Abstract
Background
Nasal (airway) epithelial methylation profiles have been associated with asthma, but the effects of such profiles on expression of distant cis-genes are largely unknown.
Research Question
To identify genes whose expression is associated with proximal and distal CpG probes (within 1 Mb), and to assess whether and how such genes are differentially expressed in atopic asthma.
Study Design and Methods
Genome-wide expression quantitative trait methylation (eQTM) analysis in nasal epithelium from Puerto Rican subjects (aged 9-20 years) with (n = 219) and without (n = 236) asthma. After the eQTM analysis, a Gene Ontology Enrichment analysis was conducted for the top 500 eQTM genes, and mediation analyses were performed to identify paths from DNA methylation to atopic asthma through gene expression. Asthma was defined as physician-diagnosed asthma and wheeze in the previous year, and atopy was defined as at least one positive IgE to allergens. Atopic asthma was defined as the presence of both atopy and asthma.
Results
We identified 16,867 significant methylation-gene expression pairs (false-discovery rate-adjusted P < .01) in nasal epithelium from study participants. Most eQTM methylation probes were distant (average distance, ∼378 kb) from their target genes, and also more likely to be located in enhancer regions of their target genes in lung tissue than control probes. The top 500 eQTM genes were enriched in pathways for immune processes and epithelial integrity and were more likely to have been previously identified as differentially expressed in atopic asthma. In a mediation analysis, we identified 5,934 paths through which methylation markers could affect atopic asthma through gene expression in nasal epithelium.
Interpretation
Previous epigenome-wide association studies of asthma have estimated the effects of DNA methylation markers on expression of nearby genes in airway epithelium. Our findings suggest that distant epigenetic regulation of gene expression in airway epithelium plays a role in atopic asthma.
Key Words: airway epithelium, asthma, eQTM, gene expression, methylation
Abbreviations: DEG, differentially expressed genes; eQTL, expression quantitative trait loci; eQTM, expression quantitative trait methylation; EVA-PR, Epigenetic Variation and Childhood Asthma in Puerto Ricans; EWAS, epigenome-wide association study; FDR-P, false-discovery rate-adjusted P; GWAS, genome-wide association study; TF, transcription factor; TPM, transcripts per kilobase million; TSS, transcription start site
FOR EDITORIAL COMMENT, SEE PAGE 1799
Asthma is affected not only by genetic variants but also by environmental factors such as secondhand smoke. Because DNA methylation is determined by both genetics and environment, studying methylation in relevant tissues may be key to understanding asthma pathogenesis.
A growing body of evidence suggests that abnormalities in airway epithelial integrity and function leads to interactions between injurious agents (such as pollutants and viruses) and dendritic cells, altered immune responses, and—ultimately—asthma. DNA methylation and gene expression in nasal (airway) epithelium are well correlated with those in bronchial (airway) epithelium.1
Because bronchial epithelial sampling requires a bronchoscopy (an invasive and costly procedure with nontrivial risks), nasal epithelial sampling is an attractive and safe approach for studies of the airway epithelium and childhood asthma.1 Indeed, a few epigenome-wide association studies (EWASs) have identified links between DNA methylation in nasal airway epithelium and asthma. For example, we reported 7,104 methylation CpGs associated with atopic (allergic) asthma in Puerto Rican subjects, an ethnic group disproportionately affected with this disease.2
In our prior EWAS, we estimated the effect of CpGs associated with atopic asthma on the expression of nearby genes (ie, those adjacent to or containing a CpG of interest).2 More recently, we showed that most single-nucleotide polymorphisms associated with asthma in a large meta-analysis of genome-wide association studies are not associated with expression of nearby genes, but rather with that of more distant cis-genes within 1 Mb.3 Given such findings, we were interested in examining whether methylation of specific CpG sites is associated with expression of non-nearby cis-genes. We thus conducted an expression quantitative trait methylation (eQTM) analysis in nasal airway epithelium from 455 Puerto Rican subjects ages 9 to 20 years, including 219 subjects with asthma (cases) and 236 control subjects.
Methods
Study Population and Study Procedures
Subject recruitment and study procedures for the Epigenetic Variation and Childhood Asthma in Puerto Ricans (EVA-PR) have been previously described.2 In brief, EVA-PR is a case-control study of asthma in subjects aged 9 to 20 years. See details in e-Appendix 1.
DNA and RNA were extracted from nasal specimens collected from the inferior turbinate. To account for potential effects of different cell types, we implemented a protocol in a subset of nasal samples (n = 31) to select CD326-positive nasal epithelial cells before DNA and
RNA extraction. Whole-genome methylation assays were done with HumanMethylation450 BeadChips (Illumina), as previously described.2 Beta-values, ranging from 0 to 1, were calculated to measure percentage methylation at each CpG site. We then transformed beta values to M values because M values are closer to having a normal distribution (for linear regression analysis). As previously described, RNASeq was conducted with the Illumina NextSeq 500 platform (Illumina), and reads were aligned to reference human genome (hg19), and transcripts per kilobase million (TPM) were used as proxy for gene expression level.2 We excluded genes with low expression levels (mean TPM < 1) and genes whose transcription start site (TSS) was unavailable in hg19. TPM values were transformed to log2 (TPM+1) for data analysis.
eQTM Analysis
We focused on identifying cis-eQTMs (ie, CpGs regulating transcription of neighboring genes), because of limited power to perform a trans analysis (ie, CpGs regulating distant genes).4 Thus, we only considered methylation probes within 1 Mb from the TSS of a gene. Using this criterion, we tested 8,552,964 methylation-gene expression pairs in analyses with and without adjustment for covariates. The unadjusted analysis was conducted to filter out potential false positive signals due to adjustment for batch effects.5,6 Of the 24,171 methylation-expression pairs with a false-discovery rate-adjusted P < .01 (FDR-P) in the adjusted analysis, 7,304 pairs had an FDR-P ≥ .01 in the unadjusted analysis and were thus excluded from further consideration (see later discussion). Thus, we identified 16,867 methylation-expression pairs that were significant in both unadjusted and adjusted analyses.
For the adjusted analysis, we fitted a multivariate linear regression model; , where y is gene expression, M is methylation value at a probe, T represents other covariates, and and are their regression coefficients. In this analysis, other covariates were asthma and atopy status, age, sex, the top five principal components from genotypic data, RNA sample sorting protocol (ie, whole-cells or CD326-positive nasal epithelial cells), methylation and RNA-Seq batch, and latent factors that capture data heterogeneity from methylation and RNA-seq—estimated from R package sva.7 To conduct an efficient analysis, we used a matrix expression quantitative trait loci (eQTL) package8 to obtain P values. FDR-P values were then calculated, based on all of the methylation-expression pairs tested. For the unadjusted model, we only included methylation value as .
Mediation Analysis
To understand how methylation affects asthma through gene expression as a putative mediator, we conducted mediation analyses to identify indirectly associated methylation CpGs to atopic asthma through gene expression. We used the Baron and Kenny9 approach instead of the Sobel method,10 because of differences in sample size between the eQTM analysis (including all subjects) and that for atopic asthma (including only subjects with atopic asthma and nonatopic control subjects).
To have a significant mediation of gene expression, all of the following needed to be significant: (1) the association between methylation and gene expression; (2) the association between methylation and atopic asthma; (3) the association between gene expression and atopic asthma. We only considered eQTM methylation probes and genes as candidates for the mediation tests. We recalculated the FDR-P values of the result from our prior EWAS2 only for the eQTM probes, to reduce multiple testing (e-Appendix 1). We conducted a TWAS fitting a logistic regression model: logit(P) , where P is the probability of having atopic asthma, X is a gene, is an adjusted covariate, and and are regression coefficients. The adjusted covariates included in the model were the first five principal components derived from genotypic data, age, sex, whether RNA samples were from CD326-positive nasal epithelial cells, RNA batches, and a latent factor of gene expression, calculated from the R package sva.7
Results
Location of eQTM-Methylation Probes
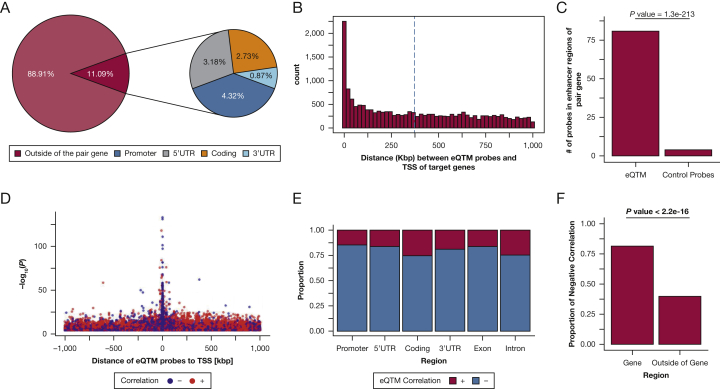
By testing associations between methylation probes within 1 Mb of TSSs of genes and gene expression, we identified 16,867 significant methylation-expression pairs (FDR-P < .01; see Methods), comprising 9,103 methylation probes associated with expression of 3,512 genes. We then investigated the position of significant methylation probes in relation to their paired genes. If a methylation probe was associated with expression of multiple genes, we counted such probe for each gene. We found that 11% and 89% of significant eQTM probes were located within and outside genes, respectively, including 4% of eQTM probes in promoter regions (Fig 1A). Most eQTM methylation probes were distant from their target genes (371,840 bp on average) (Fig 1B).
Figure 1.
Characterization and distribution of genomic location of eQTM signals for 16,867 eQTM pairs in nasal epithelium (FDR-P < 0.01). A, Chart depicting whether the probes are located inside of their paired genes. The right chart shows the specific location of probes located inside of their paired genes. B, Distance between eQTM methylation probes and transcription start sites (TSS) of their target genes in kb pairs. C, Number of probes located in enhancer regions of their target genes in lung tissue. eQTM probes vs controls (the same number of eQTM probes). Fisher exact test was conducted to calculate the P value. D, Positive/negative correlation regarding the distance between methylation and TSS and the P value in the eQTM analysis. E, The bar graph shows, within each gene region, the proportion of positive or negative correlation of the eQTM pairs. The correlation is Pearson correlation. F, The proportion of negatively correlated eQTM pairs inside (from promoter to 3’UTRs) and outside genes. The number of eQTM probes inside a gene is 1,871, and the number of the eQTM probes outside of a gene is 14,996. A χ2 test was conducted to examine the association between the region (whether the probe is located in the gene or outside of the gene) and the sign of the correlation. eQTM = expression quantitative trait methylation; FDR-P = false-discovery rate-adjusted P < .01.
Because we found distant relationships between methylation and their target genes, we assessed whether eQTM methylation probes in nasal epithelium are enriched in the enhancer regions of their paired genes. For this, we checked the enhancer database for lung tissue (http://enhanceratlas.org/),11 because nasal epithelial tissue was not available in that database, and nasal and bronchial epithelial methylation and expression are well correlated.1 We found that eQTM methylation probes are more likely to be located in enhancer regions of their paired genes than randomly selected control probes (P = 1.3 × 10-213) (Fig 1C, Table 1).
Table 1.
Top 30 eQTM Methylation CpGs That Are Located in Enhancer Regions of Their Target Genes in Lung Tissue
| Chr | Probe ID | Position | Gene ID | TSSa | eQTM P |
Distance From TSS |
|---|---|---|---|---|---|---|
| 16 | cg26259865 | 2880359 | ZG16B | 2880172 | 6.6 × 10-33 | 187 |
| 3 | cg22012981 | 58522689 | ACOX2 | 58522929 | 3.2 × 10-31 | -240 |
| 3 | cg16209444 | 58522771 | ACOX2 | 58522929 | 4.5 × 10-25 | -158 |
| 11 | cg15453278 | 67134607 | TBC1D10C | 67171383 | 1.1 × 10-22 | -36,776 |
| 11 | cg21862992 | 68658383 | MRPL21 | 68671303 | 8.6 × 10-21 | -12,920 |
| 11 | cg15453278 | 67134607 | PTPRCAP | 67205153 | 8.2 × 10-17 | -70,546 |
| 6 | cg25045942 | 33048291 | HLA-DPA1 | 33041454 | 3.9 × 10-15 | 6,837 |
| 6 | cg19053046 | 33048254 | HLA-DPA1 | 33041454 | 6.8 × 10-15 | 6,800 |
| 15 | cg10474377 | 42131658 | JMJD7 | 42120282 | 2.1 × 10-14 | 11,376 |
| 17 | cg04204452 | 1479213 | SERPINF2 | 1646129 | 5.1 × 10-14 | -166,916 |
| 11 | cg21920570 | 63766787 | FERMT3 | 63974151 | 1.7 × 10-13 | -207,364 |
| 11 | cg15995296 | 67210812 | TBC1D10C | 67171383 | 1.5 × 10-12 | 39,429 |
| 17 | cg04204452 | 1479213 | SERPINF1 | 1665218 | 1.5 × 10-12 | -186,005 |
| 15 | cg17163752 | 34729026 | GOLGA8B | 34875771 | 2.8 × 10-12 | -146,745 |
| 12 | cg21163444 | 54765670 | NCKAP1L | 54891494 | 1.3 × 10-11 | -125,824 |
| 11 | cg10161008 | 63766546 | FERMT3 | 63974151 | 3.0 × 10-11 | -207,605 |
| 6 | cg17071868 | 33047056 | HLA-DOA | 32977389 | 1.3 × 10-10 | 69,667 |
| 11 | cg21920570 | 63766787 | CCDC88B | 64107689 | 3.2 × 10-10 | -340,902 |
| 3 | cg16209444 | 58522771 | KCTD6 | 58477822 | 1.0 × 10-9 | 44,949 |
| 8 | cg20567768 | 22082066 | SLC39A14 | 22224761 | 1.3 × 10-9 | -142,695 |
| 12 | cg22824738 | 54765988 | NCKAP1L | 54891494 | 2.9 × 10-9 | -125,506 |
| 11 | cg15995296 | 67210812 | PTPRCAP | 67205153 | 4.3 × 10-9 | 5,659 |
| 6 | cg17071868 | 33047056 | HLA-DPB1 | 33043702 | 8.9 × 10-9 | 3,354 |
| 17 | cg01780984 | 79058859 | BAIAP2 | 79008946 | 1.4 × 10-8 | 49,913 |
| 6 | cg19053046 | 33048254 | HLA-DPB1 | 33043702 | 1.7 × 10-8 | 4,552 |
| 11 | cg10161008 | 63766546 | CCDC88B | 64107689 | 1.8 × 10-8 | -341,143 |
| 5 | cg23097826 | 149828748 | RPS14 | 149829319 | 2.5 × 10-8 | -571 |
| 19 | cg25264268 | 427263 | SHC2 | 460996 | 2.5 × 10-8 | -33,733 |
| 12 | cg11700959 | 7066664 | LAG3 | 6881669 | 3.0 × 10-8 | 184,995 |
| 12 | cg11700959 | 7066664 | CD4 | 6898637 | 3.0 × 10-8 | 168,027 |
The eQTM analysis was conducted in nasal airway epithelium, and the enhancer regions and their target genes were identified in lung tissue (http://www.enhanceratlas.org).11 A total of 81 eQTM CpGs that are located in enhancer regions of their target genes were found.
TSS is the transcription start site of the gene.
Although most methylation probes near TSS were negatively correlated with gene expression, more distant pairs tended to be positively correlated (Fig 1D). Of the eQTM methylation probes associated with expression of the gene they were located in, 81.9% were negatively correlated with expression (85.3% if in promoter regions) (Figs 1E, F). In contrast, only 40.2 % of eQTM methylation probes associated with expression of a distant gene (ie, methylation probes outside of the associated gene) were negatively correlated with expression levels (Fig 1F).
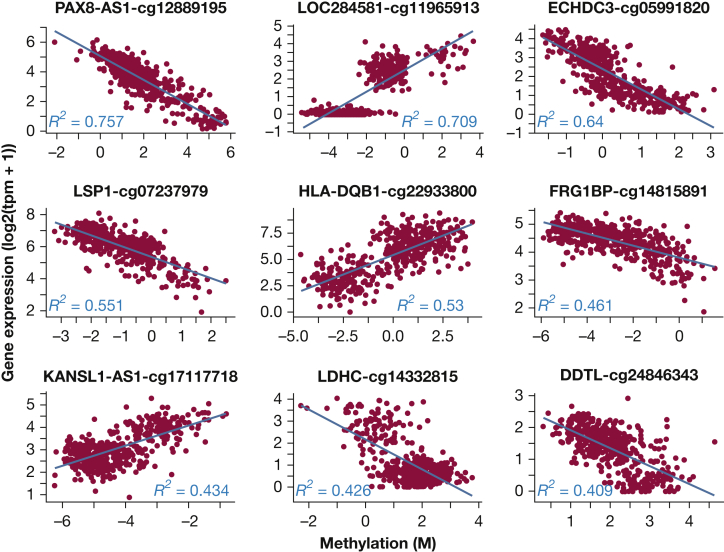
Most of the top eQTM genes (by eQTM P value) have been implicated in lung disease (Fig 2). PAX8 is associated with bronchodilator response in children with asthma,12 ECHDC3 is associated with obesity and asthma in children,13 LSP1 is associated with acute lung inflammation,14 HLA-DQB1 is associated with asthma15 and total IgE,16 FRG1B is highly mutated in lung adenocarcinoma,17 and KANSL1 is associated with pulmonary function.18
Figure 2.
Examples of the most significantly correlated gene-methylation pairs. R2 is squared Pearson correlation between methylation and gene expression. For each gene, only the most significantly associated CpG probe is plotted.
We searched for enrichment of transcription factor (TF) binding site motifs in enhancer regions associated with differentially methylated CpGs and differentially expressed genes (DEGs) in atopic asthma, using TRANSFAC MATCH19,20 software. The top 10 TF binding site motifs are shown in Figure 3. Such TFs are likely to be bound in the motifs of the enhancer regions, thus regulating expression of their target genes. Moreover, most of these TFs have been linked to asthma or respiratory disease. For example, the most enriched motif of TF HNF3 is FOXA. Experimental asthma has been associated with decreased expression of FOXA2 in murine models,21 and the loss of Hoxa5 function promotes Notch-dependent goblet cell metaplasia in the murine airway.22 Moreover, ALOX5 has been linked with reduced lung function and asthma in humans,23 downregulation of GATA-6 decreased airway inflammation via Cav-1 in a murine model of asthma24, and increased expression of the glucocorticoid receptor β-isoform has been associated with glucocorticoid resistance in subjects with asthma.25
Figure 3.
Top10 transcription factor-binding site motifs that are enriched in the enhancer regions that are associated with differential methylation CpGs and differentially expressed genes in atopic asthma. Enhancer regions were identified in lung tissue in http://www.enhanceratlas.org.11 The total number of enhancer regions that are associated with differential methylation and differentially expressed genes in atopic asthma was 26. TRANSFAC MATCH19,20 software programs were used to identify the transcription factor motifs. For each transcription factor tested, among the 26 enhancer regions, sites refer to the number of hits found across all the input sequences. Furthermore, sequences represent the number of sequences in which the hits were made. See Figure 2 legend for expansion of abbreviation.
Gene Ontology Enrichment Analysis
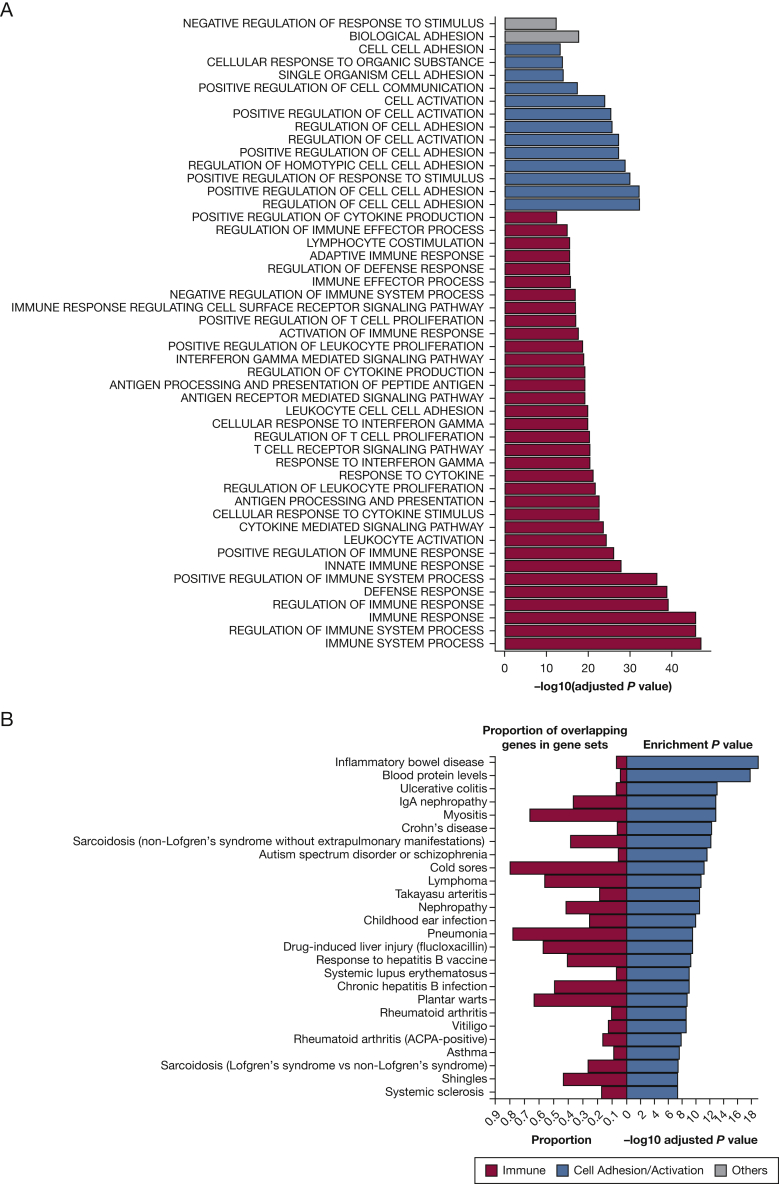
We performed a Gene Ontology enrichment analysis including the top 500 eQTM genes. In this analysis, 34 (69.4%) of the 49 most significant gene ontology categories were related to immune processes (Fig 4A)26; the second most enriched category was cell adhesion or activation.
Figure 4.
Enrichment of the top 500 eQTM genes in immune pathways/diseases. A, Gene ontology (GO) biological processes identified for the top 500 eQTM genes. B, Enrichment of top 500 eQTM genes among reported genes in GWAS catalog27 by disease. Both analyses were done through the FUMA webpage.26 GWAS = genome-wide association study. See Figure 2 legend for expansion of other abbreviation.
We then investigated whether the top 500 eQTM genes are enriched for various diseases by examining significant single-nucleotide polymorphisms from the GWAS catalog.27 Most enriched diseases were related to abnormal immunity (eg, inflammatory bowel disease and IgA nephropathy) (Fig 4B)26 and pulmonary diseases (eg, sarcoidosis, pneumonia, and asthma).
eQTM CpGs and Genes and Atopic Asthma
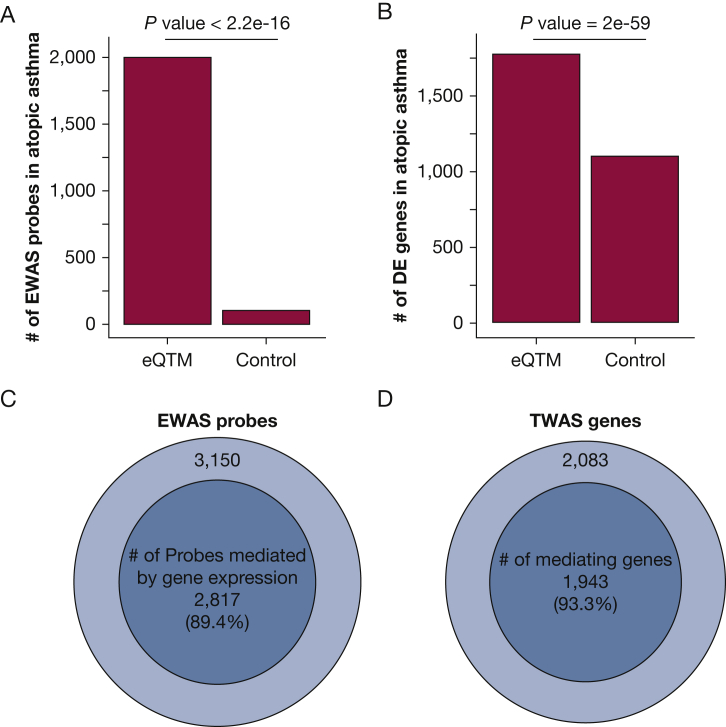
We connected our eQTM results with those from our previous EWAS of atopic asthma,2 using a genome-wide FDR-P < .01. First, we found that only 429 (6.1%) of the 7,046 CpGs that were significantly associated with atopic asthma in our prior EWAS were associated with expression of nearby genes in the eQTM analysis (Fig 1B). Second, CpGs that were significant in the eQTM analysis were overrepresented among CpGs that were significantly associated with atopic asthma in our prior EWAS, compared with randomly selected control CpGs (P < 2.2 × 10-16) (Fig 5A and Table 2).
Figure 5.
Association of eQTM methylation probes and eQTM genes with atopic asthma. A, Enrichment of eQTM methylation probes in epigenome-wide association studies (EWAS) of atopic asthma (genome-wide FDR-P < .01). eQTM refers to eQTM probes and control refers to the same number of randomly selected probes. B, Enrichment of eQTM genes in differentially expressed genes (DEG) in atopic asthma. DEG were identified in our previous study of EVA-PR (genome-wide FDR < 0.01).28 eQTM refers to eQTM genes, and control refers to the same number of randomly selected genes. C, A majority (89.4%) of the associations between eQTM probes and atopic asthma are mediated by gene expression. D, Most (98.3%) of the genes associated with asthma mediate the association between methylation and atopic asthma. See Figure 1 legend for expansion of abbreviations.
Table 2.
Top 30 eQTM Methylation Probes Identified in a Previous Epigenome-Wide Association Study (EWAS) of Atopic Asthma2
| Probe | Chr | Position | EWAS P |
Associated Genes Identified by eQTM (FDR < 0.01) | Nearest Gene |
|---|---|---|---|---|---|
| cg08844313 | 5 | 149240529 | 1.2 × 10-16 | CD74, SLC26A2, DCTN4, AFAP1L1, GRPEL2 | PDE6A |
| cg20372759 | 12 | 58162287 | 1.2 × 10-16 | MBD6, CYP27B1, AGAP2-AS1, MARCH9, TSFM, ATP23, MYO1A | METTL1 |
| cg07239613 | 16 | 67051005 | 2.3 × 10-16 | PSMB10, FBXL8, HSF4, NOL3, CMTM4, PSKH1, TRADD, CKLF | CES4A |
| cg15006973 | 1 | 35258933 | 3.2 × 10-16 | TMEM35B, GJB4, PSMB2, KIAA0319L, SFPQ, LOC653160 | GJA4 |
| cg10549071 | 2 | 235160451 | 4.0 × 10-16 | DGKD, UGT1A1, UGT1A4, UGT1A5, UGT1A3, SCARNA5 | SPP2 |
| cg00406211 | 10 | 121077022 | 5.6 × 10-16 | GRK5, FAM204A, PRDX3, MCMBP | GRK5 |
| cg00664723 | 5 | 15927184 | 5.7 × 10-16 | FBXL7 | FBXL7 |
| cg03875819 | 10 | 4386802 | 6.6 × 10-16 | AKR1C3 | LINC00704 |
| cg21158502 | 5 | 74348187 | 8.6 × 10-16 | GCNT4, LINC01336, GFM2, ENC1, NSA2, FAM169A, POC5 | ANKRD31 |
| cg13586696 | 22 | 29458723 | 1.1 × 10-15 | XBP1, KREMEN1, GAS2L1, RHBDD3 | C22orf31 |
| cg20790648 | 3 | 151619923 | 1.6 × 10-15 | GPR171, MBNL1, AADAC, P2RY13, P2RY1 | SUCNR1 |
| cg24707200 | 1 | 156833163 | 2.3 × 10-15 | SEMA4A, MEF2D, CCT3, ISG20L2, LAMTOR2, SLC25A44, RRNAD1, SMG5, UBQLN4, MRPL24 | NTRK1 |
| cg01859321 | 8 | 144970195 | 2.5 × 10-15 | MROH6, SLC52A2, SCRIB, DGAT1, ZNF707, MROH1, FBXL6,PLEC, ADCK5 | PLEC |
| cg01870976 | 15 | 101887154 | 3.2 × 10-15 | PCSK6, ALDH1A3, LRRK1, TM2D3 | PCSK6 |
| cg00285620 | 11 | 102147694 | 4.9 × 10-15 | DYNC2H1, TMEM123, MMP10 | BIRC3 |
| cg04132353 | 2 | 31440349 | 6.0 × 10-15 | CAPN14, DPY30, LBH, XDH | CAPN14 |
| cg06675531 | 5 | 150019123 | 6.4 × 10-15 | ZNF300, SLC26A2, DCTN4,SYNPO | SYNPO |
| cg22855021 | 14 | 81610812 | 7.5 × 10-15 | GTF2A1 | TSHR |
| cg09472600 | 1 | 183537770 | 9.3 × 10-15 | NPL, DHX9, TSEN15, APOBEC4 | NCF2 |
| cg19107578 | 5 | 493262 | 1.2 × 10-14 | SLC9A3, PP7080, CEP72, LOC100288152 | SLC9A3 |
| cg18749617 | 15 | 102028637 | 1.2 × 10-14 | PCSK6, ALDH1A3, LRRK1 | PCSK6 |
| cg10830021 | 11 | 3815589 | 1.3 × 10-14 | RRM1, TRIM21, TSSC2 | NUP98 |
| cg03387497 | 20 | 17680945 | 1.5 × 10-14 | POLR3F, SNRPB2, LINC00493, RRBP1, RBBP9 | BANF2 |
| cg20337028 | 17 | 75181836 | 1.8 × 10-14 | SEC14L1, SYNGR2, UBALD2, SEPT9, SPHK1, TNRC6C | SEC14L1 |
| cg17223698 | 15 | 39416631 | 2.0 × 10-14 | SRP14 | C15orf54 |
| cg19497511 | 2 | 238609807 | 2.7 × 10-14 | COPS8 | LRRFIP1 |
| cg08175352 | 3 | 101894206 | 3.2 × 10-14 | ZBTB11, TRMT10C, NXPE3, SENP7, RPL24, PCNP | ZPLD1 |
| cg02333649 | 22 | 19471093 | 5.5 × 10-14 | RTN4R, ARVCF, MRPL40, PRODH, LINC00896, UFD1L, TANGO2, DGCR8, ZDHHC8 | CDC45 |
| cg08956463 | 6 | 41168911 | 5.9 × 10-14 | MDFI, TREM2, FOXP4, C6orf132, UNC5CL | TREML2 |
| cg04320956 | 16 | 69143512 | 6.3 × 10-14 | HAS3, ZFP90, NQO1, NIP7, SLC7A6, CDH3, ESRP2 | HAS3 |
Probes sorted by EWAS P value. Genes shown in bold are the nearest gene.
Next, we checked whether the 3,512 significant eQTM genes identified in the current analysis are differentially expressed in atopic asthma (at genome-wide FDR-P < .01), by checking the results of our recently published TWAS28 (e-Appendix 1). Indeed, these 3,512 eQTM genes are significantly more likely to be DEGs in atopic asthma than 3,512 randomly selected genes (P = 1.53 × 10-59) (Fig 5B and Table 3).
Table 3.
Top 30 eQTM Genes Identified in a Previous Transcriptome-Wide Association Study (TWAS) of Atopic Asthma28
| Gene | Chr | TWAS P |
No. of Associated Probes | Probe | Position | eQTM P |
|---|---|---|---|---|---|---|
| CST1 | 20 | 1.1 × 10-64 | 1 | cg14928764 | 23064608 | 5.5 × 10-10 |
| CLCA1 | 1 | 1.4 × 10-47 | 5 | cg22175412 | 86063985 | 9.7 × 10-15 |
| NTRK2 | 9 | 4.3 × 10-44 | 2 | cg09926027 | 87285693 | 1.5 × 10-18 |
| FETUB | 3 | 7.5 × 10-42 | 5 | cg25735294 | 186353721 | 2.6 × 10-18 |
| CPA3 | 3 | 3.4 × 10-39 | 5 | cg13235059 | 149192304 | 3.1 × 10-15 |
| ITLN1 | 1 | 8.7 × 10-38 | 10 | cg10094191 | 160855148 | 6.3 × 10-9 |
| CDH26 | 20 | 8.6 × 10-37 | 7 | cg06943251 | 57615398 | 2.1 × 10-18 |
| CCL26 | 7 | 2.2 × 10-35 | 5 | cg13053914 | 75511260 | 1.4 × 10-8 |
| CST2 | 20 | 4.0 × 10-35 | 1 | cg14928764 | 23064608 | 3.2 × 10-10 |
| C3orf70 | 3 | 8.3 × 10-35 | 9 | cg01390445 | 185271312 | 1.6 × 10-13 |
| TPSAB1 | 16 | 3.3 × 10-34 | 26 | cg00943124 | 1705667 | 3.9 × 10-14 |
| CISH | 3 | 3.0 × 10-32 | 9 | cg23005227 | 50645426 | 8.4 × 10-24 |
| TPSB2 | 16 | 8.0 × 10-30 | 20 | cg00943124 | 1705667 | 4.3 × 10-13 |
| ALOX15 | 17 | 3.4 × 10-29 | 11 | cg23387401 | 4582204 | 9.7 × 10-24 |
| CEP72 | 5 | 1.3 × 10-28 | 42 | cg04221910 | 616842 | 5.1 × 10-21 |
| SLC5A5 | 19 | 5.5 × 10-28 | 10 | cg15734198 | 17423023 | 1.7 × 10-12 |
| POSTN | 13 | 7.7 × 10-28 | 4 | cg03071245 | 37463034 | 1.7 × 10-9 |
| HS3ST4 | 16 | 5.2 × 10-27 | 4 | cg26725397 | 25937266 | 6.2 × 10-15 |
| PCSK6 | 15 | 1.4 × 10-26 | 11 | cg18749617 | 102028637 | 1.4 × 10-27 |
| WBSCR17 | 7 | 2.5 × 10-26 | 3 | cg01349903 | 71148142 | 5.4 × 10-12 |
| KYAT1 | 9 | 2.8 × 10-26 | 12 | cg13835688 | 130859454 | 1.6 × 10-16 |
| ANO1 | 11 | 5.1 × 10-26 | 7 | cg11058904 | 69987299 | 1.3 × 10-15 |
| ABO | 9 | 3.0 × 10-25 | 6 | cg11879188 | 136149908 | 7.4 × 10-18 |
| CMYA5 | 5 | 4.5 × 10-25 | 1 | cg14978242 | 79501131 | 1.2 × 10-7 |
| SLC24A3 | 20 | 1.3 × 10-23 | 3 | cg08371391 | 19739935 | 3.0 × 10-8 |
| GCNT4 | 5 | 1.1 × 10-22 | 2 | cg21158502 | 74348187 | 2.0 × 10-30 |
| SLC7A1 | 13 | 1.6 × 10-22 | 3 | cg17798847 | 30098432 | 8.3 × 10-7 |
| SLC45A4 | 8 | 1.8 × 10-22 | 7 | cg07140289 | 142299684 | 1.8 × 10-11 |
| DQX1 | 2 | 6.9 × 10-22 | 9 | cg02034222 | 74753281 | 4.4 × 10-17 |
| GSN | 9 | 9.4 × 10-22 | 6 | cg13928417 | 124498782 | 2.7 × 10-9 |
| KCNJ16 | 17 | 2.3 × 10-21 | 2 | cg13606025 | 68070495 | 7.8 × 10-10 |
| LINC01336 | 5 | 7.7 × 10-21 | 2 | cg21158502 | 74348187 | 1.6 × 10-16 |
| ZNF467 | 7 | 2.2 × 10-20 | 8 | cg07970948 | 149543165 | 3.3 × 10-19 |
| RUSC1 | 1 | 3.7 × 10-20 | 15 | cg23154272 | 154966068 | 2.3 × 10-16 |
| DHX35 | 20 | 4.7 × 10-20 | 1 | cg26604799 | 36789861 | 2.5 × 10-5 |
| DPP4 | 2 | 8.1 × 10-20 | 3 | cg22143064 | 162948592 | 5.9 × 10-27 |
| SOX13 | 1 | 8.4 × 10-20 | 3 | cg17000774 | 203154457 | 1.3 × 10-8 |
| SLC18A2 | 10 | 1.1 × 10-19 | 2 | cg03519180 | 119102524 | 1.2 × 10-6 |
| ST6GAL1 | 3 | 1.6 × 10-19 | 6 | cg25735294 | 186353721 | 1.1 × 10-20 |
| C20orf197 | 20 | 3.8 × 10-19 | 2 | cg16518142 | 58533713 | 3.9 × 10-8 |
| CA2 | 8 | 1.1 × 10-18 | 1 | cg05071334 | 86195487 | 1.1 × 10-5 |
| NPDC1 | 9 | 2.3 × 10-18 | 8 | cg13850871 | 139583773 | 2.6 × 10-11 |
| RTN4R | 22 | 3.2 × 10-18 | 3 | cg02333649 | 19471093 | 2.0 × 10-22 |
| FGF11 | 17 | 3.2 × 10-18 | 12 | cg22637538 | 7348327 | 3.1 × 10-11 |
| LOC100288152 | 5 | 5.0 × 10-18 | 15 | cg22572362 | 501938 | 1.6 × 10-8 |
| ELOVL5 | 6 | 5.8 × 10-18 | 1 | cg26516974 | 52475065 | 3.5 × 10-9 |
| CMIP | 16 | 1.0 × 10-17 | 1 | cg16583186 | 81526361 | 6.7 × 10-6 |
| ADAMTS9 | 3 | 1.4 × 10-17 | 11 | cg08765100 | 64211659 | 1.4 × 10-11 |
| CCK | 3 | 1.8 × 10-17 | 2 | cg07886398 | 42131702 | 1.0 × 10-8 |
Significance for both differential expression and differential methylation defined as FDR-P < .01. Genes shown sorted by TWAS P value. Only the most significantly associated probe per gene is presented.
To test whether methylation affects atopic asthma through regulation of gene expression, we conducted a mediation analysis. In this analysis, we found 5,934 paths in which methylation of CpGs affects atopic asthma through gene expression, consisting of 2,817 methylation probes and 1,943 genes (Table 4). Of all the associations between eQTM methylation probes and atopic asthma, 89.4% were mediated by gene expression (Fig 5C). Likewise, 93.3% of the eQTM genes associated with atopic asthma mediate the association between methylation and atopic asthma (Fig 5D).
Table 4.
Top 30 Mediation Paths From Methylation to Gene Expression to Atopic Asthma
| Chr | Probe | Position | Gene | TSS | eQTM P |
EWAS P |
TWAS P |
|---|---|---|---|---|---|---|---|
| 20 | cg14928764 | 23064608 | CST1 | 23731574 | 5.5 × 10-10 | 6.0 × 10-4 | 3.2 × 10-15 |
| 3 | cg01390445 | 185271312 | C3orf70 | 184870802 | 1.5 × 10-13 | 9.6 × 10-8 | 7.0 × 10-13 |
| 5 | cg14978242 | 79501131 | CMYA5 | 78985658 | 1.2 × 10-7 | 2.0 × 10-7 | 1.4 × 10-12 |
| 16 | cg00943124 | 1705667 | TPSAB1 | 1290677 | 3.9 × 10-14 | 9.0 × 10-9 | 3.3 × 10-12 |
| 16 | cg26725397 | 25937266 | HS3ST4 | 25703346 | 6.2 × 10-15 | 6.6 × 10-8 | 6.2 × 10-12 |
| 17 | cg23387401 | 4582204 | ALOX15 | 4544960 | 9.7 × 10-24 | 2.1 × 10-13 | 6.8 × 10-12 |
| 11 | cg11058904 | 69987299 | ANO1 | 69924407 | 1.3 × 10-15 | 3.9 × 10-13 | 6.8 × 10-12 |
| 7 | cg11303839 | 75405967 | CCL26 | 75419064 | 7.3 × 10-7 | 1.1 × 10-7 | 8.9 × 10-12 |
| 5 | cg21158502 | 74348187 | GCNT4 | 74326724 | 2.0 × 10-30 | 8.5 × 10-16 | 1.1 × 10-11 |
| 9 | cg04236137 | 123655887 | GSN | 124030379 | 3.7 × 10-8 | 8.8 × 10-4 | 1.4 × 10-11 |
| 5 | cg21158502 | 74348187 | LINC01336 | 74348468 | 1.6 × 10-16 | 8.5 × 10-16 | 1.5 × 10-11 |
| 15 | cg18749617 | 102028637 | PCSK6 | 102030187 | 1.4 × 10-27 | 1.1 × 10-14 | 1.6 × 10-11 |
| 3 | cg13235059 | 149192304 | CPA3 | 148583042 | 3.0 × 10-15 | 1.7 × 10-8 | 1.8 × 10-11 |
| 20 | cg14928764 | 23064608 | CST2 | 23807312 | 3.2 × 10-10 | 6.0 × 10-4 | 1.9 × 10-11 |
| 5 | cg01181940 | 478916 | CEP72 | 612404 | 1.6 × 10-20 | 2.2 × 10-11 | 2.8 × 10-11 |
| 9 | cg09926027 | 87285693 | NTRK2 | 87283372 | 1.5 × 10-18 | 4.5 × 10-12 | 3.2 × 10-11 |
| 1 | cg23154272 | 154966068 | RUSC1 | 155290639 | 2.2 × 10-16 | 2.0 × 10-8 | 3.4 × 10-11 |
| 9 | cg11879188 | 136149908 | ABO | 136150630 | 7.3 × 10-18 | 1.9 × 10-7 | 3.8 × 10-11 |
| 3 | cg23005227 | 50645426 | CISH | 50649262 | 8.3 × 10-24 | 4.0 × 10-12 | 3.8 × 10-11 |
| 6 | cg14178895 | 11778902 | ADTRP | 11779280 | 6.8 × 10-23 | 3.6 × 10-8 | 4.0 × 10-11 |
| 3 | cg25735294 | 186353721 | ST6GAL1 | 186648314 | 1.0 × 10-20 | 3.3 × 10-10 | 5.2 × 10-11 |
| 8 | cg07140289 | 142299684 | SLC45A4 | 142238673 | 1.8 × 10-11 | 2.4 × 10-4 | 6.3 × 10-11 |
| 2 | cg04132353 | 31440349 | CAPN14 | 31440411 | 4.6 × 10-20 | 5.9 × 10-15 | 6.4 × 10-11 |
| 5 | cg14978242 | 79501131 | SERINC5 | 79551901 | 1.3 × 10-9 | 2.0 × 10-7 | 7.7 × 10-11 |
| 1 | cg03058346 | 91275170 | LRRC8D | 90286572 | 1.4 × 10-6 | 6.9 × 10-4 | 8.4 × 10-11 |
| 1 | cg01062020 | 162382848 | SH2D1B | 162381928 | 1.7 × 10-6 | 2.0 × 10-6 | 9.8 × 10-11 |
| 20 | cg26604799 | 36789861 | DHX35 | 37590980 | 2.5 × 10-5 | 2.5 × 10-3 | 1.0 × 10-10 |
| 16 | cg00943124 | 1705667 | TPSB2 | 1280185 | 4.2 × 10-13 | 9.0 × 10-9 | 1.1 × 10-10 |
| 2 | cg22143064 | 162948592 | DPP4 | 162931052 | 5.8 × 10-27 | 5.2 × 10-8 | 1.5 × 10-10 |
| 15 | cg09407660 | 59910436 | GCNT3 | 59903981 | 1.3 × 10-19 | 9.8 × 10-10 | 1.5 × 10-10 |
Of the 2,817 methylation probes identified in the mediation analysis, 143 were located in the promoter regions of the associated genes (Table 5). Of these 143 probes, 130 (90%) were associated with reduced gene expression, and 29 were located in enhancer regions of the associated genes in lung tissue. In a secondary analysis, we examined whether these probes are associated with the activation of transcription in an additional lung H3K27ac ChIP-seq dataset from ENCODE (GSM1013123). In this analysis, all but two (cg20813462 and cg24437859) of the CpGs listed in Table 5 can be mapped to H3K27ac ChIP-seq peaks, suggesting that these may be active enhancer regions. In addition, we examined possible 3D gene interaction using the 3D Genome Browser,29 in which the resolution of a lung dataset is limited to 10 kb. Interestingly, we observed possible interactions for cg15453278- TBC1D10C, cg21862992- MRPL21, and cg15453278- PTPRCAP, using the Virtual 4C method; these three CpGs are within enhancer regions.
Table 5.
Top Methylation Probes (CpGs)∗ That Are in Transcription Regulatory Elements (TREs) in the Promoter or Enhancer Regions of Genes That Are Differentially Expressed in Atopic Asthma
| Chr | Probe | Pos | Gene | TRE Start |
TRE End |
Effect size | eQTM P |
EWAS P |
TWAS P |
|---|---|---|---|---|---|---|---|---|---|
| Promoter | |||||||||
| 6 | cg11210880 | 11779911 | ADTRP | 11779280 | 11781280 | -0.43 | 1.1 × 10-12 | 7.6 × 10-9 | 4.0 × 10-11 |
| 1 | cg01062020 | 162382848 | SH2D1B | 162381928 | 162383928 | -0.2 | 1.7 × 10-6 | 2.0 × 10-6 | 9.8 × 10-11 |
| 20 | cg20895028 | 58533443 | CDH26 | 58531470 | 58533470 | -0.49 | 6.0 × 10-6 | 4.2 × 10-10 | 1.5 × 10-10 |
| 17 | cg18105842 | 7341440 | FGF11 | 7339591 | 7341591 | -0.23 | 3.6 × 10-9 | 8.5 × 10-7 | 1.5 × 10-10 |
| 20 | cg01352551 | 19191994 | SLC24A3 | 19191289 | 19193289 | -0.33 | 8.1 × 10-8 | 1.5 × 10-4 | 1.7 × 10-10 |
| 2 | cg14499385 | 190446494 | SLC40A1 | 190445537 | 190447537 | 0.48 | 1.3 × 10-10 | 6.6 × 10-4 | 3.9 × 10-10 |
| 13 | cg18341491 | 38174258 | POSTN | 38172981 | 38174981 | -0.7 | 2.9 × 10-8 | 6.3 × 10-7 | 8.2 × 10-10 |
| 17 | cg13606025 | 68070495 | KCNJ16 | 68069365 | 68071365 | -0.45 | 7.8 × 10-10 | 2.5 × 10-4 | 1.9 × 10-9 |
| 3 | cg13705284 | 58523313 | ACOX2 | 58522929 | 58524929 | -0.45 | 4.0 × 10-18 | 8.8 × 10-10 | 3.8 × 10-9 |
| 10 | cg19571004 | 135340850 | CYP2E1 | 135338866 | 135340866 | -0.49 | 5.2 × 10-21 | 3.3 × 10-6 | 3.9 × 10-9 |
| 17 | cg25170091 | 202716 | RPH3AL | 202633 | 204633 | -0.21 | 7.9 × 10-15 | 4.6 × 10-6 | 5.2 × 10-9 |
| 5 | cg00049323 | 472564 | LOC100288152 | 471350 | 473350 | 0.18 | 9.5 × 10-7 | 1.0 × 10-13 | 6.0 × 10-9 |
| 7 | cg23468130 | 114562060 | MDFIC | 114560208 | 114562208 | -0.28 | 1.1 × 10-7 | 2.2 × 10-4 | 9.1 × 10-9 |
| 8 | cg10054641 | 133773093 | TMEM71 | 133772914 | 133774914 | -0.2 | 1.0 × 10-12 | 1.6 × 10-11 | 1.9 × 10-8 |
| 3 | cg08450017 | 45984838 | CXCR6 | 45982972 | 45984972 | -0.46 | 2.0 × 10-32 | 1.8 × 10-6 | 2.5 × 10-8 |
| Enhancer | |||||||||
| 3 | cg22012981 | 58522689 | ACOX2 | 58520920 | 58522840 | -0.86 | 3.1 × 10-31 | 2.0 × 10-11 | 3.8 × 10-9 |
| 16 | cg16219266 | 67433184 | FBXL8 | 67432850 | 67433250 | -0.4 | 3.3 × 10-6 | 1.1 × 10-5 | 1.2 × 10-8 |
| 17 | cg04204452 | 1479213 | MYO1C | 1479060 | 1479890 | 0.2 | 3.9 × 10-8 | 4.4 × 10-6 | 1.4 × 10-8 |
| 16 | cg16219266 | 67433184 | NOL3 | 67432850 | 67433250 | -0.34 | 1.0 × 10-5 | 1.1 × 10-5 | 2.3 × 10-8 |
| 3 | cg16209444 | 58522771 | KCTD6 | 58520920 | 58522840 | -0.29 | 1.0 × 10-9 | 1.1 × 10-8 | 3.0 × 10-8 |
| 11 | cg15453278 | 67134607 | RHOD | 67133100 | 67134930 | -0.14 | 1.7 × 10-5 | 5.1 × 10-5 | 1.9 × 10-7 |
| 7 | cg20813462 | 2646259 | TTYH3 | 2645630 | 2646750 | -0.18 | 1.5 × 10-6 | 2.4 × 10-3 | 3.9 × 10-7 |
| 17 | cg04204452 | 1479213 | ABR | 1479060 | 1479890 | 0.15 | 3.5 × 10-7 | 4.4 × 10-6 | 4.2 × 10-7 |
| 7 | cg20813462 | 2646259 | SNX8 | 2645630 | 2646750 | -0.15 | 1.5 × 10-6 | 2.4 × 10-3 | 6.3 × 10-7 |
| 16 | cg07261196 | 75601025 | KARS | 75600810 | 75601930 | 0.16 | 2.1 × 10-5 | 5.3 × 10-4 | 4.2 × 10-6 |
| 17 | cg01780984 | 79058859 | BAIAP2 | 79058690 | 79059450 | -0.26 | 1.4 × 10-8 | 7.8 × 10-4 | 1.4 × 10-5 |
| 12 | cg24437859 | 7066614 | PTMS | 7065730 | 7066810 | 0.16 | 9.0 × 10-6 | 6.9 × 10-5 | 2.1 × 10-5 |
| 17 | cg04204452 | 1479213 | SERPINF2 | 1479060 | 1479890 | 0.37 | 5.1 × 10-14 | 4.4 × 10-6 | 2.4 × 10-5 |
| 16 | cg16219266 | 67433184 | TRADD | 67432850 | 67433250 | 0.53 | 2.5 × 10-6 | 1.1 × 10-5 | 8.6 × 10-5 |
| 6 | cg19053046 | 33048254 | HLA-DPA1 | 33046460 | 33048440 | -0.34 | 6.8 × 10-15 | 3.2 × 10-4 | 8.6 × 10-5 |
The top 15 methylation-gene pairs are presented for each category (promoter or enhancer region). Results sorted by TWAS P value.29 A total of 143 mediation paths in the promoter region of the associated genes and 29 mediation paths in the enhancer region (lung) of the associated genes were identified. Only one mediation path was presented per gene. Mediation analysis was conducted using Baron and Kenny (1986).9
Replication of eQTM Results
To attempt replication of our eQTM results in EVA-PR, we used public data from GEO (GSE6520530), which includes both methylation and gene expression array data in nasal epithelium from 69 children (36 with atopic asthma and 33 healthy control subjects, mostly [91.3%] African American). Using a similar approach to that used in EVA-PR, this replication eQTM analysis was adjusted for age, sex, race/ethnicity, atopic asthma status, and unobserved batch effects.
Of the 16,867 significant associations between methylation and gene expression in EVA-PR, we were able to test 14,397 associations in GSE65205, because of differences in the platforms used to assess gene expression (RNA-Seq vs microarray). Of these 14,397 methylation-expression pairs, 12,559 (87.2%) had the same direction of association in GSE65205. Despite the small sample size of GSE65205, 6,562 (45.6%) of the significant associations in EVA-PR were replicated at FDR-P < .05, in the same direction of association (Table 6). These replicated associations include 3,992 methylation probes and 1,106 genes. Of the 3,992 replicated methylation probes, 3,222 probes were tested in our prior EWAS in EVA-PR2: 1,412 (43.8%) of these 3,222 probes are significantly associated with atopic asthma (FDR-P < .05) (Table 7).
Table 6.
Top 30 Most Significant eQTM Methylation-Gene Pairs in EVA-PR Cohort That Replicated in a Publicly Available Dataset (GSE65205)
| Associated Pairs |
EVA-PR |
GSE65205 |
|||||
|---|---|---|---|---|---|---|---|
| Probe | Gene | Beta | P | FDR | Beta | P | FDR |
| cg05991820 | ECHDC3 | -1 | 5.1 × 10-98 | 7.2 × 10-92 | -0.31 | 3.9 × 10-3 | 1.2 × 10-2 |
| cg22933800 | HLA-DQB1 | 0.75 | 1.7 × 10-76 | 1.2 × 10-70 | 1 | 7.9 × 10-20 | 5.7 × 10-16 |
| cg07237979 | LSP1 | -0.67 | 4.6 × 10-69 | 3.0 × 10-63 | -0.57 | 2.4 × 10-12 | 4.9 × 10-10 |
| cg14332815 | LDHC | -0.71 | 1.3 × 10-56 | 4.0 × 10-51 | -1.1 | 1.5 × 10-18 | 5.3 × 10-15 |
| cg10296238 | SPATC1L | -0.31 | 1.1 × 10-51 | 2.5 × 10-46 | -0.23 | 2.4 × 10-10 | 1.7 × 10-8 |
| cg17117718 | CRHR1-IT1 | 0.29 | 4.0 × 10-51 | 8.6 × 10-46 | 0.35 | 5.2 × 10-8 | 1.2 × 10-6 |
| cg16145915 | ZFAND2A | 0.48 | 2.6 × 10-50 | 5.1 × 10-45 | 0.53 | 1.1 × 10-4 | 6.0 × 10-4 |
| cg10626236 | CDK11A | 0.59 | 3.2 × 10-50 | 6.2 × 10-45 | 0.39 | 1.0 × 10-2 | 2.5 × 10-2 |
| cg03190825 | CYP4F11 | -1.3 | 2.5 × 10-48 | 4.5 × 10-43 | -0.41 | 6.7 × 10-3 | 1.8 × 10-2 |
| cg22092521 | CFD | -0.83 | 1.7 × 10-45 | 2.6 × 10-40 | -1.2 | 1.2 × 10-6 | 1.4 × 10-5 |
| cg11375102 | TMEM204 | -0.68 | 1.3 × 10-44 | 1.9 × 10-39 | -1 | 1.7 × 10-14 | 1.2 × 10-11 |
| cg24977027 | THNSL2 | -0.78 | 5.6 × 10-44 | 8.0 × 10-39 | -1.2 | 4.6 × 10-13 | 1.3 × 10-10 |
| cg05461841 | ZG16B | -0.66 | 2.0 × 10-42 | 2.6 × 10-37 | -0.71 | 1.4 × 10-4 | 7.2 × 10-4 |
| cg06851207 | PNMAL1 | -0.79 | 2.1 × 10-42 | 2.7 × 10-37 | -0.89 | 3.0 × 10-7 | 4.6 × 10-6 |
| cg01878807 | DHRS4-AS1 | -0.42 | 4.3 × 10-42 | 5.4 × 10-37 | -0.34 | 1.3 × 10-5 | 1.0 × 10-4 |
| cg02926397 | LY6D | -1.2 | 8.3 × 10-42 | 1.0 × 10-36 | -2.5 | 6.6 × 10-10 | 3.8 × 10-8 |
| cg02719634 | SLC22A18AS | -0.28 | 5.8 × 10-41 | 6.6 × 10-36 | -0.41 | 3.0 × 10-6 | 3.0 × 10-5 |
| cg06846259 | POMC | -0.5 | 5.9 × 10-41 | 6.6 × 10-36 | -1 | 1.2 × 10-16 | 2.4 × 10-13 |
| cg15176213 | COX7A1 | -0.85 | 6.3 × 10-41 | 6.9 × 10-36 | -1.3 | 2.4 × 10-14 | 1.7 × 10-11 |
| cg14815891 | FRG1BP | -0.19 | 8.0 × 10-41 | 8.7 × 10-36 | -0.26 | 1.3 × 10-4 | 7.0 × 10-4 |
| cg24846343 | GSTT2B | -0.79 | 4.2 × 10-39 | 4.2 × 10-34 | -1 | 1.1 × 10-12 | 2.6 × 10-10 |
| cg19059861 | BPIFA1 | -3.2 | 4.6 × 10-37 | 4.1 × 10-32 | -3.9 | 1.4 × 10-8 | 4.1 × 10-7 |
| cg22933800 | HLA-DQA2 | -0.49 | 9.0 × 10-37 | 7.8 × 10-32 | -0.18 | 1.7 × 10-5 | 1.3 × 10-4 |
| cg06322601 | RASA4 | 0.18 | 3.8 × 10-35 | 3.0 × 10-30 | 0.08 | 1.6 × 10-2 | 3.7 × 10-2 |
| cg05681977 | SLC39A4 | -0.37 | 6.4 × 10-34 | 4.7 × 10-29 | -0.64 | 9.1 × 10-16 | 1.4 × 10-12 |
| cg10207745 | LINC01559 | -0.86 | 7.3 × 10-34 | 5.2 × 10-29 | -0.52 | 3.0 × 10-3 | 9.5 × 10-3 |
| cg10807101 | GSTM3 | -0.43 | 3.4 × 10-33 | 2.3 × 10-28 | -0.68 | 4.9 × 10-7 | 6.8 × 10-6 |
| cg08450017 | CXCR6 | -0.46 | 2.0 × 10-32 | 1.3 × 10-27 | -0.59 | 2.4 × 10-10 | 1.7 × 10-8 |
| cg01850135 | NLRC3 | -0.34 | 8.3 × 10-32 | 5.2 × 10-27 | -0.9 | 1.5 × 10-11 | 2.0 × 10-9 |
| cg23161218 | ACAP1 | 0.31 | 1.2 × 10-31 | 7.5 × 10-27 | 0.29 | 2.1 × 10-4 | 1.0 × 10-3 |
Replication defined as FDR P < .05 with effect in the same direction as in EVA-PR.
Table 7.
Top 30 Most Significant eQTM Methylation-Gene Pairs in EVA-PR Cohort That Replicated in GSE65205
| Associated pairs |
EVA-PR |
GSE65205 |
EWAS (EVA-PR) |
|||||
|---|---|---|---|---|---|---|---|---|
| Probe | Gene | Beta | P | FDR | Beta | P | FDR | FDR |
| cg04511125 | THNSL2 | -0.9 | 1.1 × 10-36 | 9.2 × 10-32 | -0.81 | 1.2 × 10-6 | 1.4 × 10-5 | 7.3 × 10-3 |
| cg17252645 | LY6D | -1 | 6.1 × 10-36 | 5.0 × 10-31 | -1.6 | 1.5 × 10-8 | 4.4 × 10-7 | 4.1 × 10-2 |
| cg10807101 | GSTM3 | -0.43 | 3.4 × 10-33 | 2.3 × 10-28 | -0.68 | 4.9 × 10-7 | 6.8 × 10-6 | 7.6 × 10-4 |
| cg08450017 | CXCR6 | -0.46 | 2.0 × 10-32 | 1.3 × 10-27 | -0.59 | 2.4 × 10-10 | 1.7 × 10-8 | 2.6 × 10-4 |
| cg01850135 | NLRC3 | -0.34 | 8.3 × 10-32 | 5.2 × 10-27 | -0.9 | 1.5 × 10-11 | 2.0 × 10-9 | 5.6 × 10-4 |
| cg23161218 | ACAP1 | 0.31 | 1.2 × 10-31 | 7.5 × 10-27 | 0.29 | 2.1 × 10-4 | 1.0 × 10-3 | 2.3 × 10-3 |
| cg22012981 | ACOX2 | -0.86 | 3.2 × 10-31 | 1.9 × 10-26 | -0.35 | 1.1 × 10-2 | 2.6 × 10-2 | 3.3 × 10-8 |
| cg07786657 | CD247 | -0.35 | 4.6 × 10-31 | 2.7 × 10-26 | -0.37 | 3.9 × 10-9 | 1.5 × 10-7 | 8.0 × 10-4 |
| cg03546687 | IL32 | 0.59 | 2.4 × 10-29 | 1.3 × 10-24 | 0.75 | 5.0 × 10-6 | 4.5 × 10-5 | 3.1 × 10-3 |
| cg14527029 | HGD | -0.76 | 3.2 × 10-29 | 1.6 × 10-24 | -1.1 | 9.2 × 10-8 | 1.8 × 10-6 | 1.4 × 10-7 |
| cg11453837 | PSMB9 | -0.98 | 3.3 × 10-28 | 1.6 × 10-23 | -0.78 | 6.0 × 10-4 | 2.5 × 10-3 | 1.7 × 10-4 |
| cg27583010 | SEPT1 | -0.39 | 1.0 × 10-27 | 4.6 × 10-23 | -0.61 | 3.4 × 10-9 | 1.3 × 10-7 | 4.3 × 10-2 |
| cg18749617 | PCSK6 | -0.38 | 1.4 × 10-27 | 6.3 × 10-23 | -0.22 | 1.1 × 10-3 | 4.1 × 10-3 | 1.2 × 10-10 |
| cg08450017 | CCR5 | -0.36 | 2.8 × 10-27 | 1.2 × 10-22 | -0.54 | 3.3 × 10-9 | 1.3 × 10-7 | 2.6 × 10-4 |
| cg22143064 | DPP4 | -1.1 | 5.9 × 10-27 | 2.4 × 10-22 | -0.87 | 4.7 × 10-9 | 1.8 × 10-7 | 1.8 × 10-5 |
| cg07786657 | RCSD1 | -0.27 | 3.2 × 10-26 | 1.2 × 10-21 | -0.39 | 2.0 × 10-5 | 1.4 × 10-4 | 8.0 × 10-4 |
| cg08159663 | NLRC5 | -0.63 | 1.3 × 10-25 | 4.6 × 10-21 | -0.31 | 1.2 × 10-2 | 2.9 × 10-2 | 1.3 × 10-4 |
| cg19517476 | RASAL3 | -0.27 | 2.6 × 10-25 | 9.0 × 10-21 | -0.87 | 1.6 × 10-13 | 6.5 × 10-11 | 2.8 × 10-4 |
| cg12044599 | TBC1D10C | -0.32 | 3.5 × 10-25 | 1.2 × 10-20 | -0.71 | 3.6 × 10-7 | 5.4 × 10-6 | 3.5 × 10-3 |
| cg26833120 | LCK | 0.46 | 4.8 × 10-25 | 1.6 × 10-20 | 0.73 | 4.8 × 10-5 | 3.1 × 10-4 | 1.6 × 10-3 |
| cg02297541 | HLA-DMA | 0.49 | 5.3 × 10-25 | 1.8 × 10-20 | 0.91 | 6.6 × 10-10 | 3.7 × 10-8 | 9.2 × 10-4 |
| cg06148175 | ACY3 | -0.65 | 5.7 × 10-25 | 1.9 × 10-20 | -0.35 | 2.8 × 10-3 | 8.9 × 10-3 | 2.7 × 10-2 |
| cg00676801 | STAT1 | -0.55 | 7.0 × 10-25 | 2.2 × 10-20 | -1.3 | 8.2 × 10-8 | 1.7 × 10-6 | 8.5 × 10-4 |
| cg12911952 | SLC22A18AS | -0.39 | 1.2 × 10-24 | 3.8 × 10-20 | -1.1 | 2.7 × 10-7 | 4.3 × 10-6 | 9.4 × 10-5 |
| cg05141234 | HLA-DMB | 0.57 | 1.8 × 10-24 | 5.3 × 10-20 | 0.75 | 3.5 × 10-7 | 5.3 × 10-6 | 2.9 × 10-4 |
| cg00945209 | TMC8 | -0.54 | 2.2 × 10-24 | 6.5 × 10-20 | -1.1 | 1.8 × 10-9 | 8.2 × 10-8 | 1.7 × 10-2 |
| cg09878888 | LPXN | 0.24 | 3.2 × 10-24 | 9.1 × 10-20 | 0.35 | 6.7 × 10-3 | 1.8 × 10-2 | 1.5 × 10-4 |
| cg00676801 | STAT4 | -0.33 | 3.4 × 10-24 | 9.7 × 10-20 | -0.57 | 2.0 × 10-8 | 5.5 × 10-7 | 8.5 × 10-4 |
| cg23387401 | ALOX15 | -0.37 | 9.7 × 10-24 | 2.6 × 10-19 | -0.64 | 6.2 × 10-8 | 1.4 × 10-6 | 1.0 × 10-9 |
| cg13443575 | CCL5 | 0.54 | 1.3 × 10-23 | 3.5 × 10-19 | 0.77 | 7.8 × 10-6 | 6.6 × 10-5 | 5.5 × 10-3 |
Only eQTM probes that are associated with atopic asthma in EVA-PR cohort (FDR-P < .05) are presented. Replication defined as FDR P < .05 with effect in the same direction as in EVA-PR. Only one gene per methylation probe presented.
Discussion
To date, there have been much fewer eQTM studies than eQTL studies,31 despite probable large joint causal effects of DNA methylation and gene expression on complex diseases. Although genotype does not change as a disease progresses, both epigenetic regulation and transcriptomic activity change as a disease develops or worsens. Thus, studying eQTM may complement findings from genetic or eQTL studies and add novel insights into disease pathogenesis.
Most previous genome-wide eQTM studies have been limited to healthy subjects.32,33 In the few instances in which both subjects with asthma and healthy control subjects were included, only CpGs that were significant in an EWAS—and only genes nearby those CpGs (eg, within 10 kb)—were examined.2,34 In contrast, we assessed all genome-wide CpGs along with expression of cis-genes located within 1 Mb in the current analysis of children and adolescents with and without asthma. Moreover, we were able to replicate nearly half of our significant findings in an independent cohort of predominantly African American children.
Notably, in our analysis, most significant eQTM methylation probes were not nearby their target cis-genes, a finding that may be explained by physical contact between CpG sites and promoter/coding regions of distant target genes through looping chromatin structures.35 Significant eQTM probes were also more likely to be localized in enhancer regions of their target genes in lung tissue than control probes, suggesting that CpG sites can affect transcription of non-nearby (distant) cis-genes through enhancer activity. We also found that although most methylation probes near TSS were negatively correlated with gene expression, more distant pairs tended to be positively correlated. Consistent with our findings, methylation in promoter regions and the first intron have been negatively correlated with gene expression,36 whereas methylation of more distant CpG sites and gene expression has been positively correlated with gene expression in several types of cancer.37 Moreover, the previous study of cancers showed that distal CpGs that are negatively associated with gene expression are enriched in enhancer regions, whereas those that are positively associated with gene expression are enriched in repressor regions.38 Although our findings suggest that distal methylation-gene expression pairs may be enhancers in instances of negative correlation and repressors/insulators in instances of positive correlation, this needs to be confirmed in analyses of other epigenetic (eg, histone modification) and experimental data.
We show an overrepresentation of the top eQTM methylation probes among CpGs associated with atopic asthma. Similarly, we report an overrepresentation of the top eQTM genes among DEGs in atopic asthma. Moreover, we show that most associations between eQTM methylation probes and atopic asthma are mediated by gene expression. Through the mediation analysis, we found multiple examples in which methylation in transcription regulatory elements such as promoter or enhancer regions may affect atopic asthma by regulating gene expression.
In a secondary eQTM analysis conducted separately in 158 subjects with atopic asthma and 100 nonatopic control subjects without asthma, we identified some associations that were present in cases but not in control subjects (e-Appendix 1 and e-Fig 1). In this stratified analysis, methylation probes and genes identified in the eQTM analysis of subjects with atopic asthma were more likely to be associated with atopic asthma than those identified in the analysis of control subjects. These findings must be cautiously interpreted and could be due to differences in sample size (and thus statistical power) between subgroups. Alternatively, they may represent true differences in epigenetic regulation of gene expression by disease status (eg, new enhancers or super-enhancers have been shown to occur in subjects who develop cancer38).
We recognize several study limitations. First, we only included subjects in a high-risk population (Puerto Rican subjects). However, we have previously replicated findings from GWAS10 and EWAS12 of asthma in Puerto Rican subjects in other racial or ethnic groups, including non-Hispanic whites, African Americans, and members of other Hispanic subgroups. Moreover, approximately half of the significant eQTM pairs in the current analysis in Puerto Rican subjects were significant in African Americans, despite the small sample size of the replication cohort. Second, we cannot confirm causal relationships in this cross-sectional study, in which asthma could have led to methylation changes or vice versa. Third, enhancers are tissue-specific and disease-specific, and thus the enhancer regions that we identified in a database for lung tissue in healthy subjects may differ from those in nasal (airway) epithelium from subjects with and without asthma. Moreover, the database for lung tissue was created based on computational predictions using high-throughput data (eg, H3K4me1/H3K27ac) and not on functional work such as genome editing.
In summary, we identified significant methylation-expression pairs in an eQTM analysis of nasal airway epithelium of subjects with and without asthma. Most methylation probes were associated with expression of distant cis-genes, and eQTM genes were enriched in immune regulation and epithelial integrity. Moreover, eQTM methylation probes and eQTM genes were overrepresented among those associated with atopic asthma, further suggesting a key role of epigenetic regulation of gene expression in airway epithelium in disease pathogenesis.
Acknowledgments
Author contributions: W. C. and J. C. C. conceived and designed the study. S. K. conducted the primary data analysis. E. F., R. Z., H. P., Z. X., Q. Y., N. B., E. A.-P., and G. C. participated in data collection and data analysis. S. K., E. F., W. C., and J. C. C. prepared the first draft of the manuscript. All authors reviewed the draft for intellectual content and approved submission of the final version of the manuscript. Dr. Celedón takes responsibility for the integrity of this work, from inception to published manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: J. C. C. has received research materials from Merck and GSK (inhaled steroids) and Pharmavite (vitamin D and placebo capsules), to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work. None declared (S. K., E. F., R. Z., H. J. P., Z. X., Q. Y., N. B., E. A.-P., G. C., W. C.).
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Additional information: The e-Appendix and e-Figure can be found in the Supplemental Materials section of the online article.
Footnotes
Drs Chen and Celedón contributed equally to this manuscript.
FUNDING/SUPPORT: This work was supported by grants HL079966, HL117191, MD011764 (to J. C. C.), and U54 MD007587 (to the University of Puerto Rico) from the U.S. National Institutes of Health (NIH). Dr Kim is supported by T32 training grant HL129949 from the U.S. NIH. E. F.’s contribution was supported by NIH grant HL125666. Q. Y.’s contribution was supported by NIH grant K01 HL138098.
Supplementary Data
References
- 1.Brugha R., Lowe R., Henderson A.J. DNA methylation profiles between airway epithelium and proxy tissues in children. Acta Paediatr. 2017;106(12):2011–2016. doi: 10.1111/apa.14027. [DOI] [PubMed] [Google Scholar]
- 2.Forno E., Wang T., Qi C. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: a genome-wide study. Lancet Respir Med. 2019;7(4):336–346. doi: 10.1016/S2213-2600(18)30466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S., Forno E., Yan Q. SNPs identified by GWAS affect asthma risk through DNA methylation and expression of cis-genes in airway epithelium. Eur Respir J. 2020;55(4) doi: 10.1183/13993003.02079-2019. 1902079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strunz T., Grassmann F., Gayán J. A mega-analysis of expression quantitative trait loci (eQTL) provides insight into the regulatory architecture of gene expression variation in liver. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-24219-z. 5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goh W.W.B., Wang W., Wong L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 2017;35(6):498–507. doi: 10.1016/j.tibtech.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Nygaard V., Rødland E.A., Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2015;17(1):29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron R.M., Kenny D.A. The moderator–mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51(6):1173. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 10.Sobel M.E. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol Methodol. 1982;13:290–312. [Google Scholar]
- 11.Liu S., Gao T., Qian J., He B., Tan K., Zhu H. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics. 2016;32(23):3543–3551. doi: 10.1093/bioinformatics/btw495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan Q.L., Du R., Lasky-Su J. A polymorphism in the thyroid hormone receptor gene is associated with bronchodilator response in asthmatics. Pharmacogenomics. 2013;13(2):130–136. doi: 10.1038/tpj.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rastogi D., Suzuki M., Greally J.M. Differential epigenome-wide DNA methylation patterns in childhood obesity-associated asthma. Sci Rep. 2013;3 doi: 10.1038/srep02164. 2164-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le N.P.K., Channabasappa S., Hossain M., Liu L., Singh B. Leukocyte-specific protein 1 regulates neutrophil recruitment in acute lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2015;309(9):L995–L1008. doi: 10.1152/ajplung.00068.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demenais F., Margaritte-Jeannin P., Barnes K.C. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nature Genet. 2018;50(1):42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Movahedi M., Moin M., Gharagozlou M. Association of HLA class II alleles with childhood asthma and Total IgE levels. Iran J Allergy Asthma Immunol. 2008;7(4):215–220. [PubMed] [Google Scholar]
- 17.Kim N., Hong Y., Kwon D., Yoon S. Somatic mutaome profile in human cancer tissues. Genomics Inform. 2013;11(4):239–244. doi: 10.5808/GI.2013.11.4.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyss A.B., Sofer T., Lee M.K. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nature Commun. 2018;9(1):2976. doi: 10.1038/s41467-018-05369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingender E., Dietze P., Karas H., Knüppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24(1):238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kel A.E., Gößling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31(13):3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S.-W., Verhaeghe C., Nguyenvu L.T. Distinct roles of FOXA2 and FOXA3 in allergic airway disease and asthma. Am J Respir Crit Care Med. 2009;180(7):603–610. doi: 10.1164/rccm.200811-1768OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucherat O., Chakir J., Jeannotte L. The loss of Hoxa5 function promotes Notch-dependent goblet cell metaplasia in lung airways. Biol Open. 2012;1(7):677–691. doi: 10.1242/bio.20121701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mougey E., Lang J.E., Allayee H. ALOX5 Polymorphism associates with increased leukotriene production and reduced lung function and asthma control in children with poorly controlled asthma. Clin Exp Allergy. 2013;43(5):512–520. doi: 10.1111/cea.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang P., Shi H-y, Wu X-m. Targeted inhibition of GATA-6 attenuates airway inflammation and remodeling by regulating caveolin-1 through TLR2/MyD88/NF-κB in murine model of asthma. Mol Immunol. 2016;75:144–150. doi: 10.1016/j.molimm.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Sousa A.R., Lane S.J., Cidlowski J.A., Staynov D.Z., Lee T.H. Glucocorticoid resistance in asthma is associated with elevated in vivo expression of the glucocorticoid receptor β-isoform. J Allergy Clin Immunol. 2000;105(5):943–950. doi: 10.1067/mai.2000.106486. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-01261-5. 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GWAS Catalog https://www.ebi.ac.uk/gwas/ Accessed August 1 2019.
- 28.Forno E., Zhang R., Jiang Y. Transcriptome-wide and differential expression network analyses of childhood asthma in nasal epithelium. J Allergy Clin Immunol. 2020;146(3):671–675. doi: 10.1016/j.jaci.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.http://promoter.bx.psu.edu/hi-c/index.html Accessed March 1, 2020.
- 30.Yang I.V., Pedersen B.S., Liu A.H. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017;139(5):1478–1488. doi: 10.1016/j.jaci.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S., Park H.J., Cui X., Zhi D. Collective effects of long-range DNA methylations predict gene expressions and estimate phenotypes in cancer. Sci Rep. 2020;10(1):3920. doi: 10.1038/s41598-020-60845-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez-Arcelus M., Lappalainen T., Montgomery S.B. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2 doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutierrez-Arcelus M., Ongen H., Lappalainen T. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 2015;11(1) doi: 10.1371/journal.pgen.1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reese S.E., Xu C.-J., den Dekker H.T. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. 2019;143(6):2062–2074. doi: 10.1016/j.jaci.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mora A., Sandve G.K., Gabrielsen O.S., Eskeland R. In the loop: promoter-enhancer interactions and bioinformatics. Brief Bioinform. 2016;17(6):980–995. doi: 10.1093/bib/bbv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anastasiadi D., Esteve-Codina A., Piferrer F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin. 2018;11(1) doi: 10.1186/s13072-018-0205-1. 37-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong Y., Sun J., Wong C.F. MICMIC: identification of DNA methylation of distal regulatory regions with causal effects on tumorigenesis. Genome Biol. 2018;19(1):73. doi: 10.1186/s13059-018-1442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sur I., Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16(8):483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.