Abstract
The combination of big data and artificial intelligence (AI) is having an increasing impact on the field of electrophysiology. Algorithms are created to improve the automated diagnosis of clinical ECGs or ambulatory rhythm devices. Furthermore, the use of AI during invasive electrophysiological studies or combining several diagnostic modalities into AI algorithms to aid diagnostics are being investigated. However, the clinical performance and applicability of created algorithms are yet unknown. In this narrative review, opportunities and threats of AI in the field of electrophysiology are described, mainly focusing on ECGs. Current opportunities are discussed with their potential clinical benefits as well as the challenges. Challenges in data acquisition, model performance, (external) validity, clinical implementation, algorithm interpretation as well as the ethical aspects of AI research are discussed. This article aims to guide clinicians in the evaluation of new AI applications for electrophysiology before their clinical implementation.
Keywords: Artificial intelligence, deep learning, neural networks, cardiology, electrophysiology, ECG, big data
Clinical research that uses artificial intelligence (AI) and big data may aid the prediction and/or detection of subclinical cardiovascular diseases by providing additional knowledge about disease onset, progression or outcome. Clinical decision-making, disease diagnostics, risk prediction or individualised therapy may be informed by insights obtained from AI algorithms. As health records have become electronic, data from large populations are becoming increasingly accessible.[1] The use of AI algorithms in electrophysiology may be of particular interest as large data sets of ECGs are often readily available. Moreover, data are continuously generated by implantable devices, such as pacemakers, ICDs or loop recorders, or smartphone and smartwatch apps.[2–6]
Interpretation of ECGs relies on expert opinion and requires training and clinical expertise which is subjected to considerable inter- and intra-clinician variability.[7–12] Algorithms for the computerised interpretation of ECGs have been developed to facilitate clinical decision-making. However, these algorithms lack accuracy and may provide inaccurate diagnoses which may result in misdiagnosis when not reviewed carefully.[13–18]
Substantial progress in the development of AI in electrophysiology has been made, mainly concerning ECG-based deep neural networks (DNNs). DNNs have been tested to identify arrhythmias, to classify supraventricular tachycardias, to predict left ventricular ejection fraction, to identify disease development in serial ECG measurements, to predict left ventricular hypertrophy and to perform comprehensive triage of ECGs.[6,19–23] DNNs are likely to aid non-specialists with improved ECG diagnostics and may provide the opportunity to expose yet undiscovered ECG characteristics that indicate disease.
With this progress, the challenges and threats of using AI techniques in clinical practice become apparent. In this narrative review, recent progress of AI in the field of electrophysiology is discussed together with its opportunities and threats.
A Brief Introduction to Artificial Intelligence
AI refers to mimicking human intelligence in computers to perform tasks that are not explicitly programmed. Machine learning (ML) is a branch of AI concerned with algorithms to train a model to perform a task. Two types of ML algorithms are supervised learning and unsupervised learning. Supervised learning refers to ML algorithms where input data are labelled with the outcome and the algorithm is trained to approximate the relation between input data and outcome. In unsupervised learning, input data are not labelled and the algorithm may discover data clusters in the input data.
In ML, an algorithm is trained to classify a data set based on several statistical and probability analyses. In the training phase, model parameters are iteratively tuned by penalising or rewarding the algorithm based on a true or false prediction. Deep learning is a sub-category of ML that uses DNNs as architecture to represent and learn from data. The main difference between deep learning and other ML algorithms is that DNNs can learn from raw data, such as ECG waveforms, in an end-to-end manner with extraction and classification united in the algorithm (Figure 1a). For example, in ECG-based DNNs, a matrix containing the time-stamped raw voltage values of each lead are used as input data. In other ML algorithms, features like heart rate or QRS duration are manually extracted from the ECG and used as input data for the classification algorithm.
Figure 1: Traditional Machine Learning and Deep Learning with a Schematic Representation of Fitting a Model to a Data Set.
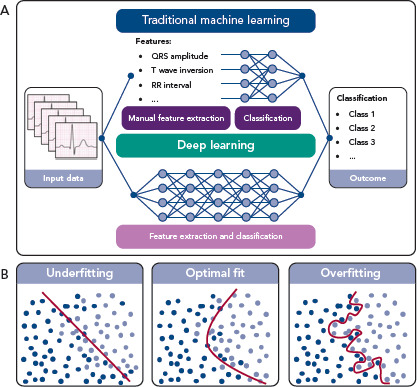
To influence the speed and quality of the training phase, the setting of hyperparameters, such as the settings of the model architecture and training, is important. Furthermore, overfitting or underfitting the model to the available data set must be prevented. Overfitting can occur when a complex model is trained using a small data set. The model will precisely describe the training data set but fail to predict outcomes using other data (Figure 1b). On the other hand, when constraining the model too much, underfitting occurs (Figure 1b), also resulting in poor algorithm performance. To assess overfitting, a data set is usually divided into a training data set, a validation data set and a test data set, or resampling methods are used, such as cross-validation or bootstrapping.[24]
To train and test ML algorithms, particularly DNNs, it is preferable to use a large data set, known as big data. Performance of highly dimensional algorithms – e.g. algorithms with many model parameters such as DNNs – depends on the size of the data set. For deep learning, more data is often required as DNNs have many non-linear parameters and non-linearity increases the flexibility of an algorithm. The size of a training data set has to reasonably approximate the relation between input data and outcome and the amount of testing data has to reasonably approximate the performance measures of the DNN.
Determining the exact size of a training and testing data set is difficult.[25,26] It depends on the complexity of algorithm (e.g. the number of variables), the type of the algorithm, the number of outcome classes and the difficulty of distinguishing between outcome classes as inter-class differences might be subtle. Therefore, size of the data set should be carefully reviewed for each algorithm. A rule of thumb for the adequate size of a validation data set is 50–100 patients per outcome class to determine overfitting. Recent studies published in the field of ECG-based DNNs used between 50,000 and 1.2 million patients.[6,19,21,27]
Prerequisites for AI in Electrophysiology
Preferably, data used to create AI algorithms is objective, as subjectivity may introduce bias in the algorithm. To ensure clinical applicability of created algorithms, ease of access to input data, difference in data quality in different clinical settings as well as the intended use of the algorithm should be considered. In this section, we mainly focus on the data quality of ECGs, as these data are easily acquired and large data sets are readily available.
Technical Specifications of ECGs
ECGs are obtained via electrodes on the body surface using an ECG device. The device samples the continuous body surface potentials and the recorded signals are filtered to obtain a clinically interpretable ECG.[28] As the diagnostic information of the ECG is contained below 100 Hz, a sampling rate of at least 200 Hz is required according to the Nyquist theorem.[29–33] Furthermore, an adequate resolution of at least 10 µV is recommended to also obtain small amplitude fluctuations of the ECG signal. In the recorded signal, muscle activity, baseline wander, motion artefacts and powerline artefacts are also present, distorting the measured ECG. To remove noise and obtain an easily interpretable ECG, a combination of a high-pass filter of 0.67 Hz and a low-pass filter of 150–250 Hz is recommended, often combined with a notch filter of 50 Hz or 60 Hz. The inadequate setting of these filters might result in a loss of information such as QRS fragmentation or notching, slurring or distortion of the ST segment. Furthermore, a loss of QRS amplitude of the recorded signal might be the result of the inappropriate combination of a high frequency cut-off and sampling frequency.[28,34] ECGs used as input for DNNs are often already filtered, thus potentially relevant information might already be lost. As DNNs process and interpret the input data differently, filtering might be unnecessary and potentially relevant information may be preserved. Furthermore, as filtering strategies differ between manufacturers and even different versions of ECG devices, the performance of DNNs might be affected when ECGs from different ECG devices are used as input data.
Apart from applied software settings, such as sampling frequency or filter settings, the hardware of ECG devices also differs between manufacturers. Differences in analogue to digital converters, type of electrodes used, or amplifiers also affect recorded ECGs. The effect of input data recorded using different ECG devices on the performance of AI algorithms is yet unknown. However, as acquisition methods may differ significantly between manufacturers, the performance of algorithms are likely to depend on the type or even version of the device.[35] Testing the performance of algorithms using ECGs recorded by different devices would illustrate the effect of these technical specifications on performance and generalisability.
ECG Electrodes
The recorded ECG is affected by electrode position with respect to the anatomical position of the heart and displacement of electrodes may result in misdiagnosis in a clinical setting.[36,37] For example, placement of limb electrodes on the trunk significantly affects the signal waveforms and lead reversal may mimic pathological conditions.[38–41] Furthermore, deviations in precordial electrode positions affect QRS and T wave morphology (Figure 2). Besides the effect of cardiac electrophysiological characteristics like anisotropy, His-Purkinje anatomy, myocardial disease and cardiac anatomy on measured ECGs, cardiac position and cardiac movement also affect the ECG.[42–45]
Figure 2: The Effect of Shifting Precordial Electrodes Upward or Downward.
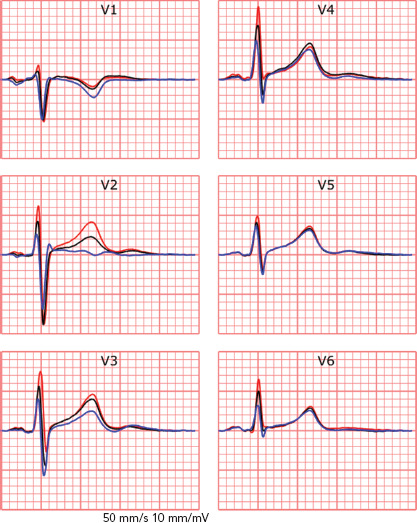
The effect of shifting precordial electrodes 4 m upward (blue) or downward (red) from standard 12-lead electrode positioning (black). Displayed signals were simultaneously recorded using a 64-electrode measurement set-up.
Conventional clinical ECGs mostly consist of the measurement of eight independent signals; two limb leads and six precordial leads (Figure 3b). The remaining four limb leads are derived from the measured limb leads. However, body surface mapping studies identified the number of signals containing unique information up to 12 for ventricular depolarisation and up to 10 for ventricular repolarisation.[46] Theoretically, to measure all information about cardiac activity from the body surface, the number of electrodes should be at least the number of all unique measurements. However, conventional 12-lead ECG is widely accepted for most clinical applications. An adjustment of a lead position is only considered when a posterior or right ventricle MI or Brugada syndrome is suspected.[27,47–50]
Figure 3: Standardised Clinical Visualised Signals.

A: Three simultaneously recorded 2.5 second measurements and raw signals. B: A 10-second measurement and median beats of all recorded leads. Displayed signals are acquired using a General Electric Healthcare MAC 5500 ECG device.
The interpretation of ECGs by computers and humans is fundamentally different and factors like electrode positioning or lead misplacement might influence algorithms. However, the effect of electrode misplacement or reversal, disease-specific electrode positions or knowledge of lead positioning on the performance on DNNs remains to be identified. A recent study was able to identify misplaced chest electrodes, implying that the effect of electrode misplacement might be able to be identified and acknowledged by algorithms.[51] Studies have suggested that DNNs can achieve similar performance when fewer leads are used.[50]
ECG Input Data Format
ECGs can be obtained from the electronic database in three formats – visualised signals (as used in standard clinical practice), raw ECG signals or median beats. Raw signals are preferable for input for DNNs as visualised signals require digitisation, which results in a loss of signal resolution. Furthermore, raw ECG signals often consist of a continuous 10-second measurement of all recorded leads, whereas visualised signals may consist of 2.5 seconds per lead with only three simultaneously recorded signals per 2.5 seconds (Figure 3). A median beat per lead can also be used, computed from measured raw ECG signals or digitised visualised signals. Using the median beat might reduce noise, as noise is expected to cancel out by averaging all beats. Therefore, subtle changes in cardiac activation, invisible due to noise might become distinguishable for the algorithm. The use of the median beat may allow for precise analysis of waveform shapes or serial changes between individuals but rhythm information will be lost.
Opportunities for Artificial Intelligence in Electrophysiology
Enhanced Automated ECG Diagnosis
An important opportunity of AI in electrophysiology is the enhanced automated diagnosis of clinical 12-lead ECGs.[8,11,12,20,52–54] Adequate computerised algorithms are especially important when expert knowledge is not readily available, such as in pre-hospital care, non-specialist departments, or facilities that have minimal resources. If high-risk patients can be identified correctly, time-to-treatment can be reduced. However, currently available computerised ECG diagnosis algorithms lack accuracy.[11] Progress has been made in using DNNs to automate diagnosis or triage ECGs to improve time-to-treatment and reduce workload.[19,55] Using very large data sets, DNNs can achieve high diagnostic performance and outperform cardiology residents and non-cardiologists.[6,19] Moreover, progress has been made in using ECG data for predictive modelling for AF in sinus rhythm ECGs or for the screening of hypertrophic cardiomyopathy.[56–58]
Combining Other Diagnostic Modalities with ECG-based DNN
Some studies have suggested the possibility of using ECG-based DNNs with other diagnostic modalities to screen for disorders that are currently not associated with the ECG. In these applications, DNNs are thought to be able to detect subtle ECG changes. For example, when combined with large laboratory data sets, patients with hyperkalaemia could be identified, or when combined with echocardiographic results, reduced ejection fraction or aortic stenosis could be identified. The created DNNs identified these three disorders from the ECG with high accuracy.[21,50,59] As a next step, supplementing ECG-based DNNs with body surface mapping data with a high spatial resolution (e.g. more than 12 measurement electrodes), inverse electrocardiography data or invasive electrophysiological mapping data, may result in the identification of subtle changes in the 12-lead ECG as a result of pathology.
Artificial Intelligence for Invasive Electrophysiological Studies
The application of AI before and during complex invasive electrophysiological procedures, such as electroanatomical mapping, is another major opportunity. By combining information from several diagnostic tools such as MRI, fluoroscopy or previous electroanatomical mapping procedures, invasive catheter ablation procedure time might be reduced through the accelerated identification of arrhythmogenic substrates. Also, new techniques such as ripple mapping may be of benefit during electroanatomical mapping studies.[60] Recent studies suggest that integration of fluoroscopy and electroanatomical mapping with MRI is feasible using conventional statistical techniques or ML, whereas others suggest the use of novel anatomical mapping systems to circumvent fluoroscopy.[61–64] Furthermore, several ML algorithms have been able to identify myocardial tissue properties using electrograms in vitro.[65]
Ambulatory Device-based Screening for Cardiovascular Diseases
One of the major current challenges in electrophysiology is the applicability of ambulatory rhythm devices in clinical practice. Several tools, such as implantable devices or smartwatch and smartphone-based devices, are becoming more widely used and continuously generate large amounts of data which would be impossible to evaluate manually.[66] Arrhythmia detection algorithms based on DNNs trained on large cohorts of ambulatory patients with a single-lead plethysmography or ECG device have shown similar diagnostic performance as cardiologists or implantable loop recorders.[2,3,6] Another interesting application of DNN algorithms are data from intracardiac electrograms before and during the activation of the defibrillator. Analysis of the signals before the adverse event might provide insight into the mechanism of the ventricular arrhythmia, providing the clinician with valuable insights. Continuous monitoring also provides the possibility of identifying asymptomatic cardiac arrhythmias or detecting post-surgery complications. Early detection might overcome serious adverse events and significantly improve timely personalised healthcare.[6,19]
A promising benefit of smartphone applications for the early detection of cardiovascular disease is in early detection of AF. As AF is a risk factor for stroke, early detection may be important to prompt adequate anticoagulant treatment.[67–69] An irregular rhythm can be accurately detected using smartphone or smartwatch-acquired ECGs. Even predicting whether a patient will develop AF in the future using smartphone-acquired ECGs recorded during sinus rhythm has been recently reported.[69,70] Also, camera-based photoplethysmography recordings can be used to differentiate between irregular and regular cardiac rhythm.[71,72] However, under-detection of asymptomatic AF is expected as the use of applications requires active use and people are likely to only use applications when they have a health complaint. Therefore, a non-contact method with facial photoplethysmography recordings during regular smartphone use may be an interesting option to explore.[70,73,74]
Apart from the detection of asymptomatic AF, the prediction or early detection of ventricular arrhythmias using smartphone-based techniques are potentially clinically relevant. For example, smartphone-based monitoring of people with a known pathogenetic mutation might aid the early detection of disease onset. In some pathogenetic mutations, this may be especially relevant as sudden cardiac death can be the first manifestation of the disease. In these patients, close monitoring to prevent these adverse events by starting early treatment when subclinical signs are detected may provide clinical benefit.
Threats of Artificial Intelligence in Electrophysiology
Data-driven Versus Hypothesis-driven Research
Data from electronic health records are almost always retrospectively collected, leading to data-driven research, instead of hypothesis-driven research. Research questions are often formulated based on readily available data, which increases the possibility of incidental findings and spurious correlations. While correlation might be sufficient for some predictive algorithms, causal relationships remain of the utmost important to define pathophysiological relationships and ultimately for the clinical implementation of AI algorithms. Therefore, big data research is argued to be in most cases solely used to generate hypotheses and controlled clinical trials remain necessary to validate these hypotheses. When AI is used to identify novel pathophysiological phenotypes, e.g. with specific ECG features, sequential prospective studies and clinical trials are crucial.[75]
Input Data
Adequate labelling of input data is important for supervised learning.[18,76,77] Inadequate labelling of ECGs or the presence of pacemaker artefacts, comorbidities affecting the ECG or medication affecting the rhythm or conduction, might influence the performance of DNNs.[13–18] Instead of true disease characteristics, ECG changes due to clinical interventions are used by the DNN to classify ECGs. For example, a DNN using chest X-rays provided insight into long-term mortality, but the presence of a thoracic drain and inadequately labelled input data resulted in an algorithm that was unsuitable for clinical decision-making.[77–80] Therefore, the critical review of computerised labels and the identification of important features used by the DNN are essential.
Data extracted from ambulatory devices consist of real-time continuous monitoring data outside the hospital. As the signal acquisition is performed outside a standardised environment, signals are prone to errors. ECGs are more often exposed to noise due to motion artefacts, muscle activity artefacts, loosened or moved electrodes and alternating powerline artefacts. To accurately assess ambulatory data without the interference of artefacts, signals should be denoised or a quality control mechanism should be implemented. For both methods, noise should be accurately identified and adaptive filtering or noise qualification implemented.[81–83] However, as filtering might remove information, rapid real-time quality reporting of the presence of noise in the acquired signal is thought to be beneficial. With concise instructions, users can make adjustments to reduce artefacts and the quality of the recording will improve. Different analysis requires different levels of data quality and through classification recorded data quality, the threshold for user notification can be adjusted per analysis.[84,85]
Generalisability and Clinical Implementation
With the increasing number of studies on ML algorithms, generalisability and implementation is one of the most important challenges to overcome. Diagnostic or prognostic prediction model research, from simple logistic regression to highly sophisticated DNNs, is characterised by three phases:
Development and internal validation.
External validation and updating for other patients.
Assessment of the implementation of the model in clinical practice and its impact on patient outcomes.[86,87]
During internal validation, the predictive performance of the model is assessed using the development data set through train-test splitting, cross-validation or bootstrapping. Internal validation is however insufficient to test generalisability of the model in ‘similar but different’ individuals. Therefore, external validation of established models is important before clinical implementation. A model can be externally validated through temporal (same institution, later period), geographical (a different institution with a similar patient group) or domain (different patient group) validation. Finally, implementation studies, such as cluster randomised trials, before and after studies or decision-analytic modelling studies, are required to assess the effect of implementing the model in clinical care.[86,87]
Most studies in automated ECG prediction and diagnosis performed some type of external validation. However, no study using external validation in a different patient group or implementation study has been published so far. A study has shown similar accuracy to predict low ejection fraction from the ECG using a DNN through temporal validation as in the development study.[88] A promising finding was a similar performance of the algorithm for different ethnic subgroups, even if the algorithm was trained on one subgroup.[89] As a final step to validate this algorithm, a cluster randomised trial is currently being performed. This might provide valuable insight into the clinical usefulness of ECG-based DNNs.[90]
Implementation studies for algorithms using ambulatory plethysmography and ECG data are ongoing. For example, the Apple Heart Study assessed the implementation of smartphone-based AF detection.[5] More than 400,000 patients who used a mobile application were included, but only 450 patients were analysed. Implementation was proven feasible as the number of false alarms was low, but the study lacks insight into the effect of smartphone-based AF detection on patient outcome. Currently, the Heart Health Study Using Digital Technology to Investigate if Early AF Diagnosis Reduces the Risk of Thromboembolic Events Like Stroke IN the Real-world Environment (HEARTLINE; NCT04276441) is randomising patients to use the smartwatch monitoring device. The need for treatment with anticoagulation of patients with device-detected subclinical AF is also being investigated.[4]
A final step for the successful clinical implementation of AI is to inform its users about adequate use of the algorithm. Standardised leaflets have been proposed to instruct clinicians when, and more importantly when not, to use an algorithm.[91] This is particularly important if an algorithm is trained on a cohort using a specific subgroup of patients. Then, applying the model to a different population may potentially result in misdiagnosis. Therefore, describing the predictive performance in different subgroups, such as different age, sex, ethnicity and disease stage, is of utmost importance as AI algorithms are able to identify these by themselves.[89,92–94] However, as most ML algorithms are still considered to be ‘black boxes’, algorithm bias might remain difficult to detect.
Interpretability
Many sophisticated ML methods are considered black boxes as they have many model parameters and abstractions. This is in contrast with the more conventional statistical methods used in medical research, such as logistic regression and decision trees, where the influence of a predictor on the outcome is clear. The trade of complexity of models and interpretability for improved accuracy is important to acknowledge; with increased complexity of the network, interpretation becomes more complicated. But interpretability remains important to investigate false positives and negatives, to detect biased or overfitted models, to improve trust in new models or to use the algorithms as a feature detector.[95] Within electrophysiology, few studies have investigated how the AI algorithms came to a certain result. For DNNs, three recent studies visualised individual examples using Guided Grad-CAM, a technique to show what the networks focus on. They showed that the DNN used the same segment of the ECG that a physician would use (Figure 4).[19,27,96–98]
Figure 4: Important Regions for the Deep Neural Network to Predict Whether an ECG is Normal, Abnormal or Acute.
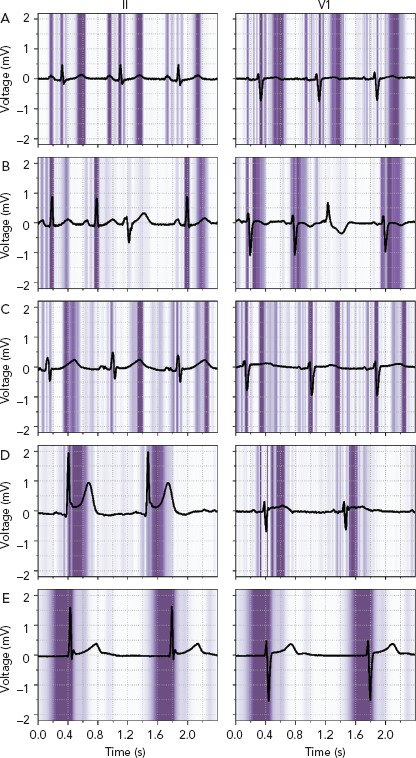
ECG leads II and V1 with a superimposed guided Grad-CAM visualisation showing regions important for the deep neural network to predict whether an ECG is normal, abnormal or acute. A and B: Normal ECGs with focus on the P wave, QRS-complex, and T wave, while correctly ignoring a premature ventricular complex. C: Abnormal ECG with a long QT interval and a focus on the beginning and end of the QT-segment. D and E: Acute ECGs with an inferior ST-segment elevation MI (D) and a focus on the ST-segment and with a junctional escape rhythm (E) and a focus on the pre-QRS-segment, where the P wave is missing. Source: van de Leur et al. 2020.[19] Reproduced from the American Heart Association, Inc., by Wiley Blackwell under a Creative Commons (CC BY-NC-ND 4.0) licence.
Visualisation techniques may provide the ECG locations which the algorithms find important, but do not identify the specific feature. Therefore, the opportunity to identify additional ECG features remains dependent on expert opinion and analysis of the data by a clinician is still required. Visualisation techniques and their results are promising and help to increase trust in DNNs for ECG analysis, but additional work is needed to further improve the interpretability of AI algorithms in clinical practice.[99,100]
Uncertainty Estimation
In contrast to physicians or conventional statistical methods, DNNs struggle to inform their users when they do not know and to give uncertainty measures about their predictions. Current models always output a diagnosis or prediction, even if they have not seen the input before. In a real-world setting, clinicians acknowledge uncertainty and consult colleagues or literature but a DNN always makes a prediction. Therefore, methods that incorporate uncertainty are essential before implementation of such algorithms is possible.[101]
Ideally, the algorithm provides results only when it reaches a high threshold of certainty, while the uncertain cases will still be reviewed by a clinician.[101] For DNNs, several new techniques are available to obtain uncertainty measures, such as Bayesian deep learning, Monte Carlo dropout and ensemble learning, but these have never been applied in electrophysiological research.[102] They have been applied to detect diabetic retinopathy in fundus images using DNNs, where one study showed that overall accuracy could be improved when uncertain cases were referred to a physician.[103] Another study suggested that uncertainty measures were able to detect when a different type of scanner was used that the algorithm had not seen before.[35] Combining uncertainty with active or online learning allows the network to learn from previously uncertain cases, which are now reviewed by an expert.[104]
Ethical Aspects
Several other ethical and legal challenges within the field of AI in healthcare are yet to be identified, such as patient privacy, poor quality algorithms, algorithm transparency and liability concerns. Data are subjected to privacy protections, confidentiality and data ownership, therefore requiring specific individual consent for use and reuse of data. However, by increasing the size of the data set, anonymisation techniques used nowadays might be inadequate and eventually result in the identification of patients.[105,106] As large data sets are required for DNNs, collaboration between institutions becomes inevitable. To facilitate data exchange, platforms have been established to allow for safe and consistent data-sharing between institutions.[107] However, these databases may still contain sensitive personal data.[54,108] Therefore, federated learning architectures are proposed that provide data-sharing while simultaneously obviating the need to share sensitive personal data. An example of this is the anDREea Consortium (andrea-consortium.org).
Another concerning privacy aspect is the continuous data acquisition through smartphone-based applications. In these commercial applications, data ownership and security are vulnerable. Security between smartphones and applications is heterogeneous and data may be stored on commercial and poorly secured servers. Clear regulations and policies should be in place before these applications can enter the clinical arena.
Data sets contain information about medical history and treatment but may also encompass demographics, religious status or socioeconomic status. Apart from medical information, sensitive personal data might be taken into account by developed algorithms, possibly resulting in discrimination in areas such as ethnicity, gender or religion.[54,108–110]
As described, DNNs are black boxes wherein input data is classified. An estimate of the competency of an algorithm can be made through the interpretation of DNNs and the incorporation of uncertainty measures. Traditionally, clinical practice mainly depends on the competency of a clinician. Decisions about diagnoses and treatments are based on widely accepted clinical standards and the level of competency is protected by continuous intensive medical training. In the case of adverse events, clinicians are held responsible if they deviated from standard clinical care. However, the medical liability of the DNN remains questionable. Incorrect computerised medical diagnoses or treatments result in adverse outcomes, thereby raising the question: who is accountable for a misdiagnosis based on an AI algorithm.
To guide the evaluation of ML algorithms, in particular DNNs, and accompanying literature in electrophysiology, a systematic overview of all relevant threats discussed in this review is presented in Table 1.
Table 1: Systematic Overview of Relevant Threats of AI Algorithms in Electrophysiology.
| Domain | Key Points | Questions |
|---|---|---|
| Algorithm input | Subjects | Is an appropriate data source used with clear inclusion and exclusion criteria? |
| Data | Is the ECG data of sufficient quality? Is the quality of ambulatory data continuously assessed? |
|
| Algorithm performance | Robustness | How does the model perform? Were there a reasonable number of subjects? Were ECGs equally sampled per subject? |
| Overfitting and optimism | Was overfitting assessed using internal validation with train-test splitting, cross-validation or bootstrapping? Was the validation data set of sufficient size (>100 participants with the outcome)? |
|
| External validation | Are there studies that provide temporal, geographical or domain validation? | |
| Subgroups | Is subgroup analysis provided to minimise the risk of poor performance in subgroups? Is there a bias based on ethnicity, gender or other demographic factors? |
|
| Algorithm implementation | Subjects | Is the population that will use the algorithm similar to the external validation population? Is the disease prevalence similar? |
| Data | Is the algorithm evaluated on the used diagnostic device of a specific manufacturer? Was data standardised according to general agreements? |
|
| Implementation studies | Have implementation studies, such as RCTs or before and after studies, been performed? Does implementation of the model positively influence patient outcomes? |
|
| Interpretation and uncertainty | Are there possibilities to check the predictions of the model in clinical practice (using visualisations)? Does the model provide uncertainty measures? How does the model deal with ECG noise or electrode misplacements? Is there a clear flowchart that allows uncertain cases to be referred to a physician? |
|
| Ethical and legal | Are the ethical and legal aspects sufficiently addressed? |
RCT = randomised controlled trial.
Conclusion
Many exciting opportunities arise when AI is applied to medical data, especially in cardiology and electrophysiology. New ECG features, accurate automatic ECG diagnostics and new clinical insights can be rapidly obtained using AI technology. In the near future, AI is likely to become one of the most valuable assets in clinical practice. However, as with every technique, AI has its limitations. To ensure the correct use of AI in a clinical setting, every clinician working with AI should be able to recognise the threats, limitations and challenges of the technique. Furthermore, clinicians and data scientists should closely collaborate to ensure the creation of clinically applicable and useful AI algorithms.
Clinical Perspective
Artificial intelligence (AI) may support diagnostics and prognostics in electrophysiology by automating common clinical tasks or aiding complex tasks through the identification of subtle or new ECG features.
Within electrophysiology, automated ECG diagnostics using deep neural networks is superior to currently implemented computerised algorithms.
Before the implementation of AI algorithms in clinical practice, trust in the algorithms must be established. This trust can be achieved through improved interpretability, measurement of uncertainty and by performing external validation and feasibility studies to determine added value beyond current clinical care.
Combining data obtained from several diagnostic modalities using AI might elucidate pathophysiological mechanisms of new, rare or idiopathic cardiac diseases, aid the early detection or targeted treatment of cardiovascular diseases or allow for screening of disorders currently not associated with the ECG.
References
- 1.Hemingway H, Asselbergs FW, Danesh J et al. Big data from electronic health records for early and late translational cardiovascular research: challenges and potential. Eur Heart J. 2018;39:1481–95. doi: 10.1093/eurheartj/ehx487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasserlauf J, You C, Patel R et al. Smartwatch performance for the detection and quantification of atrial fibrillation. Circ Arrhythm Electrophysiol. 2019;12:e006834. doi: 10.1161/CIRCEP.118.006834. [DOI] [PubMed] [Google Scholar]
- 3.Bumgarner JM, Lambert CT, Hussein AA et al. Smartwatch algorithm for automated detection of atrial fibrillation. J Am Coll Cardiol. 2018;71:2381–8. doi: 10.1016/j.jacc.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Lopes RD, Alings M, Connolly SJ et al. Rationale and design of the apixaban for the reduction of thrombo-embolism in patients with device-detected sub-clinical atrial fibrillation (ARTESiA) trial. Am Heart J. 2017;189:137–45. doi: 10.1016/j.ahj.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Perez MV, Mahaffey KW, Hedlin H et al. Large-scale assessment of a smartwatch to identify atrial fibrillation. N Engl J Med. 2019;381:1909–17. doi: 10.1056/NEJMoa1901183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannun AY, Rajpurkar P, Haghpanahi M et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nature Med. 2019;25:65–9. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadish AH, Buxton AE, Kennedy HL et al. ACC/AHA clinical competence statement on electrocardiography and ambulatory electrocardiography. A report of the ACC/AHA/ACP-ASIM Task Force on Clinical Competence (ACC/AHA Committee to Develop a Clinical Competence Statement on Electrocardiography and Ambulatory Electrocardiography). J Am Coll Cardiol. 2001;38:2091–100. doi: 10.1016/s0735-1097(01)01680-1. [DOI] [PubMed] [Google Scholar]
- 8.Salerno SM, Alguire PC, Waxman HS. Competency in interpretation of 12-lead electrocardiograms: a summary and appraisal of published evidence. Ann Intern Med. 2003;138:751–60. doi: 10.7326/0003-4819-138-9-200305060-00013. [DOI] [PubMed] [Google Scholar]
- 9.Hill AC, Miyake CY, Grady S, Dubin AM. Accuracy of interpretation of preparticipation screening electrocardiograms. J Pediatr. 2011;159:783–8. doi: 10.1016/j.jpeds.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Dores H, Santos JF, Dinis P et al. Variability in interpretation of the electrocardiogram in athletes: another limitation in pre-competitive screening. Rev Port Cardiol. 2017;36:443–9. doi: 10.1016/j.repc.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Schläpfer J, Wellens HJ. Computer-interpreted electrocardiograms benefits and limitations. J Am Coll Cardiol. 2017;70:1183–92. doi: 10.1016/j.jacc.2017.07.723. [DOI] [PubMed] [Google Scholar]
- 12.Viskin S, Rosovski U, Sands AJ et al. Inaccurate electrocardiographic interpretation of long QT: the majority of physicians cannot recognize a long QT when they see one. Heart Rhythm. 2005;2:569–74. doi: 10.1016/j.hrthm.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Willems JL, Abreu-Lima C, Arnaud P et al. The diagnostic performance of computer programs for the interpretation of electrocardiograms. N Engl J Med. 1991;325:1767–73. doi: 10.1056/NEJM199112193252503. [DOI] [PubMed] [Google Scholar]
- 14.Guglin ME, Thatai D. Common errors in computer electrocardiogram interpretation. Int J Cardiol. 2006;106:232–7. doi: 10.1016/j.ijcard.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Shah AP, Rubin SA. Errors in the computerized electrocardiogram interpretation of cardiac rhythm. J Electrocardiol. 2007;40:385–90. doi: 10.1016/j.jelectrocard.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Bae MH, Lee JH, Yang DH et al. Erroneous computer electrocardiogram interpretation of atrial fibrillation and its clinical consequences. Clin Cardiol. 2012;35:348–53. doi: 10.1002/clc.22000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anh D, Krishnan S, Bogun F. Accuracy of electrocardiogram interpretation by cardiologists in the setting of incorrect computer analysis. J Electrocardiol. 2006;39:343–5. doi: 10.1016/j.jelectrocard.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Aleexenko V, Jeevaratnam K. Computational approaches for detection of cardiac rhythm abnormalities: are we there yet? J Electrocardiol. 2020;59:28–34. doi: 10.1016/j.jelectrocard.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 19.van de Leur RR, Blom LJ, Gavves E et al. Automatic triage of 12-lead electrocardiograms using deep convolutional neural networks. J Am Heart Assoc. 2020;9:e015138. doi: 10.1161/JAHA.119.015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perlman O, Katz A, Amit G et al. Supraventricular tachycardia classification in the 12-lead ECG using atrial waves detection and a clinically based tree scheme. IEEE Biomed Health Inform. 2015;20:1513–20. doi: 10.1109/JBHI.2015.2478076. [DOI] [PubMed] [Google Scholar]
- 21.Attia ZI, Kapa S, Lopez-Jimenez F et al. Screening for cardiac contractile dysfunction using an artificial intelligence–enabled electrocardiogram. Nat Med. 2019;25:70–4. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 22.Sbrollini A, de Jongh MC, ter Haar CC et al. Serial electrocardiography to detect newly emerging or aggravating cardiac pathology: a deep-learning approach. Biomed Eng Online. 2019;18:15. doi: 10.1186/s12938-019-0630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu JM, Tsai M, Xiao S A deep neural network electrocardiogram analysis framework for left ventricular hypertrophy prediction. J Ambient Intell Human Comput. 2020. epub ahead of press. [DOI]
- 24.Hastie T, Tibshirani R, Friedman J. New York City: NY: Springer; 2009. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. [Google Scholar]
- 25.van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. 2014;14:137. doi: 10.1186/1471-2288-14-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins GS, Ogundimu EO, Altman DG. Sample size considerations for the external validation of a multivariable prognostic model: a resampling study. Stat Med. 2016;35:214–26. doi: 10.1002/sim.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghunath S, Cerna AEU, Jing L et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat Med. 2020;26:886–91. doi: 10.1038/s41591-020-0870-z. [DOI] [PubMed] [Google Scholar]
- 28.Kligfield P, Gettes LS, Bailey JJ et al. Recommendations for the standardization and interpretation of the electrocardiogram: part I: the electrocardiogram and its technology. J Am Coll Cardiol. 2007;49:1109–27. doi: 10.1016/j.jacc.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 29.Jain R, Singh R, Yamini S et al. Fragmented ECG as a risk marker in cardiovascular diseases. Curr Cardiol Rev. 2014;10:277–86. doi: 10.2174/1573403x10666140514103451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korkmaz A, Yildiz A, Demir M et al. The relationship between fragmented QRS and functional significance of coronary lesions. J Electrocardiol. 2017;50:282–6. doi: 10.1016/j.jelectrocard.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Das MK, Zipes DP. Fragmented QRS: A predictor of mortality and sudden cardiac death. Heart Rhythm. 2009;6(3 Suppl):S8–14. doi: 10.1016/j.hrthm.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 32.Thakor NV, Webster JG, Tompkins WJ. Estimation of QRS complex power spectra for design of a QRS filter. IEEE Trans Biomed Eng. 1984;31:702–6. doi: 10.1109/tbme.1984.325393. [DOI] [PubMed] [Google Scholar]
- 33.Thakor NV, Webster JG, Tompkins WJ. Optimal QRS detector. Med Biol Eng Comput. 1983;21:343–50. doi: 10.1007/bf02478504. [DOI] [PubMed] [Google Scholar]
- 34.García-Niebla J, Serra-Autonell G, de Luna AB. Brugada syndrome electrocardiographic pattern as a result of improper application of a high pass filter. Am J Cardiol. 2012;110:318–20. doi: 10.1016/j.amjcard.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 35.de Fauw J, Ledsam JR, Romera-Paredes B et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–50. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 36.Herman MV, Ingram DA, Levy JA et al. Variability of electrocardiographic precordial lead placement: a method to improve accuracy and reliability. Clin Cardiol. 1991;14:469–76. [PubMed] [Google Scholar]
- 37.Hill NE, Goodman JS. Importance of accurate placement of precordial leads in the 12-lead electrocardiogram. Heart Lung. 1987;16:561. doi: 10.1007/s11517-013-1115-9. [DOI] [PubMed] [Google Scholar]
- 38.Chanarin N, Caplin J, Peacock A. “Pseudo reinfarction”: a consequence of electrocardiogram lead transposition following myocardial infarction. Clin Cardiol. 1990;13:668–9. doi: 10.1002/clc.4960130916. [DOI] [PubMed] [Google Scholar]
- 39.Peberdy MA, Ornato JP. Recognition of electrocardiographic lead misplacements. Am J Emerg Med. 1993;11:403–5. doi: 10.1016/0735-6757(93)90177-d. [DOI] [PubMed] [Google Scholar]
- 40.Rautaharju PM, Prineas RJ, Crow RS et al. The effect of modified limb electrode positions on electrocardiographic wave amplitudes. J Electrocardiol. 1980;13:109–13. doi: 10.1016/s0022-0736(80)80040-9. [DOI] [PubMed] [Google Scholar]
- 41.Rajaganeshan R, Ludlam CL, Francis DP et al. Accuracy in ECG lead placement among technicians, nurses, general physicians and cardiologists. Int J Clin Pract. 2007;62:65–70. doi: 10.1111/j.1742-1241.2007.01390..x. [DOI] [PubMed] [Google Scholar]
- 42.van Oosterom A, Hoekema R, Uijen GJH. Geometrical factors affecting the interindividual variability of the ECG and the VCG. J Electrocardiol. 2000;33:219–28. doi: 10.1054/jelc.2000.20356. [DOI] [PubMed] [Google Scholar]
- 43.Hoekema R, Uijen GJH, van Erning L et al. Interindividual variability of multilead electrocardiographic recordings: influence of heart position. J Electrocardiol. 1999;32:137–48. doi: 10.1016/S1053-0770(99)90050-2. [DOI] [PubMed] [Google Scholar]
- 44.Mincholé A, Zacur E, Ariga R et al. MRI-based computational torso/biventricular multiscale models to investigate the impact of anatomical variability on the ECG QRS complex. Front Physiol. 2019;10:1103. doi: 10.3389/fphys.2019.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyên UC, Potse M, Regoli F et al. An in-silico analysis of the effect of heart position and orientation on the ECG morphology and vectorcardiogram parameters in patients with heart failure and intraventricular conduction defects. J Electrocardiol. 2015;48:617–25. doi: 10.1016/j.jelectrocard.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Hoekema R, Uijen G, van Oosterom A. The number of independent signals in body surface maps. Methods Inf Med. 1999;38:119–24. doi: 10.1055/s-0038-1634176. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu W, Matsuo K, Takagi M et al. Body surface distribution and response to drugs of ST segment elevation in Brugada syndrome: clinical implication of eighty-seven-lead body surface potential mapping and its application to twelve-lead electrocardiograms. J Cardiovasc Electrophysiol. 2000;11:396–404. doi: 10.1111/j.1540-8167.2000.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 48.Priori SG, Blomström-Lundqvist C, Mazzanti A et al. 2015 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur Heart J. 2015;36:2793–867. doi: 10.1093/eurheartj/ehv316. [DOI] [PubMed] [Google Scholar]
- 49.Ibánez B, James S, Agewall S et al. 2017 ESC guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation. Rev Esp Cardiol (Engl Ed) 2017;70:1082. doi: 10.1016/j.rec.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Galloway CD, Valys A v, Shreibati JB et al. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol. 2019;4:428–36. doi: 10.1001/jamacardio.2019.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rjoob K, Bond R, Finlay D et al. Data driven feature selection and machine learning to detect misplaced V1 and V2 chest electrodes when recording the 12-lead electrocardiogram. J Electrocardiol. 2019;57:39–43. doi: 10.1016/j.jelectrocard.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 52.Hong S, Zhou Y, Shang J et al. Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review. Comput Biol Med. 2020;112:103801. doi: 10.1016/j.compbiomed.2020.103801. [DOI] [PubMed] [Google Scholar]
- 53.Helbing D. Societal, economic, ethical and legal challenges of the digital revolution: from big data to deep learning, artificial intelligence, and manipulative technologies. SSRN. 2015. [DOI]
- 54.Balthazar P, Harri P, Prater A et al. Protecting your patients’ interests in the era of big data, artificial intelligence, and predictive analytics. J Am Coll Radiol. 2018;15:580–6. doi: 10.1016/j.jacr.2017.11.035. [DOI] [PubMed] [Google Scholar]
- 55.Ribeiro AH, Ribeiro MH, Paixão GMM et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat Comm. 2020;11:1760. doi: 10.1038/s41467-020-15432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Attia ZI, Noseworthy PA, Lopez-Jimenez F et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394:861–7. doi: 10.1016/S0140-6736(19)31721-0. [DOI] [PubMed] [Google Scholar]
- 57.Ko W-Y, Siontis KC, Attia ZI et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol. 2020;75:722–33. doi: 10.1016/j.jacc.2019.12.030. [DOI] [PubMed] [Google Scholar]
- 58.Attia ZI, Sugrue A, Asirvatham SJ et al. Noninvasive assessment of dofetilide plasma concentration using a deep learning (neural network) analysis of the surface electrocardiogram: a proof of concept study. PLoS ONE. 2018;13:e0201059. doi: 10.1371/journal.pone.0201059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon JM, Lee SY, Jeon KH et al. Deep learning-based algorithm for detecting aortic stenosis using electrocardiography. J Am Heart Assoc. 2020;9:e014717. doi: 10.1161/JAHA.119.014717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katritsis G, Luther V, Kanagaratnam P et al. Arrhythmia mechanisms revealed by ripple mapping. Arrhythm Electrophysiol Rev. 2018;7:261–4. doi: 10.15420/aer.2018.44.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Broek HT, Wenker S, van de Leur R et al. 3D myocardial scar prediction model derived from multimodality analysis of electromechanical mapping and magnetic resonance imaging. J Cardiovasc Transl Res. 2019;12:517–27. doi: 10.1007/s12265-019-09899-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Es R, van den Broek HT, van der Naald M et al. Validation of a novel stand-alone software tool for image guided cardiac catheter therapy. Int J Cardiovasc Imaging. 2019;35:225–35. doi: 10.1007/s10554-019-01541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zollei L, Grimson E, Norbash A . Kauai, HI. US: Institute of Electrical and Electronics Engineers: 2001. 2D-3D rigid registration of X-ray fluoroscopy and CT images using mutual information and sparsely sampled histogram estimators. In: Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. CVPR 2001 IEEE. pp. II–II. [DOI] [Google Scholar]
- 64.Walsh KA, Galvin J, Keaney J et al. First experience with zero-fluoroscopic ablation for supraventricular tachycardias using a novel impedance and magnetic-field-based mapping system. Clin Res Cardiol. 2018;107:578–85. doi: 10.1007/s00392-018-1220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantwell CD, Mohamied Y, Tzortzis KN et al. Rethinking multiscale cardiac electrophysiology with machine learning and predictive modelling. Comput Biol Med. 2019;104:339–51. doi: 10.1016/j.compbiomed.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bansal A, Joshi R. Portable out-of-hospital electrocardiography: a review of current technologies. J Arrhythm. 2018;34:129–38. doi: 10.1002/joa3.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mairesse GH, Moran P, van Gelder IC et al. Screening for atrial fibrillation: a European Heart Rhythm Association (EHRA) consensus document endorsed by the Heart Rhythm Society (HRS), Asia Pacific Heart Rhythm Society (APHRS), and Sociedad Latinoamericana de Estimulación Cardíaca y Electrofisiología (SOLAECE) Europace. 2017;19:1589–623. doi: 10.1093/europace/eux177. [DOI] [PubMed] [Google Scholar]
- 68.Freedman B, Camm J, Calkins H et al. Screening for atrial fibrillation: a report of the AF-SCREEN international collaboration. Circulation. 2017;135:1851–67. doi: 10.1161/CIRCULATIONAHA.116.026693. [DOI] [PubMed] [Google Scholar]
- 69.Wegner FK, Kochhäuser S, Ellermann C et al. Prospective blinded Evaluation of the smartphone-based AliveCor Kardia ECG monitor for Atrial Fibrillation detection: the PEAK-AF study. Eur J Intern Med. 2020;73:72–5. doi: 10.1016/j.ejim.2019.11.018. [DOI] [PubMed] [Google Scholar]
- 70.Galloway C, Treiman D, Shreibati J et al. A deep neural network predicts atrial fibrillation from normal ECGs recorded on a smartphone-enabled device. Eur Heart J. 2019;40(Suppl 1):5105. doi: 10.1093/eurheartj/ehz746.0041. [DOI] [Google Scholar]
- 71.Brasier N, Raichle CJ, Dörr M et al. Detection of atrial fibrillation with a smartphone camera: first prospective, international, two-centre, clinical validation study (DETECT AF PRO) Europace. 2019;21:41–7. doi: 10.1093/europace/euy176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McManus DD, Chong JW, Soni A et al. PULSE-SMART: pulse-based arrhythmia discrimination using a novel smartphone application. J Cardiovasc Electrophysiol. 2016;27:51–7. doi: 10.1111/jce.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Couderc JP, Kyal S, Mestha LK et al. Detection of atrial fibrillation using contactless facial video monitoring. Heart Rhythm. 2015;12:195–201. doi: 10.1016/j.hrthm.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 74.Yan BP, Lai WHS, Chan CKY et al. Contact-free screening of atrial fibrillation by a smartphone using facial pulsatile photoplethysmographic signals. J Am Heart Assoc. 2018;7:e008585. doi: 10.1161/JAHA.118.008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caliebe A, Leverkus F, Antes G et al. Does big data require a methodological change in medical research? BMC Med Res Methodol. 2019;19:125. doi: 10.1186/s12874-019-0774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hashimoto DA, Rosman G, Rus D et al. Artificial intelligence in surgery. Ann Surg. 2018;268:70–6. doi: 10.1097/sla.0000000000002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, Peng Y, Lu L ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. pp. 2097–106. [DOI]
- 78.Lu MT, Ivanov A, Mayrhofer T et al. Deep learning to assess long-term mortality from chest radiographs. JAMA Netw Open. 2019;2:e197416. doi: 10.1001/jamanetworkopen.2019.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baltruschat IM, Nickisch H, Grass M et al. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci Rep. 2019;9:6381. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oakden-Rayner L. Exploring the ChestXray14 dataset: problems. Wordpress: Luke Oakden Rayner. 2017. https://lukeoakdenrayner.wordpress.com/2017/12/18/the-chestxray14-dataset-problems/ (accessed 11 May 2020)
- 81.Moeyersons J, Smets E, Morales J et al. Artefact detection and quality assessment of ambulatory ECG signals. Comput Methods Programs Biomed. 2019;182:105050. doi: 10.1016/j.cmpb.2019.105050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clifford GD, Behar J, Li Q, Rezek I. Signal quality indices and data fusion for determining clinical acceptability of electrocardiograms. Physiol Meas. 2012;33:1419. doi: 10.1088/0967-3334/33/9/1419. [DOI] [PubMed] [Google Scholar]
- 83.Xia H, Garcia GA, McBride JC . 2011 Computing in Cardiology. Hangzhou, China: Institute of Electrical and Electronics Engineers.; 2011. Computer algorithms for evaluating the quality of ECGs in real time. In: IEEE; pp. 369–72.https://ieeexplore.ieee.org/document/6164579 [Google Scholar]
- 84.Li Q, Rajagopalan C, Clifford GD. A machine learning approach to multi-level ECG signal quality classification. Comput Methods Programs Biomed. 2014;117:435–47. doi: 10.1016/j.cmpb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 85.Redmond SJ, Xie Y, Chang D et al. Electrocardiogram signal quality measures for unsupervised telehealth environments. Physiol Meas. 2012;33:1517. doi: 10.1088/0967-3334/33/9/1517. [DOI] [PubMed] [Google Scholar]
- 86.Moons KGM, Kengne AP, Grobbee DE et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart. 2012;98:691–8. doi: 10.1136/heartjnl-2011-301247. [DOI] [PubMed] [Google Scholar]
- 87.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–9. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 88.Attia ZI, Kapa S, Yao X et al. Prospective validation of a deep learning electrocardiogram algorithm for the detection of left ventricular systolic dysfunction. J Cardiovasc Electrophysiol. 2019;30:668–74. doi: 10.1111/jce.13889. [DOI] [PubMed] [Google Scholar]
- 89.Noseworthy PA, Attia ZI, Brewer LC et al. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circ Arrhythm Electrophysiol. 2020;13:e007988. doi: 10.1161/CIRCEP.119.007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao X, McCoy RG, Friedman PA et al. ECG AI-Guided Screening for Low Ejection Fraction (EAGLE): Rationale and design of a pragmatic cluster randomized trial. Am Heart J. 2020;219:31–6. doi: 10.1016/j.ahj.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 91.Sendak MP, Gao M, Brajer N, Balu S. Presenting machine learning model information to clinical end users with model facts labels. NPJ Digit Med. 2020;3:41. doi: 10.1038/s41746-020-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Macfarlane PW, Katibi IA, Hamde ST et al. Racial differences in the ECG – selected aspects. J Electrocardiol. 2014;47:809–14. doi: 10.1016/j.jelectrocard.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 93.Rijnbeek PR, van Herpen G, Bots ML et al. Normal values of the electrocardiogram for ages 16–90 years. J Electrocardiol. 2014;47:914–21. doi: 10.1016/j.jelectrocard.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 94.Attia ZI, Friedman PA, Noseworthy PA et al. Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ Arrhythm Electrophysiol. 2019;12:e007284. doi: 10.1161/CIRCEP.119.007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carvalho D v, Pereira EM, Cardoso JS. Machine learning interpretability: a survey on methods and metrics. Electronics. 2019;8:832. doi: 10.3390/electronics8080832. [DOI] [Google Scholar]
- 96.Selvaraju RR, Cogswell M, Das A . Proceedings of the IEEE international conference on computer vision 2017. Venice, Italy: 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization. In: pp. 618–26. [Google Scholar]
- 97.Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M. International Conference on Learning Representations Workshop Track Proceedings.; San Diego, CA, US,: 7–9 May 2015. Striving for simplicity: the all convolutional net. pp. 1–14. [Google Scholar]
- 98.Strodthoff N, Strodthoff C. Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol Meas. 2019;40:015001. doi: 10.1088/1361-6579/aaf34d. [DOI] [PubMed] [Google Scholar]
- 99.Opening the black box of machine learning. Lancet Respir Med. 2018;6:801. doi: 10.1016/S2213-2600(18)30425-9. [DOI] [PubMed] [Google Scholar]
- 100.Sturmfels P, Lundberg S, Lee S-I. Visualizing the Impact of Feature Attribution Baselines. Distill. 2020;5:e22. doi: 10.23915/distill.00022. [DOI] [Google Scholar]
- 101.Filos A, Farquhar S, Gomez AN A Systematic Comparison of Bayesian Deep Learning Robustness in Diabetic Retinopathy Tasks. ArXiv. https://arxiv.org/abs/1912.10481 Available at: Published: December 2019. Accessed: June 2, 2020.
- 102.Tagasovska N, Lopez-Paz D. San Diego, CA, US: Neural Information Processing Systems,; 2019. Single-model uncertainties for deep learning. In: Wallach H. Larochelle H, Beygelzimer A, Advances in Neural Information Processing Systems 32. pp. 6414–25. [Google Scholar]
- 103.Leibig C, Allken V, Ayhan MS et al. Leveraging uncertainty information from deep neural networks for disease detection. Sci Rep. 2017;7:17816. doi: 10.1038/s41598-017-17876-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gal Y, Islam R, Ghrahramani Z. Deep Bayesian Active Learning with Image Data. Proceedings of the 34th International Conference on Machine Learning. 2017;70:1183–92. [Google Scholar]
- 105.Carter RE, Attia ZI, Lopez-Jimenez F et al. Pragmatic considerations for fostering reproducible research in artificial intelligence. NPJ Digi Med. 2019;2:42. doi: 10.1038/s41746-019-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rocher L, Hendrickx JM, de Montjoye YA. Estimating the success of re-identifications in incomplete datasets using generative models. Nat Commun. 2019;10:3069. doi: 10.1038/s41467-019-10933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mandel JC, Kreda DA, Mandl KD et al. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc. 2016;23:899–908. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vayena E, Blasimme A. Health research with big data: time for systemic oversight. J Law Med Ethics. 2018;46:119–29. doi: 10.1177/1073110518766026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vayena E, Blasimme A. Biomedical big data: new models of control over access, use and governance. J Bioeth Inq. 2017;14:501–13. doi: 10.1007/s11673-017-9809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCall B. What does the GDPR mean for the medical community? Lancet. 2018;391:1249–50. doi: 10.1016/s0140-6736(18)30739-6. [DOI] [PubMed] [Google Scholar]


