Abstract
In recent years there has been increasing advocacy for highly immunogenic gamma-irradiated vaccines, several of which are currently in clinical or pre-clinical trials. Importantly, various methods of mathematical modelling and sterility testing are employed to ensure sterility. However, these methods are designed for materials with a low bioburden, such as food and pharmaceuticals. Consequently, current methods may not be reliable or applicable to estimate the irradiation dose required to sterilize microbiological preparations for vaccine purposes, where bioburden is deliberately high. In this study we investigated the applicability of current methods to calculate the sterilizing doses for different microbes. We generated inactivation curves that demonstrate single-hit and multiple-hit kinetics under different irradiation temperatures for high-titre preparations of pathogens with different genomic structures. Our data demonstrate that inactivation of viruses such as Influenza A virus, Zika virus, Semliki Forest virus and Newcastle Disease virus show single-hit kinetics following exposure to gamma-irradiation. In contrast, rotavirus inactivation shows multiple-hit kinetics and the sterilizing dose could not be calculated using current mathematical methods. Similarly, Streptococcus pneumoniae demonstrates multiple-hit kinetics. These variations in killing curves reveal an important gap in current mathematical formulae to determine sterility assurance levels. Here we propose a simple method to calculate the irradiation dose required for a single log10 reduction in bioburden (D10) value and sterilizing doses, incorporating both single- and multiple-hit kinetics, and taking into account the possible existence of a resistance shoulder for some pathogens following exposure to gamma-irradiation.
Keywords: gamma-irradiation, inactivation curve, sterilizing dose, sterility assurance level
INTRODUCTION
Gamma (γ) radiation is widely used to sterilize materials in a variety of settings. It is used in the food [1], pharmaceutical [2] and medical industries [3, 4] due to the ability of γ-radiation to inactivate pathogens through nucleic acid damage, whilst leaving proteins and other structures largely intact. Consequently, γ-radiation has also been proposed as an inactivation method to generate highly immunogenic vaccines [5]. Several groups have demonstrated the superiority of γ-radiation to traditional methods of vaccine inactivation, including formalin and β-propiolactone [6, 7]. In addition, previous publications illustrated the development of highly immunogenic γ-irradiated vaccines against influenza A virus (IAV) [5, 7–9] and Streptococcus pneumoniae [10, 11]. Furthermore, γ-irradiated vaccines against human immunodeficiency virus (HIV) [12], and malaria [13, 14] are currently in clinical trials.
In order to ensure vaccine safety and immunogenicity, estimating the sterilizing dose (DS) under different irradiation conditions must be carefully considered. The radiosensitivity of a pathogen can be influenced by multiple factors including genome structure [15, 16], irradiation temperature [17–19], water [20] and oxygen levels [21, 22], and the presence of free-radical scavengers [23]. Importantly, resistance to γ-radiation is inversely related to genome size [15], as the chances of a single γ-ray interacting with the genome of a given pathogen is increased if the genome is larger. Accordingly, the DSs required for bacterial species are usually lower than those required for viruses [24]. In addition, it is hypothesized that viruses with more complex genomes are more radioresistant compared to viruses with simple genome structures, as a virus with a double stranded or segmented genome may require inactivation of multiple strands or segments to prevent non-damaged segments from re-assorting in a host cell. Importantly, current standard-operating procedures related to sterilization of pathogens were developed to deal with low levels of bioburden or contamination [25–27], and a dose of 50 kGy is routinely used for sterilizing pathogens that pose a biosecurity risk [28]. In this study, we investigated the effect of irradiation conditions on the irradiation dose required to sterilize highly concentrated or radioresistant pathogens, and assessed the validity of considering 50 kGy to be a widely applicable DS.
In general, DS is calculated based on the concept of a sterility assurance level (SAL). For irradiated materials, the SAL is a given probability that any single pathogen within a sample may escape inactivation following an exposure to γ-irradiation. The International Atomic Energy Agency (IAEA) recommends a SAL of 10−6 for products intended to come into contact with compromised tissues [25], and so this should be applied to γ-irradiated vaccines. A SAL of 10−6 means that there is a one in a million chance of a single infectious particle remaining following irradiation [28]. Currently, the irradiation dose required to achieve sterility at the recommended SAL of 10−6 (DSSAL) is calculated using the formula
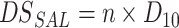 |
(1) |
where n is the number of log10 reductions in bioburden required to reach a theoretical SAL of 10−6 and D10 is the irradiation dose required for a single log10 reduction in bioburden. Equation 1 assumes a log–linear inactivation curve, which is likely observed for viruses with simple genome structure that follow one-hit kinetics (Fig. 1A). Our recent publications, however, have shown non-linear inactivation curves (Fig. 1B) for rotavirus (RV) [29] and S. pneumoniae [30], demonstrating multiple-hit kinetics for complicated pathogens. While a D10 value is usually calculated based on the linear portion of the curve [22], ignoring the shoulder of resistance could lead to miscalculation of the irradiation dose required to achieve a SAL of 10−6 (or DSSAL).
Fig. 1.
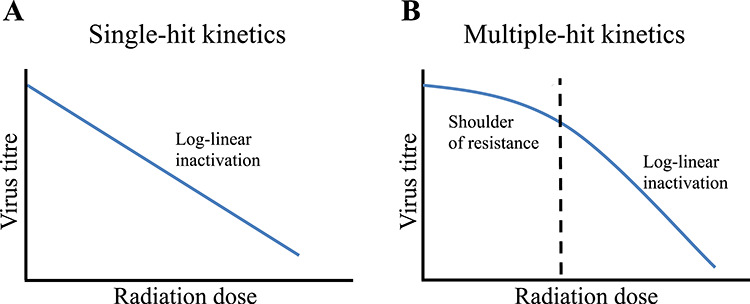
Inactivation kinetics of viruses demonstrating a model of (A) single-hit kinetics or (B) multiple-hit kinetics. Single-hit kinetics follows log–linear inactivation, whereas multiple-hit kinetics has a shoulder of resistance before damage is accumulated and log–linear inactivation occurs.
In this study we analyse the differences in D10 and DSSAL for pathogens with different genomic structures irradiated at different temperatures. Our data show both single-hit and multiple-hit inactivation kinetics and we have formulated a simple method to calculate the DSSAL. This method will ensure the shoulder of resistance is accounted for in multiple-hit inactivation models and thus allows for more accurate calculation of the SAL.
MATERIALS AND METHODS
Cells
Madin–Darby canine kidney (MDCK) and African green monkey kidney (Vero and MA104) cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% foetal bovine serum (FBS), 1% penicillin/streptomycin (P/S) and 1% 2 mM L-glutamine. For MA104 cells, 0.5% 200 mM sodium pyruvate was also added. Cells were maintained at 37°C with 5% CO2 in a humidified environment. Primary chicken embryo fibroblasts (CEF) were prepared from 10-day-old chicken embryos by removing the head, limbs and viscera. Bodies were fragmented then pushed through a 70 μm single cell strainer (BD). Cells were washed three times with phosphate buffer saline (PBS) by centrifugation at 1831 × g, then seeded into a 75cm2 tissue culture flask in DMEM +10% FBS and 1% P/S and kept at 37°C with 5% CO2 in a humidified environment. After 24 h, non-adherent cells were removed by three washes with PBS and fresh medium was added.
Viruses
Handling of all pathogens was carried out in accordance with guidelines of the biosafety committee at the University of Adelaide and all viruses were handled inside a Class II Biosafety Cabinet. Influenza A virus (IAV) A/Puerto Rico/8/1934 (A/PR8) H1N1 and Newcastle disease virus (NDV) V4 strain were grown in the allantoic cavity of 10 day old embryonated chicken eggs (ECE). Viruses were injected at 1 × 103 50% tissue culture infectious dose (TCID50)/egg in PBS containing 1% P/S. Eggs were incubated at 37°C for 48 h then chilled at 4°C overnight. Infected allantoic fluid was harvested and clarified by centrifugation at 3256 × g at 4°C for 10 min, then stored at −80°C until required.
Semliki Forest virus (SFV) A7 strain and Zika virus (ZIKV) MRC766 (Uganda 1947) strain were grown in Vero cells and rotavirus (RV) Rh452 was grown in MA104 cells. Viruses were propagated in DMEM + 1% P/S + 1% L-Glutamine RV was additionally activated by incubation at 37°C for 1 h with 10 μg/mL TPCK-trypsin (Sigma) prior to adding to cells. Viruses were all added at an multiplicity of infection (MOI) of 0.01, and infected flasks were stored at 37°C for 24–48 h until a cytopathic effect (CPE) of ~50% of the cell monolayer was observed. Virus-containing cell culture supernatants were collected and clarified by centrifugation at 3256 × g at 4°C for 10 min and stored at −80°C until required.
IAV and NDV were titrated by TCID50 in MDCK or CEF cells, respectively, in a 96-well round-bottomed microtitre plate. Virus was activated with 0.004% trypsin then 10-fold dilutions were added to confluent cell monolayers. Plates were incubated for 3 days at 37°C with 5% CO2. The presence of infectious virus was determined by agglutination of 50 μL of 0.6% chicken red blood cells (cRBC) in each well. The 50% infectious dose was determined using the method described by Reed and Muench [31] and titres were given as TCID50/mL.
SFV and ZIKV were titrated by plaque forming assay (PFA). Confluent monolayers of Vero cells were infected with serial dilutions of virus. Adsorption of virus was allowed for 1 h then a 0.9% agar overlay was added and plates were incubated for 3 days (SFV) or 5 days (ZIKV). Cells were fixed with 5% formalin for 1 h at room temperature (RT). Overlays were removed and cells were stained with 0.2% crystal violet. Plaques were enumerated and titre was calculated as plaque-forming units (PFU)/mL.
RV was titrated by focus-forming assay (FFA) as described previously [29]. Briefly, MA104 cells were seeded in 96-well flat-bottomed microtitre plates at 6.4 × 103 cells/well and plates were incubated at 37°C for 3 days until a confluent monolayer had formed. RV was activated by 10 μg/mL TPCK-trypsin for 30 min at 37°C. RV, 10-fold serially diluted, was added to wells and incubated at 37°C for 1 h to allow virus to adhere to cells. Inoculum was removed and replaced with DMEM + 1% P/S + 1% L-glutamine + 0.5% sodium pyruvate and plates were incubated for a further 18 h at 37°C. Cells were then washed, and fixed and permeabilized using acetone:methanol (1:1 ratio). RV was visualized by primary staining with a polyclonal mouse anti-RV serum for 1 h at 4°C followed by Alexa Fluor® 555 goat anti-mouse IgG (Life Technologies, USA) secondary antibody for 1 h at 4°C in the dark. Cells were also stained with 1 μg/mL DAPI (Sigma) for 10 min at RT. RV-positive cells were visualized using a Nikon Eclipse Ti fluorescent microscope and NIS-Elements AR software. Titre was calculated as focus-forming units (FFU)/mL.
Streptococcus pneumoniae
Streptococcus pneumoniae strain Rx1, a capsule-deficient derivative of D39 containing two additional mutations (ΔLytA, PdT) that has been described previously [10], was used. Streptococcus pneumoniae was inoculated into Todd Hewitt Broth supplemented with 0.5% yeast extract (THY) medium at a starting OD600 of 0.02 and then grown at 37°C + 5% CO2 until OD reached 0.65. Bacteria were centrifuged at 4000 × g for 10 min at 4°C then resuspended and washed thrice in PBS. Bacteria were then resuspended in PBS + 13% glycerol at ~1010 colony forming unit (CFU)/mL then frozen at −80°C until required. Viable titres were measured by CFU counts on blood agar plates.
Gamma-irradiation
Virus and bacteria stocks were shipped to the Australian Nuclear Science and Technology Organisation (ANSTO) whilst frozen on dry ice. Samples were thawed on ice or at RT, or kept frozen on dry ice as specified and were exposed to increasing doses (0–50 kGy) of γ-radiation at different conditions [RT (24–27°C), cold on ice water (4–8°C) or frozen on dry-ice]. Gamma-irradiation was performed using a 60Co source at the ANSTO (NSW). Radiation doses were measured using calibrated Fricke or ceric cerous dosimeters. Pathogens were then titrated to measure loss of infectivity at different radiation doses. Non-irradiated controls were treated with the same conditions (room-temperature, ice or dry ice) without exposure to γ-radiation. After irradiation all samples were stored at −80°C until required.
RESULTS
Inactivation curves
Different pathogens were exposed to incremental doses of γ-radiation and titres at each radiation dose were determined. IAV and NDV were both grown in 10-day-old ECEs and they are expected to have the same medium composition. This enabled a comparison between radiation-sensitivity of a non-segmented single-stranded RNA genome (ssRNA) genome (NDV) and a segmented ssRNA genome (IAV). Our data demonstrate log–linear inactivation for both viruses (Fig. 2), indicating single-hit inactivation kinetics.
Fig. 2.
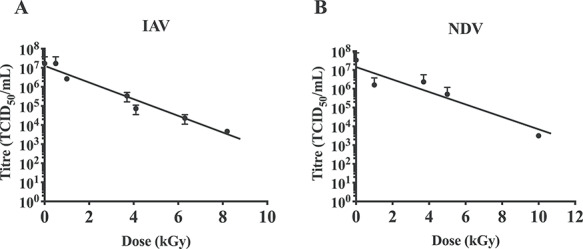
Log–linear inactivation curves of ssRNA viruses in allantoic fluid. (A) Influenza A virus and (B) Newcastle disease virus were exposed to increasing doses of γ-irradiation on dry ice. Reduced virus titre (as measured by TCID50/ml) for increasing irradiation doses helped to generate inactivation curves and log–linear inactivation was observed for both viruses. Data are expressed as mean ± SEM (n = 2). Horizontal dashed line represents background binding of virus to RBCs in the absence of a cell monolayer.
Next, we compared the inactivation curves of SFV and ZIKV under different irradiation temperatures. Both viruses have ssRNA genomes of a comparable size, and were both grown in Vero cells using DMEM with similar medium composition. Both viruses demonstrated single-hit inactivation kinetics, with increased radiosensitivity at higher temperatures, as expected (Fig. 3).
Fig. 3.
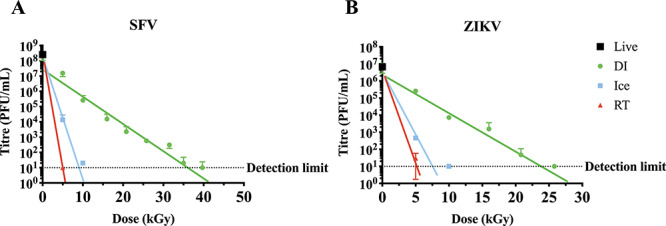
Log–linear inactivation of ssRNA viruses at different irradiation temperatures. (A) Semliki Forest virus and (B) Zika virus were exposed to increased doses of γ-irradiation on dry ice (DI) (green circles), ice (blue squares) or at room temperature (RT) (red triangles). The reduction in virus titre was estimated using plaque assay and inactivation curves were generated. Log–linear inactivation was observed for all three temperature conditions. Non-irradiated live virus was used as the starting point. Data are presented as mean ± SEM (n = 3). Horizontal dashed line represents detection limit.
We then analysed the inactivation curve of RV, a more complex virus with a segmented and double-stranded RNA genome (dsRNA) genome structure. We have previously reported that the inactivation curve for dry ice-irradiated RV is non-linear and confirmed that here using a different strain of RV (Fig. 4). The curve shows two distinct regions. A large shoulder of resistance is observed initially, with an ~2 log loss of titre occurring between 0 to 40 kGy. After this point, a rapid decline in viable titre was observed with increased radiation dose. Importantly, calculating the DS using this inactivation curve would not be possible using current mathematical models (equation 1). Interestingly, we did not detect the multiple-hit inactivation curve for RV materials irradiated on ice or at RT (Fig. 4). This could indicate that indirect damage caused by free radicals following irradiation at higher temperatures may counteract the radioresistance of pathogens with more complex genomes.
Fig. 4.
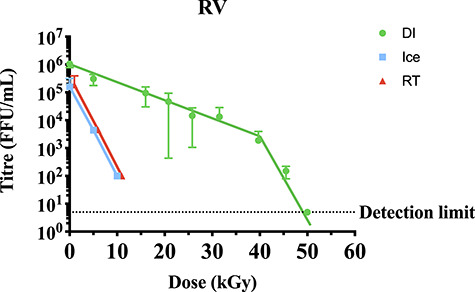
Inactivation curve of RV at different irradiation temperatures. RV was exposed to increasing doses of γ-radiation on dry ice (DI) (green circles), ice (blue squares) or at room temperature (RT) (red circles). Titre was measured by focus forming units. In contrast to both ice and RT, irradiation on DI shows an inactivation curve with multiple-hit kinetics. A shoulder of resistance appears to require an irradiation dose of 40 kGy. Data are presented as mean ± SEM (n = 2).
Calculating sterilizing doses
For viruses demonstrating single-hit kinetics, exponential lines of best fit could be determined using the equation:
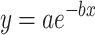 |
(2) |
where y is the titre at a given radiation dose x, a is the starting titre, and b is a constant that is determined experimentally for each individual virus under a given set of irradiation conditions. Equation (2) can then be rearranged to determine the D10 value (x), when y = 0.1a (i.e. a 90% loss of starting titre):
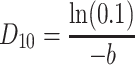 |
(3) |
Therefore, the D10 is higher where b is lower, as would be expected for more radioresistant pathogens. The line of best fit, D10 values and DSSAL were determined for IAV and NDV (Table 1), and ZIKV and SFV (Table 2). The D10 values of IAV and NDV were comparable (2.1 and 2.8 kGy, respectively), whereas SFV had a higher D10 than ZIKV for dry-ice irradiation (5.5 compared to 4.2 kGy). The D10 values were also calculated for ice and RT and were comparable, however an exact D10 value for RT-irradiated SFV could not be determined since virus was undetectable at the lowest irradiation dose used (5 kGy) in our experimental settings. Importantly, calculating a D10 value for pathogens with single-hit kinetics allowed us to calculate the DSSAL using equation (1), as shown in Tables 1 and 2. However, calculating the DS using equation (1) would not be possible for pathogens with multiple-hit kinetics as ignoring the shoulder of resistance would result in a miscalculation of the DS. Therefore, we propose a new formula to calculate the DSSAL that could accommodate both single-hit and multiple-hit inactivation kinetics:
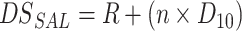 |
(4) |
where R refers to the irradiation dose required to overcome the shoulder of resistance with a value of ‘R = 0’ for pathogens that show linear inactivation curves (single-hit kinetics). This formula takes into account the distinct regions of multiple-hit curves and should allow for more accurate calculation of DSs.
Table 1.
Inactivation formulae and sterility assurance levels of NDV and IAV
| Virus | Formulaa | Starting titre (TCID50/mL) | D10 (kGy) | DS (kGy) |
|---|---|---|---|---|
| IAV | y = 2 × 107 × e−1.097x | 1.69 × 107 | 2.1 ± 0.16 | 27.77 |
| NDV | y = 2 × 107 × e−0.823x | 3.41 × 107 | 2.8 ± 0.53 | 37.86 |
aUnits for x are kGy.
Table 2.
Inactivation formulae and sterility assurance levels of ZIKV, SFV and RV
| Virus | Irradiation conditiona | Formulab | Starting titrec | D10 (kGy) | DS (kGy) |
|---|---|---|---|---|---|
| SFV | DI | y = 5 × 107 × e−0.418x | 2.55 × 108 | 5.5 ± 0.43 | 79.36 |
| Ice | y = 3 × 108 × e−1.968x | 1.2 ± 0.23 | 16.86 | ||
| RT | y = 3 × 108 × e−3.871x | <1 | 14.41 | ||
| ZIKV | DI | y = 7 × 106 × e−0.625x | 6.75 × 106 | 4.2 ± 0.35 | 54.10 |
| Ice | y = 9 × 106 × e−1.986x | 1.2 ± 0.06 | 14.87 | ||
| RT | y = 9 × 106 × e−2.533x | 0.9 ± 0.31 | 11.66 | ||
| RV | Ice | y = 1 × 105 × e−0.506x | 1.05 × 106 | 4.6 ± 1.1 | 54.71 |
| RT | y = 1 × 105 × e−0.521x | 4.4 ± 0.02 | 53.13 |
aDI = Dry ice, RT = room temperature.
bUnits for x are kGy.
cVirus titre was measured as PFU/mL for SFV and ZIKV, and FFU/mL for RV.
When considering the inactivation curve of dry-ice irradiated RV (Fig. 4), we could consider 40 kGy to be required to overcome the radioresistance (R value). We could also calculate the D10 for the radiation sensitive portion of the curve (above 40 kGy) using equation (3). The D10 for the linear portion of the curve was calculated to be 3.2 kGy (based on the formula y = 7 × 1015 × e−0.718x). To calculate DSSAL using equation (4), we need to estimate the number of log10 reduction in virus titre (n) required to achieve the internationally acceptable SAL of 10−6. For this calculation, the viable titre at x = 40 kGy was determined to be 2.4 × 103 FFU/mL. Thus, a further reduction of 9.4 log10 will be required to meet a SAL of 10−6. Hence the DSSAL for dry-ice irradiated RV could be calculated based on equation (4) as follows:
DSSAL = 40 + (9.4 × 3.2) = 70.08 kGy.
To confirm the applicability of this method, we considered the inactivation curve of the bacterial pathogen S. pneumoniae. This pathogen has a double-stranded genome, and the inactivation curve is non-linear (Fig. 5). The shoulder of resistance, or R value, was determined to be 4 kGy. At x = 4 kGy the titre was 1.7 × 109 CFU/mL, thus 15.2 log10 reductions (n = 15.2) were required to reach the accepted SAL level of 10−6. We calculated the D10 value for the log–linear curve (after 4 kGy) using the formula y = 6 × 1013 × e−2.611x, which shows a value of 0.88 kGy. Therefore, the DSSAL for S. pneumoniae irradiated on dry-ice could be calculated using equation (4) as follows:
Fig. 5.
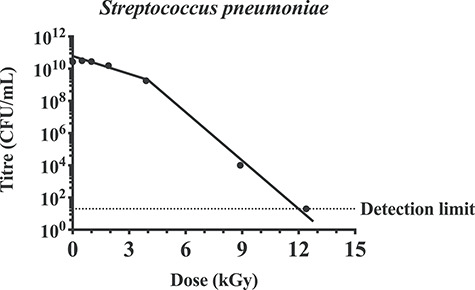
Inactivation curve of S. pneumoniae demonstrates multiple-hit kinetics. Streptococcus pneumoniae was irradiated on dry-ice (DI) at the indicated doses. Titre was measured by colony forming units and data are presented as mean ± SEM (n = 4). Inactivation curve demonstrates a multiple hit kinetics and a shoulder of resistance that require an irradiation dose of 4 kGy.
DSSAL = 4 + (15.2 × 0.88) = 17.38 kGy.
DISCUSSION
Current recommendations for calculating DSs are based on concepts and formulae generated to meet requirements to sterilize food, medical equipment and other health care products [25, 27, 32]. A dose of 25 kGy is considered the ‘gold standard’ [25] and is often substantiated for a low bioburden. In general, the contaminating species are typically bacteria, which are more sensitive to γ-radiation than viruses [24] and spores [33]. In addition, the The International Organization for Standardization (ISO) suggests that a bioburden of 106 infectious units is unusually high [27]. However, materials prepared for biomedical analysis as well as for vaccine purposes are expected to have bioburden levels much higher than 106 infectious units. Consequently, a DSSAL <25 kGy was not observed for any of the viruses irradiated on dry ice (Tables 1 and 2). Accordingly, 25 kGy should not be considered a DS for virally contaminated materials, nor for vaccine inactivation purposes, without properly addressing the inactivation curve and D10 value, particularly when frozen materials are irradiated using dry ice. For pathogens that pose a biosecurity concern a dose of 50 kGy is usually considered sufficient [28]. However, a SAL of 10−6 could not be reached following irradiation with 50 kGy on dry ice for ZIKV or SFV, or at 50 kGy using all irradiation conditions (dry ice, ice and RT) for RV (Table 2). Therefore, existing concepts that govern the use of γ-irradiation to sterilize highly infectious pathogens should be carefully considered to ensure sterility at internationally accepted levels. This will be essential for the development of highly safe and immunogenic γ-irradiated vaccines.
Inactivation curves typically follow single-hit or multiple-hit kinetics. It was expected that inactivation of single-stranded, non-segmented RNA viruses would follow single-hit kinetics. This was confirmed with NDV (Fig. 2B), SFV and ZIKV (Fig. 3), as well as previous publications [16, 34]. Interestingly, IAV also appeared to follow single-hit inactivation kinetics despite having segmented single-stranded RNA genomes (Fig. 2A). We have previously demonstrated log–linear inactivation of IAV [35]. Previous reports of inactivation curves of viruses with single-stranded segmented genomes have also demonstrated first-order kinetics [17, 36]. Conversely, the inactivation curves of RV (Fig. 4) and S. pneumoniae (Fig. 5) demonstrate multiple-hit inactivation kinetics where an accumulation of damage is required to sterilize each pathogen. Unlike other viruses used in this study, the genome of RV is comprised of 11 dsRNA segments and sufficient damage to both strands will be required to completely inactivate any genome segment. In addition, reassortment of RV is relatively frequent, and has been shown to enhance resistance in response to UV treatment [37]. Thus, incomplete inactivation of dsRNA segments accompanied by reassortment can rescue the infectivity of RV. This could explain the large shoulder of 40 kGy observed for RV. In contrast, S. pneumoniae cannot reassort, and SOS repair used by other bacterial species such as Escherichia coli [38] in response to γ-radiation do not appear to occur in S. pneumoniae [39]. However S. pneumoniae does utilize some repair mechanisms, such as excision repair [40]. It is also important to consider that S. pneumoniae has double-stranded genomes which could enhance resistance as both strands may need to be damaged to ensure inactivation. Conversely, mammalian cells are highly susceptible to γ-radiation despite having double-stranded genomes and repair mechanisms [41, 42]. This is particularly relevant to the development of γ-irradiated cancer vaccines such as GVAX, which is currently in clinical trials [43]. DSs reported are typically between 35 [44] and 100 Gy [45]. The radiosensitivity of mammalian cells is explained by a considerably larger genome than viruses and bacteria.
The ISO recommendations for calculating the DS involves setting a dose based on the calculated bioburden and a standard distribution of resistances (SDR) based on a D10 of between 2 and 3 kGy [27]. Where radioresistance is higher than the SDR (as would be the case for most viruses), the preparation is subjected to incremental increases in radiation dose and the proportion of positive samples is used to calculate the DS (i.e. at a SAL of 10−2, there should be 0, 1 or 2 positive samples out of 100 for statistically significant substantiation of the dose used). However, extrapolating this data for a SAL of 10−6 does not take into account the potential for non-linear inactivation. We have proposed an alternative method where the shoulder of resistance is calculated and accounted for as well as log–linear inactivation. To ensure the sterility and safety of irradiated materials, it is important to take into account the shape of the inactivation curve when considering the SAL, and equation (4) allows the shoulder of resistance to be incorporated when calculating the DS for pathogens that display multiple-hit inactivation kinetics. Importantly, mathematical modelling must also be coupled with rigid sterility testing.
It is important to note that γ-rays cause damage to pathogens by directly interacting with genomes to cause cross-linking, and single- and double-stranded breaks [46–49], and can interact with water or oxygen molecules to form free radicals. Oxidative damage causes most of the protein damage [20], but the formation and movement of free radicals can be reduced in frozen samples [50, 51]. In fact, irradiating frozen prions at incredibly high doses of up to 200 kGy showed minimal loss of transmission [52], demonstrating the resistance of proteins to γ-radiation at low temperatures. Thus, while irradiating at higher temperatures is more effective for sterilization (Figs 3 and 4, [16, 17, 19]), irradiating frozen samples is expected to better maintain structural integrity [35, 53]. Therefore, γ-irradiation has routinely been performed at low temperatures to obtain more effective results for both biomedical analysis and vaccine immunogenicity. However, our data clearly illustrate that sterility at an internationally accepted level based on SAL of 10−6 could not be achieved when irradiating high titres of some pathogens with 50 kGy using dry-ice conditions, and even when using room-temperature irradiation for radioresistant pathogens such as RV. Therefore, to ensure the safety of irradiated materials, the irradiation temperature, the appropriate method to calculate DSSAL and rigid sterility testing must be considered. Overall, this study highlighted a serious gap in current practices, and we propose a new mathematical formula to calculate both the D10 value and DSSAL to ensure the safety of irradiated materials for vaccine and research purposes.
ACKNOWLEDGEMENT
This work was presented at the Australian Nuclear Science and Technology Organisation (ANSTO) User Meeting, Sydney Australia, December 2019.
Contributor Information
Eve V Singleton, Research Centre for Infectious Diseases, and Department of Molecular and Biomedical Sciences, University of Adelaide, Adelaide, SA, 5005, Australia.
Shannon C David, Research Centre for Infectious Diseases, and Department of Molecular and Biomedical Sciences, University of Adelaide, Adelaide, SA, 5005, Australia.
Justin B Davies, Australian Nuclear Science and Technology Organisation, Lucas Heights, NSW, 2234, Australia.
Timothy R Hirst, Research Centre for Infectious Diseases, and Department of Molecular and Biomedical Sciences, University of Adelaide, Adelaide, SA, 5005, Australia; Gamma Vaccines Pty Ltd, Mountbatten Park, Yarralumla, ACT, 2600, Australia; GPN Vaccines Pty Ltd, Mountbatten Park, Yarralumla, ACT, 2600, Australia.
James C Paton, Research Centre for Infectious Diseases, and Department of Molecular and Biomedical Sciences, University of Adelaide, Adelaide, SA, 5005, Australia; GPN Vaccines Pty Ltd, Mountbatten Park, Yarralumla, ACT, 2600, Australia.
Michael R Beard, Research Centre for Infectious Diseases, and Department of Molecular and Biomedical Sciences, University of Adelaide, Adelaide, SA, 5005, Australia.
Farhid Hemmatzadeh, School of Animal and Veterinary Sciences, University of Adelaide, Roseworthy, SA, 5371, Australia.
Mohammed Alsharifi, Research Centre for Infectious Diseases, and Department of Molecular and Biomedical Sciences, University of Adelaide, Adelaide, SA, 5005, Australia; Gamma Vaccines Pty Ltd, Mountbatten Park, Yarralumla, ACT, 2600, Australia; GPN Vaccines Pty Ltd, Mountbatten Park, Yarralumla, ACT, 2600, Australia.
CONFLICT OF INTEREST
M.A. is head of the Vaccine Research Group at the University of Adelaide and the Chief Scientific Officer of Gamma Vaccines Pty Ltd and Director of GPN Vaccines Pty Ltd, J.C.P. is a Director of GPN Vaccines Pty Ltd, and T.R.H. is the Executive Chairman of Gamma Vaccines Pty Ltd and GPN Vaccines Pty Ltd. This does not alter adherence to policies on sharing data and materials. Both Gamma Vaccines Pty Ltd and GPN Vaccines Pty Ltd have no role in the study design, data collection and analysis, decision to publish, and preparation of the manuscript.
FUNDING
This work was supported by the following funding sources: an Australian Institute of Nuclear Science and Engineering (AINSE) Research Award (ALNGRA15517; to M.A.); an Australian Government Research Training Program (RTP) Scholarship (to E.V.S.); and a National Health and Medical Research Council Senior Principal Research Fellowship (awarded to J.C.P.).
AUTHORS’ CONTRIBUTIONS
M.A. and E.V.S. conceived and designed the study. E.V.S., S.C.D. and J.B.D. performed the experiments. E.V.S. and M.A. wrote the manuscript. F.H., T.R.H., J.C.P. and M.R.B. assisted in experimental design and preparation of the manuscript and provided reagents. M.A. supervised the study.
REFERENCES
- 1. Food and Drug Administration (FDA) . Irradiation in the production, processing and handling of food. 2017.
- 2. International Atomic Energy Agency . Manual on radiation sterilization of medical and biological materials. 1973.
- 3. Singh, R, Singh, D, Singh, A. Radiation sterilization of tissue allografts: A review. World J Radiol 2016; 8(4): 355–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doue B. Radiation doses and dose distribution during industrial sterilization by gamma rays and acclerated electron beams (part I). Medical Device Technol 2001;32–5. [Google Scholar]
- 5. Alsharifi, M, Mullbacher, A. The γ-irradiated influenza vaccine and the prospect of producing safe vaccines in general. Immunology and Cell Biology 2009; 88(2): 103. [DOI] [PubMed] [Google Scholar]
- 6. Wiktor, TJ, Aaslestad, HG, Kaplan, MM. Immunogenicity of rabies virus inactivated by β-propiolactone, acetylethyleneimine, and ionizing irradiation. Appl Microbiol 1972; 23(5): 914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuya, Y, Regner, M, Lobigs, M et al. Effect of inactivation method on the cross-protective immunity induced by whole 'killed' influenza a viruses and commercial vaccine preparations. The Journal of general virology 2010; 91(Pt 6): 1450. [DOI] [PubMed] [Google Scholar]
- 8. Alsharifi, M, Furuya, Y, Bowden, TR et al. Intranasal flu vaccine protective against seasonal and H5N1 avian influenza infections. PLoS One 2009; 4(4): e5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuya, Y, Chan, J, Wan, E-C et al. Gamma-irradiated influenza virus uniquely induces IFN-I mediated lymphocyte activation independent of the TLR7/MyD88 pathway (immune responses to inactivated influenza viruses). PLoS ONE 2011; 6(10): e25765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Babb, R, Chen, A, Hirst, TR et al. Intranasal vaccination with gamma-irradiated Streptococcus pneumoniae whole-cell vaccine provides serotype-independent protection mediated by B-cells and innate IL-17 responses. Clin Sci (Lond) 2016; 130(9): 697–710. [DOI] [PubMed] [Google Scholar]
- 11. Babb, R, Chen, A, Ogunniyi, AD et al. Enhanced protective responses to a serotype-independent pneumococcal vaccine when combined with an inactivated influenza vaccine. Clin Sci (Lond) 2017; 131(2): 169–80. [DOI] [PubMed] [Google Scholar]
- 12. Choi, E, Michalski, CJ, Choo, SH et al. First phase I human clinical trial of a killed whole-HIV-1 vaccine: Demonstration of its safety and enhancement of anti-HIV antibody responses. Retrovirology 2016; 13(1): 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seder, RA, Chang, LJ, Enama, ME et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science 2013; 341(6152): 1359–65. [DOI] [PubMed] [Google Scholar]
- 14. Ishizuka, AS, Lyke, KE, DeZure, A et al. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med 2016; 22(6): 614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jordan, RT, Kempe, LL. Inactivation of some animal viruses with gamma radiation from cobalt-60. Proc Soc Exp Biol Med 1956; 91(2): 212–5. [DOI] [PubMed] [Google Scholar]
- 16. Sullivan, R, Fassolitis, AC, Larkin, EP et al. Inactivation of thirty viruses by gamma radiation. Appl Microbiol 1971; 22(1): 61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elliott, LH, Mccormick, JB, Johnson, KM. Inactivation of Lassa, Marburg, and Ebola viruses by gamma-irradiation. Journal of Clinical Microbiology 1982; 16(4): 704–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kempner, ES, Haigler, HT. The influence of low temperature on the radiation sensitivity of enzymes. J Biol Chem 1982; 257(22): 13297–9. [PubMed] [Google Scholar]
- 19. Thayer, DW, Boyd, G. Effect of irradiation temperature on inactivation of Escherichia coli O157:H7 and Staphylococcus aureus. J Food Prot 2001; 64(10): 1624–6. [DOI] [PubMed] [Google Scholar]
- 20. Feldberg, RS, Carew, JA. Water radiolysis products and nucleotide damage in gamma-irradiated DNA. Int J Radiat Biol Relat Stud Phys Chem Med 1981; 40(1): 11–7. [PubMed] [Google Scholar]
- 21. Teoule, R, Bonicel, A, Bert, C et al. Identification of radioproducts resulting from the breakage of thymine moiety by gamma irradiation of E. coli DNA in an aerated aqueous solution. Radiation research 1974; 57(1): 46–58. [PubMed] [Google Scholar]
- 22. Gazso L. Physical, chemical and biological dose modifying factors. In: Gazso LP (ed). CCNato Sci Ser I Life. Amsterdam: IOS Press, 2005, 59–68. [Google Scholar]
- 23. Sullivan, R, Scarpino, PV, Fassolitis, AC et al. Gamma radiation inactivation of coxsackievirus B-2. Appl Microbiol 1973; 26(1): 14–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Farkas J. In: Doyle MP (ed). BLPhysical Methods of Food Preservation. 2007.
- 25. International Atomic Energy Agency . Good radiation practice (GRP). IAEA Guidelines for industrial radiation and sterilization of disposable medical products (cobalt-60 gamma irradiation). 1990.
- 26. International Atomic Energy Agency . Sterility assurance level. Guidelines for Industrial Radiation Sterilisation of Disposable Medical Products (Cobalt-60 Gamma-Irradiation). 1990Vol IAEA-TECDOC-539;39. [Google Scholar]
- 27. International Organisation for Standardization . Sterilization of health care products - radiation - part 2: Establishing the sterilization dose. 2013. ISO 11137-2:2013, 2013.
- 28. Department of Agriculture Fisheries and Forestry Biosecurity . Gamma irradiation as a treatment to address pathogens of animal biosecurity concern - final policy Review'. Canberra, CC BY 3.0 2014.
- 29. Shahrudin, S, Chen, C, David, SC et al. Gamma-irradiated rotavirus: A possible whole virus inactivated vaccine. PLoS One 2018; 13(6): e0198182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. David SC, Laan Z, Minhas V et al. Enhanced safety and immunogenicity of a psaA mutant whole-cell inactivated pneumococcal vaccine. Immunology and Cell Biology 2019. [DOI] [PubMed] [Google Scholar]
- 31. Reed, LJ, Muench, H. A simple method of estimating fifty per cent endpoints. American Journal of Epidemiology 1938; 27(3): 493–7. [Google Scholar]
- 32. Hendry JH. Radiation Sterilization of Tissue Allografts: Requirements for Validation and Routine Control : a Code of Practice. International Atomic Energy Agency, 2007. [Google Scholar]
- 33. Sommers, CH, Rajkowski, KT. Radiation inactivation of foodborne pathogens on frozen seafood products. J Food Prot 2011; 74(4): 641–4. [DOI] [PubMed] [Google Scholar]
- 34. Hume, AJ, Ames, J, Rennick, LJ et al. Inactivation of RNA viruses by gamma irradiation: A study on mitigating factors. Viruses 2016; 8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. David, SC, Lau, J, Singleton, EV et al. The effect of gamma-irradiation conditions on the immunogenicity of whole-inactivated influenza a virus vaccine. Vaccine 2017; 35(7): 1071–9. [DOI] [PubMed] [Google Scholar]
- 36. Thomas, FC, Davies, AG, Dulac, GC et al. Gamma ray inactivation of some animal viruses. Can J Comp Med 1981; 45(4): 397–9. [PMC free article] [PubMed] [Google Scholar]
- 37. McClain, ME, Spendlove, RS. Multiplicity reactivation of reovirus particles after exposure to ultraviolet light. J Bacteriol 1966; 92(5): 1422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brena-Valle, M, Serment-Guerrero, J. SOS induction by gamma-radiation in Escherichia coli strains defective in repair and/or recombination mechanisms. Mutagenesis 1998; 13(6): 637–41. [DOI] [PubMed] [Google Scholar]
- 39. Gasc, AM, Sicard, N, Claverys, JP et al. Lack of SOS repair in Streptococcus pneumoniae. Mutation research 1980; 70(2): 157–65. [DOI] [PubMed] [Google Scholar]
- 40. Sicard, N, Estevenon, AM. Excision-repair capacity in Streptococcus pneumoniae: Cloning and expression of a uvr-like gene. Mutation research 1990; 235(3): 195–201. [DOI] [PubMed] [Google Scholar]
- 41. Dolling, JA, Boreham, DR, Brown, DL et al. Modulation of radiation-induced strand break repair by cisplatin in mammalian cells. International journal of radiation biology 1998; 74(1): 61–9. [DOI] [PubMed] [Google Scholar]
- 42. Hafer, K, Iwamoto, KS, Scuric, Z et al. Adaptive response to gamma radiation in mammalian cells proficient and deficient in components of nucleotide excision repair. Radiation research 2007; 168(2): 168–74. [DOI] [PubMed] [Google Scholar]
- 43. Le, DT, Picozzi, VJ, Ko, AH et al. Results from a phase IIb, randomized, Multicenter study of GVAX pancreas and CRS-207 compared with chemotherapy in adults with previously treated metastatic pancreatic adenocarcinoma (ECLIPSE study). Clin Cancer Res 2019; 25(18): 5493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dranoff, G, Jaffee, E, Lazenby, A et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A 1993; 90(8): 3539–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baureus-Koch, C, Nyberg, G, Widegren, B et al. Radiation sterilisation of cultured human brain tumour cells for clinical immune tumour therapy. Br J Cancer 2004; 90(1): 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schulte-Frohlinde, D, Opitz, J, Gorner, H et al. Model studies for the direct effect of high-energy irradiation on DNA. Mechanism of strand break formation induced by laser photoionization of poly U in aqueous solution. Int J Radiat Biol Relat Stud Phys Chem Med 1985; 48(3): 397–408. [DOI] [PubMed] [Google Scholar]
- 47. Henner, WD, Grunberg, SM, Haseltine, WA. Sites and structure of gamma radiation-induced DNA strand breaks. J Biol Chem 1982; 257(19): 11750–4. [PubMed] [Google Scholar]
- 48. Lomax, ME, Folkes, LK, O'Neill, P. Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin Oncol (R Coll Radiol) 2013; 25(10): 578–85. [DOI] [PubMed] [Google Scholar]
- 49. Van Der Schans, GP. Gamma-ray induced double-strand breaks in DNA resulting from randomly-inflicted single-strand breaks: Temporal local denaturation, a new radiation phenomenon? Int J Radiat Biol Relat Stud Phys Chem Med 1978; 33(2): 105–20. [DOI] [PubMed] [Google Scholar]
- 50. Ormerod MG. Free-radical formation in irradiated deoxyribonucleic acid. Int J Radiat Biol Relat Stud Phys Chem Med 1965;9:291–300. [DOI] [PubMed] [Google Scholar]
- 51. Grieb, T, Forng, RY, Brown, R et al. Effective use of gamma irradiation for pathogen inactivation of monoclonal antibody preparations. Biologicals 2002; 30(3): 207–16. [DOI] [PubMed] [Google Scholar]
- 52. Gibbs, CJ, Jr., Gajdusek, DC, Latarjet, R. Unusual resistance to ionizing radiation of the viruses of kuru, Creutzfeldt-Jakob disease, and scrapie. Proc Natl Acad Sci U S A 1978; 75(12): 6268–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kitchen, AD, Mann, GF, Harrison, JF et al. Effect of gamma irradiation on the human immunodeficiency virus and human coagulation proteins. Vox sanguinis 1989; 56(4): 223–9. [DOI] [PubMed] [Google Scholar]


