Summary
NCKAP1/NAP1 regulates neuronal cytoskeletal dynamics and is essential for neuronal differentiation in the developing brain. Deleterious variants in NCKAP1 have been identified in individuals with autism spectrum disorder (ASD) and intellectual disability; however, its clinical significance remains unclear. To determine its significance, we assemble genotype and phenotype data for 21 affected individuals from 20 unrelated families with predicted deleterious variants in NCKAP1. This includes 16 individuals with de novo (n = 8), transmitted (n = 6), or inheritance unknown (n = 2) truncating variants, two individuals with structural variants, and three with potentially disruptive de novo missense variants. We report a de novo and ultra-rare deleterious variant burden of NCKAP1 in individuals with neurodevelopmental disorders which needs further replication. ASD or autistic features, language and motor delay, and variable expression of intellectual or learning disability are common clinical features. Among inherited cases, there is evidence of deleterious variants segregating with neuropsychiatric disorders. Based on available human brain transcriptomic data, we show that NCKAP1 is broadly and highly expressed in both prenatal and postnatal periods and demostrate enriched expression in excitatory neurons and radial glias but depleted expression in inhibitory neurons. Mouse in utero electroporation experiments reveal that Nckap1 loss of function promotes neuronal migration during early cortical development. Combined, these data support a role for disruptive NCKAP1 variants in neurodevelopmental delay/autism, possibly by interfering with neuronal migration early in cortical development.
Keywords: NCKAP1, autism spectrum disorder, disruptive variant, de novo variants, genotype-phenotype correlation, neurodevelopmental disorder
Introduction
Recent large-scale genome-wide sequencing studies have implicated many high-impact autism spectrum disorder (ASD) (MIM: 209850) candidate genes.1, 2, 3 However, due to the rarity of variants and the limited phenotypic data, the clinical significance and genotype-phenotype relationships for most candidate genes still remain to be determined. ASD shows a wide range of overlapping clinical features with other neurodevelopmental disorders (NDDs), such as intellectual disability (ID), language and motor developmental delay, and different psychiatric symptoms.4 As a result, it is both critical and challenging to define the particular genetic subtypes, especially among the most penetrant variants. International collaboration is key and has been pivotal in defining and establishing some recently described ASD/NDD genes, such as CHD8 (MIM: 610528),5 ADNP (MIM: 611386),6 DYRK1A (MIM: 600855),7 POGZ (MIM: 614787),8 and TANC2 (MIM: 615047).9 Although disruptive variants in any given gene explain only an extremely small proportion of individuals, defining the specific genetic subtypes not only increases the diagnostic yield of genetically defined cases but has the potential to optimize future clinical management.
We previously prioritized 58 ASD candidate genes based on gene constraint and FMRP/RBFOX binding targets9 for further consideration, including NCKAP1 (MIM: 604891). Knockout of Nckap1 in mice leads to embryonic lethal neural tube defects. Premature expression of Nckap1 retards the migration of neurons in the neocortex.10 Three de novo likely gene-disruptive (LGD) variants were originally reported in ASD probands from the Simons Simplex Collection (SSC) cohort (n = 2)2 and the Autism Clinical and Genetics Resources in China (ACGC) cohort (n = 1),11 with limited clinical information. In addition, a transmitted stop-gain variant was recently identified in a family with a multigenerational ID.12 Although previous findings implicate NCKAP1 as an ASD/ID risk gene,13 its clinical significance is undetermined and the genotype-phenotype relationships have never been demonstrated, leaving the pathogenicity of NCKAP1 variants unclear.
To address these issues, we focused on genotype-phenotype assessment of disruptive variants in NCKAP1. Based on our analysis of 21 individuals from 20 families with inherited and de novo deleterious NCKAP1 variants from a large-scale international consortium, we report a new NCKAP1-related NDD as associated with ASD.
Material and Methods
Clinical Assessment and Molecular Genetics Methods for Affected Families
The affected individuals haboring NCKAP1 variants and their family members where available were recruited to different collaborating institutes from seven countries. For each affected individual, detailed clinical information was obtained through recontact or detailed review of medical records by neurologists, psychiatrists, pediatricians, geneticists, or genetic counselors. Genomic DNA was extracted from the whole blood of the affected individuals. Family members of the probands where available were also recruited for segregation analysis and phenotyping. We collected and reviewed detailed clinical data from 21 affected individuals from 20 families. Written informed consent was obtained from study participants or their parents or legal guardians, in line with local institutional review board (IRB) requirements at the time of collection. The IRB of the Central South University approved this study. All procedures were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national). In this cohort, NCKAP1 variants were detected by different methods. In short, 16 affected individuals/families underwent exome sequencing in either a clinical or research setting, two families underwent massively parallel targeted sequencing, one family (with microdeletion) underwent array comparative genomic hybridization, and one family (with chromosome inversion) underwent karyotyping coupled with mate-pair sequencing. Detailed methods for all families are documented in the Supplemental Subjects and Methods.
Minigene Assay for Putative Splicing Variants
We designed two genomic fragments containing exons and up to 300 bp of flanking 5′ and 3′ intronic sequences from genomic DNA. The genomic fragments connect with each other using oligonucleotides containing restriction enzyme sites at their 5′ and 3′ ends. PCRs were performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) according to the manufacturer’s protocols, with primers carrying restriction sites. Inserts were cloned into the splicing vector pcDNA3.1myc-His(-)B, with the addition of T4 DNA Ligase (Thermo Scientific). The mutant plasmids were constructed by overlap-extension PCR with site-specific mutant primers. Wild-type and variant constructs were verified by Sanger sequencing.
The HEK293 cells were cultured in DMEM containing 10% fetal bovine serum, 1% nonessential amino acids, 2 mM glutamine, and 1% penicillin/streptomycin stock solution at 37°C, 5% CO2, 95% humidity. Approximately 2 × 106 of HEK293 cells were grown to 90% confluency in 6-well plates. 4 μg plasmids (wild-type or variants) were transfected into HEK293 cells using Lipofectamine 2000 Reagent (Thermo Scientific) according to standard protocols. After 36 h of culturing, cellular RNA was extracted using the Trizol reagent (Invitrogen) according to the manufacturer’s protocols. Complementary DNA synthesis was carried out with 1 μg of RNA and the Revert Aid First Strand cDNA Synthesis Kit (Life Technologies) following the standard protocol. To evaluate splicing, PCR was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific), with specialized primers. The PCR products were separated in 2% agarose gel. Transcripts were verified by Sanger sequencing.
De Novo Enrichment and Burden Analysis for NCKAP1 LGD Variants
We introduced TADA to perform enrichment analysis for NCKAP1 de novo LGD variants. The fraction of ASD risk genes which was estimated by published researches14 is 1,000 (18,271 protein-coding genes in the genome) for the sake of performing TADA analysis of de novo variants. The prior parameters (gamma.mean.dn and beta.dn) for LGD variants were confirmed as Gamma (20, 1). In addition, the background mutation rate of total genes was obtained from previous studies.15 Through simulation, we learned the distribution of Bayes factor (BF) of genes and compared this to the observed BF of genes. p values were calculated based on 1 million simulations for robustness. p values were then corrected by Bonferroni method for 18,271 protein-coding genes in the genome.
Functional Analysis for NCKAP1 Variants
Plasmid Construction
The full-length human NCKAP1 ORF (GenBank: NM_205842, No. HO205842) (YingRui Gene) was inserted into p.CAGGS-IRES-green fluorescent protein (GFP) vector, which was added a HA exogenous tag (5′-YPYDVPDYA-3′) at C-terminal of NCKAP1. Meanwhile, NCKAP1 variants were constructed into the same vector p.CAGGS-IRES-GFP via designing special primers.
Cell Culture and Transfection
HEK293 cell lines were cultured with DMEM basic (1×) (GIBCO, C11995500BT), which had been added 10% fetal bovine serum (FBS, GIBCO, 12483020), and placed into 37°C and 5% CO2 incubator. We chose Lipofectamine 2000 Reagent (Invitrogen, 11668019) to transfect exogenous plasmids into cell lines. When the cells reached 70%–80% confluent, plasmid DNA-lipid complexes were prepared and added them to cells. For detailed operating steps and components, see lipofectamine 2000 Reagent protocol at Thermo Fisher Scientific Web.
Immunofluorescence
HEK293 cells were cultured on Microscope Cover Glass (Thermo Fisher Scientific, 12-545-82) in 24-well plates and transfected for 48 h. Cells were fixed by 4% PFA for 15 min, permeabilized by 0.1% Triton X-100 (1× PBS preparation) for 10 min, and blocked by 5% bovine serum albumin (1× PBS preparation) for 1 h. Then, glasses with cells were incubated with anti-HA (Cell Signaling Technology, 3724S) primary antibodies overnight at 4°C. Then, 1× PBS was used to wash glasses three times and stained Cy3 (Jackson ImmunoResearch, 115-165-003) secondary antibodies. 4,6-diamidino-2-phenylindole (DAPI) was used to label cell nuclei. The images were collected by TCS-SP5-II confocal microscope (Leica) and analyzed with ImageJ.
Analysis of NCKAP1 Expression Pattern in the Human Brain
RNA-seq data from BrainSpan was used to illustrate the dynamic expression of NCKAP1 across brain development. The age of brain samples ranged from 8 postconceptional weeks (PCW) to 40 years old. Normalized reads per kilobase million (RPKM) expression and sample meta-data were downloaded from BrainSpan. Univariate linear regression was used to analyze the relationship between NCKAP1 expressions and development periods. The linear regression analysis was conducted in brain regions separately.
To investigate the cell-type-specific expression pattern of NCKAP1 in the developing human cerebral cortex, we introduced a single-cell RNA-seq dataset from in a previous publication.16 Briefly, the RNA-seq data were generated from 48 individuals. Processed normalized expression values were downloaded from the UCSC cell browser. Assignments of the cluster for each cell were derived from the source study. The biological interpretation for each broad cell type was derived from the previous publication. Statistical significance of enrichment or depletion for each cell type was calculated using the Wilcoxon Rank Sum Test. Bonferroni correction method was used to perform multiple testing correction. All analysis were performed in R stat (v.3.2.3).
Mouse In Utero Electroporation
The mouse Nckap1 (GenBank: NM_016965) original shRNA reference sequences were obtained from MERCK (TRCN0000112255). We changed the restriction enzyme cutting sites (XbaI and BamHI) and inserted the sequences into FUGW-H1-GFP vector.
Plasmids were deliquated to 2 μg/μL and mixed with 0.01% Fast Green (Sigma-Aldrich). Each pregnant mouse received 20 μL plasmids. We injected plasmids into lateral ventricles of embryonic 13.5/14.5-day mouse brains. Electroporation was operated using an Electro Square Porator (ECM 830) and the relevant parameter is 50 ms square pulses with 1,000 ms suspensions at 30v. The injected mouse brains were collected on embryonic 17.5/16.5 day, fixed into 4% PFA for 24 h, and gradually dehydrated into 15% sucrose for 24 h and 30% sucrose, which were compounded by 1∗PSB for 24 h until precipitated at the bottom of centrifuge tubes. Whole brains were embedded with embedding medium (the ratio of O.C.T. compound and 20% sucrose is 2:1) and frozen at −80°C. Study protocols comply with all relevant ethical regulations and were approved by the IRB of Central South University.
For immunofluorescence experiments, frozen mouse brain tissue was cut to 20–30 μm thickness along coronal planes. The neurons, which were transfected with exogenous plasmids and expressed relevant proteins, were stained by Rabbit GFP first antibody (1:500) (Invitrogen, A11122) and corrective Alex 488 secondary antibody (1:250) and emitted green fluorescence under fluorescence microscopy.
Results
Recruitment of a Large Cohort of Individuals with NCKAP1 Variants
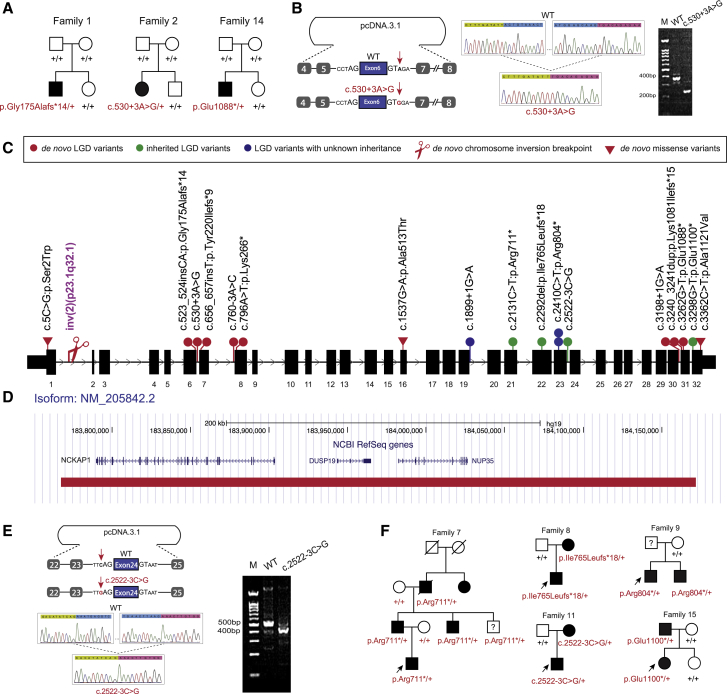
Two de novo LGD variants in NCKAP1 (GenBank: NM_205842.3) c.523_524insCA (p.Gly175Alafs∗14) and c.3262G>T (p.Glu1088∗) have been reported in the SSC cohort (Figure 1A). Through reanalysis of the SSC cohort genome-sequencing data, we identified a de novo intronic variant adjacent to the canonical splicing site (c.530+3A>G). By minigene assay, we demonstrated that this putative splicing variant affected normal splicing, leading to skipping of the 6th exon of NCKAP1 (Figure 1B). We then investigated de novo variants data from two other published large-scale ASD cohorts: the ASC cohort3 (n = 4,046) and the MSSNG cohort17 (n = 1,625); no additional de novo cases were identified in 5,671 trios, indicating the extreme rarity of NCKAP1 de novo variants in ASD.
Figure 1.
NCKAP1 Disruptive or De Novo Missense Variants and Phenotype Spectrum
(A) Three families with de novo disruptive (families 1 and 14) or intronic (family 2) variants in SSC cohort.
(B) Minigene assay shows that de novo intronic variant c.530+3A>G impairs normal splicing. Sequence highlighted by green above the Sanger trace is from exon 5. Sequence highlighted by pink is from exon 7.
(C) Distribution of NCKAP1 disruptive and de novo missense variants within the gene.
(D) A de novo deletion (red bar) removed three genes, including NCKAP1, and was identified in family 16.
(E) Minigene assay shows that the intronic variant c.2522−3C>G impairs normal splicing. Sequence highlighted by green above the Sanger trace is from exon 23. Sequence highlighted by pink is from exon 25.
(F) Pedigree plots of families with dominantly transmitted NCKAP1 disruptive variants and neuropsychiatric disorders.
Loss of Nckap1 in mice leads to neural tube and neuronal differentiation and migration defects in mouse developing brain.10 In the WAVE complex, NCKAP1 interacts directly with CYFIP2, which was recently reported to be associated with ID and autism.17, 18, 19 This interaction and the mouse knockout model warranted a more detailed search for additional individuals with potentially deleterious variants. Using GeneMatcher20 and an international network of collaborators, we recruited a total of 18 individuals with NCKAP1 putative disruptive variants (including the three individuals from the SSC cohort) (Figure 1C, Tables 1 and S1). The group consists of one individual with a de novo microdeletion involving NCKAP1 and another two adjacent genes, DUSP19 (MIM: 611437) and NUP35 (MIM: 608140) (Figure 1D); one individual with a de novo chromosome inversion for which one of the breakpoints occurred in the first intron of NCKAP1 (Figure 1C); 14 individuals with LGD variants (8 de novo, 4 inherited, 2 undetermined) (Figure 1C); and two individuals with intronic variants (c.760−3A>C and c.2522−3C>G) adjacent to splice sites (Table 1, Figure 1C). As we did for the initial splicing variant (c.530+3A>G), we applied a minigene assay to the additional two putative splicing variants. Variant c.760−3A>C is de novo and was identified in an individual with ID. However, we did not detect abnormal splicing (Figure S1), although we cannot exclude the possibility of a tissue-specific splicing effect of this variant in vivo or quantitative changes in gene expression. The pathogenic effect for this variant is still to be determined. The second putative splicing variant, c.2522−3C>G, was identified in a family where the variant was transmitted from the affected mother. Our minigene assay revealed that this variant leads to a skipping of the 24th exon of NCKAP1 (Figure 1E).
Table 1.
Summary of NCKAP1 Variants Identified in NDD-Affected Individuals
| Family Index | Cohort | Cohort Size | Methods | gDNA Change (chr2, hg19) | Function | NT Change | aa Change | Inheritance | gnomAD | Clinical Significance |
|---|---|---|---|---|---|---|---|---|---|---|
| LGD Variants | ||||||||||
| 1 | SSC | 2,508 | WES | g.183866861_183866862insTG | frameshift | c.523_524insCA | p.Gly175Alafs∗14 | de novo | 0 | P |
| 3 | GeneDx | 31,111 | WES | g.183860531_183860532insA | frameshift | c.656_657insT | p.Tyr220Ilefs∗9 | de novo | 0 | P |
| 5 | Lyon | 200 | WES | g.183859579T>A | stopgain | c.796A>T | p.Lys266∗ | de novo | 0 | P |
| 6 | GeneDx | 31,111 | WES | g.183826886C>T | splicing | c.1899+1G>A | – | unknown | 0 | LP |
| 7 | Parkville | 160 | WES | g.183821230G>A | stopgain | c.2131C>T | p.Arg711∗ | paternal | 0 | P |
| 8 | ASID | 10,927 | target | g.183817939del | frameshift | c.2292del | p.Ile765Leufs∗18 | maternal | 0 | LP |
| 9 | Poitiers | 224–350 | WES | g.183817632G>A | stopgain | c.2410C>T | p.Arg804∗ | not maternal | 0 | LP |
| 10 | Lausanne | – | WES | g.183817632G>A | stopgain | c.2410C>T | p.Arg804∗ | not maternal | 0 | LP |
| 12 | ACGC | 2,926 | target | g.183792844C>T | splicing | c.3198+1G>A | – | de novo | 0 | P |
| 13 | Lyon | 200 | WES | g.183791591_183791592dup | frameshift | c.3240_3241dup | p.Lys1081Ilefs∗15 | de novo | 0 | P |
| 14 | SSC | 2,508 | WES | g.183791570G>T | stopgain | c.3262G>T | p.Glu1088∗ | de novo | 0 | P |
| 15 | Riyadh | 105 | WES | g.183790537C>A | stopgain | c.3298G>T | p.Glu1100∗ | paternal | 0 | P |
| De Novo Intronic Variants | ||||||||||
| 2 | SSC | 2,508 | WES | g.183866852T>C | splicing | c.530+3A>G | – | de novo | 0 | P |
| 4 | Riyadh | 2,219 | WES | g.183859618T>G | splicing | c.760−3A>C | – | de novo | 0 | VUS |
| 11 | GeneDx | 31,111 | WES | g.183817233G>C | splicing | c.2522−3C>G | – | maternal | 0 | LP |
| Microdeletion | ||||||||||
| 16 | Odense | – | aCGH | g.183762482-184182761del | microdeletion | 240 kb deletion | – | de novo | 0 | P |
| Chromosome Inversion | ||||||||||
| 17 | Colorado | – | MPS | inv(2)(2pter®p23.1(30340928)::2q32.1(183896661)®p23.1(30340928)::2q32.1(183896662) ®2qter). | inversion | – | – | de novo | – | P |
| De Novo Missense Variants | ||||||||||
| 18 | GeneDx | 31,111 | WES | g.183902823G>C | missense | c.5C>G | p.Ser2Trp | de novo | 0 | VUS |
| 19 | GeneDx | 31,111 | WES | g.183832053C>T | missense | c.1537G>A | p.Ala513Thr | de novo | 1 | VUS |
| 20 | GeneDx | 31,111 | WES | g.183790473G>A | missense | c.3362C>T | p.Ala1121Val | de novo | 0 | VUS |
NCKAP1 isoform is GenBank: NM_205842.3. MPS, mate pair sequencing of inversion; P, pathogenic; LP, likely pathogenic; VUS, variants of uncertain significance.
NCKAP1 is a highly constrained gene, intolerant of variation; it is located in the top 3.9 percentile by residual variance intolerance score21 and has no reported truncating variants (the probability of being loss-of-function intolerant [pLI] = 1) in the gnomAD database.22 No LGD variant within NCKAP1, for example, has been observed in 114,704 gnomAD samples who were not ascertained for having a neurological condition in a neurological case/control study (non-neuro subset).23 Based on our larger international cohort, we assessed statistical significance of our genetic findings. We first assessed an enrichment of de novo LGD variants among probands compared with random occurrence by applying TADA.14 Six individuals with de novo NCKAP1 LGD variants were detected from four cohorts (SSC: c.523_524insCA [p.Gly175Alafs∗14], c.3262G>T [p.Glu1088∗]; GeneDx: c.656_657insT [p.Tyr220Ilefs∗9]; Lyon: c.796A>T [p.Lys266∗], c.3240_3241dup [p.Lys1081Ilefs∗15]; ACGC: c.3198+1G>A; Table 1) where a total of 36,745 individuals with NDD were tested. After combining three published ASD/NDD cohorts (ASC cohort, n = 4,046; MSSNG cohort, n = 1,625; and ASID cohort,24 n = 10,927) with no NCKAP1 de novo LGD variants identified, the TADA-denovo model reveals an exome-wide significant enrichment (p = 5.0 × 10−7, padj = 0.0091, Bonferroni correction).
Second, since almost half of the LGD variants were transmitted, we investigated whether there is a significant burden of NCKAP1 LGD variants regardless of the inheritance status by performing a Fisher’s exact test using non-neuro gnomAD subset samples as controls. Eleven individuals with NCKAP1 LGD variants (SSC: c.523_524insCA [p.Gly175Alafs∗14], c.3262G>T [p.Glu1088∗]; GeneDx: c.656_657insT [p.Tyr220Ilefs∗9], c.1899+1G>A; Parkville: c.2131C>T [p.Arg711∗]; ASID: c.2292del [p.Ile765Leufs∗18]; Poitiers: c.2410C>T [p.Arg804∗]; Lyon: c.796A>T [p.Lys266∗], c.3240_3241dup [p.Lys1081Ilefs∗15]; ACGC: c.3198+1G>A; Riyadh: c.3298G>T [p.Glu1100∗]; Table 1) were detected from eight cohorts where both de novo and inherited data are available. We observe a genome-wide significant burden of NCKAP1 LGD variants in our international NDD cohorts (11 probands in 48,206 NDD individuals versus 0 individuals in 114,704 gnomAD non-neuro samples) (p = 1.52 × 10−6, padj = 0.03, Bonferroni correction for ∼20,000 genes). Although this difference is unlikely the result of low sequence coverage considering the sequence coverage of NCKAP1 exons in gnomAD is more than 30-fold (Figure S2), the ascertainment bias in both case and control groups and the difference in terms of capture kits, sequencing methods, and variant calling pipelines between case and control subjects may also introduce inaccurate result to this analysis. Combined, these data argue for a de novo and ultra-rare deleterious variant burden of NCKAP1 in individuals with more broadly defined NDD but not yet ASD.
In addition to the high intolerance to truncating variants, NCKAP1 is also predicted to be intolerant to missense variants (missense Z-score22 = 4.27). Besides disruptive variants, we also identified three individuals with de novo missense variants (c.5C>G [p.Ser2Trp], c.1537G>A [p.Ala513Thr], c.3362C>T [p.Ala1121Val]) (Figure 1C, Tables 1 and S1). These variants are not observed among gnomAD samples. The first missense variant, c.5C>G (p.Ser2Trp), maps to the second amino acid position of the NCKAP1 protein and is predicted as damaging (SIFT,25 PolyPhen2,26 MutationTaster27) with an overall Combined Annotation Dependent Depletion (CADD) score28 of 26. The second missense variant, c.1537G>A (p.Ala513Thr), is predicted as damaging by MutationTaster, but not by SIFT or PolyPhen2. CADD score for this variant is 19. The third, c.3362C>T (p.Ala1121Val), is predicted as damaging by all three programs and has a CADD score of 31.
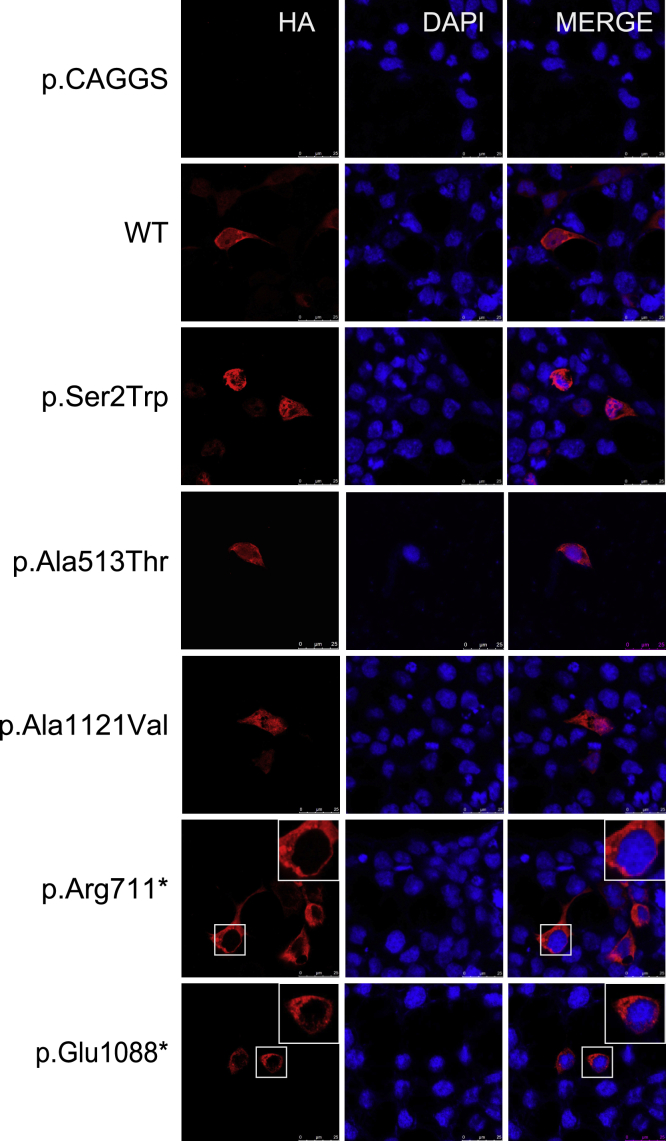
To determine the functional effects of NCKAP1 LGD and missense variants identified in our NDD cohort, we constructed the plasmids encoding HA-tagged wild-type (WT) NCKAP1, two selected LGD variants (c.2131C>T [p.Arg711∗] and c.3262G>T [p.Glu1088∗]), and three missense variants, and we expressed them into HEK293 cells. Immunoinfluence reveals that WT NCKAP1 is located at both nucleus and cytoplasm. However, we observed that both LGD variants show abnormal localization. Variants are mainly located at cytoplasm but not nucleus, indicating that the LGD variants affect nuclear entry (Figure 2). However, we did not observe this abnormal localization for the missense variants.
Figure 2.
Subcellular Localization of NCKAP1 Wild-Type and Variants Proteins in HEK293 Cells
Two LGD variants (c.2131C>T [p.Arg711∗] and c.3262G>T [p.Glu1088∗]) and three missense variants (c.5C>G [p.Ser2Trp], c.1537G>A [p.Ala513Thr], c.3362C>T [p.Ala1121Val]) were analyzed. HA-tagged NCKAP1 is shown in red and nuclei were stained by 4,6-diamidino-2-phenylindole in blue. The merged images show the subcellular localization of NCKAP1 in HEK293. Scale bar = 25 μm.
Given that our current limited functional analysis did not identity abnormal localization and that there are other alternative genetic explanations for two of the three individuals with the missense variants (a likely pathogenic variant within SCN5A [MIM: 600163] from family 18; a 375.67 kb 7q21.3 duplication and a NLGN4X [MIM: 300427] variant from family 20, Table S1), we conservatively classified the clinical significance for all three missense variants as variants of uncertain significance (VUS) (Table 1) until more genetic and/or functional data becomes available, although two of them (c.5C>G [p.Ser2Trp], c.3362C>T [p.Ala1121Val]) could be classified as likely pathogenic following the standards and guidelines for the interpretation of sequence variants from the American College of Medical Genetics and Genomics (ACMG)29 (Table S1). The putative splicing variant (c.760−3A>C), for which abnormal splicing was not detected, was also conservatively classified as VUS (Table 1). All LGD variants, structural variants, and the minigene assay validated putative splicing variants (c.530+3A>G, c.2522−3C>G) were classified as pathogenic or likely pathogenic following the ACMG guideline29 (Tables 1 and S1).
Inherited NCKAP1 Disruptive Variants in Families with a Dominant Inherited Model
In addition to de novo NCKAP1 variants, we also identified seven families with inherited disruptive variants (n = 5) or disruptive variants of unknown inheritance (n = 2). Interestingly, six of the seven families have a remarkable family history of neurodevelopmental or neuropsychiatric disease consistent with autosomal-dominant inheritance (Figure 1F, Table S1). In family 7, for example, the c.2131C>T (p.Arg711∗) variant is inherited from the father who was reported to have depression. The carrier paternal uncle has mild ID and cleft palate. The second paternal uncle also carries the variant; unfortunately, detailed clinical information is unavailable for this family member. The paternal grandfather who is deceased was reported to have had a severe psychiatric episode in adulthood, while the paternal great aunt (not tested) was diagnosed with schizophrenia. We infer that the paternal grandfather carried the variant, based on the negative results for the paternal grandmother and the presence of the variant in three of their offspring. In family 8, the c.2292del (p.Ile765Leufs∗18) variant is transmitted from an affected mother who has severe anxiety disorder and had two severe psychotic episodes around the age of 50 years. In family 9, there are two affected siblings—both have the c.2410C>T (p.Arg804∗) variant. The proband has mild ID and behavioral problems, and the brother has similar but more severe symptoms. The detection of the NCKAP1 variant in the brother was performed by Sanger sequencing; although fragile X testing, Angelman methylation analysis, and array-CGH testing were applied (Supplemental Subjects and Methods), exome sequencing was not performed, so we could not exclude additional pathogenic SNV/indel in this brother. The mother did not harbor the variant. Although the clinical information for the father could not be obtained, he was reported to be very aggressive. Behavioral problems and intellectual disability have been reported for the proband of family 10 bearing the same LGD variant discovered through exome sequencing. In family 11, the splicing variant c.2522−3C>G was inherited from the mother who has ID and a history of hyperactivity as a child and seizures (medically controlled). In family 15, the c.3298G>T (p.Glu1100∗) variant is inherited from an affected father who has ID. This family has been reported in a study with strong family history of DD (father, maternal grandmother, three paternal aunts, one paternal uncle, and two paternal cousins), albeit with limited detailed clinical information.12
Disruption of NCKAP1 Defines a NDD Subtype with Core Symptoms of ASD
Through recontact and review of the clinical information for all available affected individuals/family members from the collaborating centers, we assembled detailed phenotypic data for 17 probands (12 males and 5 females) carrying NCKAP1 pathogenic or likely pathogenic variants (Table S1). The age distribution is from 7 to 23 years old averaging 13.8 years (12 children and 5 adults). The core features are neurodevelopmental phenotypes, including ASD or autistic features, speech-language problems, childhood motor delay, and ID or learning disabilities. The most consistent neurobehavior phenotype is ASD or autistic behaviors, such as social interaction problems (11/15, 73.3%) (Table 2). Of the 15 individuals assessed for ASD, 10 individuals were formally diagnosed with ASD, two were reported to have autistic behaviors, and three individuals were reported to have no autistic phenotypes. Of the 17 individuals with language development records, 12 (73.7%) have a history of speech delay or language problems, and at least three of them exhibited absent language or have more severe language problems. Eleven out of 15 individuals (73.3%) exhibited childhood motor delay. Unlike many high-impact autism genes with which the autism diagnosis is concurrent with severe ID, only 62.5% (10/16) of individuals have diagnoses of mild to severe ID. Two individuals (2/16) were reported to have learning disabilities, and the remaining four individuals for whom this information is available were reported to have borderline to average intellectual level.
Table 2.
Summary of Genotype and Phenotype Information for NDD-Affected Individuals with NCKAP1 Pathogenic Variants
| Case Index | 1 | 2 | 3 | 5 | 6 | 7 | 8 | 9-1 | 9-2 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | Summary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation inheritance | DN | DN | DN | DN | UD | PI | MI | MI | MI | UD | MI | DN | DN | DN | PI | DN | DN | |
| Age at last examination (y) | 15 | 16 | 10 | 10 | 7 | 23 | 20 | 22 | 21 | 11 | 9 | 8 | 7 | 22 | 9 | 15 | 9 | |
| Gender | M | M | M | M | M | M | F | M | M | M | F | M | M | F | F | F | M | |
| Neurodevelopmental Problems | ||||||||||||||||||
| ASD/autistic behaviora | + | + | + | − | + | + | + | − | ND | − | + | + | + | + | ND | ± | − | 11/15 |
| Speech-language problems | − | − | + | + | + | + | + | + | + | + | + | − | + | − | + | − | + | 12/17 |
| Motor delay | − | − | − | + | + | + | + | − | ND | + | + | + | + | ND | + | + | + | 11/15 |
| ID/learning disabilityb | − | − | ND | + | + | ± | − | + | + | + | + | + | ND | − | + | ± | + | 11/15 |
| Behavior or Psychiatric Problems | ||||||||||||||||||
| Repetitive behavior | + | + | ND | + | + | + | + | − | ND | − | + | + | + | ND | ND | − | − | 9/13 |
| Aggressive behavior | + | − | ND | + | + | − | − | + | + | + | + | − | − | ND | ND | − | + | 8/14 |
| ADHD | ND | + | + | + | ND | − | − | ND | ND | − | + | + | − | + | + | − | − | 7/13 |
| Anxiety | ND | ND | ND | − | ND | + | − | + | + | − | ND | − | − | ND | ND | − | + | 4/10 |
| Depression | + | + | − | − | ND | − | − | − | ND | − | − | ND | − | + | ND | − | − | 3/13 |
| Obsessive behavior | ND | ND | ND | + | + | − | − | − | ND | − | + | − | − | ND | ND | − | − | 3/11 |
| Self-injury behavior | ND | − | − | − | − | − | − | − | + | − | − | ND | − | ND | ND | − | − | 1/13 |
| Neurological Problems | ||||||||||||||||||
| Sleep disturbances | + | + | ND | + | − | − | − | − | ND | + | + | − | + | + | + | − | + | 9/15 |
| Seizure | + | − | − | − | + | + | − | + | − | − | + | − | − | − | + | + | − | 7/17 |
| Epilepsy | ND | − | ND | − | + | + | − | − | − | − | ND | − | − | − | ND | ND | − | 2/12 |
| Brain MRI | − | − | − | − | − | + | − | − | ND | − | + | − | − | − | − | − | − | 2/16 |
| Brain EEG | − | − | − | − | − | + | − | − | ND | − | − | − | − | − | − | − | ND | 1/15 |
| Macrocephaly | − | − | − | − | − | + | − | − | − | − | ND | − | − | − | + | ND | − | 2/15 |
| Microcephaly | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | ND | − | 1/16 |
| Systemic Problems | ||||||||||||||||||
| Tall | − | − | − | − | + | + | − | + | + | − | + | ND | + | − | − | − | − | 6/16 |
| Overweightc | − | − | − | − | − | ++ | − | + | − | ++ | ++ | ND | − | − | − | − | − | 4/16 |
| Gastrointestinal disturbance | − | − | + | − | − | + | − | − | − | − | − | + | + | − | ND | − | − | 4/16 |
| Skeletal alterations | ND | ND | + | − | − | + | − | + | − | − | − | ND | + | ND | ND | − | − | 4/12 |
| Visual impairment | ND | ND | − | + | − | + | − | − | − | − | − | ND | − | ND | ND | − | + | 3/12 |
| Hearing impairment | − | − | − | − | − | + | − | − | − | + | − | ND | − | − | ND | − | − | 2/15 |
| Congenital heart defects | − | − | − | − | − | − | − | − | − | − | − | ND | − | − | ND | − | + | 1/15 |
Symbols and abbreviations: DN, de novo; MI, maternal inheritance; PI, paternal inheritance; UD, undetermined; +, present; −, absent; ND, no data or not reported.
Symbols: +, ASD diagnosis; ±, autistic features
Symbols: +, ID; ±, learning disability
Symbols: +, overweight; ++, obesity
We note a burden of neuropsychiatric behaviors in the NCKAP1-related disorders (Table 2). Repetitive behaviors, aggressive behaviors, and attention-deficit/hyperactivity disorder (ADHD) are observed in more than half of this cohort. Repetitive behaviors are observed in 9 out of 13 individuals (69.2%), aggressive behavior in 8 out of 14 individuals (57%), and ADHD in 7 out of 13 individuals (53.8%). Anxiety, depression, and obsessive behaviors are present in three or more individuals. Notably, family members with NCKAP1 variants also have psychiatric or behavioral problems (Figure 1F), as discussed above.
The most prevalent neurological phenotypes are sleeping disturbance and seizure. Nine out of 15 individuals (60%) were reported to have sleeping disturbance or have sleeping disturbance history. Seven out of 17 individuals (41.2%) were reported to have seizure. In addition to neurodevelopmental and neuropsychiatric disturbances, we observe that around one third of the individuals are tall statured (37.5%, 6/16), overweight/obese (25%, 4/16), or report gastrointestinal disturbances (25%, 4/16) (Table 2). WAVE complex has been implicated in congenital heart defects.30 Interestingly, we observed cardiac phenotypes in two unrelated families. The proband in family 17 has bicuspid aortic valve, and the proband in family 19 has dilated cardiomyopathy; left ventricular systolic dysfunction (Table S1). To further investigate whether cardiac phenotypes are related to the variants in NCKAP1, we checked the published exome-sequencing data of congenital heart disease (CHD)31 and found one CHD-affected individual with a de novo stopgain variant (GenBank: NM_013436: c.3169G>T [p.Glu1057∗]) in NCKAP1.31 Besides CHD, that individual also presents a NDD phenotype although no detailed information was provided. Those data suggest that NCKAP1 variants might be relevant to cardiac phenotypes, albeit with a very low penetrance.
In addition to the NCKAP1 variants, other variants or environmental factors (Table S1) were identified that might contribute to the phenotypes in some individuals. As discussed above, there were additional candidate variants in individuals with missense NCKAP1 variants. Of the individuals with LGD NCKAP1 variants, the proband from family 4 carried a likely pathogenic variant within GCDH that might explain the generalized chorea phenotype; glutaric aciduria type 1 was suspected as a second diagnosis for this individual. A de novo inversion (46,XX,inv(2)(p23q37.3)) in the proband from family 11 might also contribute to this individual’s NDD phenotypes, possibly through disruption of genes at inversion breakpoints. In addition, the younger brother from family 9 was reported to be severely mistreated physically and psychologically during infancy. Environmental factors may therefore have impacted neurodevelopment in this case.
Spatio-temporal and Cell-Type-Specific Expression Patterns of NCKAP1 in Developing Brain
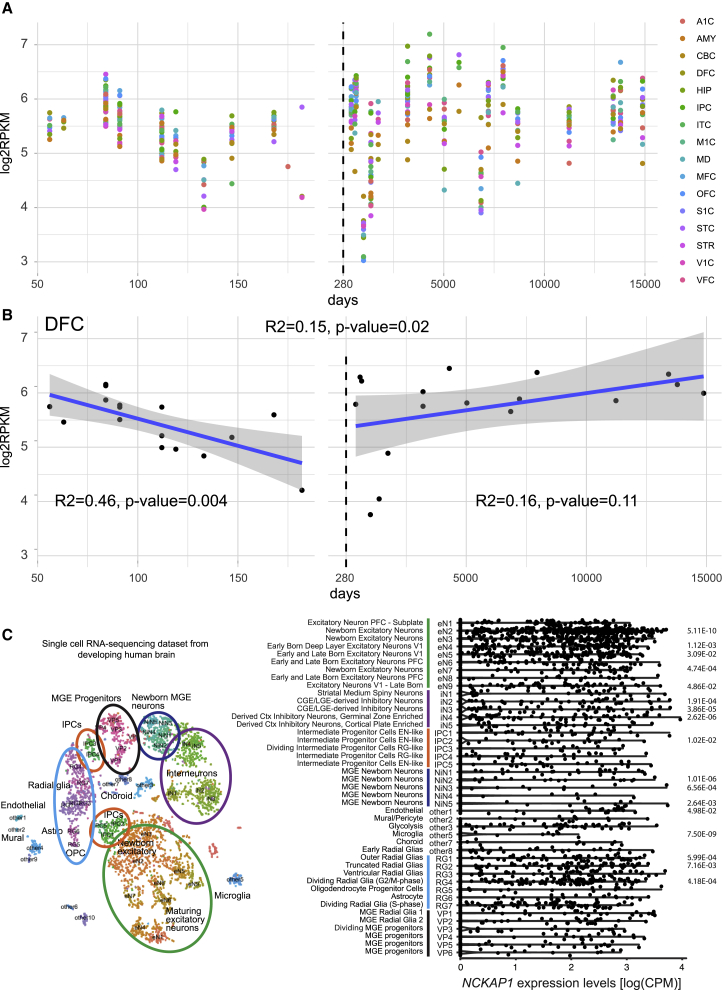
To investigate the spatio-temporal expression pattern of NCKAP1 in human fetal and postnatal brain, we analyzed the RNA sequencing data from BrainSpan (Subjects and Methods). We revealed that NCKAP1 is highly expressed throughout both fetal and postnatal brain regions (Figure 3A). To investigate whether there is a dynamic pattern of NCKAP1 throughout the development periods in specific brain regions, we performed a univariate linear regression analysis and revealed a significantly decreased expression during the prenatal period in the dorsolateral prefrontal cortex (DFC) (p = 0.004) (Figure 3B, Table S2), but no significant change in the DFC during the postnatal period (p = 0.11) (Figure 3B). These data indicate that NCKAP1 might be critical in the fetal development of DFC, which has been implicated in autism.32 We then investigated the cell-type-specific expression pattern of NCKAP1 in the developing human cerebral cortex by analyzing a single-cell RNA sequencing dataset from a recent publication.16 The transcriptomic profiles of 48 distinct clusters were broadly classified into six cell types, including excitatory neurons, inhibitory interneurons, medial ganglionic eminence (MGE) progenitors, MGE newborn neurons, intermediate progenitor cells, and radial glia (Figure 3C). Although NCKAP1 is broadly expressed across many cell types in the developing cerebral cortex, we observed enriched expression in radial glia and excitatory neurons, in particular newborn excitatory neurons (eN2) (Figure 3C, Table S3), but generally depleted expression in inhibitory neurons and MGE newborn neurons. These data indicate that NCKAP1 might have a cell-type-specific function in the development of neocortex.
Figure 3.
Expression Pattern of NCKAP1 in the Developing Human Brain
(A) Expression of NCKAP1 across development periods by brain regions. The x axis is the age of samples in days and y axis is the log2-transformed reads per kilobase million (RPKM) of NCKAP1. The dashed line shows the day of birth. A1C, primary auditory cortex; AMY, amygdaloid complex; CBC, cerebellar cortex; DFC, dorsolateral prefrontal cortex; HIP, hippocampus; IPC, inferior parietal cortex; ITC, inferolateral temporal cortex; M1C, primary motor cortex; MD, mediodorsal nucleus of thalamus; MFC, medial prefrontal cortex; OFC, orbital frontal cortex; S1C, primary somatosensory cortex; STC, superior temporal cortex; STR, striatum; V1C, primary visual cortex; VFC, ventrolateral prefrontal cortex.
(B) Expression of NCKAP1 in the DFC region. Univariate regression analysis was conducted in prenatal brains (right), postnatal brains (left), and all brains (top) separately. Blue line indicates the regression line and the gray region indicates 95% confidence interval. The text shows the R2 and p value of regression.
(C) Cell-type-specific expression of NCKAP1 in developing human brain. Left: The t-SNE plot shows the the major cell types in the developing human cerebral cortex. Cluster numbers and biological interpretations are derived from the source study. Right: Violin plot showing NCKAP1 expression across the cell types. Bonferroni adjusted p values calculated by Wilcoxon rank sum test are shown.
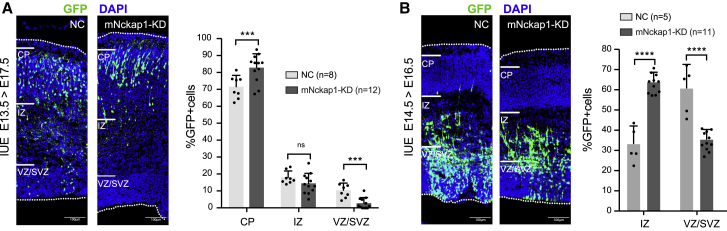
Disruption of Nckap1 Impairs Neuronal Migration in Developing Mouse Brain
The above transcriptome analysis shows that NCKAP1 is highly expressed in prenatal developing brain, and it is reported that premature expression of NCKAP1 in neocortex may inhibit neuronal migration.10 Since the pathogenic variants of NCKAP1 are loss of function, we investigated whether decreased expression of NCKAP1 affected the proliferation and radial migration of cortical neurons in prenatal developing brain. We introduced an shRNA vector that targets Nckap1 together with GFP into lateral ventricles of fetal mice at E15.5 by in utero electroporation. The distribution of GFP-labeled cells was examined at E17.5. Compared to wild-type (WT), we observed an increase of GFP-positive neurons appearing in the cortical plate, while fewer fluorescent cells were located within the intermediate and ventricular/subventricular zones (Figure 4A), suggesting that disruption of Nckap1 in the mouse may disrupt normal neuronal migration during embryonic cortical development. Reducing the interval time of harvesting (E14.5–E16.5), we observed the same effect, where more positive cells occurred in the intermediate zone in Nckap1-shRNA transferred brains compared to WT controls (Figure 4B). These results indicate that NCKAP1 loss of function may result in abnormal neuronal migration, consistent with the previous study that premature expression of Nckap1 retards neocortical neuronal migration10 although there was no significant evidence of structural abnormalities in those probands for whom neuroimaging studies were reported.
Figure 4.
Disruption of Nckap1 Promotes Neuronal Migration in Mouse Embryonic Cortical Development
(A) Wild-type mouse (B6) brains were electroporated at E13.5 with Nckap1-shRNA and collected at E17.5. More GFP-positive neurons appear in the cortical plate (CP), while fewer fluorescent cells are located at the intermediate zone (IZ) and ventricular/subventricular zones (VZ/SVZ) in Nckap1-shRNA transferred brains compared to WT controls.
(B) Reducing the interval time for harvesting (E14.5–E16.5), we find more positive cells at IZ in Nckap1-shRNA transferred brains compared to WT controls. The statistical results for the proportion of GFP-positive neurons in CP, IZ, and VZ/SVZ are shown. The bar represents standard deviation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bar = 100 μm.
Discussion
Although NCKAP1 disruptive variants have been implicated in ASD risk, the clinical or statistical significance has not yet been investigated due to the paucity of subjects. In this study, multiple lines of evidence support that disruptive variants in NCKAP1 lead to NDD, including autism. First, we identify a total of 20 NCKAP1 de novo or transmitted deleterious variants, found exclusively in affected individuals. These variants span the gamut from single-nucleotide variants/indels to chromosome-balanced rearrangements to microdeletion. We note that the microdeletion also disrupts another two genes adjacent to NCKAP1; however, both genes are tolerant to disruptive variants (pLI = 0.02 for DUSP19 and 0.33 for NUP35) and therefore the loss of these genes is unlikely to contribute to the NDD phenotypes. Of note, no LGD variant in NCKAP1 has yet been reported in gnomAD non-neurologic subset of samples consistent with NCKAP1’s extremely high intolerance to loss-of-function variant. Based on our assessment of >40,000 NDD-affected individuals, NCKAP1 achieves exome-wide significant disruptive variant burden, although ascertainment bias has likely contributed to this. Second, among the 21 individuals (20 probands and 1 affected sibling) recruited internationally across different clinical laboratories, almost all present with an NDD phenotypic profile with ASD as a core symptom. Third, families with inherited disruptive variants show segregation of the variants with NDD and/or neuropsychiatric disease in an autosomal-dominant fashion when clinical information was provided about the other family members.
Although NCKAP1 was originally highlighted due to the discovery of de novo variants, it is noteworthy that nearly half of the clinically referred samples with deleterious variants show transmission within the family. This is unlike some of the first reported high-impact autism genes, such as CHD8, SCN2A (MIM: 182390), ARID1B (MIM: 614556), SYNGAP1 (MIM: 603384), and ADNP, that predominantly occur as de novo variants. This difference may be a result of reduced penetrance and/or variability expressivity. It is interesting that affected individuals in our study with potentially pathogenic NCKAP1 variants are generally not as severely impaired intellectually as with other genes with high frequency of de novo variants. In our NCKAP1 cohort, only 63% individuals have a diagnosis of ID and most are mild even when considering the severity of co-existing conditions. Relatively higher fecundity may help explain the apparent comorbidity with other neuropsychiatric conditions. Going forward, it will be important to consider both de novo and rare transmitted variants in statistical analyses,33,34 but also longitudinal data ranging from detailed childhood developmental history to later adult neuropsychiatric data.
From the functional perspective, NCKAP1 is one of the main components of the WASP-family verprolin-homologous protein (WAVE) regulatory complex which plays important roles in activating the ARP2/3 complex to participate in actin assembly in the leading edge to drive cell migration availability.35, 36, 37 The WAVE regulatory complex consists of five core proteins, including WAVE, NCKAP1, CYFIP2, ABI2, and HSPC3000. NCKAP1 interacts directly with CYFIP2, in which variants have been recently implicated in ID, epilepsy, and ASD.18 Our human brain transcriptome analysis shows that NCKAP1 is broadly expressed in different brain regions and highly expressed in both prenatal and postnatal periods. Importantly, enriched expression is observed in radial glias and excitatory neurons implicating its cell-type-specific roles in neocortex development. Indeed, previous studies show that Nckap1 regulates neocortex neuronal cytoskeletal dynamics and loss of Nckap1 function disrupts neuronal differentiation.10 In this study, using in utero electroporation, we extend these observations by showing that knockdown of Nckap1 promotes neuronal migration. We propose that NCKAP1 is essential for neuronal mechanical changes in the cytoskeleton, which are essential for accurate timing during post-migratory differentiation. Several ASD/NDD risk genes have been implicated in promoting neuronal migration, such as PTEN (MIM: 601728), WDFY3 (MIM: 617485), and SMARCC2 (MIM: 601734). In the neocortex of Pten-heterozygous mice, for example, the number of neurons in the upper layer rises significantly.38 Wdfy3 homozygotes lead to heterotopic clusters of neurons appearing at superficial sites of cortex.39 Smarcc2/Baf170-knockout mice show enlarged cortex due to an increased number of neurons in upper layer and a decrease of such neurons in lower layer.40
In summary, our data strongly indicate heterozygous de novo and transmitted disruptive rare variants in NCKAP1 as contributing to NDD with a core ASD phenotype. Larger well-ascertained cohorts in which multigenerational familial data are available will be needed to tease apart the contribution of these variants to neurodevelopmental and neuropsychiatric disorders. Since ASD is now recognized as a group of phenotypically and genotypically heterogeneous disorders, systematically phenotyping the neurodevelopmental and neuropsychiatric features in individual families with specific ASD genetic subtypes followed up with deep functional characterization in well-established animal models will be critical not only for diagnosis and management in clinical practice but also for the development of long-term treatment strategies.
Data and Code Availability
All data needed to evaluate the paper’s conclusions are present in the paper. The data that support the findings of this study are available from the corresponding author upon request.
Declaration of Interests
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. M.J.G.S., A. Begtrup, R.E.S., S.P., I.M.W., and L.R. are employees of GeneDx, Inc. All of the remaining authors declare no conflict of interest.
Acknowledgments
We are grateful to the families involved in this study. We thank Tonia Brown for assistance in editing this manuscript. We thank Mais Hashem, Egidio Spinelli, and John J. Millichap for coordinating data collection. This work was supported by the following grants: the National Natural Science Foundation of China (81871079, 81330027, 81525007, 8173000779, 81671122) to H.G., K.X., and Z.H.; The US National Institutes of Health (NIH) grant (MH101221) to E.E.E.; and the Hunan Provincial grants (2018SK1030, 2018DK2016, 2019SK1015, 2019RS2005) to K.X., Z.H., and H.G. H.G. was also supported by the Innovation-Driven Project of Central South University (2020CX042). N.J.B., E.S., S.A., and D.E. was supported by the State Government of Victoria (Department of Health and Human Services, Melbourne Genomics Health Alliance). J.M.M. and F.S. are supported by the Swiss National Science Foundation (310030_185292). W.K.C. is supported by grants from the Simons Foundation and the JPB Foundation. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Published: November 5, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.10.002.
Contributor Information
Hui Guo, Email: guohui@sklmg.edu.cn.
Kun Xia, Email: xiakun@sklmg.edu.cn.
Web Resources
BrainSpan, https://www.brainspan.org
GeneMatcher, https://genematcher.org
OMIM, https://www.omim.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Information
References
- 1.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.-Y., Peng M., Collins R., Grove J., Klei L., Autism Sequencing Consortium. iPSYCH-Broad Consortium Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai M.C., Lombardo M.V., Baron-Cohen S. Autism. Lancet. 2014;383:896–910. doi: 10.1016/S0140-6736(13)61539-1. [DOI] [PubMed] [Google Scholar]
- 5.Barnard R.A., Pomaville M.B., O’Roak B.J. Mutations and Modeling of the Chromatin Remodeler CHD8 Define an Emerging Autism Etiology. Front. Neurosci. 2015;9:477. doi: 10.3389/fnins.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helsmoortel C., Vulto-van Silfhout A.T., Coe B.P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J.H., Marcelis C.L., Willemsen M.H., Vissers L.E. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Bon B.W.M., Coe B.P., Bernier R., Green C., Gerdts J., Witherspoon K., Kleefstra T., Willemsen M.H., Kumar R., Bosco P. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry. 2016;21:126–132. doi: 10.1038/mp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stessman H.A.F., Willemsen M.H., Fenckova M., Penn O., Hoischen A., Xiong B., Wang T., Hoekzema K., Vives L., Vogel I. Disruption of POGZ Is Associated with Intellectual Disability and Autism Spectrum Disorders. Am. J. Hum. Genet. 2016;98:541–552. doi: 10.1016/j.ajhg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H., Bettella E., Marcogliese P.C., Zhao R., Andrews J.C., Nowakowski T.J., Gillentine M.A., Hoekzema K., Wang T., Wu H., University of Washington Center for Mendelian Genomics Disruptive mutations in TANC2 define a neurodevelopmental syndrome associated with psychiatric disorders. Nat. Commun. 2019;10:4679. doi: 10.1038/s41467-019-12435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokota Y., Ring C., Cheung R., Pevny L., Anton E.S. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron. 2007;54:429–445. doi: 10.1016/j.neuron.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H., Wang T., Wu H., Long M., Coe B.P., Li H., Xun G., Ou J., Chen B., Duan G. Inherited and multiple de novo mutations in autism/developmental delay risk genes suggest a multifactorial model. Mol. Autism. 2018;9:64. doi: 10.1186/s13229-018-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anazi S., Maddirevula S., Salpietro V., Asi Y.T., Alsahli S., Alhashem A., Shamseldin H.E., AlZahrani F., Patel N., Ibrahim N. Expanding the genetic heterogeneity of intellectual disability. Hum. Genet. 2017;136:1419–1429. doi: 10.1007/s00439-017-1843-2. [DOI] [PubMed] [Google Scholar]
- 13.Ruzzo E.K., Pérez-Cano L., Jung J.-Y., Wang L.-K., Kashef-Haghighi D., Hartl C., Singh C., Xu J., Hoekstra J.N., Leventhal O. Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell. 2019;178:850–866.e26. doi: 10.1016/j.cell.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X., Sanders S.J., Liu L., De Rubeis S., Lim E.T., Sutcliffe J.S., Schellenberg G.D., Gibbs R.A., Daly M.J., Buxbaum J.D. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zweier M., Begemann A., McWalter K., Cho M.T., Abela L., Banka S., Behring B., Berger A., Brown C.W., Carneiro M., Deciphering Developmental Disorders (DDD) Study Spatially clustering de novo variants in CYFIP2, encoding the cytoplasmic FMRP interacting protein 2, cause intellectual disability and seizures. Eur. J. Hum. Genet. 2019;27:747–759. doi: 10.1038/s41431-018-0331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima M., Kato M., Aoto K., Shiina M., Belal H., Mukaida S., Kumada S., Sato A., Zerem A., Lerman-Sagie T. De novo hotspot variants in CYFIP2 cause early-onset epileptic encephalopathy. Ann. Neurol. 2018;83:794–806. doi: 10.1002/ana.25208. [DOI] [PubMed] [Google Scholar]
- 20.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruderfer D.M., Hamamsy T., Lek M., Karczewski K.J., Kavanagh D., Samocha K.E., Daly M.J., MacArthur D.G., Fromer M., Purcell S.M., Exome Aggregation Consortium Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 2016;48:1107–1111. doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stessman H.A.F., Xiong B., Coe B.P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 28.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards J.J., Rouillard A.D., Fernandez N.F., Wang Z., Lachmann A., Shankaran S.S., Bisgrove B.W., Demarest B., Turan N., Srivastava D. Systems Analysis Implicates WAVE2 Complex in the Pathogenesis of Developmental Left-Sided Obstructive Heart Defects. JACC Basic Transl. Sci. 2020;5:376–386. doi: 10.1016/j.jacbts.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawa T., Kodaira M., Oiji A., Sasayama D., Iwadare Y., Ushijima H., Usami M., Watanabe K., Saito K. Dysfunction of orbitofrontal and dorsolateral prefrontal cortices in children and adolescents with high-functioning pervasive developmental disorders. Ann. Gen. Psychiatry. 2013;12:31. doi: 10.1186/1744-859X-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.-X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilfert A.B., Turner T.N., Murali S.C., Hsieh P., Sulovari A., Wang T., Coe B.P., Guo H., Hoekzema K., Bakken T.E. Recent ultra-rare inherited mutations identify novel autism candidate risk genes. bioRxiv. 2020 doi: 10.1101/2020.02.10.932327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogdan S., Klämbt C. Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development. 2003;130:4427–4437. doi: 10.1242/dev.00663. [DOI] [PubMed] [Google Scholar]
- 36.Hummel T., Leifker K., Klämbt C. The Drosophila HEM-2/NAP1 homolog KETTE controls axonal pathfinding and cytoskeletal organization. Genes Dev. 2000;14:863–873. [PMC free article] [PubMed] [Google Scholar]
- 37.Eden S., Rohatgi R., Podtelejnikov A.V., Mann M., Kirschner M.W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Huang W.-C., Séjourné J., Clipperton-Allen A.E., Page D.T. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated β-Catenin Signaling. J. Neurosci. 2015;35:10252–10267. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orosco L.A., Ross A.P., Cates S.L., Scott S.E., Wu D., Sohn J., Pleasure D., Pleasure S.J., Adamopoulos I.E., Zarbalis K.S. Loss of Wdfy3 in mice alters cerebral cortical neurogenesis reflecting aspects of the autism pathology. Nat. Commun. 2014;5:4692. doi: 10.1038/ncomms5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuoc T.C., Boretius S., Sansom S.N., Pitulescu M.-E., Frahm J., Livesey F.J., Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the paper’s conclusions are present in the paper. The data that support the findings of this study are available from the corresponding author upon request.