Abstract
Food allergy (FA) affects 4–10% of children, especially children with atopic dermatitis (AD). During infancy the gut microbiome may determine both the course of FA and tolerance to food allergens. Analogically, the skin microbiome changes in the course of AD. Most studies have associated FA with a lower abundance and diversity of Lactobacillales and Clostridiales, but greater numbers of Enterobacterales, while AD in children has been associated with lower numbers of Staphylococcus epidermidis and S. hominis but an abundance of S. aureus and Streptococcus species. An understanding of the impact of the microbiome on the clinical course of FA and AD may allow for the development of new models of allergy treatment and prevention.
Keywords: microbiome, food allergy, atopic dermatitis, infants
The microbiome as a link between food allergy and atopic dermatitis
Although food allergy (FA) affects 4–10% of children [1], it is more frequently encountered in those with atopic dermatitis (AD), with around 15–30% of children with AD also being affected by FA [2]. AD is the most frequently observed chronic skin disease in children, affecting 15–20% of children worldwide, and is in turn more common in FA children than in the general population [1, 2]. The incidence of both FA and AD is growing [3]. Although a connection has long been recognized between the two diseases, the significance of FA in AD pathogenesis is still controversial. It has been shown that FA is connected with the exacerbation, persistence and earlier onset of AD [4], and that AD may precede FA [5]. Recent studies have shown that skin sensitization to food antigens has a significant influence on the occurrence of FA, suggesting a strong link between the skin and gut immune responses [6]. Some studies suggest that the skin and gut, which both constitute physical barriers, are linked via modulation of the immune environment via the microbiome [7, 8]. Currently, it is not clear whether modulation of the gut microbiome can impact upon the skin microbiome and vice versa. It is still not elucidated whether the skin and gut microbiomes cooperate together or can influence each other. Additionally, it is not known whether any microbiome pattern, common for skin and gut, exists and can determine sensitization to food and environmental allergens, or may predispose its host to develop an allergy.
Certain studies report that during early infancy, the microbiome may determine both the development and clinical course of food allergy as well as tolerance to food allergens [9–11]. Analogically, changes in skin microbiome composition are observed in the course of AD. A decreased abundance of Staphylococci during early infancy may predict the onset of AD until the end of the first year of life, while an increase of microbiome diversity during the course of AD treatment might indicate a high probability of remission [12, 13].
Moreover, the composition of gut microbiota is closely correlated with the development of skin inflammation. The presence of Escherichia coli in stools is associated with a higher risk of developing eczema, while Clostridium difficile is related to increasing severity of AD [14]. Alterations in the gut microbiome may be associated with AD through the action of bacterial genes that can modulate host immune cell function [15].
The gut microbiome
Development of the gut microbiome
The infant gut microbiome is composed mainly (94–98%) of bacteria coming from four phyla: Firmicutes (~64%), Bacteroidetes (~23%), Actinobacteria (~3%) and Proteobacteria (~5%). Other phyla contribute small numbers of cells and their shares vary [9, 16]. The intestines of foetuses are sterile, and colonization begins at delivery [17, 18]; however, it has been suggested that in utero colonization may be possible [19]. The initial microbiomes develop during the first 6 weeks of life, evolving to reach stability at approximately the second or third year of life [20]. Later, only environmental factors may exert any significant impact on the composition of the intestinal microbiome [17].
The precise composition of microbial strains is characteristic of a given individual. Studies on twins with similar microbiomes suggest that the genetic makeup of the host may exert a strong influence on the microbiome [21]. It was established that genetically determined physiological features of human organism, such as metabolism or immunological system, influenced colonization. It seems, however, that this relation is reciprocal, i.e. the microbiome also influences these traits in humans. On the other hand, environmental factors, such as diet, lifestyle, family size, pet owning [22–24] or medical treatment (method of delivery, antibiotic therapy, probiotic therapy) are important for shaping the microbiome and its development (Table 1) [16, 18, 25–28].
Table 1.
Factors modifying intestinal microbiome development and shaping its structure
| Factors | Commentary |
|---|---|
| Method of delivery | Natural delivery causes colonization of the infant gut by microorganisms thriving in the maternal reproductive and gastrointestinal tracts [18, 25] Caesarean section brings about colonization by bacteria dwelling on the skin of the mother and in the surrounding environment [16, 25] The composition of the gut microbiome in children delivered naturally may be different from that of children delivered by Caesarean section for at least a few months [16, 18] The stools of infants born by Caesarean delivery had lower amounts of bacteria [26] |
| Perinatal antibiotic therapy in the mother | Perinatal antibiotic therapy in the mother could be connected with dysbiosis of the gut in the child. This condition has been found to continue even until the end of the first year of life [27] |
| Probiotic treatment in the mother | Supplementation taken by the mother from 36 Hbd until the third month post-delivery (during the breastfeeding period) influences the microbiome of the infant gut, but among the supplemented bacteria (Lactobacillus (L.) rhamnosus LGG, L. acidophilus Ja-5, Bifidobacterium animalis subspecies (ssp.) lactis Bb-12) only L. rhamnosus LGG was found in increased amounts in infant stools at 10 days and 3 months post-delivery [28] However, supplementation was not found to have any effect on the infant microbiome examined at the 12th month and 24th month of life |
| Methods of feeding: breastfeeding vs. formula feeding | A link has been demonstrated between breastfeeding and diversity of the intestinal microbiome in children [19, 29] Studies comparing the microbiome of mother’s milk with that of the gut indicate the transmission of certain bacteria [30, 34] The numbers of Bacteroides, Blautia, Parabacteroides, Coprococcus, Ruminococcus and Oscillospira were elevated over the course of 5 days following the introduction of formula feeding in place of breastfeeding, while the numbers of Bifidobacterium, Lactobacillus spp., Escherichia spp. and Clostridium spp. were depressed [29] |
| Siblings | The number of siblings is correlated with the diversity of the gut microbiome at the 18th month of life [22] In another study it was found having siblings to be connected with a less diverse microbiome, particularly with a lower abundance of the Peptostreptococcaceae (Clostridium difficile) [23] |
| Pets | Pets increase the diversity of the gut microbiome in children, particularly with bacteria from the Clostridiaceae, Veillonella (in case of dogs), Peptostreptococcaceae and Coprococcus These increases are balanced with a concomitant decrease of Bifidobacteriaceae numbers (particularly in case of cats) [23] Early-life exposure to household pets has the capacity to reduce the risk of allergic disease, especially following Caesarean delivery [24] |
Of special importance for the formation of the gut microbiome is breastfeeding (Table 1) [19, 29, 30]. Breast milk contains a diverse range of bacteria, some of which are derived from the skin [31]. The number of bacterial cells per millilitre is ~106 [32]. Bacteria belong to four phyla, with the Firmicutes predominating (with the Staphylococci being most common), followed by the Actinobacteria, Proteobacteria and Bacteroidetes [31, 33]. Maternal health, body mass, atopy, diet, immunological status, duration of pregnancy, method of delivery, antibiotic therapy and lactation phase are regarded as factors influencing the microbiome of breast milk [31–34]. Grönlund et al. report that a lower abundance of Bifidobacteria in milk coming from allergic mothers is connected with lower numbers of these bacteria in the stools of their children [34]. Moreover, the breast milk microbiome positively influences digestion and absorption as well as the maturation of the intestinal barrier and pathogen defence [31]. Additionally breastfeeding has a significant impact on the infant immune system, shifting the balance between Th1 and Th2 response towards Th1, thus decreasing predisposition to allergy [29].
The gut microbiome and food allergy
Food allergy can be driven by such factors as diet and commensal microbiota, and the interactions between them determine the default state of mucosal immune tolerance [35]. Not only is the gut microbiome less diverse in allergic patients, it also varies according to the declared allergy type [11]. Studies of the gut microbiota in patients with specific food allergies shows that it is plausible that distinct types of microbiota are associated with each FA subphenotype [35]. Fujimura et al. note the presence of compositionally-distinct neonatal human gut microbiota, whose compositions are differentially related to the relative risk of childhood atopy [36]. Given differences in the presentations and natural histories of specific food allergies, it is plausible that the microbiota associated with each food allergy subphenotype are also distinct. Infants allergic to peanuts have higher levels of Bacteroides [11], and those with cow’s milk allergy (CMA) have higher levels of anaerobic bacteria [37] Ruminococcaceae and Lachnospiraceae [38].
Following allergic sensitization, changes in microbial abundance and diversity (e.g. after antibiotic exposure) decrease SCFA, IL-18, and IL-22 levels, compromising epithelial integrity and thereby facilitating the epithelial passage of microbial and food antigens. DC activation promotes inflammation, allergic sensitization and the development of Th2 cell-associated immune responses, including the production of allergen-specific IgE antibodies [39].
There are hints indicating that disturbance in microbiome composition at early stages of development may influence the onset of sensitization and FA [8]. West et al. report that Bacteroidetes diversity was lower in the stools of pregnant women who delivered children who went on to develop IgE-mediated FA and AD [40]. Azad et al. demonstrated that the composition of gut microbiome at the third month of life can predict sensitization to food allergens until the twelfth month of life [10]. They found a greater abundance of Enterobacteriaceae and lower diversity of Bacteroidetes in the gut of children with allergy to at least one food allergen. Children typified by a higher Enterobacteriaceae/Bacteroidaceae ratio tended to have a greater risk of developing sensitization [10].
Chen et al. report that children with FA aged 6–24 months displayed decreased microbiome diversity, a lower number of species of Bacteroidetes and a greater number of species of Firmicutes compared with healthy children [9]. Ling et al. showed that children with FA display lower numbers of Bacteroidetes, Proteobacteria and Actinobacteria, but greater numbers of Firmicutes. Phylotypes characteristic of FA are negatively correlated with IL-10 concentration [41]. Children with IgE-mediated FA present more Anaerobacter and Clostridium sensu stricto, and less Bacteroides and Clostridium. Thus it was shown that microbiome differentiates children with FA from healthy ones; moreover, differences between IgE-mediated and non-IgE-mediated allergy were also visible [9, 40, 41].
West et al. demonstrate that Ruminococcaceae are less abundant at the first week and the first month of age in children that developed FA than in those who did not develop any allergy until the 30th month [40]. The number of species of these bacteria was negatively correlated with the concentration of IL-6 and TNF-α induced by TLR2. A negative correlation was also found between members of the Propionibacterium genus and TNF-α induced by TLR4. At 1 year of age, children with eczema demonstrated a lower diversity of Actinobacteria compared to healthy ones [40].
Simonyté Sjödin et al. report that between infancy and school age, the gut microbiota of children with IgE-associated allergic disease were different from those of healthy children and that these differences were related to T-cell regulation [42]. Ruminococcus, Bacteroides, Prevotella and Coprococcus were found to be underrepresented in children with allergy compared to those without throughout the period of examination. At 8 years old, the microbiome of allergic children was found to be rich in Bifidobacterium but depleted of Lactobacillus, Enterococcus and Lachnospira and Faecalibacterium, which was correlated with IL-10 mRNA and Foxp3 levels (forkhead box P3). Kourosh et al. describe differences in the intestinal microbiome between children with IgE-mediated FA, their siblings without FA and non-allergic controls [43].
Bunyavanich et al. demonstrated that the acquisition of tolerance by children with CMA is connected with a greater abundance of Clostridia and Firmicutes in general [44]. Berni Canani et al. report that such acquisition is associated with changes in microbiome composition and increased butyrate levels in stools [38].
Skin microbiome
Development of skin microbiome
A square centimetre of skin may harbour even a billion bacteria, fungi and viruses, and although the majority of these microorganisms belong to the same phyla present in the gut microbiome, they are found in different quantities: Actinobacteria (52–59%), Firmicutes (24%), Proteobacteria (17%) and Bacteroidetes (7%) [45, 46]. The predominant members are Propionibacterium, Streptococcus, Staphylococcus and Corynebacterium [46, 47]. The composition of the skin microbiome varies between individuals [48]. Its acquisition begins at birth [45, 49] with the transfer of the maternal microbiome [49]. The skin microbiome evolves over the years according to the changes of skin structure and functions, becoming similar to that of adults by the age of 12–18 months [46, 50, 51].
The composition of the skin microbiome is dependent on the host, age, gender and anatomical area [45, 48, 49, 52] as well as ethnicity, climate, lifestyle (contact with other individuals, visiting different places and diet) [47, 52, 53] and medical procedures (i.e. mode of delivery) (Table 2) [49].
Table 2.
Selected factors modifying the development and shaping of the skin microbiome
| Factors | Commentary | |
|---|---|---|
| Individual | Age | The skin microbiome evolves over the years according to the changes of skin structure and functions [50], becoming similar to that of adults by the age of 12–18 months [51] |
| Skin microenvironment | The skin microenvironment is regulated by skin adnexa, so it depends on anatomical area, gender, ethnicity [52] Sebum and changes in water content are important factors shaping the environment for bacteria, which implies that differences exist among various skin areas [48, 49] Propionibacterium and staphylococci dominate in sebum-rich areas, while Corynebacterium and Staphylococci prevail in dry locations [45, 49] | |
| Environmental | Mode of delivery | The skin of an infant delivered naturally is colonized by bacteria that dominate in the vagina (Lactobacillus, Prevotella and Sneathia), while Staphylococcus, Corynebacterium, and Propionibacterium dominate in the case of Caesarean section [49] |
| Diet | Because diet impacts upon the gut microbiome, it is difficult to disentangle its direct effect on the skin microbiome. There is a lack of data suggesting a link between skin microbiome and method of feeding, including the influence of breast milk microbiome on the development of a skin microbiome | |
Skin microbiome and atopic dermatitis
Presence of healthy skin microbiome results in signalling through IL-1R and inhibits Th2 immunological response [46]. Compared to healthy subjects, the skin microbiome of AD patients has been found to be less diverse and to be dominated by Staphylococcus aureus [54, 55]. Dysbiosis of skin microbiome also can contribute to food sensitization. In the skin, Staphylococcal enterotoxin B acts as an adjuvant to promote Th2 responses and T helper cell development while cytolysins lyse epithelial cells, and serine proteases and lipases damage the skin barrier [39].
Kennedy et al. report that in infants who later developed AD, the species richness of skin microbiome was decreased at the second month of life [12]. The important role played by the skin microbiome in AD pathogenesis was confirmed in a study by Laborel-Preneron et al., who report that the S. aureus thriving on inflamed skin in children sensitized to Dermatophagoides pteronyssinus promoted inflammation through Th2 activation and regulatory T cells (Tregs) suppression, and that commensal S. epidermidis may block this effect by influencing IL-10 release from skin dendritic cells [56].
Kong et al. [13] report an increased abundance of Staphylococcus, particularly S. aureus, in children with AD. Microbiome diversity increased during treatment, with a concomitant increase in the numbers of Streptococcus, Propionibacterium and Corynebacterium. Glatz et al. [57] demonstrated changes in skin microbiota, i.e. an increased proportion of S. salivarius, associated with long-term use of emollients, and confirmed its potential preventative measure in infants at risk for AD.
There is increasing evidence that commensal skin microbes from normal skin can improve the skin barrier and augment host defence against skin pathogens, including S. aureus.
Influence of the gut and skin microbiome on the immune system
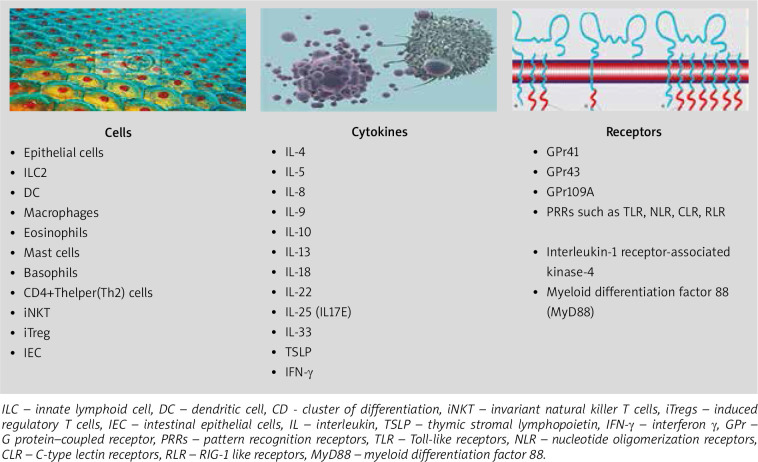
The gut microbiome serves many functions to the host, but in allergy, the influence of the gut microbiome on the immune system is of key importance [21, 58]. It is thought that the most important function of the microbiome is its influence on the development and function of the immune system, maintaining the balance between the Th1/Th2 response, by regulating various lymphocyte subpopulations, especially Tregs, responsible for immune tolerance [11, 17, 21, 58–60]. In the absence of an intact intestinal microbiome, the levels of both IgE and circulating basophils are increased [61]. Cells, cytokines, receptors involved in the regulation of type 2 immune response via the microbiome are illustrated in Figure 1.
Figure 1.
Cells, cytokines and receptors involved by microbiome in the regulation of type 2 immune response
The microbiome employs various mechanisms to influence the immune system, including direct reception and modifications at the epigenetic level (DNA methylation, histone acetylation and micro RNA activities) [21, 62, 63]. The differences in early gut microbiome composition that might exist between allergic and non-allergic subjects could be a part of causes of epigenetic modification through SCFAs, particularly butyrates as well as by vitamins of the B group and other methyl group donors that are metabolized by gut microbiome [64–67].
Microbes can modulate immunologic functions related to allergic diseases also through small molecules produced during metabolism [39, 68–70] as well as through their cell envelope or capsular components (such as DNA, RNA, proteins and cell wall components) (Table 3).Diverse microbial metabolites profoundly regulate the immune system via host receptors and other target molecules [68, 70–74].
Table 3.
Bacterial metabolites and their impact on immune responses
| Metabolites | Functions | |
|---|---|---|
| Group | Examples | |
| LCFA | HYA, conjugated linoleic acids | GPR40 ligands – suppression of TNFR2 gene expression and NF-κB |
| PPAR-γ ligands – export of NF-κB from the nucleus and decreasing NF-κB-dependent IL-8 production | ||
| PPAR-γ1 ligands – increasing the production of IL-10 | ||
| Decreased the plasma IgE levels and skin infiltration of mast cells | ||
| SCFA | Acetate | HDAC9 inhibition – acetylation of Foxp3 promoter |
| Enhance the epithelial barrier function via induction of physiological hypoxia | ||
| Diminishes the epithelial output of pro-Th2 cytokines including TSLP and IL33, via epithelial GPR43 signalling | ||
| Butyrate | Histone H3 acetylation in the Foxp3 locus – induced the differentiation of Treg cells | |
| Enhance the epithelial barrier function via induction of physiological hypoxia | ||
| Triggers CD103+ DCs to produce retinoic acid via GPR109a signalling in the presence of vitamin A, consequently resulting in the protection against FA through increased Treg differentiation by DC-derived retinoic acid | ||
| Propionate | GPR41 ligands – educate DCs to achieve high phagocytic capacity and an ability not to promote the effector function of Th2 cells | |
| GPR43 ligands – induction and function of Treg cells | ||
| Vitamins | Vitamin B9 | Folate (vitamin B9) receptor 4 on Tregs ligand – maintenance of Tregs |
| MR1-dependent antigen to MAIT cells (vitamin B9 prevents the activation of MAIT cells by competing with vitamin B2 metabolite) | ||
| Amino acids | D-tryptophan | Enhance the induction of Treg cells |
| Poly-γ-glutamic acid (γ-PGA) | Antimicrobial activity, DC activation inducing Th1 cell differentiation | |
LCFA – long-chain fatty acids, HYA – 10-hydroxy-cis-12-octadecenoic acid, TNFR – tumour necrosis factor receptor, NF-κB – nuclear factor κB, PPAR – peroxisome proliferator-activated receptor, IL – interleukin, SCFA – short-chain fatty acids, HDAC – histone deacetylases, Foxp3 – forkhead box P3, TSLP – thymic stromal lymphopoietin, GPR – G protein–coupled receptors, Treg – regulatory T cells, CD – cluster of differentiation, DCs – dendritic cells, MAIT cells mucosa-associated invariant T cells, γPGA – poly-γ-glutamic acid.
It has been found that commensal bacteria, especially lactic acid bacteria generate metabolites which decrease the levels of inflammatory cytokines, induce the production of anti-inflammatory cytokines and enhance the epithelial barrier function [69, 75].
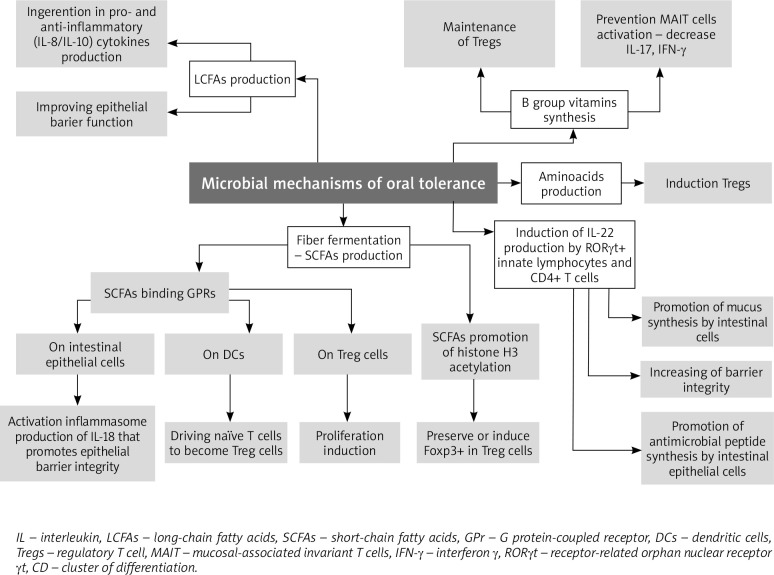
Short-chain fatty acids (SCFAs), as the most abundant microbial metabolites of the colonic lumen, play a wide range of regulatory roles in the immune system, including mediating the induction of immune tolerance and proliferation of Tregs [21, 62] (Figure 2). SCFAs act as the main energy source of epithelial cells and influence the expression of genes necessary for maintaining the epithelial barrier and defence functions. They regulate the action of innate immune cells such as macrophages, neutrophils, dendritic cells, and bidirectionally regulate antigen-specific adaptive immunity mediated by T cells and B cells.
Figure 2.
The microbial mechanisms of oral tolerance
It has been indicated that amino acids may also have a potential influence on the development, homeostasis and function of immune cells. Oral treatment with poly-γ-glutamic acid (γ-PGA) decreases the levels of serum immunoglobulin E (IgE) and Th2 cytokines, resulting in the attenuation of the clinical symptoms of AD by influencing the Th1/Th2 balance [76]. Indoxyl 3-sulfate, a derivative of indole produced from tryptophan by Lactobacillus rhamnosus GG and Lactobacillus casei W56 have the potential to enhance Treg cell induction [69].
Some commensal bacteria have the capacity to synthesize essential vitamins, especially B-family members, which have been reported to demonstrate some immunologic functions [38].
In addition to its immunologic effects, the microbiome may also influence susceptibility to FA through other mechanisms, including the control of intestinal barrier function. For example, microbe-induced IL-22 production by RORγt+ innate lymphocytes and CD4+ T cells promotes barrier integrity and intestinal epithelial cell synthesis of antimicrobial peptides and mucus [39]. Stefka et al. demonstrated that colonization with Clostridia spp. increased the expression of IL-22, a barrier-promoting cytokine, and decreased intestinal permeability [7].
The skin microbiome has been shown to promote normal skin homeostasis, maintaining skin barrier integrity and pathogen defence [46]. It works through the induction of the Th17 and T helper cell immune response and secretion of antimicrobial peptides (AMPs) [52]. It activates and assists innate immunity, and influences adaptive immunity, although these complex interactions are not completely understood. It is needed for the development of a well-functioning immune system and the modulation of the inflammatory processes [48, 52, 77, 78]. Figure 3 summarizes the relationships between the microbiome and skin and gut barriers.
Figure 3.
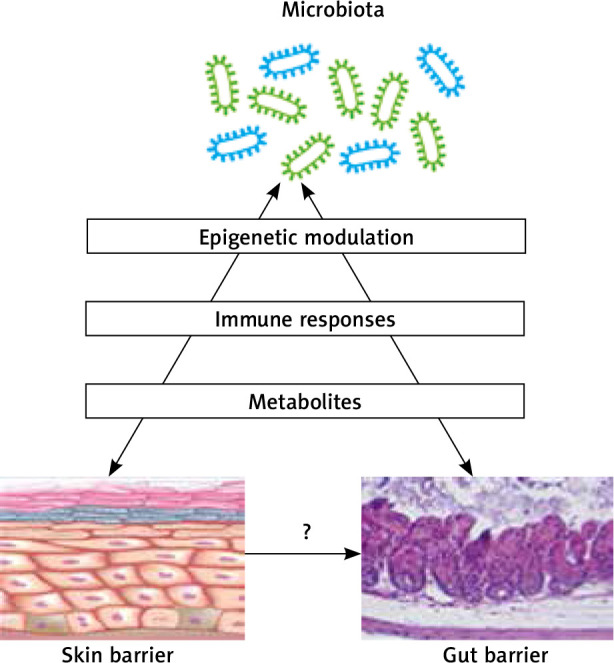
The relationships between the microbiome and skin and gut barriers
Some of commensals are capable to produce bacteriocins to inhibit the growth of harmful bacteria and to produce preventive biofilm [46].
The microbiome and probiotic therapy
A number of studies have evaluated the possibility of modifying the gut microbiome, particularly with regard to the influence of the microbiome on the risk and course of allergy. Some studies suggest that probiotic therapy may be an alternative to dietary treatment in CMA. Tolerance-promoting methylation changes were more frequently observed in patients receiving Lactobacillus GG [64, 79]. Martin et al. describe four cases of breastfed infants with allergic proctocolitis where LGG (L. rhamnosus GG) treatment, used without restrictions in the maternal diet, resulted in remission [80]. Cox et al. found that the use of LGG supplementation was associated with changes in the gut microbiome, with a lower abundance of species known to be involved in the observed pathogenesis of atopy [81].
According to Francavilla et al., the addition of lactose to an extensively hydrolysed formula positively modulated the microbiome composition, promoting the growth of bacteria observed in healthy children [82].
A previous study found that Lactobacillus probiotics are useful for the prevention, rather than treatment of AD [83]. Recently, Lactobacillus plantarum was also found to reduce the clinical index in children with AD, and this effect was associated with an increase in Treg population [84–88]. Only a few trials evaluating the use of probiotics for the prevention or treatment of challenge-proved food allergies have been published. Trials of probiotic supplementation with Lactobacillus casei and Bifidobacterium lactis for 12 months showed no effect on milk allergy resolution, although Lactobacillus rhamnosus combined with extensively hydrolysed casein formula increased rates of milk allergy resolution compared with a control group receiving formula alone [89, 90]. The probiotic Lactobacillus rhamnosus GG administered with peanut oral immunotherapy for 18 months induced desensitization compared with placebo [91]. The effects of probiotic treatment are likely strain specific, and the findings are currently insufficient to support probiotic supplementation with specific taxa for FA treatment.
A novel therapeutic strategies aimed to restore natural skin microbiome, for example application of probiotic strains/topical microbiome transplantation are being studied [46].
Conclusions
Changes in the microbiome confer protection or susceptibility to FA or AD by modulating the immune system. The potential role of exposure to microbes during early childhood in the development of FA as well as AD, and the subsequent atopic march remains to be elucidated. Early childhood appears to be critical for the colonization of a diverse microbiome. Recent research efforts have focused on the identification of bacterial strains necessary for oral tolerance and identification of the most effective probiotic strains for the prevention and treatment of FA. The role of skin microbiome in the course of FA and acquiring tolerance to food allergens by children is still unknown. Early skin topical treatment could reduce S. aureus colonization through exerting an anti-inflammatory effect. The management of skin diseases in the future may include manipulation of the gut function. Balancing the gut and skin microbiome will likely represent an effective treatment for allergy. Therefore, extensive studies are required to identify the interactions between the gut and skin microbiomes.
Acknowledgments
Project financed by National Science Centre, Poland, ul. Królewska 57, 30-081, Kraków, Grant number: 2017/25/B/NZ5/00141.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Sicherer SH, Sampson HA. Food allergy: epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol 2014; 133: 291-307. [DOI] [PubMed] [Google Scholar]
- 2.Silverberg JI, Simpson EL. Association between severe eczema in children and multiple comorbid conditions and increased healthcare utilization. Pediatr Allergy Immunol 2013; 24: 476-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lack G. Epidemiologic risks for food allergy. J Allergy Clin Immunol 2008; 121: 1331-6. [DOI] [PubMed] [Google Scholar]
- 4.Venkataraman D. Filaggrin loss-of function mutations are associated with food allergy in childhood and adolescence. J Allergy Clin Immunol 2014; 134: 876-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsakok T. Does atopic dermatitis cause food allergy? A systematic review. J Allergy Clin Immunol 2016; 137: 1071-8. [DOI] [PubMed] [Google Scholar]
- 6.Park HJ, Lee SW, Hong S. Regulation of allergic immune responses by microbial metabolites. Immune Netw 2018; 18: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefka AT, Feehley T, Tripathi P, et al. . Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA 2014; 111: 13145-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noval Rivas M, Burton OT, Wise P, et al. . A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 2013; 131: 201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CC, Chen KJ, Kong MS, et al. . Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol 2016; 27: 254-62. [DOI] [PubMed] [Google Scholar]
- 10.Azad MB, Konya T, Guttman DS, et al. . ; CHILD Study Investigators. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 2015; 45: 632-43. [DOI] [PubMed] [Google Scholar]
- 11.Hua X, Goedert JJ, Pu A, et al. . Allergy associations with the adult fecal microbita: analysis of the American Gut Project. EBioMedicine 2015; 3: 172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy EA, Connolly J, Hourihane JO, et al. . Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 2017; 139: 166-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong HH, Oh J, Deming C, et al. . Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22: 850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penders J, Thijs C, van den Brandt PA, et al. . Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 2007; 56: 661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MJ, Kang MJ, Lee SY, et al. . Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol 2018; 141: 1310-9. [DOI] [PubMed] [Google Scholar]
- 16.Rutayisire E, Huang K, Liu Y, et al. . The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterology 2016; 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gensollen T, Iyer SS, Kasper DL, et al. . How colonization by microbiota in early life shapes the immune system. Science 2016; 352: 539-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson CC, Ownby DR. Allergies and asthma: do atopic disorders result from inadequate immune homeostasis arising from infant gut dysbiosis? Expert Rev Clin Immunol 2016; 12: 379-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collado MC, Rautava S, Aakko J, et al. . Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 2016; 6: 23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu DM, Ma J, Prince AL, et al. . Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 2017; 23: 314-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D¹browska K, Witkiewicz W. Correlations of host genetics and gut microbiome composition. Front Microbiol 2016; 7: 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laursen MF, Zachariassen G, Bahl MI, et al. . Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol 2015; 15: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azad MB, Konya T, Maughan H, et al. . Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol 2013; 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tun HM, Konya T, Takaro TK, et al. . Exposure to household furry pets influences the gut microbiota of infant at 3-4 months following various birth scenarios. Microbiome 2017; 5: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, et al. . Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 2016; 22: 250-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neu J, Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol 2011; 38: 321-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azad MB, Konya T, Persaud RR, et al. . Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG 2016; 123: 983-93. [DOI] [PubMed] [Google Scholar]
- 28.Dotterud CK, Avershina E, Sekelja M, et al. . Does maternal perinatal probiotic supplementation alter the intestinal microbiota of mother and child? J Pediatr Gastroenterol Nutr 2015; 61: 200-7. [DOI] [PubMed] [Google Scholar]
- 29.Davis MY, Zhang H, Brannan LE, et al. . Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Microbiome 2016; 4: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milani C, Mancabelli L, Lugli GA, et al. . Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 2015; 81: 7078-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaTuga MS, Stuebe A Seed PC. A review of the source and function of microbiota in breast milk. Semin Reprod Med 2014; 32: 68-73. [DOI] [PubMed] [Google Scholar]
- 32.Boix-Amorós A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol 2016; 7: 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davé V, Street K, Francis S, et al. . Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr Res 2016; 79: 846-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grönlund MM, Gueimonde M, Laitinen K, et al. . Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy 2007; 37: 1764-72. [DOI] [PubMed] [Google Scholar]
- 35.Huang YJ, Marsland BJ, Bunyavanich S, et al. . The microbiome in allergic disease: current understanding and future opportunities–2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol 2017; 139: 1099-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujimura KE, Sitarik AR, Havstad S, et al. . Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016; 22: 1187-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson-Chagoyan OC, Vieites JM, Maldonado J, et al. . Changes in faecal microbiota of infants with cow’s milk protein allergy–a Spanish prospective case-control 6-month follow-up study. Pediatr Allergy Immunol 2010; 21: e394-400. [DOI] [PubMed] [Google Scholar]
- 38.Berni Canani R, Sangwan N, Stefka AT, et al. . Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 2016; 10: 742-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinthrajah RS, Hernandez JD, Boyd SD, et al. . Molecular and cellular mechanisms of food allergy and food tolerance. J Allergy Clin Immunol 2016; 137: 984-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West CE, Rydén P, Lundin D, et al. . Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy 2015; 45: 1419-29. [DOI] [PubMed] [Google Scholar]
- 41.Ling Z, Li Z, Liu X, et al. . Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol 2014; 80: 2546-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonyté Sjödin K, Hammarström ML, Rydén P, et al. . Temporal and long-term gut microbiota variation in allergic disease: a prospective study from infancy to school age. Allergy 2019; 74: 176-85. [DOI] [PubMed] [Google Scholar]
- 43.Kourosh A, Luna RA, Balderas M, et al. . Fecal microbiome signatures are different in food-allergic children compared to siblings and healthy children. Pediatr Allergy Immunol 2018; 29: 545-54. [DOI] [PubMed] [Google Scholar]
- 44.Bunyavanich S, Shen N, Grishin A, et al. . Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016; 138: 1122-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominguez Bello MG, Costello EK, Contreras M, et al. . Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010; 107: 11971-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blicharz L, Rudnicka L, Samochocki Z. Staphylococcus aureus: an understimated factor in the pathogenesis of atopic dermatitis. Adv Dermatol Allergol 2019; 36: 11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas CL, Fernández-Peńas P. The microbiome and atopic eczema: more than skin deep. Australas J Dermatol 2017; 58: 18-24. [DOI] [PubMed] [Google Scholar]
- 48.Mukherjee S, Mitra R, Maitra A, et al. . Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci Rep 2016; 6: 36062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weyrich LS, Dixit S, Farrer AG, et al. . A microbiome: associations between altered microbial communities and disease. Australas J Dermatol 2015; 56: 268-74. [DOI] [PubMed] [Google Scholar]
- 50.Baviera G, Leoni MC, Capra L, et al. . Microbiota in healthy skin and in atopic eczema. BioMed Res Int 2014; 2014: 436921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capone KA, Dowd SE, Stamatas GN, et al. . Diversity of the human skin microbi-ome early in life. J Invest Dermatol 2011; 131: 2026-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grice EA. The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Semin Cutan Med Surg 2014; 33: 98-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu TH, Zhu TR, Tran KA, et al. . Epithelial barrier dysfunctions in atopic dermatitis: a skin-gut-lung model linking microbiome alteration and immune dysregulation. Br J Dermatol 2018; 179: 570-81. [DOI] [PubMed] [Google Scholar]
- 54.Marrs T, Flohr C. The role of skin and gut microbiota in the development of atopic eczema. Br J Dermatol 2016; 175: 13-8. [DOI] [PubMed] [Google Scholar]
- 55.Sala-Cunill A, Lazaro M, Herráez L, et al. . Basic skin care and topical therapies for atopic dermatitis: beyond essential approaches. J Investig Allergol Clin Immunol 2018; 28: 379-91. [DOI] [PubMed] [Google Scholar]
- 56.Laborel-Préneron E, Bianchi P, Boralevi F, et al. . Effects of the Staphylococcus aureus and Staphylococcus epidermidis secretomes isolated from the skin microbiota of atopic children on CD4+ T cell activation. PLoS One 2015; 10: e0141067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glatz M, Jo JH, Kennedy EA, et al. . Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PLoS One 2018; 13: e0192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chunqing A, Ma N, Zhang Q, et al. . Immunomodulatory effects of different lactic acid bacteria on allergic response and its relationship with in vitro properties. PLOS One 2016; 11: e0164697.27764153 [Google Scholar]
- 59.Riiser A. The human microbiome, asthma, and allergy. Allergy Asthma Clin Immunol 2015; 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feehley T, Nagler CR. Cellular and molecular pathways through which commensal bacteria modulate sensitization to dietary antigens. Curr Opin Immunol 2014; 31: 79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill DA, Siracusa MC, Abt MC, et al. . Commensal bacteria derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med 2012; 18: 538-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arpaia N, Campbell C, Fan X, et al. . Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2014; 504: 451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet 2013; 381: 861-73. [DOI] [PubMed] [Google Scholar]
- 64.Berni Canani R, Paparo L, Nocerino R, et al. . Differences in DNA methylation profile of Th1 and Th2 cytokine genes are associated with tolerance acquisition in children with IgE-mediated cow’s milk allergy. Clin Epigenetics 2015; 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paparo L, di Costanzo M, di Scala C, et al. . The influence of early life nutrition on epigenetic regulatory mechanisms of the immune system. Nutrients 2014; 6: 4706-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canani RB, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics 2012; 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma S, Litonjua A. Asthma, allergy, and responses to methyl donor supplements and nutrients. J Allergy Clin Immunol 2014; 133: 1246-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nature Immunol 2011; 12: 5-9. [DOI] [PubMed] [Google Scholar]
- 69.Hirata S, Kunisawa J. Gut microbiome, metabolome, and allergic diseases. Allergol Int 2017; 66: 523-8. [DOI] [PubMed] [Google Scholar]
- 70.Lozupone CA. Unraveling Interactions between the microbiome and the host immune system to decipher mechanisms of disease. mSystems 2018; 3: pii: e00183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim CH. Immune regulation by microbiome metabolites. Immunology 2018; 154: 220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan J, McKenzie C, Vuillermin PJ, et al. . Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 2016; 15: 2809-24. [DOI] [PubMed] [Google Scholar]
- 73.Maglione PJ, Simchoni N, Cunningham-Rundles C. Toll-like receptor signaling in primary immune deficiencies. Ann N Y Acad Sci 2015; 1356: 1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohnmacht C, Park JH, Cording S, et al. . Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 2015; 349: 989-93. [DOI] [PubMed] [Google Scholar]
- 75.Kaikiri H, Miyamoto J, Kawakami T, et al. . Supplemental feeding of a gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, alleviates spontaneous atopic dermatitis and modulates intestinal microbiota in NC/nga mice. Int J Food Sci Nutr 2017; 68: 941-51. [DOI] [PubMed] [Google Scholar]
- 76.Lee SW, Park HJ, Park SH, et al. . Oral administration of poly-gamma-glutamic acid prevents the development of atopic dermatitis in NC/Nga mice. Exp Dermatol 2013; 22: 561-3. [DOI] [PubMed] [Google Scholar]
- 77.Knaysi G, Smith AR, Wilson JM, et al. . The skin as a route of allergen exposure: part ii. allergens and role of the microbiome and environmental exposures. Curr Allergy Asthma Rep 2017; 17: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egert M, Simmering R. The microbiota of the human skin. Adv Exp Med Biol 2016; 902: 61-81. [DOI] [PubMed] [Google Scholar]
- 79.Krogulska A, Borowiec M, Polakowska E, et al. . FOXP3, IL-10, and TGF-beta genes expression in children with IgE-dependent food allergy. J Clin Immunol 2011; 31: 205-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin VJ, Shreffler WG, Yuan Q. Presumed allergic proctocolitis resolves with probiotic monotherapy: a report of 4 cases. Am J Case Rep 2016; 17: 621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cox MJ, Huang YJ, Fujimura KE, et al. . Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One 2010; 5: e8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Francavilla R, Calasso M, Calace L, et al. . Effect of lactose on gut microbiota and metabolome of infants with cow’s milk allergy. Pediatr Allergy Immunol 2012; 23: 420-7. [DOI] [PubMed] [Google Scholar]
- 83.Lee J, Seto D, Bielory L. Meta-analysis of clinical trials of probiotics for prevention and treatment of pediatric atopic dermatitis. J Allergy Clin Immunol 2008; 121: 116-21. [DOI] [PubMed] [Google Scholar]
- 84.Prakoeswa CR, Herwanto N, Prameswari R, et al. . Lactobacillus plantarum IS-10506 Supplementation reduced SCORAD in children with atopic dermatitis. Benef Microbe 2017; 8: 833-40. [DOI] [PubMed] [Google Scholar]
- 85.Kwon HK, Lee CG, So JS, et al. . Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA 2010; 107: 2159-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ren C, Zhang Q, de Haan BJ, et al. . Identification of TLR2/TLR6 signalling lactic acid bacteria for supporting immune regulation. Sci Rep 2016; 6: 34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu L, Peng J, Zhao S, et al. . Lactic acid bacteria-specific induction of CD4(+)Foxp3(+) T cells ameliorates shrimp tropomyosin-induced allergic response in mice via suppression of mTOR signaling. Sci Rep 2017; 7: 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hacini-Rachinel F, Gheit H, Le Luduec JB, et al. . Oral probiotic control skin inflammation by acting on both effector and regulatory T cells. PLoS One 2009; 4: e4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hol J, van Leer EH, Elink Schuurman BE, et al. . The acquisition of tolerance toward cow’s milk through probiotic supplementation: a randomized, controlled trial. J Allergy Clin Immunol 2008; 121: 1448-54. [DOI] [PubMed] [Google Scholar]
- 90.Berni Canani R, Nocerino R, Terrin G, et al. . Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J Allergy Clin Immunol 2012; 129: 580-2. [DOI] [PubMed] [Google Scholar]
- 91.Tang ML, Ponsonby AL, Orsini F, et al. . Administration of a probiotic with peanut oral immunotherapy: a randomized trial. J Allergy Clin Immunol 2015; 135: 737-44. [DOI] [PubMed] [Google Scholar]