Abstract
Fanconi anemia (FA) is a rare multigenic chromosomal instability syndrome that predisposes patients to life-threatening bone marrow failure, congenital malformations, and cancer. Functional loss of interstrand cross-link (ICL) DNA repair system is held responsible, though the mechanism is not yet fully understood. The clinical and molecular findings of 20 distinct FA cases, ages ranging from perinatal stage to 32 years, are presented here. Pathogenic variants in FANCA were found responsible in 75%, FANCC, FANCE, FANCJ/BRIP1, FANCL in 5%, and FANCD1/BRCA2 and FANCN/PALB2 in 2.5% of the subjects. Altogether, 25 different variants in 7 different FA genes, including 10 novel mutations in FANCA, FANCN/PALB2, FANCE, and FANCJ/BRIP1, were disclosed. Two compound heterozygous germline cases were mosaic for one allele, revealing that the incidence of reverse mutations may not be uncommon in FA. Another case with de novo FANCD1/BRCA2 and paternally inherited FANCN/PALB2 pathogenic alleles at first glance suggested a digenic inheritance, because the presence of a second pathogenic variant in the unexamined regions of FANCD1/BRCA2 and FANCN/PALB2 were exluded by sequencing and deletion/duplication analysis. A better understanding of the complexity of the FA genotype may provide further access to undiscovered ICL components and apparently dispensable cellular pathways where FA proteins may play important roles.
Keywords: Cancer, Digenic, Fanconi anemia, Reverse mutation, Somatic mosaicism
Introduction
Fanconi anemia (FA, MIM# 227650) is a genetically heterogenous rare syndrome with variable clinical features encompassing hematologic findings of pancytopenia as a result of bone marrow failure (BMF) and diverse congenital malformations pronounced with skeletal defects of limbs with radial aplasia, hands with thumb deformities, small stature, low birth weight, microcephaly, deafness, congenital heart defects, kidney malformations, pigment changes of the skin, and predisposition to cancer [Altay et al., 1997]. While the worldwide incidence of FA is estimated to be 1:160,000–360,000 with 0.3% carrier frequency, it also is recognized to occur much more frequently among communities with consanguineous marriages [Wu, 2013].
The earliest clinical description of FA was reported in 1926 by Swiss pediatrician Guido Fanconi when he depicted birth defects and conditions resembling pernicious anemia that had afflicted 3 deceased siblings. The disease was named after Fanconi in 2006 to honor his contribution to cancer research [Lobitz and Velleuer, 2006].
Progressive BMF usually presents by the age of 8–10 years in 80% of affected individuals, with 23% of the patients encountering various forms of cancer later [Kutler et al., 2003]. Affected patients have a susceptibility to leukemia-myelodysplastic syndrome, more aggressively acute myeloid leukemia, or nonhematological solid neoplasm of squamous cell cancers in areas of the body where cells normally reproduce rapidly: head, neck, and anogenital regions [Wegman-Ostrosky and Savage, 2017]. Though regularly scheduled examinations for signs of cancer are recommended throughout a patient's lifetime, allogeneic hematopoietic stem cell transplantation has shown to be the treatment of BMF with improving protocols since 1972 [Peffault de Latour et al, 2013]. Dysregulation of DNA damage underlies the molecular pathology of FA, yielding causative chemicals to chromosomal breakage and providing a unique marker for the diagnosis before the beginning of hematologic manifestations [Esmer et al., 2004]. Diepoxybutane (DEB) or mitomycin C may be used as clastogenic agents, principally dependening upon laboratory experiences [Altay et al., 1997; Fargo et al., 2014]. Unfortunately, a small number of affected individuals are unable to benefit from this tool of chromosome breakage analysis [Auerbach, 2009]. Studies on the correlation between the clastogen hypersensitivity and the severity of the FA phenotype may suggest that genes not involved in FA may modify the cross-linking activity [Davies et al., 2005]. Although a positive cytogenetic test revealing chromatid-type aberrations is highly indicative for FA, molecular analysis is required for definitive diagnosis, assessment of prognosis, and genetic counseling. Prior to the next-generation sequencing (NGS) era, Sanger sequencing was time-consuming and burdensome. As a consequence, genetic subtypes for the limited groups were commonly predicted before mutation analysis by retrovirus-mediated complementation assays [Balta et al., 2000; Chandra et al., 2005; Navarro et al., 2006]. While advancements in sequencing technologies are today leading the genetic diagnosis of FA, limitations and gaps of the coverage, variant analysis, especially for mosaics and gross alterations, require special attention.
Presently, 20 autosomal recessive (FANCA, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG, FANCI, FANJ/BRIP1, FANCL, FANCM, FANCN/PALB2, FANCO/RAD51C, FANCP/SLX4, FANCQ/ERCC4/XPF, FANCS/BRCA1, FANCT/UBE2T, FANCU/XRCC2, FANCV/REV7, FANCW/RFWD3), 1 dominant (FANCR/RAD51), and 1 X-linked (FANCB) forms of FA are described [Ameziane et al., 2015; Knies et al., 2017; Dubois et al., 2019].
DNA cross-linking defects are new covalent bonds formed either on the same DNA strand (intrastrand) that usually occurs upon exposure to ultraviolet light or between the bases of opposite strands (interstrands) that involves the bonding of N7 nitrogen of guanines upon exposure to bifunctional alkylating anticancer agents or endogenous products of lipid peroxidation [Noll et al., 2006]. Interstrand cross-linking defects (ICLs) may develop during a DNA replication phase (GS), recognized by FA core proteins and FA-associated (FAA) proteins, and are linked to the mechanism for replication-dependent repair together with homologous recombination of cells [Deans and West, 2011; Wenzel and Singh, 2018]. In non-dividing cells, however, ICLs are repaired by transcription-coupled nucleotide excision repair [Hlavin et al., 2010]. Any lesions within the DNA structure cause stalling of the replication mechanism that enables the repair systems to function and avoid a deleterious fork collapse [Ceccaldi et al., 2016; Palovcak et al., 2017]. FA core proteins work harmoniously with 3 other DNA repair systems: nucleotide-excision repair, translesion synthesis, and homologous recombination (HR) [Xia et al., 2006; Higgs et al., 2015; Niraj et al., 2019]. Unrepaired ICLs forestall strand separation leading to breakage and viral DNA integration, which are abnormal chromosomal rearrangements that eventually may tend to convey cells into a cancerous state [Howlett et al., 2005; Deans and West, 2011]. Since FA core proteins have many interactions with mediators of cell cycle checkpoint response or DNA damage response proteins, such as ataxia telangiectasia mutated (ATM), Nijmegen breakage syndrome 1 (NBS1), DNA-repair response protein FANCR/RAD51, and in multiple breast cancer-proteins such as FANCS/BRCA1, FANCD1/BRCA2, continuity of its comprehensive trail is expressed as the FA/BRCA pathway. Alongside of acting on ICLs repair, the FA pathway also has other functionalities in the replicating cell: maintaining replication fork stability, control of chromosome segregation during mitosis, and providing genetic integrity following transcription and/or RNA-DNA hybrid formations [Zeman and Cimprich, 2014; Schwab et al., 2015]. Recent studies suggest that the FA pathway may play a role in redirecting cells from using error-prone canonical, non-homologous end joining (NHEJ) to alternative-NHEJ (alt-NHEJ) that is in the resection-dependent pathway of HR [Ceccaldi et al., 2016]. Investigations into the mechanisms of autophagy have suggested the important role of some FA genes in virophagy and Parkin-mediated autophagy of damaged mitochondria [Shyamsunder et al., 2016; Sumpter and Levine, 2017].
Here, we present clinical and molecular findings of 20 FA cases including 2 perinatal subjects.
Material and Methods
Patients
Twenty subjects were admitted for clinical genetic investigation and molecular diagnosis of FA to the Pediatric Genetic Department of Istanbul University Cerrahpasa and Medical Genetic Department of Istanbul Faculty of Medicine of Istanbul University. The patients were evaluated clinically by an experienced pediatric/medical geneticist. A review of their medical history revealed that 16 cases were reported as DEB-positive, 1 was shown negative and 1 was unknown. The remaining 2 cases were terminated fetuses due to multiple congenital anomalies assigned with a differential diagnosis of FA. The proband of one family deceased before the initiation of the molecular investigation. Peripheral blood samples from 13 subjects (Cases 2–6, 10, 12, 14, 16–20) plus the parents of one subject (Case 1), as well as skin biopsy materials from 4 bone marrow transplanted cases (Cases 7, 8, 9, 15) and from 2 fetuses (Cases 11, 13), were obtained for DNA isolation and genetic testing upon receipt of the signed consent forms from the families.
Methods
DNA isolations were performed using commercial kits (Mammalian Blood and Cells and Tissue DNA Isolation Kit, Roche). Two fetuses presented with major anomalies and were investigated genetically by chorionic villus sampling. Classical karyotyping and microarray analysis were conducted using Agilent SurePrint G3 CGH + SNP Microarray Kit (4 × 180K, Agilent Technologies, Inc., Santa Clara, CA, USA). Data analysis was performed using Agilent Genomics Workbench 5.0.2.5. Genomic linear positions were given relative to NCBI build 37 (hg19).
A targeted in-house panel-gene test was designed for radial ray defects, including 42 genes that also covered all of the coding exons and exon-intron boundaries of 17 FA-associated genes (FANCA, NM_000135.2; FANCB/FAAP95, NM_001018113.1; FANCC, NM_000136.2; FANCD1/BRCA2, NM_000059.3; FANCD2, NM_033084.3; FANCE, NM_021922.2; FANCF, NM_022725.3; FANCG/XRCC9, NM_004629.1; FANCI, NM_001113378.1; FANCJ/BRIP1, NM_032043.2, FANCL/PHF9, NM_018062.3; FANCM, NM_020937.2; FANCN/PALB2, NM_024675.3; FANCO/RAD51C, 17q22-q23, NM_058216.2; FANCP/SLX4, NM_032444.2; FANCQ/ERCC4, NM_005236.2; FANCU/XRCC2, NM_005431.1) in 785 amplicons with 99.43% coverage. Genetic analysis was performed on an Ion Torrent PGM platform by NGS. Different in silico analysis programs (MutationTaster, PolyPhen, SIFT, Human Splicer Finding, VarSEAK, and VarSome) were used for prediction of a disease-causing status of variants [Desmet et al., 2009; Kumar et al., 2009; Adzhubei et al., 2013; Schwarz et al., 2014; Kopanos et al., 2019; https://varseak.bio/]. In addition, gene level threshold values were obtained through CADD (combined annotation dependent depletion)-based MSC (mutation significance cutoff) scores providing impact predictions (http://lab.rockefeller.edu/casanova/MSC) [Itan et al., 2016]. Variant frequency was obtained from the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/about) [Karczewski et al., 2019]. Search into the phenotypic impact was obtained from ClinVar [Landrum et al., 2016] and from a literature quest. The American College of Medical Genetics and Genomics (ACMG) guidelines were used for variant classification [Richards et al., 2015] as implemented in VarSome. Uncovered and low covered regions of panel genes and all of the detected pathogenic and/or likely pathogenic variant regions were Sanger sequenced. Non-carrier and/or likely pathogenic variant carriers, or cases with novel predicted associated variant carriers and pathogenic/likely pathogenic variant carriers that do not fit to the inheritance model, were further tested by MLPA to search for deletions/duplications in FANCA, FANCD2, FANCN/PALB2, RAD50 (few exons), RAD51C, and RAD51 (MRC-Holland, MLPA P031, P032 and P260) and analyzed on Coffalyser [Coffa and Van Den Berg, 2011].
Results
Clinical Findings
Two terminated fetuses and 18 postnatal cases were evaluated. The median age of the postnatal cases were 4 years, 9 months (range: 0.8–32 years), and the male to female ratio was 1:2. Parental consanguinity was present in 65% of the cases. Four patients had similarly affected siblings, one of those patient's parents were not consanguineous. Both of the fetuses, that had been terminated (Case 11 at 22 and Case 13 at 32 gestational weeks) upon pathological ultrasound findings, revealed intrauterine growth retardation (IUGR), microcephaly, radial anomaly, bilateral ventriculomegaly, cerebellar hypoplasia, and anal atresia. Case 11 additionally presented a cleft palate at postmortem examination. Cytogenetic and microarray analysis performed in both fetuses excluded chromosomal aberrations.
The most common postnatal findings were hematologic manifestations (88%), followed by skin disorders (83%), extremity anomaly (77%), microcephaly (77%), and short stature (72%) (online suppl. Table 1; for online suppl. material, see www.karger.com/doi/10.1159/000509838). Four patients (Case 7, 8, 9, 20) had pancytopenia and/or thrombocytopenia without skeletal findings, and 2 patients (Case 16 and 17) had thumb anomalies without hematologic manifestations. One patient had been diagnosed with cancer at the age of 32 years (Case 19).
Molecular Study
All targeted bases were sequenced revealing 639 average coverage with 99.43% targeted bases covered at 20X. Pathogenic/likely pathogenic small and gross variants are presented in Table 1
Table 1.
Genotype of small and gross variants in each of our cases are presented
| Gene, transcript, peptide | Patient | Mutation |
|---|---|---|
| FANCA, NM_000135, NP_000126 | 1 | c.[(461_570)_(3937_3155)del];[(461_570)_(3937_3155)del], p.[(?)];[(?)] (Exons 6–31 homozygous deletion)a |
| 2 | c.[(461_570)_(3937_3155)del];[(461_570)_(3937_3155)del], p.[(?)];[(?)] (Exon 6–31 homozygous deletion)a | |
| 3 | c.[(?_-15)_(250_351)del];[(?_-15)_(250_351)del], p.[(?)];[(?)] (Exon 1-3 and 6–31 compound heterozygous deletion)a | |
| 4 | c.[3163C=/3163C>T];[c.3491C>T], p.[R1055 R/W];[P1164L] | |
| 5 | c.[2557C=/2557C>T];[3754G>T], p.[R853R/*];[E1252*] | |
| 6 | c.[776C>G];[3935−7G>A], p.[P259R];[(?)] | |
| 7 | c.[2938G>C];[2938G>C], p.[A980P];[A980P] | |
| 8 | c.[3026G>A];[3026G>A], p.[G1009D];[G1009D] | |
| 9 | c.[2638C>T];[2638C>T], p.[R880*];[R880*] | |
| 10 | c.[894–2A>G];[894–2A>G], p.[(?)];[(?)] | |
| 12 | c.[4261–2A>C];[4261–2A>C], p.[(?)];[(?)] | |
| 14 | c.[894–2A>G];[894–2A>G], p.[(?)];[(?)] | |
| 18 | c.[240_241delTG];[240_241delTG], p.[C80*]:[C80*] | |
| 19 | c.[2504+5G>T;2504+3G>A];[c.2504+5G>T], p.[(?)];[(?)] | |
| 20 | c.[128T>G];[2638C>T], p.[L43*];[R880*] | |
| FANCC, NM_000136,NP_000127 | 15 | c.[456+4A>T];[456+4A>T], p.[(?)];[(?)] |
| FANCD1/BRCA2, NM_000059, NP_000050 | 17 | c.[9739C>T];[9739C=], p.[Q3247*];[Q3247=] |
| FANCE, NM_021922, NP_068741 | 13 | c.[355C>T];[1509+lG>A], p.[Q119*];[(?)] |
| FANCJ/BRIP1, NM_032043, NP_114432 | 16 | c.[205+5G>T];[761_764delAGCA], p.[?];[K254Rfs*19] |
| FANCL, NM_018062, NP_060532 | 11 | c.[739_740dupAT];[739_740dupAT], p.[M247 fs*15];[M247 fs*15] |
| FANCN/PALB2, NM_024675, NP_078951 | 17 | c.[(347_2009)_(2542_2748+3)del];[(347=)_(2542=)], p.[=]:[?] (Exon 5–6 heterozygous deletion)a |
Novel variants are written in bold. References to previously published variants are provided in online suppl. Table 1.
Gross deletions are annotated according to MLPA probe-based sequence variant nomenclature suggested for positions where break points have not been sequenced [den Dunnen et al., 2016],
Fifteen patients (Cases 2–10, 12,14, 18–20) plus one of the parental samples (Case 1) carried 17 different variants in FANCA, of which 10 have previously been reported with 7 identified as novel: 2 missense: c.776C>G (p.P259R, SCV001296377) in exon 8 in Case 6 (Fig. 1c) and c.2938 G>C (p.A980P, SCV001296378) in exon 30 in Case 7 (Fig. 1d); 1 nonsense: c.128T>G (p.L43*, SCV001296382) in exon 2 in Case 20 (Fig. 1j); 1 frame-shift: c.240_241delTG (p.C80*, SCV001296379) in exon 3 in Case 18 (Fig. 1h); 3 splice site: c.3935–7G>A (p.(?), SCV001296376) in intron 39 in Case 6 (Fig. 1c), c.2504+3G>A (SCV001296380) and c.2504+5G>T (SCV001296381) intron 26 in Case 19 (Fig. 1i).
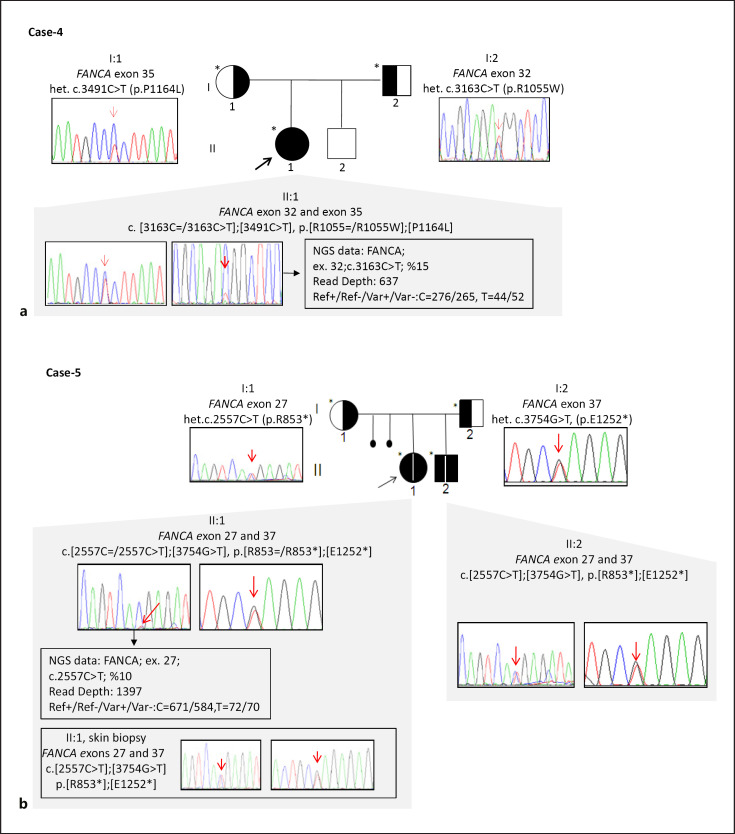
Fig. 1.
Pedigrees, Sanger sequence electropherograms, and segregation results of families with reverse mutation status (a, b) and novel pathogenic variants (c−j).
Two different mosaic pathogenic variants (c.3163C=/3163C>T, c.2557C=/2557C>T), identified in Cases 4 and 5 in compound heterozygous form in FANCA, revealed to be germline in family-based segregation (Fig. 1a, b). These mosaic alleles were delineated from IGV software with 15 and 10%, each equally represented in forward and reverse parallel sequenced amplicons. Sanger sequencing of parental alleles revealed that these variants were present in a non-mosaic heterozygous status. DNA from a skin biopsy in Case 5 also revealed a non-mosaic status.
The parents of Case 6 were first degree cousins carrying compound heterozygous novel variants in FANCA (Fig. 1c).
Five patients (Case 11, 13, 15, 16, and 17) were identified to have variants each in FANCL, FANCE, FANCC, FANCJ/BRIP1, FANCD1/BRCA2, and FANCN. Variants in FANCE (c.1509+1 G>A (p.(?), SCV001296383) in intron 9 in Case 13 (Fig. 1e), in FANCJ/BRIP1 (c.761_764delAGCA, SCV001296384) in exon 7 in Case 16 (Fig. 1f), and in FANCN (Exon 5–6 deletion, p.(?)) in case 17 (Fig. 1g) were novel. The parents of Case 1 carried a heterozygous exon 6–31 deletion in FANCA, indirectly elucidating the genetic cause of FA in the deceased proband.
An analysis of the parents of Case 17 showed that the nonsense variant (c.9739C>T, p.Q3247*) in exon 27 of FANCD1/BRCA2 was de novo, however, gonadal mosaicism could not be excluded, and a novel heterozygous gross deletion of exon 5–6 in FANCN/PALB2 was inherited from the father (Fig. 1g). Chromosomal abnormality was ruled out by karyotyping and microarray (Affymetrix, Cytoscan Optima 315K), deletions of FANCD1/BRCA2 were excluded by MLPA (SALSA MLPA Probemix P090-B1), while any other sequence variants other than FANCD1 were also excluded by whole exome sequencing (WES) performed at an external diagnostic laboratory (Centogene, Germany).
Discussion
Spontaneous errors in DNA strand, induced by exogenous exposures (anticancer agents etc.), or more commonly by endogenously originating cellular biochemical reactions (aldehydes, etc.), are corrected in majority by DNA repair systems [De Bont and Van Larebeke, 2004]. Presently, 7 different cellular repair mechanisms have been identified and described according to the type of the damage: mismatch repair for base mismatches and small insertion deletions; base-excision repair for single-base modifications or abasic formations; nucleotide-excision repair for helix-distortion lesions; variant of base excision repair mechanism for single-strand DNA breaks; HR-mediated DNA repair and NHEJ for double-strand breaks; and HR with FA/BRCA DNA repair pathway for ICLs [Mota et al., 2019]. Failure in any of those functional mechanisms, controlled by more than 50 proteins presently known, leads to genetic instability and commits an individual to a life-long risk for various somatic diseases, notably cancer, along with germline pathogenic occurrences serving as causatives for rare diseases associated with DNA repair deficiencies [Knoch et al., 2012].
Our study group of FA patients carried unique forms of variants in 7 described FA/BRCA pathway proteins: 4 encoding FA-core complex structures (FANCA, FANCC, FANCE, and FANCL) and 3 encoding downstream FA/HR proteins (FANCD1/BRCA2, FANCJ/BRIP1, and FANCN/PALB2) [Datta and Brosh, 2019]. The age of our patients at the time of their diagnosis ranged significantly from perinatal stage to 32 years, with a median age of 4 years, 9 months. This was an earlier milestone than the 2,002 cases with a median age of 6.5 years surveyed and reported by Shimamura and Alter [2010].
Individuals afflicted with FA carry an increased risk for malignancies, with an estimated 25% of patients being likely to develop a solid tumor by the age of 45 [Eiler, 2008]. A cancer risk assessment of 181 patients in the FA Registry of Germany (GEFA) reported an increase of 1.4% for acute myeloid leukemia by the age of 20; 0.7% for solid tumors by the age of 20; increasing to 5.3% by the age of 40 and to more than 10% by the age of 49, including the cumulative incidence of 50% for BMF and 28% for solid tumors similarly by the age of 49 [Rosenberg et al., 2008]. Only one patient from our cohort was diagnosed with a malignancy, which was located in her head and neck and involved her laryngeal and nasopharyngeal regions. She was the eldest patient of our series, with the next oldest subject being 13 years of age. None of the other patients in our study group has reported malignancies to-date.
Patients carrying any of 3 findings of vertebral, anal, cardiac, trachea-esophageal fistula, esophageal atresia, renal, upper limb and hydrocephalus are classified under VACTERL-H (Vertebral, Anal, Cardiac, Trachea-esophageal fistula, Esophageal atresia, Renal, upper Limb and Hydrocephalus) association [Solomon et al., 2012]. Skin pigmentation, small head, small eyes, central nervous system involvement other than hydrocephalus, otology, and short stature are commonly reported in FA patients who also had VACTERL-H [Alter and Giri, 2016]. Two perinatal cases (Case 11 and 13) were examined and evaluated through diagnostic criteria of VACTERL-H. In addition, the 2 perinatal subjects showed microcephaly and IUGR, both of which led us consider FA as a differential diagnosis upon exclusion of chromosomal abnormalities including gross copy number variations.
Though percentages may differ for certain ethnic groups in literature reports, FANCA mutations are generally kept responsible in 60% of the cases, followed by FANCC in approximately 10–15% of the instances, and by FANCG in approximately 10% of the affected individuals [Kennedy & D'Andrea, 2005]. A considerable portion of FANCA mutations are reported to be gross deletions, predominated as a result of recombination between alu-repeats in cis configurations [Castella et al, 2011; Park et al, 2013; Esmailnia et al, 2016]. In our study group, FANCA was responsible in 75% of the cases (15/20), and gross deletions were identified in 19% (3/15) of those subjects. Among the FANCA alleles, gross deletion of exon 6–31 was identified in 16% (5/30), c.894–2A>G in 13% (4/30), and c.2638C>T was found in 10% (3/30). Overall, disease responsibility of FANCC, FANCE, FANCJ, and FANCL alleles was 5%, while FANCD1 and FANCN was 2.5%. Our ratios were unlike other reported rations of different geographic regions.
Cases 4 and 5 showed a mosaic variant of FANCA (c.3163C=/3163C>T and c.2557C=/2557C>T) which had previously been described as pathogenic; the former was derived through cDNA sequencing in a Japanese FA patient with a compound heterozygous form and the latter was identified in a single chromosome upon Sanger sequencing of genomic DNA derived from patients registered with the International Fanconi Anemia Registry (IFAR), following retrovirus mediated complementation group assay. However, that particular study was inconclusive toward revealing a genetic basis since a pathogenic variant in the other allele was unidentified [Nakamura et al., 1999; Chandra et al., 2005]. Deep sequencing studies suggested that mosaic alterations in the genome could either be due to a reversal of the germline presence or post-zygotic events [Acuna-Hidalgo et al., 2016]. Family segregation revealed a germline biallelic occurrence of these variants underlining a reversal of germline existence in our cases. Mitotic recombination or gene conversion via pairing between homologous chromosomes, leading to somatic reversions, have been described in a few FA patients carrying compound heterozygous pathogenic variants [Gregory et al., 2001; Gross et al., 2002]. In our study, 28% (2/7 cases) had reverse mutations in the compound heterozygous group. It is reported that 20% of patients may benefit from genetic instability via somatic reversions of their germline pathogenic variants [Kalb et al., 2006]. Nonetheless, Case 4 suffered from BMF due to thrombocytopenia, and Case 5 had aplastic anemia, proximal replacement of her bilateral 5th finger, café au lait spots, and DEB positivity. It was observed that the anemia of Case 5 resolved spontaneously in follow-up examinations. The disease causing mutations in the second alleles of our mosaic FANCA cases (c.3491C>T in Case 4 with maternal inheritance and c.3754G>T in Case 5 with paternal inheritance, respectively) were previously described: c.3491C>T in a homozygous form in a multinational FA genetic screening study and c.3754G>T in a compound heterozygous form with another small deletion [Gille et al., 2012; Scheckenbach et al., 2012].
Two novel variants, missense (c.776C>G, p.P259R) and splice site (c.3935–7G>A) mutations, identified in FANCA in Case 6 segregated in her consanguineous parents, supporting compound heterozygosity. PolyPhen (HumVar) predicted the former as possibly damaging, but according to the ACMG classification it is predicted likely benign, as not being truncating (BP1), not having an impact on a gene product (BP2), though absent in controls (PM2). The novel splice acceptor variant identified in the other allele of this patient (c.3935–7G>A) was also predicted benign, having no impact on a splicing motif by all the prediction tools utilized (MutationTaster, Human Splicing Finder, VarSEAK and VarSome). Further segregation in her siblings was not possible since none were alive.
The homozygous novel missense variant (c.2938 G>C, p.A980P) in FANCA in Case 7 was classified as a variant of uncertain significance (VUS) according to ACMG. Nevertheless, it was predicted pathogenic by 3 in silico analysis programs, and the segregation in the family supported the inheritance model. A frame shift leading to a termination variant (c.240_241delTG, p.C80*) identified in FANCA in Case 18 is classified as likely pathogenic. Two different intronic alterations (c.2504+5G>T and c.2504+3G>A) identified in Case 19, the former homozygous and the latter de novo, were predicted with different affects: 3 in silico tools (MutationTaster, Human Splicing Finder, VarSEAK) predicted that c.2504+5G>T would strongly decrease the original donor splicing site of exon 26 of FANCA, and c.2504+3G>A would not alter the splicing motif. The compatibility of the variants within the inheritance model, being novel in disease causing genes, classified these variants (c.776C>G and c.3935–7G>A in Case 6 and c.2504+5G>T in Case 19) as possibly pathogenic, though presently their causality cannot be excluded. There are a few reported cases in the literature with secondary cis located alterations in FA genes that result in a limited recovery of protein function observed with secondary compensating mutations [Waisfisz et al., 1999]. Although Case 19 was negative for DEB and she did not have BMF and therefore did not require a bone marrow transplantation, she did exhibit multiple head and neck tumors at the age of 30. Consequently, there is no basis toward determining whether the patient's phenotype is mild or c.2504+3G>A is compensated partially to the splicing effect of c.2504+5G>T, or vice versa, or if these variants are completely irrelevant.
It is recognized that pathogenic heterozygous variants in FANCS/BRCA1, FANCD1/BRCA2, and FANCN/PALB2 are important in breast cancer predisposition [Castilla et al., 1994; Krainer et al., 1997]. FANCD1/BRCA2 mutations are reported liable to the cumulative incidence of cancer upwards to 97% by the age of 7 years and further likely to cause leukemia, brain tumor, Wilms tumor, or combinations therein with a smaller percentage developing severe BMF [Alter, 2014]. Our Case 17, with a heterozygous de novo truncating mutation in FANCD1/BRCA2, was cancer-free at the age of 6, did not develop BMF, and the child's parents were cancer-free advancing toward their middle age years (maternal age 27 and paternal age 30, respectively, at their last visit). A gross heterozygous deletion in FANCN/PALB2 was inherited from the patient's father. Although breast cancer accounts for less than 1% of all cancers in males [Ly et al., 2013], FANCN/PALB2 is classified as a moderate-penetrance gene for male breast cancers [Rizzolo et al., 2013]. Furthermore, a recently released multicenter study from Italy, using WES and targeted gene sequencing on high risk BRCA1/BRCA2 mutation-negative patients, revealed that the frequency of FANCN/PALB2 pathogenic variants in this cohort was higher than in a similarly classified cohort of females [Silvestri et al., 2017]. Since the initial breast cancer diagnosis in the study from Italy ranged from age 38–83 years, genetic counseling for the family of our Case 17 will be imperative for regular monitoring.
Investigations on peripheral blood samples of heterozygous truncating mutation carriers of FANCN/PALB2 presented impaired DNA damage response, supporting a mono-allelic negative effect of FANCN/PALB2 on genomic stability [Nikkilä et al., 2013]. A functional study, utilizing human lung cancer cells (H1299) and human 293 embryonic kidney cells, revealed that FANCS/BRCA1 and FANCD1/BRCA2, together with BRAD1 (BRCA1 associated ring domain), coexist in the same endogenous protein complex, and exposure to DNA-damaging agents expands the number of partners of foci with the inclusion of FANCR/RAD51 and p53 [Dong et al., 2003]. A knockdown experiment of FANCN/PALB2 via siRNA in HeLa S3 cells further revealed that FANCS/BRCA1 and FANCD1/BRCA2 congregation was abolished and reestablished upon removal of siRNA inhibition. These and additional investigations have shown that FANCS/BRCA1 and FANCD1/BRCA2 association is mediated by FANCN/PALB2, basically appearing at DNA double-strand break sites, however, FANCS/BRCA1 attendance was independent of FANCN/PALB2 that underlined FANCS/BRCA1's upper main regulator role in FANCD1/BRCA2 and FANCN/PALB2 foci [Zhang et al., 2009]. The functional significance of FANCD1/BRCA2 with FANCN/PALB2 investigated on multiple mouse models generated various knock-in and knock-out results [Hartford et al., 2016]. Mouse embryonic fibroblast cells with hemizygous Fancd1 (Brca2KO/+) and hemizygous Fancn (Palb2KO/+) showed a 3.4-fold increase in breaks, radials, and translocations in metaphases following mitomycin C treatment. Furthermore, these cells showed normal cell growth but decreased Fancr (Rad51) foci formation, suggesting that a heterozygous loss of FANCN/PALB2 leads to a decreased FANCR/RAD51 recruitment, important for HR and downstream function for homologous pairing and strand transfer of DNA, and further indicates the likelihood that interaction of FANCD1/BRCA2 and FANCN/PALB2 is physically important for genomic stability and that the severity of the phenotype is increased with diminishing interactions [Hartford et al., 2016]. A holistic evaluation of our Case 17 extracted from this model study is that digenic allelic null mutations of FANCD1/BRCA2 and FANCN/PALB2 would similarly generate a decrease in interaction and possibly revealing the first human presentation. Nonetheless, presently there are no sufficient functional studies that support a digenic evidence for FA. Either pathogenic sequence variants or gross alterations disabling the PALB2 gene on the maternal allele leading to biallelic functional aberration in PALB2 are a more likely disease causing scenario for our Case 17.
Early diagnosis of FA will remain extremely important in providing informed genetic counseling and contemplation of the future. Specifically, care and supervision of a patient's hematologic, oncologic and other expected complications requires life-long monitoring and treatments. Although the consanguinity rate of our cohort (65%) was mildly lower than the previous Turkish FA cohort of 52 patients (78%) [Altay et al., 1997], the consanguineous marriage rate of 21.3% in Turkey is a considerable risk for continued autosomal recessive disorders occurring in the country [Beşpınar and Beşpınar, 2017]. Furthermore, the risk of inheritance of more than one rare disease in the same family is also possible, a fact that at-risk parents need to understand and to weigh heavily [Balta et al., 2019].
A family-based approach to genetic education, diagnosis, and counseling toward such massively parallel sequences of genetically heterogeneous populations, along with continuous robust medical research, will result in life-quality improvements for affected individuals and their families, while continually achieving important steps toward a clearer understanding of the variants' pathogenic status.
Statement of Ethics
Receipt of consent forms signed by all the families preceded the study, which received approval by Istanbul University's Faculty of Medicine Ethic Council (No. 1937).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Author Contributions
G.T. contributed in constructing and conducting the bioinformatics analysis of the panel-gene; D. U.A. in designing and preparing of the clinical tables, plus performing genetic tests (panel-gene, Sanger sequencing, MLPA test) and segregation analysis in the families with major support from the team comprised of G.B., A.A., E.G.B., N.G., and S.A.. S.B. and B.K. contributed the prenatal diagnostic genetic testing, karyotyping, and arrayCGH testing; B.T., H.K., U.A., and Y.A. provided the clinical investigations and recording of the clinical findings plus oversaw all follow-up with the families. T.T.C. and H.A. assisted in the clastogenic investigations. Z.O.U. designed and managed the study, oversaw the procedures and genetic testing, and composed and edited the study manuscript.
Supplementary Material
Supplementary data
Acknowledgements
We sincerely thank the patients and their families for their partnership, their collaboration, and, ultimately, their contributions to medical science research by their involvement in this study. We wish to express our appreciation to Dr. Fatih Tepgeç for providing assistance in the organization of the reference list and to Duane DeBruyne for correcting spelling errors and providing proof reading for English language as a native speaker. We would like to convey special thanks to the Scientific Research Projects Coordination Unit of Istanbul University (Project ID 2678, Code No.: 52,556 and BEK-2017–25048), and the Turkish Pediatrics Institution (Project, No.12/2016) for their invaluable support.
References
- Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17((1)):241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7((1)):Unit7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altay C, Alikaşifoglu M, Kara A, Tunçbilek E, Özbek N, Schroeder-Kurth TM. Analysis of 65 Turkish patients with congenital aplastic anemia (Fanconi anemia and non-Fanconi anemia): Hacettepe experience. Clin Genet. 1997;51((5)):296–302. doi: 10.1111/j.1399-0004.1997.tb02477.x. [DOI] [PubMed] [Google Scholar]
- Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Clin Haematol. 2014;27((3-4)):214–21. doi: 10.1016/j.beha.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Giri N. Thinking of VACTERL-H? Rule out Fanconi Anemia according to PHENOS. Am J Med Genet A. 2016;170((6)):1520–4. doi: 10.1002/ajmg.a.37637. [DOI] [PubMed] [Google Scholar]
- Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, et al. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun. 2015;6((1)):8829. doi: 10.1038/ncomms9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balta G, de Winter JP, Kayserili H, Pronk JC, Joenje H, Joenje H. Fanconi anemia A due to a novel frameshift mutation in hotspot motifs: lack of FANCA protein. Hum Mutat. 2000;15((6)):578. doi: 10.1002/1098-1004(200006)15:6<578::AID-HUMU12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Auerbach Arleen D. Fanconi anemia and its diagnosis. Mutat. Res. 2009;668((1–2)):4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balta G, Patiroglu T, Gumruk F. Fanconi Anemia and Ataxia telangiectasia in siblings who inherited unique combinations of novel FANCA and ATM null mutations. J Pediatr Hematol Oncol. 2019;41((3)):243–6. doi: 10.1097/MPH.0000000000001336. [DOI] [PubMed] [Google Scholar]
- Beşpınar FU, Beşpınar LZ. Türkiye'de hane halki yapilari ve evlilik pratiklerinde ikili resim: geleneklerin yani sira değişimin yansimalari. Nufusbil Derg. 2017;39((1)):109–49. [Google Scholar]
- Castella M, Pujol R, Callén E, Trujillo JP, Casado JA, Gille H, et al. Origin, functional role, and clinical impact of Fanconi anemia FANCA mutations. Blood. 2011;117((14)):3759–69. doi: 10.1182/blood-2010-08-299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla LH, Couch FJ, Erdos MR, Hoskins KF, Calzone K, Garber JE, et al. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nat Genet. 1994;8((4)):387–91. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17((6)):337–49. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, et al. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol Ther. 2005;12((5)):976–84. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Coffa J, Van Den Berg J. Analysis of MLPA data using novel software Coffalyser.NET by MRC-Holland. Modern approaches to quality control. Rijeka, Croatia: InTech. 2011:125–50. [Google Scholar]
- Datta A, Brosh Jr. RM. Holding all the cards - How fanconi anemia proteins deal with replication stress and preserve genomic stability. Genes. 2019;10((2)):170. doi: 10.3390/genes10020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SM, Radloff GA, DeFor TE, Levran O, Batish SD, Hanenberg H, et al. GST genotype may modify clinical phenotype in patients with Fanconi anaemia. Br J Haematol. 2005;131((1)):118–22. doi: 10.1111/j.1365-2141.2005.05721.x. [DOI] [PubMed] [Google Scholar]
- De Bont R, Van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19((3)):169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11((7)):467–80. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37((6)):564–9. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37((9)):e67–e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12((5)):1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- Dubois EL, Guitton-Sert L, Béliveau M, Parmar K, Chagraoui J, Vignard J, et al. A Fanci knockout mouse model reveals common and distinct functions for FANCI and FANCD2. Nucleic Acids Res. 2019;47((14)):7532–47. doi: 10.1093/nar/gkz514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler ME. Fanconi Anemia: Guidelines for Diagnosis and Management. Fanconi Anemia Research Fund; 2008. [Google Scholar]
- Esmailnia G, Fadaee M, Royer R, Dehghan K, Akbari MR. Profiling Fanconi Anemia Gene Mutations among Iranian Patients. Arch Iran Med. 2016;19((4)):236–240. [PubMed] [Google Scholar]
- Esmer C, Sánchez S, Ramos S, Molina B, Frias S, Carnevale A. DEB test for Fanconi anemia detection in patients with atypical phenotypes. Am J Med Genet A. 2004;124A((1)):35–9. doi: 10.1002/ajmg.a.20327. [DOI] [PubMed] [Google Scholar]
- Fargo JH, Rochowski A, Giri N, Savage SA, Olson SB, Alter BP. Comparison of chromosome breakage in non-mosaic and mosaic patients with Fanconi anemia, relatives, and patients with other inherited bone marrow failure syndromes. Cytogenet Genome Res. 2014;144((1)):15–27. doi: 10.1159/000366251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille JJ, Floor K, Kerkhoven L, Ameziane N, Joenje H, de Winter JP. Diagnosis of Fanconi anemia: mutation analysis by multiplex ligation-dependent probe amplification and PCR-based Sanger sequencing. Anemia. 2012;2012:603253. doi: 10.1155/2012/603253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JJ, Wagner JE, Verlander PC, Levran O, Batish SD, Eide CR, et al. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci USA. 2001;98((5)):2532–7. doi: 10.1073/pnas.051609898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98((2-3)):126–35. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- Hartford SA, Chittela R, Ding X, Vyas A, Martin B, Burkett S, et al. Interaction with PALB2 is essential for maintenance of genomic integrity by BRCA2. PLoS Genet. 2016;12((8)):e1006236. doi: 10.1371/journal.pgen.1006236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, et al. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol Cell. 2015;59((3)):462–77. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Hlavin EM, Smeaton MB, Noronha AM, Wilds CJ, Miller PS. Cross-link structure affects replication-independent DNA interstrand cross-link repair in mammalian cells. Biochemistry. 2010;49((18)):3977–88. doi: 10.1021/bi902169q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14((5)):693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13((2)):109–10. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Neveling K, Nanda I, Schindler D, Hoehn H. Fanconi anemia: causes and consequences of genetic instability. In: Volff JN, Genome and Disease, editor. Genome Dyn. Vol. 1. Karger Publishers, Basel; 2006. pp. 218–42. [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxivorg. 2019:531210.. [Google Scholar]
- Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19((24)):2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Knies K, Inano S, Ramírez MJ, Ishiai M, Surrallés J, Takata M, et al. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J Clin Invest. 2017;127((8)):3013–27. doi: 10.1172/JCI92069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch J, Kamenisch Y, Kubisch C, Berneburg M. Rare hereditary diseases with defects in DNA-repair. Eur J Dermatol. 2012;22((4)):443–55. doi: 10.1684/ejd.2012.1654. [DOI] [PubMed] [Google Scholar]
- Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35((11)):1978–80. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer M, Silva-Arrieta S, FitzGerald MG, Shimada A, Ishioka C, Kanamaru R, et al. Differential contributions of BRCA1 and BRCA2 to early-onset breast cancer. N Engl J Med. 1997;336((20)):1416–21. doi: 10.1056/NEJM199705153362003. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4((7)):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101((4)):1249–56. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44((D1)):D862–8. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobitz S, Velleuer E. Guido Fanconi (1892-1979): a jack of all trades. Nat Rev Cancer. 2006;6((11)):893–8. doi: 10.1038/nrc2009. [DOI] [PubMed] [Google Scholar]
- Ly D, Forman D, Ferlay J, Brinton LA, Cook MB. An international comparison of male and female breast cancer incidence rates. Int J Cancer. 2013;132((8)):1918–26. doi: 10.1002/ijc.27841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota MBS, Carvalho MA, Monteiro ANA, Mesquita RD. DNA damage response and repair in perspective: Aedes aegypti Drosophila melanogaster and Homo sapiens. Parasit Vectors. 2019;12((1)):1–20. doi: 10.1186/s13071-019-3792-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Matsuura S, Tauchi H, Hanada R, Ohashi H, Hasegawa T, et al. Four novel mutations of the Fanconi anemia group A gene (FAA) in Japanese patients. J Hum Genet. 1999;44((1)):48–51. doi: 10.1007/s100380050106. [DOI] [PubMed] [Google Scholar]
- Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, et al. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14((4)):525–35. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Nikkilä J, Christin Parplys A, Pylkäs K, Bose M, Huo Y, Borgmann K, et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4((1)):2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niraj J, Färkkilä A, D'Andrea AD. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol. 2019;3:457–78. doi: 10.1146/annurev-cancerbio-030617-050422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106((2)):277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palovcak A, Liu W, Yuan F, Zhang Y. Maintenance of genome stability by Fanconi anemia proteins. Cell Biosci. 2017;7((1)):8. doi: 10.1186/s13578-016-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chung NG, Chae H, Kim M, Lee S, Kim Y, et al. FANCA and FANCG are the major Fanconi anemia genes in the Korean population. Clin Genet. 2013;84((3)):271–5. doi: 10.1111/cge.12042. [DOI] [PubMed] [Google Scholar]
- Peffault de Latour R, Porcher R, Dalle J, Aljurf M, Korthof ET, Svahn J, et al. FA Committee of the Severe Aplastic Anemia Working Party. Pediatric Working Party of the European Group for Blood and Marrow Transplantation Allogeneic hematopoietic stem cell transplantation in Fanconi anemia: the European Group for Blood and Marrow Transplantation experience. Blood. 2013;122((26)):4279–4286. doi: 10.1182/blood-2013-01-479733. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17((5)):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolo P, Silvestri V, Tommasi S, Pinto R, Danza K, Falchetti M, et al. Male breast cancer: genetics, epigenetics, and ethical aspects. Ann Oncol. 2013;24((Suppl l_8)):viii75–82. doi: 10.1093/annonc/mdt316. [DOI] [PubMed] [Google Scholar]
- Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica. 2008;93((4)):511–7. doi: 10.3324/haematol.12234. [DOI] [PubMed] [Google Scholar]
- Scheckenbach K, Morgan M, Filger-Brillinger J, Sandmann M, Strimling B, Scheurlen W, et al. Treatment of the bone marrow failure in Fanconi anemia patients with danazol. Blood Cells Mol Dis. 2012;48((2)):128–31. doi: 10.1016/j.bcmd.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell. 2015;60((3)):351–61. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11((4)):361–2. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24((3)):101–22. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamsunder P, Esner M, Barvalia M, Wu YJ, Loja T, Boon HB, et al. Impaired mitophagy in Fanconi anemia is dependent on mitochondrial fission. Oncotarget. 2016;7((36)):58065–74. doi: 10.18632/oncotarget.11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri V, Zelli V, Valentini V, Rizzolo P, Navazio AS, Coppa A, et al. Whole-exome sequencing and targeted gene sequencing provide insights into the role of PALB2 as a male breast cancer susceptibility gene. Cancer. 2017;123((2)):210–8. doi: 10.1002/cncr.30337. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Bear KA, Kimonis V, de Klein A, Scott DA, Shaw-Smith C, et al. Clinical geneticists' views of VACTERL/VATER association. Am J Med Genet A. 2012;158A((12)):3087–100. doi: 10.1002/ajmg.a.35638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Levine B. Emerging functions of the Fanconi anemia pathway at a glance. J Cell Sci. 2017;130((16)):2657–62. doi: 10.1242/jcs.204909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waisfisz Q, Morgan NV, Savino M, De Winter JP, Van Berkel CG, Hoatlin ME, et al. Spontaneous functional correction of homozygous fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat Genet. 1999;22((4)):379–83. doi: 10.1038/11956. [DOI] [PubMed] [Google Scholar]
- Wegman‐Ostrosky T, Savage SA. The genomics of inherited bone marrow failure: from mechanism to the clinic. Br J Haematol. 2017;177((4)):526–42. doi: 10.1111/bjh.14535. [DOI] [PubMed] [Google Scholar]
- Wenzel ES, Singh ATK. Cell-cycle checkpoints and aneuploidy on the path to cancer. In Vivo. 2018;32((1)):1–5. doi: 10.21873/invivo.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-H. The concept and practice of Fanconi Anemia: from the clinical bedside to the laboratory bench. Transl Pediatr. 2013;2((3)):112–9. doi: 10.3978/j.issn.2224-4336.2013.07.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22((6)):719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16((1)):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19((6)):524–9. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data