Summary
Plasma DNA fragmentomics is an emerging area in cell-free DNA diagnostics and research. In murine models, it has been shown that the extracellular DNase, DNASE1L3, plays a role in the fragmentation of plasma DNA. In humans, DNASE1L3 deficiency causes familial monogenic systemic lupus erythematosus with childhood onset and anti-dsDNA reactivity. In this study, we found that human patients with DNASE1L3 disease-associated gene variations showed aberrations in size and a reduction of a “CC” end motif of plasma DNA. Furthermore, we demonstrated that DNA from DNASE1L3-digested cell nuclei showed a median length of 153 bp with CC motif frequencies resembling plasma DNA from healthy individuals. Adeno-associated virus-based transduction of Dnase1l3 into Dnase1l3-deficient mice restored the end motif profiles to those seen in the plasma DNA of wild-type mice. Our findings demonstrate that DNASE1L3 is an important player in the fragmentation of plasma DNA, which appears to act in a cell-extrinsic manner to regulate plasma DNA size and motif frequency.
Keywords: liquid biopsy, cfDNA, autoimmune disease, biomarkers, systemic lupus erythematosus, circulating DNA
Introduction
The analysis of circulating nucleic acids in plasma is an emerging noninvasive diagnostic tool in many fields, including in noninvasive prenatal testing and as a liquid biopsy for cancer assessment.1, 2, 3, 4 Studies on the fragmentation of plasma DNA,5, 6, 7, 8, 9, 10, 11 referred to as fragmentomics,12, 13, 14 have received much recent attention. Plasma DNA is released upon cell death and exists in the circulation mainly as short fragments.15,16 Analysis of plasma samples from different healthy individuals reveals a characteristic plasma DNA fragmentomic profile that is observed reproducibly. For example, the size profile of plasma DNA typically shows a 166 bp major peak with 10 bp periodicities occurring from approximately 143 bp and below.6,9 Circulating fetal and tumor DNA molecules have been found to have a shorter size distribution than that of the background DNA.5,6,9 Additionally, certain sequence motifs are more frequently observed at the ends of plasma DNA molecules than others.17,18 These observations prompted our group to investigate the relationship between nucleases and plasma DNA fragmentomics.19
We have previously performed a series of studies using mouse models in which the genes for different nucleases have been knocked out.19, 20, 21 We demonstrated that different nucleases have unique DNA sequence preferences for cleavage, which result in characteristic end motif signatures of plasma DNA. Through this combination of studies, a stepwise model of plasma DNA fragmentation has been proposed.21 Plasma DNA fragments appear to be generated first intracellularly with DNA fragmentation factor subunit beta (DFFB [MIM: 601883]), intracellular deoxyribonuclease1-like 3 (DNASE1L3 [MIM: 602244]), and other nucleases. Subsequently, the fragmented DNA is released into the plasma where DNASE1L3 and DNASE1 (MIM: 125505) continue to cleave it, resulting in the typical cell-free DNA profile.
DNASE1L3, also known as DNase γ, is a member of the DNase family.22,23 It is primarily secreted by dendritic cells and macrophages and is expressed predominantly in the liver and lymphoid organs such as the spleen.24, 25, 26 Genetic deficiency of DNASE1L3 causes familial systemic lupus erythematosus (SLE [MIM: 152700]) characterized by childhood onset, anti-dsDNA antibody reactivity and lupus nephritis.27, 28, 29, 30, 31 In some cases, SLE was preceded by vascular inflammation, specifically hypocomplementemic urticarial vasculitis syndrome (HUVS), which is characterized by the presence of recurrent urticarial skin lesions, decreased plasma complement, and an array of symptoms such as arthritis, uveitis, glomerulonephritis, and abdominal pain.27,32 Similarly, the loss of DNASE1L3 in mice causes an anti-dsDNA antibody response progressing to SLE-like disease.26,33,34 In fact, DNASE1L3 and C1q (MIM: 120575) are the only genes associated with antibody-mediated monogenic SLE. Most of the others are associated with SLE-like interferonopathies such as Aicardi-Goutieres syndrome (e.g., TREX1 [MIM: 606609], DNASE2 [MIM: 126350], SAMHD1 [MIM: 606754], ADAR1 [MIM: 146920]). DNases involved in such syndromes (e.g., TREX1, DNASE2) are intracellular and their absence causes accumulation of intracellular DNA that activates cGAS/STING and interferon secretion. Thus, DNASE1L3 shows a characteristic function and represents a critical mechanism of immune tolerance to DNA, and therefore is expected to play a major role in its regulation.35
We have discovered that DNASE1L3 plays an important role in plasma DNA fragmentation in a murine model.20 Mice with homozygous inactivation of Dnase1l3 exhibit a lengthened plasma DNA size profile with increases in di-, tri-, and multi-nucleosomal plasma DNA molecules.20 Ends of plasma DNA fragments in humans and mice show high frequencies of 4-mer motifs usually starting with CC dinucleotides. Dnase1l3 deficiency in mice is associated with statistically significant reductions in the frequencies of these characteristic plasma DNA end motifs. These observations suggest that the enzyme DNASE1L3 may play an important role in the process of plasma DNA fragmentation in mice.
In this study, we provide evidence demonstrating the role of DNASE1L3 in plasma DNA fragmentation in humans. We studied the plasma DNA profile of human subjects with disease-associated variations of DNASE1L3 (Figure 1). We also investigated whether we could simulate the plasma DNA fragmentation profile using in vitro digestion of nuclei from a human cell line by DNASE1L3 (Figure 1). Finally, we employed a murine model to explore the potential normalization of plasma DNA profiles through reconstitution of DNASE1L3 in the circulation of Dnase1l3-deficient mice (Figure 1).
Figure 1.
Conceptual Diagram Showing the Design and Experimental Approaches of the Study
In part I, we aimed to study the fragmentomic characteristics of plasma DNA from human individuals with or without disease-associated variations of DNASE1L3. Plasma DNA was isolated from the blood of subjects with DNASE1L3 disease-associated variants, their parents, and control subjects for analysis and comparison. In part II, we aimed to study the fragmentation preference of DNASE1L3 and DNASE1 by in vitro digestion of DNA from a human cell line. DNA from cell nuclei and naked DNA, representing DNA with or without nucleosomal structural proteins, respectively, was subjected to enzymatic digestion by DNASE1L3 or DNASE1 prior to sequence analysis. In part III, we used a mouse model to investigate whether the fragmentomic profile of DNA in plasma of Dnase1l3-deficient mice could be corrected in vivo via DNASE1L3-expressing adeno-associated viral (AAV) transfection. Dnase1l3-deficient mice injected with empty AAV particles or PBS were used as negative controls. The DNA libraries were constructed and sequenced. The fragmentomic profiling involved the analyses of the size distribution and the 4-mer end motif frequencies of DNA fragments. The red cross denotes the inefficiency of DNASE1 in digestion of cell nuclei (refer to Figure 7).
Material and Methods
Human Subjects
We recruited 27 individuals from the Prince of Wales Hospital (Hong Kong), the Istituto Giannina Gaslini (Italy), and the Hospital for Sick Children (SickKids) (Canada) with written informed consent. In total, the cohort included 5 case subjects with DNASE1L3 disease-associated variants, 3 parents of these subjects, and 19 control subjects. Table S1 summarizes the demographics of these subjects. The five case subjects had the following sequence variations in DNASE1L3: three subjects (V1, H2, and H4) had homozygous frameshift c.290_291delCA (p.Thr97Ilefs∗2) mutation, one subject (V6) was heterozygous for c.290_291delCA (p.Thr97Ilefs∗2) and a deletion in exon 5, and one subject (A3) was found homozygous for c.563G>C (p.Gln188Arg) for DNASE1L3. A1 and A2 were the parents of A3. H1 was the parent of H2 and H4 (Figure 4). Among the control subjects, eight were healthy individuals, six were subjects with inactive SLE, two had juvenile idiopathic arthritis (JIA [MIM: 618795]), and three were subjects with kidney transplantation (due to end-stage renal failure from kidney dysplasia [MIM: 143400] and vesico-ureteral reflux [MIM: 193000]). None of these control subjects carried either c.290_291delCA (p.Thr97Ilefs∗2) or c.563G>C (p.Gln188Arg). JIA is the most common pediatric rheumatic disease and includes all forms of arthritis that begin before adulthood and are of unknown origin, which share some features, such as uveitis and presence of autoantibodies,36 with HUVS. Therefore, we enrolled patients with JIA as pediatric autoimmune disease control subjects. As subjects with DNASE1L3 disease-associated variants could develop kidney complications and require kidney transplantation, pediatric subjects, who had received kidney transplantation for non-immune related disorders, were also enrolled. One blood sample was collected from each individual, except one patient with DNASE1L3 disease-associated variant who had provided four blood samples for analysis. The study was approved by the Joint Chinese University of Hong Kong-Hospital Authority New Territories East Cluster Clinical Research Ethics Committee, the ethics committee of the Istituto Giannina Gaslini (approval BIOL 6/5/04), and the SickKids Research Ethics Board.
Figure 4.
End Motif Aberrations of Plasma DNA in Subjects with DNASE1L3 Disease-Associated Variants
Heatmap of the frequency of the top 25 ranked 4-mer end motifs in plasma DNA among the different groups. Samples V1 to V4 were taken on four separate occasions, from one patient who was homozygous for the c.290_291delCA (p.Thr97Ilefs∗2) frameshift mutation. Samples A3, H2, and H4 were taken from subjects who were also homozygous for the c.290_291delCA (p.Thr97Ilefs∗2) frameshift mutation. Sample A3 was taken from a subject who was homozygous for c.563G>C (p.Gln188Arg), and V5 was taken from a subject who was heterozygous for c.290_291delCA (p.Thr97Ilefs∗2) and a deletion in exon 5 of DNASE1L3. Sample H1 was taken from a parent who was heterozygous for c.563G>C (p.Gln188Arg), while samples H2 and H4 were taken from parents who were heterozygous for c.290_291delCA (p.Thr97Ilefs∗2) of DNASE1L3. SLE, systemic lupus erythematosus; JIA, juvenile idiopathic arthritis; transplant, subjects with a history of kidney transplantation; parent, healthy parents of subjects with DNASE1L3 disease-associated variants.
Animals
Animal experiments were performed according to a protocol approved by the Institutional Animal Care and Use Committees of New York University School of Medicine. The Dnase1l3-deficient mouse model on C57BL/6 (B6) background (Dnase1l3LacZ/LacZ) was described previously.26 The AAV8-based vector encoding human DNASE1L3 open reading frame (ORF) (GenBank: NP_004935.1) under a hepatocyte-specific plasminogen (PL1) promoter was constructed and virus particles were produced and purified by Welgen. Three Dnase1l3-deficient mice were each injected with 1 × 1011 virus genomes of AAV8-PL1-DNASE1L3 particles via the tail vein. For controls, one Dnase1l3-deficient mouse was injected with 1 × 1011 virus genomes of AAV particles encoding green fluorescent protein (GFP) under the ubiquitous cytomegalovirus (CMV) promotor (UNC Vector core) and another one was injected with sterile phosphate buffered saline (PBS) alone. After 4 weeks, the mice were euthanized and exsanguinated by cardiac puncture.
Sample Processing and DNA Extraction
The blood samples in EDTA-containing collection tubes were first centrifuged at 1,600 × g for 10 min at 4°C and the plasma portion was further subjected to centrifugation at 16,000 × g for 10 min at 4°C to pellet the residual cells and platelets.37 The resulting plasma was harvested. Circulating cell-free DNA was extracted from plasma using the DSP Blood Mini Kit (QIAGEN) as previously described.2
In Vitro Nuclease Digestion of Nuclei DNA and Naked DNA
Nuclei and genomic DNA isolated from Jurkat cells (human T lymphocyte cell line) were digested by either DNASE1 or DNASE1L3.26,38 Jurkat cell genomic DNA was purchased from ThermoFisher and nuclei were isolated as described.26 Recombinant human DNASE1 was purchased commercially (Abcam) and the DNASE1L3 was produced in-house. An 8-aa FLAG epitope was introduced into a predicted internal loop of the human full-length DNASE1L3, and the resulting protein was confirmed to retain full activity including its specific ability to digest chromatin. The resulting FLAG-DNASE1L3 ORF was stably expressed from a lentiviral vector in 293T cells, purified using anti-FLAG M2 beads (Sigma-Aldrich), eluted with FLAG peptide, and quantified by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) with bovine serum albumin (BSA) as a standard. DNases (25 μg/mL) were incubated with 2 × 105 nuclei or 200 ng of genomic DNA in Hanks’ Balanced Salt Solution (Thermo Fisher) at 37°C for 1 h in the presence of 2 mM MnCl2/CaCl2. After incubation, the digested samples were purified individually using Wizard Plus Minipreps DNA Purification System (Promega).
DNA Library Preparation and Electrophoresis
Circulating DNA libraries were constructed by using the KAPA HTP Library Preparation Kit (Roche) and purified using a MinElute Reaction Cleanup Kit (QIAGEN) according to the manufacturer’s instructions. Adaptor-ligated libraries were analyzed on an Agilent 4200 TapeStation (Agilent Technologies) using the High Sensitivity D1000 ScreenTape System (Agilent Technologies) for quality control and gel-based size determination. Before sequencing, the libraries were quantified by quantitative polymerase chain reaction (qPCR) using a KAPA Library Quantification Kit (Roche) on a LightCycler 96 instrument (Roche).
Illumina DNA Sequencing
The DNA libraries were sequenced for 75 bp for each end in a paired-end format on a NextSeq 500 instrument (Illumina). Real-time image analysis and base calling were performed using the NextSeq Control Software v.2.1.0 and Real Time Analysis (RTA) Software v.2.4.11 (Illumina). After base calling, adaptor sequences and low-quality bases (i.e., quality score < 5) on the 3′ ends of the reads were removed. The sequencing depth was 60M ± 18M unique reads per case.
For the analysis of sequencing data, the sequenced reads were aligned to the non-repeat-masked reference genomes (NCBI build 37/hg19 for human and NCBI build 37/UCSC mm9 for mouse) using the Short Oligonucleotide Alignment Program 2 as previously described.2,39 Only paired-end reads which were aligned to the same chromosome in a correct orientation with an insert size less than 600 bp were used for downstream analysis. For paired-end reads sharing the same start and end aligned genomic coordinates, only one would be kept for further analysis while the rest were deemed to be PCR duplicates and were discarded.
Single-Molecule, Real-Time Sequencing
Sequencing templates were constructed using SMRTbell Template Prep Kit 1.0-SPv3 (Pacific Biosciences). The amplicon template preparation and sequencing protocol was used, with minor modifications: DNA was purified with 1.8× AMPure PB beads, and library size was estimated using TapeStation instrument (Agilent). Sequencing primer annealing and polymerase binding conditions were calculated with SMRT Link v.5.1.0 software (Pacific Biosciences). Briefly, sequencing primer v.3 was annealed to sequencing template, then the polymerase was bound to templates using Sequel Binding and Internal Control Kit 2.1 (Pacific Biosciences). Sequencing was performed on a Sequel SMRT Cell 1M v.2 for each template. Sequencing movie was collected on the Sequel system for 10 h with Sequel sequencing kit 2.1 (Pacific Biosciences).
Size Analysis of Circulating DNA
Following paired-end sequencing, both end sequences of each DNA molecule were aligned to the human reference genome (NCBI build 37/hg19) or mouse reference genome (NCBI build 37/UCSC mm9). The genome coordinates of the aligned ends were then used to deduce the sizes of the sequenced DNA molecules.
Motif Analysis of Circulating DNA
To study whether the disease-associated variations of DNASE1L3 would alter the cleavage pattern of circulating DNA, the first 4-nucleotide sequence (i.e., a 4-mer motif) on each 5′ fragment end of plasma DNA molecules was determined. The frequency of each of the 256 possible motifs (i.e., 4 × 4 × 4 × 4) was calculated and normalized by the total number of ends. For each motif, the difference in the frequencies between subjects with DNASE1L3 disease-associated variants and healthy control subjects was tested by Wilcoxon rank-sum test and its p value was adjusted by the Holm-Bonferroni procedure because of multiple comparisons.40
Statistical Analysis
Analysis was performed using custom-built programs written in Perl and R languages. Statistical difference was calculated using the Wilcoxon rank-sum test unless otherwise specified. A p value of less than 0.05 was considered as statistically significant and all probabilities were two-tailed.
Results
Size Profiling of Plasma DNA
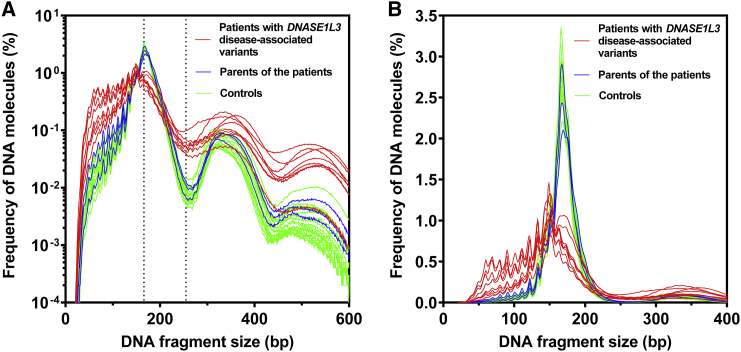
Paired-end sequencing, which allows for plasma DNA size determination at a single-nucleotide resolution, was performed for plasma DNA libraries from subjects with DNASE1L3 disease-associated variants, their parents, and control subjects. The control group consisted of both healthy subjects and subjects with inactive SLE, juvenile idiopathic arthritis, and kidney transplantation. Figure 2 shows the overall size distribution of plasma DNA molecules. Figure 2A shows the size distribution profiles plotted on a logarithmic scale to allow for better visualization of these changes in the frequencies of long DNA fragments. The plasma DNA size profile of the control subjects demonstrated the expected modal size at approximately 166 bp,6,9 which reflected the nucleosomal nature of plasma DNA. The parents, who carried one normal DNASE1L3 gene and one variant one, did not show any notable difference in the overall size distribution of plasma DNA when compared to the control subjects. In contrast, subjects carrying two DNASE1L3 disease-associated variants exhibited an aberrant size profile with an increase in the frequencies of plasma DNA molecules below 50 bp and those above 250 bp (Figure 2A). Figure S1 demonstrates that disease-associated variations of DNASE1L3 result in a near doubling in the frequency of plasma DNA molecules above 250 bp when compared to that of the control subjects (p < 0.0001, Wilcoxon rank-sum test). These observations were thus reminiscent of the plasma DNA profile of Dnase1l3-deficient mice.20 Compared to control subjects, affected individuals with DNASE1L3 disease-associated variants had an increase in the frequency of plasma DNA molecules at 250 bp or above but a reduced frequency of plasma DNA molecules at 166 bp (Figure 2A). We calculated a fragmentation ratio by dividing the sequenced read count of the DNA molecules at 166 bp by that of the DNA molecules at 250 bp. Figure 3 shows that the fragmentation indices of DNASE1L3-deficient subjects were significantly decreased by 96% (p < 0.0001, Wilcoxon rank-sum test) compared to control subjects.
Figure 2.
Size Distributions of Plasma DNA Molecules of Case Subjects, Their Parents, and Control Subjects
Subjects with DNASE1L3 disease-associated variants (red, n = 8), their parents (blue, n = 3), and control subjects (green, n = 19). The size distribution is plotted in the range (A) from 0 to 600 bp on a logarithmic scale and (B) from 0 to 400 bp on a linear scale. Each line represents one case. As one subject provided four blood collections, a total of eight plasma samples from the affected group were analyzed.
Figure 3.
Fragmentation Index of DNA in Plasma of Control Subjects and Case Subjects with DNASE1L3 Disease-Associated Variants
The fragmentation index was calculated by dividing the sequenced read count of the DNA molecules at 166 bp by that of the DNA molecules at 250 bp and plotted on a logarithmic scale. Each circle represents one case. As one subject provided four blood collections, a total of eight plasma samples from the affected group were analyzed. Multiple samples obtained from the same subject were indicated by green solid circles. One of the subjects (A3) with the missense mutation c.563G>C (p.Gln188Arg) was indicated by a yellow solid circle. The middle line and error bars indicate median ± interquartile range.
Frequency Distribution of Plasma DNA End Motifs
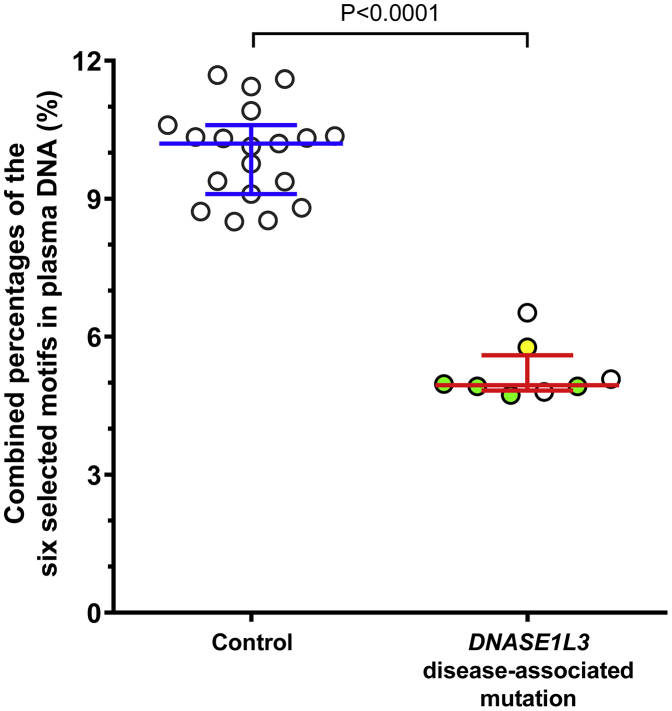
In our previous work with a murine model of Dnase1l3 gene knockout,20 we found that Dnase1l3 deletion was associated with changes in plasma DNA molecules characterized by a series of 4-mer end motifs (all starting with CC). To see whether such observations could be extrapolated to humans, we ranked the 4-mer end motifs of plasma DNA from healthy human subjects (i.e., excluding the inactive SLE, JIA, and kidney transplantation control subjects) in descending order of frequency (Data S1, tab “Controls”). The frequencies of the top 25 end motifs of each of the control subjects and case subjects with disease-associated variants are illustrated in the heatmap in Figure 4. Subjects with disease-associated variants had a distinct heatmap pattern when compared to healthy control subjects, subjects with other clinical conditions, and their parents. On the other hand, the heatmap patterns within the control subjects and the parents were similar. We then selected the end motifs that showed a significant reduction in patients with disease-associated variants from this ranked order (Data S1, tab “DNASE1L3 Def”). The top 25 end motifs, which were found by such a procedure, were all within the top 28 motifs observed in the healthy control subjects (Data S1) and are listed in Table 1. The top six selected motifs (CCCA, CCAG, CCTG, CCAA, CCCT, and CCAT) all started with CC and thus were considered together in subsequent analyses. These six selected motifs were all within the top 7 motifs observed in healthy control subjects, and together accounted for 10.64% (range 9.76%–11.69%) of plasma DNA end motifs. Figure 5 shows the frequency of the combined percentages of the six selected motifs in the plasma of control subjects compared to subjects with disease-associated variants. We observed that disease-associated variations of DNASE1L3 were associated with a significant reduction in the frequencies of these motifs. Frequencies for each of the top six motifs (Figure S2) were consistent with the combined percentages shown in Figure 5. Thus, the disease-associated variations of DNASE1L3 were associated with a reduction in the frequencies of the most common DNA end motifs in human plasma.
Table 1.
Top 25 Motifs with the Highest Frequencies in Healthy Control Subjects and Their Ranks, Frequencies in Plasma of Healthy Control Subjects and Case Subjects, as well as the Fold Change in Frequencies between Healthy Control Subjects and Case Subjects
| Motif | Rank in Healthy Control Subjects | Rank in Case Subjects | Motif Frequency of Healthy Control Subjects (a) (%) | Motif Frequency of Case Subjects (b) (%) | Fold Change (a/b) | p Value |
|---|---|---|---|---|---|---|
| CCCA | 1 | 1 | 2.29 | 0.85 | 2.69 | <0.0001 |
| CCAG | 2 | 3 | 1.91 | 0.79 | 2.43 | <0.0001 |
| CCTG | 3 | 7 | 1.89 | 0.94 | 2.01 | <0.0001 |
| CCAA | 4 | 2 | 1.64 | 0.67 | 2.45 | <0.0001 |
| CCCT | 5 | 20 | 1.51 | 0.92 | 1.64 | <0.0001 |
| CCTT | 6 | 83 | 1.48 | 1.31 | 1.13 | 0.4487 |
| CCAT | 7 | 15 | 1.41 | 0.82 | 1.72 | <0.0001 |
| CAAA | 8 | 48 | 1.41 | 1.08 | 1.30 | 0.0003 |
| CCTC | 9 | 52 | 1.23 | 0.97 | 1.27 | 0.0003 |
| CCAC | 10 | 10 | 1.12 | 0.6 | 1.85 | <0.0001 |
| TGAA | 11 | 39 | 1.04 | 0.73 | 1.42 | <0.0001 |
| TAAA | 12 | 75 | 1.03 | 0.88 | 1.16 | 0.0127 |
| CCTA | 13 | 8 | 1.02 | 0.52 | 1.97 | <0.0001 |
| CCCC | 14 | 9 | 1.01 | 0.53 | 1.90 | <0.0001 |
| TGAG | 15 | 35 | 0.99 | 0.67 | 1.48 | <0.0001 |
| TGTT | 16 | 18 | 0.98 | 0.58 | 1.69 | <0.0001 |
| CAAG | 17 | 41 | 0.97 | 0.7 | 1.40 | 0.0002 |
| CTTT | 18 | 165 | 0.96 | 1.33 | 0.73 | 0.0001 |
| AAAA | 19 | 120 | 0.95 | 0.98 | 0.96 | 0.8205 |
| TGTG | 20 | 12 | 0.91 | 0.5 | 1.82 | <0.0001 |
| CATT | 21 | 167 | 0.89 | 1.23 | 0.72 | <0.0001 |
| CACA | 22 | 45 | 0.85 | 0.65 | 1.31 | 0.0002 |
| CAGA | 23 | 57 | 0.84 | 0.68 | 1.23 | 0.0002 |
| TATT | 24 | 61 | 0.83 | 0.69 | 1.21 | 0.001 |
| CAGG | 25 | 91 | 0.81 | 0.75 | 1.08 | 0.0888 |
Figure 5.
Combined Percentages of the Six Selected Motifs in Plasma DNA of Control Subjects and Case Subjects with DNASE1L3 Disease-Associated Variants
The selected motifs CCCA, CCAG, CCTG, CCAA, CCCT, and CCAT were the top six 4-mer motifs ranked with the highest frequencies in healthy control subjects which showed a statistically significant reduction in case subjects with DNASE1L3 disease-associated variants. Each circle represents one case. As one subject provided blood samples on four different occasions, a total of eight plasma samples from the patient group were analyzed. Multiple samples obtained from the same subject were indicated by green solid circles. One of the subjects (A3) with the missense mutation c.563G>C (p.Gln188Arg) was indicated by a yellow solid circle. The middle line and error bars indicate median ± interquartile range.
In Vitro Digestion of DNA by DNASE1 and DNASE1L3
The analyses of the size distribution and end motifs of plasma DNA have suggested that the cleavage of DNA by DNASE1L3 may be inter-nucleosomal and may exhibit increased likelihood to be adjacent to cytosine dinucleotides (CC).20 To directly test this notion, we digested both cell nuclei and naked DNA by DNASE1 and DNASE1L3 and analyzed the results by DNA sequencing (Pacific Biosciences).
Table 2 shows the size metrics of the DNA molecules with and without enzymatic digestion. Digestion of cell nuclei by DNASE1L3 drastically reduced the median length of DNA fragments from 1,950 bp (no digestion) to 153 bp (after digestion), whereas DNASE1 had a much lower impact (median length after digestion: 1,640 bp). Digestion of naked DNA by DNASE1L3 and DNASE1 reduced the size of DNA fragments with median values changing from 1,871 bp to 179 bp and 217 bp, respectively. These data demonstrated that DNASE1L3 cleaved both DNA in cell nuclei and naked DNA, while DNASE1 was able to digest only naked DNA, as reported previously.41
Table 2.
Table Summarizing the Size of DNA with or without Enzyme Digestion
| Enzyme Digestion | Median DNA Size (Interquartile Range), bp | Longest DNA Size, bp |
|---|---|---|
| Cell Nuclei DNA | ||
| No digestion | 1,950 (954–3,313) | 24,100 |
| DNASE1L3 digestion | 153 (127–185) | 11,378 |
| DNASE1 digestion | 1,640 (796–2,532) | 20,010 |
| Naked DNA | ||
| No digestion | 1,871 (996–2,977) | 19,429 |
| DNASE1L3 digestion | 179 (111–308) | 4,463 |
| DNASE1 digestion | 217 (141–379) | 20,416 |
Interestingly, DNA from cell nuclei digested by DNASE1L3 exhibited a size profile similar to plasma cell-free DNA with a major peak at 153 bp followed by a series of smaller 10-bp periodic peaks (Figure 6). In contrast, this pattern was not observed after digesting naked DNA with either DNASE1L3 or DNASE1. These results suggest that these two nucleases may have different substrate preferences and that DNA-bound proteins, e.g., histones, might be a factor influencing the fragmentation patterns of plasma DNA.
Figure 6.
Size Distributions of Nuclei DNA and Naked DNA after Digestion by Nucleases In Vitro
The size distributions are in the range from 0 to 400 bp plotted on a linear scale.
We next analyzed the DNA end motifs by ranking the 256 end motifs in descending order according to their frequencies in DNASE1L3- or DNASE1-digested cell nuclei and naked DNA (Data S2). In Figures 7A and 7B, the ranking of each of the 256 DNA end motifs created by DNASE1L3 was plotted against those created by digestion with DNASE1 for nuclei and naked DNA substrates, respectively. The pink area in Figures 7A and 7B highlights motifs that were ranked within the top 10 for DNASE1L3 but ranked 11 or lower for DNASE1. Conversely, the yellow area in Figures 7A and 7B highlights motifs that were ranked within the top 10 for DNASE1 but ranked 11 or lower for DNASE1L3. Among the nine motifs in the pink area of Figure 7A (digestion of cell nuclei), six of these were also found in the pink area of Figure 7B (digestion of naked DNA). Five out of six of these started with CC. Among the five motifs starting with CC, four of them (i.e., CCTG, CCAG, CCCA, and CCAA) were previously found to be significantly reduced in the plasma of Dnase1l3-deficient mice but not Dnase1-deficient mice, implicating these motifs as specific targets for DNASE1L3.20
Figure 7.
Relationships between the Motif Rankings of In Vitro Digested DNA Substrates
Correlations between (A) the motif rankings of cell nuclei after digestion with DNASE1L3 and that of cell nuclei incubated with DNASE1 and (B) the motif rankings of naked DNA digestion by DNASE1L3 and that of naked DNA digestion by DNASE1. Each circle represents a 4-mer motif. Motifs preferentially created by the DNASE1L3 digestion of cell nuclei (A) or naked DNA (B) were indicated by open red circles. Motifs preferentially created by the DNASE1 digestion of cell nuclei (A) or naked DNA (B) were indicated by open blue circles. Two common motifs preferentially created by DNASE1 digestion of both cell nuclei and naked DNA were indicated by solid blue circles.
Conversely, among the ten motifs in the yellow area of Figure 7A (incubation of cell nuclei), only two were found in the yellow area of Figure 7B (digestion of naked DNA). Among the ten motifs in the yellow area of Figure 7B (digestion of naked DNA), nine of them started with T. DNA end motifs starting with a T were previously shown to be a signature of DNASE1 digestion in plasma.21 Previous work has indicated that DNASE1 is more efficient at digesting naked DNA compared to nucleosomal DNA.42 We hypothesized that the motifs in the yellow area in Figure 7A were located there because of their statistical frequencies within the human genome rather than due to the cutting effects mediated by DNASE1. We thus plotted the ranking of motif frequencies of cell nuclei incubated with DNASE1 versus the ranking of in silico determined motif frequencies across the human genome (Figure S3A). We observed a strong correlation between the rankings of the observed and in silico calculated frequencies (r = 0.94, p < 0.0001, Spearman’s correlation). The green area of Figure S3A denotes motifs that were highly ranked in nuclei DNA after exposure to DNASE1 and the in silico determined frequencies in the human genome. Notably, all seven motifs in the green area were found in the yellow area of Figure 7B.
In comparison, we also plotted the ranking of motif frequencies of cell nuclei digested by DNASE1L3 versus the ranking of the in silico determined motif frequencies (Figure S3B). Importantly, the correlation was not as strong (r = 0.631, p < 0.0001, Spearman’s correlation) as the correlation in DNASE1-exposed nuclei DNA; in particular, unlike in Figure S3A, there were no motifs in the top right-hand quadrant of Figure S3B. Altogether, these data suggest that DNASE1L3 directly imparts the end motif specificity on its nuclear DNA substrate.
Adeno-Associated Virus (AAV)-Mediated Reconstitution of DNASE1L3 in Dnase1l3-Deficient Mice
Our previous mouse pregnancy model demonstrated that the contribution of DNASE1L3 to the fragmentation of plasma DNA could occur at a systemic level in the circulation and within the placenta.20 To test whether DNASE1L3 may regulate plasma fragmentation in a cell-extrinsic manner, we ectopically expressed DNASE1L3 in Dnase1l3-deficient mice using AAV8 vector-based delivery. AAV8 was used as a transduction vector because of its high affinity for hepatocytes, and hepatocyte-specific expression was further reinforced by the PL1 promoter.43 Dnase1l3-deficient mice injected with PBS or with GFP-encoding AAV particles served as controls. If DNASE1L3 was expressed in Dnase1l3-deficient mice after the AAV8-Dnase1l3 transfection, then one would expect that a reversal of the phenotype associated with Dnase1l3 deletion would occur. To investigate this, we analyzed the size profile and the six selected end motifs of plasma DNA.
Figure S4A demonstrates that Dnase1l3 transduction in Dnase1l3-deficient mice partially restored the major peak at 166 bp in two out of three animals, while the controls without Dnase1l3 transduction retained a relatively more aberrant size profile of plasma DNA. Although one of the transduced Dnase1l3-deficient mice overlapped with the Dnase1l3-deficient mouse injected with PBS, an overall trend of size normalization was observed. When compared to controls, the median fragmentation index of transduced Dnase1l3-deficient mice was doubled (Figure S4B).
Figure 8 is a heatmap showing the frequencies of the top 25 end motifs of each mouse used in the AAV transfection experiment. Publicly available motif data generated from 9 wild-type and 13 Dnase1l3-deficient mice (European Genome-Phenome Archive: EGAS00001003174) were used as references. The overall motif frequency pattern of Dnase1l3-deficient mice injected with either PBS or empty AAV particles was similar to that of the reference Dnase1l3-deficient mice shown in the first two columns. In contrast, mice with Dnase1l3 transduction showed a motif frequency pattern that was more comparable to that of wild-type mice, in comparison with the reference Dnase1l3-deficient mice and those injected with PBS or empty AAV particles.
Figure 8.
Heatmap Showing the Effect of DNASE1L3 Reconstitution on the 4-mer Motifs of Plasma DNA in Dnase1l3-Deficient Mice
The top 25 4-mer motifs with the highest frequencies in the wild-type mice were compared. Publicly available data of 9 wild-type and 13 Dnase1l3-deficient mice were used as references.20
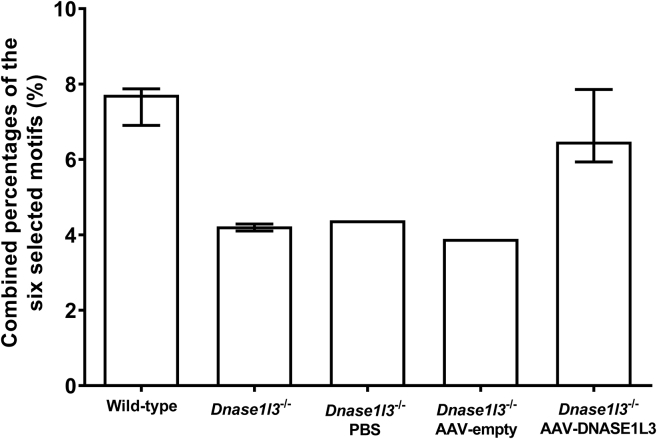
Figure 9 shows the frequency of the combined percentages of the top six motifs in the plasma DNA of these mice. After Dnase1l3 transduction, the combined percentage in Dnase1l3-deficient mice was increased and comparable to that of the reference wild-type mice. However, Dnase1l3-deficient mice injected with PBS or empty AAV particles did not show any increase in the combined percentage of the top six motifs. Frequencies for each of the top six motifs (Figures S5A–S5F) were consistent with the combined percentages shown in Figure 9. In addition to supporting the contribution of DNASE1L3 to the fragmentation of plasma DNA at the systemic level, these data suggest that Dnase1l3 transduction via AAV8 transfection could restore the DNA size and motif frequencies of plasma DNA in Dnase1l3-deficient mice to the wild-type state.
Figure 9.
Combined Percentages of the Six Selected Motifs in Plasma DNA of Mice with or without DNASE1L3 Reconstitution
The selected motifs CCCA, CCTG, CCAG, CCAA, CCAT, and CCTC were the top six 4-mer motifs ranked with the highest frequencies in wild-type mice and showed a statistically significant reduction in Dnase1l3-deficient mice. Publicly available data from 9 wild-type and 13 Dnase1l3-deficient mice were used as references.20 The bar and errors indicate median ± interquartile range.
Discussion
In this study, we found that human subjects with disease-associated variants showed a number of aberrations in the fragmentation of plasma DNA. We observed an increase in the proportion of longer DNA molecules (above 250 bp) but fewer DNA molecules at 166 bp. Hence, we used DNA fragment frequencies in these two length regions to develop a fragmentation index that could quantify such changes and even be used to identify patients with disease-associated variants (Figure 3).
In addition to the increased frequency of long DNA molecules, there was also an increase in the amount of short plasma DNA molecules below 120 bp. We have previously reported an increase in the amount of short plasma DNA molecules in patients with SLE.44 In fact, homozygous mutations of DNASE1L3 causes familial SLE characterized by childhood onset and anti-dsDNA antibody reactivity.27, 28, 29, 30, 31 The increase of short DNA fragments was associated with the presence of anti-dsDNA antibodies. One plausible explanation could be that DNASE1L3 deficiency may lead to the accumulation of the undigested circulating DNA, which could further trigger autoimmunity including production of the anti-dsDNA antibodies. Hence, the autoantibodies may bind with the short DNA molecules and result in the increased frequency of both longer and shorter DNA in plasma of the patients with DNASE1L3 mutations.26,33
Our previous study using Dnase1l3-deficient mice found that plasma DNA motifs beginning with CC appeared at relatively high levels in the plasma of wild-type mice but showed significant reductions in Dnase1l3−/− mice.20 Similar to Dnase1l3-deficient mice, the plasma DNA of human subjects with disease-associated variants exhibited a reduction of the CC end motifs (Figures 4 and S2). These results suggested that plasma DNA end motif profiling may have potential to serve as a tool for detecting diseases associated with nuclease deficiency. Indeed, we have recently demonstrated the use of plasma DNA end motif profiling in oncology, in which multiple cancer tissues have been shown to exhibit a reduction in DNASE1L3 expression.18
In our previous study on nonpregnant SLE case subjects,44 we found that the presence of high anti-dsDNA antibody levels in the active SLE subjects might contribute to the retention of the short DNA molecules in plasma due to a binding effect from IgG. As DNASE1L3 deficiency represents a subset of monogenic SLE and is associated with the presence of autoantibodies, we expect that pregnant SLE subjects with the DNASE1L3 mutation might exhibit a similar antibody-related increase in short DNA molecules in plasma.
Interestingly, the parents of the affected individuals, who carried only one copy of a mutant DNASE1L3, were clinically healthy and did not show an intermediate fragmentomic profile between the control subjects and the case subjects. These observations suggested that a single copy of DNASE1L3 might produce sufficient enzyme to maintain fragmentomic normality in the plasma DNA profile.
In ongoing projects, we have found that plasma from subjects with disease-associated variants was less effective in the digestion of native chromatin compared to healthy control subjects (J. Hartl, L.S., and B.R., unpublished data). Additionally, we found that total levels of cfDNA were not significantly changed in patients with disease-associated variants.
Both DNA from cell nuclei and naked DNA showed an increase in the frequencies of the CC end motifs after enzymatic digestion by DNASE1L3 but not by DNASE1. These findings suggested that DNASE1L3 has a strong preference for cutting DNA sequences adjacent to the CC dinucleotides regardless of the presence of nucleosomal structural proteins. DNASE1 digestion of naked DNA revealed a preponderance of high-ranking motifs starting with T. In murine models, such T end motifs have been associated with DNASE1.15 In contrast, incubation of cell nuclei with DNASE1 revealed a largely different set of high-ranking end motifs (Figure 7A), which were found to rank highly because of their statistical prevalence in the human genome (Figure S3A). Together, these results underscore the value of searching for nuclease-specific signatures in plasma DNA. A proportion of such signatures might be useful as biomarkers in the emerging field of plasma DNA fragmentomics.
By studying the fragmentomic change in plasma DNA of Dnase1l3-deficient mice where DNASE1L3 has been reconstituted in the circulation, one could further obtain insights into the site of action of DNASE1L3. AAV8 has a broad tropism in murine non-hematopoietic tissues with a distinctively strong expression in the liver,45 specifically in hepatocytes,46 and this was further reinforced with a hepatocyte-specific promoter. Conversely, DNASE1L3 is primarily expressed in hematopoietic cells including dendritic cells and macrophages, which represent a major source of the enzyme in circulation.26 If the transduced cells produce the DNASE1L3 enzyme and DNASE1L3 was able to fragment the DNA in the circulation, then one would observe a normalization of the six highest ranked motifs in plasma DNA molecules. This was indeed observed from the data shown in Figures 8, S4, and S5. Because the majority of plasma DNA was derived from hematopoietic cells and DNASE1L3 in this setting was mainly derived from the liver, these data suggest that ectopically expressed DNASE1L3 was able to regulate plasma DNA fragmentation in a cell-extrinsic manner and that the liver could play a role in plasma DNA fragmentation, at least in this experimental system. These results suggest that further studies on the replenishment of DNASE1L3 may help to explore new therapeutic approaches for treating individuals with insufficient DNASE1L3 and that plasma DNA fragmentomic profile might be a potential biomarker for monitoring the therapeutic efficacy if such an approach is developed. Due to the small number of mice in this preliminary study, further investigation with increasing sample size would be necessary to support this conclusion.
In summary, we have demonstrated that homozygous disease-associated variations of DNASE1L3 in humans are associated with aberrations in plasma DNA fragmentation. Hence, we conclude that DNASE1L3 is an important component of the mechanism for maintaining plasma DNA homeostasis in humans. Our findings suggest that a better understanding of the relationship between nuclease biology and the generation of plasma DNA might offer a new dimension of research in circulating nucleic acids and might be useful for developing fragmentomic biomarkers in plasma DNA diagnostics.
Data and Code Availability
The accession number for the sequence data reported in this paper is at the European Genome-Phenome Archive (EGA): EGAS00001004342 and EGAS00001004409.
Declaration of Interests
R.W.Y.C., M.N., P.J., A.S.H.C., W.P., D.S.C.H., K.C.A.C., R.W.K.C., and Y.M.D.L. are co-inventors on multiple patents in cell-free DNA-based diagnostics. Y.M.D.L. is a scientific co-founder, shareholder, and scientific advisor of Grail. R.W.K.C. and K.C.A.C. are shareholders of Grail. Y.M.D.L., R.W.K.C., and K.C.A.C. are co-founders and shareholders of DRA Limited and the Take2 Group of companies. Y.M.D.L. is an advisor to Decheng Capital. R.W.K.C. is an advisor to Illumina. Y.M.D.L., R.W.K.C., and K.C.A.C. receive royalties from Illumina, Sequenom, DRA, Take2, and Grail.
Acknowledgments
We thank Dr. Deborah Levy and Dr. Andrea Knight for their clinical support. We also thank Yoyo Jin, Huimin Shang, Saravanan Ramakrishnan, Paola Bocca, and Emily Leung for technical assistance. This work was supported by the Research Grants Council of the Hong Kong SAR Government under the Theme-Based Research Scheme (T12-403/15-N and T12-401/16-W), a Collaborative Research Agreement with Grail, the Lupus Research Alliance, the Colton Center for Autoimmunity, and NIH grants AR071703, AR070591, AI100853, and AR069515. Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation. L.T.H. received funding via a Canadian Institutes of Health Research (CIHR) Project Scheme grant.
Published: October 5, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.09.006.
Contributor Information
Boris Reizis, Email: boris.reizis@nyulangone.org.
Y.M. Dennis Lo, Email: loym@cuhk.edu.hk.
Web Resources
BEDTools (v.2.27.1), https://bedtools.readthedocs.io/en/latest/
European Genome-Phenome Archive, https://www.ebi.ac.uk/ega/home
Human CRCh37/hg19, https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/
Mouse mm9, https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.18/
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Chiu R.W.K., Chan K.C.A., Gao Y., Lau V.Y.M., Zheng W., Leung T.Y., Foo C.H.F., Xie B., Tsui N.B.Y., Lun F.M.F. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu R.W.K., Akolekar R., Zheng Y.W.L., Leung T.Y., Sun H., Chan K.C.A., Lun F.M.F., Go A.T.J.I., Lau E.T., To W.W.K. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan K.C.A., Woo J.K.S., King A., Zee B.C.Y., Lam W.K.J., Chan S.L., Chu S.W.I., Mak C., Tse I.O.L., Leung S.Y.M. Analysis of plasma Epstein–Barr Virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 2017;377:513–522. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 5.Chan K.C.A., Zhang J., Hui A.B.Y., Wong N., Lau T.K., Leung T.N., Lo K.W., Huang D.W.S., Lo Y.M.D. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 6.Lo Y.M.D., Chan K.C.A., Sun H., Chen E.Z., Jiang P., Lun F.M.F., Zheng Y.W., Leung T.Y., Lau T.K., Cantor C.R., Chiu R.W. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 7.Lun F.M.F., Tsui N.B.Y., Chan K.C.A., Leung T.Y., Lau T.K., Charoenkwan P., Chow K.C.K., Lo W.Y.W., Wanapirak C., Sanguansermsri T. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:19920–19925. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S.C.Y., Chan K.C.A., Zheng Y.W.L., Jiang P., Liao G.J.W., Sun H., Akolekar R., Leung T.Y., Go A.T.J.I., van Vugt J.M.G. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc. Natl. Acad. Sci. USA. 2014;111:8583–8588. doi: 10.1073/pnas.1406103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang P., Chan C.W.M., Chan K.C.A., Cheng S.H., Wong J., Wong V.W.S., Wong G.L.H., Chan S.L., Mok T.S.K., Chan H.L.Y. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam W.K.J., Jiang P., Chan K.C.A., Cheng S.H., Zhang H., Peng W., Tse O.Y.O., Tong Y.K., Gai W., Zee B.C.Y. Sequencing-based counting and size profiling of plasma Epstein-Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA. 2018;115:E5115–E5124. doi: 10.1073/pnas.1804184115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouliere F., Chandrananda D., Piskorz A.M., Moore E.K., Morris J., Ahlborn L.B., Mair R., Goranova T., Marass F., Heider K. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018;10:eaat4921. doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamyatnin A.A. Fragmentomics of natural peptide structures. Biochemistry (Mosc.) 2009;74:1575–1585. doi: 10.1134/s0006297909130100. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov M., Baranova A., Butler T., Spellman P., Mileyko V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics. 2015;16(Suppl 13):S1. doi: 10.1186/1471-2164-16-S13-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang P., Chan K.C.A., Lo Y.M.D. Liver-derived cell-free nucleic acids in plasma: Biology and applications in liquid biopsies. J. Hepatol. 2019;71:409–421. doi: 10.1016/j.jhep.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D., Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 16.Thierry A.R., El Messaoudi S., Gahan P.B., Anker P., Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrananda D., Thorne N.P., Bahlo M. High-resolution characterization of sequence signatures due to non-random cleavage of cell-free DNA. BMC Med. Genomics. 2015;8:29. doi: 10.1186/s12920-015-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang P., Sun K., Peng W., Cheng S.H., Ni M., Yeung P.C., Heung M.M.S., Xie T., Shang H., Zhou Z. Plasma DNA end motif profiling as a fragmentomic marker in cancer, pregnancy and transplantation. Cancer Discov. 2020;10:664–673. doi: 10.1158/2159-8290.CD-19-0622. [DOI] [PubMed] [Google Scholar]
- 19.Cheng T.H.T., Lui K.O., Peng X.L., Cheng S.H., Jiang P., Chan K.C.A., Chiu R.W.K., Lo Y.M.D. DNase1 does not appear to play a major role in the fragmentation of plasma DNA in a knockout mouse model. Clin. Chem. 2018;64:406–408. doi: 10.1373/clinchem.2017.280446. [DOI] [PubMed] [Google Scholar]
- 20.Serpas L., Chan R.W.Y., Jiang P., Ni M., Sun K., Rashidfarrokhi A., Soni C., Sisirak V., Lee W.S., Cheng S.H. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc. Natl. Acad. Sci. USA. 2019;116:641–649. doi: 10.1073/pnas.1815031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han D.S.C., Ni M., Chan R.W.Y., Chan V.W.H., Lui K.O., Chiu R.W.K., Lo Y.M.D. The biology of cell-free DNA fragmentation and the roles of DNASE1, DNASE1L3, and DFFB. Am. J. Hum. Genet. 2020;106:202–214. doi: 10.1016/j.ajhg.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiokawa D., Tanuma S. Characterization of human DNase I family endonucleases and activation of DNase γ during apoptosis. Biochemistry. 2001;40:143–152. doi: 10.1021/bi001041a. [DOI] [PubMed] [Google Scholar]
- 23.Keyel P.A. Dnases in health and disease. Dev. Biol. 2017;429:1–11. doi: 10.1016/j.ydbio.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiokawa D., Hirai M., Tanuma S. cDNA cloning of human DNase γ: chromosomal localization of its gene and enzymatic properties of recombinant protein. Apoptosis. 1998;3:89–95. doi: 10.1023/a:1009692807692. [DOI] [PubMed] [Google Scholar]
- 25.Liu X., Li P., Widlak P., Zou H., Luo X., Garrard W.T., Wang X. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisirak V., Sally B., D’Agati V., Martinez-Ortiz W., Özçakar Z.B., David J., Rashidfarrokhi A., Yeste A., Panea C., Chida A.S.S. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell. 2016;166:88–101. doi: 10.1016/j.cell.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozçakar Z.B., Foster J., 2nd, Diaz-Horta O., Kasapcopur O., Fan Y.S., Yalçınkaya F., Tekin M. DNASE1L3 mutations in hypocomplementemic urticarial vasculitis syndrome. Arthritis Rheum. 2013;65:2183–2189. doi: 10.1002/art.38010. [DOI] [PubMed] [Google Scholar]
- 28.Al-Mayouf S.M., Sunker A., Abdwani R., Abrawi S.A., Almurshedi F., Alhashmi N., Al Sonbul A., Sewairi W., Qari A., Abdallah E. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet. 2011;43:1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- 29.Carbonella A., Mancano G., Gremese E., Alkuraya F.S., Patel N., Gurrieri F., Ferraccioli G. An autosomal recessive DNASE1L3-related autoimmune disease with unusual clinical presentation mimicking systemic lupus erythematosus. Lupus. 2017;26:768–772. doi: 10.1177/0961203316676382. [DOI] [PubMed] [Google Scholar]
- 30.Batu E.D., Koşukcu C., Taşkıran E., Sahin S., Akman S., Sözeri B., Ünsal E., Bilginer Y., Kasapcopur O., Alikaşifoğlu M., Ozen S. Whole exome sequencing in early-onset systemic lupus erythematosus. J. Rheumatol. 2018;45:1671–1679. doi: 10.3899/jrheum.171358. [DOI] [PubMed] [Google Scholar]
- 31.Belot A., Rice G.I., Omarjee S.O., Rouchon Q., Smith E.M.D., Moreews M., Tusseau M., Frachette C., Bournhonesque R., Thielens N. Contribution of rare and predicted pathogenic gene variants to childhood-onset lupus: a large, genetic panel analysis of British and French cohorts. Lancet Rheumatol. 2020;2:e99–e109. doi: 10.1016/S2665-9913(19)30142-0. [DOI] [PubMed] [Google Scholar]
- 32.Davis M.D.P., Brewer J.D. Urticarial vasculitis and hypocomplementemic urticarial vasculitis syndrome. Immunol. Allergy Clin. North Am. 2004;24:183–213, vi. doi: 10.1016/j.iac.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Weisenburger T., von Neubeck B., Schneider A., Ebert N., Schreyer D., Acs A., Winkler T.H. Epistatic interactions between mutations of deoxyribonuclease 1-Like 3 and the inhibitory Fc gamma receptor IIB result in very early and massive autoantibodies against double-stranded DNA. Front. Immunol. 2018;9:1551. doi: 10.3389/fimmu.2018.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soni C., Perez O.A., Voss W.N., Pucella J.N., Serpas L., Mehl J., Ching K.L., Goike J., Georgiou G., Ippolito G.C. Plasmacytoid dendritic cells and type I interferon promote extrafollicular B cell responses to extracellular self-DNA. Immunity. 2020;52:1022–1038.e7. doi: 10.1016/j.immuni.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soni C., Reizis B. DNA as a self-antigen: nature and regulation. Curr. Opin. Immunol. 2018;55:31–37. doi: 10.1016/j.coi.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruschi M., Bonanni A., Petretto A., Vaglio A., Pratesi F., Santucci L., Migliorini P., Bertelli R., Galetti M., Belletti S. Neutrophil extracellular traps profiles in patients with incident systemic lupus erythematosus and lupus nephritis. J. Rheumatol. 2020;47:377–386. doi: 10.3899/jrheum.181232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu R.W.K., Poon L.L.M., Lau T.K., Leung T.N., Wong E.M.C., Lo Y.M.D. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin. Chem. 2001;47:1607–1613. [PubMed] [Google Scholar]
- 38.Nabbi A., Riabowol K. Rapid isolation of nuclei from cells in vitro. Cold Spring Harb. Protoc. 2015;2015:769–772. doi: 10.1101/pdb.prot083733. [DOI] [PubMed] [Google Scholar]
- 39.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 41.Napirei M., Wulf S., Eulitz D., Mannherz H.G., Kloeckl T. Comparative characterization of rat deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3 (Dnase1l3) Biochem. J. 2005;389:355–364. doi: 10.1042/BJ20042124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong J., Luo K., Winter P.S., Crawford G.E., Iversen E.S., Hartemink A.J. Mapping nucleosome positions using DNase-seq. Genome Res. 2016;26:351–364. doi: 10.1101/gr.195602.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sands M.S. AAV-mediated liver-directed gene therapy. Methods Mol. Biol. 2011;807:141–157. doi: 10.1007/978-1-61779-370-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan R.W.Y., Jiang P., Peng X., Tam L.-S., Liao G.J.W., Li E.K.M., Wong P.C.H., Sun H., Chan K.C.A., Chiu R.W.K., Lo Y.M. Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc. Natl. Acad. Sci. USA. 2014;111:E5302–E5311. doi: 10.1073/pnas.1421126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lisowski L., Tay S.S., Alexander I.E. Adeno-associated virus serotypes for gene therapeutics. Curr. Opin. Pharmacol. 2015;24:59–67. doi: 10.1016/j.coph.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Pei X., He T., Hall N.E., Gerber D., Samulski R.J., Li C. AAV8 virions hijack serum proteins to increase hepatocyte binding for transduction enhancement. Virology. 2018;518:95–102. doi: 10.1016/j.virol.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequence data reported in this paper is at the European Genome-Phenome Archive (EGA): EGAS00001004342 and EGAS00001004409.