Abstract
Our objective was to validate the NSABP 8-gene trastuzumab-benefit signature, developed and initially validated in NRG Oncology/NSABP B-31 in Alliance/NCCTG N9831. The B-31 and N9831 trials demonstrated the benefit of adding trastuzumab to chemotherapy in the adjuvant setting for HER2+ breast cancer patients. NSABP investigators utilized gene expression profiles of N9831 patients (N = 892) to blindly assign patients to large-, moderate-, or no-trastuzumab benefit groups and then NCCTG investigators assessed the degree of trastuzumab benefit using Cox models adjusted for age, nodes, estrogen receptor/progesterone receptor status, tumor size, and grade. Hazard ratios and 2-sided P values for recurrence-free survival of the predicted large- (n = 387), moderate- (n = 401), and no-benefit (n = 104) groups, based on the 8-gene signature were 0.47 (95% CI = 0.31 to 0.73, P < .001), 0.60 (95% CI = 0.39 to 0.92, P = .02), and 1.54 (95% CI = 0.59 to 4.02, P = .38), respectively (Pinteraction = .02), providing validation of the 8-gene signature in an independent study.
Several breast cancer clinical trials including HERA (Herceptin Adjuvant Trial), B-31, N9831, FinHer (Finland Herceptin Study), and BCIRG-006 (Breast Cancer International Research Group), demonstrated that chemotherapy-plus-trastuzumab improved outcomes for HER2+ breast cancer patients in the adjuvant setting (1–4). We previously described an 8-gene signature developed in a discovery cohort of NRG Oncology/NSABP B-31 (NCT00004067) patients to predict the degree of trastuzumab benefit, which was validated in an independent B-31 patient cohort (5). In the validation cohort, the 8-gene signature subtyped B-31 tumors into three trastuzumab-benefit groups: large- (hazard ratios [HR] = 0.28, 95% confidence interval [CI] = 0.20 to 0.41, P < .001), moderate- (HR = 0.60, 95% CI = 0.41 to 0.89, P = .01), and no-benefit (HR = 1.58, 95% CI = 0.67 to 3.69, P = .29) (Pinteraction < .001).
Our purpose was to validate the predictive ability of the 8-gene signature in Alliance/NCCTG N9831 (NCT00898898), which had three arms: Arm A (chemotherapy only), Arm B (trastuzumab after chemotherapy), or Arm C (trastuzumab given concurrently with chemotherapy). Patients from Arm B were not used for this study. There were no statistically significant differences between B-31 and N9831 Arms A and C in clinical covariates except nodal status. N9831 included node-positive and node-negative patients. B-31 only included node-positive patients (Table 1).
Table 1.
Clinical characteristics in NSABP B-31 and Alliance/NCCTG N9831
| Clinical covariates | B-31 | N9831 | P a |
|---|---|---|---|
| No. (%) | No. (%) | ||
| Age, y | |||
| <60 | 1325 (83.9) | 724 (81.2) | .09 |
| ≥60 | 254 (16.1) | 168 (18.8) | |
| Tumor size, cm | |||
| ≤2 | 584 (37) | 346 (38.8) | |
| 2.1–5 | 842 (53.3) | 466 (52.2) | .69 |
| >5 | 152 (9.6) | 80 (9.0) | |
| Unknown | 1 (0.1) | ||
| Nodal status | |||
| 0 | 0 | 118 (13.2) | <.001 |
| 1–3 | 896 (56.7) | 412 (46.2) | |
| 4–9 | 471 (29.8) | 236 (26.5) | |
| ≥10 | 212 (13.4) | 126 (14.1) | |
| ER status | |||
| Negative | 737 (46.7) | 413 (46.3) | .09 |
| Positive | 842 (53.3) | 479 (53.7) | |
| Disease-free survival | |||
| Censored | 1147 (72.6) | 603 (67.6) | .09 |
| Event | 432 (27.4) | 289 (32.4) | |
P values were determined using the χ2 test and all P values are 2-sided. ER = estrogen receptor.
N9831 cases were profiled with two custom nCounter code sets but only data for the 8 signature genes (CA12, ERBB2, ESR1, GATA3, GRB7, IGF1R, NAT1, MIEN1) and 16 housekeeping genes were sent to NSABP investigators for signature validation. Patient consent, data analysis work flow (Supplementary Figure 1, available online), and statistical methods are discussed in the Supplementary Methods (available online). All NSABP investigators were blinded to all clinical data. Patients were assigned a predicted trastuzumab-benefit group (large-, moderate-, or no-benefit) based on the 8-gene signature analysis. Details are included in the Supplementary Material (available online) and the original validation manuscript (5). Predictions were sent to N9831 investigators and prediction accuracy was tested in the chemotherapy arm (A) (n = 480) and the trastuzumab-plus-chemotherapy arm (C) (n = 412) using Cox models adjusted for age, nodes, estrogen receptor/progesterone receptor (ER/PR) status, tumor size, and grade. Only N9831 Arms A and C were analyzed because of their similarity to B-31 arms.
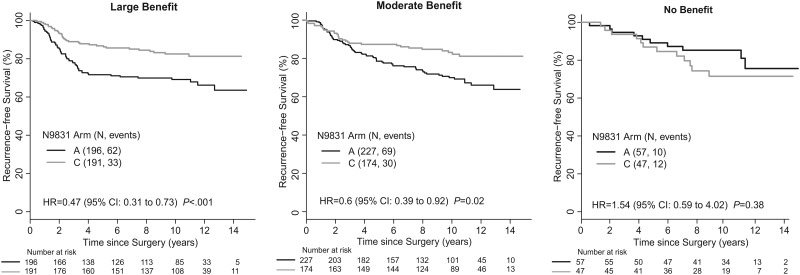
Numbers of patients assigned to the predicted large-, moderate-, and no-benefit groups were 387 (43.4%), 401 (45.0%), and 104 (11.7%), respectively. Respective recurrence-free survival (RFS) hazard ratios for trastuzumab benefit were 0.47 (95% CI = 0.31 to 0.73, P < .001), 0.6 (95% CI = 0.39 to 0.92, P = .02), and 1.54 (95% CI = 0.59 to 4.02, P = .38) in adjusted Cox models (Pinteraction = .02) (Figure 1). Ten-year RFS for trastuzumab-treated patients was 82.5% (95% CI = 77.2% to 88.2%), 83.1% (95% CI = 77.6% to 89.1%), and 71.5% (95% CI = 58.9% to 86.8%), in the large-, moderate-, and no-benefit groups, respectively.
Figure 1.
Recurrence-free survival in the 8-gene trastuzumab benefit groups of Alliance/NCCTG N9831. Cox models presented in each legend represent the hazard ratio for Arm C (adjusted for age, nodes, ER/PR, tumor size, and grade). The last follow-up date was April 25, 2015. Cox models were used to determine P values, which are 2-sided. CI = confidence interval; ER/PR = estrogen receptor/progesterone receptor.
Patients in the predicted large-, moderate- and no-benefit groups had disease-free survival (DFS) hazard ratios for trastuzumab benefit of 0.57 (95% CI = 0.40 to 0.82, P = .002), 0.66 (95% CI = 0.46 to 0.95, P = .02), and 1.13 (95% CI = 0.49 to 2.61, P = .77), respectively. The Pinteraction approached significance (P = .10) in adjusted Cox models (Supplementary Material Figure 2, available online). Ten-year DFS for trastuzumab-treated patients was 75.4% (95% CI = 69% to 82%), 76.2% (95% CI = 70% to 83%), and 71.5% (95% CI = 59% to 87%) in the large-, moderate-, and no-benefit groups, respectively.
N9831 results further validate the observation made in B-31 that the 8-gene signature may identify a relatively small, but potentially important subset of HER2+ patients who do not appear to benefit from trastuzumab. The differences between the large- and moderate-benefit groups in N9831 were less apparent than in the validation cohort of B-31, but that distinction would be of negligible clinical utility relative to identification of a subset of HER2+ patients deriving minimal, if any, benefit from trastuzumab.
Joint analysis of N9831 and B-31 established trastuzumab-plus-chemotherapy as a standard of care (SOC) for HER2+ breast cancer (2,6). Subsequently, the APHINITY (NCT01358877) trial established dual-HER2-targeted therapy with chemotherapy as the current SOC of newly diagnosed, high-risk HER2+ early-stage breast cancer, most often administered in the neoadjuvant setting, with administration of adjuvant T-DM1 for patients with residual invasive cancer following neoadjuvant chemotherapy and HER2-targeted therapy (7–9).
The 8-gene signature identifies a patient group that did not receive benefit when trastuzumab was added to chemotherapy. In B-31, the no-benefit patient group had high RNA levels of ER, intermediate levels of HER2, and included mostly luminal A and B subtypes, although HER2E was also represented (10). As expected, PIK3CA mutations were not associated with the 8-gene signature groups, because PIK3CA mutations were not associated with degree of trastuzumab benefit in the entire B-31 cohort (10). Stromal tumor-infiltrating lymphocytes (sTILs) were associated with 8-gene signature groups, with the no-benefit group showing less sTILs, although sTILs alone were not associated with trastuzumab benefit (11).
The 8-gene signature was statistically significantly associated with pathological complete response (pCR) in the neoadjuvant setting in NSABP FB-7 and NRG Oncology/NSABP B-52 using RNA-Seq data (12,13). FB-7 tested the pCR rate in locally-advanced HER2+ patients treated with chemotherapy-plus-trastuzumab, or neratinib, or the combination. The pCR rates were 22% (2/9), 53% (17/32), and 75% (9/12) in the no-, moderate-, and large-benefit groups, respectively. The pCR rate of the large group was statistically significantly different from the no-benefit group (P = .03) (12). B-52 evaluated docetaxel, carboplatin, plus trastuzumab and pertuzumab (TCHP) vs TCHP with estrogen deprivation in HER2+/hormone receptor+ breast cancer. The 8-gene signature was evaluated in the diagnostic biopsies of B-52 patients (n = 225). A total of 44 (19.6%) patients were assigned to the no-benefit group, 119 (52.9%) to the moderate-benefit group, and 62 (27.6%) to the large-benefit group. pCR rates across both arms were 6.8% in the no-benefit group compared with 54.8% in the large-benefit group (13). The low pCR rate of the no-benefit group suggests that this cohort may have derived minimal benefit from HER2-directed therapy. HER2+/hormone receptor+ early breast cancer patients have consistently shown a substantially lower pCR rate than patients with HER2+/ER- cancers (14). Additionally, differences in long-term outcomes, based on degree of pathologic response, are less pronounced in the ER+ subset of HER2+ breast cancers (14).
The 8-gene signature may identify an important subset of HER2+ breast cancers that derive minimal benefit from HER2-targeted therapy. Additional testing of this signature in other clinical trials testing dual anti-HER2 therapy such as B-41, APHINITY, KATHERINE (NCT01772472), and others, will determine whether any of the predicted trastuzumab benefit groups received differential benefit from additional anti-HER2 therapy. All information for testing the 8-gene signature is available in the Supplementary Material (available online).
Funding
The Breast Cancer Research Foundation.
Notes
Role of the funder: The study sponsor(s) played no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; nor the decision to submit the manuscript for publication.
Acknowledgments: The authors acknowledge the contributions of Barbara C. Good, PhD, Director of Scientific Publications, Christine I. Rudock, Publications and Graphics Specialist, and Wendy L. Rea, BA, Editorial Associate, all of whom are employees of NSABP. They were not compensated beyond their normal salaries for this work.
Disclosures: Dr Fumagalli reports the following grants, all to her organization, Breast International Group (BIG): Roche/Genentech, AstraZeneca, Novartis, Pfizer, Tesaro, and Servier, outside the submitted work. Dr Rastogi reports: Travel/accommodations from GNE/Roche, AZ, and Lilly. Dr Swain reports: Personal fees from Genomic Health and Tocagen; Personal fees and non-financial support from Cardinal Health, Daiichi-Sankyo, Eli Lilly & Company, Inivata, Ltd, and Pieris Pharmaceuticals; Grants from Pfizer; Grants, personal fees and non-financial support from Genentech, Inc/F. Hoffman-La Roche Ltd; Non-financial support from Novartis, Caris Life Sciences, NanoString Technologies, and Bristol-Myers Squibb; Personal fees, non-financial support and other from AstraZeneca, all outside the submitted work. Dr Mamounas reports: Advisory Board with Genomic Health, Inc, Genentech/Roche, Biotheranostics, and Daiichi Sankyo; Consultant with Merck; Speaker’s Bureau with Genentech/Roche and Genomic Health, Inc. Dr Geyer Jr reports: Supplemental funding for B-31 from Genentech, during the conduct of the study; Non-financial support from Genentech/Roche and Daiichi/Sankyo; Grants and non-financial support from Abbvie; Personal fees from Celgene and Myriad Genetics; and Grants and non-financial support from AstraZeneca and Merck, outside the submitted work. Dr Lucas reports: Stock ownership from Amgen, and Consulting (spouse) from Bayer/Loxo, outside the submitted work. Dr Paik reports: Consultancy with MedPacto; Patents related to OncotypeDx, all rights transferred to NSABP Foundation; and Stock/options with Immunoncia and Novomics. All other authors report no other conflicts of interest .
Author contributions: KLP and SP: Ideas; formulation or evolution of overarching research goals and aims. KLP, NS, and EAT: Development or design of methodology; creation of models. NS, ZL, DJS, YW, PGG, and NT: Programming, software development; designing computer programs; implementation of the computer code and supporting algorithms; testing of existing code components. NS, ZL, and DJS: Verification, whether as a part of the activity or separate, of the overall replication/ reproducibility of results/experiments and other research outputs. NS, ZL, and DJS: Application of statistical, mathematical, computational, or other formal techniques to analyze or synthesize study data. KLP-G, NS, DJS, ZL, and EAT: Conducting a research and investigation process, specifically performing the experiments, or data/evidence collection. KLP-G, NS, DJS, PGG, and EAT: Provision of study materials, reagents, materials, patients, laboratory samples, animals, instrumentation, computing resources, or other analysis tools. KLP-G, NS, DJS, and EAT: Management activities to annotate (produce metadata), scrub data and maintain research data (including software code, where it is necessary for interpreting the data itself) for initial use and later reuse. KLP-G: Preparation, creation and/or presentation of the published work, specifically writing the initial draft (including substantive translation). KLP-G, NS, DJS, YW, PGG, RK, NT, DF, YT, PR, SMS, EPM, CEG, NW, PCL, SP, and EAT: Preparation, creation and/or presentation of the published work by those from the original research group, specifically critical review, commentary or revision – including pre- or post-publication stages. NS and ZL: Preparation, creation and/or presentation of the published work, specifically visualization/ data presentation. KLP-G, SP, and EAT: Oversight and leadership responsibility for the research activity planning and execution, including mentorship external to the core team. KLP-G and EAT: Management and coordination responsibility for the research activity planning and execution. KLP-G and EAT: Acquisition of the financial support for the project leading to this publication.
Data Availability
The data underlying this article cannot be shared publicly at this time due to regulations concerning the proper consent and privacy issues of the patients. We will make an effort to make this data publicly available once regulatory issues are addressed.
Supplementary Material
Contributor Information
Katherine L Pogue-Geile, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA.
Nan Song, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA.
Daniel J Serie, Mayo Clinic Comprehensive Cancer Center, Jacksonville, FL, USA.
Ying Wang, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA.
Patrick G Gavin, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA.
Rim S Kim, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA.
Noriko Tanaka, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Tokyo Metropolitan Geriatric Medical Center, Healthy Aging Innovation Center, Department of Health Data Science Research, Tokyo, Japan.
Debora Fumagalli, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Breast International Group (BIG), Brussels, Belgium.
Yusuke Taniyama, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Tohoku University, Department of Surgery, Sendai, Japan.
Zhuo Li, Mayo Clinic Comprehensive Cancer Center, Jacksonville, FL, USA.
Priya Rastogi, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA.
Sandra M Swain, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Georgetown Lombardi Comprehensive Cancer Center, Georgetown University Medical Center, Washington, DC, USA.
Eleftherios P Mamounas, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Orlando Health UF Health Cancer Center, Orlando, FL, USA.
Charles E Geyer, Jr, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Houston Methodist Cancer Center, Houston, TX, USA.
Norman Wolmark, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Department of Surgery, Surgical Oncology, University of Pittsburgh, Pittsburgh, PA, USA.
Peter C Lucas, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; University of Pittsburgh School of Medicine, UPMC Hillman Cancer Center, Pittsburgh, PA, USA.
Soonmyung Paik, NSABP Foundation/NRG Oncology, Pittsburgh, PA, USA; Yonsei University College of Medicine, Seoul, South Korea.
E Aubrey Thompson, Mayo Clinic Comprehensive Cancer Center, Jacksonville, FL, USA.
References
- 1. Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Herceptin Adjuvant (HERA) Trial Study Team. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. [DOI] [PubMed] [Google Scholar]
- 2. Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. [DOI] [PubMed] [Google Scholar]
- 3. Joensuu H, Kellokumpu-Lehtinen PL, Bono P, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med. 2006;354(8):809–820. [DOI] [PubMed] [Google Scholar]
- 4. Slamon D, Eiermann W, Robert N, et al. Breast Cancer International Research Group. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pogue-Geile KL, Kim C, Jeong JH, et al. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105(23):1782–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perez EA, Romond EH, Suman VJ, et al. Four-year follow-up of trastuzumab plus adjuvant chemotherapy for operable human epidermal growth factor receptor 2-positive breast cancer: joint analysis of data from NCCTG N9831 and NSABP B-31. J Clin Oncol. 2011;29(25):3366–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. [Erratum in:N Engl J Med. 2017 Aug 17;377(7):702. Erratum in: N Engl J Med. 2018 Oct 18;379(16):1585] von Minckwitz G, Procter M, de Azambuja E, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N Engl J Med. 2017;377(2):122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. [DOI] [PubMed] [Google Scholar]
- 9. von Minckwitz G, Huang CS, Mano MS, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. 2019;380(7):617–628. [DOI] [PubMed] [Google Scholar]
- 10. Pogue-Geile KL, Song N, Jeong JH, et al. Intrinsic subtypes, PIK3CA mutation, and the degree of benefit from adjuvant trastuzumab in the NSABP B-31 trial. J Clin Oncol. 2015;33(12):1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim RS, Song N, Gavin PG, et al. Stromal tumor-infiltrating lymphocytes in NRG Oncology/NSABP B-31 adjuvant trial for early-stage HER2-positive breast cancer. J Natl Cancer Inst. 2019;111(8):867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. [Erratum in: Breast Cancer Res. 2020;22(1):9] Jacobs SA, Robidoux A, Abraham J, et al. NSABP FB-7: A phase II randomized neoadjuvant trial with paclitaxel + trastuzumab and/or neratinib followed by chemotherapy and postoperative trastuzumab in HER2+ breast cancer. Breast Cancer Res. 2019;21(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pogue-Geile KL, Wang Y, Feng H, et al. Association of molecular signatures, mutations, and sTILs, with pCR in breast cancer patients in NRG Oncology/NSABP B-52. AACR, 2019. Cancer Res. 2019;79(suppl 13):Abstract 4064. [Google Scholar]
- 14. [Erratum in: Lancet. 2019 Mar 9;393(10175):986] Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. Review. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly at this time due to regulations concerning the proper consent and privacy issues of the patients. We will make an effort to make this data publicly available once regulatory issues are addressed.