Abstract
The severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) causing coronavirus disease‐2019 (COVID‐19) likely has evolutionary origins in other animals than humans based on genetically related viruses existing in rhinolophid bats and pangolins. Similar to other animal coronaviruses, SARS‐CoV‐2 contains a functional furin cleavage site in its spike protein, which may broaden the SARS‐CoV‐2 host range and affect pathogenesis. Whether ongoing zoonotic infections are possible in addition to efficient human‐to‐human transmission remains unclear. In contrast, human‐to‐animal transmission can occur based on evidence provided from natural and experimental settings. Carnivores, including domestic cats, ferrets and minks, appear to be particularly susceptible to SARS‐CoV‐2 in contrast to poultry and other animals reared as livestock such as cattle and swine. Epidemiologic evidence supported by genomic sequencing corroborated mink‐to‐human transmission events in farm settings. Airborne transmission of SARS‐CoV‐2 between experimentally infected cats additionally substantiates the possibility of cat‐to‐human transmission. To evaluate the COVID‐19 risk represented by domestic and farmed carnivores, experimental assessments should include surveillance and health assessment of domestic and farmed carnivores, characterization of the immune interplay between SARS‐CoV‐2 and carnivore coronaviruses, determination of the SARS‐CoV‐2 host range beyond carnivores and identification of human risk groups such as veterinarians and farm workers. Strategies to mitigate the risk of zoonotic SARS‐CoV‐2 infections may have to be developed in a One Health framework and non‐pharmaceutical interventions may have to consider free‐roaming animals and the animal farming industry.
Keywords: SARS‐CoV‐2, COVID‐19, coronavirus, domestic animal, carnivore, farmed animal
1. INTRODUCTION
Coronavirus disease‐2019 (COVID‐19) emerged in December 2019 in Wuhan, China and quickly spread to more than 200 countries, killing over 1 million people worldwide as of October 2020. The rapid progression of the pandemic has not only taxed healthcare systems worldwide but has also wreaked havoc on the global economy and society (UN, 2020a, 2020b). COVID‐19 is caused by a newly emerged coronavirus termed severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) (Li et al., 2020). A conspecific coronavirus termed SARS‐CoV caused an outbreak in 2003–2004, infecting about 8,000 people mainly in China, of which about 10% died (Cheng et al., 2013). Control of SARS in 2003–2004 by strict non‐pharmaceutical interventions was facilitated by predominant SARS‐CoV replication in the lower respiratory tract (Gu & Korteweg, 2007), as opposed to efficient replication of SARS‐CoV‐2 in the upper respiratory tract (Wolfel et al., 2020) despite usage of the same receptor molecule angiotensin‐converting enzyme 2 (ACE2) (Hoffmann et al., 2020; Li et al., 2003). Here, we discuss the evidence for the zoonotic origins of human coronaviruses, identify epidemiologically relevant animal hosts and discuss their impact on containment strategies of COVID‐19 under a One Health approach, a concept that is gaining recognition which addresses health issues, such as zoonotic diseases, at the human‐animal‐environment interface in a collaborative effort among multiple relevant disciplines and sectors (FAO‐OIE‐WHO, 2019).
2. ZOONOTIC ORIGIN OF HUMAN CORONAVIRUSES
Endemic human coronaviruses (HCoVs) include the alphacoronaviruses HCoV‐229E and HCoV‐NL63, as well as the betacoronaviruses HCoV‐HKU1 and HCoV‐OC43, whereas emerging HCoVs include the betacoronaviruses Middle East respiratory syndrome coronavirus (MERS‐CoV), SARS‐CoV and the novel SARS‐CoV‐2 (Figure 1a). Coronavirus species are defined based upon genomic sequence distances (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020). Members of the virus species SARS‐related coronavirus that includes both SARS‐CoV and SARS‐CoV‐2 as well as multiple genetically highly diversified bat‐associated strains (summarized as SARS‐related CoVs henceforth) can infect different hosts belonging to four different mammalian orders, including bats, carnivores, pangolins and primates (Figure 1b). The large diversity of conspecific bat coronaviruses and their close genetic relationship to human viruses suggest that bats are potential animal reservoirs of the majority of HCoVs (Figure 2a,b) (Drexler et al., 2014). In the case of SARS‐related CoVs, rhinolophid bats (Rhinolophus spp., also termed horseshoe bats) are particularly relevant natural hosts, as exemplified by the extent of genetically divergent SARS‐related CoVs found in those bats across Asia, Africa and Europe (Drexler et al., 2010; Hu et al., 2017; Li et al., 2005). In the tropics, direct contact between humans and bats potentially facilitating human infection is more frequent than in temperate climates, including consumption of bats as bushmeat and their use for traditional medicine (Anti et al., 2015; Mildenstein et al., 2016). However, other animals than bats have been implicated as intermediate hosts of and source of infection with HCoVs (Figure 1a) (Corman et al., 2018). For instance, alphacoronaviruses found in camels (Camelus dromedarius) and alpacas (Vicugna pacos) share a common ancestor with HCoV‐229E (Corman et al., 2016), whereas HCoV‐OC43 shares a common ancestor with coronaviruses infecting cattle (Bos taurus) (Vijgen et al., 2006). The emerging MERS‐CoV is enzootic in camels (mainly C. dromedarius) and causes regular spill‐over infections into humans (WHO, 2020). In the case of SARS‐CoV, genetically closely related viruses were found in masked palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides) sold in a live‐animal market in the Guangdong province (Guan et al., 2003; Kan et al., 2005). Genomic comparisons of full‐length SARS‐CoV sequences from human and civets showed high nucleotide identity of approximately 99.6%. In 2005, genetically highly diverse SARS‐related CoVs were found in horseshoe bats from China (Li et al., 2005). Masked palm civets were thus likely intermediate hosts, while horseshoe bats were the likely evolutionary origin of SARS‐CoV. It has been speculated that mutations in the SARS‐CoV receptor‐binding domain may have arisen in palm civets that facilitated infection efficiency by increasing the affinity to the human ACE2 receptor (Graham & Baric, 2010; Song et al., 2005).
Figure 1.
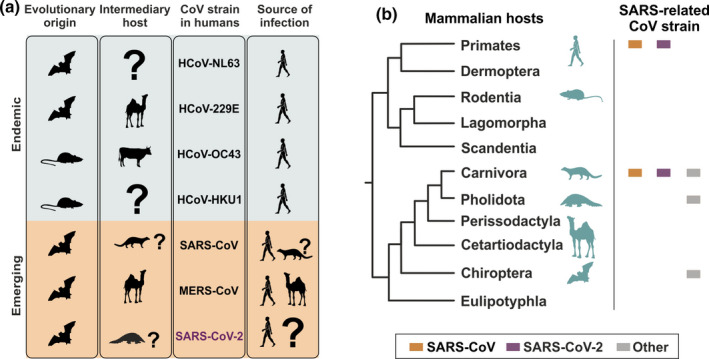
Mammals as reservoirs and intermediary hosts of endemic and emerging human coronaviruses. (a) Animal reservoirs and intermediary hosts of human coronaviruses. (b) Cladogram of mammalian orders adapted from (Foley et al., 2016). Hosts of coronaviruses are depicted by pictograms (teal). Squares depict families susceptible to SARS‐CoV (orange), SARS‐CoV‐2 (purple) and other SARS‐related CoVs (grey) [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 2.
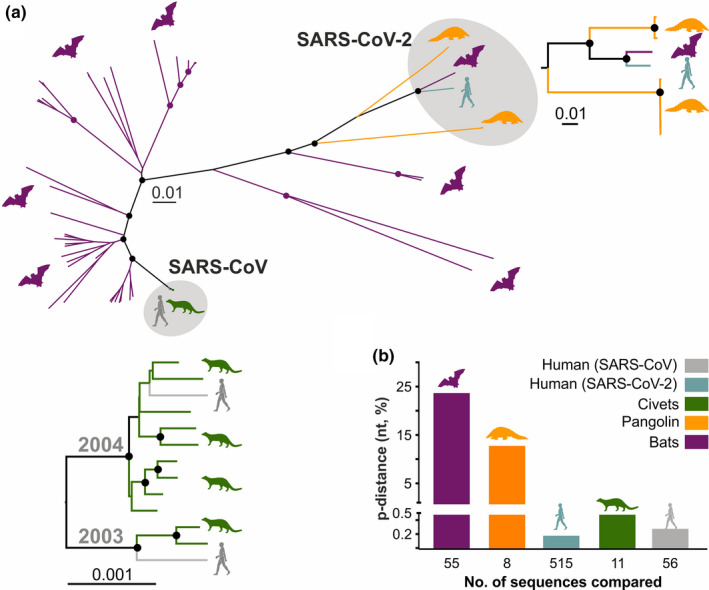
Mammalian hosts of coronaviruses. (a) Large diversity of bat‐associated SARS‐related CoVs in comparison to other hosts. The phylogeny was constructed with 77 SARS‐related CoV full‐length genomes using a neighbour‐joining method with 1,000 bootstrap replicates. Circles at nodes indicate bootstrap values ≥75%. Scale bar indicates nucleotide substitutions per site. (b) High divergence of bat SARS‐related CoVs in comparison with viruses infecting pangolin, civet and human hosts. P‐distance within SARS‐related CoVs in each host. The human SARS‐CoV‐2 and SARS‐CoV genomes included all sequences found in GenBank as of 16.04.2020, excluding identical genomes. GenBank accession numbers of all other genomes used in panel A and B: Bat SARS‐related CoVs: MN996532, MG772934, MG772933, KY938558, KU182964, KJ473811, KY770860, KJ473813, KJ473812, DQ412042, KY770859, KY770858, BKJ473816, KY417145, KF294455, DQ071615, KY417152, KY417151, KF367457, KC881006, KC881005, KY417144, KY417150, KT444582, KY417146, KY417149, KY417143, FJ588686, KY417148, KY417147, KP886809, KP886808, KJ473815, KU973692, KF569996, KJ473814, DQ648857, DQ412043, JX993987, KF294457, GQ153548, GQ153546, GQ153545, GQ153544, GQ153541, GQ153539, GQ153540, DQ084199, DQ084200, DQ022305, GQ153547, GQ153543, GQ153542, KY352407, GU190215; Human SARS‐CoV‐2: MN908947; Human SARS‐CoV: AY568539, AY390556; Civet SARS‐CoV: AY572034, AY572035, AY686863, AY686864, AY304486, AY304488, AY572038, AY686864, AY613948, AY613949, AY613950; and Pangolin SARS‐related CoVs: MT121216, MT040335, MT072865, MT072864, MT040336, MT040334, MT040333 [Colour figure can be viewed at wileyonlinelibrary.com]
Efforts to identify potential SARS‐CoV‐2 intermediate hosts have not been successful yet. The origin of the outbreak was associated with the Huanan seafood wholesale market in Wuhan during epidemiologic investigations of the first cases (Li et al., 2020), which was supported by SARS‐CoV‐2‐positive environmental samples from that market (Zhang & Holmes, 2020). However, retrospective epidemiologic analyses indicated that an earlier COVID‐19 case had no exposure to that seafood market (Huang et al., 2020). The origins of the pandemic thus remain unclear. The closest relatives to human SARS‐CoV‐2 known so far are a coronavirus coming from the intermediate horseshoe bat (R. affinis) from China with approximately 96% nucleotide identity, and two sub‐lineages of SARS‐related CoVs found in Malayan pangolins (Manis javanica) with 85.5%–92.4% nucleotide identity averaged over the whole viral genome (Figure 2a) (Lam et al., 2020; Xiao et al., 2020). Moreover, phylogenetic analyses using a large subgenomic data set of bat coronaviruses from China indicate that alike SARS‐CoV, SARS‐CoV‐2 likely originated in horseshoe bats (Latinne et al., 2020). However, whereas zoonotic infection of humans with civet‐associated SARS‐CoV strains was likely according to the genetic relatedness of those CoVs (Figure 2a, lower insert), bat‐, human‐ and pangolin‐associated SARS‐CoV‐2 strains are not as closely related and the direct ancestor of SARS‐CoV‐2 strains infecting humans remains to be determined (Figure 2a, right insert).
3. TRANSMISSION OF SARS‐COV‐2 FROM HUMANS TO ANIMALS
Human‐to‐animal transmission events during the COVID‐19 pandemic have been documented in several countries, including Hong Kong, Belgium, United States, Netherlands, Denmark, Spain, Germany and France (Barrs et al., 2020; CDC, 2020; Daly, 2020; Laudette, 2020; Oreshkova et al., 2020; ProMed‐mail, 2020a, 2020b, 2020c, 2020d; Sit et al., 2020; Sterling, 2020). Case reports on cats (Felis catus) living in the same household with COVID‐19 patients, revealed that these animals can be infected with SARS‐CoV‐2, showing either no or mild respiratory illness (CDC, 2020; Laudette, 2020; ProMed‐mail, 2020a, 2020c). A case report on two separate SARS‐CoV‐2‐infected dogs (Canis lupus familiaris), whose owners were COVID‐19 patients, showed that although both dogs tested positive by RT‐PCR and serologic methods for SARS‐CoV‐2, no apparent clinical signs were observed in the dogs (Sit et al., 2020). Other reports on SARS‐CoV‐2‐positive dogs in the Netherlands and the USA indicated that different symptoms can occur in infected dogs, ranging from mild to severe respiratory distress symptoms (Daly, 2020; Sterling, 2020). In sum, the impact of SARS‐CoV‐2 infections on domestic animal health is unclear, as both dogs and cats were reported with and without clinical signs.
A study from Wuhan showed a SARS‐CoV‐2 seroprevalence of 14.7% (15/102) in cats sampled between January and March 2020 compared with 0.0% (0/39) in cats sampled between May and March 2019 using a commercial ELISA that detects antibody reactivity against SARS‐CoV‐2 receptor‐binding domain (RBD) (Zhang et al., 2020). A study from Italy showed that 3.4% (13/388) of dogs and 3.9% (6/152) of cats living in SARS‐CoV‐2‐positive households or in regions severely affected by COVID‐19 sampled between March and May 2020, developed neutralizing antibodies against SARS‐CoV‐2 (Patterson et al., 2020). Another study, which sampled cats living in the same household with COVID‐19 patients from February to August in Hong Kong, found that 14% (6/50) were tested positive by RT‐PCR (Barrs et al., 2020). Of those positive samples, one SARS‐CoV‐2 genome was recovered and was identical to the SARS‐CoV‐2 genome recovered from the cat owner (Barrs et al., 2020). The high seroprevalence and detection rates of SARS‐CoV‐2 in cats and to some extent in dogs indicate that these animals can be infected with SARS‐CoV‐2.
Animal experimental infections can demonstrate the susceptibility of different animal species to SARS‐CoV‐2, which is on the one hand useful for the establishment of animal disease models and on the other hand provides an indication of potential animal sources that may act as animal sources of human infection. Previous infection experiments confirmed the susceptibility of ferrets (Mustela putorius), domestic cats and Syrian hamsters (Mesocricetus auratus) to SARS‐CoV (Martina et al., 2003; Roberts et al., 2005). Experimental infections of SARS‐CoV‐2 in a wide range of non‐primate animal species, including ferrets, domestic cats, raccoon dogs, Egyptian fruit bats (Rousettus aegyptiacus), Syrian hamsters, New Zealand white rabbits (Oryctolagus cuniculus) and Northern treeshrews (Tupaia belangeris), showed different degrees of susceptibility to the virus at inoculation doses of approximately 105 50% tissue culture infectious dose (TCID50) or 105 plaque‐forming units (PFU) and intranasal, intratracheal and ocular transmission routes, which may be representative of natural infections during human‐to‐human transmission (Table 1) (Freuling et al., 2020; Halfmann et al., 2020; Munoz‐Fontela et al., 2020; Mykytyn et al., 2020; Schlottau et al., 2020; Shi et al., 2020; Sia et al., 2020; Zhao et al., 2020). These animals showed viral RNA shedding in the respiratory tract and to a lesser extent or no shedding in the gastroenteric tract, development of SARS‐CoV‐2‐specific antibody responses and histopathological signs of moderate inflammation in infected respiratory tissue (Freuling et al., 2020; Halfmann et al., 2020; Munoz‐Fontela et al., 2020; Mykytyn et al., 2020; Schlottau et al., 2020; Shi et al., 2020; Sia et al., 2020; Zhao et al., 2020). Notably, although Egyptian fruit bats (belonging to the family Pteropodidae, also termed pteropodid bats) are likely not a relevant host of SARS‐related CoVs in nature, their susceptibility might be due to their close genetic relationship with the likely animal reservoir of SARS‐related CoVs, namely horseshoe bats, as both pteropodid and rhinolophid bats belong to a common suborder termed Yinpterochiroptera (Teeling et al., 2005). In contrast, SARS‐CoV‐2 replicated poorly in dogs and cattle, whereas pigs (Sus scrofa domesticus), chickens (Gallus gallus) and ducks (Anas platyrhynchos) were not susceptible (Shi et al., 2020; Ulrich et al., 2020). Apparently, limited susceptibility of dogs in experimental infection studies contrasts the data from epidemiological studies and case reports described above. These discrepancies may be due to the different susceptibility of individual dog breeds or imperfect infection settings. In addition, airborne transmission of SARS‐CoV‐2 between cats and between hamsters has also been reported (Halfmann et al., 2020; Shi et al., 2020; Sia et al., 2020). In the case of non‐human primates, given their genetic relatedness to humans and occurrences of transmission of other human respiratory viruses such as HCoV‐OC43 (Patrono et al., 2018) and respiratory syncytial virus (Grutzmacher et al., 2016), susceptibility to SARS‐CoV‐2 was not unexpected (Munster et al., 2020; Rockx et al., 2020). Beyond their use as an animal model, human‐to‐non‐human primate transmission of SARS‐CoV‐2 may be worrying due to the possible decimation of endangered non‐human primate species in the wild. In sum, surveillance strategies should include susceptible animals in close contact to humans to prevent outbreaks of SARS‐CoV‐2 in these animals and potential spillback events to humans. Whether mutations of SARS‐CoV‐2 may arise in animals other than humans that subsequently affect pathogenesis or transmissibility in humans requires urgent investigation.
Table 1.
Animal susceptibility to SARS‐CoV‐2
| Order | Species | Mode of infection | Susceptibility | Infection dose, route | Major findings | Studies |
|---|---|---|---|---|---|---|
| Primates | Rhesus macaques, Cynomologous macaques | Experimental | High | 4 × 105–106 TCID50 intranasal, intratracheal, ocular | Limited and moderate clinical signs, viral replication in upper and lower respiratory tracts, advanced age associated with increased histopathological changes, protective immune response | (Munster et al., 2020; Rockx et al., 2020) |
| Rodentia | Syrian hamster | Experimental | High | 8 × 104 TCID50, intranasal | Mild symptoms, direct contact and aerosol transmission, efficient replication in the upper and lower respiratory tract, infection of olfactory sensory neurons | (Sia et al., 2020) |
| Carnivora | Domestic cat | Natural and Experimental | High | 1‐5.2 × 105 PFU, intranasal, intratracheal, ocular |
N: 3.4%–14.7% virus seroprevalence, 8/24 seroconversion of animals roaming around SARS‐CoV‐2‐positive mink farms, 6/50 RT‐PCR‐positive cats living in households with COVID‐19‐positive patients, no or mild symptoms E: aerosol transmission confirmed, lesions in nasal and tracheal mucosa epitheliums and lungs |
(Barrs et al., 2020; Halfmann et al., 2020; Oreshkova et al., 2020; Patterson et al., 2020; Shi et al., 2020) |
| African lion | Natural | NA | NA | Mild respiratory signs | (ProMed‐mail, 2020b) | |
| Malayan tiger | Natural | NA | NA | Mild respiratory signs | (ProMed‐mail, 2020b) | |
| Amur tiger | Natural | NA | NA | Mild respiratory signs | (ProMed‐mail, 2020b) | |
| Ferret | Experimental | High | 105 PFU, 105 TCID50, intranasal | No or mild clinical signs, viral shedding in 8/9 animals and 3/3 contact animals, higher viral shedding in throat than rectum, histopathological changes including rhinitis and mild inflammation | (Schlottau et al., 2020; Shi et al., 2020) | |
| Mink | Natural | High | NA | Outbreaks in mink farms: 57 Dutch, 25 Danish, 6 USA, 1 Spain. Mild to severe respiratory symptoms, interstitial pneumonia, higher viral shedding in throat than rectum, mink‐to‐human transmission confirmed by phylogenetic analyses | (Oreshkova et al., 2020; Oude Munnink et al., 2020; ProMed‐mail, 2020d) | |
| Dog | Natural and Experimental | Inconclusive | 105 PFU, intranasal |
N: 3.4% of animals in Italy with neutralizing animals, 2/15 RT‐PCR‐positive dogs living in households with COVID‐19‐positive patients, no to severe symptoms E: faecal viral shedding was detected in 3/5 inoculated dogs but no virus isolation was possible, 2/5 inoculated and 2/2 contact dogs were seronegative |
(Patterson et al., 2020; Shi et al., 2020; Sit et al., 2020) | |
| Raccoon dog | Experimental | Moderate | 105 TCID50, intranasal | No symptoms, effective virus production and seroconversion in 6/9 animals, effective transmission to contact animals, higher viral shedding in nose and throat than rectum, mild rhinitis | (Freuling et al., 2020) | |
| Chiroptera | Egyptian fruit bat | Experimental | High | 105 TCID50, intranasal | No symptoms, viral shedding in 9/9 inoculated animals and 2/3 contact animals, low titres of neutralizing antibodies, mild rhinitis and infiltrating lymphocytes and neutrophils | (Schlottau et al., 2020) |
| Artiodactyla | Cattle | Experimental | Low | 105 TCID50, intranasal | No symptoms, viral shedding and seroconversion in 2/6 animals | (Ulrich et al., 2020) |
| Pig | Experimental | Negative | 105 PFU, 105 TCID50, intranasal | No viral replication | (Schlottau et al., 2020; Shi et al., 2020) | |
| Lagomorpha | New Zealand white rabbit | Experimental | Moderate | >105 TCID50, intranasal | No symptoms, higher viral shedding in nose and throat than rectum and seroconversion in animals infected with viral dose of >105 TCID50, mild histopathological changes | (Mykytyn et al., 2020) |
| Scandentia | Tree shrew | Experimental | Moderate | 106 PFU, intranasal | Increased body temperature in young animals, viral shedding in 16/24 animals, mild histopathological changes | (Zhao et al., 2020) |
| Galliformes | Chicken | Experimental | Negative | 105 PFU, 105 TCID50, oculo‐oronasal | No viral replication | (Schlottau et al., 2020; Shi et al., 2020) |
| Ducks | Experimental | Negative | 105 PFU | No viral replication | (Shi et al., 2020) |
Abbreviations: E, Experimental; N, Natural; NA, not applicable due to limited data; PFU, Plaque forming units; TCID50, 50% tissue culture infectious dose.
Zoonotic SARS‐CoV‐2 transmission events are not restricted to domestic animals only. In the Bronx Zoo in New York, several felids tested positive, including two Malayan tigers (Panthera tigris jacksoni), two Amur tigers (Panthera tigris altaica) and three African lions (Panthera leo), all of which developed mild respiratory signs and recovered after a week. The likely source of infection was infected zoo staff (ProMed‐mail, 2020b). Similarly, SARS‐CoV‐2‐positive minks were reported in 57 mink fur farms in the Netherlands, 25 fur farms in Denmark, six fur farms in the USA and one fur farm in Spain (ProMed‐mail, 2020d). An investigation conducted in two of the Dutch farms reported that the infected minks showed mild to severe respiratory distress (Oreshkova et al., 2020). Upon post‐mortem analyses, several minks showed interstitial pneumonia (Oreshkova et al., 2020). The source of the SARS‐CoV‐2 outbreak in minks was linked to the farmers and their family members, who either showed symptoms compatible with COVID‐19 or were tested positive for SARS‐CoV‐2. Moreover, in addition to the infected minks, 8 out of 24 stray cats surrounding two of the farms showed evidence of SARS‐CoV‐2 infection, 7 of which were confirmed by a highly specific microneutralization assay and one by PCR (Oreshkova et al., 2020). Virus transmission from infected minks back to humans was corroborated by phylogenetic comparison between human‐ and mink‐derived SARS‐CoV‐2 sequences, which grouped together (Oude Munnink et al., 2020). Zoonotic infections are common occupational hazards in individuals who are frequently exposed to animals or animal products such as veterinarians, zoo and abattoir workers, breeders and farmers (Epp & Waldner, 2012). Risk assessments should be carried out to identify occupational groups that are disproportionately exposed to potentially SARS‐CoV‐2 infected animals, as was exemplified by a novel squirrel bornavirus, which caused fatal encephalitis among squirrel breeders and a zoo worker (Tappe et al., 2018).
The combined data from natural and experimental infections together with preliminary epidemiologic investigations highlight the potential transmission of SARS‐CoV‐2 from humans to different mammalian orders, particularly members of the order Carnivora (Figure 3a, Table 1). Comparing the ACE2 residues involved in SARS‐CoV‐2 entry in humans (Shang et al., 2020) with the sequences from different carnivore species revealed only few amino acid changes, which is consistent with SARS‐CoV‐2 infection in those mammals (Table 2). It is unclear whether mutations in the SARS‐CoV‐2 spike receptor‐binding domain are needed to increase infection efficiency in different carnivore hosts or if an ACE2‐independent pathway, potentially mediated by the SARS‐CoV‐2 furin cleavage site, is employed in parallel to receptor‐mediated entry (Hoffmann et al., 2020).
Figure 3.
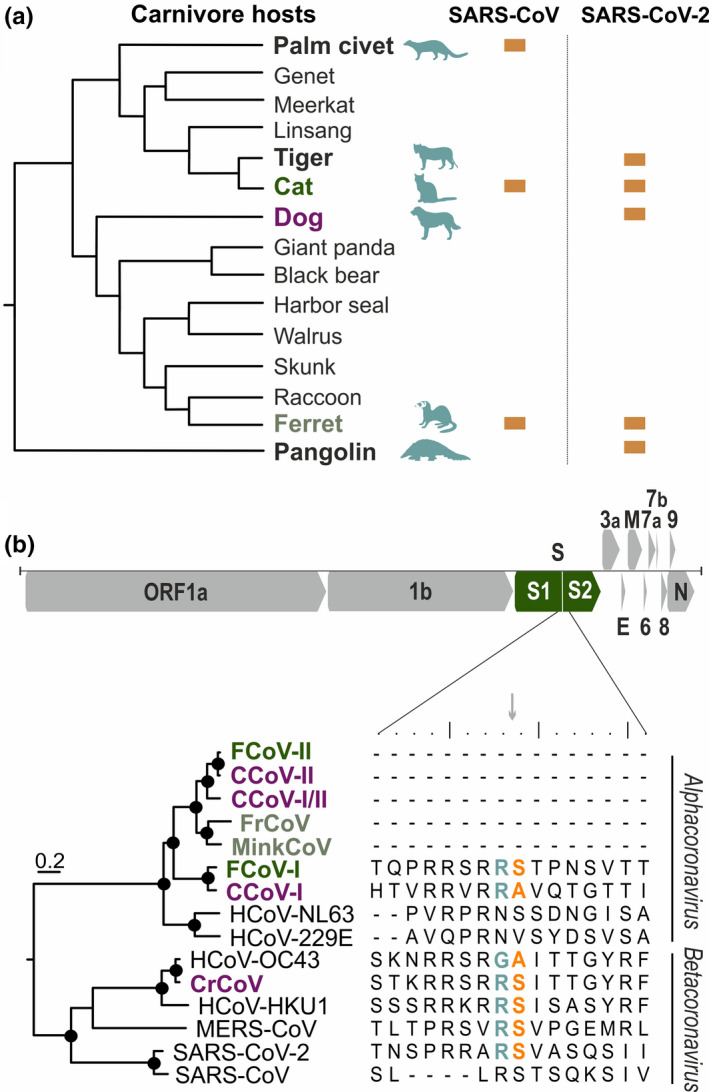
Susceptibility of carnivore hosts to coronaviruses (a) Carnivores susceptible to SARS‐related CoVs are dispersed across the family tree. Cladogram of carnivore families adapted from (Foley et al., 2016). (b) Furin cleavage site between the spike subunits S1 and S2 is predominantly present in betacoronaviruses. Scheme of SARS‐CoV‐2 genome organization with a magnified S1/S2 furin cleavage site within carnivore and human coronaviruses . Functional cleavage sites are highlighted with coloured ‘R S’ (orange and teal). Maximum‐likelihood tree of human and carnivore coronaviruses showing grouping of cat (green), dog (purple) and mustelid (grey) CoVs with human CoVs based on translated spike amino acid sequences. WAG + G + I was used as a substitution model and a complete deletion option was chosen. Scale bar indicates amino acid substitutions per site. Circle at nodes indicate bootstrap values ≥75% (1,000 bootstrap replicates). GenBank accession numbers: YP_004070194, AFG19726, AKZ66476, YP009256197, ADI80513, ACT10854, AEQ61968, YP_003767, NP_073551, AAR01015, AQT26498, YP_173238, YP_009047204, QHU36864, ACB69905 [Colour figure can be viewed at wileyonlinelibrary.com]
Table 2.
Critical ACE2 residues that interact with the receptor binding domain site of the SARS‐CoV‐2 spike protein based on human SARS‐CoV‐2 infection (Shang et al., 2020)
| Species | ACE2 residue | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 24 | 27 | 28 | 31 | 34 | 35 | 37 | 38 | 41 | 42 | 45 | 79 | 82 | 83 | 329 | 330 | 353 | 354 | 355 | 357 | |
| Human | S | Q | T | F | K | H | E | E | D | Y | Q | L | L | M | Y | E | N | K | G | D | R |
| Domestic cat | S | L | T | F | K | H | E | E | E | Y | Q | L | L | T | Y | E | N | K | G | D | R |
| Dog | S | L | T | F | K | Y | E | E | E | Y | Q | L | L | T | Y | G | N | K | G | D | R |
| Ferret | S | L | T | F | K | Y | E | E | E | Y | Q | L | H | T | Y | Q | N | K | R | D | R |
| Raccoon | S | L | T | F | N | N | E | E | E | Y | Q | L | Q | T | Y | E | N | K | G | D | R |
| Polar bear | S | L | T | F | K | Y | E | E | D | Y | Q | L | H | T | Y | E | N | K | G | D | R |
| Civet | S | L | T | F | T | Y | E | Q | E | Y | Q | V | L | T | Y | E | N | K | G | D | R |
| Meerkat | S | L | T | F | Q | H | E | Q | E | Y | L | V | R | A | Y | D | N | K | G | D | R |
| Hyena | S | L | T | F | K | Y | E | Q | E | Y | L | L | L | T | Y | E | N | K | G | D | R |
| Harbour seal | S | L | T | F | K | Y | E | E | E | Y | Q | L | Q | T | Y | E | N | K | R | D | R |
| Sea lion | S | L | T | F | K | S | E | E | E | Y | Q | L | Q | T | Y | E | N | K | H | D | R |
| Walrus | S | L | T | F | K | Y | E | E | E | Y | Q | F | Q | T | Y | E | N | K | H | D | R |
| Pangolin | S | E | T | F | K | S | E | E | E | Y | Q | L | I | N | Y | E | N | K | H | D | R |
| Horseshoe bat | S | E | M | F | K | T | K | E | D | H | Q | L | L | N | Y | N | N | K | G | D | R |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | ||||||||
Cat, dog and ferret residues identical to human ACE2 critical residues are highlighted with an asterisk.
4. FURIN CLEAVAGE SITE IN HUMAN AND CARNIVORE CORONAVIRUSES
Different from SARS‐CoV, SARS‐CoV‐2 contains a furin protease cleavage site in the viral spike protein, which is responsible for attachment and entry into cells (Coutard et al., 2020). In MERS‐CoV and influenza viruses, presence of a furin cleavage site has been associated with enhanced transmissibility, increased virulence and broader host range (Andersen et al., 2020). It has been speculated that furin‐mediated spike cleavage may also affect SARS‐CoV‐2 pathogenesis (Andersen et al., 2020). Several feline coronavirus (FCoV) strains also have a furin cleavage site in their spike proteins (Figure 3b) (Licitra et al., 2013). Interestingly, mutations in the furin cleavage site are mainly found in the feline infectious peritonitis virus (FIPV) biotype, which constitutes the virulent variant of FCoV. (Licitra et al., 2013). It is still a matter of debate if the observed mutations contribute to the acquisition of monocyte and macrophage tropism, a hallmark of FIPV (Licitra et al., 2013). SARS‐CoV‐2 studies investigating virus compartmentalization defining distinct viral populations, which may lead to different biotypes similarly to FCoV, should thus be conducted.
The bat/pangolin‐associated SARS‐CoV‐2‐related coronaviruses do not contain a functional furin cleavage site, yet a furin‐like cleavage site appears in other betacoronaviruses such as MERS‐CoV, HCoV‐HKU1 and HCoV‐OC43 (Figure 3b). The evolutionary origins of furin cleavage motifs in CoVs are not clear. Frequent recombination and high genetic diversity of bat‐associated SARS‐related CoVs (Hu et al., 2017; Latinne et al., 2020) may support an origin in the bat reservoir, but whether the motif emerged in a common coronavirus ancestor and was subsequently lost in most alphacoronaviruses, or whether the furin cleavage site was acquired independently from unknown sources remains to be determined.
5. CORONAVIRUSES NATURALLY INFECTING CARNIVORES
Besides from FCoV, which was already introduced in the section above, enzootic carnivore coronaviruses include canine coronavirus (CCoV), canine respiratory coronavirus (CrCoV), ferret coronavirus (FrCoV) and mink coronavirus (MinkCoV) (Figure 3b). Most of the mentioned carnivore CoVs belong to the genus Alphacoronavirus along with HCoV‐229E and HCoV‐NL63. CrCV is the only carnivore CoV within the genus Betacoronavirus, which includes the other HCoVs, namely HCoV‐OC43, HCoV‐HKU1, MERS‐CoV and SARS‐related CoV strains (Figure 3b).
Modified live attenuated FIPV and inactivated CCoV vaccines are commercially available. However, the use of FIPV vaccine has been controversial, as antibody‐dependent enhancement of infection has been reported in experimental settings, albeit not in the field (Takano et al., 2008). It is unclear what immunity to enzootic cat and dog CoVs elicited either by vaccination or natural infection will play in SARS‐CoV‐2 transmission between carnivores.
Currently, it is not known whether pre‐existing antibodies against human‐endemic CoVs afford cross‐ protection against or immune enhancement of SARS‐CoV‐2 infection. Despite decades of investigation into endemic HCoVs, no vaccine has been developed to prevent infections with HCoV‐229E, HCoV‐OC43, HCoV‐NL63 and HKU1, as these viruses mostly generate mild disease (Corman et al., 2018). For SARS‐CoV, translational research has been limited as the importance of the virus decreased due to the successful containment of the virus by timely public health interventions (Cheng et al., 2013). In contrast, several vaccine candidates have been tested against the recently emerged MERS‐CoV, including vectored vaccines based on adenovirus and poxvirus backbones (Alharbi, 2017). Because MERS‐CoV naturally infects camels, and human infections are almost exclusively zoonotic outside of outbreaks in hospital settings (Corman et al., 2018), MERS‐CoV vaccines were developed following a One Health approach considering applicability to both humans and camels (Alharbi, 2017). A vaccine candidate using a poxvirus vector is now in phase I trial as of December 2019 after tests in dromedary camels were successful (Alharbi et al., 2019; NLM, 2019).
Further investigation into animal coronavirus immune responses will be required, particularly into the implications of pre‐existing carnivore coronavirus immunity on SARS‐CoV‐2 infections including both cross‐protection and potential immune enhancement, which can shed light on the immune interplay in HCoV infections and potentially inform SARS‐CoV‐2 vaccine development.
6. CONCLUSION
Pandemic SARS‐CoV‐2 spread combined with over 800 million cats and dogs kept as pets worldwide (Bedford, 2020), and the large extent of the animal farming industry (Ritchie, 2017) raises concerns of domestic and farmed animals in general and carnivores in particular, becoming an epidemiologically relevant animal source for COVID‐19. Non‐pharmaceutical interventions to contain spread of SARS‐CoV‐2 may be compromised if (1) the close interaction between humans and companion or farmed animals contributes to human infections and if (2) infected domestic animals roam freely, potentially contributing to virus transmission between different households. If that was the case, strategies to mitigate and contain SARS‐CoV‐2 zoonotic transmission should be developed in a One Health framework, implementing molecular and serological surveillance as well as epidemiological assessment of SARS‐CoV‐2 occurrence in domestic and farmed animals alongside humans.
CONFLICT OF INTEREST
Authors declare no competing interests.
ETHICAL APPROVAL
No ethical approval was required as this is a review article with no original research data.
Acknowledgement
Open access funding enabled and organized by Projekt DEAL.
Jo WK, de Oliveira‐Filho EF, Rasche A, Greenwood AD, Osterrieder K, Drexler JF. Potential zoonotic sources of SARS‐CoV‐2 infections. Transbound Emerg Dis.2021;68:1824–1834. 10.1111/tbed.13872
[Correction added on 14 December 2020 after first online publication: Table 2 caption has been corrected in this version.]
DATA AVAILABILITY STATEMENT
Data sharing is not applicable as no new data was created.
References
- Alharbi, N. K. (2017). Vaccines against Middle East respiratory syndrome coronavirus for humans and camels. Reviews in Medical Virology, 27(2), e1917. 10.1002/rmv.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi, N. K. , Qasim, I. , Almasoud, A. , Aljami, H. A. , Alenazi, M. W. , Alhafufi, A. , … Balkhy, H. H. (2019). Humoral immunogenicity and efficacy of a single dose of ChAdOx1 MERS vaccine candidate in dromedary camels. Scientific Reports, 9(1), 16292. 10.1038/s41598-019-52730-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K. G. , Rambaut, A. , Lipkin, W. I. , Holmes, E. C. , & Garry, R. F. (2020). The proximal origin of SARS‐CoV‐2. Nature Medicine, 26(4), 450–452. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anti, P. , Owusu, M. , Agbenyega, O. , Annan, A. , Badu, E. K. , Nkrumah, E. E. , … Drosten, C. (2015). Human‐bat interactions in rural West Africa. Emerging Infectious Diseases, 21(8), 1418–1421. 10.3201/eid2108.142015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrs, V. R. , Peiris, M. , Tam, K. W. S. , Law, P. Y. T. , Brackman, C. J. , To, E. M. W. , … Sit, T. H. C. (2020). SARS‐CoV‐2 in quarantined domestic cats from COVID‐19 households or close contacts, Hong Kong, China. Emerg Infect Dis, 26(12). 10.3201/eid2612.202786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford, E. (2020). Global dog and cat pet population 2018 Retrieved from https://www.statista.com/statistics/1044386/dog‐and‐cat‐pet‐population‐worldwide/. [Google Scholar]
- CDC . (2020, 22.04.2020). Confirmation of COVID‐19 in Two Pet Cats in New York. Retrieved from https://www.cdc.gov/media/releases/2020/s0422‐covid‐19‐cats‐NYC.html. [Google Scholar]
- Cheng, V. C. , Chan, J. F. , To, K. K. , & Yuen, K. Y. (2013). Clinical management and infection control of SARS: lessons learned. Antiviral Research, 100(2), 407–419. 10.1016/j.antiviral.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Eckerle, I. , Memish, Z. A. , Liljander, A. M. , Dijkman, R. , Jonsdottir, H. , … Drosten, C. (2016). Link of a ubiquitous human coronavirus to dromedary camels. Proceedings of the National Academy of Sciences of the United States of America, 113(35), 9864–9869. 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Muth, D. , Niemeyer, D. , & Drosten, C. (2018). Hosts and sources of endemic human coronaviruses. Advances in Virus Research, 100, 163–188. 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of, V (2020). The species Severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nature Microbiology, 5(4), 536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard, B. , Valle, C. , de Lamballerie, X. , Canard, B. , Seidah, N. G. , & Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019‐nCoV contains a furin‐like cleavage site absent in CoV of the same clade. Antiviral Research, 176, 104742. 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, N. (2020, 04.06.2020). A German shepherd is first dog in the U.S. to test positive for the coronavirus. Retrieved from https://www.nationalgeographic.com/animals/2020/06/first‐dog‐to‐test‐positive‐in‐us‐for‐coronavirus/. [Google Scholar]
- Drexler, J. F. , Corman, V. M. , & Drosten, C. (2014). Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Research, 101, 45–56. 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler, J. F. , Gloza‐Rausch, F. , Glende, J. , Corman, V. M. , Muth, D. , Goettsche, M. , … Drosten, C. (2010). Genomic characterization of severe acute respiratory syndrome‐related coronavirus in European bats and classification of coronaviruses based on partial RNA‐dependent RNA polymerase gene sequences. Journal of Virology, 84(21), 11336–11349. 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp, T. , & Waldner, C. (2012). Occupational health hazards in veterinary medicine: zoonoses and other biological hazards. Canadian Veterinary Journal, 53(2), 144–150. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/22851775. [PMC free article] [PubMed] [Google Scholar]
- FAO‐OIE‐WHO (2019). A Tripartite Guide to Addressing Zoonotic Diseases in Countries. Retrieved from https://extranet.who.int/sph/docs/file/3866. [Google Scholar]
- Foley, N. M. , Springer, M. S. , & Teeling, E. C. (2016). Mammal madness: is the mammal tree of life not yet resolved? Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1699), 20150140. 10.1098/rstb.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling, C. M. , Breithaupt, A. , Müller, T. , Sehl, J. , Balkema‐Buschmann, A. , Rissmann, M. , Mettenleiter, T. C. (2020). Raccoon dogs are susceptible to and efficiently transmit SARS‐CoV2 and may serve as intermediate host. bioRxiv. 10.1101/2020.08.19.256800. [DOI] [Google Scholar]
- Graham, R. L. , & Baric, R. S. (2010). Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross‐species transmission. Journal of Virology, 84(7), 3134–3146. 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzmacher, K. S. , Kondgen, S. , Keil, V. , Todd, A. , Feistner, A. , Herbinger, I. , … Leendertz, F. H. (2016). Codetection of Respiratory Syncytial Virus in Habituated Wild Western Lowland Gorillas and Humans During a Respiratory Disease Outbreak. Ecohealth, 13(3), 499–510. 10.1007/s10393-016-1144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J. , & Korteweg, C. (2007). Pathology and pathogenesis of severe acute respiratory syndrome. The American Journal of Pathology, 170(4), 1136–1147. 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y. , Zheng, B. J. , He, Y. Q. , Liu, X. L. , Zhuang, Z. X. , Cheung, C. L. , … Poon, L. L. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science, 302(5643), 276–278. 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Halfmann, P. J. , Hatta, M. , Chiba, S. , Maemura, T. , Fan, S. , Takeda, M. , … Kawaoka, Y. (2020). Transmission of SARS‐CoV‐2 in Domestic Cats. New England Journal of Medicine, 383(6), 592–594. 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , & Pohlmann, S. (2020). A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Molecular Cell, 78(4), 779–784 e775. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Zeng, L. P. , Yang, X. L. , Ge, X. Y. , Zhang, W. , Li, B. , … Shi, Z. L. (2017). Discovery of a rich gene pool of bat SARS‐related coronaviruses provides new insights into the origin of SARS coronavirus. PLOS Pathogens, 13(11), e1006698. 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, B. , Wang, M. , Jing, H. , Xu, H. , Jiang, X. , Yan, M. , … Xu, J. (2005). Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus‐like virus in palm civets at an animal market and on farms. Journal of Virology, 79(18), 11892–11900. 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, T. T. , Shum, M. H. , Zhu, H. C. , Tong, Y. G. , Ni, X. B. , Liao, Y. S. , … Guan, Y. (2020). Identifying SARS‐CoV‐2 related coronaviruses in Malayan pangolins. Nature, 583(7815), 282–285. 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Latinne, A. , Hu, B. , Olival, K. J. , Zhu, G. , Zhang, L. , Li, H. , … Daszak, P. (2020). Origin and cross‐species transmission of bat coronaviruses in China. Nature Communications, 11(1), 4235. 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Laudette, C.‐L. (2020, 08.05.2020). Spanish cat tests positive for coronavirus. Retrieved from https://www.reuters.com/article/us‐health‐coronavirus‐spain‐cat/spanish‐cat‐tests‐positive‐for‐coronavirus‐idUSKBN22K1TK. [Google Scholar]
- Li, Q. , Guan, X. , Wu, P. , Wang, X. , Zhou, L. , Tong, Y. , … Feng, Z. (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. New England Journal of Medicine, 382(13), 1199–1207. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Moore, M. J. , Vasilieva, N. , Sui, J. , Wong, S. K. , Berne, M. A. , … Farzan, M. (2003). Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Shi, Z. , Yu, M. , Ren, W. , Smith, C. , Epstein, J. H. , … Wang, L. F. (2005). Bats are natural reservoirs of SARS‐like coronaviruses. Science, 310(5748), 676–679. 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Licitra, B. N. , Millet, J. K. , Regan, A. D. , Hamilton, B. S. , Rinaldi, V. D. , Duhamel, G. E. , & Whittaker, G. R. (2013). Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerging Infectious Diseases, 19(7), 1066–1073. 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina, B. E. , Haagmans, B. L. , Kuiken, T. , Fouchier, R. A. , Rimmelzwaan, G. F. , Van Amerongen, G. , … Osterhaus, A. D. (2003). Virology: SARS virus infection of cats and ferrets. Nature, 425(6961), 915. 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildenstein, T. , Tanshi, I. , & Racey, P. A. (2016). Exploitation of Bats for Bushmeat and Medicine. In Voigt C. & Kingston T. (Ed.), Bats in the Anthropocene: Conservation of Bats in a Changing World (pp. 325–375). Springer, Cham. 10.1007/978-3-319-25220-9_12. [DOI] [Google Scholar]
- Munoz‐Fontela, C. , Dowling, W. E. , Funnell, S. G. P. , Gsell, P. S. , Balta, X. R. , Albrecht, R. A. , … Barouch, D. H. (2020). Animal models for COVID‐19. Nature, –. 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster, V. J. , Feldmann, F. , Williamson, B. N. , van Doremalen, N. , Perez‐Perez, L. , Schulz, J. , … de Wit, E. (2020). Respiratory disease in rhesus macaques inoculated with SARS‐CoV‐2. Nature, 585(7824), 268–272. 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn, A. Z. , Lamers, M. M. , Okba, N. M. A. , Breugem, T. I. , Schipper, D. , van den Doel, P. B. , Haagmans, B. L. (2020). Susceptibility of rabbits to SARS‐CoV‐2. bioRxiv. 10.1101/2020.08.27.263988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NLM . (2019, 20.02.2020). A Clinical Trial to Determine the Safety and Immunogenicity of Healthy Candidate MERS‐CoV Vaccine (MERS002). Retrieved from https://clinicaltrials.gov/ct2/show/NCT04170829. [Google Scholar]
- Oreshkova, N. , Molenaar, R. J. , Vreman, S. , Harders, F. , Oude Munnink, B. B. , Hakze‐van der Honing, R. W. , Stegeman, A. (2020). SARS‐CoV‐2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance, 25(23), 2001005. 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020). Middle East respiratory syndrome coronavirus (MERS‐CoV) – United Arab Emirates. Retrieved from https://www.who.int/csr/don/31‐january‐2020‐mers‐united‐arab‐emirates/en/. [Google Scholar]
- Oude Munnink, B. B. , Sikkema, R. , Nieuwenhuijse, D. F. , Molenaar, R. J. , Munger, E. , Molenkamp, R. , Koopmans, M. (2020). Jumping back and forth: anthropozoonotic and zoonotic transmission of SARS‐CoV‐2 on mink farms. bioRxiv. 10.1101/2020.09.01.277152. [DOI] [Google Scholar]
- Patrono, L. V. , Samuni, L. , Corman, V. M. , Nourifar, L. , Rothemeier, C. , Wittig, R. M. , … Leendertz, F. H. (2018). Human coronavirus OC43 outbreak in wild chimpanzees, Cote d Ivoire, 2016. Emerging Microbes & Infections, 7(1), 118. 10.1038/s41426-018-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, E. I. , Elia, G. , Grassi, A. , Giordano, A. , Desario, C. , Medardo, M. , Decaro, N. (2020). Evidence of exposure to SARS‐CoV‐2 in cats and dogs from households in Italy. bioRxiv. 10.1101/2020.07.21.214346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ProMed‐mail . (2020a). COVID‐19 update (58): Belgium, animal, cat, clinical case, RFI Retrieved from https://promedmail.org/promed‐post/?id=20200327.7151215. [Google Scholar]
- ProMed‐mail , (2020b, 30.04.2020). COVID‐19 update (143): USA (NY) animal, zoo, tiger, lion, tests. Retrieved from https://promedmail.org/promed‐post/?id=20200430.7284183. [Google Scholar]
- ProMed‐mail (2020c). COVID‐19 update (181): Germany (BY), France (AC), cat, OIE animal case defin. Retrieved from https://promedmail.org/promed‐post/?id=20200513.7332909 [Google Scholar]
- ProMed‐mail . (2020d, 25.09.2020). COVID‐19 update (414): animal, Netherlands (LI), Denmark (ND), farm mink, spread. Retrieved from https://promedmail.org/promed‐post/?id=7813579. [Google Scholar]
- Ritchie, H. (2017). Meat and Dairy Production. Retrieved from https://ourworldindata.org/meat‐production. [Google Scholar]
- Roberts, A. , Vogel, L. , Guarner, J. , Hayes, N. , Murphy, B. , Zaki, S. , & Subbarao, K. (2005). Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. Journal of Virology, 79(1), 503–511. 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx, B. , Kuiken, T. , Herfst, S. , Bestebroer, T. , Lamers, M. M. , Oude Munnink, B. B. , … Haagmans, B. L. (2020). Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science, 368(6494), 1012–1015. 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau, K. , Rissmann, M. , Graaf, A. , Schon, J. , Sehl, J. , Wylezich, C. , … Beer, M. (2020). SARS‐CoV‐2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. The Lancet Microbe, 1(5), e218–e225. 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J. , Ye, G. , Shi, K. , Wan, Y. , Luo, C. , Aihara, H. , … Li, F. (2020). Structural basis of receptor recognition by SARS‐CoV‐2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Wen, Z. , Zhong, G. , Yang, H. , Wang, C. , Huang, B. , … Bu, Z. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science, 368(6494), 1016–1020. 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, S. F. , Yan, L. M. , Chin, A. W. H. , Fung, K. , Choy, K. T. , Wong, A. Y. L. , … Yen, H. L. (2020). Pathogenesis and transmission of SARS‐CoV‐2 in golden hamsters. Nature, 583(7818), 834–838. 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit, T. H. C. , Brackman, C. J. , Ip, S. M. , Tam, K. W. S. , Law, P. Y. T. , To, E. M. W. , … Peiris, M. (2020). Infection of dogs with SARS‐CoV‐2. Nature, 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, H. D. , Tu, C. C. , Zhang, G. W. , Wang, S. Y. , Zheng, K. , Lei, L. C. , … Zhao, G. P. (2005). Cross‐host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proceedings of the National Academy of Sciences of the United States of America, 102(7), 2430–2435. 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling, T. (2020, 15.05.2020). Dutch dog, three cats infected with coronavirus: minister. Retrieved from https://www.reuters.com/article/us‐health‐coronavirus‐netherlands‐pets/dutch‐dog‐three‐cats‐infected‐with‐coronavirus‐minister‐idUSKBN22R2EN. [Google Scholar]
- Takano, T. , Kawakami, C. , Yamada, S. , Satoh, R. , & Hohdatsu, T. (2008). Antibody‐dependent enhancement occurs upon re‐infection with the identical serotype virus in feline infectious peritonitis virus infection. Journal of Veterinary Medical Science, 70(12), 1315–1321. 10.1292/jvms.70.1315. [DOI] [PubMed] [Google Scholar]
- Tappe, D. , Schlottau, K. , Cadar, D. , Hoffmann, B. , Balke, L. , Bewig, B. , … Beer, M. (2018). Occupation‐associated fatal limbic encephalitis caused by variegated squirrel bornavirus 1, Germany, 2013. Emerging Infectious Diseases, 24(6), 978–987. 10.3201/eid2406.172027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling, E. C. , Springer, M. S. , Madsen, O. , Bates, P. , O'Brien, S.J. , & Murphy, W. J. (2005). A molecular phylogeny for bats illuminates biogeography and the fossil record. Science, 307(5709), 580–584. 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Ulrich, L. , Wernike, K. , Hoffmann, D. , Mettenleiter, T. C. , & Beer, M. (2020). Experimental infection of cattle with SARS‐CoV‐2. bioRxiv. 10.1101/2020.08.25.254474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UN (2020a). Coronavirus: the economic impact. Retrieved from https://www.unido.org/stories/coronavirus‐economic‐impact. [Google Scholar]
- UN , (2020b). The Social Impact of COVID‐19. Retrieved from https://www.un.org/development/desa/dspd/2020/04/social‐impact‐of‐covid‐19/. [Google Scholar]
- Vijgen, L. , Keyaerts, E. , Lemey, P. , Maes, P. , Van Reeth, K. , Nauwynck, H. , … Van Ranst, M. (2006). Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. Journal of Virology, 80(14), 7270–7274. 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Muller, M. A. , … Wendtner, C. (2020). Virological assessment of hospitalized patients with COVID‐2019. Nature, 581(7809), 465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xiao, K. , Zhai, J. , Feng, Y. , Zhou, N. , Zhang, X. , Zou, J. J. , … Shen, Y. (2020). Isolation of SARS‐CoV‐2‐related coronavirus from Malayan pangolins. Nature, 583(7815), 286–289. 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Zhang, H. , Gao, J. , Huang, K. , Yang, Y. , Hui, X. , … Jin, M. (2020). A serological survey of SARS‐CoV‐2 in cat in Wuhan. Emerging Microbes & Infections, 9(1), 2013–2019. 10.1080/22221751.2020.1817796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. Z. , & Holmes, E. C. (2020). A genomic perspective on the origin and emergence of SARS‐CoV‐2. Cell, 181(2), 223–227. 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Wang, J. , Kuang, D. , Xu, J. , Yang, M. , Ma, C. , Peng, X. (2020). Susceptibility of tree shrew to SARS‐CoV‐2 infection. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable as no new data was created.


