Summary
Variation in levels of the human metabolome reflect changes in homeostasis, providing a window into health and disease. The genetic impact on circulating metabolites in Hispanics, a population with high cardiometabolic disease burden, is largely unknown. We conducted genome-wide association analyses on 640 circulating metabolites in 3,926 Hispanic Community Health Study/Study of Latinos participants. The estimated heritability for 640 metabolites ranged between 0%–54% with a median at 2.5%. We discovered 46 variant-metabolite pairs (p value < 1.2 × 10−10, minor allele frequency ≥ 1%, proportion of variance explained [PEV] mean = 3.4%, PEVrange = 1%–22%) with generalized effects in two population-based studies and confirmed 301 known locus-metabolite associations. Half of the identified variants with generalized effect were located in genes, including five nonsynonymous variants. We identified co-localization with the expression quantitative trait loci at 105 discovered and 151 known loci-metabolites sets. rs5855544, upstream of SLC51A, was associated with higher levels of three steroid sulfates and co-localized with expression levels of SLC51A in several tissues. Mendelian randomization (MR) analysis identified several metabolites associated with coronary heart disease (CHD) and type 2 diabetes. For example, two variants located in or near CYP4F2 (rs2108622 and rs79400241, respectively), involved in vitamin E metabolism, were associated with the levels of octadecanedioate and vitamin E metabolites (gamma-CEHC and gamma-CEHC glucuronide); MR analysis showed that genetically high levels of these metabolites were associated with lower odds of CHD. Our findings document the genetic architecture of circulating metabolites in an underrepresented Hispanic/Latino community, shedding light on disease etiology.
Keywords: metabolomics, genome-wide association study, Hispanic/Latino community
Introduction
The metabolome is a complete set of small molecules (<1.5 kDa) in a biological sample, including biochemicals of cellular metabolism and xenobiotics from diet and environment.1,2 Metabolites play a critical role in various biological processes starting with the beginning of life,3 and variation in levels of the human metabolome reflects changes in homeostasis that provide a window into health and disease. Genome-wide association studies (GWASs) with metabolic traits have identified hundreds of genetic variants associated with levels of numerous metabolites in multiple biospecimens, including blood,4, 5, 6, 7, 8 urine,9, 10, 11 feces,12 and saliva.13 The resulted findings have illuminated mechanisms underlying human metabolism and provided insights relevant to common complex diseases.14, 15, 16 For example, in the early GWAS era, one single intronic variant, rs174548 (FADS1), was shown to affect serum glycerophospholipids, which were involved in cholesterol metabolism. Prior analyses showed that rs174548 was also associated with blood lipids—known cardio-vascular disease risk factors.14 In a recent report, sets of variants were shown to be associated with glycine levels, and genetically increased glycine levels were associated with decreased risk of coronary heart disease (CHD) potentially driven by blood pressure, providing insight in the causal pathways of CHD.15
Despite the success of previous GWAS discoveries, the genetic impact on circulating metabolites in Hispanics/Latinos, a population with unique genetic background17,18 and disproportionately high cardiometabolic disease burden,19 is largely unknown. To address this gap, we performed a GWAS investigating the effect of low frequency (1% ≤ minor allele frequency [MAF] ≤ 5%) and common genetic variants (MAF > 5%) on the circulating metabolites in 3,926 participants from the Hispanic Community Health Study/Study Of Latinos (HCHS/SOL) and replicated our findings by using a public database from TwinsUK and the data from the Atherosclerosis Risk In Communities (ARIC) study. We implemented co-localization and network analytical approaches on the identified loci to interpret the underlying biological processes. Our findings complement the catalog of genetic loci associated with metabolites with a focus on an underrepresented Hispanic/Latino population and provide candidate pathways for future research to illuminate underlying mechanisms for cardiometabolic diseases.
Subjects and Methods
Study Populations
HCHS/SOL
A comprehensive description of the community-based cohort study HCHS/SOL has previously been published.20 Participants 18–74 years of age at their baseline examination were recruited through a stratified multistage area probability sample design from four communities (San Diego, California; Chicago, Illinois; The Bronx, New York, New York; and Miami, Florida). Overall, 16,415 participants, representing major self-reported US Hispanic/Latino background groups (Mexicans, Puerto Ricans, Cubans, Central Americans, Dominicans, and South Americans), took part in baseline examinations that occurred in June 2008–July 2011. Second visits were performed in 2014–2017, and the completion of third visits began in 2020 and is ongoing. A random subset of 3,926 participants with available genetic and metabolomic measures from their first visit were included in the present analysis. The HCHS/SOL study was approved by institutional review boards at participating institutions. Written informed consent was obtained from all participants.
ARIC
The ARIC study is a prospective cohort study with 15,792 participants (73%, European Americans [EAs]; 27%, African Americans) from four US communities (Forsyth County, North Carolina; Jackson, Mississippi; Minneapolis, Minnesota; and Washington County, Maryland); participants were 45 to 64 years of age at the baseline examination (1987–1989).21 Six follow-up visits were performed in 1990–92 (visit 2), 1993–95 (visit 3), 1996–98 (visit 4), 2011–2013 (visit 5), 2016–2017 (visit 6), and 2018–2019 (visit 7). We performed replication in 1,509 ARIC EAs from the baseline examination with available genetic and metabolomics measures in the present analysis. The ARIC study was approved by the institutional review boards at each ARIC center, and participants provided informed consent.
Metabolite Measurements
In HCHS/SOL and ARIC, fasting blood samples were collected, processed, and stored at −70°C since collection. Stored serum samples at the HCHS/SOL and ARIC’s baseline examination were used for metabolomic profiling. The metabolomic profiling was conducted at Metabolon (Durham, NC) with Discovery HD4 platform in 2017 for HCHS/SOL and Discovery HD3 platform in 2014 for ARIC. Serum metabolites were quantified with untargeted, liquid chromatography-mass spectrometry (LC-MS)-based quantification protocol.22,23 In HCHS/SOL, the platform captured a total of 1,136 metabolites, including 782 known and 354 unknown metabolites. In ARIC, the platform captured a total of 1,160 metabolites, including 787 known and 373 unknown metabolites. For quality control and better understanding of the biological mechanisms underlying disease etiology, only known metabolites with missing rates ≤25% were considered in this study, including 640 metabolites in HCHS/SOL and 635 metabolites in ARIC.
In TwinsUK, the metabolites profiling was also performed at Metabolon via a platform consisting of four independent ultra-high-performance liquid chromatography–tandem mass spectrometry (UPLC–MS/MS) instruments (detailed description is available elsewhere8). A total of 901 metabolites were identified and quantified; 644 metabolites were deemed stable due to having consistent levels across three longitudinal data collections and taken forward for the association analysis.
Genotyping, Quality Control, and Imputation
The HCHS/SOL participants were genotyped on HCHS/SOL custom 15041502 B3 Illumina array, which includes Illumina Omni 2.5M array (HumanOmni2.5-8v1-1) and 150,000 custom variants.18 Quality metrics used to filter variants for the imputation basis and association testing included missing call rate (>2%), Mendelian errors (>3 in 1,343 trios or duos), duplicate-sample discordance (>2 in 291 sample pairs), and deviation from Hardy-Weinberg equilibrium (p < 1 × 10−5).18 Pre-phasing was performed with SHAPEIT2 (v.2.r644) and imputation was performed with IMPUTE2 (v.2.3.0)24 with the 1000 Genomes Project phase III reference panel. A total of 13,039,987 variants passed quality filters with MAF ≥ 1% and imputation quality (Rsq) ≥ 0.3 and were carried forward for the association analyses.
For ARIC, genotype data was completed via Affymetrix Array 6.0. Quality metrics used to filter variants for the imputation basis and association testing included sex mismatch, discordance with previously-genotyped markers, first-degree relative of an included individual, and genetic outlier based on allele sharing and principal-component analyses, insufficient call rate (>5%), and deviation from Hardy-Weinberg equilibrium (p < 1 × 10−5).25 Imputation was performed on the data that passed quality filters in two steps: (1) pre-phasing with ShapeIt (v1.r532) and (2) imputation with IMPUTE2 with the 1000 Genomes Project phase I reference panel.25 A total of 38,038,545 variants had Rsq ≥ 0.3 and were considered in the replication analyses for which MAF-based variant exclusion was not performed.
For both HCHS/SOL and ARIC, population structure was estimated with principal-component analysis as implemented in EIGENSTRAT.26 We considered only autosomal variants.
For TwinsUK, whole-genome sequencing was performed on an Illumina HiSeqX sequencer with a 150-base-paired-end single-index-read format. Data from twins with European descent (verified with ADMIXTURE) was used for genomic data analysis. Genomes with a ratio of heterozygous to homozygous variants >2.5 were excluded. Overall, 1,960 subjects (383 monozygotic twins and 522 dizygotic twins) passed quality control and association tests were performed on ∼11,350,000 variants (further details on sample preparation and quality control are available elsewhere8).
Statistical Analysis
Genotype-Phenotype Analyses
In order to address any deviation of metabolites from normality, we applied a two-stage procedure for rank normalization in genotype-metabolite association analyses, which was shown to be superior to reduce type I errors and to improve statistical power.27 We analyzed a total of 640 (Table S1) known metabolites in HCHS/SOL, and missing values for the included metabolites were imputed to half of the lowest value in the present analysis.28 For each of the 640 metabolites, residuals were first obtained by regressing age, sex, estimated glomerular filtration rate (eGFR),29 recruitment center, the first five principal components, and genetic analysis group by origin (Cuban, Dominican, Puerto Rican, Mexican, Central American, and South American); residuals were then inverse normal transformed to be used in the genetic analyses adjusting the same aforementioned covariates. In addition, we used three random effects terms18 to account for genetic relatedness (kinship) and environmental correlations (shared household and census block group) by using a linear mixed-effect model. Overall, 13,039,987 variants (MAF ≥ 1% and Rsq ≥ 0.3) were analyzed individually with each metabolite. Analyses were performed in GMMAT30 with additive genetic models.
Out of 640 metabolites, we identified 333 independent metabolites; the independence was defined as pairwise Pearson correlation coefficient, r ≤ 0.7.31 The remaining 307 metabolites were grouped into 75 metabolite sets (Table S2), and all metabolites in one set correlated with at least one other metabolite in the same set (r > 0.7); that is, metabolites within a particular set did not correlate (r > 0.7) with any metabolites outside of the set. Significance for single variant analysis was defined as p value ≤ 1.23 × 10−10 (accounting for ∼1,000,000 independent variants and 408 independent metabolite sets [including 333 independent metabolites and 75 metabolite sets]).
We obtained the heritability of each of the 640 metabolites based on the genotype-metabolite summary statistics by using LDSC v1.0.0.32 For each metabolite, summary statistics were used to perform single-trait linkage disequilibrium (LD) score regression with genomic control correction with genetic information only from 3,289 unrelated individuals (kinship coefficient > 0.022).18 The slope of the LD score regression was used to estimate the heritability explained by all variants used in the LD score estimation.32
Conditional Analysis
Among the metabolites and metabolite sets that reached genome-wide significance, we identified 497 genetic locus-metabolite pairs containing statistically significant variants (Supplemental Methods and Table S3). There were 462 out of 497 identified genetic locus-metabolite pairs that had more than one statistically significant variant. Therefore, we applied genome-wide complex trait analysis (GCTA v1.91.4)33 to identify independent genetic variants within each of the selected regions. Variants that were both statistically significant in the primary analysis (p value ≤ 1.23 × 10−10) and genome-wide statistically significant (p valueconditional < 5 × 10−8) within the GCTA joint model were considered conditionally independent associations. Overall, we identified 608 conditionally independent variant-metabolite associations (Tables S4 and S5).
To annotate the identified 608 independent variant-metabolite associations, we obtained reports from the Metabolomic GWAS Server,4 TwinsUK study,8 GWAS Catalog, GRASP Search, PhenoScanner, and our previous reports5, 6, 7 and performed manual search through published papers to detect known loci that overlap with our findings. If a variant belonging to a region-metabolite pair was previously associated with any of the metabolites in its respective metabolite set, the region-metabolite pair was considered known, and otherwise, it was considered previously unreported.
Replication Analysis
We conducted replication analysis in ARIC EAs and additionally used TwinsUK (up to 1,960 twins of European ancestry, mostly female, recruited as volunteers by media campaigns8) published summary statistics. In the replication populations, 107 variant-metabolite associations were represented in 1,509 ARIC EAs and 73 metabolites were present in the TwinsUK dataset.8 Although the TwinsUK dataset only provided variant-metabolite associations with p value < 1 × 10−5 (i.e., 24 pairs were available), we assumed that the dataset contained all 95 variant-metabolite pairs to minimize type I error rate. Because discovery was performed in a Hispanic/Latino population, whereas replication was sought in individuals of European Ancestry, we estimated the generalized effect across these two populations. Significant generalized effects were defined as follows: (1) had p value ≤ 2.48 × 10−4 in either of the European Ancestry datasets (accounting for 202 associations; 95 variant-metabolite pairs in TwinsUK and 107 variant-metabolite pairs in ARIC EA, and 53 potential pairs were represented in both datasets; Table S5) and (2) had consistent direction of effect in both discovery and replication dataset(s).
For variant-metabolite association with generalized effect, we obtained information from publicly available data by using PhenoScanner34 to identify diseases, intermediate phenotypes, expression quantitative trait loci (eQTLs), protein quantitative trait loci (pQTLs), and DNA methylation quantitative trait loci (methQTLs) associated with the previously unreported variants (Tables S6–S9). Results were filtered to include associations reaching a widely used genome-wide significance threshold p value < 5 × 10−8.
Co-localization of Metabolites with eQTLs
To evaluate whether the identified 497 genetic locus-metabolite associations share genetic loci with any gene expression levels, we performed co-localization analysis with gene eQTLs summary by using HyPrColoc.35 HyPrColoc can identify subsets of traits co-localizing at distinct causal variants in the genomic locus.35 All metabolites associated with variants belonging to the same genetic locus were analyzed simultaneously. Analysis was performed on variants present in both datasets, taking into account all 48 tissues available in GTEx V736,37 (prior structure, = 0.0001, = 0.98, Table S10).
Locus-Specific Investigations
We investigated associations between selected identified variants with generalized effect and clinical outcomes of interest in HCHS/SOL and ARIC EAs (Supplemental Methods). To assess the association between rs2328895, N-acetyltryptophan, and prevalent CHD, as well as between rs324420, N-oleoyltaurine, and alcohol and tobacco use, we used general linear models with binomial link (R packages survey v3.35.1 in HCHS/SOL and stats v3.5.2 in ARIC). To assess the relationship between the N-acetyltryptophan and rs2328895 and incident CHD and heart failure (HF) in ARIC, we used Cox proportional hazards models as implemented in R package survival v2.43.3. A landmark analysis approach was applied to investigate the relationship between rs2328895 and incident CHD in older ARIC EAs.38
Assessing Tissue Specificity of Metabolites
To assess the tissue specificity of genes associated with those metabolites, we first calculated gene-based p values from the genotype-metabolite summary statistics for each of the 39 metabolites, which were associated with previously unreported findings with generalized effect. Specifically, we mapped variants to genes if they are located in the gene body or 50 kb upstream/downstream of the gene. We used the method Pascal to calculate a sum of chi-square statistics for each gene, requiring a gene to have at least two variants for the analyses.39 The gene-based score was adjusted for the gene length and the local LD, which was estimated with the admixed American (AMR) population from the 1000 Genomes Project phase I.40 For each metabolite, we defined the associated genes as those with gene p value from Pascal less than 5.79 × 10−8 (0.05/39 metabolites/22,129 genes) (Table S11). To identify in which tissues the metabolite-associated genes were most specifically expressed, we conducted tissue-specific enrichment analysis by using the tool deTS.41 deTS uses Fisher’s exact test to assess whether a query list of genes (e.g., metabolite-associated genes) is overrepresented with tissue-specific genes in a particular tissue. For the deTS analyses, we used 47 tissues from GTEx V7, after exclusion of EBV cells and tissues with <30 samples. The p value from the enrichment test was further adjusted by the Benjamini-Hochberg (BH) method for 39 metabolites.42
Pathway Analysis
We conducted a network-based analysis to identify modules of genes whose combined effect was overrepresented in GWASs by using the previously developed dense module searching for GWASs (dmGWAS, version 2.7).43 Specifically, we performed our analysis by using two protein-protein interaction networks: one from PathwayCommons (PC)44 (16,332 genes and 369,895 interactions) and the other from in-house curation with 21,137 genes and 413,492 interactions. In brief, dmGWAS implements a dense module searching algorithm to search for modules in the reference network with a goal of achieving a maximum module score.43 For each metabolite, the top ten most significant modules were identified (Table S12) and combined as metabolite-related subnetworks. We also performed gene set enrichment analysis by using DAVID45 to assess the functions of the subnetworks. Gene Ontology biological process (GOBP), KEGG, and BIOCARTA (Table S13) were examined. Significant gene sets were defined as those with false discovery rate (FDR) < 0.05 via the BH method.
Mendelian Randomization
For each of the previously unreported variants with generalized effect, we performed a lookup in the cardiovascular disease knowledge portal (Web Resources) for the GWAS associations with disease (p valueGWAS < 4.07 × 10−4, accounting for 41 variants and three diseases: CHD-CARDIoGRAMplusC4D, HF-HERMES, and type 2 diabetes [T2D]-DIAGRAM). We then conducted two-sample Mendelian randomization (MR) analyses to detect any potential causal effects of the levels of these metabolites and aforementioned diseases. For each selected metabolite, we considered all statistically independent variants (Tables S4 and S5). For each independent variant, we obtained a causal estimate as the ratio of the association of the variant with disease with published GWAS summary statistics46, 47, 48 to the association of the variant with metabolite with the summary statistics generated in the present study. If a metabolite was associated with more than one independent variant, we performed a fixed effect inverse variance meta-analysis by using R package meta v4.10-0 to obtain the overall estimates. Associations with p valueMR < 1.23 × 10−3 (accounting for 39 metabolites) were considered statistically significant.
Results
Genotype-Phenotype Analysis
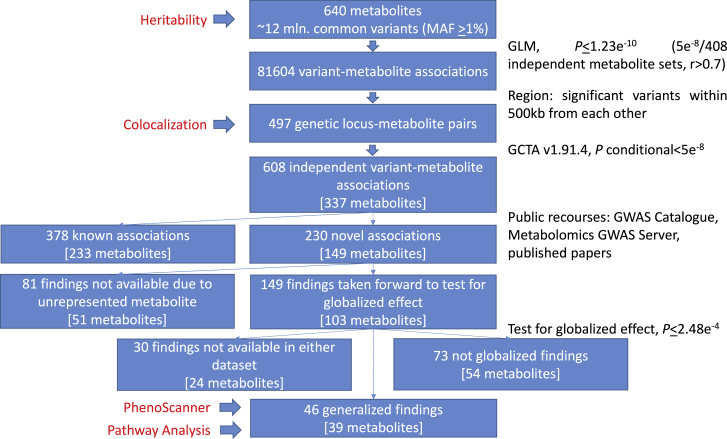
We performed a GWAS of genotyped and imputed variants with 640 circulating metabolites in 3,926 HCHS/SOL participants with mean age at 46 years old and 43% males. The demographics of study participants and the biochemical name and distribution for each metabolite are summarized in Table S1. Our study design, statistical and functional analyses performed, and an overview of the known and previously unreported findings are presented in Figure 1.
Figure 1.
Study Design
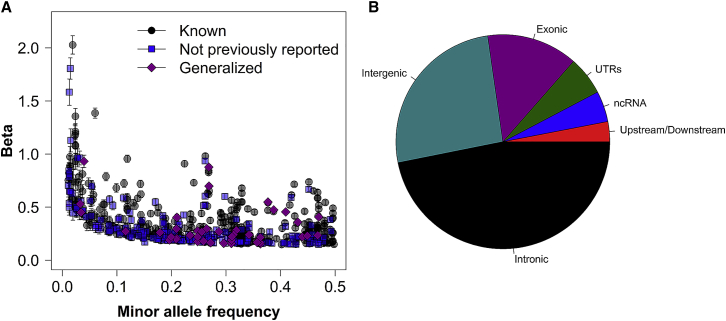
We identified 81,604 variant-metabolite associations reaching the p value ≤ 1.23 × 10−10. Among these, 608 significant single variant-metabolite pairs were independent (p value ≤ 1.23 × 10−10, p valueconditional ≤ 5 × 10−8), including 429 unique variants (∼12% were low frequency, 1% ≤ MAF ≤ 5%) with an average variance explained at 3.5%, where 378 pairs were known and 230 pairs were previously unreported. As expected, assessed low-frequency variants had a 2.5× larger effect on metabolites levels compared to common variants (mean effect at 0.77 SD and 0.30 SD change per minor allele for low-frequency and common variants, respectively, Figure 2A). Around 60% of detected variants belonged to genes, harboring 14% exonic variants (Figure 2B).
Figure 2.
Known and Previously Unreported Independent Associations
(A) Minor allele frequency against absolute effect estimates for 608 variant-metabolite associations with 95% confidence intervals.
(B) Functional consequences of 429 unique variants associated with metabolites levels.
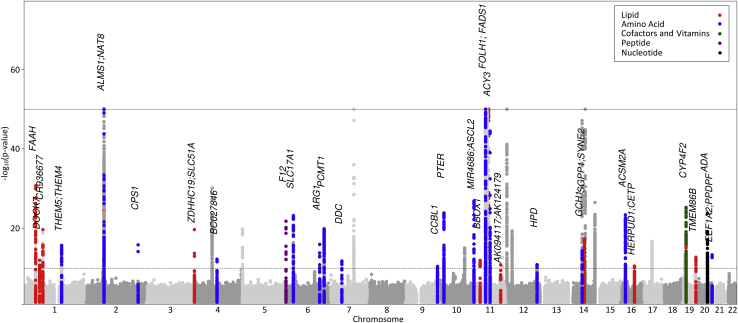
Among 378 known variant-metabolite pairs, we reproduced 171 previously reported variant-metabolite associations; 207 additional independent variant-metabolite associations were located within the known metabolite loci (Table S4). In the remaining 230 previously unreported variant-metabolite pairs, 46 pairs of 41 unique variants and 39 metabolites were successfully replicated in the ARIC and/or TwinsUK study with generalized effects (Figure 3), and the explained variances ranged from 1% to 22% (Table 1). The strongest associations were detected for N-acetyl amino acids at NAT8 (N2-acetyllysine, p value = 5.98 × 10−252; N-acetylleucine, p value = 5.66 × 10−160) and TPRKB (N-acetylarginine, p value = 3.46 × 10−123). Six loci (NAT8, FOLH1, ACY3, CPS1, SLC51A, and UCA1-CYP4F2) affected more than one metabolite. Among the 46 previously unreported pairs with generalized effects, we report genetic effect on twelve metabolites and six loci (PCMT1, PTER, FOLH1, TMEM86B, and EEF1A2), which were not shown previously to be associated with any other human metabolites.
Figure 3.
Manhattan Plot of 39 Metabolites Associated with 46 Previously Unreported Findings with Generalized Effect
For each of the corresponding super-pathways, a color scheme for the previously unreported loci with generalized effect is represented in the legend, whereas all other signals are shown in gray.
Table 1.
Previously Unreported Independent Statistically Significant Single Variant-Metabolite Associations with Generalized Effect in HCHS/SOL, ARIC, and TwinsUK
| Phenotype |
Variant Information |
HCHS/SOL (n = 3,926) |
ARIC (n = 1,509) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Gene | Consequence | M/O | MAF | Beta | p Value | MAF | Beta | p Value | |
| N-oleoyltaurinea | rs324420 | FAAH | nonsynonymous | A/C | 0.319 | 0.28 | 2.05E−31 | 0.202 | 0.19 | 5.84E−05 |
| Caprylate (8:0) | rs12760091 | CR936677 | ncRNA_intronic | C/T | 0.44 | −0.22 | 2.35E−20 | 0.699 | −0.22 | 1.03E−05 |
| Indoleacetylglutamine | rs7541453 | THEM5; THEM4 | intergenic | C/T | 0.36 | 0.18 | 2.21E−16 | 0.407 | 0.21 | 3.51E−08 |
| N-acetylleucinea | rs28879089 | ALMS1 | intronic | A/G | 0.269 | 0.70 | 5.66E−160 | 0.241 | 0.55 | 2.48E−40 |
| N2-acetyllysine | rs13409366 | ALMS1; NAT8 | intergenic | G/A | 0.268 | 0.88 | 5.98E−252 | 0.241 | 1.02 | 6.42E−162 |
| 2-hydroxyoctanoate | rs111540621 | ALMS1; NAT8 | intergenic | G/C | 0.264 | −0.30 | 2.08E−30 | 0.232 | −0.35 | 2.94E−16 |
| N2-acetyllysine | rs10200762 | ALMS1P; NAT8B | intergenic | T/C | 0.387 | 0.47 | 5.93E−92 | 0.388 | 0.58 | 5.84E−59 |
| N-acetylargininea | rs12713794 | TPRKB | downstream | A/G | 0.377 | 0.55 | 3.46E−123 | 0.36 | 0.56 | 5.18E−54 |
| Isobutyrylglycinea | rs1047891 | CPS1 | nonsynonymous | A/C | 0.31 | 0.16 | 6.98E−11 | 0.326 | 0.40 | 1.66E−22 |
| Isovalerylglycinea | 0.19 | 1.58E−16 | 0.326 | 0.20 | 8.86E−07 | |||||
| Androsterone sulfate | rs376277540 | ZDHHC19; SLC51A | intergenic | T/TG | 0.363 | 0.16 | 6.09E−11 | 0.422 | 0.17 | 1.80E−05 |
| Epiandrosterone sulfate | 0.16 | 6.36E−11 | 0.422 | 0.18 | 3.50E−06 | |||||
| 16a-hydroxy DHEA 3 -sulfate | 0.22 | 2.29E−20 | 0.422 | 0.15 | 0.000115 | |||||
| Tiglylcarnitine (C5:1-DC) | rs7678928 | BC027846 | ncRNA_intronic | T/C | 0.362 | −0.17 | 6.75E−13 | 0.462 | −0.15 | 6.41E−05 |
| Methionine sulfone | rs151067661 | BC032469; SLC6A19 | intergenic | C/inc | 0.31 | 0.22 | 1.68E−20 | 0.286 | 0.17 | 3.49E−05 |
| Leucylglycinea | rs1801020 | F12 | UTR5 | A/G | 0.342 | −0.24 | 1.74E−22 | 0.253 | −0.23 | 1.68E−05 |
| N-acetyltryptophana | rs2328895 | SLC17A1 | intronic | C/T | 0.314 | −0.24 | 6.60E−24 | 0.445 | −0.14 | 0.000137 |
| S-adenosylhomocysteinea | rs2095375b | PCMT1 | intronic | C/A | 0.363 | −0.22 | 1.43E−20 | 0.637 | −0.21 | 2.21E−08 |
| 3-methoxytyrosine | rs57835901 | DDC | intronic | A/T | 0.034 | 0.45 | 1.88E−12 | 0.015 | 0.77 | 3.29E−07 |
| N-acetyltaurine | rs6602116b | PTER | intronic | G/A | 0.449 | 0.23 | 1.44E−24 | 0.535 | 0.16 | 2.28E−05 |
| Deoxycarnitine | rs2304913 | BBOX1 | intronic | C/T | 0.034 | 0.45 | 1.33E−12 | 0.044 | 0.43 | 1.26E−06 |
| N-acetyl-aspartyl-glutamatea | rs4929895b | FOLH1 | intronic | G/A | 0.409 | −0.46 | 3.30E−84 | 0.419 | −0.39 | 6.10E−25 |
| N-acetyl-aspartyl-glutamatea | rs61885293b | FOLH1; LOC440040 | intergenic | C/A | 0.04 | −0.93 | 3.99E−58 | 0.048 | −1.01 | 5.73E−34 |
| N-acetyl-aspartyl-glutamatea | rs9704992 | TRIM48 | intergenic | T/C | 0.432 | −0.36 | 6.78E−55 | 0.531 | −0.32 | 1.03E−13 |
| N-acetyltryptophana | rs2290958 | ACY3 | intronic | T/C | 0.21 | −0.40 | 3.28E−45 | 0.178 | −0.47 | 4.05E−20 |
| N-acetylkynurenine (2)a | rs948445 | ACY3 | nonsynonymous | C/T | 0.209 | −0.21 | 4.72E−13 | 0.18 | −0.26 | 1.53E−07 |
| 3-methoxytyramine sulfatea | rs7141433 | GCH1 | intronic | T/C | 0.116 | −0.27 | 4.82E−15 | 0.125 | −0.27 | 8.04E−07 |
| Indoleacetylglutamine | rs146233716 | ACSM2A | nonsynonymous | A/G | 0.179 | −0.26 | 1.89E−17 | 0.246 | −0.30 | 9.43E−09 |
| Gamma-CEHC | rs79400241 | UCA1; CYP4F2 | intergenic | G/C | 0.248 | −0.28 | 5.93E−26 | 0.279 | −0.24 | 4.81E−09 |
| Gamma-CEHC glucuronidea | −0.18 | 1.34E−15 | 0.279 | −0.31 | 2.01E−14 | |||||
| Phenotype | Variant Information | HCHS/SOL (n = 3,926) | TwinsUK (n = 1,293–1,959) | |||||||
| 1-stearoyl-2-arachidonoyl-GPI (18:0/20:4) | rs10889335 | DOCK7 | synonymous | G/A | 0.357 | −0.17 | 1.19E−12 | 0.377 | −0.24 | 1.71E−08 |
| Dopamine 3-O-sulfate | rs7129483 | MIR4686; ASCL2 | intergenic | T/C | 0.287 | 0.27 | 1.52E−27 | 0.236 | 0.18 | 2.84E−06 |
| 1-arachidonylglycerol (20:4) | rs102274 | TMEM258 | intronic | C/T | 0.47 | −0.41 | 7.52E−62 | 0.354 | −0.25 | 3.13E−10 |
| 1-palmitoyl-2-stearoyl-GPC (16:0/18:0) | rs174554 | FADS1 | intronic | G/A | 0.468 | −0.24 | 4.76E−21 | 0.354 | −0.39 | 4.90E−08 |
| Palmitoyl dihydrosphingomyelin (d18:0/16:0) | rs8008068 | SGPP1; SYNE2 | intergenic | G/A | 0.234 | −0.24 | 5.09E−18 | 0.152 | −0.22 | 1.18E−08 |
| Indoleacetylglutamine | rs7185111 | ACSM2A | intronic | G/C | 0.311 | 0.22 | 4.30E−24 | 0.328 | 0.27 | 3.70E−09 |
| 1-(1-enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0) | rs247617 | HERPUD1; CETP | intergenic | A/C | 0.296 | 0.16 | 3.57E−11 | 0.333 | 0.14 | 4.31E−07 |
| 1-(1-enyl-palmitoyl)-2-arachidonoyl-GPE (P-16:0/20:4) | rs3826884b | TMEM86B | UTR3 | A/G | 0.208 | −0.20 | 2.16E−13 | 0.171 | −0.19 | 2.52E−06 |
| Arginine | rs17788484 | ARG1 | UTR5 | T/C | 0.033 | −0.53 | 1.23E−16 | 0.017 | −0.73 | 5.44E−07 |
| 2-hydroxyphenylacetate | rs7849982 | CCBL1 | intronic | T/C | 0.272 | −0.17 | 4.40E−11 | 0.261 | −0.23 | 4.79E−08 |
| N-acetyltyrosine | rs948445 | ACY3 | nonsynonymous | C/T | 0.209 | −0.30 | 3.15E−24 | 0.19 | −0.17 | 1.99E−07 |
| Taurocholenate sulfate | rs10488763 | AK094117; AK124179 | ncRNA_intronic | T/A | 0.259 | −0.18 | 1.74E−11 | 0.131 | −0.28 | 1.84E−06 |
| 2-hydroxyphenylacetate | rs11614623 | HPD | intronic | T/C | 0.196 | −0.19 | 1.56E−11 | 0.114 | −0.28 | 1.87E−07 |
| Octadecanedioate | rs2108622 | CYP4F2 | nonsynonymous | T/C | 0.248 | −0.22 | 6.12E−17 | 0.308 | −0.21 | 1.50E−06 |
| N1-methyladenosine | rs406383 | ADA | intronic | G/C | 0.256 | −0.27 | 1.75E−24 | 0.231 | −0.19 | 3.64E−06 |
| Guanidinoacetate | rs2314639b | EEF1A2; PPDPF | intergenic | T/C | 0.199 | 0.22 | 5.37E−14 | 0.141 | 0.25 | 6.45E−06 |
RsID, reference SNP ID; MA/OA, minor allele/other allele; MAF, minor allele frequency; n, number of participants.
Metabolite has never been reported to be statistically significantly associated with genetic variants.
Variant belongs to a locus that has never been reported to be associated with other human metabolome metabolites.
Out of the 640 metabolites considered in this study, 366 metabolites had a positive estimated heritability (h2), with the mean at 0.111, ranging from 0 to 0.539 in HCHS/SOL participants (Table S14); heritability estimates and their summary by super-pathway are presented in Figure S2. Heritability explained >20% of inter-individual variation in 50 metabolites, and amino-acid-related metabolites, such as N-acetyl-asparagine (h2 = 0.539) and N-acetyl-aspartyl-glutamate (h2 = 0.464), tended to have the highest heritability estimates.
Co-localization of Metabolites with eQTLs
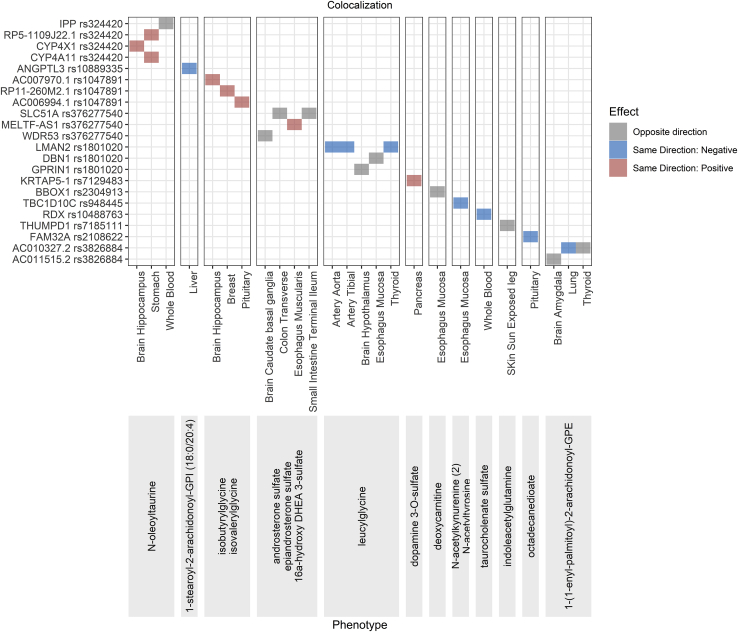
To better describe the potential genetic contribution of the identified GWAS loci, we performed co-localization analysis on 158 metabolite loci with gene expression in GTEx V7 (Table S10) to understand whether they might also affect gene expression levels in various tissues. Variants belonging to 78 genetic loci (136 variants, 339 variant-metabolite pairs) were found to have evidence of co-localization (posterior probability, PPr > 0.6). There were 54 loci where a single potential causal variant underlies both the expression of a single gene and the metabolite(s). Twelve previously unreported independent variants with generalized effect were identified as co-localizing for the association with 45 metabolites and 22 gene eQTLs in various tissues (Figure 4), suggesting that the expression of these genes may be responsible for the variation of metabolite levels in this locus. Among 22 previously unreported generalized variant-gene eQTL pairs, 41% was attributed to four nonsynonymous variants (rs324420-FAAH, rs1047891-CPS1, rs948445-ACY3, and rs2108622-CYP4F2), and two intronic variants were co-localizing with the same gene to which they were annotated (rs376277540-SLC51A and rs2304913-BBOX1).
Figure 4.
Previously Unreported Variant-Metabolite Pairs with Generalized Effect Co-localizing for the Association with Gene eQTLs
The direction of the effect of the minor allele on metabolite levels and gene expression is shown in the legend.
Functional Annotation of GWAS Loci
We further used PhenoScanner34 to determine whether 41 previously unreported independent variants with generalized effect detected in the current study were also associated with other traits and diseases to improve the functional annotation of our GWAS loci. Twelve variants were genome-wide significant for 65 phenotypes (Figure S3), including blood lipids levels, blood cell characteristics, factors related to coagulation, and various diseases, such as gout, asthma, and chronic kidney disease (Table S7). We correlated the metabolites with those reported phenotypes that are available in the HCHS/SOL study and found 24 significant metabolite-phenotype associations, including eight metabolite-lipid and three metabolite-blood cell characteristics (p valuecor < 7.81 × 10−4, accounting for 64 tests, Table S15).
Assessing Tissue Specificity of Metabolites
To improve biological insight of our findings that were previously unreported, we assessed tissue specificity of metabolites. Pascal analysis identified 376 statistically significant gene-metabolite pairs, including 313 pairs located in loci containing previously unreported variants with generalized effect (Table S11). For the tissue-specific enrichment analysis, top “one” ranked tissue-metabolite pairs included nine liver-metabolite pairs and three kidney-metabolite pairs (Figure S4).
Pathway Analysis
In genetic loci containing previously unreported variants with generalized effect, the top ten significant modules of both protein-protein interaction networks identified included 70 gene-metabolite pairs, where 41 gene-metabolite pairs were also selected for DAVID modules (metabolite-specific FDR < 0.05), including three genes to which three variant-metabolite pairs with generalized effect were annotated (Tables S11–S13). Among the latter, two genes were selected by both approaches in several identical pathways: the EEF1A2-guanidinoacetate pair is involved in regulation of metabolic process and transferase activity, whereas the F12-leucylglycine pair is involved in blood coagulation, according to KEGG and BIOCARTA.
Locus-Specific Investigations
Changes in metabolites levels may be reflecting the changes in homeostasis and, possibly, an underlying disease process; therefore, studying relationships between the identified variants, the respective metabolites levels, and the disease outcomes can contribute to our understanding of the latter. Out of 39 metabolites associated with variants with generalized effect, 12 were previously reported to be associated with various health conditions (Table S16).
We examined metabolites without known evidence for disease conditions. For example, a nonsynonymous variant, rs324420 (FAAH), was associated with increased levels of an endocannabinoid N-oleoyltaurine (MAF = 32%, b = 0.28). Fatty acid amide hydrolase (FAAH) plays role in drug addiction (MIM: 606581), which led us to selected smoking and drinking status as outcomes of interest. Each SD increase in N-oleoyltaurine was associated with lower odds of current smoking (odds ratio [OR] = 0.86, 95% confidence interval [CI] = 0.79–0.93) in 3,595 HCHS/SOL participants as well as with lower odds of current alcohol drinking (OR = 0.82, 95% CI = 0.71–0.93) in 3,925 ARIC EAs (p value < 0.006, Supplemental Methods). However, we did not observe statistically significant associations between rs324420 and smoking or drinking status (Table S17).
Intronic variant rs2328895 (SLC17A1) was associated with decreased N-acetyltryptophan in our study and was associated with decreased risk of self-reported gout in previous a GWAS (see “Rapid GWAS of Thousands of Phenotypes for 337,000 Samples in the UK BioBank” in Web Resources). Gout increases the risk of CHD and HF, therefore, we investigated both the variant and the metabolite with CHD and HF. Each SD increase in N-acetyltryptophan levels was associated with increased odds of prevalent CHD in both HCHS/SOL participants (OR = 1.24, 95% CI = 1.06–1.46) and ARIC EAs (OR = 1.75, 95% CI = 1.16–2.63) and with 231% increase in odds of prevalent (OR = 3.31, 95% CI = 2.12–5.16) and 58% increase risk of incident (HR = 1.58, 95% CI = 1.25–1.99) HF among ARIC EAs (Table S18). Rs2328895 was suggestively associated with decreased risk of incident CHD in ARIC EAs (p value = 0.09), where the risk-decreasing effect was predominantly seen in the elderlies (>60 years of age, HR = 0.86, 95% CI = 0.77–0.96, Table S18 and Figure S5).
Mendelian Randomization
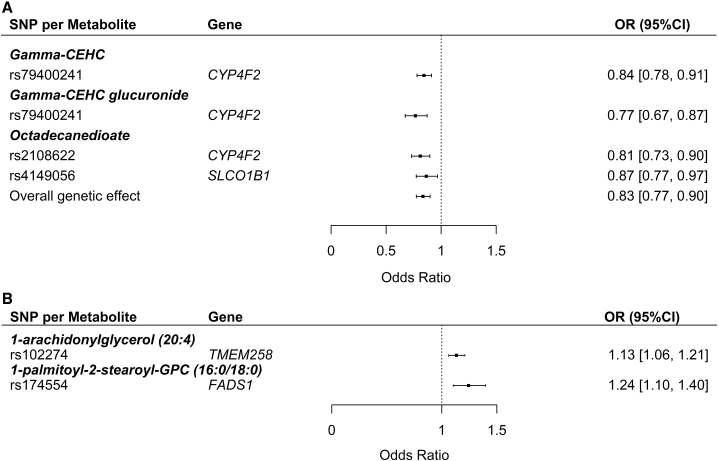
In addition to non-genetic observational analyses, we performed MR analyses to elucidate the possible causal pathways relevant for diseases of heart (CHD and HF) and diabetes mellitus (T2D), both of which are among the ten leading causes of death in US.49 Genetically regulated higher levels of five metabolites, which were not previously associated with any applicable health traits, including known risk factors for CHD and T2D (according to the human metabolome database, Table S16), had a statistically significant causal effect on CHD and T2D (Figures 5A and 5B and Table S19). We estimated that each SD increase in the levels of octadecanedioate, gamma-CEHC, and gamma-CEHC glucuronide was associated with 17% (OR = 0.83, 95% CI = 0.77–0.90), 16% (OR = 0.84, 95% CI = 0.78–0.91), and 23% (OR = 0.77, 95% CI = 0.67–0.87) lower odds of CHD, respectively. Additionally, each SD increase in 1-arachidonylglycerol (20:4) and 1-palmitoyl-2-stearoyl-GPC (16:0/18:0) was associated with increased odds of T2D (OR = 1.13, 95% CI = 1.06–1.21 and OR = 1.24, 95% CI = 1.10–1.40, respectively).
Figure 5.
Forest Plots for Mendelian Randomization Analysis Results
(A) Coronary heart disease.
(B) Type 2 diabetes.
Discussion
We conducted a GWAS based on 1000 Genomes panel to detect genetic loci associated with 640 circulating metabolites in a Hispanic population and identified 46 previously unreported generalized metabolite-genetic locus associations; seven associations were driven by nonsynonymous variants. Five loci harboring six variants with generalized effect have never been reported with circulating metabolites levels. We demonstrated that levels of five metabolites were genetically associated with the risk of CHD and/or T2D. Our study represents the GWAS of the metabolome in an underrepresented Hispanic/Latino community, which provides additional insights beyond previous GWASs in other ethnicities.
The 46 previously unreported generalized associations consist of metabolites from five super pathways, including amino acids, lipids, cofactors and vitamins, nucleotides, and peptides. To understand the potential function of those findings, we applied a series of complementary methods. By using co-localization analyses, we identified twelve unique loci where the previously unreported generalized variant co-localized with the eQTL for 22 unique genes in GTEx tissues, highlighting the biologically plausible genes. Previously unreported generalized variants in or near genes DOCK7, CPS1, F12, and CYP4F2, which co-localize with gene expression in seven specific tissues, are also associated with clinically relevant phenotypes. For example, synonymous DOCK7 variant rs10889335, associated with decreased levels of phosphatidylinositol 1-stearoyl-2-arachidonoyl-GPI (18:0/20:3) (HMDB09815), co-localized with decreased ANGPTL3 levels in the liver (Table S10). ANGPTL3 participates in lipid metabolism and causes familial hypobetalipoproteinemia type 2 (MIM: 605019); rs10889335 was previously associated with decreased level of triglycerides, low density lipoproteins, and total cholesterol.50
To generate new hypotheses, we explored the potential pathways by using knowledge-based approaches. A nonsynonymous variant, rs324420, belonging to a degrading enzyme of endocannabinoids, FAAH, was associated with increased levels of an endocannabinoid N-oleoyltaurine. rs324420 increases sensitivity of FAAH to proteolytic degradation51 and was previously associated with several other endocannabinoids.52 FAAH plays a role in drug addiction (MIM: 606581), smoking cessation in humans,53 and alcohol consumption in mice.54 We tested and identified that increased levels of N-oleoyltaurine were associated with lower odds of current smoking and drinking status, providing supporting evidence for the importance of this endocannabinoid in the addiction pathways (Table S17). Additionally, rs324420 was identified as a causal variant, underlying the effect on both expression of CYP4X1 (a cytochrome P450 family member participating in oxidation of endocannabinoids) in the hippocampus, a part of the endocannabinoid brain signaling system (Table S10),55, 56, 57 and it was associated with several endocannabinoids and derivatives, including palmitoylethanolamide, linoleoylethanolamide, N-oleoyltaurine, and N-oleoylserine. Although we did not observe statistical significance in both HCHS/SOL and ARIC for the associations between N-oleoyltaurine with drinking and smoking, the directionality of the effects was consistent for the N-oleoyltaurine-drinking association in both studies (Table S17). N-oleoyltaurine can be synthesized from taurine and oleic acid, both of which are common in the human diet. The lack of reproducibility of statistically significant associations might be due to the differences between the cohorts, such as culture and lifestyles, because taurine and oleic acid are mainly obtained from diet58 and it has been shown that N-oleoyltaurine can affect food intake in mice.59 We were also underpowered to detect the effect of current smoking observed in HCHS/SOL in 1,553 ARIC EAs. Further investigation is warranted to elucidate the relationship between FAAH, N-oleoyltaurine, and addiction behavior.
Intergenic variant rs376277540, located upstream of SLC51A, encoding a subunit of a bidirectional transporter for steroid-derived molecules, OSTa–OSTb, is associated with increased levels of three androgenic sulfated steroids (androsterone sulfate, epiandrosterone sulfate, and ahydroxy-DHEA3 sulfate).60 OSTa–OSTb can be inhibited by sulfate-conjugates of steroids in transport experiments,61 supporting the importance of this transporter in relation to the detected steroids. Additionally, rs376277540 co-localizes with decreased expression levels of SLC51A in transverse colon and terminal ileum (Table S10).
A previously unreported intronic variant rs2328895, belonging to SLC17A1 and encoding a sodium-dependent phosphate transport protein located in the proximal convoluted renal tubule, is associated with decreased N-acetyltryptophan levels. rs2328895 was previously reported to be associated with several anthropometric characteristics, blood cell traits,62 DNA methylation in whole blood, expression levels of several surrounding genes, and with decreased risk for disorders of mineral metabolism and self-reported gout (Figure S3 and Table S7)(see “Rapid GWAS of Thousands of Phenotypes for 337,000 Samples in the UK BioBank” in Web Resources). Gout, which is characterized by hyperuricemia, increases the risk of CHD63 and HF,64 and high N-acetyl-tryptophan levels were found to be associated with increased odds of prevalent CHD and HF and increased risk of HF (Table S18). Moreover, we observed that rs2328895 was associated with decreased risk of CHD in older ARIC participants. N-acetyltryptophan has not been previously reported for CHD and HF. It belongs to tryptophane metabolism65 with anti-oxidant properties.66 Increased urine levels of N-acetyltryptophan have recently been reported to be associated with hyperlipidemia.67 Interestingly, for the same genetic locus, DAVID analysis with both PPI and PC modules suggested enrichment of N-acetyltryptophan-related genes with chromatin assembly and organization, the latter playing a key role in determining cellular fate and identity (Table S13).68 Further studies are needed to confirm the roles of rs2328895 and N-acetyltryptophan in CHD and HF.
With the emerging large-scale GWAS discoveries of complex traits, the MR analyses have been widely used to explore the causal relationship between traits. We utilized three large GWAS summary statistics and identified two causal pathways for CHD and one causal pathway for T2D. Allele C of an intergenic variant, rs79400241, located downstream of CYP4F2 involved in catabolism of vitamin E,69 is associated with increased levels of gamma-CEHC and gamma-CEHC glucuronide. Both metabolites are vitamin E derivatives,70 and gamma-CEHC possesses antioxidant and natriuretic properties.71,72 C allele of a nonsynonymous variant, rs2108622, located in CYP4F2, is associated with increased levels of octadecanedioate. Increased octadecanedioate levels have previously been associated with decreased odds of preeclampsia.73 DAVID analysis with PPI and PC interaction networks suggested enrichment of octadecanedioate-related genes with apoptosis (Table S13). Our MR analyses show that increase in octadecanedioate, gamma-CEHC, and gamma-CEHC glucuronide levels are all associated with decreased odds of CHD (Table S19), suggesting that these metabolites might help to prevent CHD and improve CHD risk prediction. CYP4F2 disruption is known to lead to accumulation of vitamin E and to decreased production of CEHCs,74 therefore, our results are consistent with previous MR analyses showing that genetically regulated high levels of vitamin E are increasing the odds of CHD.75
Allele A of an intronic variant, rs174554 (FADS1, an inflammation initiation and resolution regulator involved in hepatic lipogenesis76), is associated with increased levels of 1-palmitoyl-2-stearoyl-GPC (16:0/18:0)—a phosphatidylcholine. FADS1 is thought to modulate the hepatic accumulation of phosphatidylcholines.77 Phosphatidylcholine species are suggested to play a role in insulin resistance,78 which is further supported by our MR results, according to which increase in 1-palmitoyl-2-stearoyl-GPC (16:0/18:0) is associated with increased odds of T2D (Table S19). Allele T of another intronic variant, rs102274 of TMEM258, a central regulator of intestinal inflammation and endoplasmic reticulum stress responses79 located in the same cluster of genes on chromosome 11, is associated with increased levels of 1-arachidonylglycerol (20:4). 1-arachidonylglycerol (20:4) is a derivative of arachidonic acid (which plays a key role in inflammation) and is also associated with increased odds of T2D in our MR analysis.
Furthermore, genes associated with several metabolites discussed above are most specifically expressed in liver, including androsterone sulfate, epiandrosterone sulfate, ahydroxy-DHEA3 sulfate, and palmitoyl-2-stearoyl-GPC (16:0/18:0) (concordant with the known metabolites synthesis organ), and more importantly, N-acetyltryptophan, octadecanedioate and gamma-CECH glucuronide (Figure S4), which do not have known tissue locations in the human metabolome database.80 Our findings pinpoint the biochemical synthesis “factory” in humans related to the latter metabolites.
Previous GWASs largely focus on European ancestry,4,8,9 and a few considered African ancestry.5, 6, 7 Our metabolite heritability estimates in Hispanics ranged from 0%–54%, which are comparable with other ancestries.4,81 We also reproduced 171 previously reported variant-metabolite associations and generalized 46 previously unreported variant-metabolite associations in Hispanics to European ancestry, implying that the genetic effects of many metabolites are consistent across ancestries. Out of 149 metabolites (forming 230 variant-metabolite pairs) associated with previously unreported variants and considered for replication, about half (73 metabolites and 107 variant-metabolite associations) were available for replication in ARIC EAs. 69 (50 metabolites and 63 variants) associations were available in ARIC EAs but were not replicated. Among these 63 variants, nine variants had imputation quality < 0.6 and six had MAF < 1.4% and would require a larger sample size for their replication. Future studies with a broad measure of metabolites and greater samples may provide additional insight into those associations that were not replicated in our study, especially given that some of the previously unreported findings that were not generalized in ARIC EAs are biologically plausible.
For example, intergenic variant rs12416738 was enriched in Hispanics/Latinos (HCHS/SOL MAF = 15%, ARIC EA MAF = 0.3%) and is associated with increased levels of several metabolites participating in arachidonate-phospholipid remodeling cycle (also known as CoA-independent transacylation system, involved in cell proliferation), including arachidonate, 1-arachidonoyl-GPC (20:4n6), 1-stearoyl-2-arachidonoyl-GPC (18:0/20:4), and 1-palmitoyl-2-arachidonoyl-GPC (16:0/20:4n6) (Table S5). Notably, no genes or corresponding enzymes responsible for the CoA-independent transacylation have been identified so far,82 making this finding a particularly interesting candidate for future follow-up. Prior GWASs suggest distinct genetic effects on complex traits in the Hispanic population.83 The lack of generalizability in the present study may be in part due to the unique Hispanic genetic architecture, which warrants future investigation.
In summary, our study demonstrated the genetic architecture of circulating metabolites in an underrepresented Hispanic/Latino community and improved the functional annotation of GWAS loci. This work was further strengthened by identification of causal relationships between selected genetically regulated metabolites and disease outcomes. Our results provide a unique resource and interesting insights for follow-up studies in basic science and clinical medicine to further unravel disease etiology.
Data and Code Availability
The genotype data used in this paper are available under accession number dbGaP: phs000880.v1.p1. The metabolomics data used in this paper are available on request through the HCHS/SOL web portal (Web Resources). We comply with NIH summary data sharing policy. The summary statistics are available from the corresponding author or Dr. Robert Kaplan (robert.kaplan@einsteinmed.org) on request.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by R01HL141824 (to B.Y.). B.Y. was supported in part by R01HL141824, R01HL142003, and the JLH Foundation (Houston, Texas). M.G. and E.B. were supported in part by the JLH Foundation (Houston, Texas). Q.Q. was supported by R01DK119268, R01DK120870, R01HL060712, and R01HL140976. The authors thank the staff and participants of the ARIC and HCHS/SOL studies for their important contributions. The ARIC study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I), R01HL087641, R01HL059367, and R01HL086694, National Human Genome Research Institute (NHGRI) contract U01HG004402, and NIH contract HHSN268200625226C. Infrastructure was partly supported by grant number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Metabolomics measurements were sponsored by NHGRI (3U01HG004402-02S1). The HCHS/SOL is a collaborative study supported by contracts from the NHLBI to the University of North Carolina (HHSN268201300001I / N01-HC-65233), University of Miami (HHSN268201300004I / N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I / N01-HC-65235), University of Illinois at Chicago (HHSN268201300003I / N01-HC-65236 Northwestern University), and San Diego State University (HHSN268201300005I / N01-HC-65237). The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Support for metabolomics data was graciously provided by the JLH Foundation (Houston, Texas).
Published: October 7, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.09.003.
Web Resources
Cardiovascular disease knowledge portal, http://broadcvdi.org/home/portalHome
HCHS/SOL web portal, https://sites.cscc.unc.edu/hchs/
“Rapid GWAS of Thousands of Phenotypes for 337,000 Samples in the UK BioBank,” http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank
Supplemental Information
and Supplemental Methods
References
- 1.Nicholson J.K., Lindon J.C. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 2.Ryals J., Lawton K., Stevens D., Milburn M. Metabolon, Inc. Pharmacogenomics. 2007;8:863–866. doi: 10.2217/14622416.8.7.863. [DOI] [PubMed] [Google Scholar]
- 3.Botros L., Sakkas D., Seli E. Metabolomics and its application for non-invasive embryo assessment in IVF. Mol. Hum. Reprod. 2008;14:679–690. doi: 10.1093/molehr/gan066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin S.-Y., Fauman E.B., Petersen A.-K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., Yang T.-P., Multiple Tissue Human Expression Resource (MuTHER) Consortium An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B., de Vries P.S., Metcalf G.A., Wang Z., Feofanova E.V., Liu X., Muzny D.M., Wagenknecht L.E., Gibbs R.A., Morrison A.C., Boerwinkle E. Whole genome sequence analysis of serum amino acid levels. Genome Biol. 2016;17:237. doi: 10.1186/s13059-016-1106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries P.S., Yu B., Feofanova E.V., Metcalf G.A., Brown M.R., Zeighami A.L., Liu X., Muzny D.M., Gibbs R.A., Boerwinkle E., Morrison A.C. Whole-genome sequencing study of serum peptide levels: the Atherosclerosis Risk in Communities study. Hum. Mol. Genet. 2017;26:3442–3450. doi: 10.1093/hmg/ddx266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feofanova E.V., Yu B., Metcalf G.A., Liu X., Muzny D., Below J.E., Wagenknecht L.E., Gibbs R.A., Morrison A.C., Boerwinkle E. Sequence-Based Analysis of Lipid-Related Metabolites in a Multiethnic Study. Genetics. 2018;209:607–616. doi: 10.1534/genetics.118.300751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long T., Hicks M., Yu H.C., Biggs W.H., Kirkness E.F., Menni C., Zierer J., Small K.S., Mangino M., Messier H. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 9.Suhre K., Wallaschofski H., Raffler J., Friedrich N., Haring R., Michael K., Wasner C., Krebs A., Kronenberg F., Chang D. A genome-wide association study of metabolic traits in human urine. Nat. Genet. 2011;43:565–569. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- 10.Raffler J., Friedrich N., Arnold M., Kacprowski T., Rueedi R., Altmaier E., Bergmann S., Budde K., Gieger C., Homuth G. Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality. PLoS Genet. 2015;11:e1005487. doi: 10.1371/journal.pgen.1005487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlosser P., Li Y., Sekula P., Raffler J., Grundner-Culemann F., Pietzner M., Cheng Y., Wuttke M., Steinbrenner I., Schultheiss U.T., GCKD Investigators Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nat. Genet. 2020;52:167–176. doi: 10.1038/s41588-019-0567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zierer J., Jackson M.A., Kastenmüller G., Mangino M., Long T., Telenti A., Mohney R.P., Small K.S., Bell J.T., Steves C.J. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018;50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nag A., Kurushima Y., Bowyer R.C.E., Wells P.M., Weiss S., Pietzner M., Kocher T., Raffler J., Völker U., Mangino M. Genome-wide scan identifies novel genetic loci regulating salivary metabolite levels. Hum. Mol. Genet. 2020;29:864–875. doi: 10.1093/hmg/ddz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gieger C., Geistlinger L., Altmaier E., Hrabé de Angelis M., Kronenberg F., Meitinger T., Mewes H.W., Wichmann H.E., Weinberger K.M., Adamski J. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittemans L.B.L., Lotta L.A., Oliver-Williams C., Stewart I.D., Surendran P., Karthikeyan S., Day F.R., Koulman A., Imamura F., Zeng L. Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat. Commun. 2019;10:1060. doi: 10.1038/s41467-019-08936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L., Kiryluk K. Insights into CKD from Metabolite GWAS. J. Am. Soc. Nephrol. 2018;29:1349–1351. doi: 10.1681/ASN.2018030291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González Burchard E., Borrell L.N., Choudhry S., Naqvi M., Tsai H.J., Rodriguez-Santana J.R., Chapela R., Rogers S.D., Mei R., Rodriguez-Cintron W. Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am. J. Public Health. 2005;95:2161–2168. doi: 10.2105/AJPH.2005.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernández-Rhodes L., Justice A.E., Graff M. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daviglus M.L., Pirzada A., Talavera G.A. Cardiovascular disease risk factors in the Hispanic/Latino population: lessons from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) Prog. Cardiovasc. Dis. 2014;57:230–236. doi: 10.1016/j.pcad.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Lavange L.M., Kalsbeek W.D., Sorlie P.D., Avilés-Santa L.M., Kaplan R.C., Barnhart J., Liu K., Giachello A., Lee D.J., Ryan J. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol. 2010;20:642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(1989). The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol. 129, 687–702. [PubMed]
- 22.Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 23.Ohta T., Masutomi N., Tsutsui N., Sakairi T., Mitchell M., Milburn M.V., Ryals J.A., Beebe K.D., Guo L. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol. Pathol. 2009;37:521–535. doi: 10.1177/0192623309336152. [DOI] [PubMed] [Google Scholar]
- 24.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhaaren B.F.J., Debette S., Bis J.C., Smith J.A., Ikram M.K., Adams H.H., Beecham A.H., Rajan K.B., Lopez L.M., Barral S. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet. 2015;8:398–409. doi: 10.1161/CIRCGENETICS.114.000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Sofer T., Zheng X., Gogarten S.M., Laurie C.A., Grinde K., Shaffer J.R., Shungin D., O’Connell J.R., Durazo-Arvizo R.A., Raffield L., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium A fully adjusted two-stage procedure for rank-normalization in genetic association studies. Genet. Epidemiol. 2019;43:263–275. doi: 10.1002/gepi.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei R., Wang J., Su M., Jia E., Chen S., Chen T., Ni Y. Missing Value Imputation Approach for Mass Spectrometry-based Metabolomics Data. Sci. Rep. 2018;8:663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levey A.S., Stevens L.A., Schmid C.H., Zhang Y.L., Castro A.F., 3rd, Feldman H.I., Kusek J.W., Eggers P., Van Lente F., Greene T., Coresh J., CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H., Wang C., Conomos M.P., Stilp A.M., Li Z., Sofer T., Szpiro A.A., Chen W., Brehm J.M., Celedón J.C. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. Am. J. Hum. Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukaka M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 32.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staley J.R., Blackshaw J., Kamat M.A., Ellis S., Surendran P., Sun B.B., Paul D.S., Freitag D., Burgess S., Danesh J. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foley C.N., Staley J.R., Breen P.G., Sun B.B., Kirk P.D.W., Burgess S., Howson J.M.M. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. bioRxiv. 2019 doi: 10.1038/s41467-020-20885-8. 101101/592238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dafni U. Landmark analysis at the 25-year landmark point. Circ. Cardiovasc. Qual. Outcomes. 2011;4:363–371. doi: 10.1161/CIRCOUTCOMES.110.957951. [DOI] [PubMed] [Google Scholar]
- 39.Lamparter D., Marbach D., Rueedi R., Kutalik Z., Bergmann S. Fast and Rigorous Computation of Gene and Pathway Scores from SNP-Based Summary Statistics. PLoS Comput. Biol. 2016;12:e1004714. doi: 10.1371/journal.pcbi.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei G., Dai Y., Zhao Z., Jia P. deTS: tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics. 2019;35:3842–3845. doi: 10.1093/bioinformatics/btz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 43.Jia P., Zheng S., Long J., Zheng W., Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loley C., Alver M., Assimes T.L., Bjonnes A., Goel A., Gustafsson S., Hernesniemi J., Hopewell J.C., Kanoni S., Kleber M.E. No Association of Coronary Artery Disease with X-Chromosomal Variants in Comprehensive International Meta-Analysis. Sci. Rep. 2016;6:35278. doi: 10.1038/srep35278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott R.A., Scott L.J., Mägi R., Marullo L., Gaulton K.J., Kaakinen M., Pervjakova N., Pers T.H., Johnson A.D., Eicher J.D., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heron M. Division of Vital Statistics; Hyattsville, MD, USA: 2019. Deaths: leading causes for 2017. National Center for Health Statistics; p. 68. [PubMed] [Google Scholar]
- 50.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang K.P., Gerber A.L., Sipe J.C., Cravatt B.F. Reduced cellular expression and activity of the P129T mutant of human fatty acid amide hydrolase: evidence for a link between defects in the endocannabinoid system and problem drug use. Hum. Mol. Genet. 2004;13:2113–2119. doi: 10.1093/hmg/ddh216. [DOI] [PubMed] [Google Scholar]
- 52.Sipe J.C., Scott T.M., Murray S., Harismendy O., Simon G.M., Cravatt B.F., Waalen J. Biomarkers of endocannabinoid system activation in severe obesity. PLoS ONE. 2010;5:e8792. doi: 10.1371/journal.pone.0008792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Justinova Z., Panlilio L.V., Moreno-Sanz G., Redhi G.H., Auber A., Secci M.E., Mascia P., Bandiera T., Armirotti A., Bertorelli R. Effects of Fatty Acid Amide Hydrolase (FAAH) Inhibitors in Non-Human Primate Models of Nicotine Reward and Relapse. Neuropsychopharmacology. 2015;40:2185–2197. doi: 10.1038/npp.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y., Huang T., Lee F., Kreek M.J. Involvement of Endocannabinoids in Alcohol “Binge” Drinking: Studies of Mice with Human Fatty Acid Amide Hydrolase Genetic Variation and After CB1 Receptor Antagonists. Alcohol. Clin. Exp. Res. 2016;40:467–473. doi: 10.1111/acer.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snider N.T., Walker V.J., Hollenberg P.F. Oxidation of the endogenous cannabinoid arachidonoyl ethanolamide by the cytochrome P450 monooxygenases: physiological and pharmacological implications. Pharmacol. Rev. 2010;62:136–154. doi: 10.1124/pr.109.001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cascio M.G. PUFA-derived endocannabinoids: an overview. Proc. Nutr. Soc. 2013;72:451–459. doi: 10.1017/S0029665113003418. [DOI] [PubMed] [Google Scholar]
- 57.Wilson R.I., Nicoll R.A. Endocannabinoid signaling in the brain. Science. 2002;296:678–682. doi: 10.1126/science.1063545. [DOI] [PubMed] [Google Scholar]
- 58.Wójcik O.P., Koenig K.L., Zeleniuch-Jacquotte A., Costa M., Chen Y. The potential protective effects of taurine on coronary heart disease. Atherosclerosis. 2010;208:19–25. doi: 10.1016/j.atherosclerosis.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grevengoed T.J., Trammell S.A.J., McKinney M.K., Petersen N., Cardone R.L., Svenningsen J.S., Ogasawara D., Nexøe-Larsen C.C., Knop F.K., Schwartz T.W. N-acyl taurines are endogenous lipid messengers that improve glucose homeostasis. Proc. Natl. Acad. Sci. USA. 2019;116:24770–24778. doi: 10.1073/pnas.1916288116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeifer N.D., Hardwick R.N., Brouwer K.L.R. Role of hepatic efflux transporters in regulating systemic and hepatocyte exposure to xenobiotics. Annu. Rev. Pharmacol. Toxicol. 2014;54:509–535. doi: 10.1146/annurev-pharmtox-011613-140021. [DOI] [PubMed] [Google Scholar]
- 61.Fang F., Christian W.V., Gorman S.G., Cui M., Huang J., Tieu K., Ballatori N. Neurosteroid transport by the organic solute transporter OSTα-OSTβ. J. Neurochem. 2010;115:220–233. doi: 10.1111/j.1471-4159.2010.06920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.e1419. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muiesan M.L., Agabiti-Rosei C., Paini A., Salvetti M. Uric Acid and Cardiovascular Disease: An Update. Eur. Cardiol. 2016;11:54–59. doi: 10.15420/ecr.2016:4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishnan E. Gout and the risk for incident heart failure and systolic dysfunction. BMJ Open. 2012;2:e000282. doi: 10.1136/bmjopen-2011-000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pavlova T., Vidova V., Bienertova-Vasku J., Janku P., Almasi M., Klanova J., Spacil Z. Urinary intermediates of tryptophan as indicators of the gut microbial metabolism. Anal. Chim. Acta. 2017;987:72–80. doi: 10.1016/j.aca.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 66.Dion M.Z., Leiske D., Sharma V.K., Zuch de Zafra C.L., Salisbury C.M. Mitigation of Oxidation in Therapeutic Antibody Formulations: a Biochemical Efficacy and Safety Evaluation of N-Acetyl-Tryptophan and L-Methionine. Pharm. Res. 2018;35:222. doi: 10.1007/s11095-018-2467-5. [DOI] [PubMed] [Google Scholar]
- 67.Yang L., Li Z., Song Y., Liu Y., Zhao H., Liu Y., Zhang T., Yuan Y., Cai X., Wang S. Study on urine metabolic profiling and pathogenesis of hyperlipidemia. Clin. Chim. Acta. 2019;495:365–373. doi: 10.1016/j.cca.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Yadav T., Quivy J.-P., Almouzni G. Chromatin plasticity: A versatile landscape that underlies cell fate and identity. Science. 2018;361:1332–1336. doi: 10.1126/science.aat8950. [DOI] [PubMed] [Google Scholar]
- 69.Kiyose C., Saito K., Yachi R., Muto C., Igarashi O. Changes in the concentrations of vitamin E analogs and their metabolites in rat liver and kidney after oral administration. J. Clin. Biochem. Nutr. 2015;56:143–148. doi: 10.3164/jcbn.14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmölz L., Birringer M., Lorkowski S., Wallert M. Complexity of vitamin E metabolism. World J. Biol. Chem. 2016;7:14–43. doi: 10.4331/wjbc.v7.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murray E.D., Jr., Wechter W.J., Kantoci D., Wang W.H., Pham T., Quiggle D.D., Gibson K.M., Leipold D., Anner B.M. Endogenous natriuretic factors 7: biospecificity of a natriuretic gamma-tocopherol metabolite LLU-alpha. J. Pharmacol. Exp. Ther. 1997;282:657–662. [PubMed] [Google Scholar]
- 72.Saito H., Kiyose C., Yoshimura H., Ueda T., Kondo K., Igarashi O. Gamma-tocotrienol, a vitamin E homolog, is a natriuretic hormone precursor. J. Lipid Res. 2003;44:1530–1535. doi: 10.1194/jlr.M300061-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Ross K.M., Baer R.J., Ryckman K., Feuer S.K., Bandoli G., Chambers C., Flowers E., Liang L., Oltman S., Dunkel Schetter C., Jelliffe-Pawlowski L. Second trimester inflammatory and metabolic markers in women delivering preterm with and without preeclampsia. J. Perinatol. 2019;39:314–320. doi: 10.1038/s41372-018-0275-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Traber M.G. Mechanisms for the prevention of vitamin E excess. J. Lipid Res. 2013;54:2295–2306. doi: 10.1194/jlr.R032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang T., Xu L. Circulating Vitamin E Levels and Risk of Coronary Artery Disease and Myocardial Infarction: A Mendelian Randomization Study. Nutrients. 2019;11:2153. doi: 10.3390/nu11092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gromovsky A.D., Schugar R.C., Brown A.L., Helsley R.N., Burrows A.C., Ferguson D., Zhang R., Sansbury B.E., Lee R.G., Morton R.E. Δ-5 Fatty Acid Desaturase FADS1 Impacts Metabolic Disease by Balancing Proinflammatory and Proresolving Lipid Mediators. Arterioscler. Thromb. Vasc. Biol. 2018;38:218–231. doi: 10.1161/ATVBAHA.117.309660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han L., Liu P., Wang C., Zhong Q., Fan R., Wang L., Duan S., Zhang L. The interactions between alcohol consumption and DNA methylation of the ADD1 gene promoter modulate essential hypertension susceptibility in a population-based, case-control study. Hypertens. Res. 2015;38:284–290. doi: 10.1038/hr.2014.172. [DOI] [PubMed] [Google Scholar]
- 78.Chang W., Hatch G.M., Wang Y., Yu F., Wang M. The relationship between phospholipids and insulin resistance: From clinical to experimental studies. J. Cell. Mol. Med. 2019;23:702–710. doi: 10.1111/jcmm.13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graham D.B., Lefkovith A., Deelen P., de Klein N., Varma M., Boroughs A., Desch A.N., Ng A.C.Y., Guzman G., Schenone M. TMEM258 Is a Component of the Oligosaccharyltransferase Complex Controlling ER Stress and Intestinal Inflammation. Cell Rep. 2016;17:2955–2965. doi: 10.1016/j.celrep.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., Sajed T., Johnson D., Li C., Karu N. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hagenbeek F.A., Pool R., van Dongen J., Draisma H.H.M., Jan Hottenga J., Willemsen G., Abdellaoui A., Fedko I.O., den Braber A., Visser P.J., BBMRI Metabolomics Consortium Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat. Commun. 2020;11:39. doi: 10.1038/s41467-019-13770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamashita A., Hayashi Y., Matsumoto N., Nemoto-Sasaki Y., Koizumi T., Inagaki Y., Oka S., Tanikawa T., Sugiura T. Coenzyme-A-Independent Transacylation System; Possible Involvement of Phospholipase A2 in Transacylation. Biology (Basel) 2017;6:23. doi: 10.3390/biology6020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L. The PAGE Study: How Genetic Diversity Improves Our Understanding of the Architecture of Complex Traits. bioRxiv. 2018 101101/188094. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and Supplemental Methods
Data Availability Statement
The genotype data used in this paper are available under accession number dbGaP: phs000880.v1.p1. The metabolomics data used in this paper are available on request through the HCHS/SOL web portal (Web Resources). We comply with NIH summary data sharing policy. The summary statistics are available from the corresponding author or Dr. Robert Kaplan (robert.kaplan@einsteinmed.org) on request.