Summary
PRKACA and PRKACB code for two catalytic subunits (Cα and Cβ) of cAMP-dependent protein kinase (PKA), a pleiotropic holoenzyme that regulates numerous fundamental biological processes such as metabolism, development, memory, and immune response. We report seven unrelated individuals presenting with a multiple congenital malformation syndrome in whom we identified heterozygous germline or mosaic missense variants in PRKACA or PRKACB. Three affected individuals were found with the same PRKACA variant, and the other four had different PRKACB mutations. In most cases, the mutations arose de novo, and two individuals had offspring with the same condition. Nearly all affected individuals and their affected offspring shared an atrioventricular septal defect or a common atrium along with postaxial polydactyly. Additional features included skeletal abnormalities and ectodermal defects of variable severity in five individuals, cognitive deficit in two individuals, and various unusual tumors in one individual. We investigated the structural and functional consequences of the variants identified in PRKACA and PRKACB through the use of several computational and experimental approaches, and we found that they lead to PKA holoenzymes which are more sensitive to activation by cAMP than are the wild-type proteins. Furthermore, expression of PRKACA or PRKACB variants detected in the affected individuals inhibited hedgehog signaling in NIH 3T3 fibroblasts, thereby providing an underlying mechanism for the developmental defects observed in these cases. Our findings highlight the importance of both Cα and Cβ subunits of PKA during human development.
Keywords: PRKACA, PRKACB, PKA, cAMP signaling, hedgehog signaling, GLI transcritpion factors, Ellis-van Creveld syndrome, congenital heart defects, postaxial polydactyly, mosaicism
Main Text
Protein kinase A (PKA) can be found as an inactive tetrameric holoenzyme formed by the association of two catalytic (C) subunits with a regulatory (R) subunit dimer. Activation is achieved through binding of two molecules of cyclic AMP (cAMP) to each R-subunit and subsequent unleashing of the C-subunits to engage substrates. PRKACA (MIM: 601639) and PRKACB (MIM: 176892) code for the highly homologous Cα- and Cβ-subunits, respectively, and the four functionally non-redundant R-subunits (RIα, RIβ, RIIα, and RIIβ) are encoded by four genes (PRKAR1A [MIM: 188830], PRKAR1B [MIM: 176911], PRKAR2A [MIM: 176910], and PRKAR2B [MIM: 176912]). A-kinase anchoring proteins (AKAPs) and PKA inhibitor proteins (PKI) contribute to PKA subcellular localization and function by binding to R-subunits and C-subunits, respectively.1
PKA functions as an intracellular mediator of a variety of G-protein coupled receptor (GPCR) ligands, including specific hormones. Signaling from GPCRs coupled to protein Gαs stimulates adenylate cyclase, leading to increased levels of cAMP and consequently to higher PKA activity. The cAMP/PKA pathway is known to play a central role in the endocrine system because, in addition to mediating the effects of various hormones, it regulates hormone secretion and the proliferation of endocrine cells.2
In vertebrates, PKA also works to restrain hedgehog (Hh) signaling through phosphorylation of GLI transcription factors.3,4 PKA-mediated phosphorylation of full-length GLI3 (GLI3-FL [MIM: 165240]) promotes the conversion of this factor into a strong transcriptional repressor (GLI3R) of Hh-target genes by inducing the proteolytic processing of its C-terminus. GLI3 has a dual function, and uncleaved GLI3FL can be transformed into a transcriptional activator (GLI3A). Hh ligands counteract the activity of PKA by de-repressing the main Hh signal transducer Smoothened (SMO [MIM: 601500]), which is classified as a Frizzled-class GPCR, and recruiting it into the primary cilium. Although the mechanism by which SMO regulates PKA is not fully elucidated, activated SMO suppresses PKA activity, at least partially, by removing from cilia the GPCR GPR161 [MIM: 612250], which presumably operates by increasing the levels of cAMP.4, 5, 6
PRKACA germline copy number gains have previously been associated with cortisol-producing bilateral adrenal hyperplasias and Cushing's syndrome (CS [MIM: 615830]),7,8 and PRKACA somatic mutations are also found in tumors: cortisol-producing adrenal adenomas of CS individuals, hypothalamic hamartomas, and cardiac myxomas.7,9,10 Similarly, a PRKACB somatic mutation was detected in tumor DNA from a CS individual,11 and a 1p31.1 triplication encompassing PRKACB was described in another individual who had a specific form of Carney complex (CNC [MIM: 160980]) characterized by skin pigmentation, acromegaly, and myxomas, but not CS.12
Herein, we studied seven unrelated individuals of different ancestries (P1–P7; Figure 1A–B), all born to non-consanguineous healthy parents, who presented with congenital defects. Two individuals had offspring with the same condition, and the other five were simplex cases. All probands had limb abnormalities consisting of postaxial polydactyly of the hands (6/7 bilateral; 1/7 unilateral) and feet (4/7 bilateral; 1/7 unilateral) and brachydactyly (4/7). Congenital heart defects comprising common atrium or an atrioventricular septal defect (AVSD) were observed in 5/7 individuals. The two probands (P1, P2) without a heart condition had offspring with AVSD. Additionally, short stature/length, short limbs, narrow chest, abnormal teeth, oral frenula, nail dysplasia, and intellectual disability were features present in more than one affected individual. One proband had a history of unusual tumors. Affected individuals were initially diagnosed as having either Ellis-van Creveld syndrome (EvC; MIM: 225500), Weyers acrodental dysostosis (WAD; MIM: 193530), or an undiagnosed syndrome, depending on the presence and severity of chondroectodermal features (Table 1). Serum levels of hormones and bone metabolic markers were assessed in four affected individuals (P1, P2, P4, P7). Endocrine investigations did not show hypercortisolism or an overt endocrine dysfunction. Adrenal imaging in the same four probands (P1, P2, P4, P7) was also negative for adrenal abnormalities. An extended clinical description of the affected individuals is available as Supplemental Information (Supplemental Case Reports). The study was conducted in accordance with the declaration of Helsinki for medical research involving human subjects and was approved by the corresponding institutional ethics committees of the participant institutions. All affected individuals or their legal guardians and family members provided written informed consent for their participation in the study and publication of photographs.
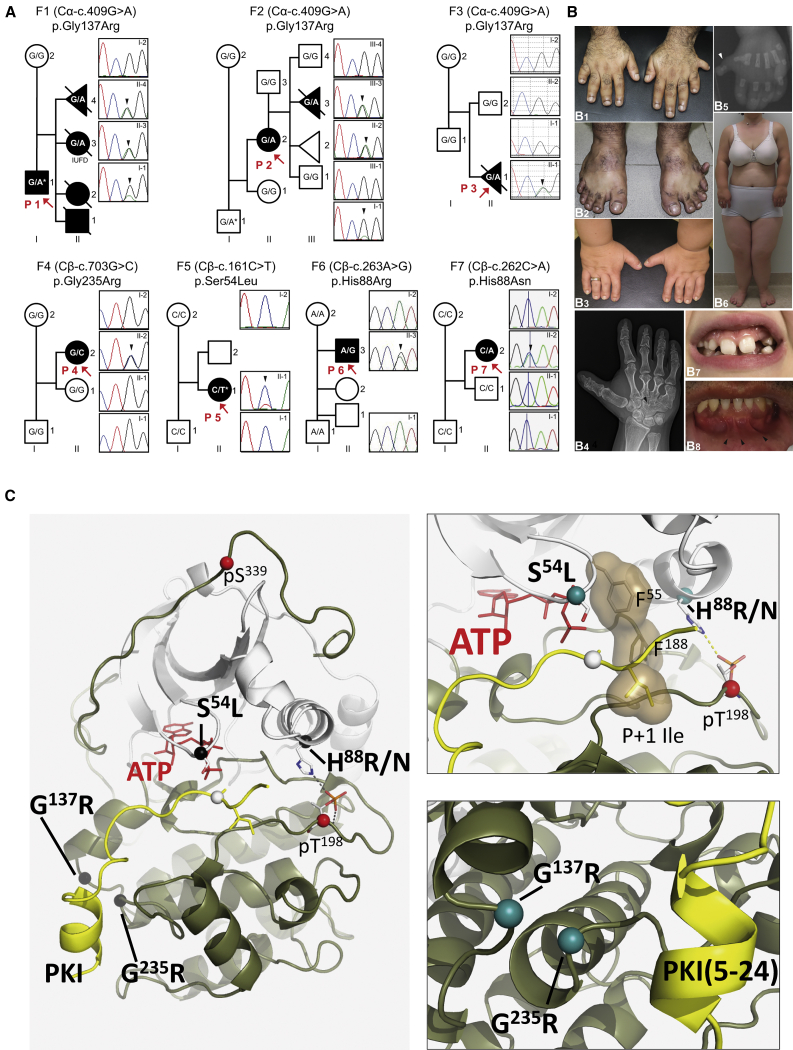
Figure 1.
Affected Individuals and Mutations
(A) Family (F) pedigrees of the seven probands (P; red arrows) of this study and DNA sequence electropherograms illustrating mutations (black arrowheads) and their co-segregation with the disease phenotype. Asterisks denote mosaic state of the corresponding mutation in P1, the father of P2 (I-1) and in P5. IUFD: intrauterine fetal death.
(B) Clinical images. Hands and feet of P1 with bilateral postaxial polydactyly and wide sandal gap. The extra digit of the right hand and foot were surgically removed (B1 and B2). Hands of P2 demonstrating brachydactyly and nail dysplasia. Postaxial polydactyly had been previously corrected (B3). Radiograph of the right hand of P2 with carpal bone fusion (arrowhead) and brachydactyly (B4). Hand radiograph of III-3 (F2) showing postaxial polydactyly (arrowhead) (B5). Clinical image of P2 demonstrating short stature with short limbs (B6). Diastema and abnormal teeth in P4 at age 9 years (B7). Orodental features of P1 with diastema and multiple lower lingual frenula (arrowheads) (B8).
(C) Sites of mutations in the catalytic subunit of PKA. In the full-length C-subunit (left), the mutations are black spheres. p.Ser54Leu and p.His88Arg/Asn are near the active site (top right, teal spheres) and p.Gly137Arg and p.Gly235Arg (bottom right) are at a tethering surface that interacts with partner proteins, in this case the PKA inhibitor (PKI) peptide (yellow), whose tethering helix docks onto this site.
Table 1.
Summary of Clinical and Molecular Findings of Affected Individuals
| Clinical features (ethnic origin) | P1 (Egypt) | P2 (Belgium) | P3 (Italy) | P4 (Denmark) | P5 (France) | P6 (France) | P7 (Australia) |
|---|---|---|---|---|---|---|---|
| Age, gender | 33 years, male | 42 years, female | fetus (23 weeks), female | 18 years, female | 15 years, female | 20 years, male | 41 years, female |
| Height | 165 cm (−1.61 SD) | 139 cm (−5 SD); disproportionate short stature | fetus length: 27 cm (<3%) | 163 cm (−1 SD) | 148.5 cm (−1.8 SD) | 175 cm (0 SD) | 165 cm (−0.19 SD) |
| Weight | 97 kg (+1.74 SD) | 61,5 kg (+0.46 SD) | 467 g (25% < p < 50%) | 47.3 kg (−2.8 SD) | 47 kg (−1 SD) | 53 kg (−1.5 SD) | 51.2 kg (−1.03 SD) |
| Head circumference | 57 cm (+1.32 SD) | 52,6 cm (−1.6 SD) | not available | 51 cm (−3 SD) | 55 cm (M) | 56,5 cm (+ 0.5 SD) | 56 cm (+ 1.18 SD) |
| Congenital heart abnormalities | no, but present in the affected offspring of the proband | no, but present in the affected offspring of the proband | yes, AVSD with myocardial hypertrophy | yes, AVSD and left cava superior entering into the coronary sinus | yes, single atrium, mitral anomaly | yes, single atrium, mild mitral valve regurgitation | yes, single atrium, surgically corrected in infancy; atrial fibrillation in adulthood with persistent incompetence of the valves |
| Postaxial polydactyly of the hands | yes, bilateral | yes, bilateral | yes, bilateral | yes, bilateral | yes, bilateral | yes, bilateral | yes, unilateral (right hand) |
| Postaxial polydactyly of the feet | yes, bilateral | no | yes, unilateral (hexadactyly of the left foot) | yes, bilateral | no | yes, bilateral | yes, bilateral |
| Other hands/feet anomalies | brachydactyly | brachydactyly; fusion of hamate and capitate in right hand | not reported | short and broad with shortening of middle and distal phalanges and toes | brachydactyly and large great toe | fifth finger clinodactyly (unilateral) | fifth finger clinodactyly, broad toes, and mild digital clubbing |
| Long trunk | yes | yes, in childhood | not reported | yes | yes, moderate | no | no |
| Narrow thorax | no, but present in the affected offspring of the proband | yes, in childhood | yes, short ribs | yes | yes, moderate | no | no |
| Upper/lower limb shortening | no (arm span 162 cm), but present in the affected offspring of the proband | yes (arm span 121 cm) | micromelia | yes | no | no | no |
| Genu valgum | yes | yes | not available | yes, genu valgum and previous surgery for coxa vara | yes | no | no, but recurrent dislocated patellae |
| Teeth abnormalities | yes, congenitally missing upper lateral incisors bilateral and lower right lateral incisor. Diastema | yes, conical teeth; early decay | not available | small central maxillary incisors, conical right canine, and hypodontia, invagination, agenesis, and supernumerary teeth of lateral mandibular incisors | yes, hypodontia | no | no |
| Nail dysplasia | no, but present in the affected offspring of the proband | yes | not available | yes, especially on the toes, also broad nails on both thumbs | no | no | no |
| Facial/lip abnormalities | long face with mid face hypoplasia, short philtrum, overhanging nasal tip | notched upper lip | not available | long face, short and deep philtrum, tented upper lip | long face | no | broad forehead, hypertelorism, prognathism, prominent nasal tip |
| Multiple frenula or abnormal gum-lip attachment | yes, multiple upper and lower lingual frenula, hypoplastic maxilla with cross bite | multiple oral frenula at lower lip present at birth | not available | yes, abnormal gum-lip attachment | multiple oral frenula at lower lip | no | no |
| Intellectual disability | no | no | not applicable, fetus with brain edema | no, in childhood a period with mild developmental delay including mild language delay, gross motor difficulties, balance problems, and concentration problems; later diagnosed with dyslexia | no | yes, mild intellectual disability, reading and writing acquired, severe anxiety | yes, severe intellectual disability with autistic features. Medically refractory focal epilepsy |
| Neoplastic lesions | absent at age 33 years | absent at age 42 years | not reported | absent at age 18 years | absent at age 15 years | absent at age 20 years | yes, grade 1 borderline mucinous ovarian tumor, liver haemangioma, renal cell carcinoma |
| Clinical diagnosis | WAD | EvC | EvC | EvC | EvC | common atrium and polydactyly | common atrium and polydactyly |
| Affected gene variant description (GRCh37/hg19) |
PRKACA chr19: 14211648 C>T NM_002730.4: c.409 G>A p.Gly137Arg |
PRKACA chr19: 14211648 C>T NM_002730.4: c.409 G>A p.Gly137Arg |
PRKACA chr19: 14211648 C>T NM_002730.4: c.409 G>A p.Gly137Arg |
PRKACB chr1: 84668426 G>C NM_002731.3: c.703G>C p.Gly235Arg |
PRKACB Chr1: 84647935 C>T NM_002731.3: c.161C>T p.Ser54Leu |
PRKACB Chr1: 84649745 A>G NM_002731.3: c.263A>G p.His88Arg |
PRKACB Chr1: 84649744 C>A NM_002731.3: c.262C>A p.His88Asn |
| Inheritance | mosaic | inherited | de novo | de novo | mosaic | de novo | de novo |
| NGS; altered allele reads/total read depth | 0.28 (811/2,858)a | 0.55 (41/74) | detected via Sanger sequencing; equal representation of altered and reference alleles in sequencing chromatograms | 0.42 (102/239) | 0.32 (39/122) | 0.54 (20/37) | 0.31 (4/13) (the mutant allele was demonstrated to be in the heterozygous state in blood-derived DNA of P7 [~59% mutant allele frequency] and absent in both parents via droplet digital PCR [ddPCR]) |
| Other affected family members | yes, two offspring with postaxial polydactyly of both hands, short limbs, and congenital heart septal defects; both died early after birth; an IUFD at 33 weeks of gestation with bilateral postaxial polydactyly and congenital heart disease and a fetus with similar manifestations | yes, one affected fetus with short limbs, narrow thorax, postaxial polydactyly of both hands, and complete AVSD | no | no | no | no | no |
| Other information | surgery for a lobar emphysema in the left lung at the age of 2 years | the fetus presented with bicornuate and didelphys uterus; lungs with immature parenchyma at canalicular stage were also observed | dural ectasia and osteoporosis with multiple fractures |
P: Proband. M: median or 50th percentile. AVSD: atrioventricular septal defect. WAD: Weyers acrodental dysostosis. EvC: Ellis-van Creveld syndrome. IUFD: Intrauterine fetal death. NGS: next-generation sequencing.
To further confirm the mosaic state of the mutation detected by initial standard WES, additional deep WES was carried out in P1. Deep WES was also performed in the father of P2 (altered allele reads (508)/total read depth (3,097) = 0.16).
After we excluded mutations in the EvC genes (EVC [MIM: 604831 and EVC2 [MIM: 607261]), we conducted whole-exome sequencing (WES) in families 1 and 2. This analysis identified the same heterozygous missense variant in PRKACA (GenBank: NM_002730.4), c.409G>A (p.Gly137Arg), in both unrelated families. Remarkably, this mutation was also found in individual P3. The c.409G>A variant was mosaic in the unaffected father of P2 (variant allele fraction [VAF] = 0.16; altered allele read depth = 508/total read depth = 3,097), and was germline-transmitted in P2 (VAF = 0.55) and her affected offspring (VAF = 0.46). P1 was also mosaic for the same PRKACA variant (VAF = 0.28; 811/2,858), and his two affected offspring from whom there was available DNA (II-3 and II-4 in Figure 1A); both carried the variant in the heterozygous state. In P3, the mutation was identified as de novo. Next-generation sequencing (NGS) data for the c.409G>A variant and pedigree segregation were confirmed via Sanger sequencing in each family (Figure 1A, Table 1). Trio-WES in P4, P6, and P7 did not detect changes in PRKACA but revealed different de novo heterozygous missense variants predicted to be damaging in PRKACB (GenBank: NM_002731.3) in the three affected individuals (P4: c.703G>C [p.Gly235Arg], P6: c.263A>G [p.His88Arg], and P7: c.262C>A [p.His88Asn]). WES analysis of P5 also identified a pathogenic change in PRKACB (c.161C>T [p.Ser54Leu]). All four PRKACB variants were proved to be de novo through the use of Sanger sequencing. In P5, the mutation was present in 32% of NGS reads (VAF = 0.32; 39/122) and P5’s electropherograms were consistent with this individual also being mosaic (Figure 1A, Table 1). All PRKACA and PRKACB variants were absent in gnomAD v2.1.1/v313 and involved evolutionarily conserved residues (Figure S1). Detailed WES results, including other variants detected and analysis pipelines used, are provided in the Supplemental Information (Figure S2). The p.Ser54Leu variant was previously identified as a somatic mutation in a cortisol-producing adenoma from an individual with CS.11
We next confirmed expression of Cα and Cβ transcripts in dermal fibroblasts through the use of RT-PCR (Figure S3A). Sequencing of the resulting RT-PCR fragments demonstrated expression of both normal and mutant PRKACA or PRKACB alleles in fibroblasts from affected individuals. We also observed the levels and localization of EvC proteins to be similar between cells from control and affected individuals (Figure S3B–S3C). Similarly, localization of PKA-C was found to be unaffected in PRKACA- or PRKACB-mutant fibroblasts (Figure S3D). In addition, because defects in one PKA subunit can lead to expression changes in other components of the holoenzyme,14, 15, 16 we used qRT-PCR and immunoblotting to study PKA-C and -R expression in dermal fibroblasts. Compared to control cells, fibroblasts from individuals with PRKACB mutations showed a slight increase in the mRNA levels of PRKACA. PKA-C protein levels were also found to be increased in these cells, although statistical significance was only reached respecting one of the two controls included in the analysis. Furthermore, PRKACB mutant cells showed decreased PRKAR1B transcript levels. No significant differences, neither at the mRNA nor at the protein levels, of PKA-C or -R subunits were identified in fibroblasts from individuals with the PRKACA mutation with respect to controls. Changes in RIIβ protein levels were present in cells from both control and affected individuals and therefore cannot be attributed to the mutations (Figure S3E–S3F).
Analysis of the tertiary structure of the C-subunit revealed that mutations cluster in two groups with Cβ-Ser54 and Cβ-His88 being located in or near the Glycine-rich loop (G-Loop) at the active site and Cα-Gly137 and Cβ-Gly235 in a shared pocket at the end of the D and F helices (Figure 1C). Ensemble models, generated for each mutation, showed that both Cβ-p.His88Arg and Cβ-p.His88Asn altered the dynamics of the G-Loop, predictably affecting synergistic ATP and substrate peptide binding.17,18 These mutations and the previously described Cβ-p.Ser54Leu11 likely disrupt ATP-dependent regulation in the G-Loop. Cα-p.Gly137Arg and Cβ-p.Gly235Arg do not affect ATP binding, but they share an interface that forms interactions with regulatory proteins that include PKI, RIα, and RIIβ (Figure 2A–2D and Figure S4).
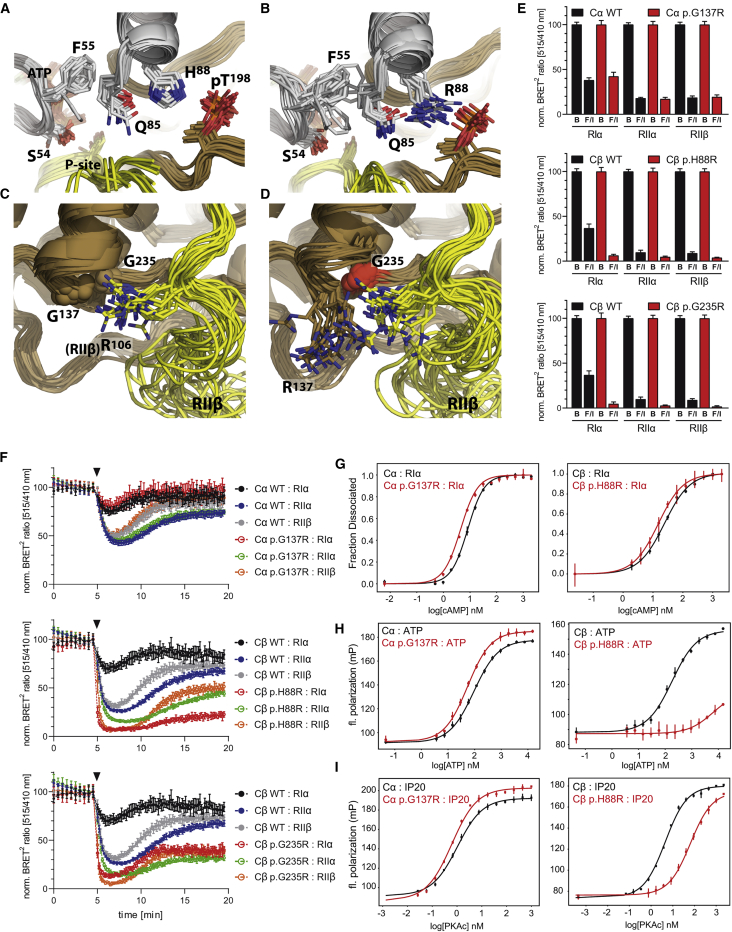
Figure 2.
Structural and Functional Assessment of Mutations
(A–D.) Ensemble model of mutations: (A) A stabilizing four-residue network that involves salt bridges between the activation loop phosphate (phospho-Thr198), the C-helix (His88 and Gln85), and hydrophobic packing of Phe55 of the G-Loop onto Gln85 is found in WT PKA. This interaction modulates the conformational dynamics of the G-Loop.
(B) The p.His88Arg (R88) mutation pulls Gln85 away from Phe55 and releases the G-Loop, likely leading to reduced synergistic binding of ATP and reduced affinity for the ATP-dependent RIα subunit. p.Ser54Leu, in the G-Loop, likely similarly affects ATP-dependent regulation by disrupting the G-Loop dynamics.
(C) In WT PKA, a pocket is formed by the D and F helices, and that pocket is accessed in an RIIβ-specific manner by Arg106 in the inhibitor segment (PDB:3TNP) and by the tethering helix in PKI(5-24).
(D) p.Gly137Arg and p.Gly235Arg disturb this RIIβ-specific interaction. This pocket is also at the RIα cAMP-binding domain-B interface (PDB:6NO7) and at the interface with the tethering helix in PKI (Figure S4B–S4C).
(E) In BRET2 experiments, Cα-p.Gly137Arg shows comparable dissociation to that of Cα-WT upon full cAMP-stimulation by Forskolin/IBMX (F/I) for RIα-, RIIα, and RIIβ-holoenzymes, whereas Cβ-p.His88Arg- and Cβ-p.Gly235Arg-holoenzymes fully dissociate. BRET2 data from unstimulated cells treated with buffer only are designated by the letter B. Normalized data are shown as means ± SD of three independent experiments with n = 6 replicates each (total n = 18).
(F) Kinetic BRET2 analyses demonstrate full dissociation upon addition of the physiological β-adrenergic agonist isoproterenol (100 nM, triangle) for Cβ-p.His88Arg and Cβ-p.Gly235Arg and identical behavior of Cα-WT and Cα-p.Gly137Arg. Data shown are means ± SD of n = 6 replicates showing one of three (two for RIIα) independent experiments. For expression levels of GFP-C-subunits used in BRET, see Figure S5B.
(G) FPA analysis: RIα holoenzyme activation by cAMP shows increased sensitivity compared to WT with both Cα-p.Gly137Arg and Cβ-p.His88Arg (n = 3).
(H–I) FPA: Synergistic binding of ATP (H) with PKI 5-24 (IP20) peptide (I) to the C-subunit shows slightly increased binding affinity compared to WT with Cα-p.Gly137Arg and strongly decreased cooperativity with Cβ-p.His88Arg (n = 3). To illustrate differences in total binding, raw fluorescence polarization is expressed as millipolarization units (mP), and otherwise mP has been converted to fraction dissociated. Graphs show the mean ± SD.
The effect of the identified mutations in PKA holoenzymes was analyzed by using bioluminescence resonance energy transfer (BRET2), which provides in cellulae analysis of holoenzyme dissociation. This study showed a dramatic increase in the sensitivity to cAMP of Cβ-p.His88Arg and Cβ-p.Gly235Arg PKA holoenzymes formed with RIα, RIIα and RIIβ upon Forskolin/IBMX (Figure 2E) or isoproterenol (Figure 2F) stimulation in comparison to the corresponding Cβ-wild-type (Cβ-WT) PKA holoenzymes. Cβ-p.His88Asn showed almost full dissociation upon Forskolin/IBMX (Figure S5A) but a lower response to 100 nM isoproterenol compared to Cβ-p.His88Arg (Figure S5C and Figure 2F). Higher sensitivity to cAMP of the Cβ-p.His88Arg:RIα holoenzyme was additionally demonstrated through the use of fluorescence polarization assays (FPA). Using these assays, we also proved that the reduction in the stability of the Cβ-p.His88Arg:RIα holoenzyme can be attributed to loss of the synergistic effect of ATP binding, which is also true for the PKI peptide (PKI5-24) (Figure 2G–2I; Table S1). In contrast to the Cβ mutations, BRET2 assays showed the dissociation kinetics for Cα-p.Gly137Arg and Cα-WT holoenzymes with RIα, RIIα, and RIIβ to be comparable (Figures 2E-2F). However, using FPA of purified holoenzymes (both RIα and RIIβ), we found greater sensitivity of Cα-p.Gly137Arg to lower cAMP concentration than the wild-type (WT) protein (Figure 2G and Figure S5D). Cα-p.Gly137Arg was additionally characterized with slightly increased cooperative binding for ATP and PKI peptide substrate (Figure 2H–2I; Table S1). Reduced association of Cα-p.Gly137Arg and Cβ-p.Gly235Arg with both RIα and RIIβ compared to their corresponding control C-WT proteins was also observed via co-immunoprecipitation (Figure S5E–S5F). Consistently, the kinase activity of Cα-p.Gly137Arg and Cβ-p.Gly235Arg determined through the use of the PepTag assay, which uses a fluorescent-labeled Kemptide substrate, in extracts from HEK293T co-transfected with both PKA-C and -RIα subunits, was found to be higher than that of their respective WT proteins at low cAMP concentrations (Figure S6).
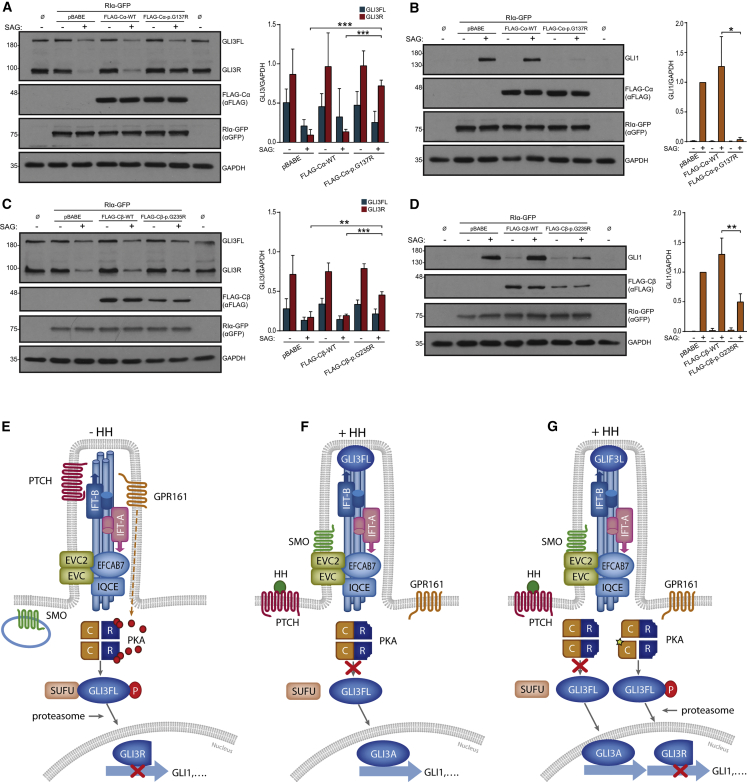
Subsequently, we assessed the effect of mutations in the Hh pathway by ectopically expressing normal or mutant (Cα-p.Gly137Arg or Cβ-p.Gly235Arg) FLAG-tagged C-subunits together with RIα-GFP in NIH 3T3 via retroviral delivery. Notably, after stimulation of the pathway with the SMO-agonist SAG, the cells that were retrotransduced with the mutant C-subunits showed increased levels of GLI3R and reduced expression of the readout of the Hh pathway GLI1 compared to the control cultures, indicating that both Cα- and Cβ-mutations impair SAG-mediated inactivation of PKA in NIH 3T3 (Figure 3A–3D). Results were similar in cells treated only with FLAG-tagged C-subunit retroviruses (Figure S7). A model explaining the pathological mechanism of C-mutations in Hh signaling is shown in Figure 3E–3G.
Figure 3.
PRKACA and PRKACB Mutations Impair Hh Signaling in NIH 3T3
(A–D) Analysis of GLI3 and GLI1 protein levels in NIH 3T3 co-infected with human FLAG-Cα-WT or FLAG-Cα-p.Gly137Arg and RIα-GFP retroviral vectors (A–B) or alternatively with FLAG-Cβ-WT or FLAG-Cβ-p.Gly235Arg and RIα-GFP retroviruses (C–D), exposed to SAG (+) or its vehicle DMSO (-). Non-infected cells are indicated with Ø. Expression levels of FLAG-C and RIα-GFP are shown in the underneath panels. After incubation with SAG, Cα-p.Gly137Arg and Cβ-p.Gly235Arg retrotransduced cells showed increased GLI3R protein levels and reduced expression of GLI1 compared to cells retrotransduced with pBABE (empty vector) or with FLAG-Cα-WT or FLAG-Cβ-WT. Representative immunoblots are on the left and histograms show densitometric quantification of the levels of GLI3R and GLI3FL referred to GAPDH in (A) and (C), or GLI1/GAPDH levels normalized to the value of SAG-pBABE cells in (B) and (D). Data are expressed as mean ± SD from three experiments corresponding to three independent retroviral infections (n = 3). ∗ = p < 0.05; ∗∗ = p < 0.01; ∗∗∗ = p < 0.001. Student’s t-test.
(E–G) Model of action of PKA-Cα/β mutations. (E) In the absence of signal, PTCH (the receptor of Hh ligands [HH]) is in the cilium and represses SMO. GPR161 is also located in the cilium membrane and negatively regulates Hh signaling by promoting adenylyl-cyclase-dependent cAMP synthesis (red spheres). Consequently, PKA holoenzymes are active and their C-subunits (C) are free from R-subunits (R) to phosphorylate GLI3FL, which is bound to the inhibitory protein SUFU. Phosphorylated GLI3FL undergoes C-terminal proteolytic processing by the proteasome and is transformed into GLI3R, leading to reduced expression of Hh targets such as GLI1. (F) The interaction of Hh ligands with PTCH disables this protein to continue repressing SMO and PTCH-HH complexes exit from cilia. De-repressed SMO accumulates into the cilium and interacts with the EvC ciliary complex, which is retained at the base of this organelle through binding of the C-terminal of EVC2 to the EFCAB7-IQCE complex.19 In this manner, SMO signaling is enriched at the EvC region. SMO and the EvC proteins promote GLI3FL-SUFU dissociation and stimulate the recruitment of GLI3 to cilia tips.20, 21, 22 Active SMO additionally causes GPR161 to abandon the cilium, and this, in combination with other not fully understood SMO-mediated mechanisms, results in decreased levels of cAMP and the inactivation of PKA.4,23 Accordingly, GLI3FL phosphorylation is stopped and the production of GLI3R discontinued while GLI3FL is converted into a functional transcriptional activator (GLI3A). (G) The same situation as in (F), but in an individual with a PRKACA or PRKACB mutation. Due to higher cAMP sensitivity of the mutant PKA holoenzymes, the mutant PKA C-subunits (star) remain active following downregulation of cAMP levels associated with the activation of the Hh pathway, thus leading to abnormally increased levels of GLI3R and reduced Hh pathway activity. Affected individuals are expected to have holoenzymes containing two normal or two mutant C-subunits and holoenzymes composed of one normal and one mutant C-subunit. Intraflagellar transport protein complexes (IFT-A and IFT-B) which are also involved in Hh signaling are indicated in (E–G).
In summary, we describe a syndrome involving multiple congenital anomalies caused by germline or mosaic mutations in PRKACA or PRKACB. Affected individuals had a constellation of features with the major shared findings being common atrium/AVSD and postaxial polydactyly. The association of these two features without other defects was postulated as an independent syndrome in a number of reported affected individuals.24,25 Common atrium/AVSD and polydactyly are also part of the clinical spectrum of several ciliopathies, and their co-morbidity is often thought to be a consequence of abnormal Hh signaling.26 Accordingly, germline or mosaic PRKACA or PRKACB mutations may explain the phenotype in other undiagnosed individuals with common atrium/AVSD-polydactyly alone or as part of more complex phenotypes.
Five of the seven probands in this report (P1–P5) showed phenotypic overlap with EvC or its less-severe dominantly inherited allelic form, WAD.27 Biallelic loss-of-function mutations in EVC or EVC2, which encode the two subunits of the EvC ciliary complex (EVC and EVC2), are the primary cause of EvC, whereas specific heterozygous C-terminal truncating mutations in EVC2 are responsible for WAD.28, 29, 30 The EvC complex, which localizes at the base of primary cilia, is required downstream of SMO for complete inhibition of GLI3FL processing in response to Hh ligands.20, 21, 22 Consequently, PKA and EVC-EVC2 act at the same level in the Hh pathway, but in an opposing manner. A scaffolding role for concentrating SMO signaling to the cilium base has been proposed for the EVC-EVC2 complex.4,21 Given the overlap between EvC/WAD and PRKACA/B phenotypes, EVC-EVC2 could specifically link SMO signaling to PKA acting as scaffold, or be involved in a biochemical reaction to prevent the phosphorylation of GLI3 by PKA. Intriguingly, RIα is known to bind specifically to GPR161.31 The ciliopathy-like phenotype of the individuals of this report is in agreement with the negative effect caused by the identified PRKACA and PRKACB variants on Hh signaling. However, we cannot rule out the possibility that these mutations could also alter additional molecular pathways regulated by PKA that may be contributing to the phenotype. Skeletal defects have been reported in association with variants in other PKA subunits or other components of cAMP/PKA signaling. Specific variants in PRKAR1A lead to acrodysosotosis type 1 (MIM: 101800),32 and variants in the cAMP phosphodiesterase encoded by PDE4D lead to acrodysostosis type 2 (MIM 614613).33,34 The skeletal phenotype of acrodysostosis (brachydactyly, short stature, facial dysostosis, and nasal hypoplasia) is similar to that of Albright hereditary osteodystrophy and does not typically resemble a ciliopathy. Loss-of-function mutations in PRKAR1A resulting in unrestricted PKA activity cause CNC,35 which is a condition characterized by skin pigmentary abnormalities, endocrine tumors or overactivity, and other tumors such as myxomas or schwannomas. However, polydactyly, common atrium/AVSD, and skeletal and ectodermal defects are not considered to be part of the CNC diagnostic criteria.36 Prkar1a+/− mice have also been shown to be prone to developing bone lesions.14,15 Considering the individuals reported here, only P7 had tumors, but she did not have evidence of CS, and to date, no adrenal, pituitary, or thyroid tumors have been found on imaging. She did not have the typical skin manifestations of CNC, either. Whether the presence of tumors in P7 is due to the Cβ-p.His88Asn variant needs to be clarified through further investigations. The hormonal profile in the four affected individuals analyzed did not show signs of overt endocrine alterations, and until now, bone tumors have not been identified in any of the affected individuals.
We show that the mutations reported here affect the interaction of C- and R-subunits through an ATP-dependent mechanism (for p.His88Arg, p.His88Asn, and p.Ser54Leu) or through disruption of interfacial surfaces (for p.Gly137Arg and p.Gly235Arg), creating holoenzymes that are more sensitive to cAMP for different reasons. This implies that the mutant Cα- and Cβ-subunits remain more active following the downregulation of cAMP levels associated with Hh signaling,23 thus decreasing the strength of this pathway. Indeed, diminished Hh signaling activity was observed in NIH 3T3 ectopically expressing mutant C-subunits. Of note, using random mutagenesis in a plasmid containing the mouse Cα subunit, Orellana and McKnight described a p.His87Gln variant (p.His88Gln using our variant nomenclature) that compared to the WT protein retained partial activity in the presence of an excess of RIα subunit.37
In our assays, Cα-p.Gly137Arg caused a less severe impact in PKA holoenzymes than the Cβ mutations did. Because Cα is the major PKA C-subunit and is ubiquitously expressed, whereas Cβ is mainly expressed in brain and lymphoid tissues,38 mutations in Cβ may need to be more damaging than in Cα in order to cause a phenotype in tissues with low Cβ expression. Nonetheless, we cannot discard the possibility that Cα-p.Gly137Arg could also alter an unknown Hh-specific regulatory mechanism of PKA inactivation. During the preparation of this manuscript, we became aware of a large-scale clinical exome sequencing study compiling WES results from >2,200 Saudi families; in this study, the Cα-p.Gly137Arg variant was observed to be de novo in one affected individual with clinical suspicion of EvC. The WES result of this individual was stated as ambiguous, and the case was considered not solved because of the unknown causality of the change, which is now demonstrated by our data.39 This observation further reinforces the recurrent character of the Cα-p.Gly137Arg mutation. While much is known about the Cα-subunit, surprisingly little is known about the Cβ-subunit. Our discovery of these mutations underscores the need to now distinguish between the structural and functional differences of Cβ splice variants that remain as an unexplored frontier. Our findings demonstrate a critical role of both Cα- and Cβ-subunits of PKA in human development.
Data and Code Availability
Specific datasets supporting this article or additional information not subjected to ethical restrictions can be obtained from the corresponding author upon request.
Declaration of Interests
I.E.S has served on scientific advisory boards for UCB, Eisai, GlaxoSmithKline, BioMarin, Nutricia, and Xenon Pharmaceuticals and on editorial boards of the Annals of Neurology, Neurology and Epileptic Disorders; may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound, a patent for SCN1A testing held by Bionomics Inc. and licensed to various diagnostic companies; has received speaker honoraria from GlaxoSmithKline, Athena Diagnostics, UCB, BioMarin, Biocodex, Eisai, and Transgenomics; and has received funding for travel from Athena Diagnostics, UCB, Biocodex, GlaxoSmithKline, Biomarin, and Eisai. The remaining authors declare no competing interests.
Acknowledgments
We thank all affected individuals, their siblings, and their parents or legal guardians for their participation in this study. Some individuals were included after a GeneMatcher match.40 This work was partially supported by funding from the Spanish Ministry of Science, Innovation and Universities (SAF2016-75434-R [AEI/FEDER, UE] and PID2019-105620RB-I00/AEI/10.13039/501100011033) to V.L.R.-P. S.S.T. was supported by NIH grant R03TR002947, E.M.F.M. by Kassel graduate school “Clocks”, and A.D.L. by the Italian Ministry of Health (RC-2019). The University of Antwerp supported G.M. and W.V.H. with Methusalem funding (FFB190208) and S.P. with a predoctoral grant. E.B. was supported by The Research Foundation Flanders with a postdoctoral grant (12A3814N). The study was also funded by a National Health and Medical Research Council Program Grant (1091593) to I.E.S., a Practitioner Fellowship (1006110) to I.E.S., a Senior Research Fellowship (1102971) to M.B., and an R.D. Wright Career Development Fellowship (1063799) to M.S.H. B.S.S. and G.L. were supported by Throne Holst Foundation UiO (2019-2021) and Strategic PhD fund by UiO/IMB.
Published: October 14, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.09.005.
Web Resources
gnomAD Browser, https://gnomad.broadinstitute.org/
OMIM, https://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Søberg K., Skålhegg B.S. The Molecular Basis for Specificity at the Level of the Protein Kinase a Catalytic Subunit. Front. Endocrinol. (Lausanne) 2018;9:538. doi: 10.3389/fendo.2018.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peverelli E., Mantovani G., Lania A.G., Spada A. cAMP in the pituitary: an old messenger for multiple signals. J. Mol. Endocrinol. 2013;52:R67–R77. doi: 10.1530/JME-13-0172. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y., Roelink H., McKnight G.S. Protein kinase A deficiency causes axially localized neural tube defects in mice. J. Biol. Chem. 2002;277:19889–19896. doi: 10.1074/jbc.M111412200. [DOI] [PubMed] [Google Scholar]
- 4.Kong J.H., Siebold C., Rohatgi R. Biochemical mechanisms of vertebrate hedgehog signaling. Development. 2019;146:dev166892. doi: 10.1242/dev.166892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S.J., Jackson P.K. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 6.Nachury M.V., Mick D.U. Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 2019;20:389–405. doi: 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beuschlein F., Fassnacht M., Assié G., Calebiro D., Stratakis C.A., Osswald A., Ronchi C.L., Wieland T., Sbiera S., Faucz F.R. Constitutive activation of PKA catalytic subunit in adrenal Cushing’s syndrome. N. Engl. J. Med. 2014;370:1019–1028. doi: 10.1056/NEJMoa1310359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carney J.A., Lyssikatos C., Lodish M.B., Stratakis C.A. Germline PRKACA amplification leads to Cushing syndrome caused by 3 adrenocortical pathologic phenotypes. Hum. Pathol. 2015;46:40–49. doi: 10.1016/j.humpath.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hildebrand M.S., Griffin N.G., Damiano J.A., Cops E.J., Burgess R., Ozturk E., Jones N.C., Leventer R.J., Freeman J.L., Harvey A.S. Mutations of the Sonic Hedgehog Pathway Underlie Hypothalamic Hamartoma with Gelastic Epilepsy. Am. J. Hum. Genet. 2016;99:423–429. doi: 10.1016/j.ajhg.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng I.C., Huang W.J., Jhuang Y.L., Chang Y.Y., Hsu H.P., Jeng Y.M. Microinsertions in PRKACA cause activation of the protein kinase A pathway in cardiac myxoma. J. Pathol. 2017;242:134–139. doi: 10.1002/path.4899. [DOI] [PubMed] [Google Scholar]
- 11.Espiard S., Knape M.J., Bathon K., Assié G., Rizk-Rabin M., Faillot S., Luscap-Rondof W., Abid D., Guignat L., Calebiro D. Activating PRKACB somatic mutation in cortisol-producing adenomas. JCI Insight. 2018;3:e98296. doi: 10.1172/jci.insight.98296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forlino A., Vetro A., Garavelli L., Ciccone R., London E., Stratakis C.A., Zuffardi O. PRKACB and Carney complex. N. Engl. J. Med. 2014;370:1065–1067. doi: 10.1056/NEJMc1309730. [DOI] [PubMed] [Google Scholar]
- 13.Karczewski K.J.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 14.Liu S., Saloustros E., Mertz E.L., Tsang K., Starost M.F., Salpea P., Faucz F.R., Szarek E., Nesterova M., Leikin S., Stratakis C.A. Haploinsufficiency for either one of the type-II regulatory subunits of protein kinase A improves the bone phenotype of Prkar1a+/- mice. Hum. Mol. Genet. 2015;24:6080–6092. doi: 10.1093/hmg/ddv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang K.M., Starost M.F., Nesterova M., Boikos S.A., Watkins T., Almeida M.Q., Harran M., Li A., Collins M.T., Cheadle C. Alternate protein kinase A activity identifies a unique population of stromal cells in adult bone. Proc. Natl. Acad. Sci. USA. 2010;107:8683–8688. doi: 10.1073/pnas.1003680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z., Pringle D.R., Jones G.N., Kelly K.M., Kirschner L.S. Differential role of PKA catalytic subunits in mediating phenotypes caused by knockout of the Carney complex gene Prkar1a. Mol. Endocrinol. 2011;25:1786–1793. doi: 10.1210/me.2011-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor S.S., Yang J., Wu J., Haste N.M., Radzio-Andzelm E., Anand G. PKA: a portrait of protein kinase dynamics. Biochim. Biophys. Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 18.Lew J., Coruh N., Tsigelny I., Garrod S., Taylor S.S. Synergistic binding of nucleotides and inhibitors to cAMP-dependent protein kinase examined by acrylodan fluorescence spectroscopy. J. Biol. Chem. 1997;272:1507–1513. doi: 10.1074/jbc.272.3.1507. [DOI] [PubMed] [Google Scholar]
- 19.Pusapati G.V., Hughes C.E., Dorn K.V., Zhang D., Sugianto P., Aravind L., Rohatgi R. EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Dev. Cell. 2014;28:483–496. doi: 10.1016/j.devcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caparrós-Martín J.A., Valencia M., Reytor E., Pacheco M., Fernandez M., Perez-Aytes A., Gean E., Lapunzina P., Peters H., Goodship J.A., Ruiz-Perez V.L. The ciliary Evc/Evc2 complex interacts with Smo and controls Hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum. Mol. Genet. 2013;22:124–139. doi: 10.1093/hmg/dds409. [DOI] [PubMed] [Google Scholar]
- 21.Dorn K.V., Hughes C.E., Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell. 2012;23:823–835. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C., Chen W., Chen Y., Jiang J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell Res. 2012;22:1593–1604. doi: 10.1038/cr.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore B.S., Stepanchick A.N., Tewson P.H., Hartle C.M., Zhang J., Quinn A.M., Hughes T.E., Mirshahi T. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc. Natl. Acad. Sci. USA. 2016;113:13069–13074. doi: 10.1073/pnas.1602393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin S.E., Dansky R., Milner S., Benatar A., Govendrageloo K., du Plessis J. Atrioventricular septal defect and type A postaxial polydactyly without other major associated anomalies: a specific association. Pediatr. Cardiol. 1995;16:242–246. doi: 10.1007/BF00795716. [DOI] [PubMed] [Google Scholar]
- 25.Onat T. Post-axial hexodactily and single atrium: a new syndrome? Hum. Genet. 1994;94:104–106. doi: 10.1007/BF02272854. [DOI] [PubMed] [Google Scholar]
- 26.Digilio M.C., Pugnaloni F., De Luca A., Calcagni G., Baban A., Dentici M.L., Versacci P., Dallapiccola B., Tartaglia M., Marino B. Atrioventricular canal defect and genetic syndromes: The unifying role of sonic hedgehog. Clin. Genet. 2019;95:268–276. doi: 10.1111/cge.13375. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz-Perez V.L., Goodship J.A. Ellis-van Creveld syndrome and Weyers acrodental dysostosis are caused by cilia-mediated diminished response to hedgehog ligands. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:341–351. doi: 10.1002/ajmg.c.30226. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Perez V.L., Ide S.E., Strom T.M., Lorenz B., Wilson D., Woods K., King L., Francomano C., Freisinger P., Spranger S. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat. Genet. 2000;24:283–286. doi: 10.1038/73508. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Perez V.L., Tompson S.W., Blair H.J., Espinoza-Valdez C., Lapunzina P., Silva E.O., Hamel B., Gibbs J.L., Young I.D., Wright M.J., Goodship J.A. Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am. J. Hum. Genet. 2003;72:728–732. doi: 10.1086/368063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valencia M., Lapunzina P., Lim D., Zannolli R., Bartholdi D., Wollnik B., Al-Ajlouni O., Eid S.S., Cox H., Buoni S. Widening the mutation spectrum of EVC and EVC2: ectopic expression of Weyer variants in NIH 3T3 fibroblasts disrupts Hedgehog signaling. Hum. Mutat. 2009;30:1667–1675. doi: 10.1002/humu.21117. [DOI] [PubMed] [Google Scholar]
- 31.Bachmann V.A., Mayrhofer J.E., Ilouz R., Tschaikner P., Raffeiner P., Röck R., Courcelles M., Apelt F., Lu T.W., Baillie G.S. Gpr161 anchoring of PKA consolidates GPCR and cAMP signaling. Proc. Natl. Acad. Sci. USA. 2016;113:7786–7791. doi: 10.1073/pnas.1608061113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linglart A., Menguy C., Couvineau A., Auzan C., Gunes Y., Cancel M., Motte E., Pinto G., Chanson P., Bougnères P. Recurrent PRKAR1A mutation in acrodysostosis with hormone resistance. N. Engl. J. Med. 2011;364:2218–2226. doi: 10.1056/NEJMoa1012717. [DOI] [PubMed] [Google Scholar]
- 33.Lee H., Graham J.M., Jr., Rimoin D.L., Lachman R.S., Krejci P., Tompson S.W., Nelson S.F., Krakow D., Cohn D.H. Exome sequencing identifies PDE4D mutations in acrodysostosis. Am. J. Hum. Genet. 2012;90:746–751. doi: 10.1016/j.ajhg.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michot C., Le Goff C., Goldenberg A., Abhyankar A., Klein C., Kinning E., Guerrot A.M., Flahaut P., Duncombe A., Baujat G. Exome sequencing identifies PDE4D mutations as another cause of acrodysostosis. Am. J. Hum. Genet. 2012;90:740–745. doi: 10.1016/j.ajhg.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirschner L.S., Carney J.A., Pack S.D., Taymans S.E., Giatzakis C., Cho Y.S., Cho-Chung Y.S., Stratakis C.A. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat. Genet. 2000;26:89–92. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- 36.Mateus C., Palangié A., Franck N., Groussin L., Bertagna X., Avril M.F., Bertherat J., Dupin N. Heterogeneity of skin manifestations in patients with Carney complex. J. Am. Acad. Dermatol. 2008;59:801–810. doi: 10.1016/j.jaad.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 37.Orellana S.A., McKnight G.S. Mutations in the catalytic subunit of cAMP-dependent protein kinase result in unregulated biological activity. Proc. Natl. Acad. Sci. USA. 1992;89:4726–4730. doi: 10.1073/pnas.89.10.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cadd G., McKnight G.S. Distinct patterns of cAMP-dependent protein kinase gene expression in mouse brain. Neuron. 1989;3:71–79. doi: 10.1016/0896-6273(89)90116-5. [DOI] [PubMed] [Google Scholar]
- 39.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Specific datasets supporting this article or additional information not subjected to ethical restrictions can be obtained from the corresponding author upon request.