To the Editor,
Coronavirus disease 2019 (COVID‐19) is a primary acute respiratory infection caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). It has become a worldwide pandemic and public health emergency and is associated with considerable morbidity and mortality. SARS‐CoV‐2 utilizes angiotensin‐converting enzyme II (ACE2) as its cellular receptor, 1 and high ACE2 expression has been shown to increase cell susceptibility to SARS‐CoV‐2 in vitro. 1 In addition, elevated ACE2 expression has been reported in high‐risk patient groups including smokers and those with diabetes and hypertension. 2 Thus, understanding the expression and regulation of ACE2 under different pathological conditions may help to predict patient's susceptibility to COVID‐19 and ultimately their clinical outcomes. 2
Chronic rhinosinusitis (CRS) is a chronic inflammatory disease of the sinonasal mucosa that affects 5%–15% of the adult population. 3 Understanding the associations between CRS and COVID‐19 is important for the management of CRS in the era of COVID‐19. Using single‐cell RNA‐sequencing analysis, Ziegler et al recently reported that ACE2 mRNA expression level is lower in the sinus mucosa of patients with CRS than in the inferior turbinate tissues (IT) of non‐CRS controls. 4 However, the sample size in that study was small and the RNA data need to be verified at the protein level. Moreover, IT may not be a proper control for diseased sinus mucosa as a previous study demonstrated significant differences in the expression of several innate host defense molecules between ethmoid sinus tissues (ET) and IT. 5 Therefore, it is necessary to evaluate ACE2 expression in different regions of the nasal cavity.
Here, we evaluated ACE2 expression in repository sinonasal mucosal samples from 39 control subjects without CRS, 55 patients with CRS and nasal polyps (CRSwNP), and 42 patients with CRS without nasal polyps (CRSsNP), which were collected before the COVID‐19 outbreak. Samples were evaluated by RT‐PCR and immunohistochemistry. This study was approved by the ethics committee of the Tongji Hospital, and written informed consent was obtained from each patient. More information regarding the subjects, the different types of sinonasal mucosal specimen and the methods used in their evaluation are provided in this article's Online Supplement, Tables S1‐S3.
We found that ACE2 mRNA expression in the tissue and protein expression in the epithelium was lower in ET samples from CRSsNP patients and nasal polyps (NP) from CRSwNP patients than in ET samples from non‐CRS control subjects, with a more profound decrease in the NP group (Figure 1A,B). Since IT is the proximal point of contact for inhaled air, it is critical for nasal immunity against airborne pathogens; consequently, we also studied ACE2 expression in IT. In contrast to the sinus mucosa, we found that ACE2 expression was comparable in IT samples from non‐CRS controls, CRSsNP patients, and CRSwNP patients (Figure 1C,D). Immunohistochemistry revealed that ACE2 was primarily expressed in the epithelial cells of the sinonasal mucosa (Figure 1B‐D). In addition, ACE2 expression level was lower in the ET and NP samples of the CRSsNP and CRSwNP patients than in their IT samples. However, ACE2 expression level was comparable in the ET and IT samples from the control subjects (Figure S1A,B).
FIGURE 1.
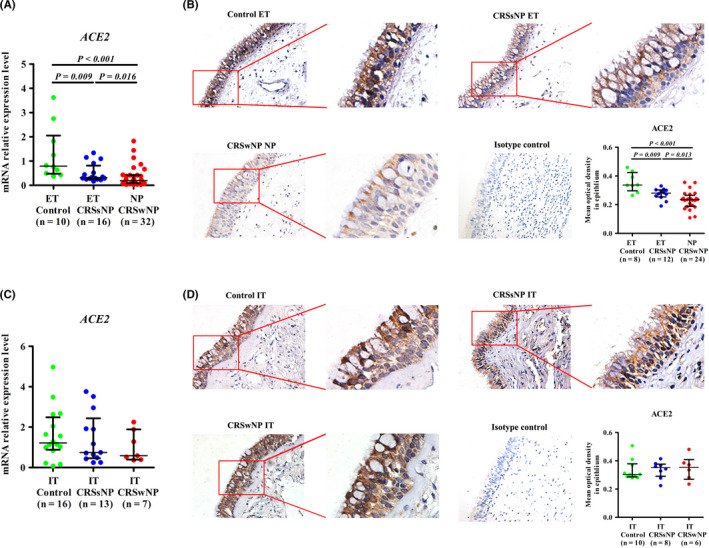
ACE2 expression in the sinus mucosa and IT samples from patients with CRS. A, The mRNA expression levels of ACE2 in normal ET from control subjects, diseased ET from CRSsNP patients, and NP from CRSwNP patients as detected by quantitative RT‐PCR. B, The immunoreactivity of ACE2 in sinus mucosal samples as detected by immunohistochemistry. C, The mRNA expression levels of ACE2 in IT samples from control subjects, CRSsNP patients and CRSwNP patients as detected by quantitative RT‐PCR. D, The immunoreactivity of ACE2 in IT samples as detected by immunohistochemistry. For B and D, the representative photomicrographs are shown (original magnification × 400). A higher magnification of the outlined area is shown as an inset. The staining intensity in the epithelium was quantified and the results are presented as the average optical density value per unit area. Data are presented as the median and interquartile range. The Kruskal‐Wallis H test was used to assess significant intergroup variability. The Mann‐Whitney U 2‐tailed test was used for between‐group comparisons. Significance was set at a P value of less than 0.017. ACE2, angiotensin‐converting enzyme II; CRSsNP, chronic rhinosinusitis without nasal polyps; CRSwNP, chronic rhinosinusitis with nasal polyps; IT, inferior turbinate tissues; ET, ethmoid sinus tissues; NP, nasal polyps
We further investigated the relationship between ACE2 mRNA expression and differential inflammatory patterns. We found no differences in ACE2 mRNA and protein expression between eosinophilic and non‐eosinophilic NP (Figure S2). ACE2 expression showed no correlation with interleukin (IL)‐5, IL‐17A, or interferon (IFN)‐γ expression in sinus mucosal tissues (Figure S3), or the expression of various type 2 response genes including beta‐galactoside alpha‐2,6‐sialyltransferase (ST6GAL) 1, chemokine (C‐C motif) ligand 26 (CCL26), and periostin (POSTN) (Figure S4). We found that the expression of various IFN response genes including chemokine (C‐X‐C motif) ligand (CXCL) 10, and CXCL11 were significantly lower in the ET samples from CRSsNP patients and NP samples from CRSwNP patients than in the ET samples from control groups, with a further decrease in NP (Figure S5A). CXCL10 and CXCL11 expression did not differ between eosinophilic and non‐eosinophilic NP (Figure S5B). Notably, ACE2 mRNA expression was positively correlated with CXCL10 and CXCL11 mRNA expression in sinus tissues (Figure 2A).
FIGURE 2.
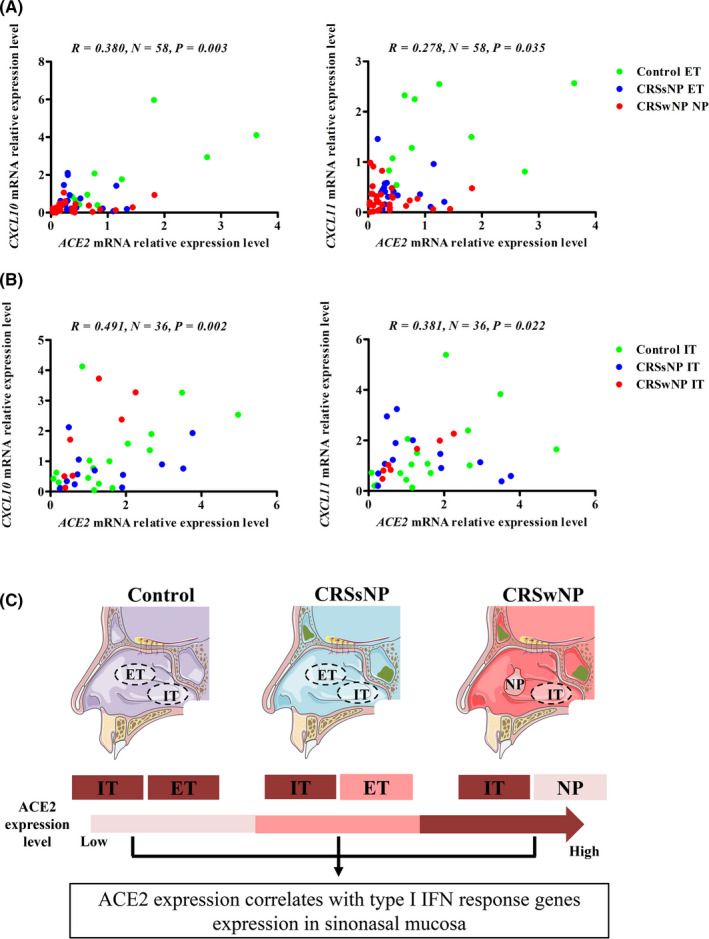
Correlations between ACE2 gene expression and IFN response gene expression in the sinus mucosa and IT. A, ACE2 mRNA expression positively correlates with CXCL10 and CXCL11 mRNA expression in the sinus mucosa. B, ACE2 mRNA expression positively correlates with CXCL10 and CXCL11 mRNA expression in IT. C, A graphical summary of ACE2 expression in the sinonasal mucosa. Spearman rank correlation analysis was used to evaluate correlation, and significance was set at a P value of less than 0.05. ACE2, angiotensin‐converting enzyme II; IFN, interferon; CXCL, chemokine (C‐X‐C motif) ligand; CRSsNP, chronic rhinosinusitis without nasal polyps; CRSwNP, chronic rhinosinusitis with nasal polyps; IT, inferior turbinate tissues; ET, ethmoid sinus tissues; NP, nasal polyps
We found that CXCL10 and CXCL11 mRNA levels in IT did not differ between the various groups; this finding was consistent with the ACE2 expression data (Figure S6A). CXC10 and CXCL11 mRNA expression levels were lower in the sinus mucosa of patients with CRS than in their IT samples, but these levels were comparable between the ET and IT samples from the control subjects (Figure S6B,C). In the IT samples, ACE2 mRNA expression was also positively correlated with the expression of CXCL10 and CXCL11 (Figure 2B) but not with the expression of IL‐5, IL‐17A, IFN‐γ, ST6GAL1, CCL26, or POSTN (Figures S7 and S8).
Here, where we used ET samples from non‐CRS subjects as a control, we were able to demonstrate that ACE2 mRNA and protein expression levels decreased in diseased ET samples from CRSsNP patients and NP samples from CRSwNP patients. Recent studies reported that ACE2 mRNA expression in airway epithelial cells was upregulated by the type I interferon (IFN) response and downregulated by type 2 inflammation. 4 , 6 We found no differences in ACE2 expression between eosinophilic and non‐eosinophilic NP and no correlations between ACE2 expression and type 2 cytokine or type 2 response gene expression, countermanding the dominant role of type 2 response‐mediated regulation of ACE2 expression in the sinus mucosa. In contrast, we found that type I IFN responses were decreased in ET or NP samples from patients with CRS and that these changes correlated with ACE2 expression. The reduced expression of IFN‐stimulated genes in the sinus mucosa of patients with CRS has also been reported by other groups, indicating that patients with CRS may have a deficient local anti‐viral defense. 7 IFN response deficits may limit the entry of SARS‐CoV‐2 by downregulating ACE2 expression. However, IFN response‐mediated elimination of the virus is also impaired. Therefore, the contribution of the complicated interactions between ACE2 and the IFN response during the development of COVID‐19 in patients with CRS warrants further investigation.
Unlike the sinus mucosa, there were no changes in ACE2 expression in the IT samples from any of the evaluated groups, which again correlated with the type I IFN response gene expression. Since both IT and sinus mucosa are important parts of the nasal cavity mucosal lining, our study suggests that it is necessary to consider each of the different regions of the sinonasal mucosa independently when assessing the function of the nasal cavity, particularly in patients with CRS. Our study shows that ACE2 expression in the nasal cavity may not increase the risk for developing COVID‐19 in patients with CRS. This is consistent with our recent data that CRS prevalence in hospitalized COVID‐19 patients in Wuhan (6.1%) was close to the CRS prevalence in the general population of China (8%), and CRS comorbidity was not associated with severity of COVID‐19. 8 Chhiba et al reported that COVID‐19 patients with rhinosinusitis presented a lower risk of hospitalization than those without rhinosinusitis. 9 However, Chhiba et al did not differentiate between acute and chronic rhinosinusitis. 9
Our study has several limitations. First, there are no available data regarding the correlations between ACE2 expression and the use of corticosteroids or biologics in our cohort, suggesting that the influence of steroid‐ or biologics‐based treatments on the risk of COVID‐19 requires further study. Second, there are overlaps in ACE2 mRNA expression in the ET samples from the control and CRSsNP subjects and the NP samples from the CRSwNP patients. These data should be considered with caution. Moreover, since ACE2 is primarily expressed in the epithelium of sinonasal tissues, examining ACE2 mRNA expression in nasal epithelial cells could provide more precise information. Third, ACE2 expression was observed in ciliated and goblet cells in a previous single‐cell RNA‐sequencing study. 4 It would be of great interest to explore whether the regional differences in ACE2 expression in CRS are the result of intrinsic differences in ACE2 expression in specific cell types or the change in the abundance of specific cell types. Fourth, since it is very difficult to obtain tissue samples from patients with CRS who experience a good response to medical treatment without surgical intervention, whether these patients have similar ACE2 expression profiles as those undergoing surgery remain an open question. Fifth, beyond being a receptor for SARS‐CoV‐2, ACE2 plays a crucial role in maintaining the balance of the renin‐angiotensin system by countering the activities of angiotensin II. Whether ACE2 plays an additional role in the nasal cavity warrants further study.
In summary, our study provides the first evidence of regional differences in ACE2 expression in the sinonasal mucosa of patients with CRS and that ACE2 expression in CRS tissues is associated with type I IFN response and not type 2 response (Figure 2C).
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Supplementary Material
Wang and Song these authors contributed equally to this work.
Funding information This study was supported by grants from the National Natural Science Foundation of China (NSFC) [81630024 and 81920108011 (ZL), and 81900925 (JS)], and the Natural Science Foundation of Hubei Province, China [2018CFB602 (MZ)].
Contributor Information
Ming Zeng, Email: zmsx77@163.com.
Zheng Liu, Email: zhengliuent@hotmail.com.
REFERENCES
- 1. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Structure VD. Function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐292.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li Y, Zhou W, Yang L, You R. Physiological and pathological regulation of ACE2, the SARS‐CoV‐2 receptor. Pharmacol Res. 2020;157:104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol. 2017;12:331‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 Is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016‐1035.e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seshadri S, Rosati M, Lin DC, et al. Regional differences in the expression of innate host defense molecules in sinonasal mucosa. J Allergy Clin Immunol. 2013;132(5):1227‐1230.e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sajuthi SP, DeFord P, Jackson ND, et al. Type 2 and interferon inflammation strongly regulate SARS‐CoV‐2 related gene expression in the airway epithelium. medRxiv preprint. 2020. 10.1101/2020.04.09.034454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peng Y, Zi XX, Tian TF, et al. Whole‐transcriptome sequencing reveals heightened inflammation and defective host defence responses in chronic rhinosinusitis with nasal polyps. Eur Respir J. 2019;54(5):1900732. 10.1183/13993003.00732-2019 [DOI] [PubMed] [Google Scholar]
- 8. Wang H, Song J, Pan LI, et al. The characterization of chronic rhinosinusitis in hospitalized patients with COVID‐19. J Allergy Clin Immunol Pract. 2020. 10.1016/j.jaip.2020.09.013. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chhiba KD, Patel GB, Vu THT, et al. Prevalence and characterization of asthma in hospitalized and non‐hospitalized patients with COVID‐19. J Allergy Clin Immunol. 2020;146(2):307‐314.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Supplementary Material


