Summary
To facilitate scientific collaboration on polygenic risk scores (PRSs) research, we created an extensive PRS online repository for 35 common cancer traits integrating freely available genome-wide association studies (GWASs) summary statistics from three sources: published GWASs, the NHGRI-EBI GWAS Catalog, and UK Biobank-based GWASs. Our framework condenses these summary statistics into PRSs using various approaches such as linkage disequilibrium pruning/p value thresholding (fixed or data-adaptively optimized thresholds) and penalized, genome-wide effect size weighting. We evaluated the PRSs in two biobanks: the Michigan Genomics Initiative (MGI), a longitudinal biorepository effort at Michigan Medicine, and the population-based UK Biobank (UKB). For each PRS construct, we provide measures on predictive performance and discrimination. Besides PRS evaluation, the Cancer-PRSweb platform features construct downloads and phenome-wide PRS association study results (PRS-PheWAS) for predictive PRSs. We expect this integrated platform to accelerate PRS-related cancer research.
Keywords: polygenic risk scores, genome-wide association studies, GWAS, PRS, phenome-wide association studies, PheWAS, electronic health records, EHR, complex traits, cancer genetics
Introduction
Since 2005, genome-wide association studies (GWASs) have successfully uncovered many common genetic variants associated with a plethora of complex traits and disorders.1, 2, 3 Translation of these findings into clinical practice to improve pre-symptomatic screening and patient care is a major aspiration in the research community. However, genetic risk factors for complex diseases like cancer usually have relatively small risk effects and/or low frequencies and thus have only limited ability as individual predictors of risk in the overall population. Alternatively, the integration of all common risk variants into a single biomarker, called a polygenic risk score (PRS), represents a widely used approach for potentially identifying high-risk individuals at the highest levels of a PRS.4, 5, 6 For example, it was shown that PRSs for five common complex diseases (coronary artery disease [MIM: 608320], atrial fibrillation, type 2 diabetes, inflammatory bowel disease [MIM: 266600], and breast cancer [MIM: 114480]) have the potential to detect individuals at significantly higher genetic risk4 who might benefit from intensified screening efforts, prophylactic prevention, or earlier treatment. Several challenges have to be overcome for constructing a PRS that incorporates state of the art scientific knowledge: one needs (1) summary statistics from an independent discovery GWAS with phenotype and ancestry matching the target study;7 (2) individual-level genetic data of a sufficiently large cohort to adjust for linkage disequilibrium (LD) between genetic variants; and (3) a computationally efficient method to calculate each PRS and to find the best PRS construct for the target cohort.
The gold standards for GWASs to define PRS constructs are independent, large GWAS analyses or GWAS meta-analyses. Full summary statistics enable exploration of the complete spectrum of PRS construction methods, e.g., those that determine the optimal inclusion p value threshold of risk variants for prediction, which often deviates from the standard threshold for genome-wide significance (p value ≤ 5 × 10−8). So far, several cancer GWAS research groups and consortia have openly shared their full GWAS summary statistics with the research community: ovarian carcinoma (MIM: 167000),8,9 breast cancer,10,11 prostate cancer (MIM: 176807),12 colorectal cancer (MIM: 608812),13 and cervical carcinoma (MIM: 603956).14 Other groups have released variants that reached an arbitrarily chosen p value threshold below genome-wide significance (e.g., p value < 10−5).15 In addition to complete or partial GWAS summary statistics, lists of genome-wide significant hits are available for nearly all published GWAS results. The NHGRI-EBI GWAS Catalog2 curates and stores published risk variants for a wealth of traits in a structured database, offering a convenient and efficient way to extract GWAS hits for automated processing.
Alternative and growing sources for publicly available GWAS summary statistics across a large ensemble of diseases use UK Biobank genotype and phenotype data,16 adjusting for population stratification and/or relatedness between individuals17 (see Web Resources). These biobank-based approaches accessed thousands of phenotypes and traits that were defined in efficient automated fashion, e.g., by ICD10 diagnosis category, with specific phenotype defining algorithms like PHESANT18 or PheWAS Codes (PheCodes),19 or even with consortium-based curated phenotype constructs using the content of the electronic health records (EHR) (FINNGEN, see Web Resources).
Another important aspect of finding a suitable set of GWAS summary statistics for a PRS is the mapping of the discovery GWAS trait, here the cancer phenotype, with the trait of interest in the target study. GWAS efforts usually balance specificity and sample size to maximize power for discovery. Consequently, the analyzed phenotype definition might not necessarily represent an ideal match to the phenotype of the target study. Also, differences in diagnosis coding practices in EHR systems, e.g., the preference for certain diagnoses due to billing purposes, might limit the transferability of phenotype definitions across cohorts, even if the same coding systems were used.20
The simplest form of PRS construction requires two things: a selected set of independent risk variants with estimated or weighted risk effect sizes (say ) and genotype data of individuals genotyped at the selected sites (say Gi where . A PRS can then be calculated for each individual as the sum of the weighted risk increasing alleles, namely .
PRS construction methods and their underlying variant selection procedures can roughly be categorized into four groups: (1) fixed p value thresholds of independent risk variants, e.g., “GWAS hits,” variants that reached genome-wide significance (with p < 5 × 10−8); (2) LD pruning (actually clumping) / p value thresholding (“P&T”) of summary statistics that increases power by determining the most predictive p value cut-off that can be above or below genome-significance;6 (3) genome-wide PRS that consider the full GWAS summary statistics after modeling LD, applying shrinkage or Bayesian approaches, e.g., lassosum and LDPred;21, 22, 23, 24, 25 and (4) methods that use individual-level data from a GWAS to determine an optimal set of independent predictors through Bayesian spike and slab or mixture priors.26 The first two approaches typically use the originally reported effect sizes for weighting, while the latter two approaches model LD and/or shrink effect sizes. All methods require a reference panel for LD estimation that ideally resembles or matches the genotype data underlying the discovery GWAS source. Since most only have summary statistics and not individual-level data of the discovery study, we will use only the first three approaches for PRS construction, i.e., fixed p value thresholds, LD pruning / p value thresholding, and lassosum.
PRSs have increasingly been used in cancer risk prediction and stratification. A brief survey of PRS-related literature in PubMed shows that ∼15% of all PRS articles are related to cancer, with 67% of cancer PRS papers focusing on common cancers (defined by the US National Cancer Institute [NCI] as estimated incidence of 40,000 or more in the United States in 2019). As of November 9, 173 PubMed articles on PRSs and cancer have been published in 2019, more than double the previous high of 86 set in 2018, indicating the rapid growth in collection, curation, and generation of genetic data. These studies typically employ construction methods (1) and (2) as described above, although joint variant models are becoming more common because they generally outperform methods (1) and (2) and advanced software has made joint modeling more computationally efficient for large sample sizes.27,28 Several publications constructed PRSs for cancer traits using different methods29, 30, 31 and described their PRS methodology. However, very few share the variants selected and their corresponding weights, making it a challenge to compare or replicate PRS results in different cohorts. The Polygenic Score Catalog (see Web Resources) is a resource under active development to help researchers share, apply, and evaluate PRSs. This resource primarily relies on external PRS sources and currently includes 97 traits; however, no validation is carried out in large biobanks.32
The primary goal of this study is the generation of PRS constructs for common groupings of cancer by using published, freely available cancer GWAS summary statistics and established PRS methods and genetic data from two large biobanks: the Michigan Genomics Initiative (MGI) and the UK Biobank (UKB). We explore hundreds of PRS constructs and offer optimized predictive PRSs (in terms of maximal increase in an R2-type metric) for 35 cancers. The resulting repository of cancer PRSs is made available online via an interactive platform, called Cancer PRSweb (see Web Resources). In this platform, we accompany each GWAS source / PRS method combination with its downloadable constructs and performance metrics (like area under the receiver operating curve, tail enrichment, and Brier score), and we offer insights into secondary trait associations through screening of hundreds of cancer and non-cancer phenotypes of the EHR-derived phenomes of MGI and UKB. We also make the summary statistics for the phenome-wide association study (PheWAS) available. Thus, this centralized and unified platform is a timely attempt to accelerate cancer research related to PRSs.
Our repository contributes to the new and necessary work of democratizing PRS constructions and applications for several cancers under a uniform analytic framework to eventually develop transferable risk scores with clinical utility. We also offer phenome-wide exploration of PRS association through PRS-PheWAS, a tool previously introduced by this group.33,34
Subjects and Methods
Evaluation Cohorts
MGI Cohort
Adult participants aged between 18 and 101 years at enrollment were recruited through the Michigan Medicine health system between 2012 and 2018 while awaiting diagnostic or interventional procedures either during a preoperative visit prior to the procedure or on the day of the procedure that required anesthesia. In addition to coded biosamples and secure, protected health information, participants understood that all EHR, claims, and national data sources linkable to the participant may be incorporated into the MGI databank. Each participant donated a blood sample for genetic analysis, underwent baseline vital sign testing, and completed a comprehensive history and physical assessment (also see ethics statement below). We report results obtained from 38,360 unrelated, genotyped patients of inferred recent European ancestry with available integrated EHR data (∼90% of all MGI participants were inferred to be of recent European ancestry).33 The data used in this study included diagnoses coded with the Ninth and Tenth Revision of the International Statistical Classification of Diseases (ICD9 and ICD10) with clinical modifications (ICD9-CM and ICD10-CM), sex, precomputed principal components (PCs), genotyping batch, and age. Data were collected according to the Declaration of Helsinki principles.35 MGI study participants’ consent forms and protocols were reviewed and approved by the University of Michigan Medical School Institutional Review Board (IRB ID HUM00099605 and HUM00155849). Opt-in written informed consent was obtained. Additional details about MGI can be found online (see Web Resources). A detailed comparison of the MGI versus UKB cohort (see below) can be found in Beesley et al.36
UK Biobank Cohort (UKB)
UKB is a population-based cohort collected from multiple sites across the United Kingdom and includes more than 500,000 participants aged between 40 and 69 years when recruited in 2006–2010.16 The open-access UK Biobank data used in this study included genotypes, ICD9 and ICD10 codes, inferred sex, inferred white British ancestry, kinship estimates down to third degree, birth year, genotype array, and precomputed principal components of the genotypes. Table 1 provides some descriptive statistics of the MGI and UK Biobank samples.
Table 1.
Demographics and Clinical Characteristics of the Analytic Datasets
| Characteristic | MGI | UKB |
|---|---|---|
| Total participants | 38,360 | 408,595 |
| Females, n (%) | 20,141 (52.5%) | 220,896 (54.1%) |
| Mean age, years (SD) | 56.8 (16.2) | 56.9 (8.0) |
| Median number of visits per participant | 45 | not available |
| Median time (years) between first and last visit | 5.5 | not available |
| Median number of unique ICD9 codes | 36a | 2 |
| Median number of unique ICD10 codes | 31a | 6 |
| Number of PheCodes with more than 50 cases | 1,689 | 1,419 |
| Any cancer diagnosis | 20,751 (54.1%) | 69,190 (16.9%) |
| 20 Most Common Cancer Traits in MGI (PheCode) | ||
| Basal cell carcinoma (172.21)b | 2,988 (7.79%) | not available |
| Melanomas of skin, dx or hx (172.1) | 2,701 (7.04%) | 2,682 (0.66%) |
| Breast cancer [female] (174.1) | 2,605 (12.93%) | 12,483 (5.65%) |
| Cancer of prostate (185) | 2,432 (13.35%) | 5,977 (3.18%) |
| Squamous cell carcinoma (172.22)b | 1,917 (5.00%) | not available |
| Cancer of bladder (189.2) | 1,575 (4.11%) | 2,413 (0.59%) |
| Colorectal cancer (153) | 1,196 (3.12%) | 4,585 (1.12%) |
| Non-Hodgkins lymphoma (202.2) | 1,141 (2.97%) | 1,810 (0.44%) |
| Cancer of connective tissue (170.2) | 1,097 (2.86%) | 331 (0.08%) |
| Malignant neoplasm of kidney, except pelvis (189.11) | 1,083 (2.82%) | 1,033 (0.25%) |
| Colon cancer (153.2) | 941 (2.45%) | 3,108 (0.76%) |
| Myeloproliferative disease (200) | 886 (2.31%) | 992 (0.24%) |
| Cancer of bronchus; lung (165.1) | 874 (2.28%) | 2,232 (0.55%) |
| Thyroid cancer (193) | 798 (2.08%) | 347 (0.08%) |
| Malignant neoplasm of rectum, rectosigmoid junction, and anus (153.3) | 669 (1.74%) | 2,167 (0.53%) |
| Malignant neoplasm of uterus (182) | 643 (3.19%) | 1,285 (0.58%) |
| Nodular lymphoma (202.21) | 632 (1.65%) | 365 (0.09%) |
| Cancer of tongue (145.2) | 550 (1.43%) | 310 (0.08%) |
| Leukemia (204) | 545 (1.42%) | 1,665 (0.41%) |
| Cancer of brain (191.11) | 483 (1.26%) | 525 (0.13%) |
The provided characteristics are based on the European subjects in MGI and white British subjects in UKB for which phenotype and imputed genotype data were available. SD, standard deviation.
ICD9/10-CM codes
Skin cancer sub-types
Genotyping, Sample Quality Control, and Imputation
MGI
DNA from 47,364 blood samples was genotyped on customized Illumina Infinium CoreExome-24 bead arrays and subjected to various quality-control filters, resulting in a set of 392,323 polymorphic variants. Principal components and ancestry were estimated by projecting all genotyped samples into the space of the principal components of the Human Genome Diversity Project reference panel using PLINK (938 individuals).37,38 Pairwise kinship was assessed with the software KING,39 and the software FastIndep was used to reduce the data to a maximal subset that contained no pairs of individuals with 3rd or closer degree relationship.40 We removed participants without EHR data and participants not of recent European descent from the analysis, resulting in a final sample of 38,360 unrelated subjects. Additional genotypes were obtained using the Haplotype Reference Consortium reference panel of the Michigan Imputation Server41 and included more than 24 million imputed variants with R2 ≥ 0.3 and minor allele frequency (MAF) ≥ 0.01%. Genotyping, quality control, and imputation are described in detail elsewhere.33
UK Biobank
We used the UK Biobank Imputed Dataset (v3) and limited analyses to the documented 408,961 white British42 individuals and 47,836,001 variants with imputation information score ≥ 0.3 and MAF ≥ 0.01% of which 22,846,729 overlapped with the imputed MGI data (see above). Two random subsets of 5,000 and 10,000 unrelated, white British individuals were used for LD analyses of UKB-based summary statistics.
Phenome Generation
MGI
The MGI phenome was based on ICD9-CM and ICD10-CM code data for 38,360 unrelated, genotyped individuals of recent European ancestry. Longitudinal time-stamped diagnoses were recoded to indicators for whether a patient ever had given a diagnosis code recorded by Michigan Medicine. These ICD9-CM and ICD10-CM codes were aggregated to form up to 1,857 PheCodes using the PheWAS R package (as described in detail elsewhere33,43). For each trait, we identified case and control samples by using the PheCode system where case subjects had at least one observed diagnosis code of the trait while control subjects (reference in fitted models) were individuals who did not have any diagnosis codes belonging to the trait and/or to the trait-specific PheCode exclusion list (see example in Figure S1). To minimize differences in age and sex distributions, avoid extreme case-control ratios, and reduce the computational burden, we matched up to 10 control subjects to each case subject using the R package “MatchIt.”44 Nearest neighbor matching was applied for age and the first four principal components of the genotype data (PC1-4) using Mahalanobis distance with a caliper/width of 0.25 standard deviations. Exact matching was applied for sex and genotyping array. A total of 1,689 case-control studies with >50 cases were used for our analyses of the MGI phenome.
UK Biobank
The UK Biobank phenome was based on ICD9 and ICD10 code data of 408,961 white British,42 genotyped individuals that were similarly aggregated to PheCodes as MGI (as described elsewhere17). In contrast to MGI, there were many pairwise relationships reported for UKB participants.
To retain a larger effective sample size for each phenotype, we first selected a maximal set of unrelated case subjects for each phenotype (defined as no pairwise relationship of 3rd degree or closer11,40) before selecting a maximal set of unrelated control subjects unrelated to these case subjects. Similar to MGI, we matched up to 10 control subjects to each case subject using the R package “MatchIt.”44 Nearest neighbor matching was applied for birth year (as proxy for age, because age at diagnosis was not available to us) and PC1-4 (Mahalanobis-metric matching; matching window caliper/width of 0.25 standard deviations), and exact matching was applied for sex and genotyping array. A total of 1,419 case-control studies with >50 cases each were used for our analyses of the UK Biobank phenome.
On average, we were able to match 9 control subjects per case subject in the MGI phenome and 9.9 control subjects per case subject in the UKB phenome. Additional phenotype information for MGI and UK Biobank is included in Table S1.
PRS Structure
PRSs combine information across a defined set of genetic loci, incorporating each locus’s association with the target trait. The PRS for patient j takes the form PRSj = where i indexes the included loci for that trait, weight is the log odds ratios retrieved from the external GWAS summary statistics for locus i, and Gij is a continuous version of the measured dosage data for the risk allele on locus i in subject j. In order to construct a PRS, one must determine which genetic loci to include in the PRS and their relative weights. Below, we obtain GWAS summary statistics from several different sources, resulting in several sets of weights for each trait of interest. For each set of weights, we consider several strategies for determining which genetic loci to include in the PRS construction.
Sources of GWAS Summary Statistics
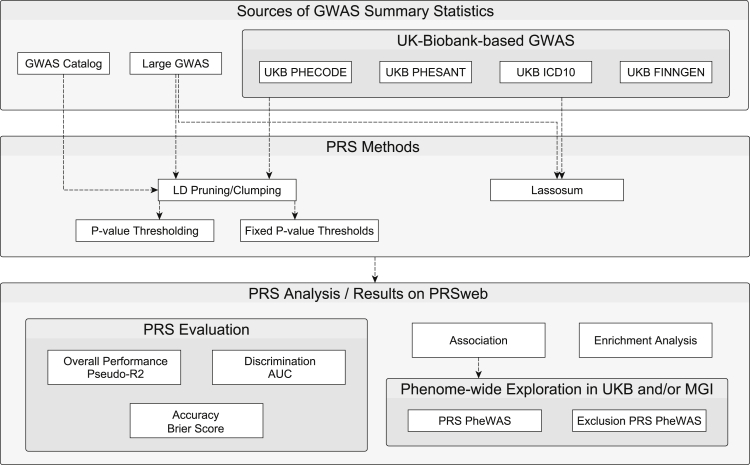
For each of 68 cancers of interest, we collected GWAS summary statistics from up to three different sources: (1) merged genome-wide significant association signals published in the NHGRI EBI GWAS Catalog45 if available; (2) large cancer GWAS meta-analysis if available; and (3) publicly available GWAS summary statistics of phenome × genome screening efforts of the UK Biobank data17 (see Web Resources; Figure 1). If needed, we used LiftOver to convert coordinates of GWAS summary statistics to human genome assembly GRCh37 (UCSC Genome Browser Store, see Web Resources).
Figure 1.
Schematic Overview of PRS Generation and Analysis
GWAS Catalog
We downloaded previously reported GWAS variants from the NHGRI-EBI GWAS Catalog (file version: r2019-05-03).45,46 Single nucleotide polymorphism (SNP) positions were converted to GRCh37 using variant IDs from dbSNP (build 151; UCSC Genome Browser) after updating outdated dbSNP IDs to their merged dbSNP IDs.
Entries with missing risk alleles, risk allele frequencies, or SNP-disease odds ratios were excluded. If a reported risk allele did not match any of the reported forward strand alleles of a non-ambiguous SNP (not A/T or C/G) in the imputed MGI genotype data (which correspond to the alleles of the imputation reference panel), we assumed minus-strand designation and corrected the effect allele to its complementary base of the forward strand. Entries with a reported risk allele that did not match any of the alleles of an ambiguous SNP (A/T and C/G) in our data were excluded at this step. We only included entries with broad European ancestry (as reported by the NHGRI-EBI GWAS Catalog) to match ancestries of discovery GWAS and target cohorts (MGI and UKB). As a quality-control check, we compared the GWAS Catalog reported risk allele frequencies (RAF) with the RAF in MGI individuals. We then excluded entries whose RAF deviated more than 15%. This chosen threshold is subjective and was based on clear differentiation between correct and likely flipped alleles on the two diagonals (Figure S2), as noted frequently in GWAS meta-analyses quality-control procedures.47 For SNPs with multiple entries, we kept the SNP with the most recent publication date (and smaller p value, if necessary) and excluded the others.
Large GWAS Meta-analyses
We downloaded full GWAS summary statistics made available by the Breast Cancer Association Consortium (BCAC),11 the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL),12 and the Ovarian Cancer Association Consortium (OCAC).2,9 In addition, we extracted partial GWAS summary statistics that accompanied recent publications but were incomplete, i.e., reporting only SNPs below a certain p value threshold.15,48, 49, 50 GWAS summary statistics were harmonized and, if needed, lifted over to human genome assembly GRCh37. In this paper, this source is referred to as Large GWAS.
UK Biobank-Based GWAS
We downloaded UK Biobank-based GWAS summary statistics from two public repositories.
The first set of UK Biobank GWAS summary statistics were based on the analysis of up to 408,961 white British European-ancestry samples (UKB GWAS Lee Lab, see Web Resources). SNP-disease odds ratios were estimated using logistic mixed modeling adjusting for sample relatedness, and p values were estimated using saddlepoint approximations (SAIGE method)17 to calibrate the distribution of score test statistics and, thus, control for unbalanced case-control ratios. The underlying phenotypes were auto-curated phenotypes based on the PheCodes of the PheWAS R package17,33,43 similar to the phenomes used in our study and in the following are referred to as UKB PHECODE (Table S2).
The second set of UK Biobank GWAS summary statistics were based on a linear regression model of up to 361,194 unrelated white British samples adjusting for relevant covariates (UKB GWAS Neale Lab, see Web Resources). Three phenotype models were used in their analyses: (1) PHESANT, auto-curated phenotypes using PHEnome Scan ANalysis Tool; (2) ICD10, individuals with the same ICD10 category code (first three characters, e.g., “C50”) were used as case subjects while all non-coded individuals were treated as control subjects, and (3) FINNGEN, curated phenotypes/endpoints based on definitions of the Finngen consortium. In addition to the UKB PHECODE (described above), these three latter sources are referred to as UKB PHESANT, UKB ICD10, and UKB FINNGEN, respectively (Table S2; see Web Resources).
PRS Construction
For each set of GWAS summary statistics from the above-mentioned sources and each cancer, we develop up to seven different PRSs using three different construction methods (Figure 1). Our goal of this approach was to compare multiple PRS methods and find the method that works best for the various types of GWAS summary statistics.
For the first two construction strategies, we performed LD clumping/pruning of variants with p values below 10−4 by using the imputed allele dosages of 10,000 randomly selected samples and a pairwise correlation cut-off at r2 < 0.1 within 1 Mb window. Using the resulting loci, we defined up to five sub-sets of variants with p values below different thresholds (<5 × 10−9 to <5 × 10−5). These were used to construct a PRS tied to each threshold, where the PRS associated with p values less than 5 × 10−8 is sometimes denoted as “GWAS hits.” For the second PRS construction method, we construct many different PRSs across a fine grid of p value thresholds. The p value threshold with the highest cross-validated pseudo-R2 (see PRS Evaluation below) was used to define the more optimized “Pruning and Thresholding (P&T)” PRS.
As an alternative to the p value thresholding and P&T PRS construction strategies, we also used the software package “lassosum”24 to define a third type of PRS for GWAS sources with full summary statistics. Lassosum obtains PRS weights by applying elastic net penalization to GWAS summary statistics and incorporating LD information from a reference panel. Here, we used 5,000 randomly selected, unrelated samples as the LD reference panel. We applied a MAF filter of 1% and, in contrast to the other two approaches, only included autosomal variants that overlap between summary statistics, LD reference panel, and target panel. Each lassosum run resulted in up to 76 combinations of the elastic net tuning parameters s and l, and consequently, in 76 SNP sets with corresponding weights used to construct 76 PRS. We then selected the PRS with the highest pseudo-R2 to define the lassosum PRS (see PRS Evaluation below).
For each cancer and set of GWAS summary statistics, this approach resulted in up to seven PRSs, where PRSs with less than 5 included variants were excluded and the available GWAS summary statistics limited the available PRS construction techniques in some cases. Using the R package Rprs (see Web Resources), the value of each PRS was then calculated for each MGI participant and, if the GWAS source was not based on UKB, also for each UKB participant. For comparability of association effect sizes corresponding to the continuous PRS across cancer traits and PRS construction methods, we centered PRS values in MGI and UKB to their mean and scaled them to have a standard deviation of 1.
PRS Evaluation
For the PRS evaluations, except for when computing the pseudo-R2 (which is a measure of marginal association of the PRS with the outcome), we fit the following model for each PRS and cancer phenotype adjusting for covariates:
| (Equation 1) |
We used Nagelkerke’s pseudo-R2 to select the tuning parameters within the P&T and lassosum construction methods (p value for P&T SNP sets; s and λ for lassosum) and kept the PRS with the highest pseudo-R2 for further analyses. For each PRS derived for each GWAS source/method combination, we assessed the following performance measures relative to observed disease status in MGI and UKB:
-
(1)
overall performance with Nagelkerke’s pseudo-R2 using R package “rcompanion” (see Web Resources)
-
(2)
accuracy with Brier score using R package “DescTools” (see Web Resources)
-
(3)
ability to discriminate between case and control subjects as measured by the area under the covariate-adjusted receiver operating characteristic (AROC; semiparametric frequentist inference55) curve (denoted AAUC) using R package “ROCnReg” (see Web Resources)51.
For cross-validation purposes, we split the data corresponding to each trait in a phenome into training and test set. To retain case-control matching (see Phenome Generation above), we randomly and equally distributed unique strata from matching and thus obtained a 50%/50% split of cases where their matched control subjects were assigned to the same subset. We used the training set to determine the PRS tuning parameter(s) with the highest pseudo-R2 and used the testing set to obtain performance metric for that PRS. Firth’s bias reduction method was used to resolve the problem of separation in logistic regression (see R package brglm2 in Web Resources).52
PRS Association Testing
Next, we assessed the strength of the relationship between these PRSs and the traits they were designed for. To do this we fit the model of Equation 1 for each PRS and cancer phenotype adjusting for various covariates, where the PCs were the first four principal components obtained from the principal component analysis of the genotyped GWAS markers, where “age” was the age at last observed diagnosis in MGI and birth year in UKB and where “array” represents the genotyping array. Our primary interest is , while the other factors (age, sex, and PC) were included to address potential residual confounding and do not provide interpretable estimates due to the preceding application of case-control matching. Firth’s bias reduction method was used to resolve the problem of separation in logistic regression (R package brglm2; see Web Resources).53,52
To study the ability of the PRS to identify high-risk patients, we fit the above model but replacing the PRS with an indicator for whether the PRS value was in the top 1%, 2%, 5%, 10%, or 25% (defined in control subjects) among the matched case control cohort.
Phenome-wide Exploration of PRS Associations
We selected PRSs that were strongly associated with the cancer trait they were designed for phenome-wide association exploration in the phenomes of MGI and UKB for (p value ≤ (0.05 / [#phenotypes in corresponding phenome]); see below).
We conducted PheWAS in MGI and also UKB (if the GWAS source was not based on UKB) to identify additional, secondary phenotypes associated with the PRS.33 To evaluate PRS-phenotype associations, we conducted Firth bias-corrected logistic regression by fitting model of Equation 1 above for each PRS and each phenotype of the corresponding phenome. To adjust for multiple testing, we applied the conservative phenome-wide Bonferroni correction according to the total number of analyzed PheCodes (MGI: 1,689 phenotypes; UKB: 1,419 phenotypes; Table S1). In Manhattan plots, we present –log10 (p value) corresponding to tests of . Directional triangles on the PheWAS plot indicate whether a phenome-wide significant trait was positively (pointing up) or negatively (pointing down) associated with the PRS.
To investigate the possibility of the secondary trait associations with PRS being completely driven by the primary trait association, we performed a second set of PheWAS after excluding individuals affected with the primary or related cancer traits for which the PRS was constructed, referred to as Exclusion-PRS-PheWAS as described previously.33
Online Visual Catalog: PRSweb
The online open access visual catalog PRSweb was implemented using Grails, a Groovy- and Java-based backend logic, to integrate interactive visualizations and MySQL databases. Interactive PheWAS plots are drawn with the JavaScript library “LocusZoom.js” which is maintained by the UM Center for Statistical Genetics (Locuszoom, see Web Resources) and offers dynamic plotting, automatic plot sizing, and label positioning. Additional data-driven visualizations (e.g., temporal relationship plots) were implemented with the JavaScript library “D3.js.”
Unless otherwise stated, analyses were performed using R 3.6.1.54
Results
PRS Construction
We screened the GWAS Catalog, PubMed, and UK Biobank GWAS efforts for any cancer GWAS summary statistics that were reported for European ancestry, to match the predominantly European cohorts of MGI and UKB, and that were openly available, i.e., did not require contacting the main authors or any form of written approval process. We identified 232 source sets that reported complete information for each tested single nucleotide polymorphisms (SNP) (position [and/or dbSNP ID], effect allele, effect estimate, p value, and, ideally, effect allele frequency). We obtained 188 SNP sets based on UKB GWAS, 24 based on excerpts from the GWAS Catalog, and 20 from large GWAS or GWAS meta-analyses (Tables S2 and S3).
We manually matched the traits of the identified cancer GWAS to cancer traits of MGI and UKB PheCodes and analyzed each GWAS source separately, generating PRS for each. The discovery GWAS traits of the 232 source sets approximated 68 cancer PheCodes of the MGI phenome and 21 PheCodes in the UKB phenome (Tables S2 and S3). Following the scheme in Figure 1 and Table 2, we generated PRSs using the P&T and/or lassosum approach and also generated PRSs using fixed p value thresholds after LD clumping (p value ≤ 5 × 10−5, 5 × 10−6, 5 × 10−7, 5 × 10−8 [“GWAS Hits”], or 5 × 10−9). Using these methods and the available GWAS sources, we generated a total of 1,307 PRSs (1,077 PRSs for the MGI cohort and 230 PRSs for the UKB cohort) (Table S4).
Table 2.
Overview of GWAS Sources and PRS Construction Methods
| Source of Summary Statistics |
PRS Construction Method |
|||
|---|---|---|---|---|
| Fixed p Value Thresholdsa | P&Tb | Lassosum | ||
| GWAS Catalog | yes | yes | no | |
| Large GWAS | yes | yes | yes, if full GWAS | |
| UKB GWAS | PHECODE | yes | yes | yes |
| FINNGEN | yes | yes | yes | |
| ICD10 | yes | yes | yes | |
| PHESANT | yes | yes | yes | |
Multiple PRSs were constructed per trait of interest depending on availability of GWAS summary statistics.
Uncorrelated variants with p value ≤ 5 × 10−5, 5 × 10−6, 5 × 10−7, 5 × 10−8 (“GWAS Hits”), or 5 × 10−9
LD pruning and p value thresholding
PRS Evaluation
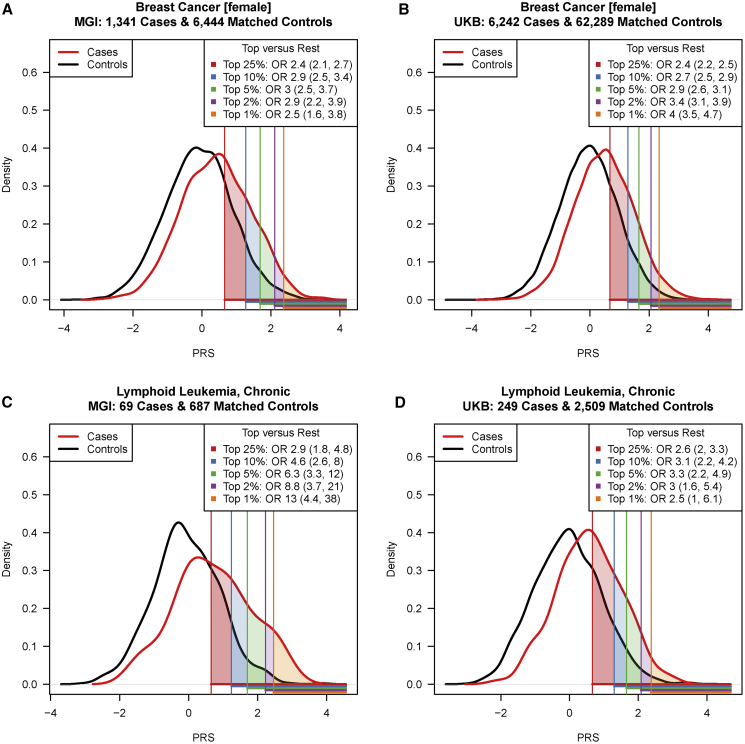
We tested the association between each PRS and its corresponding cancer trait and evaluated each PRS in terms of performance (pseudo-R2), accuracy (Brier score), and discrimination (covariate-adjusted area under the receiver operating characteristic curve [AAUC]). Finally, we tested their utility for risk stratification, i.e., their ability to enrich cases in five selected top percentiles (1%, 2%, 5%, 10%, and 25%) versus the rest of the PRS distribution (Figure 2).
Figure 2.
Distribution of Breast Cancer and Chronic Lymphoid Leukemia PRSs in MGI and UKB
Breast cancer (A, B) and chronic lymphoid leukemia (C, D) PRSs in matched case-controls samples in MGI (A, C) and UKB (B, D) are shown. The five top PRS percentiles (1%, 2%, 5%, 10%, and 25% [defined in control subjects]) are indicated by the shaded areas under the density curves while corresponding odds ratios (OR) and their 95% confidence intervals are given in the top right corner of each plot. PRSs were standardized.
As an initial filtering step, we removed 760 PRSs (57% of total PRSs considered) that were not significantly (751 PRSs with p > 0.05) or negatively (252 PRSs) associated with their corresponding cancer trait in MGI and/or UKB. The majority of these filtered PRSs were either based on discovery GWAS with small sample sizes that often did not identify any genome-wide significant hits or were evaluated for diseases with few cases or both, indicating a potential lack of power in our analysis. A total of 547 PRSs for 35 different cancer traits were positively and significantly associated with their corresponding cancers in MGI (354 PRSs; 31 cancer traits) and UKB (193 PRSs; 20 cancer traits) (Table S4).
Comparison of Performance Metrics
In general, we found that the ranking by pseudo-R2 ensured strong performance across other metrics related to discrimination, accuracy, and overall association of PRS constructs for their specific cancers. Conversely, the enrichment analyses in the extreme PRS percentiles (e.g., top 5% versus rest) was not always concordant with the selection of optimal PRSs based on pseudo-R2, showing that performance in the extreme tails could be optimized by a modified criterion that focuses on extremes of the risk distribution.55
An example evaluation is shown in Table 3. Here we compare PRSs across seven construction methods (lassosum, P&T, and five fixed p value thresholds) that were all based on a single summary statistics source, a large GWAS on overall breast cancer.11 In MGI, we observed that the lassosum-based PRS (118,388 SNPs) had the best performance (highest pseudo-R2 = 0.059), the highest accuracy (Brier score = 0.134), the best discrimination between breast cancer case and control subjects (AAUC = 0.641 [95% confidence interval (CI): 0.625,0.656]), and showed the strongest association with breast cancer itself (odds ratio [OR] continuous PRS = 1.70 [95% CI: 1.59,1.81]). In this scenario, modeling LD information with lassosum retained more information than LD clumping,24 even though, unlike the other methods, lassosum only considered autosomal variants.
Table 3.
Comparison of PRS Methods on Breast Cancer PRS Performance in MGI and UKB
| Method Tuning Parameter | # SNPs | Pseudo-R2 | Brier Score | AAUC (95% CI) | Odds Ratio Continuous PRS (95% CI) | Odds Ratio Top 1% (95% CI) |
|---|---|---|---|---|---|---|
| MGI Cohort | ||||||
| Lassosum: s = 0.5, λ = 0.0043 | 118,388 | 0.0592 | 0.134 | 0.641 (0.625,0.656) | 1.70 (1.59,1.81) | 2.48 (1.63,3.77) |
| P&T: p ≤ 4 × 10−4 | 3,038 | 0.0532 | 0.135 | 0.635 (0.618,0.651) | 1.64 (1.54,1.75) | 2.84 (1.88,4.28) |
| Fixed threshold: p ≤ 5 × 10−5 | 1,307 | 0.0521 | 0.135 | 0.634 (0.618,0.65) | 1.63 (1.53,1.74) | 2.75 (1.81,4.17) |
| Fixed threshold: p ≤ 5 × 10−6 | 712 | 0.0484 | 0.135 | 0.629 (0.612,0.645) | 1.60 (1.50,1.70) | 3.06 (2.04,4.59) |
| Fixed threshold: p ≤ 5 × 10−7 | 464 | 0.0476 | 0.135 | 0.627 (0.611,0.644) | 1.60 (1.50,1.70) | 3.38 (2.28,5.02) |
| Fixed threshold: p ≤ 5 × 10−8 | 334 | 0.0462 | 0.136 | 0.625 (0.609,0.641) | 1.58 (1.49,1.69) | 3.32 (2.24,4.93) |
| Fixed threshold: p ≤ 5 × 10−9 | 264 | 0.0455 | 0.136 | 0.624 (0.608,0.64) | 1.58 (1.48,1.68) | 2.56 (1.68,3.90) |
| UKB Cohort | ||||||
| Lassosum: s = 0.9, λ = 0.0043 | 286,144 | 0.0487 | 0.0807 | 0.643 (0.637,0.65) | 1.70 (1.65,1.75) | 4.02 (3.46,4.67) |
| P&T: p ≤ 1 × 10−4 | 1,682 | 0.0401 | 0.0811 | 0.63 (0.623,0.637) | 1.61 (1.57,1.66) | 3.69 (3.16,4.31) |
| Fixed threshold: p ≤ 5 × 10−6 | 712 | 0.0402 | 0.0811 | 0.628 (0.62,0.635) | 1.61 (1.57,1.65) | 3.32 (2.83,3.90) |
| Fixed threshold: p ≤ 5 × 10−5 | 1,307 | 0.0392 | 0.0812 | 0.627 (0.62,0.634) | 1.60 (1.56,1.64) | 3.49 (2.98,4.08) |
| Fixed threshold: p ≤ 5 × 10−7 | 464 | 0.0384 | 0.0812 | 0.626 (0.618,0.633) | 1.59 (1.55,1.63) | 3.81 (3.27,4.44) |
| Fixed threshold: p ≤ 5 × 10−8 | 334 | 0.0361 | 0.0813 | 0.622 (0.615,0.63) | 1.57 (1.53,1.61) | 3.69 (3.16,4.31) |
| Fixed threshold: p ≤ 5 × 10−9 | 264 | 0.0347 | 0.0813 | 0.62 (0.612,0.627) | 1.55 (1.51,1.59) | 3.28 (2.79,3.86) |
PRSs are based on the BCAC Consortium GWAS on overall breast cancer.11 Italic values indicate best performing PRSs according to the corresponding metrics for MGI or UKB.
The enrichment of cases in the top 1% compared to the rest was more pronounced for the PRSs with a fixed p value threshold (p ≤ 5 × 10−7; 464 SNPs; ORTop1% 3.38 [95% CI: 2.28,5.02]) than for the lassoum PRS (OR Top1% = 2.48 [95% CI: 1.63,3.77]) (Table 3). In UKB, we observed a similar ranking of PRS methods in terms of pseudo-R2 and AAUC, but we noted several differences with MGI. First, the tuning parameters of the lassosum PRS and the P&T PRS differed between MGI and UKB, resulting in a different number of included variants (lassosum: MGI 118,388 variants versus UKB 286,144 variants; P&T: MGI 3,038 variants versus UKB 1,682 variants) (Table 3). Closer inspection of the underlying tuning parameter optimization revealed comparable parameter ranking for lassosum and P&T, suggesting that optimizations seem cohort specific but stable, i.e., tuning parameters for PRSs established in UKB might perform similarly well in MGI and vice versa (Spearman’s rank correlation rho > 0.982) (Figure S3).
Comparison across GWAS Sources
We also explored the influence of various GWAS sources on the predictive performance of PRSs. As an illustrative example, we again focus on breast cancer PRSs, but now consider PRSs constructed from different breast cancer GWAS sources, using for each source the method that yielded the highest pseudo-R2 (Table 4). In MGI, the PRS (lassosum) of the largest available GWAS (122,977 case subjects and 105,974 control subjects) yielded the best performance across most PRS metrics (e.g., pseudo-R2 = 0.0592, AAUC = 0.641 [0.625,0.656]). The GWAS Catalog PRS (P&T), which included 62 top hits from 18 different GWASs,11,56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73 was ranked second (pseudo-R2 = 0.0346) and showed inferior discrimination ability (AAUC 0.607 [0.589,0.623]). The case enrichment in the top 1% was pronounced but not significantly different from the top-ranked PRS (ORTop1%[GWAS Catalog] = 3.06 [2.04,4.60] versus ORTop1%[Large GWAS] = 2.48 [1.63,3.77]). The four UKB GWAS-based PRSs (all based on lassosum) followed next and showed similar performances (pseudo-R2: 0.034–0.020; AAUC between 0.609 and 0.579 with overlapping confidence intervals) and could be ranked according to their effective sample sizes. Most interestingly, the UKB PheCode PRS (6,977 variants) could differentiate case and control subjects as well as the GWAS Catalog PRS, which was based on 62 independent risk variants with p ≤ 3.2 × 10−9 reported in 18 GWASs (AAUC[UKB PheCode] 0.609 [0.592,0.624] and AAUC[GWAS Catalog] 0.607 [0.589,0.623]). This suggested that biobank-based PRSs can be a viable alternative for PRS construction, especially if summary statistics from a large disease-specific GWASs are unavailable (Table 4) though UKB-GWAS-based PRSs underperformed compared to PRSs based on GWASs from much larger meta-analysis efforts. A detailed comparison of GWAS sources across the 31 cancer traits in MGI is available in Table S5.
Table 4.
Influence of GWAS Sources on Breast Cancer PRS Performance in MGI
| GWAS Source (Effective Sample Size)a | Method Tuning Parameter | # SNPs | Pseudo-R2 | Brier Score | AAUC (95% CI) | Odds Ratio Continuous PRSb | Odds Ratio Top 1%c |
|---|---|---|---|---|---|---|---|
| MGI Cohort | |||||||
| Large GWAS Michailidou et al.11 (113,845) | Lassosum: s = 0.5, λ = 0.0043 | 118,388 | 0.0592 | 0.134 | 0.641 (0.625,0.656) | 1.70 (1.59,1.81) | 2.48 (1.63,3.77) |
| GWAS Catalog (N/A) | P&T: p ≤ 3.2 × 10−9 | 62 | 0.0346 | 0.136 | 0.607 (0.589,0.623) | 1.49 (1.40,1.58) | 3.06 (2.04,4.6) |
| UKB GWAS PHECODE (23,839) | Lassosum: s = 0.5, λ = 0.014 | 6,977 | 0.0340 | 0.137 | 0.609 (0.592,0.624) | 1.48 (1.39,1.57) | 1.94 (1.21,3.11) |
| UKB GWAS FINNGEN (18,376) | Lassosum: s = 0.5, λ = 0.018 | 2,267 | 0.0300 | 0.137 | 0.600 (0.584,0.616) | 1.44 (1.35,1.53) | 2.37 (1.54,3.65) |
| UKB GWAS ICD10 (15,792) | Lassosum: s = 0.5, λ = 0.018 | 4,047 | 0.0264 | 0.137 | 0.595 (0.579,0.610) | 1.40 (1.32,1.49) | 2.05 (1.30,3.24) |
| UKB GWAS PHESANT (15,282) | Fixed threshold: p ≤ 5 × 10−8 | 22 | 0.0204 | 0.138 | 0.579 (0.561,0.597) | 1.34 (1.27,1.43) | 2.32 (1.49,3.61) |
| UKB Cohort | |||||||
| Large GWAS Michailidou et al.11 (113,845) | Lassosum: s = 0.9, λ = 0.0043 | 286,144 | 0.0487 | 0.0807 | 0.643 (0.637,0.65) | 1.70 (1.65,1.75) | 4.02 (3.46,4.67) |
| GWAS Catalog (N/A) | P&T: p ≤ 2.5 × 10−8 | 79 | 0.0226 | 0.0819 | 0.598 (0.59,0.605) | 1.43 (1.39,1.46) | 2.68 (2.25,3.18) |
Italic values indicate best performing PRS according to the corresponding metrics for MGI or UKB.
Effective sample size: 4 / (1/#cases + 1/#controls); n/a: not available; references of studies contributing to GWAS Catalog PRS are listed in Table S4.
PRSs were scaled to mean = 0 and SD = 1.
Top 1% versus rest.
Comparison of Performance across Methods
First, we explored the benefit of p value thresholding for the pre-filtered risk variants of the GWAS Catalog. Compared to the GWAS hits only approach, i.e., only perform LD-clumping of risk variants with p ≤ 5 × 10−8, the p value thresholding step of the P&T PRS construction overall was very similar but we observed for several UKB PRSs improved performance, as previously reported.74 This implied that p value thresholding might be beneficial for some of the relatively sparse sets of GWAS hits reported in the GWAS Catalog (Figure S4).
The P&T approach will, by definition, also cover fixed p value thresholds in its tuning parameter optimization; therefore, we limited our next comparison of PRS methods for full summary statistics to P&T and lassosum PRSs. We assessed both methods for different GWAS sources in MGI (37 PRSs) and UKB (10 PRSs). We found that both methods ranked comparably, i.e., a GWAS source that produced a lassosum PRS with high pseudo-R2 also produced a P&T PRS with high pseudo-R2 and vice versa (Spearman’s rank correlation: rho > 0.907; Figure S5).
Comparison of Performance across Cancers
Next, we were interested in comparisons between PRSs across traits to assess overall performance and general differences between cancer traits. Table 5 shows the top-ranked PRSs for the 20 most common cancer traits in MGI and highlights the different properties of the generated PRSs. The PRSs vary in their numbers of included SNPs and their abilities to distinguish case from control subjects or to enrich cases in the top percentiles. The AUC of the presented PRSs was highest for the chronic lymphoid leukemia PRS (AUC = 0.696 [0.621,0.764]) and lowest for the lung cancer PRS (0.529 [95% CI:0.503,0.558]). Significant enrichment of cases in the top 1% ranged from OR of 12.9 (95% CI: 4.45,37.6; chronic lymphoid leukemia) to 2.48 (95% CI: 1.63,3.77; breast cancer [female]). Table 6 shows that similar trends were observed for traits in UKB. Due to limited sample sizes in the top percentiles, we could not detect significant enrichment for most of the rarer cancers.
Table 5.
Top PRSs for the 20 Most Common Cancer Traits in MGI
| PRS Cancer Trait (PheCode) | GWAS Source | Method Tuning Parameter | # SNPs | Brier Score | AAUC (95% CI) | Odds Ratio Continuous PRS (95% CI)a | Odds Ratio Top 1% (95% CI)b |
|---|---|---|---|---|---|---|---|
| Basal cell carcinoma (172.21) | Large GWAS: Chahal et al.49 | P&T: p ≤ 4 × 10−8 | 27 | 0.106 | 0.632 (0.616,0.647) | 1.66 (1.57,1.76) | 3.79 (2.68,5.35) |
| Melanomas of skin (172.1) | UKB GWAS PHECODE | P&T: p ≤ 2 × 10−7 | 15 | 0.0952 | 0.604 (0.587,0.62) | 1.49 (1.4,1.57) | 2.97 (2.04,4.34) |
| Breast cancer [female] (174.1) | Large GWAS: Michailidou et al.11 | Lassosum: s = 0.5, λ = 0.0043 | 118,388 | 0.134 | 0.641 (0.625,0.656) | 1.70 (1.59,1.81) | 2.48 (1.63,3.77) |
| Cancer of prostate (185) | Large GWAS: Schumacher et al.12 | Lassosum: s = 0.5, λ = 0.007 | 26,418 | 0.145 | 0.665 (0.647,0.684) | 1.91 (1.77,2.05) | 4.92 (3.21,7.55) |
| Squamous cell carcinoma (172.22) | GWAS Catalog | P&T: p ≤ 1 × 10−11 | 7 | 0.0977 | 0.593 (0.573,0.613) | 1.45 (1.36,1.55) | 3.74 (2.46,5.68) |
| Cancer of bladder (189.2) | GWAS Catalog | P&T: p ≤ 5 × 10−8 | 13 | 0.0917 | 0.572 (0.55,0.594) | 1.29 (1.2,1.39) | 1.47 (0.779,2.77) |
| Colorectal cancer (153) | Large GWAS: Huyghe et al.15 | P&T: p ≤ 4 × 10−7 | 81 | 0.0828 | 0.553 (0.525,0.577) | 1.21 (1.12,1.32) | 3.04 (1.79,5.17) |
| Colon cancer (153.2) | UKB GWAS PHECODE | Lassosum: s = 0.2, λ = 0.038 | 150 | 0.083 | 0.567 (0.54,0.594) | 1.25 (1.13,1.37) | 1.17 (0.477,2.87) |
| Cancer of bronchus/lung (165.1) | GWAS Catalog | P&T: p ≤ 1 × 10−10 | 14 | 0.0827 | 0.529 (0.503,0.558) | 1.12 (1.01,1.24) | 1.75 (0.796,3.85) |
| Thyroid cancer (193) | GWAS Catalog | P&T: p ≤ 3.2 × 10−10 | 8 | 0.0812 | 0.618 (0.587,0.647) | 1.57 (1.41,1.74) | 5.14 (2.94,8.99) |
| Nodular lymphoma (202.21) | UKB GWAS FINNGEN | Lassosum: s = 1, λ = 0.018 | 2,209,179 | 0.0825 | 0.538 (0.504,0.573) | 1.15 (1.02,1.29) | 1.48 (0.538,4.05) |
| Cancer of brain (191.11) | UKB GWAS ICD10 | Lassosum: s = 0.9, λ = 0.1 | 522 | 0.0824 | 0.546 (0.504,0.587) | 1.20 (1.04,1.37) | 1.42 (0.453,4.47) |
| Cancer of esophagus (150) | UKB GWAS ICD10 | Lassosum: s = 1, λ = 0.078 | 2,001 | 0.0826 | 0.551 (0.51,0.588) | 1.20 (1.04,1.39) | 1.81 (0.56,5.82) |
| Cancer of larynx (149.4) | UKB GWAS ICD10 | Lassosum: s = 0.9, λ = 0.1 | 25,920 | 0.0822 | 0.570 (0.522,0.618) | 1.28 (1.09,1.51) | 2.14 (0.649,7.06) |
| Cancer of other male genital organs (187) | UKB GWAS FINNGEN | P&T: p ≤ 4 × 10−6 | 97 | 0.083 | 0.558 (0.506,0.606) | 1.23 (1.03,1.46) | 1.04 (0.183,5.95) |
| Lymphoid leukemia (204.1) | UKB GWAS FINNGEN | P&T: p ≤ 1 × 10−6 | 6 | 0.0819 | 0.578 (0.517,0.642) | 1.36 (1.11,1.66) | 3.69 (1.01,13.4) |
| Multiple myeloma (204.4) | UKB GWAS ICD10 | P&T: p ≤ 7.9 × 10−6 | 27 | 0.0823 | 0.547 (0.479,0.613) | 1.24 (1,1.53) | 2.6 (0.593,11.4) |
| Cancer of testis (187.2) | UKB GWAS PHESANT | Lassosum: s = 0.9, λ = 0.078 | 771 | 0.084 | 0.656 (0.593,0.717) | 1.67 (1.3,2.14) | 2.72 (0.568,13.1) |
| Hodgkin’s disease (201) | GWAS Catalog | P&T: p ≤ 1 × 10−6 | 20 | 0.0821 | 0.620 (0.559,0.688) | 1.48 (1.15,1.89) | 2.64 (0.572,12.2) |
| Lymphoid leukemia, chronic (204.12) | GWAS Catalog | P&T: p ≤ 7 × 10−6 | 44 | 0.0776 | 0.696 (0.621,0.764) | 2.12 (1.65,2.74) | 12.9 (4.45,37.6) |
Cancer traits are sorted by observed case counts in MGI; references of studies contributing to GWAS Catalog PRSs are listed in Table S4.
PRSs were scaled to mean = 0 and SD = 1.
Top 1% versus rest.
Table 6.
Best Performing PRSs for the 20 Analyzed Cancer Traits in UKB
| PRS Cancer Trait (PheCode) | GWAS Source | Method Tuning Parameter | # SNPs | Brier Score | AAUC (95% CI) | Odds Ratio Continuous PRS (95% CI)a | Odds Ratio Top 1% (95% CI)b |
|---|---|---|---|---|---|---|---|
| Breast cancer [female] (174.1) | Large GWAS: Michailidou et al.11 | Lassosum: s = 0.9, λ = 0.0043 | 286,144 | 0.0807 | 0.643 (0.637,0.65) | 1.70 (1.65,1.75) | 4.02 (3.46,4.67) |
| Cancer of prostate (185) | Large GWAS: Schumacher et al.12 | Lassosum: s = 0.9, λ = 0.0055 | 178,259 | 0.0794 | 0.699 (0.690,0.710) | 2.13 (2.04,2.22) | 5.88 (4.85,7.14) |
| Colorectal cancer (153) | Large GWAS: Huyghe et al.15 | P&T: p ≤ 7.8 × 10−6 | 87 | 0.0812 | 0.617 (0.605,0.630) | 1.55 (1.48,1.62) | 4.00 (3.11,5.13) |
| Melanomas of skin (172.1) | GWAS Catalog | P&T: p ≤ 5 × 10−7 | 27 | 0.0812 | 0.619 (0.603,0.634) | 1.56 (1.48,1.66) | 3.12 (2.18,4.47) |
| Cancer of bladder (189.2) | GWAS Catalog | P&T: p ≤ 7 × 10−7 | 15 | 0.0821 | 0.571 (0.555,0.588) | 1.30 (1.23,1.38) | 2.91 (1.99,4.24) |
| Cancer of other lymphoid, histiocytic tissue (202) | GWAS Catalog | P&T: p ≤ 5 × 10−7 | 5 | 0.0822 | 0.490 (0.476,0.505) | 1.15 (1.09,1.21) | 1.97 (1.25,3.12) |
| Cancer of bronchus/lung (165.1) | GWAS Catalog | P&T: p ≤ 2.5 × 10−8 | 19 | 0.0824 | 0.552 (0.534,0.569) | 1.22 (1.15,1.30) | 1.94 (1.22,3.10) |
| Non-Hodgkins lymphoma (202.2) | GWAS Catalog | P&T: p ≤ 1 × 10−9 | 10 | 0.082 | 0.547 (0.527,0.566) | 1.24 (1.16,1.32) | 2.05 (1.24,3.40) |
| Cancer of uterus (182) | GWAS Catalog | P&T: p ≤ 1 × 10−7 | 20 | 0.082 | 0.572 (0.549,0.596) | 1.30 (1.20,1.41) | 2.60 (1.50,4.51) |
| Cancer of kidney, except pelvis (189.11) | GWAS Catalog | P&T: p ≤ 5 × 10−8 | 12 | 0.0825 | 0.517 (0.492,0.540) | 1.15 (1.06,1.25) | 2.17 (1.13,4.14) |
| Cancer of ovary (184.11) | Large GWAS: Phelan et al.9 | P&T: p ≤ 1.3 × 10−9 | 12 | 0.0824 | 0.558 (0.530,0.586) | 1.23 (1.12,1.35) | 1.55 (0.71,3.38) |
| Pancreatic cancer (157) | GWAS Catalog | P&T: p ≤ 5 × 10−9 | 10 | 0.0822 | 0.579 (0.548,0.611) | 1.34 (1.20,1.50) | 1.64 (0.655,4.12) |
| Cancer of brain and nervous system (191.1) | GWAS Catalog | P&T: p ≤ 3.2 × 10−9 | 19 | 0.0812 | 0.622 (0.590,0.653) | 1.56 (1.40,1.75) | 2.93 (1.34,6.41) |
| Multiple myeloma (204.4) | GWAS Catalog | P&T: p ≤ 2.5 × 10−8 | 21 | 0.0818 | 0.576 (0.536,0.616) | 1.32 (1.16,1.50) | 2.20 (0.854,5.66) |
| Cancer of brain (191.11) | GWAS Catalog | P&T: p ≤ 5 × 10−29 | 5 | 0.0813 | 0.606 (0.568,0.642) | 1.52 (1.34,1.71) | 4.15 (2.04,8.41) |
| Lymphoid leukemia, chronic (204.12) | GWAS Catalog | P&T: p ≤ 2.5 × 10−8 | 27 | 0.0796 | 0.672 (0.637,0.703) | 1.85 (1.62,2.11) | 2.52 (1.04,6.08) |
| Thyroid cancer (193) | GWAS Catalog | P&T: p ≤ 1 × 10−16 | 5 | 0.0804 | 0.628 (0.582,0.675) | 1.61 (1.38,1.88) | 4.41 (1.81,10.7) |
| Cancer of testis (187.2) | GWAS Catalog | P&T: p ≤ 5 × 10−6 | 44 | 0.0793 | 0.703 (0.659,0.745) | 2.11 (1.73,2.56) | 4.60 (1.75,12.1) |
| Basal cell carcinoma (172.21) | GWAS Catalog | P&T: p ≤ 5 × 10−9 | 24 | 0.0813 | 0.615 (0.608,0.623) | 1.53 (1.48,1.57) | 3.05 (2.55,3.64) |
| Squamous cell carcinoma (172.22) | Large GWAS: Chahal et al.50 | P&T: p ≤ 1.6 × 10−8 | 9 | 0.0819 | 0.571 (0.563,0.579) | 1.33 (1.30,1.37) | 1.93 (1.56,2.39) |
Cancer traits are sorted by observed case counts in UKB; references of studies contributing to GWAS Catalog PRSs are listed in Table S4.
PRSs were scaled to mean = 0 and SD = 1.
Top 1% versus rest.
Our observed variations between these cancer PRSs likely recapitulates the different genetic architectures of cancers in combination with their prevalences in the discovery and evaluation cohorts. First, the prevalence impacted the ability to identify true associations in the discovery GWASs and also affected our capacity to observe significant effects in the PRS performance evaluation.
Comparison of Performance across Cohorts
The two evaluation cohorts, MGI and UKB, varied in, among other things, their sample sizes, their use of diagnosis code systems, and their recruitment mechanisms, with UKB representing a population-based cohort and MGI an EHR-based, cancer-enriched cohort. We limited a comparison of the cancer PRSs to the top-ranked PRSs for 13 cancers that were present for both cohorts. We selected the top PRS for each cancer within each cohort, i.e., their GWAS source and method might be different between MGI and UKB (Table S6).
We noticed the similar ranking of AAUC values for most cancer PRSs but found significantly higher estimates for cancer of brain, colorectal cancer, and prostate cancer in UKB than in MGI (Figure S6). The former estimate might reflect the different underlying GWAS sources, while the latter two might be inflated in UKB due to overlapping samples between their discovery GWAS meta-analyses and the UKB cohort.12,15 The other ten cancers showed a similar ranking of AAUC estimates in both cohorts that ranged between “cancer of bronchus/lung” (AAUCMGI: 0.520, AAUCUKB: 0.552) and highest for “chronic lymphoid leukemia” (AAUCMGI: 0.696, AAUCUKB: 0.672). AAUC values tended to be slightly higher for UKB than for MGI, while confidence intervals were mostly smaller in UKB corresponding to their (often) larger observed effective sample sizes.
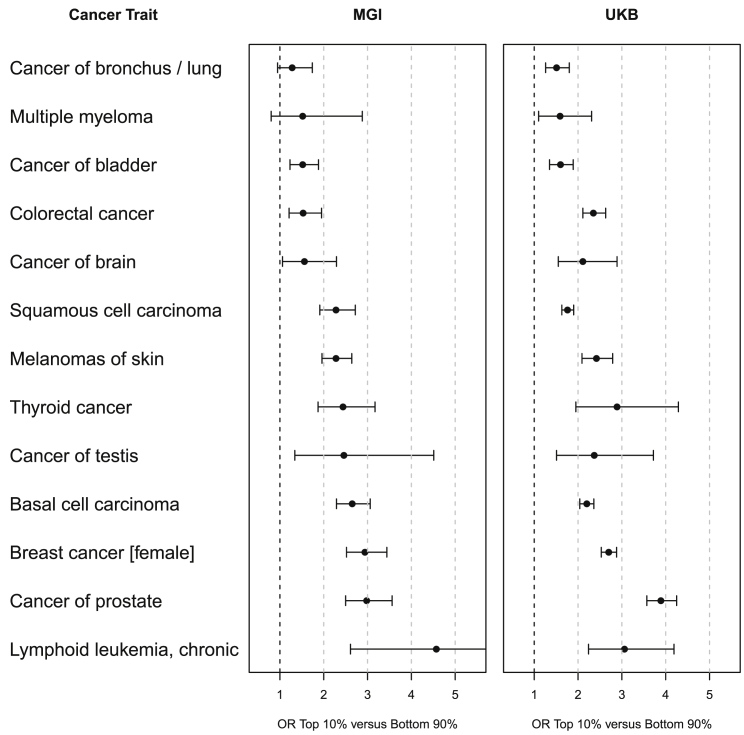
A similar comparison of the enrichment of cases in the top 10% versus bottom 90% revealed a lack of power for two cancer PRSs in MGI, but a relatively consistent ranking from PRSs for bladder cancer (MGI ORTop10%: 1.52 and UKB ORTop10%: 1.60) to chronic lymphoid leukemia (MGI ORTop10%: 4.57 and UKB ORTop10%: 3.06). Overall enrichment effects were often stronger in UKB compared to MGI, reflecting the larger sample sizes of these cancers, but also indicated a disparity between population- and hospital-based control subjects (Tables 1, S1, and S6; Figure 3). However, when comparing the enrichment of cases for two PRSs that were well powered in both cohorts (PRSs for breast cancer and chronic lymphoid leukemia), we found it to be strikingly comparable across all tested percentiles (1%, 2%, 5%, 10%, and 25% versus rest; Figure 2).
Figure 3.
Case Enrichment of the Top Ranked PRSs for 13 Cancers that Were Available in MGI and UKB
Odds ratios (OR, top 10% versus bottom 90% of PRS distribution [defined in control subjects]) and their 95% confidence intervals are shown for MGI (left) and UKB (right).
Phenome-wide Association Analyses
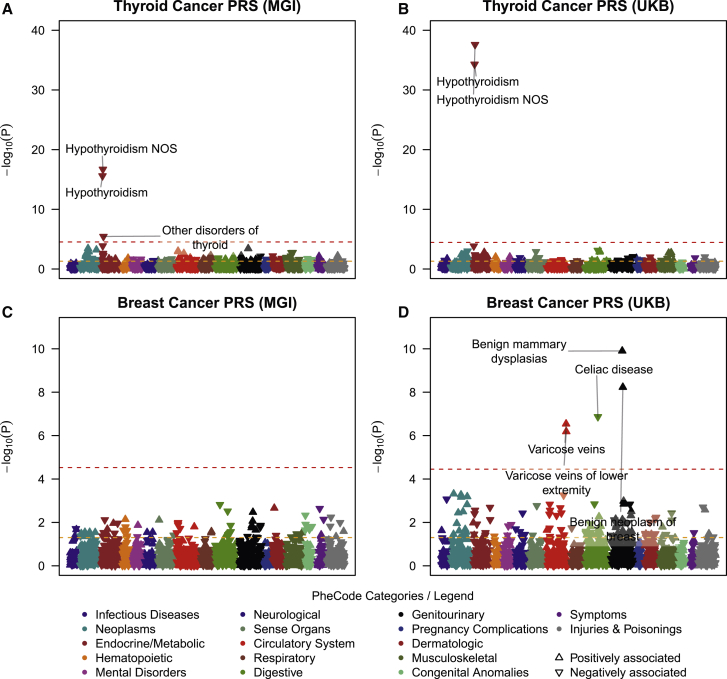
Beyond case enrichment and risk stratification, PRSs can also be used in phenome-wide screenings to uncover secondary trait associations through shared genetic risk factors.33,34 These secondary traits might uncover features in the EHR that occur before cancer diagnosis and thus could represent important predictors for cancer outcomes. From the generated PRSs for 35 cancer traits, we selected 14 PRSs in MGI and 19 PRSs in UKB (whose association with their corresponding cancer traits reached phenome-wide significance) for phenome-wide screens of PRS associations. In total, we observed phenome-wide significant associations between 19 cancer PRSs and 143 different secondary traits (Table S7). We performed Exclusion-PRS-PheWAS (i.e., removed primary cancer cases and repeated the phenome-wide analysis) to assess whether the identified secondary associations were mainly driven by the primary cancer trait, e.g., through intensified screening or represent post-treatment effects.33 While the exclusion of case subjects markedly decreased case counts of secondary traits, we still identified 63 secondary traits that remained significantly associated with the corresponding cancer PRS (Table S7). Most of the secondary traits in MGI that remained phenome-wide significant in the Exclusion-PRS-PheWAS, e.g., skin cancer PRS associated with actinic keratosis or thyroid cancer PRS associated with hypothyroidism, were reported in our previous studies.34,75 Due to the larger sample sizes for most traits in UKB compared to MGI (Table S1), we observed more and stronger secondary trait associations in UKB PRS-PheWAS. Several secondary trait associations were seen in both cohorts (e.g., hypothyroidism associated with thyroid cancer PRS after exclusion of thyroid cancer cases: ORMGI = 0.883 [0.858,0.910] and ORUKB = 0.896 [0.881,0.912]; Figures 4A and 4C). We also observed several secondary trait associations exclusively in UKB. Some of these associations, e.g., hyperplasia of prostate associated with cancer of prostate PRS (Exclusion-PRS-PheWAS in UKB: OR 1.07 [95% CI: 1.05,1.09], p = 2.16 × 10−10), represent known risk factors or presentation features of primary cancers.76,77 However, we also observed traits where the cancer relevance was less clear, e.g., varicose veins associated with breast cancer PRS (Exclusion-PRS-PheWAS in UKB: OR 1.05 [95% CI: 1.03,1.07], p = 2.88 × 10−7) (Figures 4C and 4D; Table S7). Deeper explorations and replications are needed to understand these observed associations and to distinguish between spurious and genuine associations.
Figure 4.
Example Exclusion PRS-PheWAS in the MGI and UKB Phenomes
The plots show Exclusion PheWAS for the thyroid cancer PRS (A, B) and for breast cancer [female] (C, D). The horizontal line indicates phenome-wide significance. Only the strongest and phenome-wide significantly associated traits within a category are labeled. Directional triangles indicate whether a phenome-wide significant trait was positively (pointing up) or negatively (pointing down) associated with the PRS.
Online Visual Catalog: Cancer PRSweb and R package Rprs
In our current study, we compared three PRS construction methods for 68 cancer traits using more than 232 sets of GWAS summary statistics. By doing so, we created a large number of PRSs in which predictive or enrichment properties differed between GWAS source, PRS method, and/or evaluation cohort. After assessing 1,307 constructed PRSs, we found PRSs for 35 different cancer traits that we deemed to have predictive value. In our explorations, we established that it could be beneficial to select PRSs with certain predictive properties for a specific application instead of using one PRS for all applications. Also, it could be computationally more convenient to use a slightly less powerful PRS based on a fewer number of SNPs than to use a PRS that is based on a few hundred thousand variants. To allow the user the option to explore various PRS constructs, we created PRSweb (see Web Resources), an interactive and intuitive web interface, to explore the available PRS constructs for 35 different cancer traits as well as their performance metrics and suitability for risk stratification, association studies, or other PRS applications.
After an initial selection menu for cancer trait and evaluation cohort (MGI or UKB), PRSweb provides tabularized information about all available PRSs, their evaluation metrics (performance, discrimination, calibration, and accuracy) and case enrichment capabilities in five top percentiles of their distributions. The tables, similar to Tables 3 and 4, can be sorted, filtered, or downloaded in full. These tables contain detailed information about the underlying GWAS source(s) and LD reference panels and are directly linked to downloadable PRS constructs. The PRS construct files include headers with information about the PRS construction (source, version, method, and references) and lists its underlying risk variants, their physical positions, effect/non-effect alleles (forward strand orientation for a given genome assembly), and its weights. Together with the Rprs R package we developed (see Web Resources), the construct file will enable the reproduction of PRS association in MGI or UKB and allow a straightforward generation of comparable PRS in external datasets using imputed dosage data in VCF or BCF format.
For phenome-wide predictive PRSs (association PPrimaryCancer ≤ 0.05 / [# phenotypes in phenome]), PRSweb also links to PRS-PheWAS results for their evaluation cohort. The PRS-PheWAS result page includes interactive Manhattan plots for PRS-PheWAS and Exclusion-PRS-PheWAS with mouseover information for each tested association. The PheWAS plots can be exported as scalable vector graphics (SVG) and are accompanied by interactive and downloadable result tables that provide PheWAS summary statistics plus sample counts per analyzed phenotype.
We also implemented a search interface for each phenotype/PheCode to provide insights into the ICD-codes underlying the primary cancers as well as the traits of our EHR-derived MGI and UKB phenomes. A methods section describes the approaches we applied.
Discussion
In our study, we constructed and evaluated a large set of cancer PRSs using more than 200 different sources of GWAS summary statistics. We applied three common PRS construction methods: GWAS hits, LD pruning/p value thresholding, and lassosum. While doing so, we created an online repository called PRSweb with more than 500 PRSs for 35 cancer traits.
We observed that construction and resulting performance of PRSs depend on multiple factors, including GWAS source, PRS method, and evaluation cohort. Researchers who plan to apply PRSs in their projects are often faced with an agony of choice from a set of PRSs in the current literature or might not find predictive PRS at all. Furthermore, if PRSs are available, a direct comparison of multiple constructs is often not feasible, as their performance can be cohort specific and limited by available sample size.
To alleviate this situation, we generated PRSweb that could serve as a central hub for standardized PRSs. PRSweb so far offered a selection and exploration of PRSs based on publicly available cancer GWAS data. The platform integrated the evaluation of the rich EHR data of two independent biobanks, MGI and UKB. In our initial version of PRSweb, we focused on cancer traits because MGI is enriched for cancer.
There are several remaining challenges in developing PRSs, and we will discuss the following four: access to independent GWAS summary statistics, mapping of trait definitions between discovery and evaluation cohorts, power limitations, and finally, transferability of PRSs across cohorts and ancestries.
Access to Independent GWAS Summary Statistics
Limited accessibility to full summary statistics for cancer GWASs in the published literature resulted in a lack of PRS constructs for many cancers. By systematically integrating openly available cancer GWAS summary statistics, we can also openly share PRS constructs, some with millions of markers, with the research community. However, there are large cancer GWAS datasets used in the cancer research community that are not yet integrated into PRSweb. For example, a recent study analyzed 14 different cancer types based on summary-level association statistics from larger cancer GWAS consortia.78 To our knowledge, only the full summary statistics on breast cancer,11 ovarian cancer,9 and prostate cancer12 were openly shared. We are confident that future versions of PRSweb will be able to integrate summary statistics from other large GWAS consortia, e.g., on chronic lymphocytic leukemia, glioma, melanoma, esophageal, testicular, oropharyngeal, pancreatic, renal, colorectal, endometrial, or lung cancer, some with substantially larger samples sizes than the GWASs used in our current analysis.
With the tendency to form large consortia and to integrate available biobank data comes another challenge, namely the potential overlap between the discovery and evaluation cohorts and, thus, potential overfitting. For our current study, we used GWASs that are (to the best of our knowledge) independent from MGI. Since UKB is a popular and widely used resource, the assumption of independence of large GWAS efforts from UKB does not always hold true as we have seen for the large colorectal cancer GWAS.15 In the future, the assessment of independence of GWASs from PRS construction will become more challenging, especially when relying on GWAS databases (e.g., the GWAS Catalog), where the distinction of contributing cohorts might not be obvious from a database entry alone. If the performance of optimized PRSs is promising in both MGI and UKB, we would recommend the use of the MGI-optimized PRSs to alleviate concerns about potential overlap between the discovery GWAS that led to the summary data used for PRS construction and the cohort used for PRS evaluation. The risk of such overlap is minimal with MGI and substantial with UKB. An alternative solution, especially for consortia joining large GWASs, is leave-one-out meta-analysis where in addition to the full meta-analysis results, a separate set of meta-analysis results will be provided for each contributing cohort so that each resulting leave-one-out meta-analysis can be shared and used for PRS generation in that cohort to avoid overfitting. Until such leave-one-out meta analyses summary statistics become publicly available, we recommend users interested in applying PRSs to UKB to use UKB- independent PRS constructs, e.g., the MGI-based PRS constructs that are shared through PRSweb.
We anticipate a more accessible landscape of high-quality full GWAS results in the near future, not only for cancer. First, funding agencies, e.g., the US National Institutes of Health (NIH), are updating their policy regarding access to GWAS summary statistics of funded projects.79 Second, biobank studies are growing in numbers and size and, when connected to EHR data, enable GWASs for thousands of traits each.75 In addition, global efforts are forming that will enable even more powerful phenome × GWAS meta-analyses through collaboration, likely reaching sample sizes that can compete with classical disease-specific consortia.80
Mapping of Trait Definitions
One of the premises for PRS utility is the resemblance of the original trait in the discovery GWAS with the trait of the evaluation cohort.
For our current study, we relied on EHR-based cohorts and defined cancer via PheCodes that are adopted from ICD codes. It is important to bear in mind that we used EHR-based diagnosis data that per se were not collected for research. Besides misclassification, EHR-derived phenomes might be prone to selection and recruitment biases that can negatively impact power or result in false-positive associations.36 ICD codes usually serve administrative and billing purposes and often lack the specificity found in the discovery GWAS. Due to the difference in trait definitions, we often had to fall back to the broad phenotype definition in the EHR cohorts and, by doing so, might have negatively influenced the predictive power for PRS.41 For example, we had only one definition for ovarian cancer in MGI and UKB (PheCode 184.11 “malignant neoplasm of ovary”) that was defined by ICD9 codes 183.0 and V10.43 as well as by ICD10:C56 and their sub-codes. In contrast, the large GWAS on ovarian cancer included results for nine more refined cancer subtypes: invasive epithelial, low-grade serous, high-grade serous, serous invasive, endometrioid, epithelial, mucinous, low-grade serous and serous borderline ovarian cancer, and ovarian clear cell cancer. For our PRS generation, we used all nine GWAS as separate sources and tested each resulting PRS against the single PheCode 184.11. Consequently, the best performing PRS might represent the combination where the discovery GWAS’s trait specificity and the cohort’s trait composition maximized predictive power.
While we restricted our analysis to PheCode definitions, future PRS explorations and evaluations with growing EHR data should include more refined cancer phenotypes by integrating cancer registry data, pathology results, and/or natural language processing of clinical notes. The currently chosen phenotype definitions represent valid and common cancer groupings that are frequently used in clinical and research applications.81 A broader phenotype definition may lead to a larger sample size but may also lump genetically distinct phenotypes together. This heterogeneity can dilute the specificity of the PRS to molecular sub-types of cancer and consequently lower the predictive power.
Power Limitations
For our project, we used data from MGI, a medical center-based cohort, and UKB, a population-based cohort. Due to MGI’s recruitment mechanism through surgery, the observed case counts in MGI reflected the numbers of adult (18+) patients that underwent a surgical procedure and had at least one corresponding cancer diagnosis in their medical records. The case counts in UKB, a rather healthy subset of the older (40–69) British population,82 might be even lower than the population’s cancer prevalence. We observed an enrichment of many cancers in MGI compared to UKB, especially for rarer cancers like thyroid cancer, but generally registered more case subjects in the UKB because its cohort is ten times larger (Table S1). In addition, MGI’s recruitment through surgical procedures likely resulted in a relative depletion of blood cancers (e.g., leukemia, lymphoma, and myeloma), since affected patients undergo surgery less frequently than somatic cancer patients. As a consequence, we often had sufficient power to evaluate and analyze PRSs for these diseases in UKB but not MGI.
We also recognize that each cancer we consider is heterogeneous and there are molecular subtypes that behave very differently (for example estrogen receptor status for breast cancer). In these situations, using a broader phenotype definition will entail larger sample sizes but might lead to an inclusion of genetically distinct phenotypes that increases heterogeneity of the disease and consequently lower the predictive power.34 Similarly, a more refined phenotype might increase homogeneity but consequently reduce sample size and lead to a loss of power. Moreover, GWAS data for subtypes may be limited and based on smaller studies, making summary statistics less reliable in some cases. Considering established sub-types of a given cancer and optimizing between phenotype definition and sample size will be critical as cancer PRS research continues to grow.
One may be interested in defining a combined phenotype of “any cancer” for a composite cancer PRS with a maximal sample size. We defined this phenotype in UKB (with 69,190 cases of any cancer), performed a GWAS that revealed known risk variants for numerous cancers, and created an “any cancer” PRS using our established methods (Tables S1 and S8, Figures S7 and S8). The lassosum PRS with a choice for 179 variants performed best among the constructs (Table S9). However, while defining such a composite phenotype, we have to remember that the endpoint is a heterogeneous mix of various cancers, and the discovery will be driven by the cancers with larger numbers of cases or strong risk effects in the discovery (UKB) and evaluation (MGI) cohort. In the PRS PheWAS in MGI, we saw many related traits associated with the overall PRS. No secondary trait reached phenome-wide significance in the exclusion PRS-PheWAS (Table S10; Figure S9). We incorporated this overall PRS construct in Cancer-PRSweb.
Besides accessible sample sizes, the ability to create predictive PRSs depends on the cancers’ “chip heritability,” i.e., the variance explained through polygenic variants of genotyped and imputed datasets. A previous study on six common cancers found that chip heritability estimates can vary substantially for cancers (e.g., estimated heritability for prostate cancer: 27%, breast cancer 12%, and pancreatic cancer 7%).83 Thus, indicating that even if the most powerful cancer PRS can be generated, other factors play a bigger role, emphasizing the limitations of PRSs for personal risk prediction if used on its own without considering other risk factors.84
Also, genetic architecture affects the choice of PRS construction methods. A recent study estimated the heritability explained by genome-wide significant variants for 14 common cancers and found a wide variability of explained heritability estimates among the analyzed cancer types. For some cancers like testicular cancer, chronic lymphocytic leukemia, prostate, and breast cancer, GWAS hits could explain a large fraction of the chip heritability, while GWAS hits for other cancers like esophageal, colorectal, endometrial, ovarian, or lung cancer explained only moderate to very low fractions.78 Consequently, approaches that only consider GWAS hits might work better for the former, while less conservative p value thresholds or genome-wide PRS methods might work better for the latter cancer traits.
Finally, we realize that a k-fold cross-validation will be more ideal than the single 50:50 split we have adopted to define our training and test sets. Our choice was governed by computational consideration, ease of presentation, and the fact that with larger sample sizes the selection of tuning parameters and the optimized pseudo-R2 values remained relatively stable across multiple random splits.
Transferability of PRS across Cohorts
In our current study, we constructed and evaluated PRSs in individuals of broadly European ancestry. However, we recognize the need to also construct and share PRSs for non-European ancestry groups, especially because of the limited transferability of PRSs across ancestries and ethnicities.7 The integration of PRSs for non-European individuals into our platform PRSweb so far is hampered by the scarcity of GWAS data for diverse ancestry groups85 and by the limited diversity in MGI and UKB, both encompassing predominantly European ancestry individuals. The next largest ancestry groups are Blacks/African Americans (5.4% in MGI; 1.6% in UKB) and Asian (1.6% in MGI, 2.0% in UKB), but even the most common cancers like breast or prostate cancer had less than 250 cases in these groups and thus limited power for PRS evaluations. Moreover, lack of publicly available GWAS summary statistics made it very difficult to construct ancestry-specific PRSs for Blacks.
Differences in genotyping and sequencing strategies can also negatively impact comparability between studies. Ideally, genotype data in the discovery GWAS, the LD reference panel, and the evaluation cohort should be comparable in quality, density, and LD structure for ultimate compatibility. GWASs usually rely on genotyping arrays that can differ in composition and density of variants. Phasing and imputation methods are constantly improving thanks to growing reference panels and refined methods86 and are essential in harmonizing genotype data across cohorts. However, the achievable accuracy is dependent on the study’s sample size and variant density. Consequently, a PRS that was constructed from a large and marker-dense GWAS might not be directly transferable to smaller, sparser genotype data.
In our current analysis of two genotype datasets that differed in genotype density and sample size, we found that the tuning parameters of PRSs established separately for MGI and UKB were ranked similarly in terms of their resulting predictive performance. This indicated that sharing of PRS constructs might represent a feasible and convenient alternative to computationally expensive PRS methods and evaluations.
Conclusions
By generating PRSs from a large collection of freely available cancer GWAS summary statistics and by evaluating them in two independent biobanks, we created the analytical backbone of PRSweb, an online repository for cancer PRSs offering detailed constructs and comparisons. So far, we included PRS constructs and analyses for 35 different cancer traits that showed promising performance in MGI and/or UKB.
We designed PRSweb with the following scientific goals in mind. (1) Expedite and accelerate research with cancer PRSs by curating the freely available GWAS summary statistics. Researchers can access GWAS and PheWAS summary data and optimized constructs without exhaustive computational work. (2) Provide PRS PheWAS results in two biobanks with interactive plots to open up exploring association of cancer with other phenotypes through underlying common genetic susceptibilities. (3) Share a comprehensive evaluation framework for selecting PRSs with an ensemble of metrics that can be adopted in other studies. (4) Distribute PRS constructs and an R package Rprs (see Web Resources) to generate dosage-based PRSs in external dataset so that the PRSs can be easily generated and used as a covariate in research studies or used as an instrumental variable in research related to Mendelian randomization. Our long-term goal is to integrate PRSs with the subject’s EHR to enable translational PRS research in MGI. For now, we plan to simply flag which percentile of PRS distribution the subject falls into for risk stratification. The goal is for the physician to have easy access to this information as a potential tool to inform their cancer screening decisions. We are also actively working on protocols for the return of PRSs results through the University of Michigan Precision Health Initiative. The ultimate goal is to have absolute risk metrics for each individual in the EHR, translating GWAS findings to informed patient care.
The next logical step using the PRS constructs will be to proceed toward absolute risk prediction at an individual level that will require auxiliary data beyond summary statistics from case-control studies. We anticipate the inclusion of additional PRS constructs and methods in an upcoming version of PRSweb that also will expand our focus beyond cancers. There are several Bayesian methods using continuous shrinkage priors that have been proposed for PRS construction (LDPred, PRS-CS, DBSLMM).21, 22, 23 In our initial exploration with a limited number of traits, we found the predictive performance of PRS-CS to be better than the Lassosum method (Table S11) but at a higher computational cost. Operationalizing these three methods to the PRSweb platform requires massive computational resources, especially because MGI is a growing resource with ongoing recruitment. We are currently working on implementing these choices to the PRSweb menu.
Several challenges remain in PRS research in terms of access, power, and transferability. Nevertheless, PRSs have proven to be a valuable tool for risk stratification, especially if combined with non-genetic risk factors.87, 88, 89 PRSs will likely become more powerful with growing sample sizes, better tools, and more diverse resources.
Data Availability
The PRS constructs, evaluations, and PheWAS summary statistics generated during this study are available on the Cancer PRSweb site (see Web Resources).
There are restrictions to the availability of individual-level data of the MGI study due to patient confidentiality; however, researchers who meet the criteria for access to confidential data can apply for access through the University of Michigan Medical School Central Biorepository and from the UK Biobank (see Web Resources).
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors acknowledge the Michigan Genomics Initiative participants, Precision Health at the University of Michigan, and the University of Michigan Medical School Data Office for Clinical and Translational Research, the University of Michigan Medical School Central Biorepository, and the University of Michigan Advanced Genomics Core for providing data storage, management, processing, and distribution services, and the Center for Statistical Genetics in the Department of Biostatistics at the School of Public Health for genotype data curation, imputation, and management in support of the research reported in this publication.
Part of this research has been conducted using both the UK Biobank Resource under application number 24460 and also using results and data generated by previous researchers who have used the UK Biobank Resource.
This material is based in part upon work supported by the National Institutes of Health/NIH (NCI P30CA046592 [L.G.F., L.J.B., M.S., B.M.], NHGRI R01 HG008773 [B.M.], and T32 CA83654 [R.B.P.]), by the University of Michigan (UM-Precision Health Investigators Award U063790 [L.G.F., S.P., Y.M., B.M.]), and by the National Science Foundation under grant number DMS-1712933. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Additional acknowledgments of GWAS sources are listed in Supplemental Acknowledgments.
Published: September 28, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.025.
Contributor Information
Lars G. Fritsche, Email: larsf@umich.edu.
Bhramar Mukherjee, Email: bhramar@umich.edu.
Web Resources
Breast Cancer Association Consortium, http://bcac.ccge.medschl.cam.ac.uk
brglm2, https://cran.r-project.org/web/packages/brglm2/index.html
Cancer PRSweb, https://prsweb.sph.umich.edu
DescTools, https://andrisignorell.github.io/DescTools/
FINNGEN Clinical Endpoints, https://www.finngen.fi/en/researchers/clinical-endpoints
Liftover, https://genome-store.ucsc.edu
Locuszoom, https://github.com/statgen/locuszoom
Michigan Genomics Initiative, https://precisionhealth.umich.edu/our-research/michigangenomics/
NCI Common Cancers Statistics, https://www.cancer.gov/types/common-cancers
NHGRI-EBI GWAS Catalog, https://www.ebi.ac.uk/gwas
OMIM, http://www.omim.org/
Ovarian Cancer Association Consortium, http://ocac.ccge.medschl.cam.ac.uk
PHESANT, https://github.com/MRCIEU/PHESANT
Polygenic Score Catalog, http://www.pgscatalog.org
Rcompanion, https://rdrr.io/cran/rcompanion/
ROCnReg, https://cran.r-project.org/web/packages/ROCnReg/index.html
UCSC Genome Browser, http://genome.ucsc.edu
UK Biobank, https://www.ukbiobank.ac.uk
UK Biobank dataset, https://www.ebi.ac.uk/ega/datasets/EGAD00010001474
UKB GWAS Lee Lab, https://www.leelabsg.org/resources
UKB GWAS Neale Lab, http://www.nealelab.is/uk-biobank
University of Michigan Medical School Central Biorepository, https://research.medicine.umich.edu/our-units/central-biorepository/get-access
Supplemental Information
References
- 1.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee N., Wheeler B., Sampson J., Hartge P., Chanock S.J., Park J.H. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet. 2013;45:400–405. doi: 10.1038/ng.2579. e1–e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrenson K., Song F., Hazelett D.J., Kar S.P., Tyrer J., Phelan C.M., Corona R.I., Rodríguez-Malavé N.I., Seo J.H., Adler E., Australian Ovarian Cancer Study Group Genome-wide association studies identify susceptibility loci for epithelial ovarian cancer in east Asian women. Gynecol. Oncol. 2019;153:343–355. doi: 10.1016/j.ygyno.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phelan C.M., Kuchenbaecker K.B., Tyrer J.P., Kar S.P., Lawrenson K., Winham S.J., Dennis J., Pirie A., Riggan M.J., Chornokur G., AOCS study group. EMBRACE Study. GEMO Study Collaborators. HEBON Study. KConFab Investigators. OPAL study group Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017;49:680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.Y., Kim J., Kim S.W., Park S.K., Ahn S.H., Lee M.H., Suh Y.J., Noh D.Y., Son B.H., Cho Y.U. BRCA1/2-negative, high-risk breast cancers (BRCAX) for Asian women: genetic susceptibility loci and their potential impacts. Sci. Rep. 2018;8:15263. doi: 10.1038/s41598-018-31859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., NBCS Collaborators. ABCTB Investigators. ConFab/AOCS Investigators Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., Profile Study. Australian Prostate Cancer BioResource (APCB) IMPACT Study. Canary PASS Investigators. Breast and Prostate Cancer Cohort Consortium (BPC3) PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium. Cancer of the Prostate in Sweden (CAPS) Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility Loci (PEGASUS) Genetic Associations and Mechanisms in Oncology (GAME-ON)/Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanikawa C., Kamatani Y., Takahashi A., Momozawa Y., Leveque K., Nagayama S., Mimori K., Mori M., Ishii H., Inazawa J. GWAS identifies two novel colorectal cancer loci at 16q24.1 and 20q13.12. Carcinogenesis. 2018;39:652–660. doi: 10.1093/carcin/bgy026. [DOI] [PubMed] [Google Scholar]
- 14.Leo P.J., Madeleine M.M., Wang S., Schwartz S.M., Newell F., Pettersson-Kymmer U., Hemminki K., Hallmans G., Tiews S., Steinberg W. Defining the genetic susceptibility to cervical neoplasia-A genome-wide association study. PLoS Genet. 2017;13:e1006866. doi: 10.1371/journal.pgen.1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huyghe J.R., Bien S.A., Harrison T.A., Kang H.M., Chen S., Schmit S.L., Conti D.V., Qu C., Jeon J., Edlund C.K. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millard L.A.C., Davies N.M., Gaunt T.R., Davey Smith G., Tilling K. Software Application Profile: PHESANT: a tool for performing automated phenome scans in UK Biobank. Int. J. Epidemiol. 2017;47:29–35. doi: 10.1093/ije/dyx204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P., Gifford A., Meng X., Li X., Campbell H., Varley T., Zhao J., Carroll R., Bastarache L., Denny J.C. Developing and Evaluating Mappings of ICD-10 and ICD-10-CM Codes to PheCodes. bioRxiv. 2019 doi: 10.1101/462077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X., Pashova H., Heagerty P.J. Comparing healthcare utilization patterns via global differences in the endorsement of current procedural terminology codes. Ann. Appl. Stat. 2017;11:1349–1374. [Google Scholar]
- 21.Ge T., Chen C.Y., Ni Y., Feng Y.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S., Zhou X. Accurate and Scalable Construction of Polygenic Scores in Large Biobank Data Sets. Am. J. Hum. Genet. 2020;106:679–693. doi: 10.1016/j.ajhg.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilhjálmsson B.J., Yang J., Finucane H.K., Gusev A., Lindström S., Ripke S., Genovese G., Loh P.R., Bhatia G., Do R., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mak T.S.H., Porsch R.M., Choi S.W., Zhou X., Sham P.C. Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol. 2017;41:469–480. doi: 10.1002/gepi.22050. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd-Jones L.R., Zeng J., Sidorenko J., Yengo L., Moser G., Kemper K.E., Wang H., Zheng Z., Magi R., Esko T. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 2019;10:5086. doi: 10.1038/s41467-019-12653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Privé F., Aschard H., Ziyatdinov A., Blum M.G.B. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics. 2018;34:2781–2787. doi: 10.1093/bioinformatics/bty185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Privé F., Aschard H., Blum M.G.B. Efficient Implementation of Penalized Regression for Genetic Risk Prediction. Genetics. 2019;212:65–74. doi: 10.1534/genetics.119.302019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavaddat N., Michailidou K., Dennis J., Lush M., Fachal L., Lee A., Tyrer J.P., Chen T.H., Wang Q., Bolla M.K., ABCTB Investigators. kConFab/AOCS Investigators. NBCS Collaborators Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Z., Hopp H., Ingles S.A., Huff C., Sheng X., Weaver B., Stern M., Hoffmann T.J., John E.M., Van Den Eeden S.K. A genome-wide association study of prostate cancer in Latinos. Int. J. Cancer. 2020;146:1819–1826. doi: 10.1002/ijc.32525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shieh Y., Fejerman L., Lott P.C., Marker K., Sawyer S.D., Hu D., Huntsman S., Torres J., Echeverry M., Bohorquez M.E. A polygenic risk score for breast cancer in U.S. Latinas and Latin-American women. J. Natl. Cancer Inst. 2020;112:590–598. doi: 10.1093/jnci/djz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert S.A., Gil L., Jupp S., Ritchie S.C., Xu Y., Buniello A., Abraham G., Chapman M., Parkinson H., Danesh J. The Polygenic Score Catalog: an open database for reproducibility and systematic evaluation. medRxiv. 2020 doi: 10.1101/2020.05.20.20108217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritsche L.G., Gruber S.B., Wu Z., Schmidt E.M., Zawistowski M., Moser S.E., Blanc V.M., Brummett C.M., Kheterpal S., Abecasis G.R., Mukherjee B. Association of Polygenic Risk Scores for Multiple Cancers in a Phenome-wide Study: Results from The Michigan Genomics Initiative. Am. J. Hum. Genet. 2018;102:1048–1061. doi: 10.1016/j.ajhg.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fritsche L.G., Beesley L.J., VandeHaar P., Peng R.B., Salvatore M., Zawistowski M., Gagliano Taliun S.A., Das S., LeFaive J., Kaleba E.O. Exploring various polygenic risk scores for skin cancer in the phenomes of the Michigan genomics initiative and the UK Biobank with a visual catalog: PRSWeb. PLoS Genet. 2019;15:e1008202. doi: 10.1371/journal.pgen.1008202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 36.Beesley L.J., Salvatore M., Fritsche L.G., Pandit A., Rao A., Brummett C., Willer C.J., Lisabeth L.D., Mukherjee B. The emerging landscape of health research based on biobanks linked to electronic health records: Existing resources, statistical challenges, and potential opportunities. Stat. Med. 2020;39:773–800. doi: 10.1002/sim.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C., Zhan X., Bragg-Gresham J., Kang H.M., Stambolian D., Chew E.Y., Branham K.E., Heckenlively J., Fulton R., Wilson R.K., FUSION Study Ancestry estimation and control of population stratification for sequence-based association studies. Nat. Genet. 2014;46:409–415. doi: 10.1038/ng.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 39.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham K.J., Diaz C. Identifying large sets of unrelated individuals and unrelated markers. Source Code Biol. Med. 2014;9:6. doi: 10.1186/1751-0473-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 2017 doi: 10.1101/166298. [DOI] [Google Scholar]
- 43.Carroll R.J., Bastarache L., Denny J.C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho D.E., Imai K., King G., Stuart E.A. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J. Stat. Softw. 2011;42:1–28. [Google Scholar]
- 45.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R., Ferreira T., Fall T., Graff M., Justice A.E., Genetic Investigation of Anthropometric Traits (GIANT) Consortium Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ransohoff K.J., Wu W., Cho H.G., Chahal H.C., Lin Y., Dai H.J., Amos C.I., Lee J.E., Tang J.Y., Hinds D.A. Two-stage genome-wide association study identifies a novel susceptibility locus associated with melanoma. Oncotarget. 2017;8:17586–17592. doi: 10.18632/oncotarget.15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chahal H.S., Wu W., Ransohoff K.J., Yang L., Hedlin H., Desai M., Lin Y., Dai H.J., Qureshi A.A., Li W.Q. Genome-wide association study identifies 14 novel risk alleles associated with basal cell carcinoma. Nat. Commun. 2016;7:12510. doi: 10.1038/ncomms12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chahal H.S., Lin Y., Ransohoff K.J., Hinds D.A., Wu W., Dai H.J., Qureshi A.A., Li W.Q., Kraft P., Tang J.Y. Genome-wide association study identifies novel susceptibility loci for cutaneous squamous cell carcinoma. Nat. Commun. 2016;7:12048. doi: 10.1038/ncomms12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janes H., Pepe M.S. Adjusting for covariate effects on classification accuracy using the covariate-adjusted receiver operating characteristic curve. Biometrika. 2009;96:371–382. doi: 10.1093/biomet/asp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosmidis I., Clovis Kenne Pagui E., Sartori N. Mean and median bias reduction in generalized linear models. arXiv. 2018 1804.04085. [Google Scholar]
- 53.Heinze G. A comparative investigation of methods for logistic regression with separated or nearly separated data. Stat. Med. 2006;25:4216–4226. doi: 10.1002/sim.2687. [DOI] [PubMed] [Google Scholar]
- 54.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2016. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 55.Song M., Kraft P., Joshi A.D., Barrdahl M., Chatterjee N. Testing calibration of risk models at extremes of disease risk. Biostatistics. 2015;16:143–154. doi: 10.1093/biostatistics/kxu034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., SEARCH collaborators. kConFab. AOCS Management Group Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 59.Gold B., Kirchhoff T., Stefanov S., Lautenberger J., Viale A., Garber J., Friedman E., Narod S., Olshen A.B., Gregersen P. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl. Acad. Sci. USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., SEARCH. GENICA Consortium. kConFab. Australian Ovarian Cancer Study Group Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., Breast Cancer Susceptibility Collaboration (UK) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., EMBRACE. GEMO Study Collaborators. HEBON. kConFab. SWE-BRCA. MOD SQUAD. GENICA A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J., Humphreys K., Heikkinen T., Aittomäki K., Blomqvist C., Pharoah P.D., Dunning A.M., Ahmed S., Hooning M.J., Martens J.W. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res. Treat. 2011;126:717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- 65.Fletcher O., Johnson N., Orr N., Hosking F.J., Gibson L.J., Walker K., Zelenika D., Gut I., Heath S., Palles C. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl. Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 66.Sehrawat B., Sridharan M., Ghosh S., Robson P., Cass C.E., Mackey J.R., Greiner R., Damaraju S. Potential novel candidate polymorphisms identified in genome-wide association study for breast cancer susceptibility. Hum. Genet. 2011;130:529–537. doi: 10.1007/s00439-011-0973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rinella E.S., Shao Y., Yackowski L., Pramanik S., Oratz R., Schnabel F., Guha S., LeDuc C., Campbell C.L., Klugman S.D. Genetic variants associated with breast cancer risk for Ashkenazi Jewish women with strong family histories but no identifiable BRCA1/2 mutation. Hum. Genet. 2013;132:523–536. doi: 10.1007/s00439-013-1269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., Breast and Ovarian Cancer Susceptibility Collaboration. Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) kConFab Investigators. Australian Ovarian Cancer Study Group. GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361, e1–e2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia-Closas M., Couch F.J., Lindstrom S., Michailidou K., Schmidt M.K., Brook M.N., Orr N., Rhie S.K., Riboli E., Feigelson H.S., Gene ENvironmental Interaction and breast CAncer (GENICA) Network. kConFab Investigators. Familial Breast Cancer Study (FBCS) Australian Breast Cancer Tissue Bank (ABCTB) Investigators Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013;45:392–398, e1–e2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaudet M.M., Kuchenbaecker K.B., Vijai J., Klein R.J., Kirchhoff T., McGuffog L., Barrowdale D., Dunning A.M., Lee A., Dennis J., KConFab Investigators. Ontario Cancer Genetics Network. HEBON. EMBRACE. GEMO Study Collaborators. GENICA Network Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Couch F.J., Wang X., McGuffog L., Lee A., Olswold C., Kuchenbaecker K.B., Soucy P., Fredericksen Z., Barrowdale D., Dennis J., kConFab Investigators. SWE-BRCA. Ontario Cancer Genetics Network. HEBON. EMBRACE. GEMO Study Collaborators. BCFR. CIMBA Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M., BOCS. kConFab Investigators. AOCS Group. NBCS. GENICA Network Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Milne R.L., Kuchenbaecker K.B., Michailidou K., Beesley J., Kar S., Lindström S., Hui S., Lemaçon A., Soucy P., Dennis J., ABCTB Investigators. EMBRACE. GEMO Study Collaborators. HEBON. kConFab/AOCS Investigators. NBSC Collaborators Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017;49:1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Euesden J., Lewis C.M., O’Reilly P.F. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beesley L., Salvatore M., Fritsche L., Pandit A., Rao A., Brummett C., Willer C.J., Lisabeth L.D., Mukherjee B. 2018. The Emerging Landscape of Epidemiological Research Based on Biobanks Linked to Electronic Health Records: Existing Resources, Analytic Challenges and Potential Opportunities. Preprints.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ørsted D.D., Bojesen S.E., Nielsen S.F., Nordestgaard B.G. Association of clinical benign prostate hyperplasia with prostate cancer incidence and mortality revisited: a nationwide cohort study of 3,009,258 men. Eur. Urol. 2011;60:691–698. doi: 10.1016/j.eururo.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 77.Dai X., Fang X., Ma Y., Xianyu J. Benign Prostatic Hyperplasia and the Risk of Prostate Cancer and Bladder Cancer: A Meta-Analysis of Observational Studies. Medicine (Baltimore) 2016;95:e3493. doi: 10.1097/MD.0000000000003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y.D., Hurson A.N., Zhang H., Choudhury P.P., Easton D.F., Milne R.L., Simard J., Hall P., Michailidou K., Dennis J., Breast Cancer Association Consortium (BCAC) Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON) Colon Cancer Family Registry (CCFR) Transdisciplinary Studies of Genetic Variation in Colorectal Cancer (CORECT) Endometrial Cancer Association Consortium (ECAC) Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) Melanoma Genetics Consortium (GenoMEL) Glioma International Case-Control Study (GICC) International Lung Cancer Consortium (ILCCO) Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium. International Consortium of Investigators Working on Non-Hodgkin’s Lymphoma Epidemiologic Studies (InterLymph) Ovarian Cancer Association Consortium (OCAC) Oral Cancer GWAS. Pancreatic Cancer Case-Control Consortium (PanC4) Pancreatic Cancer Cohort Consortium (PanScan) Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) Renal Cancer GWAS. Testicular Cancer Consortium (TECAC) Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat. Commun. 2020;11:3353. doi: 10.1038/s41467-020-16483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.National Institutes of Health (NIH) 2018. Update to NIH Management of Genomic Summary Results Access.https://grants.nih.gov/grants/guide/notice-files/NOT-OD-19-023.html [Google Scholar]
- 80.Zhou W., Neale B.M., Daly M.J., Global Biobank Meta-analysis Initiative . Houston; Tx, USA: 2019. Global Biobank Meta-analysis Initiative: Powering genetic discovery across human diseases. In 69th Annual Meeting of the American Society of Human Genetics. [Google Scholar]
- 81.Wei W.Q., Bastarache L.A., Carroll R.J., Marlo J.E., Osterman T.J., Gamazon E.R., Cox N.J., Roden D.M., Denny J.C. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE. 2017;12:e0175508. doi: 10.1371/journal.pone.0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flint E., Cummins S. Active commuting and obesity in mid-life: cross-sectional, observational evidence from UK Biobank. Lancet Diabetes Endocrinol. 2016;4:420–435. doi: 10.1016/S2213-8587(16)00053-X. [DOI] [PubMed] [Google Scholar]
- 83.Lindström S., Finucane H., Bulik-Sullivan B., Schumacher F.R., Amos C.I., Hung R.J., Rand K., Gruber S.B., Conti D., Permuth J.B., PanScan, GECCO and the GAME-ON Network: CORECT, DRIVE, ELLIPSE, FOCI, and TRICL-ILCCO Quantifying the Genetic Correlation between Multiple Cancer Types. Cancer Epidemiol. Biomarkers Prev. 2017;26:1427–1435. doi: 10.1158/1055-9965.EPI-17-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 85.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maas P., Barrdahl M., Joshi A.D., Auer P.L., Gaudet M.M., Milne R.L., Schumacher F.R., Anderson W.F., Check D., Chattopadhyay S. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA Oncol. 2016;2:1295–1302. doi: 10.1001/jamaoncol.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garcia-Closas M., Gunsoy N.B., Chatterjee N. Combined associations of genetic and environmental risk factors: implications for prevention of breast cancer. J. Natl. Cancer Inst. 2014;106(11) doi: 10.1093/jnci/dju305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Das J.K., Choudhury P.P., Chaturvedi N., Tayyab M., Hassan S.S. Ranking and clustering of Drosophila olfactory receptors using mathematical morphology. Genomics. 2019;111:549–559. doi: 10.1016/j.ygeno.2018.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PRS constructs, evaluations, and PheWAS summary statistics generated during this study are available on the Cancer PRSweb site (see Web Resources).
There are restrictions to the availability of individual-level data of the MGI study due to patient confidentiality; however, researchers who meet the criteria for access to confidential data can apply for access through the University of Michigan Medical School Central Biorepository and from the UK Biobank (see Web Resources).