprises (1) patients with MNA, (2) patients of 18 months of age or older and M stage and (3) patients under 18 months, Ms stage, 11q deletion and no MNA [3]. Heterogeneous MNA (hetMNA) was distinguished from homogeneous MNA (homMNA), as there is evidence that they define distinct clinico-biological patient groups [20]. MNA heterogeneity was defined as coexistence of non-MNA cells and a variable percentage of MNA cells. The cases analyzed here belong to one of these 5 categories: <1,1-5, 6-10,11-50, and >50% [20,21]. Clinical and biological characteristics of the entire cohort are shown in Supplementary Table 1.
Calculation of indexes to evaluate genomic instability derived from SCAs
SNPa genomic profiles were analyzed with CNAG beta 3.0 (for GeneChip Human mapping 250k, from Affymetrix, Santa Clara, CA, USA), ChAS v4.0 (for CytoScanHD and Oncoscan CNV, from Affymetrix), KaryoStudio v1.4 (for Human Cyto SNP-12 bead array from Illumina, San Diego, CA, USA), and BlueFuse Multi v4.5 (for Illumina arrays) to visualize gain and loss SCAs. Increases in partial copy number status (for Affymetrix arrays) or logR ratio (for Illumina arrays) in chromosome arms compared to the whole chromosome were defined as gains, while decreases in these parameters represented losses. Cutoffs for these parameters described elsewhere [22] guided the definition of gains (+0.15) and losses (−0.25), together with a variation in the number of tracks of data points in the B allele frequency.
For each sample, we annotated the length and the number of copies gained or lost of all SCAs. The somy for each chromosome was determined individually, assuming that SCA event may also occur after whole chromosome CNA. When available, fluorescence in situ hybridization data (for 1p and 11q regions and MYCN status with their corresponding chromosome controls) were helpful to better establish the different chromosome somies.
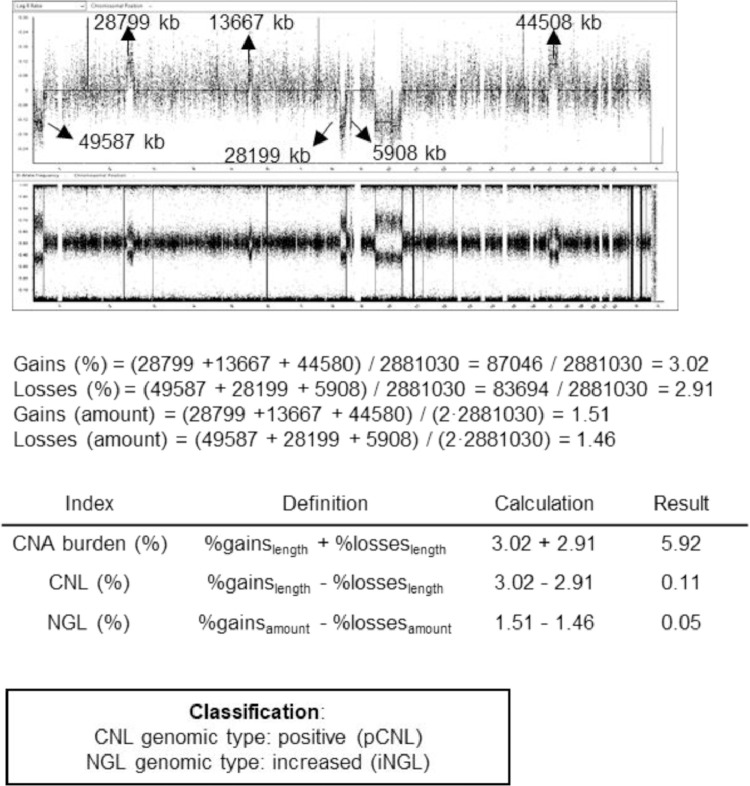
With the above data, 3 genomic instability (GI) indexes were calculated: (1) the CNA burden represented the overall percentage of the genome length (from chromosomes 1 to 22) affected by SCAs and measured the grade of genome alteration (%gainslength+%losseslength), (2) the CNL was indicative of the extent of imbalance between length of gained and lost genome (%gainslength-%losseslength), and (3) the NGL quantified the contribution of the SCAs to gain or loss in total amount of genetic material in the tumor, considering the somy of each SCA (%gainsamount-%lossesamount). Therefore, GI indexes CNL and NGL could be divided in 2 genomic subtypes each. Henceforth, tumors with positive CNL (pCNL) had a higher proportion of the genome gained than lost, and tumors with negative CNL (nCNL) had a higher proportion of the genome lost than gained. Likewise, in tumors with an increased NGL (iNGL) or a decreased NGL (dNGL) SCAs contributed to a higher or lower amount of genetic material, respectively (See Figure 1 for more details). Focal segmental aberrations (FSCAs, <3Mb) were not considered in calculating genomic instability indexes, except for FSCAs in TERT and ATRX which were annotated as they define subgroups of patients with worse prognosis [9,11].
Figure 1.
Example of calculation of genomic instability indexes in a sample. The genomic profile is visualized in BlueFuse Multi v4.5. All chromosomes are diploid. Segmental aberrations are indicated with arrows for gains (above the horizontal line) and losses (under the line). Note that for the calculus of NGL we consider that in a diploid sample without chromosomal aberrations the quantity of genetic material is 2-fold the genome length (i.e., 2•2,881,030 kb in this case). CNA, copy number aberration; CNL, copy number load; iNGL, increased net genomic load; NGL, net genomic load; pCNL, positive copy number load.
Ploidy estimation
Ploidy was estimated on the basis of SNPa data for consistency with CNL and NGL computing and was calculated as follows:
Where chr = chromosome length (kb), CN = copy number (somy) of each chromosome, CNX = 1 (XY/X0) or 2 (XX) and CNY = 1 (XY) or 0 (XX).
Tumors were classified according to the value obtained as diploid (1–1.25), triploid (1.26–1.80), tetraploid (1.8–2.26), and pentaploid or higher (≥2.27).
Statistics
Means (±SD) and medians were compared with parametric tests (Student's t, one-way ANOVA) or nonparametric tests (Mann-Whitney U, Kruskal-Wallis) according to data distribution. Chi-squared test was used to find differences in clinical and biological data and SCA frequencies between genomic subtypes. Correlations were determined with Spearman's correlation coefficient (rs) for continuous variables and with Cramer's V for categorical variables. Stepwise forward logistic regression was performed to assess the contribution of clinical and biological factors to the genomic subtypes.
Survival analysis was performed with univariate and multivariate Cox regression for GI indexes and clinical and biological features. Survival curves were generated using the Kaplan-Meier method and the curves were compared using the log-rank test. Overall survival (OS) time was calculated from diagnosis until death or last follow-up for censored patients. EFS time was calculated from diagnosis to event (relapse or death) or last follow-up. OS and EFS are presented as estimate ± SE. For censored patients, a cut-off of 9 months’ follow-up was established to be included in the survival analysis, leaving a cohort of 111 patients. Level of significance was established at P < 0.05. Tests are two-sided. Statistical analyses were performed using SPSS statistical analysis software (IBM, version 24).
Results
Exploratory analysis of genomic instability indexes
All tumors showed both segmental chromosomal gains and losses, except for 4 tumors that only harbored gains and 3 tumors that only harbored losses. Maximum gained genome was 19.4% and maximum lost genome was 14.6%.
The CNA burden ranged from 0.67% to 27.70%, with a mean of 10.71% ± 5.83% and median of 9.96%. Length of both gains and losses correlated with CNA burden (rs = 0.855 and rs = 0.835 respectively, P < 0.001). We then measured the grade of genomic imbalance in the cohort calculating CNL, which represents extra length of either gained or lost genome. The CNL ranged from 0.01% to 17.69%, with a mean of 2.90% ± 3.08% and median of 2.08%. To evaluate SCA contribution to a gain or a loss of genetic material in the tumor, we calculated NGL. Tumors showed an NGL range from 0.01% to 14.35%, mean of 1.88% ± 2.29% and median of 1.17%.
Tumor classification according to CNL genomic subtype reported a slightly higher frequency of nCNL than pCNL tumors (56% vs 44%). We found a high correlation between CNL and NGL genomic types (i.e., nCNL and dNGL; pCNL and iNGL, Cramer's V = 0.856, P < 0.001), which means that even though more than one copy of the same region can be gained or lost, this is not sufficient to trigger a net gain of genetic material in nCNL tumors or a net loss of genetic material in pCNL tumors.
Degree of genomic instability is related to MYCN status and 11q loss
MNA and 11q loss are genetic features that are present inversely in neuroblastoma tumors [23]. This was reproduced in the cohort, where 75.6% of the tumors harbored either 11q loss (42.5% of the entire cohort) or MNA (33.1%), 12.6% had combinations of MNA and 11q loss and only 11.8% did not show these aberrations. Although the representation of these profiles may be skewed by the election criteria of the cases included, the cohort is representative for high-risk neuroblastoma referent to patterns of genetic aberrations, as further described. As CNA burden, CNL and NGL were variable across tumors; we assessed their relation to clinical and biological factors. The factors related to the GI indexes were age (only for CNA burden), MYCN status (for CNA burden and CNL), and 11q loss (for the 3 indexes) in the entire cohort. All these associations were influenced by CNL genomic subtypes, as the 3 indexes were only related to 11q loss in nCNL tumors, while they were related to both 11q loss and MYCN status in pCNL tumors (Supplementary Table 2). We analyzed the GI indexes according to a classification of tumors by all possible combinations of 11q loss and MYCN status and determined that in both CNL genomic subtype groups, non-MNA tumors harboring 11q loss had higher GI indexes than homMNA tumors without 11q loss (Supplementary Figure 1).
The copy number load genomic background influences MYCN impact on survival
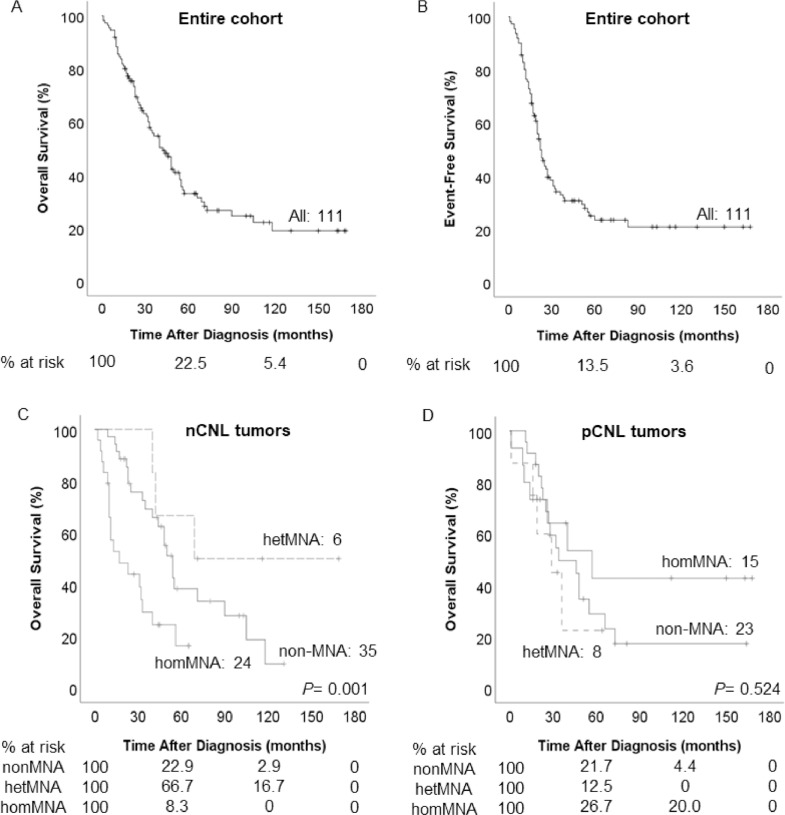
Our main objective was to determine whether GI indexes are useful to identify HR-NB patient subgroups with worse outcome. Total OS in the cohort was 19.9% ± 5.2%, 5-year OS was 33.0% ± 5.0%, total EFS was 20.8% ± 4.7% and 5-year EFS was 23.4% ± 4.6% (Figures 2A and B). We performed univariate Cox regression analyses for the GI indexes, CNL and NGL genomic subtypes, finding no link to survival in the entire cohort (Table 1). We then divided the cohort into 2 groups corresponding to CNL and NGL genomic subtypes and examined the impact of clinical and biological factors and GI indexes in each group. We found that in the nCNL subset, the MYCN status was statistically associated with OS (Table 1), while it had no impact within the pCNL subgroup, as depicted in Figures 2C and D. Thus, within the nCNL subgroup, patients with homMNA had 5-year OS of 16.3% ± 9.1%, inferior to those with hetMNA (66.7% ± 19.2%) and non-MNA tumors (38.4% ± 9.4%). We found no difference in OS between non-MNA and MNA tumors considering hetMNA and homMNA in the same MNA group (Table 1).
Figure 2.
Survival curves illustrate the influence of the CNL genomic background over the impact of MYCN status over survival. (A and B) OS and EFS of HR-NB patients without any stratification (C and D) The impact of MYCN status on the OS of HR-NB patients depends on the CNL genomic subtype. (C) Patients with nCNL tumors and homMNA have a markedly low OS in comparison to patients with non-MNA and hetMNA, while no difference is observed for the different MYCN status in the OS of patients with pCNL tumors (D). CNL, copy number load; hetMNA, heterogeneous MYCN amplification; homMNA, homogeneous MYCN amplification; MNA, MYCN amplification; nCNL, negative copy number load; OS, overall survival; pCNL, positive copy number load.
Table 1.
Cox regression analyses for overall survival.
| HR | 95%CI | P | |
|---|---|---|---|
| Entire cohort (univariate) | |||
| CNA burden (%) | 0.99 | 0.95 to 1.03 | 0.667 |
| CNL (%) | 1.00 | 0.94 to 1.07 | 0.999 |
| NGL (%) | 0.97 | 0.87 to 1.08 | 0.566 |
| CNL genomic subtype | 0.99 | 0.62 to 1.59 | 0.971 |
| NGL genomic subtype | 0.77 | 0.48 to 1.24 | 0.285 |
| nCNL subgroup (univariatea) | |||
| MYCN status | 0.003 | ||
| non-MNA | ref | ref | ref |
| hetMNA | 0.49 | 0.14 to 1.64 | 0.244 |
| homMNA | 2.72 | 1.41 to 5.26 | 0.003 |
| MYCN status | |||
| non-MNA | ref | ref | ref |
| MNA | 1.61 | 0.88 to 2.94 | 0.120 |
| nCNL value (continuous) | |||
| All patients | 1.11 | 1.01 to 1.22 | 0.048 |
| non-MNA patients | 1.14 | 1.01 to 1.29 | 0.033 |
| MNA patients (hetMNA + homMNA) | 1.05 | 0.91 to 1.20 | 0.498 |
| homMNA patients | 1.05 | 0.90 to 1.22 | 0.544 |
| hetMNA patients | 1.23 | 0.84 to 1.82 | 0.293 |
| nCNL subgroup (multivariatea,b) | |||
| MYCN status | 0.002 | ||
| non-MNA | ref | ref | ref |
| hetMNA | 0.33 | 0.07 to 1.42 | 0.136 |
| homMNA | 2.77 | 1.40 to 5.49 | 0.004 |
| nCNL value (continuous) | 1.13 | 1.04 to 1.24 | 0.006 |
Age, stage, histology, ploidy, MYCN status, 11q loss, nCNL value (continuous). Only statistically significant variables are reported.
Stepwise forward. CI, confidence interval; CNA, copy number aberration; CNL, copy number load; hetMNA, heterogeneous MYCN amplification; homogeneous MYCN amplification; HR, hazard ratio; MNA, MYCN amplification; NGL, net genomic load; OS, overall survival; nCNL, negative copy number load; pCNL, positive copy number load.
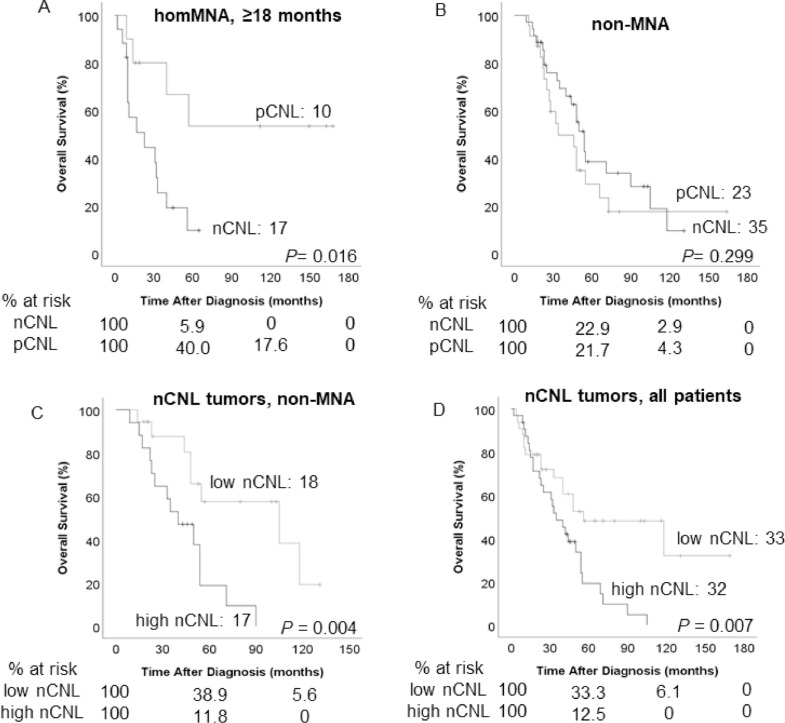
We then studied the impact of the CNL genomic subtype on survival for each MYCN status patient group, determining that worst outcomes were seen in patients ≥18 months with homMNA nCNL tumors (5-year OS of 9.5 ± 8.3%, Figure 3A). For patients ≥18 months with homMNA, the CNL genomic subtype was the only factor impacting on OS (hazard ratio = 3.40), in multivariate analysis regarding other factors (stage, histology, ploidy and 11q loss).
Figure 3.
Impact of the CNL genomic subtype on overall survival of patients with different MYCN status. (A) The nCNL genomic background is unfavorable for patients ≥18 months. (B) The CNL genomic background has not a direct impact over the survival of non-MNA patients. (C) High values of nCNL (≥2.24%) are related to low OS in non-MNA patients. (D) A slightly difference in OS is observed for patients with high nCNL (≥1.74%), including non-MNA and MNA. CNL, copy number load; hetMNA, heterogeneous MYCN amplification; homMNA, homogeneous MYCN amplification; MNA, MYCN amplification; nCNL, negative copy number load; OS, overall survival; pCNL, positive copy number load.
The CNL genomic subtype had no direct impact on OS in patients with non-MNA tumors (Figure 3B). Nevertheless, high nCNL values were related to worse outcomes when considering nCNL as a continuous variable (Table 1) and above the median (>2.24%). Thus, for patients with non-MNA tumors, 5-year OS was 18.8% ± 11.4% for high nCNL values and 57.6% ± 13.4% for patients with low nCNL values (Figure 3C). We also report that the nCNL value was related to OS considered as a continuous variable (Table 1) and as high values above the median (1.74% ± 2.88%) for all the nCNL cases, although the curves crossed at the beginning (Figure 3D). Interestingly, the nCNL value was not related to OS in patients with MNA tumors (Table 1).
The low number of patients allocated in homMNA <18 months and hetMNA groups limited the statistical analysis. No difference between CNL genomic subtypes was observed for patients <18 months with homMNA (Supplementary Figure 2A), nor for all homMNA patients. Patients with hetMNA pCNL tumors seemed to have worse prognostic (5-year OS of 22.5 ± 18.5%, Supplementary Figure 2B). Similar survival curves were observed for EFS as well, although only the effect of CNL on EFS of patients ≥18 months homMNA was statistically significant (Supplementary Figure 3).
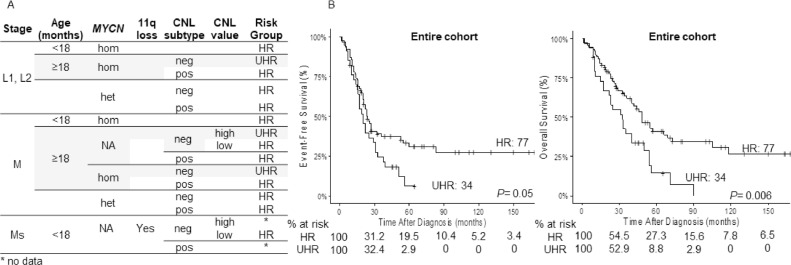
Combining CNL genomic subtype, MYCN status and age (Figure 4A), and excluding very small groups of patients as explained above, we typified 31% of the cohort as UHR patients, with a 5-year OS of 14.3% ± 7.18% and 5-year EFS of 6.1% ± 5.5% (Figure 4B).
Figure 4.
Overall identification of ultra-high-risk patients in the cohort. (A) Classification of NB patients into HR and UHR groups used into his work. (B) The log-rank test reports statistically significant differences between survival curves of the patients classified as UHR in comparison to the rest of the cohort for overall survival (right). CNL, copy number load; het, heterogeneous MYCN amplification; hom, homogeneous MYCN amplification; HR, high-risk; L1,L2, localized; Ms, metastatic special; neg, negative copy number load; non-MNA, not MYCN-amplified; pos, positive copy number load; UHR, ultra-high-risk.
Number, size, and pattern of SCAs differ between copy number load genomic subtypes
We searched for differences between nCNL and pCNL tumors that could explain the results of the survival analysis dependent on the CNL genomic subtype. We found no differences in the value of CNA burden, CNL and NGL between CNL genomic subtypes (Supplementary Figure 4A). Interestingly, the genome of pCNL tumors was more fragmented than nCNL tumors, as they harbored a higher number of SCAs (8.0 ± 5.7 vs 6.0 ± 4.1, P < 0.01). The fraction of gained genome was higher in pCNL tumors, which was due to a higher number of SCA gain than nCNL tumors, and not to the size of the SCAs, which had a similar mean size (Supplementary Figure 4B). The fraction of lost genome was higher in nCNL tumors, which owed not to the number of SCAs lost, but rather to the larger mean size of these SCAs (Supplementary Figure 4C). Therefore, the CNL genomic types differed in the higher number of gains for pCNL and in the larger size of losses for nCNL. We also assessed correlations between GI indexes in both CNL subgroups (Supplementary Figure 4D). As expected, CNA burden and number of SCAs were strongly correlated, as were CNL and NGL. Unexpectedly, we found other weaker but still positive correlations suggesting that CNA burden, CNL, NGL and number of SCAs all contribute to genomic instability. These correlations were slightly weaker in nCNL tumors than in the entire cohort or pCNL tumors.
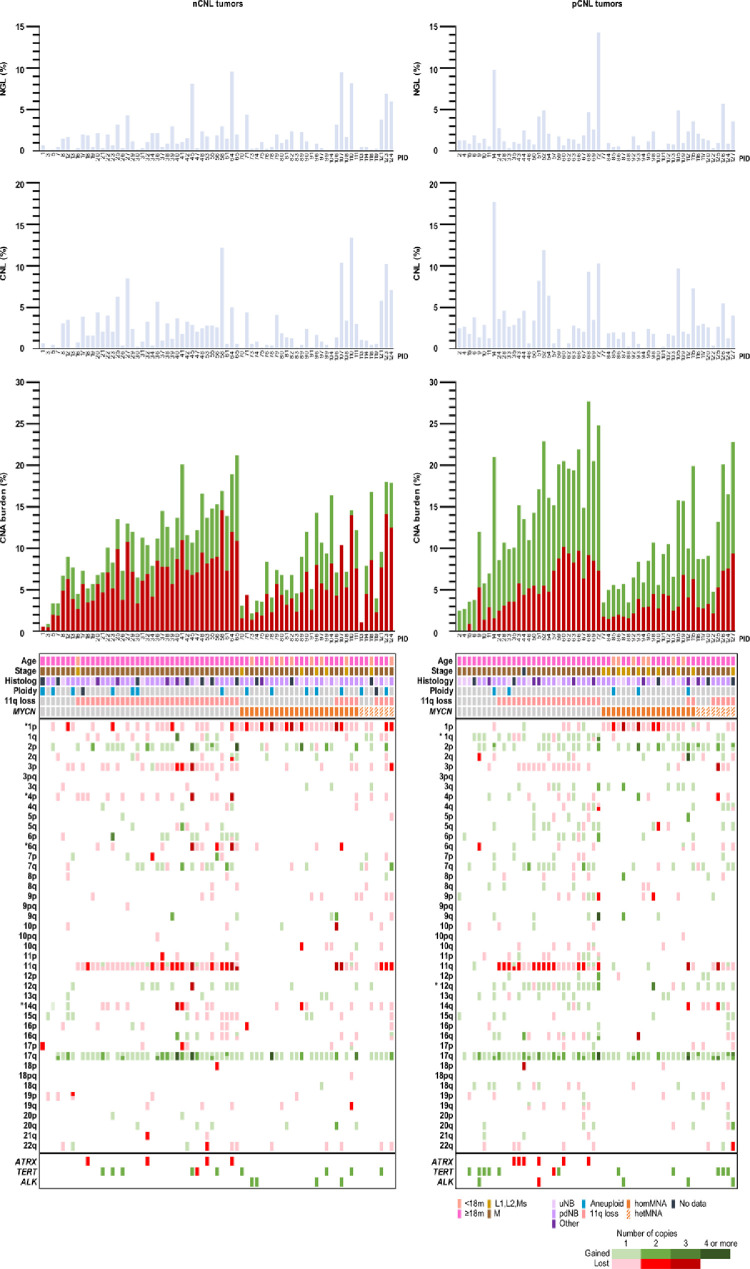
Next, we compared the distribution of age, stage, histology, ploidy, 11q loss and MYCN status in contingency tables and performed logistic regression including these factors to compare CNL genomic subtypes, which revealed no differences between them. Going one step further, we selected the most frequent SCAs apart from MNA and 11q loss to compare CNL genomic subtypes: gain for 17q (84%), 2p (47%), 12q (27%), 7q (22%) and 1q (20%), and loss for 1p (58%), 3p (35%), 4p (18%), 14q (17%) and 6q (15%). Gain of 1q, 2p and 17q, and loss of 1p, 3p, 4p are typical SCAs together with 11q loss. The MNA and the 11q loss define distinct genetic NB subgroups related to different SCAs [23]. Known associations between MYCN status and other SCAs were observed in the cohort, as 1p loss was more frequent in MNA tumors and 3p and 4p loss and 1q gain were more frequent in non-MNA tumors. However, these frequencies were skewed by CNL genomic subtype for 1p and 4p loss and for 1q gain (Supplementary Figure 5). We detected a higher frequency of 1p loss in non-MNA tumors in the nCNL subgroup in comparison to pCNL (48% vs 14%, P = 0.003). Besides this, 8 out of the 19 cases in non-MNA nCNL tumors were 1p heterozygous losses, while the 4 cases of 1p loss in non-MNA pCNL tumors were all homozygous losses. The frequency of 1q gain in non-MNA tumors was higher in the pCNL subgroup than in nCNL (52% vs 7.5%, P < 0.001), thus in nCNL tumors the 1q gain was not related to MYCN status. Also, for 4p loss, the relation with non-MNA tumors was lost in pCNL tumors, as we observed that it was less frequent than in nCNL tumors (14% vs 33%), although it was not statistically significant. In non-MNA tumors, comparing the nCNL and pCNL subsets we also observed a tendency to loss of the 6q (25% vs 7%) and 14q (25% vs 3%) chromosomal regions. A higher frequency of 12q gain (52% vs 23%, P = 0.005) was associated with pCNL. In homMNA and hetMNA tumor groups we observed that the SCAs were practically confined to 1p loss and 2p and 17q gains, and although the number of SCAs increased in the pCNL compared to the nCNL group, any region was significantly differentially represented in any of them. Focal SCAs in ATRX and TERT were related to MYCN status but not to the CNL genomic subtype. Focal ATRX deletions (N = 10, 14%) occurred only in non-MNA tumors, as described [9]. Focal gain SCAs affecting TERT were detected in non-MNA (N = 10, 14%) and in MNA tumors (N = 9, 15%). Comparison between the CNL genomic types is visualized in the summarizing graph provided in Figure 5.
Figure 5.
Distribution of clinical and biological features according to copy number load genomic subtypes. The 127 HR-NB cases are classified according to CNL genomic subtype (left: nCNL tumors, 71 patients; right: pCNL tumors, 56 patients) and ranged by patterns of SCAs defined by MYCN status, from least to greatest number of SCAs to facilitate comparison of clinical and biological data. The SCAs in the different chromosomal regions are represented with a sliding scale from light to dark according to number of copies gained or lost with respect to the somy of the chromosome inferred from SNPa (green: gains, red: losses). When there are two SCAs in the same chromosome arm (p or q), they are different colored in the same region (bottom region in p arms and upper region in q arms are closer to centromere). For non-MNA tumors, loss of 1p, 4p, 6q and 14q is more frequent in nCNL tumors and gain of 1q and 12q is more frequent in pCNL tumors (marked with a star). Focal deletions in ATRX in non-MNA tumors and focal gains and losses in TERT and ALK are independent of the CNL genomic subtype. No difference was found in genomic instability indexes shown in the histograms nor in clinical and biological characteristics represented in the chart below. Patient identifiers (PID) are shown on the x-axes.
Discussion
In this study we have improved identification of HR-NB patients with especially unfavorable outcomes based on the genomic instability of their primary tumors. The vast majority of tumors from HR-NB patients harbor several gain and loss SCAs. However, studies that address how whole SCAs contribute to genomic instability are scarce, and limited to exploring the number of SCAs [13]. In this study we paid special attention to the imbalance between gains and losses and explored the predominance of length (measured by the CNL) and amount (termed NGL) of either gained or lost chromosomal material as forms of genomic instability. We also evaluated the CNA burden, which has prognostic value in other CNA-driven cancers [14], [15], [16]. To our knowledge, this is an unprecedentedly comprehensive study about genomic instability related to SCAs in HR-NB, considering the imbalance between gains and losses.
Our first significant conclusion is that CNL genomic background influences the impact of MNA on survival in HR-NB patients. MNA is the best-characterized genetic aberration in NB and is linked to aggressive tumor behavior in the overall population of NB patients [3], but there is evidence both for [24] and against [5,8] its prognostic value in HR-NB (discussed in [5]). In our study, MYCN status had an impact on survival of patients with tumors where losses predominated (nCNL), indicating that homMNA was more closely related to worse outcomes than non-MNA and hetMNA, regardless of other clinicobiological factors. However, MYCN status was not associated with survival in the group with an excess of gains (pCNL). We have demonstrated that differences in genomic background between the CNL genomic subtypes basically refer to a greater size of losses for nCNL tumors and a higher number of gains for pCNL tumors, which accounts for the higher fraction of genome lost in the former and higher fraction of genome gained in the latter. Here the question arises as to how the CNL genomic background could shed light on the uncertain prognostic value of MNA in HR-NB. Copy number aberrations affect gene dosage, which correlates with gene expression in several types of cancer [25,26]. Expression levels of candidate oncogenes and tumor suppressor genes are associated with recurrent gains and losses in NB tumors [27,28]. As the fraction of genome gained or lost alone did not correlate with survival, we hypothesize that the CNL genomic background reflects a disequilibrium between several oncogenes and tumor suppressor genes. It is likely that a nCNL scenario, where gene loss of function may predominate over gene activation, is favorable for the homMNA to confer tumor aggressiveness, whereas in a pCNL scenario activation of genes other than MYCN might mask the impact of the homMNA. Gene expression and mutation data are needed to test this hypothesis. Copy number aberrations are also related to ploidy. In NB we demonstrated that the SCAs of nCNL tumors contribute to a net loss in the amount of DNA, while in pCNL tumors they cause an increase in the amount of DNA. However, even the highest value of NGL in the tumors (14.35% increase) was not enough to explain a change in ploidy as a consequence of SCA acquisition. It is not known whether small changes in DNA index could contribute to a more aggressive or favorable genomic background.
Our second observation is that the CNL genomic subtype is not a prognostic marker itself in the entire cohort, but is in combination with MYCN status. So far, potential genetic prognostic markers for HR-NB patients have been suggested for certain MYCN status patient groups. This is because MYCN status determines patterns of genetic aberrations, so non-MNA tumors show loss of 11q, 3p and 4p, gain of 1q and aberrations in telomere maintenance genes ATRX and TERT, while MNA tumors often harbor 1p loss and TERT upregulation mediated by MYCN, and do not show alterations in ATRX and TERT [23,29]. The degree of genomic instability in HR-NB is also related to MYCN status. Our results are consistent with a higher genomic instability in non-MNA than in MNA tumors previously reported by a higher number of SCAs and breakpoints [13,30]. Non-MNA tumors not only show a higher number of SCAs, which correlates well with CNA burden, but also have a higher degree of imbalance between fraction and amount of genome gained and lost. 11q deletion also contributes to higher genomic instability, as previously described [31]. Although tumor classification according to MYCN status must be still considered when studying genomic instability, identification of patients with worse outcome based on the CNL genomic subtype is available for all MYCN status groups. In overall, we classified 31% of the cohort analyzed as UHR, with 5-year OS under 20%. This percentage is not far from the classification of UHR patients with RAS and p53 pathway gene mutations [32], and therefore further collaborative studies would be useful to pinpoint whether UHR patients share both characteristics.
For patients with homMNA, differentiating between younger and older patients was decisive to analyze the impact of the CNL genomic subtype. Age is strongly associated with prognosis and younger patients generally have a better outcome; cut-offs of 12 and 18 months at diagnosis are commonly used [3]. Age was not related to survival within the homMNA patient group. Nevertheless, we observed that in the ≥18 months patient group, the nCNL subgroup had a 5-year survival probability of 9.5%, while it had no impact on survival in patients <18 months. This shows that factors other than SCAs may drive tumor aggressiveness in younger homMNA patients. Tumors with homMNA harbor few SCAs apart from 1p loss and 17q gain, which are present in the vast majority of tumors; it is therefore challenging to identify specific SCAs related to more adverse outcomes. Apart from the presence of amplifications not encompassing the MYCN region [8], no SCAs have been related to prognosis of homMNA patients. Therefore, characterization of the CNL genomic subtype opens up an interesting opportunity for therapeutic stratification of these patients.
Patients presenting with hetMNA constitute a clinico-biological category distinct from non-MNA and homMNA [20]. Beyond the mere presence of MNA clones, age and genomic background seem to play an important role in hetMNA tumor aggressiveness [33]. In the group of 15 hetMNA cases we analyzed, the correlation between age and outcome could not be proven, as 2 patients were 14 and 17 months of age who would be classified as older or younger depending on whether the cut-off was 12 or 18 months; and the rest were ≥18 months old. Tumors from older hetMNA patients are di-tetraploid and harbor fewer MNA clones and more SCAs than younger patients [33]. We report that for older hetMNA patients, the CNL genomic background also differentiates patients with worse and better outcomes, as the pCNL subgroup had a 5-year OS of 22.5%. As the 14- and 17-month-old patients belonged to the nCNL group we cannot rule out an age-related influence regarding the superior prognosis of this group, although at >12 months and close to the 18-month cut-off we considered them older patients. In contrast to homMNA, where all tumoral cells are MNA, in hetMNA tumors the CNL genomic background can influence competition between MNA and non-MNA cells. Thus, the pCNL background may encourage proliferation of subclones harboring aberrations favorable for tumor aggressiveness. The effect of CNL genomic subtype on survival in older hetMNA patients compared with homMNA patients also underlines that hetMNA and homMNA tumors behave differently, as the nCNL subgroup is favorable for hetMNA patients but unfavorable for homMNA patients. The importance of genomic background in the different behavior of homMNA and hetMNA tumors has been previously discussed [21,33]. Also, the fact that there was no difference in survival between non-MNA and hetMNA patients for both CNL genomic subtypes, indicates that hetMNA tumors may behave more similarly to non-MNA than to homMNA tumors. These results must be confirmed in a larger or independent patient cohort, as the number of patients allocated to individual groups in the survival analysis was small.
Lastly, for patients with non-MNA tumors, there was no survival difference in patients with nCNL and pCNL tumors, which reinforces the idea of an underlying relationship between MYCN and the CNL background. Nevertheless, a high percentage imbalance toward loss of genetic material, considering the nCNL as a continuous variable and values above the median (>2.24%), was related to a worse prognosis in these patients. Of interest, all patients with nCNL values over 4% (N = 5) died from disease, 4 of them within 5 years after diagnosis. High nCNL values might correspond to an intermediate degree of genomic instability tolerated by tumoral cells with therapeutic implications [34]. The inference is that patients with high nCNL values could benefit from alternative therapies. High genomic instability measured by a high number of SCAs (≥3) has been related to worse prognosis in non-MNA HR-NB patients [13]. We obtained discordant results, as the number of SCAs was not predictive of survival, either as a continuous variable or as a dichotomized variable. In fact, only 7 (10%) non-MNA tumors showed 3 SCAs or less.
We also observed that the CNL genomic subtype is related to certain SCA patterns in non-MNA tumors. Intriguingly, in nCNL tumors we detected a higher frequency of 1p loss, which is linked to worse prognosis if appearing together with MNA, while it is an uncommon aberration in non-MNA tumors [35]. In contrast, 1q gain, which often appears in non-MNA tumors [23], was scarce in the nCNL background. Therefore, segmental aberrations in chromosome 1 are not only related to MYCN status, but also to the CNL genomic background. Other SCAs frequent in non-MNA tumors that were related to the CNL genomic subtype were loss of 4p, 6q and 14q and gain of 12q. Oncogenes have been identified in 12q [36] and tumor suppressor genes in 1p [27,37] and 6q [37]. Distal 6q loss has recently been related to worse prognosis in non-MNA HR-NB patients [8]. 1p and 6q losses were more frequent in nCNL tumors while 12q gain was more frequent in pCNL tumors, suggesting that oncogenes and tumor suppressor genes contained in these regions may play an important role in CNL genomic subtypes. The correlation between the CNL genomic background and survival might then lead to discovery of genes related to UHR disease which could be potential biomarkers or therapeutic targets. Whereas specific SCAs are related to CNL in non-MNA tumors containing relevant suppressor tumors, as mentioned above, for homMNA tumors more extensive in-depth work must be done to find genomic regions that are differentially represented in the CNL groups with different prognoses.
In conclusion, we report that the imbalance between gains and losses in HR-NB tumors defines distinct genomic backgrounds with prognostic impact on MNA and non-MNA patient groups. Further in-depth characterization of HR-NB tumors according to CNL genomic type would pave the way to enhanced genetic characterization and identification of HR-NB patients with worse outcomes. Validation of these results in a larger cohort is warranted.
Author's contribution
B F-B analyzed the data, performed the statistical analysis, and wrote the original draft of the manuscript. AP carried out the genetic analyses, provided data analysis support and suggested draft changes. S M-V carried out the genetic analyses. VC provided the pediatric clinical data used for this study. SN performed the histopathological analyses of the samples. RN conceived and designed the study, reviewed the manuscript, and suggested draft changes. All authors reviewed the draft and approved the final version of the manuscript.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors would like to thank Juan Carbonell for statistical assistance and Kathryn for English correction. We also thank the Spanish Society of Pediatric Haemato-Oncology (SEHOP) for data patient management.
Footnotes
Funding: This work was supported by grants from ISCIII and ERDF (PI17/01558), CIBERONC and ERDF (CB16/12/00484); Fundación Científica de la Asociación Española contra el Cáncer (FAECC2015/006) and NEN Association (Nico contra el cancer infantil 2017 – PVR00157). The funders had no involvement in the research process nor in the preparation and submission of the article.
Conflict of interest: The authors have declared that no conflict of interest exists.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.11.001.
Appendix. Supplementary materials
References
- 1.Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O'Leary M, Smith FO, Reaman GH. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;28:2625. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trigg RM, Turner SD. ALK in neuroblastoma: biological and therapeutic implications. Cancers (Basel) 2018;10(4) doi: 10.3390/cancers10040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgenstern DA, Bagatell R, Cohn SL, Hogarty MD, Maris JM, Moreno L, Park JR, Pearson AD, Schleiermacher G, Valteau-Couanet D. The challenge of defining "ultra-high-risk" neuroblastoma. Pediatr Blood Cancer. 2019;66:e27556. doi: 10.1002/pbc.27556. [DOI] [PubMed] [Google Scholar]
- 6.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 8.Depuydt P, Boeva V, Hocking TD, Cannoodt R, Ambros IM, Ambros PF, Asgharzadeh S, Attiyeh EF, Combaret V, Defferrari R. Genomic amplifications and distal 6q loss: novel markers for poor survival in high-risk neuroblastoma patients. JNCI. 2018;110:1084–1093. doi: 10.1093/jnci/djy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann JM. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 11.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 12.Hwang WL, Wolfson RL, Niemierko A, Marcus KJ, DuBois SG, Haas-Kogan D. Clinical impact of tumor mutational burden in neuroblastoma. J Natl Cancer Inst. 2019;111:695–699. doi: 10.1093/jnci/djy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stigliani S, Coco S, Moretti S, Oberthuer A, Fischer M, Theissen J, Gallo F, Garavent A, Berthold F, Bonassi S. High genomic instability predicts survival in metastatic high-risk neuroblastoma. Neoplasia. 2012;14:823–832. doi: 10.1593/neo.121114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso MH, Aussó S, Lopez-Doriga A, Cordero D, Guinó E, Solé X, Barenys M, de Oca J, Capella G, Salazar R. Comprehensive analysis of copy number aberrations in microsatellite stable colon cancer in view of stromal component. Br J Cancer. 2017;117:421–431. doi: 10.1038/bjc.2017.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Feizi N, Chi C, Hu P. Association analysis of somatic copy number alteration burden with breast cancer survival. Front Genet. 2018;9:421. doi: 10.3389/fgene.2018.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hieronymus H, Murali R, Tin A, Yadav K, Abida W, Moller H, Berney D, Scher H, Carver B, Scardino P. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife. 2018:7. doi: 10.7554/eLife.37294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siaw JT, Javanmardi N, Van den Eynden J, Lind DE, Fransson S, Martinez-Monleon A, Djos A, Sjöberg RM, Östensson M, Carén H. 11q deletion or ALK activity curbs DLG2 expression to maintain an undifferentiated state in neuroblastoma. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108171. [DOI] [PubMed] [Google Scholar]
- 18.Tang YC, Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemente-Ruiz M, Murillo-Maldonado JM, Benhra N, Barrio L, Pérez L, Quiroga G, Nebreda AR, Milán M. Gene dosage imbalance contributes to chromosomal instability-induced tumorigenesis. Dev Cell. 2016;36:290–302. doi: 10.1016/j.devcel.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Berbegall AP, Bogen D, Pötschger U, Beiske K, Bown N, Combaret V, Defferrari R, Jeison M, Mazzocco K, Varesio L. Heterogeneous MYCN amplification in neuroblastoma: a SIOP Europe Neuroblastoma Study. Br J Cancer. 2018;118:1502–1512. doi: 10.1038/s41416-018-0098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berbegall AP, Villamón E, Piqueras M, Tadeo I, Djos A, Ambros PF, Martinsson T, Ambros IM, Cañete A, Castel V. Comparative genetic study of intratumoral heterogenous MYCN amplified neuroblastoma versus aggressive genetic profile neuroblastic tumors. Oncogene. 2016;35:1423–1432. doi: 10.1038/onc.2015.200. [DOI] [PubMed] [Google Scholar]
- 22.Depuydt P, Koster J, Boeva V, Hocking TD, Speleman F, Schleiermacher G, De Preter K. Meta-mining of copy number profiles of high-risk neuroblastoma tumors. Sci Data. 2018;5 doi: 10.1038/sdata.2018.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandesompele J, Baudis M, De Preter K, Van Roy N, Ambros P, Bown N, Brinkschmidt C, Christiansen H, Combaret V, Lastowska M. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol. 2005;23:2280–2299. doi: 10.1200/JCO.2005.06.104. [DOI] [PubMed] [Google Scholar]
- 24.Kushner BH, Modak S, Kramer K, LaQuaglia MP, Yataghene K, Basu EM, Roberts SS, Cheung NK. Striking dichotomy in outcome of MYCN‐amplified neuroblastoma in the contemporary era. Cancer. 2014;120:2050–2059. doi: 10.1002/cncr.28687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehrmann RS, Karjalainen JM, Krajewska M, Westra HJ, Maloney D, Simeonov A, Pers TH, Hirschhorn JN, Jansen RC, Schultes EA. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 26.Menezes RX, Boetzer M, Sieswerda M, van Ommen GJ, Boer JM. Integrated analysis of DNA copy number and gene expression microarray data using gene sets. BMC Bioinformatics. 2009;10:203. doi: 10.1186/1471-2105-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama H, Zhuang T, Light JE, Kolla V, Higashi M, McGrady PW, London WB, Brodeur GM. Mechanisms of CHD5 inactivation in neuroblastomas. Clin Cancer Res. 2012;18:1588–1597. doi: 10.1158/1078-0432.CCR-11-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–623. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- 29.Hertwig F, Peifer M, Fischer M. Telomere maintenance is pivotal for high-risk neuroblastoma. Cell Cycle. 2016;15:311–312. doi: 10.1080/15384101.2015.1125243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, Klijanienko J, Couturier J, Pierron G, Mosseri V, Valent A, Auger N, Plantaz D. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–3130. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 31.Carén H, Kryh H, Nethander M, Sjöberg RM, Träger C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci U S A. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackermann S, Cartolano M, Hero B, Welte A, Kahlert Y, Roderwieser A, Bartenhagen C, Walter E, Gecht J, Kerschke L. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362:1165–1170. doi: 10.1126/science.aat6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogen D, Brunner C, Walder D, Ziegler A, Abbasi R, Ladenstein RL, Noguera R, Martinsson T, Amann G, Schilling FH. The genetic tumor background is an important determinant for heterogeneous MYCN-amplified neuroblastoma. Int J Cancer. 2016;139:153–163. doi: 10.1002/ijc.30050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andor N, Maley CC, Ji HP. Genomic instability in cancer: teetering on the limit of tolerance. Cancer Res. 2017;77:2179–2185. doi: 10.1158/0008-5472.CAN-16-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attiyeh EF, London WB, Mossé YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 36.Schnepp RW, Khurana P, Attiyeh EF, Raman P, Chodosh SE, Oldridge DA, Gagliardi ME, Conkrite KL, Asgharzadeh S, Seeger RC. A LIN28B-RAN-AURKA signaling network promotes neuroblastoma tumorigenesis. Cancer Cell. 2015;28:599–609. doi: 10.1016/j.ccell.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Jr Diaz LA, Papadopoulos N. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.