Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a new member of the coronavirus family that can cause coronavirus disease 2019 (COVID‐19). COVID‐9 has become a global pandemic with severe health issues around the world. Identifying the accurate immunopathogenesis of the COVID‐19 and the immune response against SARS‐CoV‐2 is necessary for the development of therapeutic approaches and rational drug design. This paper aims to overview the updated clinical data on the immunopathogenesis of the COVID‐19 and review the innate and adaptive immune response to SARS‐CoV‐2. Also, challenges of the immune response to SARS‐CoV‐2 leading to dysfunctional immune response and their contribution to the progression of the disease have been discussed. To achieve a more efficient immune response, multiple methods could be applied, including regulation of the immune response, augmentation of the immune system against the virus, inhibition of the dysfunctional immune checkpoints, and inhibition of the viral replication/infection. Based on the immune response against SARS‐CoV‐2 and its dysfunction, we introduce potential immunotherapies as well as reviewing recruiting/completed clinical trials of COVID‐19.
Keywords: clinical trial, COVID‐19, immunopathogenesis, immunotherapy, SARS‐CoV‐2
Understanding the immunology of coronavirus disease 2019 (COVID‐19) is necessary for rational drug and vaccine design against it. In this paper, we have reviewed the updated data on immunobiology and immunotherapeutic‐based approaches for COVID‐19.
1. INTRODUCTION
Multiple cases of pneumonia of unknown origin were reported last December 2019, in Wuhan, Hubei province, China (Phelan et al., 2020; Tahmasebi et al., 2020; Zhang, Litvinova et al., 2020). Analysis of the bronchoalveolar lavage fluids of the patients by next‐generation sequencing (NGS) identified a novel viral RNA genome incompatible with previous viral sources. Further studies identified this novel virus to be a member of the beta coronaviruses. The phylogenetic study demonstrated the virus to be a novel zoonotic virus that belonged to the subgenus Sarbecovirus of the beta‐coronavirus genus (Wang, Horby, et al., 2020; N. Zhu et al., 2020). Coronaviruses are enveloped positive‐stranded RNA viruses. Up to now, seven coronaviruses have been reported to cross species and infect humans. The previous members of the coronaviruses infecting humans were 229E, OC43, NLG3, and HKU1. These coronaviruses can cause minor symptoms of respiratory tract infection in immune‐competent individuals. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the seventh member of the betacoronavirus family that has crossed species and infected humans. Similar to the SARS viruses and Middle East Respiratory Syndrome (MERS), SARS‐CoV‐2 causes an acute respiratory syndrome that is represented by pneumonia‐associated symptoms. The whole‐genome analysis demonstrated that the SARS‐CoV‐2 genome is 96% identical to a bat‐origin coronavirus, introducing bat as a probable source for COVID‐19 (P. Zhou et al., 2020).
Initially, the virus spread was considered to be only through direct contact with the seafood market in Wuhan; however, further research by Chan et al. (2020) reported a familial cluster of COVID‐19‐caused pneumonia. This indicated the human‐to‐human transmission of the virus, which was supported by further research. The rapid distribution of the virus in China and other countries lead COVID‐19 to be a public health emergency of international concern and convinced the World Health Organization (WHO) to announce the COVID‐19 outbreak as a global health emergency on January 30, 2020. Also, on March 11, 2020, COVID‐19 was announced global pandemic by the WHO. By the end of September 6, 2020, this virus has reported being spread to most of the countries around the world, infecting approximately 27 million individuals, of which 900,000 have died (https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200907-weekly-epi-update-4.pdf?sfvrsn=f5f607ee_2).
Considering the impacts of the COVID‐19 on global health around the world, there is an urgent need for the development of effective therapeutic approaches and vaccine design (Lim et al., 2020). Understanding the immunopathogenesis of SARS‐COV‐2 and the host‐virus interaction is necessary for the development of novel insights into the treatment and management of COVID‐19. In this paper, we aim to review the immunopathogenic characteristics of the COVID‐19 and the innate and adaptive immune response against the virus. We also introduce immunotherapeutic potentials and approaches for the treatment of this disease, based on recently identified data on SARS‐COV‐2. The immunotherapeutic approaches for COVID‐19 aim to inhibit the viral infection or modify the hyperactivated immune response against SARS‐CoV‐2. Moreover, an updated overview of the prospective clinical trials targeting the COVID‐19 has been discussed. This could pave the way to identify novel treatments against this pandemic and reduce disease burden and deaths caused by COVID‐19.
2. CLINICAL SIGNS AND SYMPTOMS OF COVID‐19
SARS‐CoV‐2 is mainly transmitted through close contact by respiratory droplets of the infected patients. Also, positive fecal specimens of the infected patients have increased the possibility of fecal‐oral transmission. Bare contact with infected surfaces has also been suggested as a potential route of infection of COVID‐19 (Meselson, 2020). The incubation period of the COVID‐19 is estimated 1–14 days after exposure. A study has reported the incubation period up to 27 days after exposure; however, most patients exhibit symptoms approximately after 5 days (Lauer et al., 2020). There is a wide range of variability in clinical symptoms of the patients with COVID‐19, with most of the patients remaining asymptomatic. In symptomatic patients, initial clinical symptoms include fever, myalgia, pharyngalgia, sore throat, dry cough, dyspnea (shortness of breath), fatigue, and malaise. Diarrhea and loss of appetite can also be among the early signs of the disease. Despite SARS and MERS that do not accompany gastrointestinal symptoms, COVID‐19 can cause gastrointestinal tract symptoms such as diarrhea (Holshue et al., 2020; Zhang, Wang et al., 2020). Also, headache and hemoptysis have been reported in some patients (Z. Xu et al., 2020).
Most of the patients show mild symptoms of infection with SARS‐CoV‐2; however, in a small proportion of the patients, the respiratory symptoms can worsen and lead to a severe respiratory syndrome that needs intensive care. According to documents, most of the mortalities were reported to happen in elderly patients or patients with multiple comorbidities, including cardiovascular diseases, respiratory diseases, diabetes mellitus, hypertension, and immune‐compromised patients, such as cancer. Also, the mortality rate varies based on age with most of the deaths occurring in older male patients (Jung et al., 2020).
COVID‐19 has some nonspecific laboratory signs. The laboratory signs of the COVID‐19 include leukocytosis, lymphopenia, increased C‐reactive protein, increased d‐dimer, and increased lactate dehydrogenase. Procalcitonin is not reported being increased except in patients with severe disease who needed intensive care (C. Huang et al., 2020). In the chest X‐ray, patchy infiltrations with diffuse ground‐glass patchy shadow can be observed. In the chest computed tomography (chest‐CT), bilateral ground‐glass opacification and diffuse consolidation are the most specific signs for COVID‐19‐related pneumonia (Bernheim et al., 2020). Multiple tests can be used to confirm the diagnosis. The most common definitive test for COVID‐19 is the viral RNA detecting technique, real‐time quantitative polymerase chain reaction (RT‐qPCR). Despite the high sensitivity of the RT‐qPCR test for SARS‐COV‐2, the false‐negative results have been reported in patients with typical clinical symptoms and chest‐CT results (Yelin et al., 2020). Thus, CT and clinical symptoms must be attended in the evaluation of the patients with negative RT‐qPCR results. A recent study reported the sensitivity of chest‐CT to be higher in comparison to RT‐qPCR. Thus, in clinically highly suspicious patients with negative RT‐qPCR test, chest‐CT and repeating RT‐qPCR has been proposed. Moreover, antiviral Immunoglobulin M (IgM)/IgG‐detecting kits have been studied and produced by multiple companies.
3. IMMUNOPATHOGENESIS OF THE COVID‐19
The SARS‐CoV‐2 is transmitted through droplets, which are distributed by coughing and sneezing from infected patients. Also, the positive‐viral tested neonates born from infected pregnant women have suggested the vertical transmission of the virus to the infants; however, unlike SARS and MERS, COVID‐19 does not seem to cause maternal mortality or intrauterine growth retardation in humans (L. Wang et al., 2020).
After being transmitted, the first location that the virus starts to replicate is the airway epithelial cells. As the disease progresses, the virus transmits to the lower sections of the airway. The main host cells for SARS‐CoV‐2 are the type II pneumocytes of the lung and the enterocytes of the gut. Similar to SARS, SARS‐CoV‐2 utilizes the angiotensin‐converting enzyme‐2 (ACE‐2) receptor to enter the host cell; however, the presence of the ACE‐2 receptor is not the sole factor that determines the infection of the tissue with SARS‐CoV‐2 (W. Li et al., 2003; P. Zhou et al., 2020). A recent study has demonstrated that SARS‐COV‐2 possesses 10–20 fold more affinity for the ACE‐2 receptor (X. T. Xu et al., 2020). Through the spike (S) glycoprotein of its receptor‐binding domain (RBD), SARS‐CoV‐2 binds to the ACE‐2 receptor. Spike glycoprotein of the SARS‐CoV‐2 is composed of two sections, S1 and S2. S1 is responsible for the binding of the viral RBD to the host cell receptor, and S2 is responsible for the fusion of the virus to the host cell membrane. Since the ACE‐2 receptor is highly expressed by the epithelium of the small intestine, upper respiratory tract, and alveolar pneumocytes, COVID‐19 can enter the cells of these two organs and cause upper respiratory and gastrointestinal symptoms (X. Li et al., 2020). After the infection with SARS‐CoV, the expression of the ACE‐2 receptor by lung cells has reported being decreased (Kuba et al., 2006). The downregulation of the ACE‐2 expression by lung cells is associated with acute lung injury (Imai et al., 2005). Therefore, ACE‐2 downregulation in lung cells is another pathologic mechanism of SARS‐CoV leading to acute lung injury and acute respiratory distress syndrome (ARDS). Considering the application of the same receptor for cell entrance and the similarity of the pathogenesis of SARS‐COV with SARS‐CoV‐2, the downregulation of ACE‐2 could be another possible mechanism of acute lung injury in the pathogenesis of SARS‐CoV‐2. All other organs of the body that express the ACE‐2 receptor could also be infected with SARS‐COV‐2. As an example, a study (Jia et al., 2020) has investigated the infection of the adipose tissue by SARS‐COV‐2. This study indicated that since adipose tissue expresses the ACE‐2 receptor, adipose cells can be infected with SARS‐COV‐2.
As mentioned, pulmonary pneumocytes are the most common lung cells infected by SARS‐CoV‐2. The cytotoxic effects of the virus on pneumocytes stimulate the release of inflammatory mediators that trigger a local immune response. Additionally, resident alveolar macrophages exert an inflammatory immune response against the virus by producing proinflammatory cytokines and chemokines, such as interferon (IFN)‐γ, IP‐10, monocyte chemotactic protein‐1 (MCP‐1), tumor necrosis factor (TNF)‐α, interleukin (IL)‐6, and IL‐1β. The release of these proinflammatory cytokines and chemokines to the blood stimulates an inflammatory response in the lung by recruiting blood monocytes and T‐lymphocytes. The inflammatory response can vary from mild inflammation to severe inflammation. In most cases, the immune response is capable of eliminating the virus; however, deficient immune response or the respiratory hyperinflammation can lead to severe respiratory failure in severe COVID‐19 cases. Severe pulmonary inflammation also increases capillary leakage that can cause ARDS (Rockx et al., 2020). Thus, the immune response against SARS‐CoV‐2 and the severity of the inflammation are two major factors that define the outcome in patients with COVID‐19.
The analysis of the lung autopsy samples of the patients with COVID‐19 has exhibited the patients to have cellular fibromyxoid exudates and desquamation of the pneumocytes. Also, hyaline membrane formation and pulmonary edema were seen, which identifies the progression of the ARDS as the leading cause of death. Similar to SARS‐CoV, the uncontrolled infiltration of the immune cells and unrestrained production of inflammatory cytokines are the main factors that contribute to the progression of cytokine storm and ARDS in COVID‐19. ARDS reduces the oxygen‐exchange capacity of the lungs and causes respiratory failure, which can lead to patient death (Z. Xu et al., 2020). Multiple‐organ failure is one of the consequences of the cytokine storm that can lead to dysfunction of the kidneys, liver, heart, and other end organs. In conclusion, the immune response is the factor that defines whether the infection with SARS‐CoV‐2 is going to be promptly cleaned, leading to mild disease, or it is going to develop severe disease and respiratory failure (F. Zhang et al., 2020).
In addition to the immune system, the ABO blood group is another factor that could affect the susceptibility of the individuals to COVID‐19. In a recent study, Zhao et al. (2020) inspected the relationship between the ABO blood and COVID‐19 infection. This study demonstrated that individuals with A blood group were at an increased risk for COVID‐19 (OR = 1.45), while O blood group individuals had reduced risk for COVID‐19 infection (OR = 45; Ellinghaus et al., 2020). The lower susceptibility of O blood group patients can be explained by the presence of the cross‐neutralizing anti‐blood group antibodies against SARS‐CoV‐2 in their serum. These anti‐blood group antibodies, and especially anti‐A antibodies, have shown neutralizing activity against the SARS‐CoV‐2, and can thus reduce the risk of infection with SARS‐CoV‐2 in patients with O blood group (Zhao et al., 2020).
One of the most common laboratory characteristics of the COVID‐19 is lymphopenia (Z. Wu & McGoogan, 2020), which is commonly accompanied by an increase in the number of neutrophils. There are several reasons for lymphopenia in COVID‐19. The pulmonary infiltration of the lymphocytes, apoptosis/pyroptosis of the lymphocytes, lateral margination, and infection of the lymphocytes by SARS‐CoV‐2, are the most common reasons leading to lymphopenia (Tay et al., 2020). Studies have demonstrated that the lymphopenia mostly results from the reduced number of CD8+ cells, rather than CD4+ T cells, B cells, or NK cells (Wan et al., 2020). Since CD8+ cells have an essential role in the antiviral immune response against SARS‐COV‐2‐infected cells, the reduction in their number weakens the antiviral immunity of the patients. Thus, lymphopenia has been reported to be more common in severe cases, occurring in 63%–70.3% of critically ill patients (Tavakolpour et al., 2020). Accordingly, lymphopenia has been proposed as a prognostic marker for COVID‐19. Since the number of lymphocytes is reduced and the number of neutrophils is increased, a study has identified the neutrophil‐lymphocyte ratio (NLR, the proportion of the neutrophil count to lymphocyte count) to be increased in COVID‐19. The NLR was reported to be more increased in severe COVID‐19 patients, and thus, the increased NLR could be used as a prognostic criterion for the COVID‐19. This study also demonstrated that as the number of lymphocytes reduces, the inflammatory cytokines increase and the disease becomes more critical, which emphasizes the severe inflammatory status as the leading cause of respiratory failure in severe COVID‐19 patients (J. Liu et al., 2020).
A study in Wuhan on 99 COVID‐19 patients (N. Chen et al., 2020) demonstrated that in severe cases who needed intensive care in the intensive care unit (ICU), the serum levels of inflammatory cytokines, including TNF‐α, IP‐10, macrophage inflammatory protein 1A (MIP‐1A), and MCP‐1, were higher than other patients. This emphasizes the role of hyperinflammatory status in the progression and severity of the COVID‐19. Also, the first study on 41 COVID‐19 patients in Wuhan demonstrated that severe cases had higher levels of inflammatory cytokines including MIP‐1A, IL‐7, IL‐2, IP‐10, G‐CSF, IL‐10, MCP‐1, and TNF‐α (C. Huang et al., 2020). Severe COVID‐19 patients were also reported to have an increased percentage of custer of differentiation 14 (CD14+)CD16+ monocytes in their peripheral blood, in comparison to mild COVID‐19 patients. The inflammatory CD14+CD16+ monocytes contribute to the hyperinflammatory status and the cytokine storm by producing multiple inflammatory cytokines (Y. Zhou et al., 2020). These findings exhibit the role of the dysfunctional immune response leading to hyperinflammatory status and the cytokine storm in severely ill patients. Identification of the proper function of the immune system against COVID‐19 and managing immune dysfunction could pave the way through discovering effective treatments and reducing the mortality rate of COVID‐19. To do so, it is critical to understand the exact mechanisms of the innate and adaptive immune response against SARS‐CoV‐2. Here, we aim to discuss the innate and adaptive immune responses to the SARS‐COV‐2 based on recent data. Also, we aim to address the dysfunctional immune response and the challenges of the immune response against SARS‐CoV‐2.
3.1. Innate immunity against SARS‐COV‐2
To exert an innate immune response against SARS‐COV‐2, it should be first sensed by the innate immunity as a pathogen. The recognization of the coronavirus family is usually exerted through the identification of the pathogen‐associated molecular patterns (PAMPs), viral single‐stranded RNA, and viral double‐stranded RNA, by pathogen‐related receptors of the macrophages. Then, the Toll‐like receptors 3 and 7, cytosolic RNA sensor, and RIG1/MDA5 contribute to the identification of the coronavirus. This mechanism leads to the activation of the downstream inflammatory signaling pathways, such as the phosphorylation of nuclear factor‐κB (NF‐κB), phosphoinositide 3‐kinase, mitogen‐activated protein kinase, and IFN regulatory factor‐3 (IRF‐3). Activation of these inflammatory transcription factors finally leads to the induction of innate immune response, production of type 1 interferon (IFN‐1), and the release of proinflammatory cytokines such as IL‐1β, TNF‐α, IL‐6, and IL‐18 (X. Li et al., 2020).
The macrophage‐derived cytokines and type I IFNs also participate to activate the natural killer (NK) cells. NK cells are members of the innate immune system that contribute to the initial immune response against SARS‐CoV‐2. NK cells exert a major histocompatibility complex (MHC)‐independent immune response against SARS‐CoV‐2 and can restrict the pathogenesis of the virus at the initial steps of infection (Florindo et al., 2020).
The respiratory airway epithelial cells are also among the first immune effector cells that recruit immune cells to the lung tissue by producing immune cytokines and expression of adhesive molecules. Type 1 macrophages (M1), members of the innate immunity with proinflammatory properties, produce antiviral immune response after recognizing the virus. The immune response by macrophages is exerted by the production of multiple immune cytokines, such as type I IFNs, and their phagocytosis (Vabret et al., 2020).
The respiratory mucosa contains many immune effector cells, such as dendritic cells (DCs). DCs inhibit the viral infection as members of the innate immune system. After infection of the respiratory system, DCs start an immediate immune response by producing type I IFNs and IL‐6. DCs also stimulate the adaptive immune response by acting as antigen‐presenting cells (APCs.) Although the type I IFN‐based immune response is initially critical to restrict viral infection, the excess production of type I IFNs by DCs leads to severe inflammation and ARDS in severe patients (Chiappelli et al., 2020).
One of the essential innate immune signals that have a significant antiviral function is IFN‐1, including IFN‐α, IFN‐ β, and IFN‐λ. After the recognition of the virus, the activation of the IRF‐3 in the macrophage leads to the production of IFN‐1. Through its downstream signaling pathways, IFN‐1 induces an adaptive immune response against the virus. One of the mechanisms of antiviral immunity by type I IFN is the activation of the IFN‐induced transmembrane family proteins, which inhibit the entrance of the virus to the host cell. This inhibits the replication of the virus and the infection of the further cells by SARS‐CoV‐2 (Mosaddeghi et al., 2020). IFN‐1 has shown to have an important role in the immune response against SARS‐CoV and thus, is one of the most important immunotherapeutic potentials in the treatment of COVID‐19 (Mantlo et al., 2020).
3.2. Adaptive immunity against SARS‐COV‐2
APCs, including monocytes and DCs, distinguish the viral antigen on infected cells. Then, APCs introduce the antigen to the helper T (Th) cells. The mechanism of antigen presentation of SARS‐COV‐2 is not entirely identified yet; however, since the pathogenesis of SARS‐COV‐2 is similar to SARS‐CoV, the same pathway might be applied for the antigen presentation, which is through the MHC‐1 (K. Yang et al., 2009). APCs also produce cytokines that direct the antiviral immune response of the T cells. The cytokines produced by APCs and helper T cells drive the cellular immune response to SARS‐CoV‐2. Lymphopenia, the reduced number of lymphocytes in the peripheral blood, following mononuclear infiltrations in the COVID‐19 autopsy samples, demonstrate the activation of the lymphocytes and their recruitment to the lung tissue to inhibit the infection by SARS‐CoV‐2.
Helper T cells are the directors of the cellular immunity against SARS‐CoV‐2. In COVID‐19 patients, the serum levels of the Th1‐associated cytokines are reported to be increased. Th1 cells release cytokines, including IFN‐γ, TNF‐α, and IL‐2, that activate the cytotoxic T cells (CTLs). CTLs attack the viral‐contaminated cells and destroy them by the producing perforin and granzyme. Also, Th2 cells present the viral antigen to the B lymphocytes, which subsequently produce neutralizing antibodies against the spike (S) protein of the virus. The neutralizing antibodies inhibit the replication of the virus inside the body and produce humoral immunity, which is one of the main concepts for vaccine design against SARS‐CoV‐2 (X. Li et al., 2020).
Considering SARS‐CoV, cellular memory immunity by T cells has been reported to be present for 6 years after SARS (Oh et al., 2011); however, there are no reports of how long can the cellular immunity maintain memory‐immunity against SARS‐COV‐2. Th17 cells are another subgroup of the helper T cells that have been reported to be increased in COVID‐19. The production of IL‐17, a proinflammatory cytokine, by Th17 cells, promotes the inflammatory response. The exact mechanism of the antiviral function of the Th17 cells, another group of cellular immunity, has not been determined yet. Th17 cells could have antiviral function through the production of inflammatory cytokines; however, further study is required to understand the exact antiviral role of Th17 cells (Prompetchara et al., 2020; D. Wu & Yang, 2020).
As well as T cells, humoral immunity, mostly through B cells, has a significant role in the induction of adaptive immunity against coronaviruses. The activation of the B cells and the plasma cells leads to the production of the neutralizing antibodies that prevent further contamination by the virus. In most patients, the neutralizing antibodies are present 3 weeks after infection (Bernheim et al., 2020). Firstly, the B cells start their antibody responses against nucleocapsid (N) of the SARS‐CoV‐2. Furthermore, the B cells produce anti‐spike (S) antibodies. The anti‐SARS‐CoV‐2 antibodies include two subclasses: IgM and IgG antibodies. SARS‐CoV‐2‐IgM has been reported being present in the serum of the COVID‐19 patients 9 days after the onset of the disease. SARS‐CoV‐2‐IgG is present in patient serum 2 weeks after exposure (Totura & Baric, 2012). Appropriate antiviral antibodies prevent the patient from being reinfected by the virus; however, it has been recently reported that inadequate serum antibody levels could expose the patients to reinfection. Besides, some cases of reinfection have been reported, which questions the humoral‐memory immunity against SARS‐COV‐2 (L. Zhou et al., 2020). Further research is needed to be inducted for understanding the accurate immune response mechanism to the SARS‐COV‐2. SARS‐CoV‐2‐specific T cells and B cells have major roles in the immune response in COVID‐19 and thus, must be considered as promising approaches in rational drug and vaccine design against COVID‐19.
During the cytokine storm, monocytes and Th1 cells produce granulocyte‐monocyte colony‐stimulating factor (GM‐CSF) and IL‐6. GM‐CSF and IL‐6 have a major role in the progression of mild to severe respiratory inflammation and ARDS (X. Li et al., 2020; Y. Zhou et al., 2020). A recent systematic review study has exhibited that higher serum levels of IL‐6 are associated with poor clinical outcomes and severe pulmonary complications (Russell et al., 2020). Another important factor that contributes to the dysfunction of the cellular immune response in COVID‐19 is the exhaustion of the T cells. T cells separated from COVID‐19 patients have an increased expression of programmed death‐1 (PD‐1) and T‐cell immunoglobulin and mucin‐domain containing‐3 (Tim‐3), two major surface markers of the T cell exhaustion. The expression of the PD‐1 and Tim‐3 is associated with disease severity in highly symptomatic patients (Diao et al., 2020).
Human leukocyte antigens (HLAs) are critical genetic compounds in APCs that affect the expression of the pathogen‐antigens to the T cells through MHC‐1. Since MHC‐1 has an essential role in the induction of the cellular immune response against SARS‐CoV‐2, HLA‐polymorphisms could have an important role in the susceptibility or resistance of the individuals to COVID‐19. Certain HLAs have proved to be related to the susceptibility of the patients to multiple viral infections such as SARS and MERS. Discussing the role of HLAs in COVID‐19 infection is beyond the goal of this article (Shi et al., 2020).
3.3. Challenges related to the immune response against SARS‐CoV‐2
The immune system is the main system defending the body against SARS‐CoV‐2 infection; however, some challenges dampen the antiviral immune against SARS‐CoV‐2 and lead to dysfunctional immune activity. Both immune‐suppressing challenges and severe inflammatory response can lead to dysfunction of the immune system in COVID‐19 (Chang et al., 2020).
3.3.1. Insufficient immune response in immunocompromised patients
The insufficient immune response in immune‐compromised patients is one of the significant events that prevent the immune system from producing sufficient immunity against the SARS‐COV‐2. Old patients with multiple comorbidities, cancer patients, patients on immunosuppressive drugs, and other immune‐compromised patients are highly susceptible to severe infection with COVID‐19. Since their immune system is compromised, they are unable to inhibit the replication of the virus. Thus, SARS‐CoV‐2 can infect the lower respiratory tract faster and cause pneumonia that can be followed by severe inflammation and ARDS. In conclusion, the insufficient immune response in immune‐compromised patients is the underlying cause of higher mortality in old‐aged and comorbid patients (Monti et al., 2020).
3.3.2. SARS‐CoV‐2 immune evasion
The second challenge that depletes appropriate antiviral immune response is the capability of the coronaviruses to evade identification by the immune system. This immune‐evasion mechanism could be the underlying reason for the prolonged incubation period of the SARS‐COV‐2 in comparison to influenza or other respiratory viruses. The immune‐evasion mechanism of SARS‐COV‐2 is similar to the immune‐evasion mechanism of the SARS and MERS (Park, 2020; Sariol & Perlman, 2020). Two mechanisms have been identified in SARS and MERS: disturbance in RNA‐sensing and type I IFN producing pathways. Accordingly, recent research has reported the production of type I IFN to be decreased in COVID‐19 patients (Prompetchara et al., 2020). Since type I IFN (both α and β subtypes) are important antiviral components of the immune system, the antagonization of the type I interferon signaling in COVID‐19 patients reduces the power of the immune system to battle the virus. Also, the type I IFN signature is remarkably disturbed in critically ill patients. It has been demonstrated that SARS‐CoV‐2 restricts the production of type I IFN through inhibiting IFN‐signal transduction pathways, such as IRF‐3. This study supports the immune‐evasion mechanism of SARS‐CoV‐2 to be similar to SARS‐CoV, through a reduction in the production of the type I IFN (Prompetchara et al., 2020). The mechanism of action of the IFNs in the COVID‐19 is time dependent. In the first stages, IFNs inhibit the progression of the disease by inhibiting the infection of new host cells. However, in the late stages of the disease, IFNs might promote the progression of the disease by inducing the upregulation of ACE‐2 by airway epithelial cells, which increases the chance infection of these cells (Vabret et al., 2020).
Another plausible immune evasion mechanism of COVID‐19 is the escaping of the virus from the MHC‐dependent presentation of its antigens. The MHC‐dependent presentation of viral antigens by APCs triggers the adaptive immune response. Thus, the escape of the virus from MHC‐dependent antigen presentation restricts the adaptive immune response against SARS‐CoV‐2, which must be addressed as an immune response challenges against it (Kumar et al., 2020).
3.3.3. Cytokine storm
The leading mechanism of respiratory failure in COVID‐19 is the lung damage caused by exaggerated immune response, severe inflammation, ARDS, and further fibrosis, rather than the direct cytopathic effects of the virus on epithelial cells. The uncontrolled production of the immune cytokines in COVID‐19 leads to cytokine storm. Similar to SARS, the cytokine storm is a significant mechanism in the pathogenesis of the SARS‐COV‐2 in critical ICU patients. The cytokine storm caused by SARS‐COV‐2 finally leads to the inflammation of the lungs and the formation of hyaluronan (Liu et al., 2019). The accumulation of the hyaluronan in the lung leads to the hyaloid membrane formation and further ARDS (Harrison, 2010; W. Wang et al., 2020).
Damaged lung cells and M1 macrophages of the lung produce chemotactic factors that recall innate immune cells and induce an uncontrolled inflammatory response in the lung. Then, the inflammatory cells start an uncontrolled production of proinflammatory cytokines and chemokines. CXCL‐8, 9, 10, and CCL‐2, 3, and 5 are the major produced chemokines in the process of the cytokine storm. Also, TGFβ, TNF‐α, IFN‐γ, IFN‐α, IL‐1β, 6, 12, 18, and 33, are the cytokines that contribute to the cytokine storm. Also, similar to SARS‐CoV1, SARS‐CoV‐2 induces the expression of IL‐6 and IL‐8 by its NSP‐9 and NSP‐10. The upregulation in the expression of IL‐6 and IL‐8 leads to the progression of mild inflammation to severe inflammation in critical patients (Vabret et al., 2020).
Cytokine storm, the uncontrolled production of immune cytokines, causes lung‐tissue damage by activating the immune‐inflammatory cells to attack the alveoli and produce fibrotic tissue in the lung, which could finally induce ARDS. Also, the leakage of the fluid to the alveoli and accumulation of the inflammatory exudates lead to respiratory failure (Park, 2020; Wan et al., 2020). The cytokine storm can also lead to multiple‐organ failure that disturbs the function of the kidneys, liver, and heart. Multiple‐organ failure is diagnosed by an increase in the level of creatinine, blood urea, AST, ALT, and other end‐organ enzymes in severe COVID‐19 patients (Tjendra et al., 2020).
According to the destructive role of the cytokine storm in the pathogenesis of the COVID‐19, the management of the cytokine storm can reduce lung tissue damage and lead to a better outcome in COVID‐19 patients (Ye et al., 2020). Multiple strategies, such as the injection of immunomodulatory drugs and mesenchymal stem cells, have applied to inhibit lung damage and multiple‐organ failure in severe patients with cytokine storm. Also, targeting inflammatory cytokines could reduce the severity of the cytokine storm. Interventions that lead the immune response through Th2‐mediated immune responses are also among other options. These strategies have been discussed in further sections.
3.3.4. T cell exhaustion
T cells separated from COVID‐19 patients have shown to exhibit T cell exhaustion markers. PD‐1 and Tim‐3 are two T cell exhaustion markers that were reported to be highly expressed by T cells of the COVID‐19 patients. Since T cells are major cellular immunity cells that control the infection of SARS‐CoV‐2, the exhaustion of the T cells is associated with severe infection and poor antiviral immune response (Diao et al., 2020). Identification and inhibition of the mechanisms that contribute to exhaustion of the T cells could increase the efficacy of the immune response against SARS‐CoV‐2 and lead to improved clinical results.
4. IMMUNOTHERAPY OF COVID‐19
Similar to MERS and SARS, no specific treatment has been approved for COVID‐19. The first‐line treatment for COVID‐19 is supportive treatment, including oxygen therapy, mechanical ventilator support for patients with respiratory failure, antibiotics for prevention of secondary bacterial infection, and body fluid management (K. Xu et al., 2020). Also, some drugs have shown promising results in the treatment of COVID‐19. Since the outbreak of the COVID‐19, clinicians have started to assess the antiviral functions of existing drugs on this disease, and multiple preclinical and clinical trials have been launched (Dong et al., 2020; Stebbing et al., 2020).
Viral targeted inhibitors were among the first studied drugs. Adenosine‐analogs such as Remdisivir block the viral RNA synthesis process. Also, Remdesivir has shown promising results in the treatment of COVID‐19 patients in the clinical setting (Chang et al., 2020). Other nucleoside analogs such as ribavirin and favipiravir are among the antiviral drugs that could be effective in the treatment of COVID‐19 patients; however, no reports have been published about the efficacy of these drugs (Chang et al., 2020).
Immunotherapy uses the potentials of the patient's immune system to fight diseases. Immunotherapy has shown considerable results in the treatment of many diseases such as cancer (Tahmasebi et al., 2019) and viral infections (Boeckh & Corey, 2017). Amplification and reinforcement of the immune system using immune‐reinforcing material could have benefits in the treatment of COVID‐19.
Inadequate antiviral immune response and severe inflammation induced by dysfunctional immune response are the two main challenges in COVID‐19. Developing novel immune‐based therapeutics that target viral infection and dysfunctional immune response can improve the clinical outcome of patients with COVID‐19. This novel approach is named as “immunotherapy of COVID‐19,” and is among the novel treatments being developed. Multiple approaches can be administered as immunotherapeutic treatments for COVID‐19 (Figure 1), which are discussed below.
Figure 1.
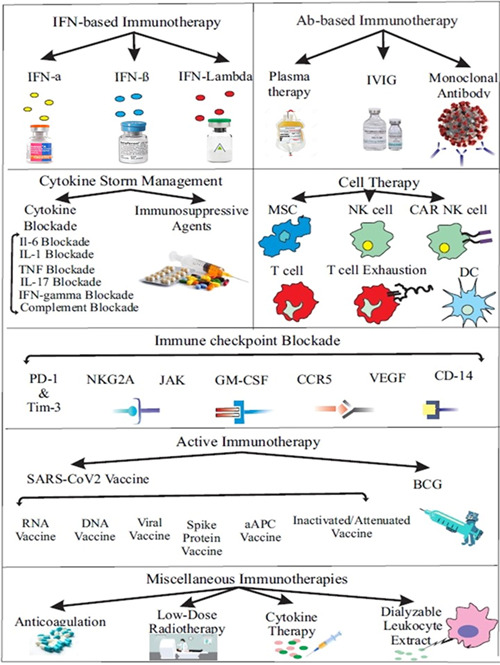
Immunotherapeutic approaches for COVID‐19. (Created by Esmaeilzadeh et al.) aAPC, artificial antigen‐presenting cell; BCG, Bacille Calmette–Guérin; CAR NK cell, chimeric antigen receptor natural killer cell; CCR5, CC chemokine receptor 5; CD14, cluster of differentiation 14; DC, dendritic cell; DNA, deoxyribonucleic acid; GM‐CSF, granulocyte‐macrophage colony‐stimulating factor; IFN‐γ, interferon‐γ; IL, interleukin; IVIG, intravenous immunoglobulin; JAK, Jasus kinase; MSC, mesenchymal stem cell; NK cell, natural killer cell; PD‐1, programmed death‐1; RNA, ribonucleic acid; Tim‐3, T‐cell immunoglobulin and mucin‐domain containing‐3; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor
4.1. Interferon‐based immunotherapy
4.1.1. Type I IFNs
Interferons, as components of the immune system, have a critical role in defending the body against viruses. SARS‐CoV‐2 can hinder the production of IFN‐1 by impairing IFN‐signaling pathways. This effect is conducted through the inhibition of the signal transducer and activator of transcription 1 (STAT‐1) and IRF‐3, which are intracellular pathways necessary for the production of the IFN (Vabret et al., 2020). IFN‐based immune responses are exerted against the virus; however, the threshold of the stimulation for the production of IFN is different based on age. The low‐fatality rate of the COVID‐19 in children has been attributed to the low IFN‐producing threshold of IFN, which causes the early induction of IFN, and can finally inhibit the SARS‐CoV‐2 infection. In contrast, the higher mortality rate in elderly patients is at least partly attributed to the higher threshold for IFN production, leading to the delay in IFN production and inadequate immune response. The inhibition of the IFN‐producing pathway by the virus and inadequate IFN‐production is one of the major challenges of the immune response against COVID‐19 (Mosaddeghi et al., 2020).
In an in vitro study, in comparison to many other pathogenic viruses, SARS‐CoV‐2 was more sensitive to treatment with IFN‐α and β (Mantlo et al., 2020). In the clinical setting, early tripple treatment of COVID‐19 with IFN‐β1b, lopinavir, and ritonavir, was reported to be more effective than lopinavir‐ritonavir alone, inhibit the infection more effectively, and reduce the progression of the disease to severe stages (Hung et al., 2020). According to in vitro and clinical results supporting the efficacy of treatment with type I IFNs, IFN‐based immunotherapy has been considered in clinical trials of COVID‐19 patients (Sallard et al., 2020). Multiple clinical trials are evaluating the efficacy of IFN‐α1β, IFN‐β, recombinant human IFN‐α, IFN‐β1a, and IFN‐β1b, in the treatment of the COVID‐19 in early stages. The clinical trials related to IFN‐based immunotherapy have been shown in Table 1.
Table 1.
Immunotherapeutic clinical trials on COVID‐19 registered in clinicaltrials.gov
| Treatment | Phase | Method/drug | NCT |
|---|---|---|---|
| Ab‐based immunotherapy | |||
| Convalescent plasma | 2/3 | Comparison of treatment with convalescent plasma and anti‐COVID19 human immunoglobulin | NCT04395170 |
| 2/3 | Hyperimmune (Convalescent) Plasma | NCT04385043 | |
| 3 | Convalescent plasma | NCT04372979 | |
| 2 | Convalescent plasma | NCT04345991 | |
| 3 | Convalescent plasma | NCT04372979 | |
| 1 | Convalescent plasma | NCT04353206 | |
| 2 | Convalescent plasma | NCT04343755 | |
| 2/3 | Convalescent plasma | NCT04388410 | |
| 1/2 | Convalescent plasma | NCT04384497 | |
| – | Immunoglobulin of cured patients + γ‐Globulin | NCT04264858 | |
| – | Anti‐corona VS2 immunoglobulins prepared from COVID19 convalescent Plasma | NCT04383548 | |
| – | Immunoglobulin of cured patients | NCT04264858 | |
| Monoclonal Ab | – | Generation of human monoclonal antibodies neutralizing SARS‐Cov‐2 from B cells of convalescent patients | NCT04354766 |
| IVIG | 3 | Polyvalent immunoglobulin (IVIG) | NCT04350580 |
| 2/3 | IVIG | NCT04261426 | |
| 3 | OCTAGAM 10% | NCT04400058 | |
| 1/2 | IVIG | NCT04521309 | |
| 2 | High‐dose IVIG | NCT04432324 | |
| 2 | High‐dose IVIG | NCT04480424 | |
| 4 | IVIG | NCT04411667 | |
| 3 | IVIG | NCT04500067 | |
| IFN‐based immunotherapy | |||
| IFN‐α | 1 | Recombinant human interferon α1β | NCT04293887 |
| 3 | Recombinant human interferon α‐1b | NCT04320238 | |
| 1/2 | Recombinant interferon α‐2b | NCT04379518 | |
| IFN‐β | 2 | Inhaled SNG001 (IFN‐β1) | NCT04385095 |
| 4 | IFNβ‐1A + lopinavir/ritonavir | NCT04350671 | |
| 2 | IFNβ‐1A + IFNβ‐1B + lopinavir/ritonavir + hydroxychloroquine | NCT04343768 | |
| 4 | Interferon‐β 1a + umifenovir + lopinavir/ritonavir + hydroxychloroquine | NCT04350684 | |
| 2 | Interferon β‐1B + hydroxychloroquine | NCT04350281 | |
| 2 | Interferon β‐1B + lopinavir/ritonavir + ribavirin | NCT04276688 | |
| IFN‐λ | 2 | Peginterferon IFN‐lambda | NCT04343976 |
| 2 | Peginterferon IFN‐lambda‐1A | NCT04354259 | |
| 2 | Peginterferon Lambda‐1A | NCT04388709 | |
| 2 | Peginterferon lambda α‐1a subcutaneous injection | NCT04344600 | |
| Cell therapy | |||
| MSC | 2/3 | MSCs | NCT04366063 |
| 1/2 | Umbilical Cord‐derived MSCs | NCT04333368 | |
| 2 | Hope Biosciences allogeneic adipose‐derived MSCs (HB‐adMSCs) | NCT04348435 | |
| 1/2 | MSCs | NCT04392778 | |
| 1/2 | Human embryonic stem cells derived M cells (CAStem) | NCT04331613 | |
| 1/2 | Allogenic pooled olfactory mucosa‐derived mesenchymal stem cells | NCT04382547 | |
| 1/2 | Bone marrow‐derived MSCs | NCT04346368 | |
| 1/2 | Allogeneic MSCs derived from adipose tissue | NCT04366323 | |
| 1 | Dental pulp‐derived MSCs | NCT04302519 | |
| – | CAP‐1002 allogeneic cardiosphere‐derived cells | NCT04338347 | |
| 1 | Inhalation of MSCs‐derived exosomes | NCT04276987 | |
| NK cell | 1/2 | Allogeneic NK cells | NCT04344548 |
| 1/2 | CYNK‐001 | NCT04365101 | |
| 1 | NK cells | NCT04280224 | |
| 1 | FT516 (an off‐the‐shelf cryopreserved NK cell product) | NCT04363346 | |
| T cell | – | Adoptive cellular therapy With SARS‐CoV‐2 specific T cells from convalescent donors | NCT04351659 |
| 1 | COVID‐19‐specific T Cell‐derived exosomes (CSTC‐Exo) | NCT04389385 | |
| 1/2 | Antigen‐specific CTLs + LV‐SMENP‐DC vaccine | NCT04276896 | |
| CAR NK cell | 1/2 | Universal Off‐the‐shelf NKG2D‐ACE2 CAR‐NK cells | NCT04324996 |
| DC | – | – | – |
| Macrophage | – | – | – |
| Management of cytokine storm | |||
| IL‐6 blockade | 2 | Clazakizumab | NCT04348500 |
| 4 | Tocilizumab | NCT04377750 | |
| 2 | Tocilizumab | NCT04370834 | |
| 1 | Sarilumab | NCT04386239 | |
| 2 | Sarilumab | NCT04357808 | |
| 2 | Thalidomide | NCT04273581 | |
| IL‐1 blockade | 2/3 | Emapalumab + anakinra | NCT04324021 |
| 2 | Tocilizumab + anakinra | NCT04339712 | |
| – | Canakinumab | NCT04348448 | |
| 2 | Anakinra | NCT04341584 | |
| IFN‐γ blockade | 2/3 | Emapalumab + anakinra | NCT04324021 |
| TNF blockade | 2 | XPro1595 | NCT04370236 |
| IL‐17 blockade | – | – | – |
| GM‐CSF blockade | 3 | Lenzilumab | NCT04351152 |
| 1/2 | TJ003234 | NCT04341116 | |
| 2 | Otilimab | NCT04376684 | |
| 2 | Gimsilumab | NCT04351243 | |
| 2 | Mavrilimumab | NCT04397497 | |
| 4 | Sargramostim | NCT04326920 | |
| Complement C5 | 4 | Baricitinib + ravulizumab | NCT04390464 |
| 2 | Eculizumab | NCT04346797 | |
| 2 | Avdoralimab (anti‐C5aR antibody) | NCT04371367 | |
| 2 | Zilucoplan | NCT04382755 | |
| Complement C3 | 2 | AMY‐101 (complement 3 inhibitor) | NCT04395456 |
| Immunosuppressive agents | 3 | Methylprednisolone pulse + tacrolimus | NCT04341038 |
| 2 | Fingolimod | NCT04280588 | |
| 2 | Sirolimus | NCT04341675 | |
| 2 | Ozanimod | NCT04405102 | |
| 2 | Tofacitinib | NCT04415151 | |
| 1/2 | Methotrexate‐loaded nanoparticles | NCT04352465 | |
| 2 | Low‐doses of melphalan inhalation | NCT04380376 | |
| Polymerase type I collagen | 1 | Collagen‐polyvinylpyrrolidone | NCT04517162 |
| Checkpoint inhibition | |||
| PD‐1 inhibition | 2 | PD‐1 blocking antibody | NCT04268537 |
| 2 | Nivolumab | NCT04356508 | |
| JAK inhibition | 4 | Baricitinib + ravulizumab | NCT04390464 |
| 1/2 | Ruxolitinib | NCT04334044 | |
| 2/3 | Baricitinib | NCT04320277 | |
| 2 | Ruxolitinib + simvastatin | NCT04348695 | |
| NKG2A inhibition | – | – | – |
| VEGF | 2 | Bevacizumab | NCT04344782 |
| – | Bevacizumab | NCT04305106 | |
| 2/3 | Bevacizumab | NCT04275414 | |
| CCR5 blockade | 2 | Leronlimab (PRO 140) | NCT04343651 |
| 2 | Leronlimab | NCT04347239 | |
| CD14 blockade | – | IC14 (monoclonal antibody against CD14) | NCT04346277 |
| – | IC14 (monoclonal antibody against CD14) | NCT04391309 | |
| Active immunotherapy | |||
| BCG | 4 | BCG vaccination | NCT04369794 |
| 3 | BCG vaccination | NCT04362124 | |
| 3 | BCG vaccination | NCT04379336 | |
| 3 | BCG vaccination | NCT04350931 | |
| 3 | BCG vaccination | NCT04327206 | |
| 3 | BCG vaccination | NCT04373291 | |
| DC vaccine | 1/2 | Vaccine consisting of autologous dendritic cells loaded with antigens from SARS‐CoV‐2 | NCT04386252 |
| 1/2 | Antigen‐specific CTLs + LV‐SMENP‐DC vaccine | NCT04276896 | |
| aAPC vaccine | 1 | Pathogen‐specific aAPC | NCT04299724 |
| RNA vaccine | 1 | mRNA‐1273 | NCT04283461 |
| ½ | BNT162a1 + BNT162b1 + BNT162b2 + BNT162c2 | NCT04368728 | |
| DNA vaccine | 1 | INO‐4800 | NCT04336410 |
| Viral vector vaccine | 2 | Recombinant novel coronavirus vaccine (Ad5‐nCoV) | NCT04341389 |
| ½ | Recombinant novel coronavirus vaccine (adenovirus type 5 vector) | NCT04398147 | |
| Inactivated/attenuated SARS‐CoV‐2 vaccine | ½ | Low‐dose Inactivated SARS‐CoV‐2 vaccine | NCT04383574 |
| Spike protein vaccine | 1 | Oral bacTRL‐Spike | NCT04334980 |
| Miscellaneous immunotherapies | |||
| VIP analog | 2 | IV Aviptadil | NCT04311697 |
| Low‐dose radiotherapy | – | Low dose anti‐inflammatory radiotherapy | NCT04380818 |
| Low‐dose IL‐2 | – | ILT101 (low‐dose IL‐2) | NCT04357444 |
| Dialyzable leukocyte extract | 2 | Dialyzable leukocyte extract | NCT04379479 |
| IL‐15 agonist | 1 | N‐803 (recombinant human superagonist IL‐15) | NCT04385849 |
| Anticoagulation | 3 | Therapeutic anticoagulation | NCT04362085 |
| 3 | Enoxaparin | NCT04359277 | |
| 2 | Enoxaparin | NCT04377997 | |
| 4 | Enoxaparin + heparin | NCT04367831 | |
| 2 | Tinzaparin + unfractionated Heparin | NCT04345848 | |
Abbreviations: γ‐globulin, γ‐globulin; aAPC, artificial antigen‐presenting cell; Ab‐based immunotherapy, antibody‐based immunotherapy; BCG, Bacille Calmette–Guérin; CAR NK cell, chimeric antigen receptor natural killer cell; CCR5, CC chemokine receptor 5; CD14, cluster of differentiation 14; COVID‐19, coronavirus disease 2019; DC, dendritic cell; GM‐CSF, granulocyte monocyte‐colony‐stimulating factor; IL, interleukin; IFN, interferon; IFN‐based immunotherapy, interferon‐based immunotherapy; peginterferon, pegylated interferon; IVIG, intravenous immunoglobulin; JAK, Janus kinase; MSC, mesenchymal stem cell; NK cell, natural killer cell; PD‐1, programmed‐death 1; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor; VIP, vasoactive intestinal peptide.
4.1.2. Type III IFNs
Also named as IFN‐λ (lambda), type III IFNs are involved in the antiviral immune response against viral infections. IFN‐λ triggers the Janus kinase (JAK)‐STAT signaling pathway, which finally activates inflammatory transcription factors that lead to the expression of IFN‐related genes. IFN‐λ has shown to induce a robust immune response in chronic viral hepatitis, leading to better clinical outcomes (Phillips et al., 2017). Considering COVID‐19, treatment with IFN‐λ could stimulate a stronger immune response against the virus. Thus, Peginterferon IFN‐λ, a form of IFN‐λ, has been used in COVID‐19‐treating clinical trials (Table 1).
At the first stages of the disease, IFNs are critical for inducing an adequate immune response against the virus. At late and severe stages, however, the main immune response against the virus is mediated by granulocytes, lymphocytes, monocytes, and macrophages. The uncontrolled activation of the monocytes and macrophages leads to the production of excess amounts of proinflammatory cytokines. Thus, in the IFN‐based treatment of the COVID‐19, the stage of the disease must be considered. Application of IFNs in the earlier stages of the COVID‐19 could induce a stronger antiviral response and inhibit the infection. However, at later stages of the disease, injection of the IFNs could induce a hyperactivated immune response and worsen the cytokine storm. Thus, severe patients are not candidates if immunotherapy and suppression of the excessive immune response through immunomodulatory approaches are indicated in these patients (Mosaddeghi et al., 2020).
4.2. Antibody‐based treatments
Inadequate anti‐SARS‐CoV antibody levels are associated with increased mortality and a high risk of recurrence (Jacofsky et al., 2020). Antibody‐based treatments are a kind of passive immunotherapy that can improve the immune response and inhibit SARS‐CoV‐2 infection in COVID‐19 patients (Long et al., 2020). In this section, antibody‐based immunotherapies for COVID‐19 have been described.
4.2.1. Convalescent plasma therapy
Passive immunity in COVID‐19 patients can be achieved by the infusion of the SARS‐CoV‐2 convalescent plasma from recovered patients. Convalescent plasma therapy has been previously used for the treatment of multiple diseases, such as influenza (X.‐X. Wu et al., 2015). It has also shown clinical benefits in patients with SARS (Cheng et al., 2005) and MERS (Ko et al., 2017). This approach has shown effective results in treating patients with acute and severe COVID‐19 patients, as well (Cheraghali et al., 2020; Focosi et al., 2020).
Convalescent plasma can inhibit the progression of the infection and reverse the inflammatory process in COVID‐19 patients by multiple mechanisms. First is the presence of neutralizing antibodies in the plasma. Neutralizing IgM/IgG antibodies, produced via a competent immune response in recovered patients, restrict the progression of the infection by inhibiting the entrance of the virus to the host cell and viral amplification, activating the complement system, inducing phagocytosis of the virus, and activating antibody‐dependent cellular toxicity (Rojas et al., 2020). Also, the convalescent plasma contains immune‐modulatory cytokines and autoantibodies that control the hyperinflammatory process and the cytokine storm (Bloch et al., 2020). Using these mechanisms, convalescent plasma restricts viral infection and modulates the cytokine storm, which finally improves the respiratory function and outcome in COVID‐19 patients.
In a study, Shen et al. (2020) reported the injection of donated convalescent plasma of recovered COVID‐19 patients to five patients with severe COVID‐19 with respiratory failure. The patients received concomitant antiviral therapy and IFN‐therapy, as well. Plasma‐treated patients were reported to have a normal temperature and improved respiratory function after 1 week. Also, 1–12 days following infusion, the respiratory samples were reported to be negative regarding SARS‐CoV‐2 RT‐PCR.
In another study, 10 severe COVID‐19 patients with positive RT‐PCR received 200 ml of convalescent plasma. The titer of the neutralizing antibodies was greater than 1:640. Compared to the control group, the patients who received convalescent plasma had improved clinical symptoms, reduced lung lesions, and increased lymphocyte count. Also, seven patients who received convalescent plasma had undetectable viral RNA 7 days after infusion (Duan et al., 2020). Despite promising results of treatment with convalescent plasma in COVID‐19 patients, larger scale clinical trials with a higher number of patients are still required to define the dosage and efficacy of this approach in the treatment of COVID‐19. Another possible application of the convalescent plasma could be the prophylaxis with convalescent plasma in high‐risk populations. This could reduce the risk of infection and mortality, especially in comorbid patients (Sridevi et al., 2020).
United States Food and Drug Administration (FDA) has already approved the infusion of convalescent plasma for the treatment of severely ill COVID‐19 patients (Tanne, 2020); however, multiple challenges still need to be concerned. Low antibody levels in some recovered patients, limited access to large doses of convalescent plasma for large groups of patients, the possibility of adverse reactions after the infusion, and the donor‐dependent nature of the antibody subtypes, are the most important challenges that need further considerations. Some clinical trials have been launched to investigate the efficacy of convalescent plasma therapy in COVID‐19 patients which are shown in Table 1.
4.2.2. Monoclonal antibody
Monoclonal antibodies (mAbs) are a group of antibodies that are specifically produced against a specific epitope of an antigen by a specific group of B cells. mAbs are used for the treatment of multiple diseases, including cancers and infectious diseases (Shanmugaraj et al., 2020). Also, treatment with mAb against previous coronaviruses has shown clinical efficacy in SARS and MERS patients (Modjarrad, 2016). Since the S1 subunit has an important role in the immunogenicity of coronaviruses, most mAbs target the S1 subunit. Considering SARS, the application of SARS‐CoV‐specific neutralizing mAbs, m396 and CR3022, has shown promising results (Prabakaran et al., 2009). Since the RBD of the SARS‐CoV and SARS‐CoV‐2 are extremely identical, Tian et al. (2020) thought to study the efficacy of CR3022 in COVID‐19 patients. In this study, CR3022, a SARS‐CoV‐specific neutralizing mAb, was reported to bind the SARS‐CoV‐2 RBD and inhibit it significantly. In contrast, m396 and CR3014, which are among other SARS‐CoV‐specific mAbs, failed to bind to the SARS‐CoV‐2‐spike protein. In conclusion, this study reported the potent cross‐efficacy of CR3022 in inhibition of SARS‐CoV‐2, alone, or in combination with other treatments (Tian et al., 2020).
SARS‐CoV‐2‐specific neutralizing antibodies are also under investigation and primary results have been published in the preclinical setting (Ju et al., 2020; Wang, Li et al., 2020). Further research is still required to translate the production and application of the monoclonal neutralizing antibodies to the clinical setting. To achieve this, recent knowledge of mAb therapy for SARS and MERS should be taken into consideration. Moreover, further studies are going to define whether SARS‐CoV‐2‐specific monoclonal neutralizing antibodies could be applied to prevent the infection in high‐risk patients.
4.2.3. Intravenous immunoglobin
Intravenous immunoglobin (IVIG) is a biological product that is produced by gathering polyclonal IgGs from the serum of hundreds of donors. IVIG is routinely used for the treatment of multiple systemic inflammatory diseases, such as autoimmune thrombocytopenic purpura and Kawasaki disease (Jawhara, 2020). Several mechanisms contribute to the anti‐inflammatory effects of IVIG treatment. Examples are the inhibition of Fcγ receptors, neutralizing cytokines and antibodies, suppressing the activation of lymphocytes, and inhibiting the function of the complement system (X. Li et al., 2020). In a recent study, Cao et al. (2020) investigated the therapeutic efficacy of high‐dose IVIG in three patients with severe COVID‐19 pneumonia. This study reported the application of high‐dose IVIG, at a dosage of 0.3–0.5g/kg daily, and the repetition of injection for 5 days, to significantly improve the fever and dyspnea in the patients. Also, no adverse effects were observed in any of the patients (Cao et al., 2020).
4.3. Management of the cytokine storm
The cytokine storm is the main cause that induces the progression of mild inflammation to ARDS and multiple‐organ failure in COVID‐19. The activation of the immune cells and the release of inflammatory cytokines is the underlying mechanism leading to severe inflammation in the cytokine storm. Management of the cytokine storm can improve respiratory failure in severe cases with hyperinflammation. This can be achieved using two approaches. First is the inhibition of the inflammatory mechanisms. To inhibit the inflammatory pathways, one could block the pathways of the inflammatory cytokines. Second is the administration of immunosuppressive drugs in severely ill patients with cytokine storm (Bhaskar et al., 2020). Here, we discuss different approaches to manage the cytokine storm in COVID‐19.
4.3.1. Blockade of the cytokine pathways
In response to the cytokines, chemokines, and immune receptors expressed by infected cells and alveolar macrophages, inflammatory immune cells such as macrophages, neutrophils, and monocytes infiltrate into the respiratory tissue. In most cases, the recruitment of these immune cells leads to the clearance of the pathogen through a mild immune response. However, in severe cases, these inflammatory immune cells exert an intense immune response that leads to the uncontrolled production of inflammatory cytokines, which is known as the cytokine storm. IL‐1β, IL‐6, and TNF‐α are the most important cytokines that contribute to the cytokine storm (Tanaka et al., 2016).
4.3.1.1. IL‐6 blockade
The excessive uncontrolled production of IL‐6 induces ARDS, leads to the destruction of the alveoli membrane, and causes hemorrhage in the lung. These events finally lead to pulmonary fibrosis. Higher serum IL‐6 levels are associated with the severity of the COVID‐19 infection. Therefore, blocking IL‐6 could reverse the inflammatory mechanism and reduce the severity of COVID‐19. Tocilizumab is an anti‐IL‐6 mAb that can inhibit the inflammatory pathway induced by IL‐6. The application of tocilizumab has shown promising clinical benefits in some COVID‐19 patients (B. Liu et al., 2020). Tocilizumab inhibits the destructive effects of the SARS‐COV‐2 on the respiratory system; however, this drug does not have a direct antiviral function on SARS‐CoV‐2. Similar to other immune‐restraining drugs, uncontrolled or unnecessary application of tocilizumab could be associated with some adverse effects, including an increase in hepatic enzymes, hypercholesterolemia, skin allergy, and opportunistic fungal infections (Mehta et al., 2020). Tocilizumab (ACTEMRA) and Sarilumab are two IL‐6 mAbs that have been approved to be studied on COVID‐19 patients in phase III clinical trial by the FDA (https://www.cancernetwork.com/news/fda-approves-phase-iii-clinical-trialtocilizumab-COVID-19-pneumonia; Cascella et al., 2020). Clazakizumab is another anti‐IL6 mAb that is under investigation in current clinical trials (Table 1).
4.3.1.2. IL‐1 blockade
IL‐1β is a proinflammatory cytokine that has an important role in the progression of respiratory inflammation in many viral infections (Conti et al., 2020). In response to alveolar infection by SARS‐CoV‐2, macrophages secrete IL‐1β, which induces fever and stimulates the mechanisms that lead to respiratory fibrosis in the lung. Higher levels of IL‐1β are associated with the severity of COVID‐19 infection in critically ill patients (Price et al., 2020). Thus, inhibition of the IL‐1β in severe stages of the disease could be an effective approach to reduce the progression of the cytokine release syndrome and ARDS. Anakinra is a recombinant IL‐1 receptor antagonist that has been considered in the clinical trial of COVID‐19 (NCT04341584). Canakinumab is another mAb against IL‐1β that has been proposed to be investigated in the COVID‐19 clinical trials (Table 1).
4.3.1.3. TNF‐α blockade
TNF‐α is an inflammatory mediator that is produced by the innate immune cells in response to the SARS‐CoV‐2. TNF‐α is among the first cytokines that are produced by the innate immune cells in COVID‐19 and has shown to have an important role in the induction of the immune response against coronaviruses. This effect is conducted by inducing the recruitment of the leukocytes to the infection site and differentiation of the DCs (Price et al., 2020). According to the role of the TNF‐α in the progression of the cytokine storm, inhibiting TNF‐α could be considered as a promising approach to reduce the hyperinflammation in severe COVID‐19 infection. XPro1595 is a soluble TNF‐α‐neutralizing protein that inhibits the interaction between soluble TNF‐α and its receptor. XPro1595 has shown appropriate clinical results in chronic inflammatory disorders, such as Alzheimer's disease, and is being investigated in the clinical trial of COVID‐19 (Table 1).
4.3.1.4. IL‐17 blockade
IL‐17 is an inflammatory cytokine that has shown to have a significant role in the progression of the ARDS in multiple in vivo and clinical studies (Crowe et al., 2009). Produced by innate lymphoid cells, IL‐17 contributes to the excessive inflammation in the lung by inducing the activation of Th17 cells, excretion of proinflammatory mediators, and stimulating the recruitment of inflammatory neutrophils (Muir et al., 2016). No studies have reported the role of IL‐17 in the progression of the COVID‐19‐induced pulmonary inflammation; however, similar to the ARDS due to other inflammatory conditions, IL‐17 might be contributing to ARDS and lung tissue damage in COVID‐19 (Q. Li et al., 2016). Thus, inhibiting IL‐17 could be introduced as a potential approach to inhibit the progression of ARDS and lung tissue damage in COVID‐19 (Pacha et al., 2020).
4.3.1.5. IFN‐γ blockade
IFN‐γ is a soluble cytokine and a member of the type II interferons. Produced by NK, CD4+, and CD8+ lymphocytes, IFN‐γ exerts an important role in the immune response against viruses. However, the excessive production of IFN‐γ promotes the progression of severe inflammation and lung tissue damage (L. Chen et al., 2018; Meissner et al., 2010). The inhibition of IFN‐γ has previously shown to alleviate acute lung injury in H1N1 influenza in in vivo studies (B. Liu et al., 2019). In clinical studies, IFN‐γ has shown to be an important biomarker promoting the pathogenesis of ARDS and acute lung injury (Spadaro et al., 2019). Thus, the inhibition of IFN‐γ, using anti‐IFN‐γ antibodies, could be considered as an immunotherapeutic method to inhibit the progression of the ARDS in severe COVID‐19 infection. An example is the application of Emapalumab, an anti‐IFN‐γ Ab, for the treatment of COVID‐19 in a clinical trial (NCT04324021).
4.3.1.6. Complement blockade
As members of the innate immune system, complements are inactive proteins in the serum. After being activated, the complement system assists the antimicrobial function of the immune system and stimulates the activation of neutrophils. Although complements are not cytokines, some members of the complement system have some immune functions that are similar to cytokines. Through these mechanisms, the complement system exerts an important role in the progression of the inflammation (Risitano et al., 2020). C3 and C5 are two major members of the complement system. Inhibition of the complement system, using avdoralimab, zilucoplan, and ravulizumab, as C5 inhibitors, and AMY‐101, as a C3 inhibitor, has been considered in the clinical trials of COVID‐19 (Table 1).
4.3.2. Immunosuppressive/cytotoxic agents
Considering the immunomodulatory effects of corticosteroids, dexamethasone, and other oral/IV corticosteroids, were among the first drugs that were proposed for the treatment of cytokine storm in severe COVID‐19. Corticosteroids exert their anti‐inflammatory effect by inhibiting the expression of proinflammatory transcription factors in the nuclei of inflammatory cells (Ramesh et al., 2015). The application of dexamethasone is not indicated in mild to moderate patients; however, in severe COVID‐19 infection with hyperinflammatory status, the injection of corticosteroids could alleviate the inflammation and ARDS (Azimi et al., 2020). Multiple clinical trials are investigating the efficacy of treatment with steroids in severe COVID‐19, which are described in Table 1. As immune suppressor drugs, corticosteroid application is accompanied by some known side effects, such as vascular necrosis and diabetes, which must be taken into consideration (Tang et al., 2020).
4.4. Active immunotherapy
The antibody‐based immunity against SARS‐CoV‐2 can be achieved by both passive and active immunotherapeutic methods. Plasma therapy and treatment with IVIG are two examples of passive immunotherapy. Considering the challenges of treatment with passive immunotherapy, such as limited resources and high cost, active immunotherapy using antiviral vaccines has been considered from the first steps of the pandemic (Roback & Guarner, 2020). Here, we aim to discuss the recent progress in the production of SARS‐CoV‐2‐specific vaccines as novel immunotherapeutic approaches.
4.4.1. SARS‐CoV‐2 vaccination
Viral vaccines are bioproducts that can induce active immunity by introducing the virus‐specific antigen, inactivated viral antigen, virus‐specific APCs, adenoviral vectors, viral DNA/RNA vaccine, or attenuated virus, to the immune system. Soon after the COVID‐19 outbreak, multiple studies have started to design SARS‐CoV‐2‐specific vaccines. Most vaccine‐based studies have targeted the spike (S) glycoprotein of the virus (C. Chen et al., 2020). Here, we aim to review different kinds of vaccine design against SARS‐CoV‐2.
4.4.1.1. RNA vaccines
RNA vaccines include viral RNA/mRNA, carried by a vector, that is, lipid nanoparticles, to the host body. After transduction, mRNA induces the expression of the viral antigen in the target cells. The expression of the viral antigen stimulates the adaptive immune system to produce neutralizing antibodies against the target antigen. mRNA vaccines have shown promising results against infectious diseases and malignancies in previous studies and were among the first vaccines being developed during the COVID‐19 pandemic. Multiple drug‐developing countries have started to design RNA vaccines for COVID‐19. Anti‐SARS‐CoV‐2 RNA vaccines could be produced to encode multiple antigens, such as the spike (S) protein and RBD. Despite great immune potentials, RNA vaccines could be associated with adverse effects, as well. By inducing the production of type I IFNs, RNA vaccines can lead to autoimmune disorders. Also, since RNA vaccines contain a non‐human polynucleotide, they can stimulate an unintended immune response which can lead to severe immune reactions and undesirable outcomes.
mRNA‐1273 is a biological product that is composed of the lipid nanoparticles (LNPs) as the vector and mRNA‐1273, as the viral antigen producing mRNA. mRNA‐1273 encodes the viral spike (S) protein. Expression of the viral spike protein induces the production of spike‐neutralizing antibodies by adaptive immune cells. A clinical trial has been launched that has been designed to investigate the efficacy of this vaccine in preventing COVID‐19 (NCT04283461). Moreover, BNT162a1, BNT162b1, BNT162b2, and BNT162c2, are nucleoside modified mRNAs that can induce the expression of the viral RBD. Carried by an LNP vector, these modified mRNAs express the SARS‐CoV‐2‐RBD, which leads to the secretion of RBS‐neutralizing antibodies from the adaptive immune cells. A clinical trial has been designed to investigate the safety, immunogenicity, and efficacy of these mRNA vaccines in COVID‐19 (NCT04368728).
4.4.1.2. DNA vaccine
DNA vaccines include an antigen‐expressing plasmid, and a vector responsible for carrying the plasmid into the host cell. The inserted plasmid is genetically engineered to encode a pathogen‐specific antigen. The expression of the pathogen‐specific antigen activates the adaptive immune system and leads to the production of antigen‐specific antibodies. DNA vaccines have been studied in a wide range of infectious diseases and have also been considered in the COVID‐19 pandemic. Some preclinical studies are designed to develop DNA vaccines against SARS‐CoV‐2. INO‐4800 is a synthetic DNA vaccine against SARS‐CoV‐2 that is being studied in phase I clinical trial. This clinical trial has enrolled 40 healthy volunteers to be injected with two doses of INO‐4800 within four weeks (NCT04336410). Being genetically engineered, DNA vaccines are also associated with some limitations and side effects. Only protein antigens can be used as immunogens in DNA vaccines. Also, the possibility of anti‐DNA antibody production and developing dysplasia by inducing mutations in the host genome, are two examples of the adverse effects accompanied by DNA vaccines.
4.4.1.3. Spike protein vaccine
Spike (S) protein is the main immunogenic protein of the SARS‐CoV‐2 structure. Similar to SARS, the spike protein seems to be an ideal target for developing antigen‐based vaccines against SARS‐CoV‐2 (L. Du et al., 2009). Also, called subunit vaccines, the Spike protein vaccines are capable of inducing a strong immune response against the virus. Spike protein vaccines have shown promising potential to induce a virus‐specific immune response in SARS (McPherson et al., 2016). Multiple preclinical and clinical studies are investigating the immunogenicity and efficacy of spike protein vaccine design against SARS‐CoV‐2.
4.4.1.4. Viral live vaccine
Live vector vaccines use a virus as the platform vector that contains a genetic sequence encoding the viral antigen. Live nonproliferative viral vectors can be constructed using retroviruses, lentiviruses, adenoviruses, and adeno‐associated viruses. The live virus is capable of integrating its genome into the host cell genome, which finally leads to the expression of the target antigen and stimulation of the adaptive immune response against it (Amanat & Krammer, 2020). This approach has been used to design vaccines against SARS‐CoV‐2, which is named as Ad5‐nCoV vaccine. To the best of our knowledge, two clinical and several preclinical studies have been started to evaluate the safety and efficacy of the live vector vaccines against SARS‐CoV‐2 (NCT04341389, NCT04341389).
4.4.1.5. Artificial APC vaccine
Artificial APCs (aAPCs) contribute to the antiviral immune response by introducing the pathogen‐associated antigen to the adaptive immune cells. The presentation of the antigen triggers the adaptive immune response against the pathogen. aAPCs are lentivirally modified APCs that express a pathogen‐specific structural and/or protease protein. The expression of the virus‐associated proteins leads to the activation of the T cells and the induction of a strong immune response against the virus (H. Li et al., 2017). The artificial APC vaccine design has been considered for the development of the anti‐SARS‐CoV‐2 vaccine in preclinical and clinical studies (L. Huang et al., 2020).
4.4.1.6. Inactivated/attenuated vaccine
Inactivated/attenuated viral vaccines are among the oldest methods for vaccine design. This kind of vaccine has been successfully designed for different viral infections, including influenza and measles. Inactivated/attenuated vaccine includes the injection of the inactivated/attenuated virus/viral particles to the host. The injection induces diverse immune responses against the virus and thus can trigger strong immunity against the virus (Nachbagauer et al., 2018). Several institutions have launched related research to study this kind of vaccine in preclinical and clinical studies (Pandey et al., 2020).
4.4.2. Bacille Calmette–Guérin vaccine
Bacille Calmette–Guérin (BCG) is a routinely used vaccine for protection against tuberculosis. In addition to the long‐time active immunization against tuberculosis, BCG vaccination has shown nonspecific immune response against other diseases, such as viral upper respiratory infections. Possibly, this protection is exerted by a nonspecific innate immune response to BCG (Miller et al., 2020). To investigate the probable association between BCG vaccination and COVID‐19 mortality, Shet et al. (2020) used a simple log‐linear regression model. This study demonstrated that BCG‐vaccinating countries had shown much lower COVID‐19‐associated mortality in comparison with non‐BCG‐using countries (Shet et al., 2020). These data demonstrated the protective role of BCG vaccination against COVID‐19 through a nonspecific immune response. Thus, BCG vaccination could be introduced as nonspecific immunotherapy to induce nonspecific innate immune response against COVID‐19; however, further research is required to evaluate the efficacy of this approach. Several clinical trials have been started to compare the infection and mortality in BCG‐vaccinated individuals compared to non‐vaccinated individuals (Table 1).
4.5. Cell therapy
Cell therapy is an immunotherapeutic approach that has recently gained much attention for the treatment of viral respiratory infections, as well as cancers (Elahi et al., 2018). Showing therapeutic efficacy against other viral respiratory infections, cell therapy has been developed as an attractive treatment strategy in COVID‐19 (J. Du et al., 2020). In this section, we describe different cell therapy methods developed for the treatment of COVID‐19.
4.5.1. Mesenchymal stem cell therapy
Mesenchymal stem cells (MSCs) are a group of stem cells that possess intense immunomodulatory effects. Due to their immunomodulatory impacts, MSCs are widely utilized in the treatment of multiple chronic inflammatory diseases (Marofi et al., 2017). Also, MSCs possess regenerative effects on target tissue by producing multiple growth factors, such as glial cell line‐derived neurotrophic factor, vascular endothelial growth factor (VEGF), hepatocyte growth factor (HGF), and keratinocyte growth factor (KGF; Hofer & Tuan, 2016). MSCs exert their immunomodulatory effects by inhibiting the production of proinflammatory cytokines, including IFN‐γ, TNF‐α, IL‐6, and IL‐12, suppressing the overactivation of immune cells including macrophages and T lymphocytes, and stimulating regulatory T cells and M2 macrophages. By inhibiting proinflammatory cytokines and immune cells, MSCs can inhibit the cytokine storm and its harmful effects on the lung tissue. Also, by producing IL‐10 and multiple growth factors, such as HGF and KGF, MSCs improve lung tissue repair and inhibit alveolar fibrosis (J. Chen et al., 2020).
MSC therapy has previously shown to reduce influenza‐associated acute lung injury in multiple in vivo and clinical studies (J. Chen et al., 2020; Y. Li et al., 2016). The immunomodulatory effects of MSCs, along with the previously published data on the clinical efficacy and safety of MSC therapy on viral respiratory infections, have suggested the application of MSCs in the treatment of acute lung injury in severe COVID‐19 patients (Golchin et al., 2020).
Considering the harmful effects of the cytokine storm and uncontrolled immune response in severe COVID‐19 patients, Leng et al. (2020) transplanted ACE2‐negative MSCs to 7 COVID‐19‐pneumonia patients in Beijing YouAn Hospital, China. The patients were followed‐up for 14 days after injection. This study reported the symptoms and pulmonary function of the patients to be remarkably ameliorated 2 days after injection. One of the patients had severe COVID‐19 that was significantly improved and was discharged 10 days after injection. Also, after transplantation of MSCs, the serum levels of the inflammatory markers were decreased, the number of inflammatory cytokine‐secreting cells was decreased, and the number of peripheral lymphocytes were increased. In conclusion, this study reported the transplantation of the ACE2‐negative MSCs to be a safe and effective treatment approach for COVID‐19. Other ongoing clinical trials are designed to study the safety and efficacy of MSC‐transplantation in COVID‐19‐associated pneumonia patients (Table 1).
Another MSC‐therapy method is using MSC‐derived secretome. The MSCs secrete their extracellular components to the cell culture media via extracellular vesicles and exosomes, which are named as secretome. The secretome of the MSCs includes MSC‐derived cytokines and growth factors. MSC‐derived secretome has shown to have similar MSC‐associated results, including immunomodulatory, regenerative, and anti‐inflammatory effects (Bari et al., 2020). Thus, similar to the transplantation of the MSCs, treatment with MSC‐derived secretomes has also been suggested as a potential therapeutic approach in COVID‐19. The secretome can be administered in two ways, first is the inhaled spray form of the secretome, and the second is the intravenous prescription (Deffune et al., 2020). Based on this, some clinical trials have been launched to assess the safety and efficacy of MSC‐derived secretome in the treatment of COID‐19 (Table 1).
4.5.2. NK cell therapy
NK cells are a subset of lymphocytes and members of the innate immunity. NK cells are among the first immune components contributing to the initial immune response against virus‐infected and cancer cells (Hammer et al., 2018). After being activated by macrophage‐derived cytokines and type I IFNs, NK cells attack the virus‐infected cells. The most specific characteristic of the NK‐derived immune response is its MHC‐independency and rapidity (Paul & Lal, 2017). Since the COVID‐19 outbreak, scientists have considered the application of NK cells as a potential novel treatment for COVID‐19. Multiple clinical trials have been designed to assess the therapeutic efficacy of COVID‐19 treatment with NK cells that are described in Table 1.
4.5.3. Chimeric antigen receptor NK cell therapy
Chimeric antigen receptor (CAR) is a genetically engineered receptor that is widely applied in the treatment of multiple cancers (Tahmasebi et al., 2019). Engineered cells expressing CAR can specifically target the antigen‐expressing cells. Since NK cells have an important role in the antiviral immune response against SARS‐CoV‐2, engineered CAR NK cells have been suggested as a novel approach for the treatment of COVID‐19. ACE‐2 is the antigen that could be utilized for designing CAR NK cells against SARS‐CoV‐2. NCT04324996 is a phase I/II clinical trial that has been started to assess the therapeutic efficacy of universal Off‐the‐shelf NKG2D‐ACE2 CAR NK cells in the treatment of COVID‐19 pneumonia.
4.5.4. SARS‐CoV‐2‐specific T cells and T cell‐derived exosomes
T cells are the major members of the adaptive immunity that have an important role in eliminating SARS‐CoV‐2 infection. Cytotoxic T cells and helper T cells exert their antiviral immune response after being recruited to the lung tissue. T cells could be applied to induce a stronger immune response in COVID‐19 patients (L. Zhu et al., 2020). This concept is named as “adoptive cell therapy of COVID‐19,” an emerging field of immunotherapy in COVID‐19. Adoptive cell therapy could be applied using many approaches. One method is the transplantation of the SARS‐CoV‐2‐specific T cells from convalescent donors. Since convalescent donors have overcome the COVID‐19 infection, the T cells separated from them have higher antiviral potential against SARS‐CoV‐2 (Bachanova et al., 2020). Thus, the transplanted virus‐specific T cells from treated donors could eliminate SARS‐CoV‐2 more specifically and aggressively.
Another approach is to transfer antigen‐specific cytotoxic T lymphocytes. This approach includes exposing the T cells to viral antigens in the culture media and transferring the virus‐specific T cells to the COVID‐19 patients. The T cells could either be autologous (separated from the patients), or allogeneic (separated from other individuals). The viral‐antigen cultured T cells can recognize the viral antigen faster and lead to a faster and stronger immune response against SARS‐CoV‐2 (Tu et al., 2020).
Similar to MSCs, the virus‐specific T cell culture media contain T cell‐derived extracellular exosomes that include antiviral materials (Golchin et al., 2020). The T cell‐derived extracellular exosomes could improve the antiviral immune response by activating immune cells (Fu et al., 2019). Thus, SARS‐CoV‐2‐specific T cell‐derived exosome transfer is a novel therapeutic method for the treatment of COVID‐19. Corresponding clinical trials have been launched to investigate the therapeutic efficacy of adoptive cell therapy in COVID‐19, which have been described in Table 1.
4.5.5. Inhibiting T cell exhaustion
Lymphopenia is one of the most common laboratory signs in COVID‐19. In addition to the reduction in the number of cytotoxic CD8+ T cells, two studies have also reported the exhaustion of the T cells in COVID‐19 patients. PD‐1 is an exhaustion marker of the T cells. The higher levels of PD‐1 expression by T cells is associated with higher exhaustion of these cells. Exhausted T cells have a reduced antiviral function and thus have lower antiviral potency. Also, Tim‐3, another T cell exhaustion surface marker, is highly expressed by the T cells in COVID‐19 patients (Diao et al., 2020). From this standpoint, some clinical trials have targeted the inhibition of PD‐1 and Tim‐3 in the treatment of COVID‐19. Clinical trials investigating the efficacy of PD‐1 blockade in COVID‐19 patients are described in Table 1. NKG2A, an exhaustion marker expressed by NK cells and cytotoxic T lymphocytes, has been reported to be upregulated in severe COVID‐19 patients. The upregulated expression of NKG2A reduces the production of proinflammatory cytokine by NK cells and T lymphocytes (Zheng et al., 2020). Inhibition of the NKG2A could also be considered in the treatment of the COVID‐19 patients, which has been discussed in further sections.
By producing immune‐stimulating cytokines (CD4+ cells) and attacking the virus‐infected cells (CD8+) cells, T cells exert their immune response against SARS‐CoV‐2 (Bernheim et al., 2020). Similar to CAR‐engineered NK cells, T cells engineered to express CAR (CAR T cells) could also be suggested to be applied in the treatment of COVID‐19‐associated pneumonia. Although no clinical trials have been designed, the antiviral potential of the CAR T cell therapy could open a promising window in the treatment of COVID‐19 and might be considered in further clinical trials.
4.5.6. DC therapy
As members of the innate immune system, type 1 DCs (DC‐1) exert their antiviral immune response by producing IL‐6 and type I IFN, and acting as APCs (Vabret et al., 2020). DCs also activate the NK cells by expressing NKG2D (Draghi et al., 2007). Moreover, the hypersecretion of IL‐6 by DC‐1 cells is considered as one of the important mechanisms that contribute to the progression of respiratory inflammation and lung tissue damage in ARDS (Rajaei & Dabbagh, 2020). To manage the hyperinflammatory process in severe COVID‐19 patients, it could be useful to inhibit the proinflammatory effects of DCs, either by application of DC‐blocking agents or using engineered DCs (Lega et al., 2020).
4.5.7. Macrophage therapy
Two types of macrophages have been identified. Type 1 macrophages (M1) with proinflammatory functions, and type 2 macrophages (M2) with anti‐inflammatory properties. In COVID‐19, M1 macrophages contribute to the severe inflammation by secreting proinflammatory cytokines, such as IL‐6 and IL‐1β (Merad & Martin, 2020). To suppress the hyperinflammatory condition, macrophages can be modified in two ways. The first method is the modulation of the M1 macrophages to secrete lower levels of proinflammatory cytokines. The second approach could be the application of M2 macrophages to suppress the inflammation of the lungs (Lega et al., 2020). Considering the application of macrophages for the treatment of COVID‐19 could be considered in further research.
4.6. Immune checkpoint inhibition
Two main challenges reduce the optimal antiviral immune response of the lymphocytes in COVID‐19. First is lymphocytopenia which is decrease in the number of T and NK cells. Second is the exhaustion of T cells which is identified by overexpression of NKG2A, PD‐1, and Tim‐3, on NK cells and cytotoxic T lymphocytes (Diao et al., 2020; Zheng et al., 2020). Immune checkpoint inhibition is a developing approach that has been widely studied in the concept of cancer treatment (Decazes & Bohn, 2020). Here, we aim to review recent data on checkpoint inhibition as a developing method for the treatment of COVID‐19.
4.6.1. PD‐1 and Tim‐3 inhibition
PD‐1 and Tim‐3 are surface markers of T cell exhaustion expressed by exhausted T lymphocytes (Luo et al., 2020). Previous studies have reported the upregulated expression of PD‐1 and Tim‐3 on T lymphocytes in COVID‐19 ICU‐admitted patients compared to non‐ICU patients. Thus, the reduction in the number of lymphocytes (lymphopenia) is accompanied by the upregulated expression of PD‐1 and Tim‐3 on residual T lymphocytes in severe COVID‐19 cases (Diao et al., 2020). Inhibition of the PD‐1 could reduce the exhaustion of the lymphocytes and increase the immune response potency in COVID‐19 patients (López‐Collazo et al., 2020). Therefore, inhibition of PD‐1 has been considered in the clinical trial design in COVID‐19 that has been described in Table 1.
Higher levels of proinflammatory cytokines have been shown to be associated with higher PD‐1 and Tim‐3 expression by T cells in severe COVID‐19 cases. This supports the role of the cytokine storm in the exhaustion of T lymphocytes (Moon, 2020). Therefore, management of the cytokine storm not only reduces lung tissue damage, but also leads to an improved immune response by limiting T cell exhaustion.
4.6.2. NKG2A inhibition
NKG2A is an immune‐inhibitory receptor expressed by NK cells and cytotoxic T lymphocytes. The higher expression of NKG2A is associated with lower immune functionality of the NK and cytotoxic T cells, which is conducted by reducing the capability of these cells to secrete granzymes and release cytokines (Antonioli et al., 2020). Considering the immune‐suppressive state in tumors, inhibiting NKG2A has shown to overcome the immune resistance by increasing the immune function of the NK cells (Kamiya et al., 2019). Serum samples separated from COVID‐19 patients have also shown upregulated expression of NKG2A by NK and cytotoxic T cells. The upregulated expression of NKG2A was associated with lower secretion of TNF‐α, IL‐2, IFN‐γ, granzyme B, and CD107a (Zheng et al., 2020). As a result, the upregulated expression of NKG2A is sought to have an important role in the compromised immune response against SARS‐CoV‐2 in COVID‐19 patients. Therefore, inhibition/downregulation of NKG2A could reduce the exhaustion of T cells and produce a stronger immune response by T and NK cells.
4.6.3. JAK inhibition
JAK is a member of the intracellular cytokine signaling pathway, named as the JAK‐STAT pathway. JAK stimulates the production of proinflammatory cytokines by mediating the phosphorylation of the STAT family. Phosphorylation of the STAT leads to the production of several proinflammatory cytokines, including IL‐6. The uncontrolled production of the proinflammatory cytokines leads to severe inflammation and cytokine storm, which causes severe damage to the lung and develops multiorgan failure. Accordingly, inhibition of the JAK could reduce the production of proinflammatory cytokines and can thus be suggested in the treatment of the cytokine storm in severe COVID‐19 patients (Favalli et al., 2020).
Several JAK‐inhibitors could be used in the treatment of COVID‐19. Recent literature has introduced Baricitinib, a JAK inhibitor, as a potential treatment for acute viral pneumonia in COVID‐19. As well as reducing inflammation by inhibiting JAK, Barticinib limits the entrance of the virus to the host cells by inhibiting adaptor‐associated protein kinase 1 receptor (Richardson et al., 2020). Thus, barticinib could be introduced as a therapeutic agent to be studied in further clinical trials of COVID‐19.
At the first stages of COVID‐19, SARS‐CoV‐2‐associated reduction in type I IFN is mediated by the inhibition of the JAK‐STAT pathway. Thus, time must be noted in the prescription of JAK‐inhibitors. Treatment with JAK‐inhibitors in the first stages can worsen the infection by reducing the production of type I IFN and limiting its antiviral potency. However, treatment with JAK‐inhibitors in severe cases with cytokine could be beneficial by reducing proinflammatory cytokines (Fleming, 2016). Currently, some clinical trials have been launched to evaluate the efficacy of JAK inhibitors in the treatment of COVID‐19 that have been discussed in Table 1.
4.6.4. GM‐CSF inhibition
GM‐CSF is a glycoprotein secreted by monocytes, macrophages, and Th1 cells. GM‐CSF acts on the bone marrow and stimulates the production of granulocytes (Merad & Martin, 2020). It also increases the migration of the neutrophils and monocytes to the inflammatory site. GM‐CSF, in companion with IL‐6, contributes to the progression of the severe inflammation during ARDS. The inhibition of GM‐CSF has previously shown to improve ARDS (Goodman et al., 1999). From this standpoint, inhibiting GM‐CSF could also reduce the severity of ARDS and lung tissue damage in COVID‐19. Multiple clinical trials have considered GM‐CSF blockade using lenzilumab, otilimab, gimsilumab, mavrilimumab, and sargramostim, as a potential therapeutic method in COVID‐19 (Table 1).
4.6.5. CCR5 inhibition
C‐C chemokine receptor type 5 (CCR5) is a chemokine receptor for CCL3, CCL4, CCL5, and CCL3L1. CCR5 is expressed by multiple leukocytes, including macrophages, T cells, and DCs (Wei & Nielsen, 2019) and has shown to have an important role in the direction of the leukocytes to the inflammatory site. Therefore, CCR5 is one of the major checkpoints promoting inflammation in COVID‐19 (Merad & Martin, 2020). Inhibiting CCR5 is one of the recently proposed methods to inhibit the severe inflammation in COVID‐19. To the best of our knowledge, a CCR5‐specific mAb, lenrulimab, is being used to inhibit CCR5 in two recent clinical trials (NCT0434365 and NCT04347239).
4.6.6. VEGF inhibition
VEGF is a subset of the growth factor family that contributes to the angiogenesis, formation of new vessels in the tissue, and induction of the migration and mitosis of the endothelial cells (Niu & Chen, 2010). Recent studies have reported the serum levels of VEGF to be increased in severe COVID‐19 patients (X. Yang et al., 2020). VEGF contributes to the progression of acute lung injury by increasing the permeability of the vessels that leads to respiratory edema, a major factor in ARDS. Thus, VEGF is a potent therapeutic target in ARDS and acute lung injury in COVID‐19. Clinical trials have astarted to investigate the therapeutic efficacy of VEGF‐inhibition, using a VEGF‐specific mAb, Bevacizumab (Table 1).
4.6.7. CD14 inhibition
CD14 is expressed on the surface of the macrophages and monocytes. As part of the innate immune system, CD14 contributes to the recognition of the PAMPs. Since macrophages play a major role in the inflammatory process, inhibition of CD14 attenuates macrophage‐associated signaling pathways, and suppresses inflammation (da Silva et al., 2017). Thus, CD14 could be considered as a potential therapeutic target in COVID‐19‐associated inflammation and ARDS. IC14, an anti‐CD14 mAb, can appropriately inhibit CD14, and thus, has been used to inhibit CD14 in corresponding clinical trials (Table 1).
4.7. Miscellaneous immunotherapies
4.7.1. Anticoagulation
There is a proven interaction between inflammation and coagulation. An example is the increased risk of diffuse intravascular coagulation in severe sepsis. Chronic severe inflammation increases the chance of developing intravascular thrombosis. Proinflammatory cytokines, mostly IL‐6, stimulate coagulation markers such as the tissue factor, and suppress anticoagulatory factors (Levi & van der Poll, 2010), which lead to the increased production of thrombin and fibrin. On the other hand, coagulatory factors can increase inflammation by acting on specific cell receptors. Coagulopathy has also shown to have an important role in the progression of multiple‐organ failure by causing small thrombosis in different organs (Levi & van der Poll, 2005). Considering the two‐way crosstalk between coagulation and inflammation, inhibiting coagulation could reduce mortality in COVID‐19 by decreasing the risk of intravascular small thrombosis, as well as attenuating the severe inflammation. Prophylaxis of coagulation is being studied in COVID‐19 clinical trials (Table 1).
4.7.2. Anti‐inflammatory radiotherapy
Low‐dose radiotherapy has shown to have anti‐inflammatory effects and is being used for the treatment of multiple chronic inflammatory disorders (Arenas et al., 2012). Low‐dose ionizing radiation exerts its anti‐inflammatory effect by acting on the endothelial cells. Ionizing radiation also increases the adhesion of the monocytes to the wall of the vessels (Schröder et al., 2018). In conclusion, low‐dose radiotherapy could be used to suppress the inflammation in COVID‐19. Corresponding clinical trials regarding anti‐inflammatory radiation therapy of COVID‐19 have been described in Table 1.
4.7.3. Immune‐stimulating cytokine therapy
Immune‐enhancing therapies can improve the antiviral immune response in the early stages of the infection with SARS‐CoV‐2. IL‐2 is an immune‐stimulating cytokine that is produced by the helper and cytotoxic T cells. IL‐2 exerts its immunogenic effect by inducing the differentiation of the T cells to effector and memory cells (Buchbinder et al., 2019). The application of IL‐2 could increase the immune system potential to fight infections, such as COVID‐19. Correspondingly, the therapeutic efficacy of low‐dose IL‐2 is being investigated in a clinical trial on COVID‐19 patients (NCT04357444).
IL‐15 is another member of the cytokine family that shares high structural similarity with IL‐2 (Berger et al., 2019). IL‐15 has an important role in the immune response against viral infections. Following infection with the virus, IL‐15 is secreted by mononuclear leukocytes. IL‐15 induces the differentiation, activation, and proliferation of the antiviral NK cells. Induction of the IL‐15 could induce a robust immune response against SARS‐CoV‐2 (Knudson et al., 2019). N‐803 is a recombinant human IL‐15 superagonist that is being investigated in a clinical trial in COVID‐19 (NCT04385849).
4.7.4. Dialyzable leukocyte extract
Dialyzable leukocyte extract (DLE) is a mixture of several low‐molecular peptides that are produced from released cytokines of disrupted leukocytes (Castrejón Vázquez et al., 2019). DLE has shown to have anti‐inflammatory and immunomodulatory effects in several studies (Pérez‐Alvarado et al., 2017). To suppress the hyperinflammatory status in COVID‐19, DLE could be injected into the patients. Thus, the immunomodulatory effect of DLE in the cytokine storm is being investigated in a clinical trial (NCT04379479).
In attention to the potential capabilities of immunotherapy against SARS‐CoV‐2, the development of novel immune‐based approaches could have a critical role in fighting viral epidemics, such as COVID‐19. Despite promising advances, further studies are required to develop and evaluate the efficacy of immunotherapeutic methods against COVID‐19 and other viral epidemics.
5. CONCLUSION
In this review, we have discussed the mechanisms underlying COVID‐19 immunopathogenesis and immune response against SARS‐CoV‐2. Understanding the immunopathogenesis of the COVID‐19 is critical to control the infection. As well as viral infection, the management of the inflammatory response is vital to inhibit the progression of ARDS in severe cases. Targeting viral infection by inhibiting the restriction of type I interferon‐producing pathways is an example of controlling the COVID‐19 infection. Also, modulation of the severe inflammatory response by using immunomodulatory approaches is among the methods introduced for inflammatory response management. The potentials of immunotherapy could help to overcome the challenges and develop efficient treatments for COVID‐19 patients. Moreover, the induction of active immunity by viral vaccines can inhibit the spread of the virus among communities, especially in patients with severe comorbidities. Further research is still required to determine the host‐viral interaction and the efficacy of current immunotherapeutic treatments for COVID‐19.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Abdolreza Esmaeilzadeh contributed to the hypothesis, data gathering, final manuscript edition, and scientific support for this review. Reza Elahi contributed to data gathering, preparing the primary manuscript, manuscript edition, and table/figure design.
ACKNOWLEDGMENTS
The authors would like to acknowledge COVID‐19 patients and health workers (physicians, nurses, laboratory staff, etc.) around the world for their precious efforts fighting COVID‐19.
Esmaeilzadeh A, Elahi R. Immunobiology and immunotherapy of COVID‐19: A clinically updated overview. J Cell Physiol. 2021;236:2519–2543. 10.1002/jcp.30076
REFERENCES
- Amanat, F. , & Krammer, F. (2020). SARS‐CoV‐2 vaccines: Status report. Immunity, 52, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonioli, L. , Fornai, M. , Pellegrini, C. , & Blandizzi, C. (2020). NKG2A and COVID‐19: Another brick in the wall. Cellular & Molecular Immunology, 17, 672–674. 10.1038/s41423-020-0450-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas, M. , Sabater, S. , Hernández, V. , Rovirosa, A. , Lara, P. , Biete, A. , & Panes, J. (2012). Anti‐inflammatory effects of low‐dose radiotherapy. Strahlentherapie und Onkologie, 188(11), 975–981. [DOI] [PubMed] [Google Scholar]
- Azimi, S. , Sahebnasagh, A. , Sharifnia, H. , & Najmeddin, F. (2020). Corticosteroids administration following COVID‐19‐induced acute respiratory distress syndrome. Is it harmful or life‐saving? Advanced Journal of Emergency Medicine, 4(2s):e43. [Google Scholar]
- Bachanova, V. , Bishop, M. R. , Dahi, P. , Dholaria, B. , Grupp, S. A. , Hayes‐Lattin, B. , Janakiram, M. , Maziarz, R. T. , McGuirk, J. P. , Nastoupil, L. J. , Oluwole, O. O. , Perales, M. A. , Porter, D. L. , Riedell, P. A. , & CAR T‐cell Consortium (2020). CAR T cell therapy during the COVID‐19 pandemic. Biology of Blood and Marrow Transplantation, 26, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari, E. , Ferrarotti, I. , Saracino, L. , Perteghella, S. , Torre, M. L. , & Corsico, A. G. (2020). Mesenchymal stromal cell secretome for severe COVID‐19 infections: Premises for the therapeutic use. Cells, 9(4):924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, A. , Colpitts, S. J. , Seabrook, M. S. S. , Furlonger, C. L. , Bendix, M. B. , Moreau, J. M. , McKillop, W. M. , Medin, J. A. , & Paige, C. J. (2019). Interleukin‐15 in cancer immunotherapy: IL‐15 receptor complex versus soluble IL‐15 in a cancer cell‐delivered murine leukemia model. Journal for Immunotherapy of Cancer, 7(1):355. 10.1186/s40425-019-0777-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim, A. , Mei, X. , Huang, M. , Yang, Y. , Fayad, Z. A. , Zhang, N. , Diao, K. , Lin, B. , Zhu, X. , Li, K. , Li, S. , Shan, H. , Jacobi, A. , & Chung, M. (2020). Chest CT findings in coronavirus disease‐19 (COVID‐19): Relationship to duration of infection. Radiology, 295, 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar, S. , Sinha, A. , Banach, M. , Mittoo, S. , Weissert, R. , Kass, J. S. , Rajagopal, S. , Pai, A. R. , & Kutty, S. (2020). Cytokine storm in COVID‐19—Immunopathological mechanisms, clinical considerations, and therapeutic approaches: The REPROGRAM Consortium position paper. Frontiers in Immunology, 11, 1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, E. M. , Shoham, S. , Casadevall, A. , Sachais, B. S. , Shaz, B. , Winters, J. L. , van Buskirk, C. , Grossman, B. J. , Joyner, M. , Henderson, J. P. , Pekosz, A. , Lau, B. , Wesolowski, A. , Katz, L. , Shan, H. , Auwaerter, P. G. , Thomas, D. , Sullivan, D. J. , Paneth, N. , … Tobian, A. A. (2020). Deployment of convalescent plasma for the prevention and treatment of COVID‐19. The Journal of Clinical Investigation, 130(6), 2757–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckh, M. , & Corey, L. (2017). Adoptive immunotherapy of viral infections: Should infectious disease embrace cellular immunotherapy? Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Buchbinder, E. I. , Dutcher, J. P. , Daniels, G. A. , Curti, B. D. , Patel, S. P. , Holtan, S. G. , Miletello, G. P. , Fishman, M. N. , Gonzalez, R. , Clark, J. I. , Richart, J. M. , Lao, C. D. , Tykodi, S. S. , Silk, A. W. , & McDermott, D. F. (2019). Therapy with high‐dose Interleukin‐2 (HD IL‐2) in metastatic melanoma and renal cell carcinoma following PD1 or PDL1 inhibition. Journal for Immunotherapy of Cancer, 7(1):49. 10.1186/s40425-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, W. , Liu, X. , Bai, T. , Fan, H. , Hong, K. , Song, H. , Han, Y. , Lin, L. , Ruan, L. , & Li, T. (2020). High‐dose intravenous immunoglobulin as a therapeutic option for deteriorating patients with coronavirus disease 2019. Open Forum Infectious Diseases, 7(3):ofaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M. , Rajnik, M. , Cuomo, A. , Dulebohn, S. C. , & Di Napoli, R. (2020). Features, evaluation and treatment coronavirus (COVID‐19). StatPearls Publishing. [PubMed] [Google Scholar]
- Castrejón Vázquez, M. I. , Reséndiz‐Albor, A. A. , Ynga‐Durand, M. A. , Martínez, A. , Maciel, I. , Orellana‐Villazon, V. I. , García López, C. A. , Laue Noguera, M. L. , & Vargas Camaño, M. E. (2019). Dialyzable leukocyte extract (Transferon™) administration in sepsis: Experience from a single referral pediatric intensive care unit. BioMed Research International, 2019, 8980506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.‐W. , Yuan, S. , Kok, K.‐H. , To, K. K.‐W. , Chu, H. , Yang, J. , Xing, F. , Liu, J. , Yip, C. C‐Y. , Poon, R. W.‐S. , Tsoi, H. W. , Lo, S. K.‐F. , Chan, K.‐H. , Poon, V. K.‐M. , Chan, W. M. , Ip, J.D. , Cai, J.‐P. , Cheng, V. C.‐C. , Chen, H. , Hui, C. K.‐M. , & Yuen, K.‐Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. , Tung, Y. , Lee, K. , Chen, T. , Hsiao, Y. , Chang, H. , Hsieh, T. , Su, C. , Wang, S. , Yu, J. , Shih, S. , Lin, Y. , Lin, Y. , Tu, Y.E. , Tung, C. , & Chen, C. (2020). Potential therapeutic agents for COVID‐19 based on the analysis of protease and RNA polymerase docking. Preprints, 2020020242. 10.20944/preprints202002.0242.v1 [DOI] [Google Scholar]
- Chen, L. , Deng, H. , Cui, H. , Fang, J. , Zuo, Z. , Deng, J. , Li, Y. , Wang, X. , & Zhao, L. (2018). Inflammatory responses and inflammation‐associated diseases in organs. Oncotarget, 9(6):7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Hu, C. , Chen, L. , Tang, L. , Zhu, Y. , Xu, X. , Chen, L. , Gao, H. , Lu, X. , Yu, L. , Dai, X. , Xiang, C. , & Li, L. (2020). Clinical study of mesenchymal stem cell treatment for acute respiratory distress syndrome induced by epidemic Influenza A (H7N9) infection: A hint for COVID‐19 treatment. Engineering. 10.1016/j.eng.2020.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Zhang, X. , Ju, Z. , & He, W. (2020). Advances in the research of cytokine storm mechanism induced by Corona Virus Disease 2019 and the corresponding immunotherapies. Zhonghua shao shang za zhi = Zhonghua shaoshang zazhi = Chinese journal of burns, 36, E005. [DOI] [PubMed] [Google Scholar]
- Chen, N. , Zhou, M. , Dong, X. , Qu, J. , Gong, F. , Han, Y. , Qiu, Y. , Wang, J. , Liu, Y. , Wei, Y. , Xia, J. , Yu, T. , Zhang, X. , & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Wong, R. , Soo, Y. O. , Wong, W. S , Lee, C. K. , Ng, M. H. , Chan, P. , Wong, K. C. , Leung, C. B. , & Cheng, G. (2005). Use of convalescent plasma therapy in SARS patients in Hong Kong. European Journal of Clinical Microbiology and Infectious Diseases, 24(1), 44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheraghali, A. M. , Abolghasemi, H. , & Eshghi, P. (2020). Management of COVID‐19 virus infection by convalescent plasma. Iranian Journal of Allergy, Asthma and Immunology, 19(S1). [DOI] [PubMed] [Google Scholar]
- Chiappelli, F. , Khakshooy, A. , & Greenberg, G. (2020). CoViD‐19 Immunopathology and Immunotherapy. Bioinformation, 16(3), 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, P. , Ronconi, G. , Caraffa, A. , Gallenga, C. , Ross, R. , Frydas, I. , & Kritas, S. (2020). Induction of pro‐inflammatory cytokines (IL‐1 and IL‐6) and lung inflammation by Coronavirus‐19 (COVI‐19 or SARS‐CoV‐2): Anti‐inflammatory strategies. Journal of Biological Regulators and Homeostatic Agents, 34(2), 327–331. [DOI] [PubMed] [Google Scholar]
- Crowe, C. R. , Chen, K. , Pociask, D. A. , Alcorn, J. F. , Krivich, C. , Enelow, R. I. , Ross, T. M. , Witztum, J. L. , & Kolls, J. K. (2009). Critical role of IL‐17RA in immunopathology of influenza infection. The Journal of Immunology, 183(8), 5301–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decazes, P. , & Bohn, P. (2020). Immunotherapy by immune checkpoint inhibitors and nuclear medicine imaging: Current and future applications. Cancers, 12(2):371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffune, E. , Prudenciatti, A. , & Moroz, A. (2020). Mesenchymal stem cell (MSc) secretome: A possible therapeutic strategy for intensive‐care COVID‐19 patients. Medical Hypotheses, 142, 109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao, B. , Wang, C. , Tan, Y. , Chen, X. , Liu, Y. , Ning, L. , Chen, L. , Li, M. , Liu, Y. , Wang, G. , Yuan, Z. , Feng, Z. , Zhang, Y. , Wu, Y. , & Chen, Y. (2020). Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID‐19). Frontiers in Immunology, 11, 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L. , Hu, S. , & Gao, J. (2020). Discovering drugs to treat coronavirus disease 2019 (COVID‐19). Drug Discoveries & Therapeutics, 14(1), 58–60. [DOI] [PubMed] [Google Scholar]
- Draghi, M. , Pashine, A. , Sanjanwala, B. , Gendzekhadze, K. , Cantoni, C. , Cosman, D. , Moretta, A. , Valiante, N. M. , & Parham, P. (2007). NKp46 and NKG2D recognition of infected dendritic cells is necessary for NK cell activation in the human response to influenza infection. The Journal of Immunology, 178(5), 2688–2698. [DOI] [PubMed] [Google Scholar]
- Du, L. , He, Y. , Zhou, Y. , Liu, S. , Zheng, B.‐J. , & Jiang, S. (2009). The spike protein of SARS‐CoV—a target for vaccine and therapeutic development. Nature Reviews Microbiology, 7(3), 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J. , Li, H. , Lian, J. , Zhu, X. , Qiao, L. , & Lin, J. (2020). Stem cell therapy: A potential approach for treatment of influenza virus and coronavirus‐induced acute lung injury. Stem Cell Research Therapy, 11(1):192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, K. , Liu, B. , Li, C. , Zhang, H. , Yu, T. , Qu, J. , Zhou, M. , Chen, L. , Meng, S. , Hu, Y. , Peng, C. , Yuan, M. , Huang, J. , Wang, Z. , Yu, J. , Gao, X , Wang, D , Yu, X , Li, L. , … Yang, X. (2020). Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proceedings of the National Academy of Sciences of the United States of America, 117(17), 9490–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi, R. , Khosh, E. , Tahmasebi, S. , & Esmaeilzadeh, A. (2018). Immune cell hacking: Challenges and clinical approaches to create smarter generations of chimeric antigen receptor T cells. Frontiers in Immunology, 9, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus, D. , Degenhardt, F. , Bujanda, L. , Buti, M. , Albillos, A. , Invernizzi, P. , Fernandez, J. , Prati, D. , Baselli, G. , Asselta, R. , Grimsrud, M. M. , Milani, C. , Aziz, F. , Kassens, J. , May, S. , Wendorff, M. , Wienbrandt, L. , Uellendahl‐Werth, F. , Zheng, T. , … Karlsen, T. H. (2020). The ABO blood group locus and a chromosome 3 gene cluster associate with SARS‐CoV‐2 respiratory failure in an Italian‐Spanish genome‐wide association analysis. medRxiv. 10.1101/2020.05.31.20114991 [DOI] [Google Scholar]
- Favalli, E. G. , Biggioggero, M. , Maioli, G. , & Caporali, R. (2020). Baricitinib for COVID‐19: A suitable treatment? The Lancet Infectious Diseases, 20, 1012–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, S. B. (2016). Viral inhibition of the IFN‐induced JAK/STAT signalling pathway: Development of live attenuated vaccines by mutation of viral‐encoded IFN‐antagonists. Vaccines, 4(3):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florindo, H. F. , Kleiner, R. , Vaskovich‐Koubi, D. , Acúrcio, R. C. , Carreira, B. , Yeini, E. , Tiram, G. , Liubomirski, Y. , & Satchi‐Fainaro, R. (2020). Immune‐mediated approaches against COVID‐19. Nature Nanotechnology, 15, 630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focosi, D. , Tang, J. , Anderson, A. , & Tuccori, M. (2020). Convalescent plasma therapy for COVID‐19: State of the art. Preprints, 2020040097. 10.20944/preprints202004.0097.v1 [DOI] [PMC free article] [PubMed]
- Fu, W. , Lei, C. , Liu, S. , Cui, Y. , Wang, C. , Qian, K. , Li, T. , Shen, Y. , Fan, X. , Lin, F. , Ding, M. , Pan, M. , Ye, X. , Yang, Y. , & Hu, S. (2019). CAR exosomes derived from effector CAR‐T cells have potent antitumour effects and low toxicity. Nature Communications, 10(1):4355. 10.1038/s41467-019-12321-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golchin, A. , Seyedjafari, E. , & Ardeshirylajimi, A. (2020). Mesenchymal stem cell therapy for COVID‐19: Present or future. Stem Cell Reviews and Reports, 16(3), 427–433. 10.1007/s12015-020-09973-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, E. , Stricker, P. , Velavicius, M. , Fonseca, R. , Kleinstein, E. , Lavery, R. , Deitch, E. A. , Hauser, C. J. , & Simms, H. (1999). Role of granulocyte‐macrophage colony‐stimulating factor and its receptor in the genesis of acute respiratory distress syndrome through an effect on neutrophil apoptosis. Archives of Surgery, 134(10), 1049–1054. [DOI] [PubMed] [Google Scholar]
- Hammer, Q. , Rückert, T. , & Romagnani, C. (2018). Natural killer cell specificity for viral infections. Nature Immunology, 19(8), 800–808. [DOI] [PubMed] [Google Scholar]
- Harrison, C. (2010). Calming the cytokine storm. Nature Reviews Drug Discovery, 9(5), 360–361. [DOI] [PubMed] [Google Scholar]
- Hofer, H. R. , & Tuan, R. S. (2016). Secreted trophic factors of mesenchymal stem cells support neurovascular and musculoskeletal therapies. Stem Cell Research & Therapy, 7(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue, M. L. , DeBolt, C. , Lindquist, S. , Lofy, K. H. , Wiesman, J. , Bruce, H. , Spitters, C. , Ericson, K. , Wilkerson, S. , Tural, A. , Diaz, G. , Cohn, A. , Fox, L. , Patel, A. , Gerber, S.I. , Kim, L. , Tong, S. , Lu, X. , Lindstrom, S. , … Washington State 2019‐nCoV Case Investigation Team (2020). First case of 2019 novel coronavirus in the United States. New England Journal of Medicine, 382(10), 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. , Rong, Y. , Pan, Q. , Yi, K. , Tang, X. , Zhang, Q. , Wang, W. , Wu, J. , & Wang, F. (2020). SARS‐CoV‐2 vaccine research and development: Conventional vaccines and biomimetic nanotechnology strategies. Asian Journal of Pharmaceutical Sciences. 10.1016/j.ajps.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , Zhang, L. , Fan, G. , Xu, J. , Gu, X. , Cheng, Z. , Yu, T. , Xia, J. , Wei, Y. , Wu, W. , Xie, X. , Yin, W. , Li, H. , Liu, M. , … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, I. F.‐N. , Lung, K.‐C. , Tso, E. Y.‐K. , Liu, R. , Chung, T. W.‐H. , Chu, M.‐Y. , Ng, Y.‐Y. , Lo, J. , Chan, J. , Tam, A. R. , Shum, H.‐P. , Chan, V. , Wu, A.K.‐L. , Sin, K.M. , Leung, W.‐S. , Law, W.‐L. , Lung, D. C. , Sin, S. , Yeung, P. , … Yuen, K.‐Y. (2020). Triple combination of interferon beta‐1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID‐19: An open‐label, randomised, phase 2 trial. The Lancet, 395, 1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, Y. , Kuba, K. , Rao, S. , Huan, Y. , Guo, F. , Guan, B. , Yang, P. , Sarao, R. , Wada, T. , Leong‐Poi, H. J. N. , Crackower, M. A. , Fukamizu, A. , Hui, C.‐C. , Hein, L. , Uhlig, S. , Slutsky, A. S. , Jiang, C. , & Penninger, J. M. (2005). Angiotensin‐converting enzyme 2 protects from severe acute lung failure. Nature, 436(7047), 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacofsky, D. , Jacofsky, E. M. , & Jacofsky, M. (2020). Understanding Antibody Testing for COVID‐19. The Journal of Arthroplasty, 35, S74–S81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhara, S. (2020). Could intravenous immunoglobulin collected from recovered coronavirus patients protect against COVID‐19 and strengthen the immune system of new patients? International Journal of Molecular Sciences, 21(7):2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, X. , Yin, C. , Lu, S. , Chen, Y. , Liu, Q. , Bai, J. , & Lu, Y. (2020). Two things about COVID‐19 might need attention. Preprints. 10.20944/preprints202002.0315.v1 [DOI] [Google Scholar]
- Ju, B. , Zhang, Q. , Ge, X. , Wang, R. , Yu, J. , Shan, S. , Zhou, B. , Song, S. , Tang, X. , Yu, J. , Ge, J. , Lan, J. , Yuan, J. , Wang, H. , Zhao, J. , Zhang, S. , Wang, Y. , Shi, X. , Liu, L. , … Zhang, L. (2020). Potent human neutralizing antibodies elicited by SARS‐CoV‐2 infection. bioRxiv. 10.1101/2020.03.21.990770 [DOI] [PubMed] [Google Scholar]
- Jung, S.‐M , Akhmetzhanov, A. R. , Hayashi, K. , Linton, N. M. , Yang, Y. , Yuan, B. , Kobayashi, T. , Kinoshita, R. , & Nishiura, H. (2020). Real‐time estimation of the risk of death from novel coronavirus (covid‐19) infection: Inference using exported cases. Journal of Clinical Medicine, 9(2):523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, T. , Seow, S. V. , Wong, D. , Robinson, M. , & Campana, D. (2019). Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. The Journal of Clinical Investigation, 129(5), 2094–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson, K. M. , Hicks, K. C. , Alter, S. , Schlom, J. , & Gameiro, S. R. (2019). Mechanisms involved in IL‐15 superagonist enhancement of anti‐PD‐L1 therapy. Journal for Immunotherapy of Cancer, 7(1):82. 10.1186/s40425-019-0551-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J.‐H. , Müller, M. , Seok, H. , Park, G. E. , Lee, J. Y. , Cho, S. Y. , Ha, Y. E. , Baek, J. Y. , Kim, S. H. , Kang, J.‐M. , Kim, Y.‐J , Jo, I. J. , Chung, C. R. , Hahn, M.‐J. , Drosten, C. , Kang, C.‐I. , Chung, D. R. , Song, J.‐H. , Kang, E.‐S. , … Peck, K. R. (2017). Suggested new breakpoints of anti‐MERS‐CoV antibody ELISA titers: Performance analysis of serologic tests. European Journal of Clinical Microbiology & Infectious Diseases, 36(11), 2179–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba, K. , Imai, Y. , & Penninger, J. M. (2006). Angiotensin‐converting enzyme 2 in lung diseases. Current Opinion in Pharmacology, 6(3), 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Nyodu, R. , Maurya, V. K. , & Saxena, S. K. (2020). Host immune response and immunobiology of human SARS‐CoV‐2 infection. Coronavirus Disease 2019 (COVID‐19): Epidemiology, Pathogenesis, Diagnosis, and Therapeutics (pp. 43–53). 10.1007/978-981-15-4814-7_5 [DOI] [Google Scholar]
- Lauer, S. A. , Grantz, K. H. , Bi, Q. , Jones, F. K. , Zheng, Q. , Meredith, H. R. , Azman, A. S. , Reich, N. G. , & Lessler, J. (2020). The incubation period of coronavirus disease 2019 (COVID‐19) from publicly reported confirmed cases: Estimation and application. Annals of Internal Medicine, 172, 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lega, S. , Naviglio, S. , Volpi, S. , & Tommasini, A. J. V. (2020). Recent insight into SARS‐CoV2 immunopathology and rationale for potential treatment and preventive strategies in COVID‐19. Vaccines, 8, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, Z. , Zhu, R. , Hou, W. , Feng, Y. , Yang, Y. , Han, Q. , Shan, G. , Meng, F. , Wang, S. , Fan, J. , Wang, W. , Deng, L. , Shi, H. , Li, H. , Hu, Z. , Zhang, F. , Gao, J. , Liu, H. , Li, X. , … Zhao, R. C. (2020). Transplantation of ACE2‐Mesenchymal Stem Cells Improves the Outcome of Patients with COVID‐19 Pneumonia. Aging and Disease, 11(2), 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi, M. , & van der Poll, T. (2005). Two‐way interactions between inflammation and coagulation. Trends in Cardiovascular Medicine, 15(7), 254–259. [DOI] [PubMed] [Google Scholar]
- Levi, M. , & van der Poll, T. (2010). Inflammation and coagulation. Critical Care Medicine, 38, S26–S34. [DOI] [PubMed] [Google Scholar]
- Li, X. , Geng, M. , Peng, Y. , Meng, L. , & Lu, S. (2020). Molecular immune pathogenesis and diagnosis of COVID‐19. Journal of Pharmaceutical Analysis, 10, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Gu, Y. , Tu, Q. , Wang, K. , Gu, X. , & Ren, T. (2016). Blockade of interleukin‐17 restrains the development of acute lung injury. Scandinavian Journal of Immunology, 83(3), 203–211. [DOI] [PubMed] [Google Scholar]
- Li, W. , Moore, M. J. , Vasilieva, N. , Sui, J. , Wong, S. K. , Berne, M. A. , Somasundaran, M. , Sullivan, J. L. , Luzuriaga, K. , Greenough, T. C. , Choe, H. , & Farzan, M. (2003). Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Shao, S. , Cai, J. , Burner, D. , Lu, L. , Chen, Q. , Minev, B. , & Ma, W. (2017). Artificial human antigen‐presenting cells are superior to dendritic cells at inducing cytotoxic T‐cell responses. Immunology, 152(3), 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Xu, J. , Shi, W. , Chen, C. , Shao, Y. , Zhu, L. , Lu, W. , & Han, X. (2016). Mesenchymal stromal cell treatment prevents H9N2 avian influenza virus‐induced acute lung injury in mice. Stem Cell Research & Therapy, 7(1):159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. , Jeon, S. , Shin, H.‐Y. , Kim, M. J. , Seong, Y. M. , Lee, W. J. , Choe, K. W. , Kang, Y. M. , Lee, B. , & Park, S.‐J. (2020). Case of the index patient who caused tertiary transmission of Coronavirus disease 2019 in Korea: The application of lopinavir/ritonavir for the treatment of COVID‐19 pneumonia monitored by quantitative RT‐PCR. Journal of Korean Medical Science, 35(6):e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Bao, L. , Wang, L. , Li, F. , Wen, M. , Li, H. , Deng, W. , Zhang, X. , & Cao, B. (2019). Anti‐IFN‐γ therapy alleviates acute lung injury induced by severe influenza A (H1N1) pdm09 infection in mice. Journal of Microbiology, Immunology and Infection. 10.1016/j.jmii.2019.07.009 [DOI] [PubMed] [Google Scholar]
- Liu, B. , Li, M. , Zhou, Z. , Guan, X. , & Xiang, Y. (2020). Can we use interleukin‐6 (IL‐6) blockade for coronavirus disease 2019 (COVID‐19)‐induced cytokine release syndrome (CRS)? Journal of Autoimmunity, 111, 102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Liu, Y. , Xiang, P. , Pu, L. , Xiong, H. , Li, C. , Zhang, M. , Tan, J. , Xu, Y. , Song, R. , Song, M. , Wang, L. , Zhang, W. , Han, B. , Yang, L. , Wang, X. , Zhou, G. , Zhang, T. , Li, B. , … Wang, X. (2020). Neutrophil‐to‐lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. Journal of Translational Medicine, 1. 10.1186/s12967-020-02374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, Q.‐X. , Liu, B.‐Z. , Deng, H.‐J. , Wu, G.‐C. , Deng, K. , Chen, Y.‐K. , Liao, P. , Qiu, J.‐F. , Lin, Y. , Cai, X.‐F. , Wang, D.‐Q. , Hu, Y. , Ren, J.‐H. , Tang, N. , Xu, Y.‐Y. , Yu, L.‐H. , Mo, Z. , Gong, F. , Zhang, X.‐L. , … Huang, A.‐L. (2020). Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nature Medicine, 26, 845–848. [DOI] [PubMed] [Google Scholar]
- López‐Collazo, E. , Avendaño‐Ortiz, J. , Martín‐Quirós, A. , & Aguirre, L. A. (2020). Immune response and COVID‐19: A mirror image of Sepsis. International Journal of Biological Sciences, 16(14), 2479–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Rizvi, H. , Egger, J. V. , Preeshagul, I. R. , Wolchok, J. D. , & Hellmann, M. D. (2020). Impact of PD‐1 blockade on severity of COVID‐19 in patients with lung cancers. Cancer Discovery, 10, 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo, E. , Bukreyeva, N. , Maruyama, J. , Paessler, S. , & Huang, C. (2020). Antiviral activities of type I interferons to SARS‐CoV‐2 infection. Antiviral Research, 179, 104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marofi, F. , Vahedi, G. , Biglari, A. , Esmaeilzadeh, A. , & Athari, S. S. (2017). Mesenchymal stromal/stem cells: A new era in the cell‐based targeted gene therapy of cancer. Frontiers in Immunology, 8, 1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, C. , Chubet, R. , Holtz, K. , Honda‐Okubo, Y. , Barnard, D. , Cox, M. , & Petrovsky, N. (2016). Development of a SARS coronavirus vaccine from recombinant spike protein plus delta inulin adjuvant. Vaccine Design, 1403, 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, P. , McAuley, D. F. , Brown, M. , Sanchez, E. , Tattersall, R. S. , & Manson, J. J. (2020). COVID‐19: Consider cytokine storm syndromes and immunosuppression. The Lancet, 395, 1033–1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner, N. , Swain, S. , McInnerney, K. , Han, S. , & Harmsen, A. G. (2010). Type‐I IFN signaling suppresses an excessive IFN‐γ response and thus prevents lung damage and chronic inflammation during Pneumocystis (PC) clearance in CD4 T cell‐competent mice. American Journal of Pathology, 176(6), 2806–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad, M. , & Martin, J. C. (2020). Pathological inflammation in patients with COVID‐19: A key role for monocytes and macrophages. Nature Reviews Immunology, 20, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson, M. (2020). Droplets and aerosols in the transmission of SARS‐CoV. The New England Journal of Medicine, 382, 2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. , Reandelar, M. J. , Fasciglione, K. , Roumenova, V. , Li, Y. , & Otazu, G. H. (2020). Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID‐19: An epidemiological study. medRxiv. [Google Scholar]
- Modjarrad, K. (2016). Research and development activities for Middle East respiratory syndrome: The current landscape . World Health Organization.
- Monti, S. , Balduzzi, S. , Delvino, P. , Bellis, E. , Quadrelli, V. S. , & Montecucco, C. (2020). Clinical course of COVID‐19 in a series of patients with chronic arthritis treated with immunosuppressive targeted therapies. Annals of the Rheumatic Diseases, 79(5), 667–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, C. (2020). Fighting COVID‐19 exhausts T cells. Nature Reviews Immunology, 20, 277. 10.1038/s41577-020-0304-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosaddeghi, P. , Negahdaripour, M. , Dehghani, Z. , Farahmandnejad, M. , Moghadami, M. , Nezafat, N. , & Masoompour, S. M. (2020). Therapeutic approaches for COVID‐19 based on the dynamics of interferon‐mediated immune responses.
- Muir, R. , Osbourn, M. , Dubois, A. V. , Doran, E. , Small, D. M. , Monahan, A. , O'Kane, C. M. , McAllister, K. , Fitzgerald, D. C. , Kissenpfennig, A. , McAuley, D. F. , & Ingram, R. J. (2016). Innate lymphoid cells are the predominant source of IL‐17A during the early pathogenesis of acute respiratory distress syndrome. American Journal of Respiratory and Critical Care Medicine, 193(4), 407–416. [DOI] [PubMed] [Google Scholar]
- Nachbagauer, R. , Krammer, F. , & Albrecht, R. A. (2018). A live‐attenuated prime, inactivated boost vaccination strategy with chimeric hemagglutinin‐based universal influenza virus vaccines provides protection in ferrets: A confirmatory study. Vaccines, 6(3):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, G. , & Chen, X. (2010). Vascular endothelial growth factor as an anti‐angiogenic target for cancer therapy. Current Cancer Drug Targets, 11(8), 1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, H.‐L. J. , Chia, A. , Chang, C. X. L. , Leong, H. N. , Ling, K. L. , Grotenbreg, G. M. , Gehring, A. J. , Bertoletti, A. , Tan, Y. J. , & Bertoletti, A. (2011). Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. Journal of Virology, 85(20), 10464–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacha, O. , Sallman, M. A. , & Evans, S. E. (2020). COVID‐19: A case for inhibiting IL‐17? Nature Reviews Immunology, 20, 345–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S. C. , Pande, V. , Sati, D. , Upreti, S. , & Samant, M. J. L. S. (2020). Vaccination strategies to combat novel corona virus SARS‐CoV‐2. Life Sciences, 256, 117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M. D. (2020). Immune evasion via SARS‐CoV‐2 ORF8 protein? Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S. , & Lal, G. (2017). The molecular mechanism of natural killer cells function and its importance in cancer immunotherapy. Frontiers in Immunology, 8, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Alvarado, C. , Gómez, C. , Reyes, M. , García, M. , Pérez, E. , Pérez de la Mora, C. , Sanchez, V. , & Pérez Ishiwara, D. G. (2017). Anti‐inflammatory effect of dialyzable leukocyte extract in autoimmune prostatitis: Evaluation in animal model. BioMed Research International, 2017, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan, A. L. , Katz, R. , & Gostin, L. O. (2020). The novel coronavirus originating in Wuhan, China: challenges for global health governance. Journal of the American Medical Association, 323(8), 709–710. [DOI] [PubMed] [Google Scholar]
- Phillips, S. , Mistry, S. , Riva, A. , Cooksley, H. , Hadzhiolova‐Lebeau, T. , Plavova, S. , Katzarov, K. , Simonova, M. , Zeuzem, S. , Woffendin, C. , Chen, P. J. , Peng, C. Y. , Chang, T. T. , Lueth, S. , De Knegt, R. , Choi, M. S. , Wedemeyer, H. , Dao, M. , Kim, C. W. , … Chokshi, S. (2017). Peg‐interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Frontiers in Immunology, 8, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran, P. , Zhu, Z. , Xiao, X. , Biragyn, A. , Dimitrov, A. S. , Broder, C. C. , & Dimitrov, D. S. (2009). Potent human monoclonal antibodies against SARS CoV, Nipah and Hendra viruses. Expert Opinion on Biological Therapy, 9(3), 355–368. 10.1517/14712590902763755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, K. N. , Frew, J. W. , Hsiao, J. L. , & Shi, V. Y. (2020). COVID‐19 and immunomodulator/immunosuppressant use in dermatology. Journal of the American Academy of Dermatology, 82(5), e173–e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara, E. , Ketloy, C. , & Palaga, T. (2020). Immune responses in COVID‐19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific Journal of Allergy and Immunology, 38, 1–9. [DOI] [PubMed] [Google Scholar]
- Rajaei, S. , & Dabbagh, A. (2020). The immunologic basis of COVID‐19: A clinical approach. Journal of Cellular & Molecular Anesthesia, 5(1), 37–42. [Google Scholar]
- Ramesh, G. , Meisner, O. C. , & Philipp, M. T. (2015). Anti‐inflammatory effects of dexamethasone and meloxicam on Borrelia burgdorferi‐induced inflammation in neuronal cultures of dorsal root ganglia and myelinating cells of the peripheral nervous system. Journal of Neuroinflammation, 12(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, P. , Griffin, I. , Tucker, C. , Smith, D. , Oechsle, O. , Phelan, A. , & Stebbing, J. (2020). Baricitinib as potential treatment for 2019‐nCoV acute respiratory disease. Lancet, 395(10223), e30–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risitano, A. M. , Mastellos, D. C. , Huber‐Lang, M. , Yancopoulou, D. , Garlanda, C. , Ciceri, F. , & Lambris, J. D. (2020). Complement as a target in COVID‐19? Nature Reviews Immunology, 20(6), 343–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roback, J. D. , & Guarner, J. (2020). Convalescent plasma to treat COVID‐19: Possibilities and challenges. Journal of the American Medical Association, 323, 1561. [DOI] [PubMed] [Google Scholar]
- Rockx, B. , Kuiken, T. , Herfst, S. , Bestebroer, T. , Lamers, M. M. , Oude Munnink, B. B. , de Meulder, D. , van Amerongen, G. , van den Brand, J. , Okba, N. M. A. , Schipper, D. , van Run, P , Leijten, L. , Sikkema, R. , Verschoor, E. , Verstrepen, B. , Bogers, W. , Langermans, J. , Drosten, C. , … Haagmans, B. L. (2020). Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science, 368(6494), 1012–1015. 10.1126/science.abb7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, M. , Rodríguez, Y. , Monsalve, D. M. , Acosta‐Ampudia, Y. , Camacho, B. , Gallo, J. E. , Rojas‐Villarraga, A. , Ramírez‐Santana, C. , Díaz‐Coronado, J. C , Manrique, R. , Mantilla, R. D. , Shoenfeld, Y. , & Anaya, J. M. (2020). Convalescent plasma in Covid‐19: Possible mechanisms of action. Autoimmunity Reviews, 19, 102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, B. , Moss, C. , George, G. , Santaolalla, A. , Cope, A. , Papa, S. , & Van Hemelrijck, M. (2020). Associations between immune‐suppressive and stimulating drugs and novel COVID‐19—A systematic review of current evidence. Ecancermedicalscience, 14, 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallard, E. , Lescure, F.‐X. , Yazdanpanah, Y. , Mentre, F. , & Peiffer‐Smadja, N. (2020). Type 1 interferons as a potential treatment against COVID‐19. Antiviral Research, 178, 104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol, A. , & Perlman, S. (2020). Lessons for COVID‐19 immunity from other coronavirus infections. Immunity, 53, 248–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, S. , Kriesen, S. , Paape, D. , Hildebrandt, G. , & Manda, K. (2018). Modulation of inflammatory reactions by low‐dose ionizing radiation: Cytokine release of murine endothelial cells is dependent on culture conditions. Journal of Immunology Research, 2018, 2856518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugaraj, B. , Siriwattananon, K. , Wangkanont, K. , & Phoolcharoen, W. (2020). Perspectives on monoclonal antibody therapy as potential therapeutic intervention for coronavirus disease‐19 (COVID‐19). Asian Pacific Journal of Allergy and Immunology, 38(1), 10–18. [DOI] [PubMed] [Google Scholar]
- Shen, C. , Wang, Z. , Zhao, F. , Yang, Y. , Li, J. , Yuan, J. , Wang, F. , Li, D. , Yang, M. , Xing, L. , Wei, J. , Xiao, H. , Yang, Y. , Qu, J. , Qing, L. , Chen, L. , Xu, Z , Peng, L. , Li, Y. , … Liu, L. (2020). Treatment of 5 critically ill patients with COVID‐19 with convalescent plasma. Journal of the American Medical Association, 323, 1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shet, A. , Ray, D. , Malavige, N. , Santosham, M. , & Bar‐Zeev, N. (2020). Differential COVID‐19‐attributable mortality and BCG vaccine use in countries. medRxiv. 10.1101/2020.04.01.20049478 [DOI] [Google Scholar]
- Shi, Y. , Wang, Y. , Shao, C. , Huang, J. , Gan, J. , Huang, X. , Bucci, E. , Piacentini, M. , Ippolito, G. , & Melino, G. (2020). COVID‐19 infection: The perspectives on immune responses. Cell Death & Differentiation, 27, 1451–1454. 10.1038/s41418-020-0530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, T. A. , Zorzetto‐Fernandes, A. L. , Cecílio, N. T. , Sardinha‐Silva, A. , Fernandes, F. F. , & Roque‐Barreira, M. C. (2017). CD14 is critical for TLR2‐mediated M1 macrophage activation triggered by N‐glycan recognition. Scientific Reports, 7(1):7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro, S. , Park, M. , Turrini, C. , Tunstall, T. , Thwaites, R. , Mauri, T. , Ragazzi, R. , Ruggeri, P. , Hansel, T. T. , Caramori, G. , & Volta, C. A. (2019). Biomarkers for Acute Respiratory Distress syndrome and prospects for personalised medicine. Journal of Inflammation, 16(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridevi, K. , Munjal, A. , Chandran, A. , Nachiappan, S. , Raman, P. , Bhalla, S. , Kumar, S. , & Nayyar, A. S. (2020). Convalescent plasma therapy for prophylaxis and treatment of COVID‐19: A systematic research of facts and files. A Narrative Review, 8(2):314. [Google Scholar]
- Stebbing, J. , Phelan, A. , Griffin, I. , Tucker, C. , Oechsle, O. , Smith, D. , & Richardson, P. (2020). COVID‐19: Combining antiviral and anti‐inflammatory treatments. The Lancet Infectious Diseases, 20(4), 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi, S. , Elahi, R. , & Esmaeilzadeh, A. (2019). Solid tumors challenges and new insights of car t cell engineering. Stem Cell Reviews and Reports, 15(5), 619–636. [DOI] [PubMed] [Google Scholar]
- Tahmasebi, S. , Khosh, E. , & Esmaeilzadeh, A. (2020). The outlook for diagnostic purposes of the 2019‐novel coronavirus disease. Journal of Cellular Physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T. , Narazaki, M. , & Kishimoto, T. (2016). Immunotherapeutic implications of IL‐6 blockade for cytokine storm. Immunotherapy, 8(8), 959–970. 10.2217/imt-2016-0020 [DOI] [PubMed] [Google Scholar]
- Tang, C. , Wang, Y. , Lv, H. , Guan, Z. , & Gu, J. (2020). Caution against corticosteroid‐based COVID‐19 treatment. The Lancet, 395, 1759–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne, J. H. (2020). Covid‐19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ, 368, m1256. [DOI] [PubMed] [Google Scholar]
- Tavakolpour, S. , Rakhshandehroo, T. , Wei, E. X. , & Rashidian, M. J. (2020). Lymphopenia during the COVID‐19 infection: What it shows and what can be learned. Immunology Letters, 225, 31–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, M. Z. , Poh, C. M. , Rénia, L. , MacAry, P. A. , & Ng, L. F. (2020). The trinity of COVID‐19: Immunity, inflammation and intervention. Nature Reviews Immunology, 20, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. , Li, C. , Huang, A. , Xia, S. , Lu, S. , Shi, Z. , Lu, L. , Jiang, S. , Yang, Z. , Wu, Y. , & Ying, T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerging Microbes & Infections, 9(1), 382–385. 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjendra, Y. , Al Mana, A. F. , Espejo, A. P. , Akgun, Y. , Millan, N. C. , Gomez‐Fernandez, C. , & Cray, C. (2020). Predicting disease severity and outcome in COVID‐19 patients: A review of multiple biomarkers. Archives of Pathology & Laboratory Medicine. 10.5858/arpa.2020-0471-SA [DOI] [PubMed] [Google Scholar]
- Totura, A. L. , & Baric, R. S. (2012). SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Current Opinion in Virology, 2(3), 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y.‐F. , Chien, C.‐S. , Yarmishyn, A. A. , Lin, Y.‐Y. , Luo, Y.‐H. , Lin, Y.‐T. , Lai, W. Y. , Yang, D. M. , Chou, S. J. , Yang, Y. P. , Wang, M. L. , & Chiou, S. H. (2020). A review of SARS‐CoV‐2 and the ongoing clinical trials. International Journal of Molecular Sciences, 21(7):2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret, N. , Britton, G. J. , Gruber, C. , Hegde, S. , Kim, J. , Kuksin, M. , Levantovsky, R. , Malle, L. , Moreira, A , Park, M. D. , Pia, L. , Risson, E , Saffern, M. , Salomé, B. , Esai Selvan, M , Spindler, MP , Tan, J. , van der Heide, V. , Gregory, J. K. , … Sinai Immunology Review Project (2020). Immunology of COVID‐19: Current state of the science. Immunity, 52, 910–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, S. , Yi, Q. , Fan, S. , Lv, J. , Zhang, X. , Guo, L. , Lang, C. , Xiao, Q. , Xiao, K. , Yi, Z. , Qiang, M. , Xiang, J. , Zhang, B. , & Chen, Y. (2020). Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv. 10.1101/2020.02.10.20021832 [DOI] [Google Scholar]
- Wang, C. , Horby, P. W. , Hayden, F. G. , & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Liu, X. , Wu, S. , Chen, S. , Li, Y. , Nong, L. , Lie, P. , Huang, L. , Cheng, L. , Lin, Y. , & He, J. (2020). Definition and risks of Cytokine Release Syndrome in 11 critically ill COVID-19 patients with pneumonia: Analysis of disease characteristics. The Journal of Infectious Diseases. jiaa387. Advance online publication. 10.1093/infdis/jiaa387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Li, W. , Drabek, D. , Okba, N. M. A. , van Haperen, R. , Osterhaus, A. D. M. E. , van Kuppeveld, F. J. M. , Haagmans, B. L. , Grosveld, F. , & Bosch, B.‐J. (2020). A human monoclonal antibody blocking SARS‐CoV‐2 infection. Nature Communications, 11, 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Shi, Y. , Xiao, T. , Fu, J. , Feng, X. , Mu, D. , Feng, Q. , Hei, M. , Hu, X. , Li, Z. , Lu, G. , Tang, Z. , Wang, Y. , Wang, C. , Xia, S. , Xu, J. , Yang, Y. , Yang, J. , Zeng, M. , … Working Committee on Perinatal and Neonatal Management for the Prevention and Control of the 2019 Novel Coronavirus Infection (2020). Chinese expert consensus on the perinatal and neonatal management for the prevention and control of the 2019 novel coronavirus infection. Annals of Translational Medicine, 8(3):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X. , & Nielsen, R. (2019). CCR5‐∆ 32 is deleterious in the homozygous state in humans. Nature Medicine, 25(6), 909–910. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu, X.‐X. , Gao, H.‐N. , Wu, H.‐B. , Peng, X.‐M. , Ou, H.‐L. , & Li, L.‐J. (2015). Successful treatment of avian‐origin influenza A (H7N9) infection using convalescent plasma. International Journal of Infectious Diseases, 41, 3–5. [DOI] [PubMed] [Google Scholar]
- Wu, Z. , & McGoogan, J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Journal of the American Medical Association, 323, 1239. [DOI] [PubMed] [Google Scholar]
- Wu, D. , & Yang, X. O. (2020). TH17 responses in cytokine storm of COVID‐19: An emerging target of JAK2 inhibitor fedratinib. Journal of Microbiology, Immunology and Infection, 53, 368–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Cai, H. , Shen, Y. , Ni, Q. , Chen, Y. , Hu, S. , Li, J. , Wang, H. , Yu, L. , Huang, H. , Qiu, Y. , Wei, G. , Fang, Q. , Zhou, J. , Sheng, J. , Liang, T. , & Li, L. (2020). Management of corona virus disease‐19 (COVID‐19): The Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban, 49(1), 147–157. 10.3785/j.issn.1008-9292.2020.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. T. , Chen, P. , & Wang, J. F. (2020). Evolution of the novel corona. Virus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63, 457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Shi, L. , Wang, Y. , Zhang, J. , Huang, L. , Zhang, C. , Liu, S. , Zhao, P. , Liu, H. , Zhu, L. , Tai, Y. , Bai, C. , Gao, T , Song, J , Xia, P , Dong, J , Zhao, J , & Wang, F. S. (2020). Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. The Lancet Respiratory Medicine, 8, 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K. , Sun, K. , Srinivasan, K. , Salmon, J. , Marques, E. , Xu, J. , & August, J. (2009). Immune responses to T‐cell epitopes of SARS CoV‐N protein are enhanced by N immunization with a chimera of lysosome‐associated membrane protein. Gene Therapy, 16(11), 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Yu, Y. , Xu, J. , Shu, H. , Liu, H. , Wu, Y. , Zhang, L. , Yu, Z. , Fang, M. , Yu, T. , Wang, Y. , Pan, S. , Zou, X. , Yuan, S. , & Shang, Y. (2020). Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: A single‐centered, retrospective, observational study. The Lancet Respiratory Medicine, 8, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Q. , Wang, B. , & Mao, J. (2020). Cytokine storm in COVID‐19 and treatment. Journal of Infection, 80(6). 10.1016/j.jinf.2020.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin, I. , Aharony, N. , Shaer Tamar, E. , Argoetti, A. , Messer, E. , Berenbaum, D. , Shafran, E. , Kuzli, A. , Gandali, N. , Shkedi, O. , Hashimshony, T. , Mandel‐Gutfreund, Y. , Halberthal, M. , Geffen, Y. , Szwarcwort‐Cohen, M. , & Kishony, R. (2020). Evaluation of COVID‐19 RT‐qPCR test in multi‐sample pools. Clinical Infectious Diseases , ciaa531. Advance online publication. 10.1093/cid/ciaa531 [DOI] [PMC free article] [PubMed]
- Zhang, F. , Gan, R. , Zhen, Z. , Hu, X. , Li, X. , Zhou, F. , Liu, Y. , Chen, C. , Xie, S. , Zhang, B. , Wu, X. , & Huang, Z. (2020). Adaptive immune responses to SARS‐CoV‐2 infection in severe versus mild individuals. Signal Transduction and Targeted Therapy, 5(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Litvinova, M. , Wang, W. , Wang, Y. , Deng, X. , Chen, X. , Li, M. , Zheng, W. , Yi, L. , Chen, X. , Wu, Q. , Liang, Y. , Wang, X. , Yang, J. , Sun, K. , Longini, I. M., Jr. , Halloran, M. E. , Wu, P. , Cowling, B. J. , … Yu, H. (2020). Evolving epidemiology of novel coronavirus diseases 2019 and possible interruption of local transmission outside Hubei Province in China: A descriptive and modeling study. medRxiv: The Preprint Server for Health Sciences . 10.1101/2020.02.21.20026328 [DOI]
- Zhang, J. , Wang, S. , & Xue, Y. (2020). Fecal specimen diagnosis 2019 novel coronavirus‐infected pneumonia. Journal of Medical Virology, 92, 680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Yang, Y. , Huang, H. , Li, D. , Gu, D. , Lu, X. , Zhang, Z. , Liu, L. , Liu, T. , Liu, Y. , He, Y , Sun, B. , Wei, M. , Yang, G. , Wang, X. , Zhang, L. , Zhou, X. , Xing, M. , & Wang, P. G. (2020). Relationship between the ABO blood group and the COVID‐19 susceptibility. Clinical Infectious Diseases, ciaa1150. 10.1093/cid/ciaa1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M. , Gao, Y. , Wang, G. , Song, G. , Liu, S. , Sun, D. , Xu, Y. , & Tian, Z. (2020). Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cellular & Molecular Immunology, 17, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Fu, B. , Zheng, X. , Wang, D. , & Zhao, C. (2020). Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID‐19 patients. National Science Review, 7, 998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Liu, K. , & Liu, H. (2020). Cause analysis and treatment strategies of "recurrence" with novel coronavirus pneumonia (covid‐19) patients after discharge from hospital. Zhonghua Jie He He Huxi Za Zhi, 43, E028. [DOI] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X.‐L. , Wang, X.‐G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C.‐L. , Chen, H.D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R.‐D. , Liu, M.‐Q. , Chen, Y. , Shen, X.‐R. , Wang, Y.‐Y. , … Shi, Z.‐L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Yang, P. , Zhao, Y. , Zhuang, Z. , Wang, Z. , Song, R. , Zhang, J. , Liu, C. , Gao, Q. , Xu, Q. , Wei, X. , Sun, H.X. , Ye, B. , Wu, Y , Zhang, N. , Lei, G. , Yu, L. , Yan, J. , Diao, G. , … Liu, W. J. (2020). Single‐cell sequencing of peripheral blood mononuclear cells reveals distinct immune response landscapes of COVID‐19 and influenza patients. Immunity, 53, 685–696.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , Zhao, X. , Huang, B. , Shi, W. , Lu, R. , Niu, P. , Zhan, F. , Ma, X. , Wang, D. , Xu, W. , Wu, G. , Gao, G. F. , & Tan, W. , for the China Novel Coronavirus Investigating and Research Team . (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine, 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]


