Abstract
Background: The pathogenic role of Wilms tumor 1 gene (WT1) is well known in renal cancer. However, recently, its over expression is been documented in cases of acute myeloid leukaemia (AML), acute lymphoblastic leukaemia (ALL) and myelodysplastic syndrome (MDS). WT1 mutations is found in about 6%-15% of cases of AML affecting mainly hotspot exon 7 and 9, and less frequently in other exon such as 1, 2, 3, and 8. Different studies have shown equivocal findings with few of them depicting poorer prognosis, while others suggesting lack of any significant clinical impact. Objective: This study was planned to evaluate prevalence of WT1 gene mutation on exon 7 & 9 in de novo cases of AML and its correlation with their clinical features and disease course. Methodology: A total of newly diagnosed and treatment naive 100 cases of AML, having blast count of ≥20% in peripheral blood or bone marrow were enrolled. Genomic DNA of all participants was extracted from blood/bone marrow sample using Qiagen® DNA extraction kit. Haematological workup for counts and flow cytometry based immunophenotypes was done. Mutation on exon 7 & 9 were detected with the help of Sanger sequencing. Results: WT1 mutations were detected in both types of cases having normal vs. abnormal cytogenetics. The overall prevalence of WT1 mutation of 2% was found. We have reported one novel mutation on exon 9 of WT1 gene. Twelve cases (12%) among all analyzed AMLs were found to have synonymous single nucleotide polymorphism (SNPs) on exon 7 which has been previously reported in SNP database (rs16754). Conclusion: In our study, presence of synonymous SNP was not associated with any change at protein level. We also evaluated mutational status with deaths during induction remission and concluded that presence of WT1 gene mutation was associated with death during induction therapy.
Keywords: WT1 gene, mutation, sequencing, AML
Introduction
Acute myeloid leukaemia (AML) is a complex multisystem hematopoietic disorder having system specific symptoms and complications [1]. It is second most common type of leukaemia being diagnosed among adults and children [2,3]. However, the incidence of AML rises with increasing age [4]. The disease is an outcome of various multistep mutational insults leading to phenomenon such as uncontrolled proliferation, hampered maturation and reduction in apoptosis of myeloid lineage specific cancer stem cells [5,6]. With advancement of therapeutic interventions, there has been a slight improvement in survival figures. However, most of the diagnosed cases of AML usually succumb to disease within time frame of approximately two years [7]. The survival is an outcome of on genomic profile of mutational burden along with other clinical factors. In addition, posttranscriptional and post-translational modification after mutations may also contribute significantly promoting malignant cell proliferation [8,9].
Wilms Tumor 1 (WT1) gene was initially identified as a tumor suppressor gene linked with nephroblastoma [10,11]. Many recent studies [12-16] have shown involvement of WT1 gene in cell survival, proliferation and differentiation in various other neoplasm like leukaemia, lung cancer, colon cancer and pancreatic cancer emphasizing its strong oncogenic potential. However, its mechanism is not so well understood and remains elusive till date. It is expressed in different tissues in a specific sequential manner. During embryogenesis, it is found predominantly in urogenital system, while, in adults it is also present in central nervous system and haemato-lymphoid tissues along with urogenital system [12]. Despite of its behaviour as tumor suppressor gene, it has been functioning as tumor oncogene in leukemia and breast cancer [17].
WT1 gene is at 11p13 and encodes for 10 exons to generate approximately 3 kb mRNA (Table 1) from ncbi.nlm.nih.gov/nuccore/NM_024426.4?report = fasta. The gene undergoes alternative splicing at exon 7 and exon 9 to form four different protein isoforms [18]. WT1 gene is proposed to play a predominant oncogenic role in contrast to its tumor suppressor role in cases of AML [19]. It controls important biological processes such as transcription, translation and RNA metabolism [20,21]. In addition to Wilms tumor, mutations of WT1 gene also leads to disorders such as Denys-Drash syndrome [22]. The expression of WT gene has been high in normal blast cell and down regulates with normal maturation of cell [23,24], however it has been seen persistently high among cases of AML and in human cell lines [25,26].
Table 1.
Sequence of WT1 gene
| Parameters | Age/Karyotype |
|---|---|
| Cytogenetics | 11p13 |
| Base Pairs | 32,387,775 to 32,435,539 |
| mRNA | 3 KB |
| No of Exons | 10 |
| Sequence | |
| >NM_024426.4 Homo sapiens Wilms tumor 1 (WT1), transcript variant D, mRNA | |
| AGCTGGGGTAAGGAGTTCAAGGCAGCGCCCACACCCGGGGGCTCTCCGCAACCCGACCGCCTGTCCGCTC | |
| CCCCACTTCCCGCCCTCCCTCCCACCTACTCATTCACCCACCCACCCACCCAGAGCCGGGACGGCAGCCC | |
| AGGCGCCCGGGCCCCGCCGTCTCCTCGCCGCGATCCTGGACTTCCTCTTGCTGCAGGACCCGGCTTCCAC | |
| GTGTGTCCCGGAGCCGGCGTCTCAGCACACGCTCCGCTCCGGGCCTGGGTGCCTACAGCAGCCAGAGCAG | |
| CAGGGAGTCCGGGACCCGGGCGGCATCTGGGCCAAGTTAGGCGCCGCCGAGGCCAGCGCTGAACGTCTCC | |
| AGGGCCGGAGGAGCCGCGGGGCGTCCGGGTCTGAGCCGCAGCAAATGGGCTCCGACGTGCGGGACCTGAA | |
| CGCGCTGCTGCCCGCCGTCCCCTCCCTGGGTGGCGGCGGCGGCTGTGCCCTGCCTGTGAGCGGCGCGGCG | |
| CAGTGGGCGCCGGTGCTGGACTTTGCGCCCCCGGGCGCTTCGGCTTACGGGTCGTTGGGCGGCCCCGCGC | |
| CGCCACCGGCTCCGCCGCCACCCCCGCCGCCGCCGCCTCACTCCTTCATCAAACAGGAGCCGAGCTGGGG | |
| CGGCGCGGAGCCGCACGAGGAGCAGTGCCTGAGCGCCTTCACTGTCCACTTTTCCGGCCAGTTCACTGGC | |
| ACAGCCGGAGCCTGTCGCTACGGGCCCTTCGGTCCTCCTCCGCCCAGCCAGGCGTCATCCGGCCAGGCCA | |
| GGATGTTTCCTAACGCGCCCTACCTGCCCAGCTGCCTCGAGAGCCAGCCCGCTATTCGCAATCAGGGTTA | |
| CAGCACGGTCACCTTCGACGGGACGCCCAGCTACGGTCACACGCCCTCGCACCATGCGGCGCAGTTCCCC | |
| AACCACTCATTCAAGCATGAGGATCCCATGGGCCAGCAGGGCTCGCTGGGTGAGCAGCAGTACTCGGTGC | |
| CGCCCCCGGTCTATGGCTGCCACACCCCCACCGACAGCTGCACCGGCAGCCAGGCTTTGCTGCTGAGGAC | |
| GCCCTACAGCAGTGACAATTTATACCAAATGACATCCCAGCTTGAATGCATGACCTGGAATCAGATGAAC | |
| TTAGGAGCCACCTTAAAGGGAGTTGCTGCTGGGAGCTCCAGCTCAGTGAAATGGACAGAAGGGCAGAGCA | |
| ACCACAGCACAGGGTACGAGAGCGATAACCACACAACGCCCATCCTCTGCGGAGCCCAATACAGAATACA | |
| CACGCACGGTGTCTTCAGAGGCATTCAGGATGTGCGACGTGTGCCTGGAGTAGCCCCGACTCTTGTACGG | |
| TCGGCATCTGAGACCAGTGAGAAACGCCCCTTCATGTGTGCTTACCCAGGCTGCAATAAGAGATATTTTA | |
| AGCTGTCCCACTTACAGATGCACAGCAGGAAGCACACTGGTGAGAAACCATACCAGTGTGACTTCAAGGA | |
| CTGTGAACGAAGGTTTTCTCGTTCAGACCAGCTCAAAAGACACCAAAGGAGACATACAGGTGTGAAACCA | |
| TTCCAGTGTAAAACTTGTCAGCGAAAGTTCTCCCGGTCCGACCACCTGAAGACCCACACCAGGACTCATA | |
| CAGGTAAAACAAGTGAAAAGCCCTTCAGCTGTCGGTGGCCAAGTTGTCAGAAAAAGTTTGCCCGGTCAGA | |
| TGAATTAGTCCGCCATCACAACATGCATCAGAGAAACATGACCAAACTCCAGCTGGCGCTTTGAGGGGTC | |
| TCCCTCGGGGACCGTTCAGTGTCCCAGGCAGCACAGTGTGTGAACTGCTTTCAAGTCTGACTCTCCACTC | |
| CTCCTCACTAAAAAGGAAACTTCAGTTGATCTTCTTCATCCAACTTCCAAGACAAGATACCGGTGCTTCT | |
| GGAAACTACCAGGTGTGCCTGGAAGAGTTGGTCTCTGCCCTGCCTACTTTTAGTTGACTCACAGGCCCTG | |
| GAGAAGCAGCTAACAATGTCTGGTTAGTTAAAAGCCCATTGCCATTTGGTGTGGATTTTCTACTGTAAGA | |
| AGAGCCATAGCTGATCATGTCCCCCTGACCCTTCCCTTCTTTTTTTATGCTCGTTTTCGCTGGGGATGGA | |
| ATTATTGTACCATTTTCTATCATGGAATATTTATAGGCCAGGGCATGTGTATGTGTCTGCTAATGTAAAC | |
| TTTGTCATGGTTTCCATTTACTAACAGCAACAGCAAGAAATAAATCAGAGAGCAAGGCATCGGGGGTGAA | |
| TCTTGTCTAACATTCCCGAGGTCAGCCAGGCTGCTAACCTGGAAAGCAGGATGTAGTTCTGCCAGGCAAC | |
| TTTTAAAGCTCATGCATTTCAAGCAGCTGAAGAAAAAATCAGAACTAACCAGTACCTCTGTATAGAAATC | |
| TAAAAGAATTTTACCATTCAGTTAATTCAATGTGAACACTGGCACACTGCTCTTAAGAAACTATGAAGAT | |
| CTGAGATTTTTTTGTGTATGTTTTTGACTCTTTTGAGTGGTAATCATATGTGTCTTTATAGATGTACATA | |
| CCTCCTTGCACAAATGGAGGGGAATTCATTTTCATCACTGGGAGTGTCCTTAGTGTATAAAAACCATGCT | |
| GGTATATGGCTTCAAGTTGTAAAAATGAAAGTGACTTTAAAAGAAAATAGGGGATGGTCCAGGATCTCCA | |
| CTGATAAGACTGTTTTTAAGTAACTTAAGGACCTTTGGGTCTACAAGTATATGTGAAAAAAATGAGACTT | |
| ACTGGGTGAGGAAATCCATTGTTTAAAGATGGTCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTTG | |
| TGTTGTGTTTTGTTTTTTAAGGGAGGGAATTTATTATTTACCGTTGCTTGAAATTACTGTGTAAATATAT | |
| GTCTGATAATGATTTGCTCTTTGACAACTAAAATTAGGACTGTATAAGTACTAGATGCATCACTGGGTGT | |
| TGATCTTACAAGATATTGATGATAACACTTAAAATTGTAACCTGCATTTTTCACTTTGCTCTCAATTAAA | |
| GTCTATTCAAAAGGAAAAAAAAAAAAA | |
WT1 gene expression has been extensively studied on pre-malignant and malignant haematological conditions. Majority of studies have found increased WT gene expression in various subsets of AML and have correlated with their respective outcomes and survivals. A comparative evaluation of WT1 expression among cases of myelodysplastic syndromes (MDS) and AML on pre and post-transplant occasions was done for assessment of future relapse [26]. It has also been studied in AML cases along with other associated co-existing genetic abnormality [27]. There is a study from china in bone marrow aspiration samples of 195 AML cases [28] to validate WT1 gene expression as predictive biomarker for detection of minimal residual disease. The same has been studied on AML cases undergoing allogenic stem cell transplant to predict post transplant relapses [29]. On the basis of available medical studies published till date, it can be concluded that WT gene expression has been found in the range of 70-90% in patients with AML [30-32].
However, published data on WT1 mutational pattern exhibits relative paucity when compared with its expression related studies. Mutation in WT1 gene and modifications of down-regulating pathway is potentially playing a critical role for malignant blast proliferation and impaired differentiation of neoplastic blast cell. WT1 mutations have been seen in about 6%-15% of cases of AML clustered mainly around exon 7 and 9, and less frequently in exon 1, 2, 3, and 8 [33-37]. Mutation in WT1 gene causes conformational changes in binding capacity of WT1 protein leading to its deficient tumor suppressing activity and creates pro-tumor environment [33]. One study has reported deranged TET2 function as a consequence to WT1 mutation [38]. Different studies have showed variable conflicting results with few of them depicting a poorer prognosis in cases of AML due to WT1 over expression or mutation, while other few suggests lack of any significant impact [35,40-43].
There has been only one study [36] carried out in Indian population which shows prevalence of 6.7% of WT1 mutation among cases of AML. In view of the above equivocal conflicting scenario, the current study was planned to evaluate prevalence of WT1 gene hotspots mutation in de novo cases of AML and its correlation with clinical features and prognosis.
Materials and methods
Design of study: Prospective, diagnostic study evaluation.
Sample size calculation
The sample size to be studied was as per the review of literature, the prevalence of WT1 mutation is approximately 6.7% AML [36]. In view of the cost of experiment, benchmark of 7% was considered. Accordingly, at 95% of confidence and absolute precision as 5% the required sample size came around 99. According to formula n = 3.84 pq/d2 where p = prevalence, Q = 1-p and “d” is precision rate.
Number of samples = 3.84 × 7 × 93/5 × 5 = 99 round off to 100.
The proposed sample size is likely to provide reasonably better preliminary findings on the topic.
Sample collection, diagnosis and treatment protocol
This study was performed at our institute after obtaining ethical approval from institute ethics committee in accordance with the Declaration of Helsinki. After informed consent was granted then samples were collected. The diagnosis of AML was made on basis of clinical features, morphology, cytogenetics and immunophenotyping [44]. Blasts ≥20% in bone marrow aspirate or peripheral blood was considered as a cut off for final diagnosis of AML. The diagnosis were confirmed by flowcytometry based immunophenotyping using Beckman coulter flow cytometer (Gallios & FC 500) with cMPO, CD13, CD33, CD45 and CD117, CD14, CD36, CD11b, CD64, CD34, CD65, CD38, CD19, CD2, CD9, CD15, cCD79a and cCD3 as markers for diagnosing and differentiating AML blasts from other haemato-lymphoid neoplasm. Clinical details of cases were noted from records file of the patient and they were regularly followed up for a year. Cytogenetic results and other molecular markers (AML-ETO, FLT3-ITD, CBFB-MYH11, and NPM1 etc.) were also recorded. All cases of AML included in this study received uniform treatment of 3+7 intensive induction chemotherapy and High dose cytarabine (HiDAC) as consolidation therapy according to standard AML treatment protocol regimen [45].
Cell samples and DNA isolation
Bone marrow aspirate/peripheral blood samples were collected from the AML patients at diagnosis in EDTA vials. Peripheral blood mononuclear cells were separated using RBCs lysis method to isolate lymphocytes and stored in phosphate buffer saline (PBS) at -80°C. DNA was extracted using Qiagen® DNA extraction kit according to the manufacturer’s protocol. DNA was further quantified by spectrophotometry and its quality was checked by agarose gel electrophoresis.
Amplification of WT1 gene
High molecular weight genomic DNA was amplified for exons 7 and 9 of WT1 gene using the following primer sets: WT-Ex-7F: 5-GACCTACGTGAAT GTTCACATG-3; WT-Ex-7R: 50-ACCAACACCTGGATCAGACCT-30; WT-Ex-9F: 50-TGCAGACATT GC AGGCATGGCAGG-30; and WT-Ex-9R: 50-GCACTATTCCTTCTCTCAACTGAG-30, as per a previous report [36]. Briefly, the polymerase chain reaction (PCR) was performed in a 50 µL volume containing 50 ng of genomic DNA, 1.5 mmol/L MgCl2, 0.2 mM dNTPs, 20 pmol of each oligonucleotide primer, and 5 unit of Taq polymerase (commercially available). The PCR conditions consisted of an initial denaturation step at 94°C for 5 min followed by 34 cycles at 94°C for 30 s, 60°C for 1 min, 72°C for 1 min, and a final step at 72°C for 10 min. The bands of amplified products on gel were captured using UV light gel imaging system. The base pair size of both amplified product size was same as 349 bp for exon 9 & 7 respectively (Figure 1A and 1B).
Figure 1.
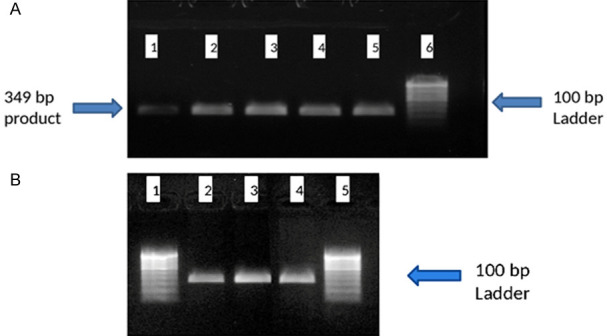
PCR products on 1% agarose gel electrophoresis. A. Lane 1-5; Amplified product of 349 bp of exon 9. Lane 6; Molecular ladder of 100 bp. B. Lane 2-4; Amplified product of 349 bp of exon 7. Lane 1 & 5; Molecular ladder of 100 bp.
Sequencing of WT1 gene
As per manufacturer’s protocol, PCR amplified products were subjected to the clean-up process for removal of leftover dNTPs, primers and other unwanted/inhibitory components acquired during PCR reaction using QI quick® PCR purification kit (Qiagen, Germany). After purification step, amplified PCR products were subjected to direct sanger sequencing to detect any nucleotide sequence variation in targeted region by using capillary electrophoresis on genetic analyser 3500 (Applied bio-system, USA). The targeted gene sequence results were analysed with reference sequence of gene using software Chromas© 2.6 tool. The altered gene sequence results were then reconfirmed again by a repeat PCR amplification with both primers (forward & reverse) followed by re-sequencing of the amplified products.
Statistical analysis
The statistical analysis (STATA:14 software) of data was analyzed using Pearson’s chi-squared test (χ2) and Fisher’s exact test to evaluate significant association of WT1 gene mutations among AML cases for disease risk stratification and clinical management. The qualitative value was represented in form of numbers and percentage while quantitative value in median and range. The p-value of results was calculated for all parameters and p-value ≤0.05 was considered as significant.
Results
Prevalence of hotspots in Exon 7 & 9 of WT1 gene mutation
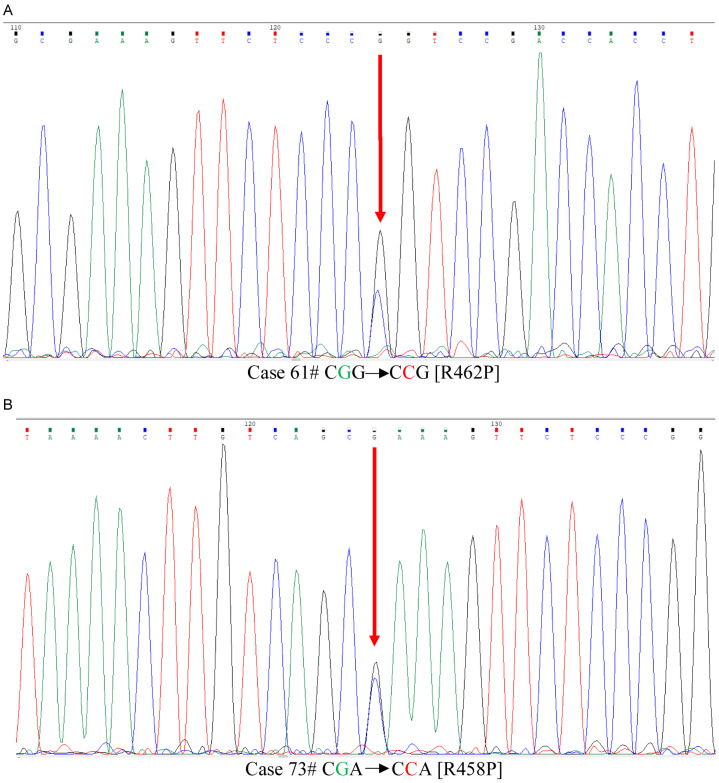
WT1 gene mutation was analysed for coding region and intron-exon flanking region of exon 7 & 9 using Sanger sequencing in a total of 100 cases. Two (2) out of 100 cases i.e. 2% showed point mutation i.e. single base substitution (Figure 2A, 2B) only in exon 9. Interestingly, we observed a novel mutation in case id 73 in exon 9, which was due to single base substitution of nucleotide resulting change in amino acid sequence of protein (Table 2A). The nucleotide and protein variation of was c.1373 G>C; p.Arg458Pro. This novel mutation finding was submitted to ClinVar database of NCBI. We also observed an already reported point mutation in case id 61 in exon 9. The altered nucleotide and protein sequence of this case was c.1385 G>C; p.Arg462Pro. Another 12 case (12%) had synonymous single nucleotide polymorphism (SNP) exclusively in exon 7 (Table 2B), which have also been previously reported in SNP database (dbSNP -rs16754). Six (6) out 100 i.e. 6% had intronic single nucleotide variant (iSNV) (Table 2C) which were found in exon 7 (n = 2) as well as in exon 9 (n = 4).
Figure 2.
Sanger basedsequence result of exon 9 of WT1 gene. A. Case 61 shows G>C nucleotide change at c.1385/G>C. B. Case 73 G>C shows novel nucleotide change c.1373 G>C.
Table 2A.
Brief profile of point mutation in Exon 9 of WT1 gene in AML patients with novel reported change in protein
| Serial No | Age/Karyotype | Nucleotide change | Protien Change |
|---|---|---|---|
| 1 | 28/46xy | c.1385/G>C | p.R462P |
| 2 | 15/46xx | c.1385/G>C | p.R458P |
Table 2B.
Brief profile of point mutation in Exon 7 of WT1 gene in AML patients
| Serial No | Age/Karyotype | Nucleotide change | Protien Change |
|---|---|---|---|
| 1 | 0.8/46, XX | 1293 A>G | No change |
| 2 | 39/46, XY | 1293 A>G | No change |
| 3 | 37/46, XY | 1293 A>G | No change |
| 4 | 7/45, del (Xq), -Y, add(1p)/46, XY | 1293 A>G | No change |
| 5 | 54/46, XY | 1293 A>G | No change |
| 6 | 62/46, XX | 1293 A>G | No change |
| 7 | 21/46, XY | 1293 A>G | No change |
| 8 | 12/46, XY | 1293 A>G | No change |
| 9 | 19/46, XX | 1293 A>G | No change |
| 10 | 24/46, XX | 1293 A>G | No change |
| 11 | 04/58XY, +X, (+4, +10), +22/46, XY | 1293 A>G | No change |
| 12 | 28/46, XX | 1293 A>G | No change |
| 0.8/46, XX |
Table 2C.
Intronic variation found in Exon 7 & 9 of WT1 gene in AML patients
| Serial No | Exon no. | Nucleotide change | Nucleotide seq variants |
|---|---|---|---|
| 1 | Exon 9 | upstream -87G>A | GCTGG G>A CTCC |
| 2 | Exon 9 | upstream -87G>A | GCTGG G>A CTCC |
| 3 | Exon 9 | upstream -87G>A | GCTGG G>A CTCC |
| 4 | Exon 9 | upstream -87G>A | GCTGG G>A CTCC |
| 5 | Exon 7 | upstream -20T>C | CCT TCC>T TCT |
| 6 | Exon 7 | upstream -09T>A | TCT CT>AG CCT |
Correlation of WT1 hotspots mutation finding with demographic & clinical characteristics
The demographic and clinical characteristics of AML patients with wild-type & mutant-type of WT1 gene has been summarized in (Table 3). In this study, 60 cases were males and 40 were females with median age of 17.5 year (range-3 months - 69 years). The WT1 mutation was identified in two patients aged 15 and 28 years respectively, which was found in younger adult age group. There were no statistically significant differences in clinical parameters like haemoglobin levels, white blood cells count (WBCs), platelets count, & blast cell percentage at primary diagnosis between wild type and mutant-type cases. Both the cases having WT1 mutation died during induction remission therapy was statistically significant.
Table 3.
Clinical and demographic characteristics cases of acute myeloid leukemia
| Characteristics (n) | Wild type | Mutant type | p value |
|---|---|---|---|
| Gender (100) | |||
| Male (60) | 59 | 1 | 0.851 |
| Female (40) | 39 | 1 | |
| AgeMed.17.5 (0.3-69) | |||
| ≤18 years (52) | 51 | 1 | 0.802 |
| >18 years (48) | 20 | 1 | |
| Blast % | |||
| <20-30 (8) | 8 | 0 | 0.161 |
| 31-50 (19) | 19 | 0 | |
| 51-79 (35) | 34 | 1 | |
| >80 (38) | 37 | 1 | |
| Remission Status | |||
| Under Remission (27) | 27 | 0 | 0.05 |
| Relapse (42) | 41 | 1 | |
| Unknown (31) | 30 | 1 | |
| Follow-up status | |||
| Active | 38 | 0 | 0.04 |
| Death | 14 | 2 | |
| Lost | 46 | 0 |
Correlation of hotspots WT1 mutation with cytogenetic findings
Cytogenetic findings were available for only 39 (39%) out of 100 patients including WT1 mutations as mentioned in (Table 4). According to cytogenetic findings, patients were categorised into two main groups one with normal karyotype (n = 19); and other with abnormal karyotype (n = 20). Cases with abnormal karyotypes were further sub-grouped as t (8; 21), t (15; 17) having (n = 9), deletion (n = 6), other complex cytogenetic (n = 5). The karyotypes of rest of the 61 cases were not available for analysis. The WT1 mutation result showed no significant differences with respect to these cytogenetic results (p value = 0.649).
Table 4.
WT1 mutation and its correlation with other cytogenetic abnormalities
| Characteristics (n) | Wild type | Mutant type | p value |
|---|---|---|---|
| Kayotype (100) | |||
| Unknown (61) | 61 | 0 | |
| Known (39) | 37 | 2 | 0.851 |
| Cytogenetics (39) | |||
| Normal | 19 | 0 | |
| Abnormal | 18 | 2 | |
| Abnormal Cytogenetics (20) | |||
| t (8; 21), t (15; 17) | 8 | 1 | 0.649 |
| Deletions | 6 | 0 | |
| Other complex abnormality | 4 | 1 |
Discussion
In this study, we have analyzed prevalence of mutations on hotspots locations on exon 7 & 9 of WT1 gene and its clinical correlation among treatment naïve newly diagnosed primary cases of AML. Few studies of WT1 gene mutation in AML have been published till date mostly representing western countries [32-36]. A most recent study [37] from Pakistan has reported WT1 gene to be associated with recruitment of three independent germ line mutations in AML cases. There has been paucity of such data from India; however, a single study [36] has reported prevalence of 6.7% of WT1 mutation among cases from southern India. The current study has been conducted in the northern part of India, though caters to medical services to whole country. In our study, WT1 mutation was found only in 2 out of 100 cases (2%) including one novel mutation onexon 9, which was due to single base substitution of nucleotide resulting change in amino acid sequence of protein (Table 2A). The nucleotide and protein variation of was c.1373 G>C; p.Arg458Pro. This novel mutation finding was submitted to ClinVar database (accession IDVCV000625171.1) of NCBI as per Genome Reference Consortium Human Build 37. We also observed an already reported point mutation on exon 9. The altered nucleotide and protein sequence of this case was c.1385 G>C; p.Arg462Pro. There were 12 cases (12%) having synonymous SNP (Table 2B) which has already been reported as SNP rs16754 in dbSNP database. Six out of 100 cases (6%) had intronic variants (Table 2C) on intron-exon flanking region of both exon 7 & 9. Prevalence of WT1 mutation was found to be very low (2%) as compared to the reported data of 6-13% [34-37] from western European and middle east countries. The reported prevalence of 2% is even lower than already reported data of 6.7% from a previous Indian study [36]. This disparity in our study can be due to the fact, that, we have analyzed WT1 mutation cluster only on exon 7 & 9 considering them as hotspots gene. While, other studies have examined whole of WT1 gene which also included exons 1, 2, 3 & 8, apart from exon 7 and 9. Though, sample size was calculated as per benchmark of 7% adopted from Indian study [36], the overall prevalence was expected to be within the range of reported data. The possible hypothesis to explain this lower prevalence figure is that previous study was done predominantly on south Indian population, which shares a different gene pool [36]. Other possible potential cause could also be limited number of samples analyzed in our study. The sanger sequencing analysis revealed, that WT1 mutation was found mainly in exon 9 (n = 2), and not in exon 7 which is different from some of the other previous published studies [41]. Both were single base substitution mutation (point mutation) in exon 9, where one nucleotide was changed to other thus resulting in a change in amino acid sequence. This may ultimately lead to a variation in protein conformation and hence in protein function [34].
In correlation with age and sex between WT1 mutant & wild-type cases, there was no statistically significant differences and p Value for age group less than 18 & age group more than 18 was P = 0.743 and between male & female cases was (P = 0.851) (Table 3). There were also no statistically significant differences in other clinical parameters as hemoglobin level, WBCs count and platelet count between WT1 mutant-type and wild-type cases, which was in concordance with other published studies [41]. A correlation was noticed between WT1 mutant-type patients and high percentage (%) of blast counts at presentation but it was also not statistically significant (P = 0.161), may be due to a lesser number of mutant when compared with wild cases. This finding was different in comparison to a previously published studies [39,40] which stated that WT1 gene mutation might induce high proliferation capacity of blast cells and that may affect its count and distribution.
Considering the critical impact of WT1 gene in cell survival and its role as oncogene in various neoplasm, increased WT1 expression levels is considered to have a prognostic significance and is associated with a poor response to therapy [46,47]. In a study [47], WT1 transcript levels were evaluated in blast cells of 139 AML cases who were treated with standard chemotherapy regimen. Probability of 3-year overall survival was found to be 59% in patients with low WT1 expression levels compared to 21% in patients with high WT1 levels (P<0.046). These findings contrasted with another observation of [45] where another research group evaluated WT1 RNA expression in 125 de novo cases of AML and did not find any correlation with their disease-free survival or clinical remission. This discrepancy in observation may be explained due to differences in protocol methodology in measuring WT1 gene expression and differences in selection of target sample size. Further, in the intensity of the treatment regimens used would also have their confounding effect on survival parameters. In our study, both cases having WT1 mutation died during their course of induction therapy and this finding was statistically significant (P<0.04) suggesting a overall poor prognostic effect of WT1 mutation. However, we cannot conclude with confidence due to a small percentage of cases harboring mutational signature and a fairly limited sample size of the study.
In karyotype analysis, WT1 mutation was found in both, cases with normal as well as abnormal cytogenetics (Table 4) but there was no statistically significant difference among both groups. This finding in our study was not in consensus with a previous published study which reported that WT1 mutation are more prevalent in cases with normal cytogenetics compared with abnormal cytogenetics [41]. This could be attributed to the fact that in our study we had cytogenetic results available for limited no of cases (39/100). Twelve (12%) among all analyzed cases of AML were found to have synonymous single nucleotide polymorphism (SNPs) in exon 7 which has been previously reported in SNP database (rs16754) and it is heterozygous in nature (WT1 A→G) (Table 2B). Synonymous SNPs causes a change in nucleotide sequence of gene without altering protein sequence. Such synonymous SNPs were thought to be insignificant earlier but recent studies have reported that WT1 SNP rs16754 is an independent prognostic marker in certain cases of AML [48-51]. A recent study [51] showed that these synonymous SNPs may also directly alter the gene function and phenotype by mechanisms such as alteration of miRNA binding, protein folding, or by affecting mRNA splicing, stability or SNP expression. In our study presence of synonymous SNP was not associated with any protein change and was not statistically significant with prognosis or for any other clinical parameters. The correlation among the demographic data and clinical parameters was not statistically significant among the two groups. This is may be explained due to low number of cases among mutant positive category.
Conclusion
The current study concluded that exon 7 & 9 WT1 mutations are not so prevalent among cases of AML in Indian population as compared to western population. WT1 mutation was found in AML with both normal as well as abnormal cytogenetics. It was also evaluated that WT1 gene mutation corresponded with bad prognosis and showed poorer clinical outcome in the form of early induction deaths. Presence of synonymous SNP in cases of AML was not statistically significant. The limitation of study was analysis of only mutational hotspot region i.e. exon 7 & 9 of WT1 gene along with relatively limited sample size. Therefore, an elaborate study on NGS based sequencing of WT1 covering all exons with greater number of cases may give better evaluation of its clinical impact on disease causation, prognosis and overall survival. Molecular studies in cases of AML with mRNA expression of WT1 gene using real time PCR might also be incorporated in future research. WT1 gene mutation and its expression have potential of being a effective minimal residual disease marker and further needs to be deciphered.
Acknowledgements
We are thankful to Research Section, AIIMS, New Delhi and Science and Engineering Research Board, Department of Science and Technology, Government of India through Grant EEQ/2016/000318 for supporting resources in this study.
Disclosure of conflict of interest
None.
References
- 1.Goryainova NV. The clinical significance of genetic mutations in acute myeloid leukemia. Lik Sprava. 2014;12:10–18. [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;1:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Sill H, Olipitz W, Zebisch A, Schulz E, Wolfler A. Therapy-related myeloid neoplasms: pathobiology and clinical characteristics. Br J Pharmacol. 2011;4:792–805. doi: 10.1111/j.1476-5381.2010.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, Anderson JE, Petersdorf SH. Age and acute myeloid leukemia. Blood. 2006;9:3481–3485. doi: 10.1182/blood-2005-09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagunas-Rangel FA, Chavez-Valencia V, Gomez-Guijosa MA, Cortes-Penagos C. Acute myeloid leukemia-genetic alterations and their clinical prognosis. Int J Hematol Oncol Stem Cell Res. 2017;4:328–339. [PMC free article] [PubMed] [Google Scholar]
- 6.Pulte D, Redaniel MT, Jansen L, Brenner H, Jeffreys M. Recent trends in survival of adult patients with acute leukemia: overall improvements, but persistent and partly increasing disparity in survival of patients from minority groups. Haematologica. 2013;2:222–229. doi: 10.3324/haematol.2012.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlen M, Klusmann JH, Hoell JI. Molecular approaches to treating pediatric leukemias. Front Pediatr. 2019;7:368. doi: 10.3389/fped.2019.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;7756:345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- 9.Janin M, Coll-SanMartin L, Esteller M. Disruption of the RNA modifications that target the ribosome translation machinery in human cancer. Mol Cancer. 2020;1:1–13. doi: 10.1186/s12943-020-01192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiyama H. Wilms’ tumor gene WT1: its oncogenic function and clinical application. Int J Hematol. 2001;73:177–187. doi: 10.1007/BF02981935. [DOI] [PubMed] [Google Scholar]
- 11.Huff V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;2:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Han Y, Suarez FS, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 13.Williams RD, Chagtai T, Alcaide-German M, Apps J, Wegert J, Popov S, Vujanic G, van Tinteren H, van den Heuvel-Eibrink MM, Kool M, de Kraker J, Gisselsson D, Graf N, Gessler M, Pritchard-Jones K. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget. 2015;6:7232–7243. doi: 10.18632/oncotarget.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rampal R, Figueroa ME. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica. 2016;6:672–679. doi: 10.3324/haematol.2015.141796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida S, Hosen N, Shirakata T, Kanato K, Yanagihara M, Nakatsuka SI, Hoshida Y, Nakazawa T, Harada Y, Tatsumi N, Tsuboi A. AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1. Blood. 2006;107:3303–3312. doi: 10.1182/blood-2005-04-1656. [DOI] [PubMed] [Google Scholar]
- 16.Ariyaratana S, Loeb DM. The role of the Wilms tumour gene (WT1) in normal and malignant haematopoiesis. Expert Rev Mol Med. 2007;14:1. doi: 10.1017/S1462399407000336. [DOI] [PubMed] [Google Scholar]
- 17.Artibani M, Sims AH, Slight J, Aitken S, Thornburn A, Muir M, Brunton VG, Del-Pozo J, Morrison LR, Katz E, Hastie ND. WT1 expression in breast cancer disrupts the epithelial/mesenchymal balance of tumour cells and correlates with the metabolic response to docetaxel. Sci Rep. 2017;7:45255. doi: 10.1038/srep45255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlton J, Pritchard-Jones K. In: WT1 mutation in childhood cancer. methods in molecular biology. Hastie N, editor. New York: Humana Press; 2016. pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 19.Lindstedt I, Lindgren MA, Andersson E, Engstrom W. The WT1 gene--its role in tumourigenesis and prospects for immunotherapeutic advances. In vivo. 2014;5:675–681. [PubMed] [Google Scholar]
- 20.Toska E, Roberts SG. Mechanisms of transcriptional regulation by WT1 (Wilms’ tumour 1) Biochem J. 2014;1:15–32. doi: 10.1042/BJ20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lv L, Chen G, Zhou J, Li J, Gong J. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J Exp Clin Cancer Res. 2015;1:1–9. doi: 10.1186/s13046-015-0233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto H, Zhang X, Zheng Y, Wilson GG, Cheng X. Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications. Nucleic Acids Res. 2016;21:10165–10176. doi: 10.1093/nar/gkw766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosen N, Sonoda Y, Oji Y, Kimura T, Minamiguchi H, Tamaki H, Kawakami M, Asada M, Kanato K, Motomura M, Murakami M. Very low frequencies of human normal CD34+ haematopoietic progenitor cells express the Wilms’ tumour gene WT1 at levels similar to those in leukaemia cells. Br J Haematol. 2002;2:409–420. doi: 10.1046/j.1365-2141.2002.03261.x. [DOI] [PubMed] [Google Scholar]
- 24.Ellisen LW, Carlesso N, Cheng T, Scadden DT, Haber DA. The Wilms tumor suppressor WT1 directs stage-specific quiescence and differentiation of human hematopoietic progenitor cells. EMBO J. 2001;8:1897–1909. doi: 10.1093/emboj/20.8.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird PN, Simmons PJ. Expression of the Wilms’ tumor gene [WT1] in normal hemopoiesis. Exp Hematol. 1997;4:312–320. [PubMed] [Google Scholar]
- 26.Rautenberg C, Pechtel S, Hildebrandt B, Betz B, Dienst A, Nachtkamp K, Kondakci M, Geyh S, Wieczorek D, Haas R, Germing U. Wilms tumor 1 gene expression using a standardized European leukemia net-certified assay compared to other methods for detection of minimal residual disease in myelodysplastic syndrome and acute myelogenous leukemia after allogeneic blood stem cell transplantation. Biol Blood Marrow Transplant. 2018;11:2337–2343. doi: 10.1016/j.bbmt.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Hidaka D, Onozawa M, Hashiguchi J, Miyashita N, Kasahara K, Fujisawa S, Hayase E, Okada K, Shiratori S, Goto H, Sugita J. Wilms tumor 1 expression at diagnosis correlates with genetic abnormalities and polymorphism but is not independently prognostic in acute myelogenous leukemia: a Hokkaido leukemia net study. Clin Lymphoma Myeloma Leuk. 2018;11:469–479. doi: 10.1016/j.clml.2018.07.291. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Wang X, Zhang H, Wang J, Chen Y, Ma T, Shi J, Kang Y, Xi J, Wang M, Zhang M. Dynamic changes in the level of WT1 as an MRD marker to predict the therapeutic outcome of patients with AML with and without allogeneic stem cell transplantation. Mol Med Report. 2019;3:2426–2432. doi: 10.3892/mmr.2019.10440. [DOI] [PubMed] [Google Scholar]
- 29.Cho BS, Min GJ, Park SS, Shin SH, Yahng SA, Jeon YW, Yoon JH, Lee SE, Eom KS, Kim YJ, Lee S. WT1 measurable residual disease assay in patients with acute myeloid leukemia who underwent allogeneic hematopoietic stem cell transplantation: optimal time points, thresholds, and candidates. Biol Blood Marrow Transplant. 2019;10:1925–1932. doi: 10.1016/j.bbmt.2019.05.033. [DOI] [PubMed] [Google Scholar]
- 30.Pronier E, Bowman RL, Ahn J, Glass J, Kandoth C, Merlinsky TR, Whitfield JT, Durham BH, Gruet A, Hanasoge Somasundara AV, Rampal R. Genetic and epigenetic evolution as a contributor to WT1-mutant leukemogenesis. Blood. 2018;12:1265–1278. doi: 10.1182/blood-2018-03-837468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramzi M, Moghadam M, Cohan N. Wilms tumor-1 (WT1) rs16754 polymorphism and clinical outcome in acute myeloid leukemia. Turk J Haematol. 2019;1:67–68. doi: 10.4274/tjh.galenos.2018.2018.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauth MT, Alpermann T, Bacher U, Eder C, Dicker F, Ulke M, Kuznia S, Nadarajah N, Kern W, Haferlach C, Haferlach T. WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia. 2015;3:660–667. doi: 10.1038/leu.2014.243. [DOI] [PubMed] [Google Scholar]
- 33.Aref S, El Sharawy S, Sabry M, Azmy E, Raouf DA. Prognostic relevance of Wilms tumor 1 (WT1) gene Exon 7 mutations in-patient with cytogenetically normal acute myeloid leukemia. Indian J Hematol Blood Transfus. 2014;4:226–230. doi: 10.1007/s12288-013-0288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen C, Fitzgibbon J, Paschka P. The clinical relevance of Wilms Tumour 1 (WT1) gene mutations in acute leukaemia. Hematol Oncol. 2010;1:13–19. doi: 10.1002/hon.931. [DOI] [PubMed] [Google Scholar]
- 35.Renneville A, Boissel N, Zurawski V, Llopis L, Biggio V, Nibourel O, Philippe N, Thomas X, Dombret H, Preudhomme C. Wilms tumor 1 gene mutations are associated with a higher risk of recurrence in young adults with acute myeloid leukemia: a study from the Acute Leukemia French Association. Cancer. 2009;16:3719–3727. doi: 10.1002/cncr.24442. [DOI] [PubMed] [Google Scholar]
- 36.Ahmad F, Dsouza W, Mandava S, Ranjan Das B. Molecular analysis of WT1 and KIT mutations in patients from an Indian population with de novo acute myeloid leukemia: determination of incidence, distribution patterns, and report of a novel KIT mutation. Leuk Lymphoma. 2011;52:865–876. doi: 10.3109/10428194.2011.552137. [DOI] [PubMed] [Google Scholar]
- 37.Shahid S, Shakeel M, Siddiqui S, Ahmed S, Sohail M, Khan IA, Abid A, Shamsi T. Novel genetic variations in acute myeloid leukemia in Pakistani population. Front Genet. 2020;11:560. doi: 10.3389/fgene.2020.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, Lu C. DNA hydroxymethylation profiling reveals thatWT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;5:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panuzzo C, Signorino E, Calabrese C, Ali MS, Petiti J, Bracco E, Cilloni D. Landscape of tumor suppressor mutations in acute myeloid leukemia. J Clin Med. 2020;3:802. doi: 10.3390/jcm9030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buelow DR, Pounds SB, Wang YD, Shi L, Li Y, Finkelstein D, Shurtleff S, Neale G, Inaba H, Ribeiro RC, Palumbo R. Uncovering the genomic landscape in newly diagnosed and relapsed pediatric cytogenetically normal FLT3-ITD AML. Clin Transl Sci. 2019;6:641–647. doi: 10.1111/cts.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho PA, Zeng R, Alonzo TA, Gerbing RB, Miller KL, Pollard JA, Stirewalt DL, Heerema NA, Raimondi SC, Hirsch B, Franklin JL. Prevalence and prognostic implications of WT1 mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood. 2010;116:702–710. doi: 10.1182/blood-2010-02-268953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abbas S, Erpelinck-Verschueren CA, Goudswaard CS, Löwenberg B, Valk PJ. Mutant Wilms’ tumor 1 (WT1) mRNA with premature termination codons in acute myeloid leukemia (AML) is sensitive to nonsense-mediated RNA decay (NMD) Leukemia. 2010;24:660–663. doi: 10.1038/leu.2009.265. [DOI] [PubMed] [Google Scholar]
- 43.Hou HA, Huang TC, Lin LI, Liu CY, Chen CY, Chou WC, Tang JL, Tseng MH, Huang CF, Chiang YC, Lee FY. WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood. 2010;115:5222–5231. doi: 10.1182/blood-2009-12-259390. [DOI] [PubMed] [Google Scholar]
- 44.Harris NL, Arber DA, Campo E, Jaffe ES, Orazi A, Pileri SA, Stien H, Swerdlow SH, Thiele J, Vardiman JW. Introduction to the WHO classification of tumours of haematopoietic and lymphoid tissue. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stien H, Thiele JJ, editors. WHO Classification of Tumours of the Haematopoietic and Lymphoid Tissues. Geneva: WHO Press; 2017. pp. 13–27. [Google Scholar]
- 45.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heesch S, Goekbuget N, Stroux A, Tanchez JO, Schlee C, Burmeister T, Schwartz S, Blau O, Keilholz U, Busse A, Hoelzer D. Prognostic implications of mutations and expression of the Wilms tumor 1 (WT1) gene in adult acute T-lymphoblastic leukemia. Haematologica. 2010;6:942–949. doi: 10.3324/haematol.2009.016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assem M, Osman A, Kandeel E, Elshimy R, Nassar H, Ali R. Clinical impact of overexpression of FOXP3 and WT1 on disease outcome in egyptian acute myeloid leukemia patients. Asian Pac J Cancer Prev. 2016;10:4699–4711. doi: 10.22034/APJCP.2016.17.10.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayatollahi H, Sadeghian MH, Naderi M, Jafarian AH, Shams SF, Motamedirad N, Sheikhi M, Bahrami A, Shakeri S. Quantitative assessment of Wilms tumor 1 expression by real-time quantitative polymerase chain reaction in patients with acute myeloblastic leukemia. J Res Med Sci. 2017;22:54. doi: 10.4103/jrms.JRMS_448_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damm F, Heuser M, Morgan M, Yun H, Grobhennig A, Gohring G, Schlegelberger B, Dohner K, Ottmann O, Lubbert M, Heit W. Single nucleotide polymorphism in the mutational hotspot of WT1 predicts a favorable outcome in patients with cytogenetically normal acute myeloid leukemia. J. Clin. Oncol. 2010;4:578–85. doi: 10.1200/JCO.2009.23.0342. [DOI] [PubMed] [Google Scholar]
- 50.Ho PA, Kuhn J, Gerbing RB, Pollard JA, Zeng R, Miller KL, Heerema NA, Raimondi SC, Hirsch BA, Franklin JL, Lange B. WT1 synonymous single nucleotide polymorphism rs16754 correlates with higher mRNA expression and predicts significantly improved outcome in favorable-risk pediatric acute myeloid leukemia: a report from the children’s oncology group. J. Clin. Oncol. 2011;29:704–711. doi: 10.1200/JCO.2010.31.9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;66:110635–110649. doi: 10.18632/oncotarget.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]