Abstract
Viral infections cause high morbidity and mortality, threaten public health, and impose a socioeconomic burden. We have seen the recent emergence of SARS‐CoV‐2 (Severe Acute Respiratory Syndrome Coronavirus 2), the causative agent of COVID‐19 that has already infected more than 29 million people, with more than 900 000 deaths since its identification in December 2019. Considering the significant impact of viral infections, research and development of new antivirals and control strategies are essential. In this paper, we summarize 96 antivirals approved by the Food and Drug Administration between 1987 and 2019. Of these, 49 (51%) are used in treatments against human immunodeficiency virus (HIV), four against human papillomavirus, six against cytomegalovirus, eight against hepatitis B virus, five against influenza, six against herpes simplex virus, 17 against hepatitis C virus and one against respiratory syncytial virus. This review also describes future perspectives for new antiviral therapies such as nanotechnologies, monoclonal antibodies and the CRISPR‐Cas system. These strategies are suggested as inhibitors of viral replication by various means, such as direct binding to the viral particle, blocking the infection, changes in intracellular mechanisms or viral genes, preventing replication and virion formation. We also observed that a large number of viral agents have no therapy available and the majority of those approved in the last 32 years are restricted to some groups, especially anti‐HIV. Additionally, the emergence of new viruses and strains resistant to available antivirals has necessitated the formulation of new antivirals.
Keywords: antivirals, CRISPR‐Cas, monoclonal antibodies, nanoparticles
Introduction
Viruses are aetiological agents of several chronic and severe diseases and have a global presence. Some viruses can lead to epidemics or pandemics, as observed throughout history such as those caused by the influenza virus [1], human immunodeficiency virus (HIV) – the aetiological agent of acquired immunodeficiency syndrome (AIDS) – and recently COVID‐19, the disease caused by the new coronavirus SARS‐CoV‐2. It is estimated that 1.8 million people are infected with HIV worldwide, with Africa and Southeast Asia being the most affected regions [2]. There were about 12 800 deaths globally due to the H1N1 Influenza A pandemic in 2009, and the highest mortality rate was registered in the South American continent, with 76.9 deaths per 10 000 inhabitants [3]. Since its emergence in December 2019, COVID‐19 has infected more than 29 million people with more than 900 000 deaths worldwide until August 2020, characterising the largest pandemic of the current century [4]. Due to their ease of transmission and wide dissemination, along with the possibility of potentially lethal complications, respiratory viral infections account for high rates of morbidity and mortality [5]. Central nervous system (CNS) tropism, shown by some viruses such as herpes simplex virus types 1 and 2 (HSV‐1 and HSV‐2) and varicella zoster virus (VZV), can also result in severe acute infections, such as meningitis and encephalitis or microcephaly, as seen in Zika virus (ZIKV) infection [6, 7]. In addition to viruses causing chronic diseases, there exist oncogenic viruses such as hepatitis B virus (HBV), which has infected approximately 250 million people, and human papillomavirus (HPV), the aetiological agent of sexually transmitted infection and cervical cancer [8, 9]. Viruses are also associated with socioeconomic ratios and longevity and affect the quality of life in a population. Some African countries have suffered recurrent epidemics caused by the Ebola virus, linked to low health development rates and serious public health problems [10].
Considering the high prevalence of viral infections, the limited number of antivirals available, the emergence of new viruses, and the re‐emergence of some viruses, research on new antivirals has become extremely important. Moreover, high genetic variation presented by some viruses necessitates constant monitoring for possible pandemics in the form of updating available vaccines that have become obsolete due to emergence of new viral strains [11]. The search for and development of new antiviral drugs is quite challenging; however, new and encouraging antiviral therapies have been proposed. In this study, we review on the antiviral drugs approved in recent years and new proposals for antiviral therapies.
Antiviral Therapies
Antiviral drugs are still quite limited in terms of number and variety. Since viruses are obligatory intracellular pathogens, they are essentially dependent on the machinery and metabolism of a host cell. Therefore, selective toxicity and minimization of major side effects to the host are necessary during the development of new antivirals [12, 13]. In addition to the possibility of resistance presented by certain viral strains, limitations of in vitro laboratory techniques and availability of animal models that mimic human infections in vivo also hinder the discovery of new candidate molecules for antivirals [9].
Antiviral drug therapy started with iododeoxyuridine (IDU), a nucleoside analogue synthesized in 1959, initially as a potential anticancer drug [14]. In the late 1970s, the development of acyclovir (ACV) and the subsequent addition of its prodrugs represented a major advance in the treatment of viral infections. ACV acts on HSV‐infected cells, where its active form is effectively acquired through phosphorylation by a viral enzyme (thymidine kinase), demonstrating low toxicity and potent selectivity of this drug [15].
By definition, antivirals are compounds that inhibit the formation of new viruses and do so by blocking essential stages of viral replication. Thus, understanding these steps is essential for the development of new antiviral agents [16]. A generalized summary of the viral replication steps is presented in Figure 1 and comprises the adsorption of the virion onto the host cell, penetration into the intracellular environment, uncoating and release of viral nucleic acid, replication of genetic material, synthesis of viral components, and virion assembly and release.
Figure 1.
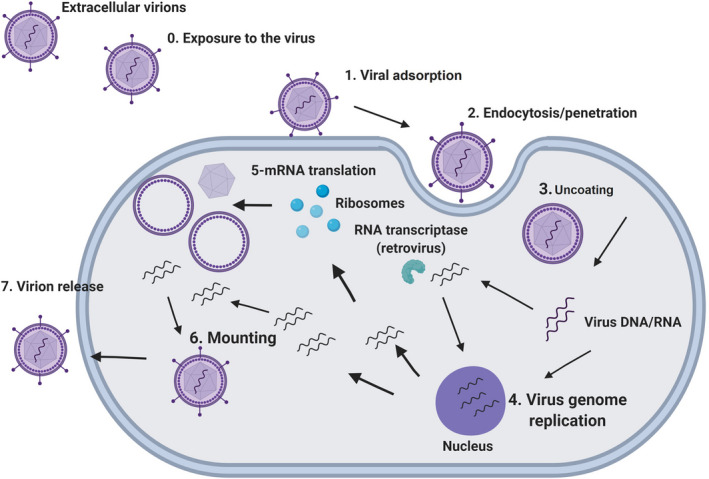
Steps of viral replication: exposure to the virus (0); viral adsorption onto the host cell surface (1); penetration of the virus into the intracellular environment (2); uncoating and disassembly (3); viral genome replication and transcription (4); viral mRNA translation (5); mounting (6); and release of the viral progeny (7).
Adsorption and penetration are excellent targets for antiviral activity. For example, the anti‐HIV drug enfuvirtide blocks the fusion of the viral envelope with the cell membrane, specifically binding to the glycoprotein (gp‐41) of the envelope [17]. After penetration, some viruses need to carry their genetic material to the host cell nucleus using microtubules, suggesting a potential target for therapeutic intervention [18]. Blocking replicative enzymes such as viral polymerases is another targetable viral replication step for drugs like zidovudine, which inhibits HIV reverse transcriptase [19].
Due to the complexity of the viral replication steps associated with the mutagenic capacity of some viruses, antiviral therapies occasionally involve the use of two or more drugs in order to increase the effectiveness of the treatment, featuring antiviral polytherapy or combination therapy. Polytherapy seems to be a viable alternative treatment for some viruses such as HIV, especially against resistant viral strains [20]. In addition to HIV, in a study by Beigel et al. [21], the synergistic effect of oseltamivir, amantadine and ribavirin against H1N1 was demonstrated to be more efficacious than that of monotherapy with oseltamivir.
In addition to antivirals that work by interfering in the replication cycle, other compounds that control the viral infection by modulating the host immune response have also been investigated. This group includes interferons which represent a family of cytokines that activate the innate immune response against viral infections [22]. Briefly, interferons can act on both, infected and healthy cells, imparting resistance to viral infection, called an antiviral state. The interferons secreted by infected cells prevent viral replication by binding to cell receptors, especially on adjacent cells, activating the transcription of genes encoding antiviral proteins, such as ribonucleases, thereby blocking viral replication [23]. The antiviral imiquimod, which acts as an antagonist to toll‐like receptors, is an example of such immune response modifiers [12]. Toll‐like receptors are crucial in the immune response against infections, since they recognize conserved regions of pathogens and induce the production of cytokines [24].
Food and Drug Administration (FDA)‐approved substances and drugs which have other therapeutic indications have also been evaluated as possible antivirals by expanding their pharmacological action. Currently, this strategy has been widely used in emergency attempts to control the COVID‐19 pandemic, taking advantage of the knowledge of pharmacokinetics, pharmacodynamics, along with awareness of side effects of previously available drugs. However, the inhibitory effects found in vitro can only be confirmed in vivo in long‐term studies. Currently, around 900 clinical trials are underway worldwide, including those for antivirals, corticosteroids and antibiotics, among others, against COVID‐19 [4]. However, only few candidate agents, such as hydroxychloroquine (HCQ), and chloroquine and remdesivir and lopinavir‐ritonavir, have been included in the World Health Organization (WHO) SOLIDARITY clinical trial. By enrolling patients from several countries, the SOLIDARITY trial aims to quickly determine whether any of these drugs slow the progression of the disease or improve patient survival.
Chloroquine or Hydroxychloroquine – indicated for the treatment of malaria and rheumatoid arthritis – has already been evaluated for treatment of viruses such as hepatitis A, HIV, flavivirus [11] and coronavirus [25]. Its antiviral mechanisms are based on its capacity to increase the endosomal pH. This prevents enveloped viruses, such as those belonging to the Coronaviridae family (e.g. SARS‐CoV‐2), from entering and releasing their genetic material into the host cells [26]. The anti‐SARS‐CoV‐2 activity of hydroxychloroquine has been tested in different countries through randomized multicenter studies. Although the studies varied in the number of patients evaluated, period of pharmacological intervention (outpatients or inpatients), and design, that is, monotherapy or combination therapy (azithromycin/oseltamivir), hydroxychloroquine treatment does not seem to show significant improvement in mortality, in addition to showing possible adverse events such as heart failure, increased hospital stay, and risk of progression to invasive mechanical ventilation or death [27, 28, 29, 30, 31, 32, 33, 34]. Another randomized study evaluated the use of lopinavir‐ritonavir in the treatment of COVID‐19 in critically ill patients hospitalized in Wuhan but did not identify any benefit in relation to the control [35].
Remdesivir, an adenosine analogue that interferes with viral RNA polymerase activity, has a broad antiviral spectrum. Its activity against the MERS and SARS coronaviruses has been demonstrated in vitro and in vivo. Recently, two clinical practice guidelines recommended the administration of remdesivir [36, 37], based on the findings of the randomized clinical trial (RCT) ACTT‐1. This trial enrolled 1 063 patients from 13 countries, selecting patients with severe as well as mild/moderate illness. It reported a considerable reduction in recovery time for COVID‐19 patients treated with remdesivir compared with that for placebo treatment; however, the trial revealed no major differences in mortality or the need for ventilation [38]. Another randomized trial carried out on 237 patients with severe COVID‐19 revealed no significant clinical improvement, neither in the mortality rate (14%) nor in the time for elimination of the virus in the group treated with remdesivir compared with that for placebo treatment. However, all patients included in this study had pre‐existing illnesses [39]. These trials both addressed the critical outcomes for COVID‐19 treatment (defined by the panel), including mortality, mechanical ventilation, time to clinical improvement, duration of hospitalization and adverse events related to drug administration. Another cohort study on patients hospitalized for severe COVID‐19 showed 68% clinical improvement upon remdesivir treatment; however, the mortality rate observed in this study corroborated with that reported by Wang et al. [39]. Moreover, 60% of the patients experienced one or more adverse drug events, of which 23% had serious conditions. The most common adverse events were abnormal liver function, diarrhoea, rash, renal impairment and hypotension [40]. Thus far, the ongoing trials examining remdesivir include WHO SOLIDARITY, DISCOVERY (NCT04315948) and SIMPLE (NCT04292899).
A large number of natural compounds have also demonstrated antiviral effects in vitro and in vivo, such as those derived from the Digitalis lanata plant are already being used as cardiotonic drugs due to the presence of cardiac glycosides and have shown promising anti‐HIV‐1 activity by inducing splicing during the viral replication process [41]. In addition to the compounds (synthetic or natural) proposed as antivirals, many studies have demonstrated the potential of metallic nanoparticles, monoclonal antibodies and the CRISPR system in controlling viral infections. The antiviral activity of nanoparticles has already been demonstrated against H1N1 influenza using iron oxide [42] and against hepatitis C virus (HCV) using copper nanoparticles [43]. Monoclonal antibodies have been shown to be effective in neutralising several viruses and can mount an immune defence by acting in a way similar to that of natural antibodies [44]. In addition to their therapeutic role, monoclonal antibodies can also have a prophylactic function due to their persistence in the serum [45]. The CRISPR‐Cas9 technique is based on a gene‐editing system which uses CRISPR sequences – grouped and regularly spaced short palindromic repetitions – that are naturally present in prokaryotes [46]. It has been investigated experimentally for viral infections such as HIV infections wherein it acts directly on the viral genetic material, especially on the provirus, in patients in whom the disease is under control [47]. These techniques are described below.
FDA‐approved antivirals between 1987 and 2019
In this review, antivirals approved by the FDA between 1987 and 2019 have been analysed. These drugs are being used for therapeutic use against RNA viruses, including HCV, HIV, influenza virus (INFLU), respiratory syncytial virus (RSV) and against DNA virus, including cytomegalovirus (CMV), HBV, HPV and herpes simplex virus (HSV). The year of approval, generic and commercial name, and targeted viral component or mechanism is mentioned in Table I [17, 48].
Table I.
FDA approved antivirals between 1987 and 2019 for HCV, HIV, INFLU, RSV, CMV, HBV, HPV and HSV.
| Virus | Year | Drugs | Commercial | Route of administration | Action location |
|---|---|---|---|---|---|
| Generic | |||||
| HCV | 1991 | Interferon Alfa‐2B | Intron A (Merck, USA) |
Subcutaneous/ Intramuscular |
Host |
| 1997 | Interferon Alfacon‐1 | Infergen (Amgen, USA) |
Subcutaneous/ Intramuscular |
Host | |
| 1998 | Ribavirin | Rebetol (Merck, USA) | Oral | Host | |
| 2001 | Peginterferon Alfa‐2B | Pegintron (Merck, USA) | Subcutaneous | Host | |
| 2002 | Peginterferon Alfa‐2A | Pegasys (Roche, SWI) | Subcutaneous | Host | |
| 2011 |
Boceprevir Telaprevir |
Victrelis (Merck, USA) Incivek (Vertex P.Inc, USA) |
Oral Oral |
Pr Pr |
|
| 2013 |
Simeprevir sodium Sofosbuvir |
Olysio (Janssen‐Cilag, BE) Sovaldi (Gilead Sc., USA) |
Oral Oral |
Pr Pol |
|
| 2014 |
Ledipasvir/sofosbuvir DasabuvirSodium/ombitasvir/paritaprevir/ritonavir |
Harvoni (Gilead Sc., USA) Viekira Pak (AbbVie, USA) |
Oral Oral |
NS5A/Pol Pr/Pr/NsS5A/Pol |
|
| 2015 |
Daclatasvir dihydrochloride Ombitasvir/paritaprevir/ritonavir |
Daklinza (Bristol‐Myers S., USA) Technivie (AbbVie, USA) |
Oral Oral |
NS5A Pr/Pr/NS5A |
|
| 2016 |
Elbasvir/grazoprevir Sofosbuvir/velpatasvir |
Zepatier (Merck, USA) Epclusa (Gilead Sc., USA) |
Oral Oral |
Pr/NS5A Pol/NS5A |
|
| 2017 |
Sofosbuvir/velpatasvir/voxilaprevir Glecaprevir/pibrentasvir |
Vosevi (Gilead Sc., USA) Mavyret (AbbVie, USA) |
Oral Oral |
Pr/Pol/NS5A Pr/NS5A |
|
| 1987 | Zidovudine | Retrovir (GSK, UK) | Oral | Pol | |
| 1991 | Didanosine | Videx (Bristol‐Myers S., USA) | Oral | Pol | |
| 1992 | Zalcitabine | Hivid (Roche, SWI) | Subcutaneous/ Intramuscular | Pol | |
| 1994 | Stavudine | Zerit (Bristol‐Myers S., USA) | Oral | Pol | |
| 1995 |
Lamivudine Saquinavir mesylate |
Epivir (ViiV Healthcare, UK) Invirase (Genentech, USA) |
Oral Oral |
Pol Pr |
|
| HIV | 1996 |
Ritonavir Indinavir sulphate Nevirapine |
Norvir (AbbVie, USA) Crixivan (Merck, USA) Viramune (Boehringer Ing., DE) |
Oral Oral Oral |
Pr Pr Pol |
| 1997 |
Nelfinavir mesylate Delavirdine mesylate Lamivudine/zidovudine |
Viracept (Pfizer, USA) Rescriptor (Pfizer, USA) Combivir (GSK, UK) |
Oral Oral Oral |
Pr Pr Pol/Pol |
|
| 1998 |
Efavirenz Abacavir sulphate |
Sustiva (Bristol‐Myers S., USA) Ziagen (GSK, UK) |
Oral Oral |
Pol Pol |
|
| 1999 | Amprenavir | Agenerase (GSK, UK) | Oral | Pr | |
| 2000 | Lopinavir/ritonavir | Kaletra (AbbVie, USA) | Oral | Pr/Pr | |
| 2001 | Tenofovir disoproxil fumarate | Viread (Gilead Sc., USA) | Oral | Pol | |
| 2002 | Abacavir/ sulphate/ lamivudine/ zidovudine | Trizivir (GSK, UK) | Oral | Pol/Pol/Pol | |
| 2003 |
Enfuvirtide (T20) Atazanavir sulphate Emtricitabine Fosamprenavir calcium |
Fuzeon (Genentech, USA) Reyataz (Bristol‐Myers S., USA) Emtriva (Gilead Sc., USA) Lexiva (GSK, UK) |
Subcutaneous Oral Oral Oral |
Fusion Pr Pol Pr |
|
| 2004 |
Abacavir/lamivudine/emtricitabine/tenofovir Disoproxil fumarate |
Epzicom (ViiV Healthcare, USA) Truvada (Gilead Sc., USA) |
Oral Oral |
Pol Pol |
|
| 2005 | Tipranavir |
Aptivus (Boehringer Ing., DE) |
Oral | Pr | |
| 2006 |
Darunavir ethanolate/efavirenz/emtricitabine/tenofovir Disoproxil fumarate |
Prezista (Janssen‐Cilag, BE) Atripla (Gilead Sc., USA) |
Oral Oral |
Pr Pol/Pol/Pol |
|
| HIV | 2007 |
Maraviroc Raltegravir potassium |
Selzentry (Pfizer, USA) Isentress (Merck, USA) |
Oral Oral |
Host Int |
| 2008 | Etravirine | Intelence (Janssen‐Cilag, BE) | Oral | Pol | |
| 2011 |
Rilpivirine Emtricitabine/rilpivirine/tenofovir disoproxil/fumarate |
Edurant (Janssen‐Cilag, BE) Complera (Gilead Sc., USA) |
Oral Oral |
Pol Pol/Pol/Pol |
|
| 2012 | Cobicistat/elvitegravir/emtricitabine/tenofovir disoproxil/fumarate | Stribild (Gilead Sc., USA) | Oral | Host/Int/Pol/Pol | |
| 2013 | Dolutegravir Sodium | Tivicay (ViiV Healthcare, USA) | Oral | Int | |
| 2014 |
Elvitegravir Abacavir sulphate/dolutegravir sodium/lamivudine |
Vitekta (Gilead Sc., USA) Triumeq (ViiV Healthcare, USA) |
Oral Oral |
Int Pol/Int/Pol |
|
| 2015 |
Cobicistat/darunavir Atazanavir/cobicistat Lamivudine/raltegravir Cobicistat/elvitegravir/emtricitabine/tenofovir alafenamide Emtricitabine/tenofovir alafenamide |
Prezcobix (Janssen‐Cilag, BE) Evotaz (Bristol‐Myers S., USA) Dutrebis (Merck, USA) Genvoya (Gilead Sc., USA) Descovy (Gilead Sc., USA |
Oral Oral Oral Oral Oral |
Pr Pr Pol/Int Host/Int/Pol/Pol Pol |
|
| 2016 |
Emtricitabine/rilpivirine/tenofovir alafenamide |
Odefsey (Gilead Sc., USA) |
Oral |
Pol |
|
| 2017 | Dolutegravir/rilpivirine | Juluca (ViiV Healthcare, USA) | Oral | Pol/Int | |
| HIV | 2018 |
Ibalizumab‐uiyk Doravirine Bictregavir/emtricitabine/tenofovir alafenamide Cobicistat/darunavir/emtricitabine/tenofovir alafenamide Doravirine/lamivudine/tenofovir disoproxil |
Trogarzo (TaiMed Biologics, USA) Pifeltro (Merck, USA) Biktarvy (Gilead Sc., USA) Symtuza (Janssen‐Cilag, BE) Delstrigo (Merck, USA) |
Intravenous Oral Oral Oral Oral |
Host Pol Pr/Pol/Pol Host/Pr/Pol/Pol Pol/Pol/Pol |
| 2019 | Dolutegravir sodium/lamivudine | Dovato (ViiV Healthcare, USA) | Oral | Pr/Pol | |
| 1993 | Rimantadine | Flumadine (Sun pharma, IN) | Oral | Uncoating | |
| INFLU | 1999 |
Zanamivir Oseltamivir |
Relenza (GSK, UK) Tamiflu (Genentech, USA) |
Inhalation Oral |
Neu Neu |
| 2014 | Peramivir |
Rapivab (BioCryst Pharma, USA) |
Intravenous | Neu | |
| 2018 | Baloxavir marboxil | Xofluza (Genentech, USA) | Oral | Pol | |
| RSV | 1998 | Palivizumab | Synagis (AstraZeneca, USA) | Intramuscular | Fusion |
| 1989 | Ganciclovir sodium | Cytovene (Roche, SWI) | Intravenous | Pol | |
| 1991 | Foscarnet sodium | Foscavir (AstraZeneca, USA) | Intravenous | Pol | |
| CMV | 1996 | Cidofovir | Vistide (Gilead Sc., USA) | Intravenous | Pol |
| 1998 | Fomivirsen sodium | Vitravene (Isis Pharma., USA) | Intravenous | NA | |
| 2001 | Valganciclovir hydrochloride | Valcyte (Genentech, USA) | Oral | Pol | |
| 2017 | Letermovir | Prevymis (Merck, USA) | Oral or Intravenous | Rep | |
| 1992 | Interferon Alfa‐2B | Intron A (Merck, USA) |
Subcutaneous/ Intramuscular |
Host | |
| 1998 | Lamivudine | Epivir‐HBV (GSK, UK) | Oral | Pol | |
| 2002 | Adefovir dipivoxil | Hepsera (Gilead Sc., USA) | Oral | Pol | |
| HBV | 2005 |
Entecavir Peginterferon Alfa‐2A |
Baraclude Bristol‐Myers S., USA) Pegasys (Genentech, USA) |
Oral Subcutaneous |
Pol Host |
| 2006 | Telbivudine | Tyzeka (Novartis Pharm., SWI) | Oral | Pol | |
| 2008 | Tenofovir disoproxil fumarate | Viread (Gilead Sc., USA) | Oral | Pol | |
| 2015 | Tenofovir alafenamide fumarate | Vemlidy (Gilead Sc., USA) | Oral | Pol | |
| 1988 | Interferon Alfa‐2B | Intron A (Merck, USA) | Subcutaneous/Intramuscular | Host | |
| HPV | 1989 | Interferon Alfa‐N3 | Alferon N (AIM ImmunoTech,, USA) | intralesional | Host |
| 1997 | Imiquimod | Aldara (Graceway Pharma., USA) | Topical | Host | |
| 2006 | Sinecatechins |
Veregen (Bradley Pharma., USA) |
Topical | Host | |
| 1994 | Famciclovir | Famvir (Novartis Pharm., SWI) | Oral | Pol | |
| 1995 | Valacyclovir hydrochloride | Valtrex (GSK, UK) | Oral | Pol | |
| HSV | 1996 | Penciclovir | Denavir (Prestium Pharma., USA) | Oral | Pol |
| 1998 | Trifluridine | Viroptic (Monarch Pharma., MY) | Ophthalmic Solution | Pol | |
| 2000 | Docosanol | Abreva (Avanir Pharma., USA) | Topical | Fusion | |
| 2009 | Acyclovir/hydrocortisone |
Xerese (Valeant Pharma., CA) |
Topical | Pol |
CMV, cytomegalovirus; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HPV, human papilloma virus; HSV, herpes simplex virus; INFLU, influenza; Int, integrase; NA, nucleic acid; Neu, neuraminidase; NS5A, non‐structural protein 5A; Pol, polymerase; Pr, protease; Rep, replication; RSV, respiratory syncytial virus.
There are 49 HIV antivirals available so far, with the first one, zidovudine approved in 1987. Some drugs are used in polytherapy, while some are used in combination, especially those approved in the last decade. HIV treatment usually includes different medications with different targets so that in case of resistance to one, the others are still effective [49]. Overall, we have listed 96 FDA‐approved antivirals between 1987 and 2019, of which 49 are anti‐HIV, with the most number of approved drugs (51%), followed by 17 anti‐HCV, five anti‐influenza, one anti‐RSV, six anti‐CMV, eight anti‐HBV, four anti‐HPV and six anti‐HSV drugs. The years with the most approvals were 1998 and 2015 with seven, followed by 2018 with six and 1996, 1997, 2014, with five approvals each (Figure 2 ).
Figure 2.
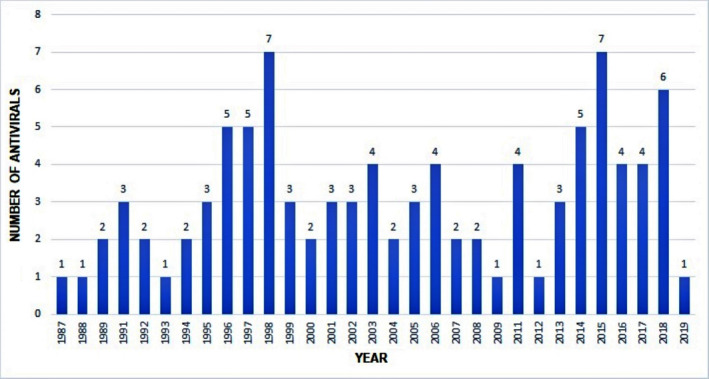
FDA‐approved antivirals between 1987 and 2019 by year of approval.
New Perspectives for the Treatment of Viral Diseases
Nanotechnology applied to antivirals
Scientific interest in nanoparticles is on the rise, due to their versatility and especially their wide applicability. Owing to the conversion of pure metals into nanoparticles, nanotechnology has become a powerful research technique in medical sciences and is used in the diagnosis of diseases, for carrying drugs, and even for their antimicrobial and anticancer activities [42, 50, 51]. These infinitesimal particles, produced using varying methods and used in different formats and compositions, are capable of transporting substances into the body, improving therapeutic efficacy and decreasing toxicity of the substance carried [43, 44, 52]. Nanostructured delivery systems have numerous advantages for drug delivery, including the abilities to protect the active molecules against degradation in physiological medium and to release the active substance in a controlled manner at the site of action. This biotechnology, therefore, assists in conducting active principles to their target locations with more specificity, bypassing problems associated with commercially available drugs. Further, features such as easy synthesis, biocompatibility and optical properties make them suitable for biological applications.
Depending on the materials with which they are prepared, nanoparticles are classified as polymeric (usually prepared using biodegradable polymers to provide controlled release of encapsulated therapeutic substances), solid lipids, magnetics or metallics [53]. Regarding their mechanism of action, although not confirmed or outlined, it is assumed that nanoparticles act directly on the virus or that they induce structural changes in the virus, preventing it from penetrating the target cell. Some studies suggest the intracellular action of nanoparticles through protein interference. Thus, the possible mechanisms of action of nanoparticles still need to be elucidated. Table II shows data gathered from some recently published studies investigating nanoparticle antiviral activity, proving their effectiveness and wide use [12].
Table II.
Update on research of nanoparticles with antiviral activity.
| Year | Reference | Nanoparticle | Virus | Effect | ||
|---|---|---|---|---|---|---|
| 2019 | [51] | Iron oxide | Influenza A | Alteration of viral RNA transcripts, with an 8‐fold reduction in viral RNA | ||
| 2018 | [44] | Chitosan/siRNA | Influenza A | Inhibition of viral replication | ||
| 2018 | [42] | Tannic/Silver Acid | HSV‐1 e HSV‐2 | Inhibition of viral adsorption | ||
| 2018 | [54] | Dentritic polyglycerol sulphate | HSV‐1 | Inhibition of viral entry | ||
| 2018 | [52] | Gold coated with polymers (MES & MUS) | HSV‐1, HSV‐2, HPV‐16, RSV, LV‐VSV‐G e DENV‐2 | Virucidal effect | ||
| 2018 | [55] | Diphyline and bafilomycin | Influenza A | Increased antiviral activity in vitro, but in vivo diphyline nanoparticles reduced pulmonary viral charge in mice | ||
| 2017 | [56] | Ritonavir carrier lipids | HIV | Maintained ritonavir activity and modulated drug release | ||
| 2016 | [57] | Zinc oxide tetrapods | HSV‐2 | Inhibition of viral entry and virustatic effect | ||
| 2016 | [58] | Siallylactosamine‐carrying glyco‐nanoparticles | Influenza A | Inhibition of virus binding to cell | ||
| 2016 | [59] | Galactosylated conjugated amines | HCV | Inhibition of virus replication through siRNA release by nanoparticles | ||
| 2016 | [60] | Dapivirine carriers | HIV | Improvement in distribution, retention, and modulation of drug release by nanoparticles in vaginal gel formulation | ||
| 2015 | [61] | Silver using aqueous extract of Bruguiera cylindrica | DENV‐2 | Significant reduction in viral RNA | ||
| 2015 | [62] | Silver modified with curcumin | RSV | Effective virus inactivation before entering the host cell | ||
| 2015 | [63] | Cuprous oxide | HCV | Inhibition of viral entry | ||
Nanotechnology was recently suggested for combatting SARS‐CoV‐2 infection. Results of successful studies on respiratory viruses, including SARS‐CoV [64], could also be applied to this new coronavirus through polymeric inorganic nanoparticles, using self‐assembling and peptide‐based proteins [65]. This method will enable not only faster mucosal penetration and dissemination but also formulation of stable biodegradable products with minimal levels of pulmonary toxicity during the treatment. The activity against SARS‐CoV‐2 could be explained on the basis of a study by Schmitt et al. [66], in which virus‐like nanoparticles containing a fusion protein conferred a protective effect against RSV in mice. This effect is suggested to be stimulated by the induction of defence cells as natural killers, tumour necrosis factor (TNFα) and interferon gamma (IFN‐γ) (+) in the pulmonary and bronchiolar airways during the infection stage and the absence of harmful plasmacytoid dendritic cells (pDCs) and effector T cells.
Nanoparticles offer many advantages, especially in the treatment and eradication of infectious diseases. As antivirals, they show improved effectiveness and overcome limitations such as low bioavailability, adverse side effects, frequency of ingestion and treatment time. However, this new technology still needs complementary studies that seek the characterization of nanomaterials and development of highly biocompatible, biodegradable, and non‐cytotoxic nanocarrier systems as well as nano‐transporters to act specifically on viral infection without affecting healthy cells and tissues. These nanomaterials, in addition to their role in drug delivery, should be able to exert their expected antiviral therapeutic characteristics. Thus, studies investigating the interactions of nanomaterials with the immune system, which plays the most important role in controlling viral infections, are required.
Monoclonal antibodies against viral infections
The production and isolation of monoclonal antibodies (mAbs) began in the 1970s by stimulating antibody production in animals via myeloma tumour cells using a technique known as hybridoma. Currently, this production can be optimized using chimeric monoclonal antibodies from camelids, by ‘phage display’, isolation of memory B cells, and direct cloning of the secretory plasmacytic or the antibody itself, via genetic and molecular coding [67]. mAbs have high specificity with direct and rapid viral repression [68]. They neutralize virions by identifying viral surface antigens which are essential for recognition and binding to host cells (Figure 3 ). Moreover, viral dissemination can be reduced by the effector functions of the Fc fragment) and recognition of the complement system [69].
Figure 3.
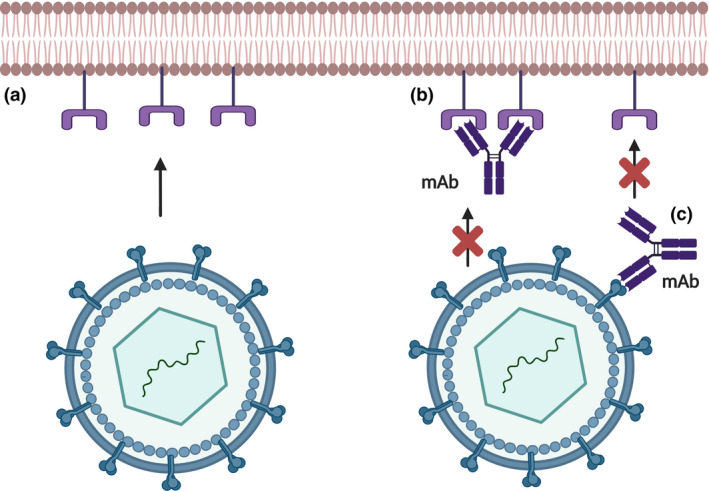
Without the presence of the monoclonal antibody (mAbs), the virion glycoproteins have the ability to bind to the receptors of host cells (a). In therapy, mAbs may show affinity for viral receptors in host cells (b) or directly to the virus (c), preventing viral adsorption and consequently, infection.
The first FDA‐approved mAb was ibalizumab (Theratechnologies Inc.) and was used to treat HIV in adult patients refractory to conventional antiviral treatment [70]. It acts by blocking the entry of HIV‐1 into host cells by non‐competitive binding to TCD4+ lymphocytes [71]. It induces a conformational change in the gp120 binding site, preventing the virus from fusing with the cell membrane [72]. Other mAbs such as 3BNC117, 3BNC117‐LS and VRC01 also function by blocking the TCD4 binding site, which is necessary for viral envelope adsorption; however, they are still in the clinical trial phase [73, 74].
mAbs are also beneficial in prophylactic treatment, especially against emerging viruses such as ZIKV in pregnant women, preventing the impairment of foetal development. In this context, three antibodies have already been formulated and are undergoing laboratory testing: SMZAb1, SMZAb2 and SMZAb5, which prevented viral replication in monkeys [74]. Sapparapu et al. [75] demonstrated a decrease in infection in pregnant mice and foetal tissues with the use of mAb ZIKV‐117. The first human mAb to proceed to the clinical trial stage will be Z021, which has significant neutralising potential after recognising an envelope domain (EDIII) epitope of ZIKV. INO‐A002 is the most recently announced mAb, which uses DNA decoding technology for the production of antibodies (dmAb). When released into the body, dmAb is capable of genetically instructing the immune system cells to produce modified antibodies. dmAbs exhibit improved and more stable treatment kinetics, providing greater protection to experimental models and representing a major advance over conventional approaches [76].
Zmapp is an experimental drug that has shown therapeutic benefits in Ebola infections and was formulated by combining three mAbs (13C6, 2G4 and 4G), produced in plants such as tobacco (Nicotiana benthamiana) [77, 78]. There are four reports describing the exorable use of ZMAb and Zmapp in humans infected with Ebola [79]. To date, no clinical study has been conducted for mAb114, MB‐003, ZMAb and MIL‐77E and the data obtained are based on in vitro studies and those in non‐human primates. These mAbs have different origins: mAb114 is a human mAb, recently isolated from a survivor of the 1995 Ebola outbreak in the Democratic Republic of Congo [80]; MB‐003 and ZMAb are cocktails of mAbs isolated from mice which were injected with replicas of the Venezuelan equine encephalitis virus that encoded the Ebola virus glycoprotein [81]; MIL77E is a cocktail containing two mAbs (13C6 and 2G4 from ZMapp) and is produced in modified Chinese hamster ovary cells [82]. However, the problem with the use of mAbs for the Ebola virus is that high doses are required to ensure efficacy. Additionally, it is necessary to keep developing new mAb mixtures due to the possibility of emergence of new epitopes in outbreaks [79].
Although it is an innovative and a very recent advancement, mAbs have promising therapeutic potential due to their high specificity and few adverse effects. To date, two mAbs have been approved by the FDA: palivizumab for RSV and ibalizumab for HIV. Twelve mAbs are in phase I, 19 in phase II and two in phase III of clinical research, covering nine different viruses: RSV, Epstein‐Barr virus (EBV), HIV, influenza, CMV, ZIKV, HBV, HCV and rabies virus [67].
During the SARS‐CoV‐2 pandemic, new mAbs have been suggested as a therapeutic option in infection control. Several mAbs have been proposed for coronaviruses responsible for severe acute respiratory syndrome (SARS‐CoV) and Middle East respiratory syndrome (MERS‐CoV) [83]. Due to the genetic similarity with these other coronaviruses, especially with SARS‐CoV, an mAb with the potential for reactivity or cross‐neutralization could also be tested for SARS‐CoV‐2 [84]. Some studies have concluded that the human mAb CR3022, specific for SARS‐CoV, could bind to SARS‐CoV‐2 with high affinity and recognize the receptor‐binding domain (RBD) binder [85]. Serum containing polyclonal antibodies from SARS‐CoV convalescent patients and animals could also have therapeutic potential [86]. Hwang et al. [87] reported that four out of 51 mAb hybridomas, derived from transgenic H2L2 mice immunized against SARS‐CoV, showed cross‐reactivity observed through the ELISA test, but only mAb 47D11 showed neutralising cross‐reactivity for SARS‐CoV‐2 S1. The chimeric antibody 47D11 H2L2 was recombined with a fully human immunoglobulin, through the cloning of human variable regions and light chains in human IgG1 isotype. Complementary in vivo studies and preclinical and clinical trials should be performed to ascertain its therapeutic and protective response and future clinical applications [88].
Thus far, only a small number of mAbs have been approved as antivirals, and despite their advantages (specificity, speed and little adverse effect), some challenges need to be overcome for better use of this technology. It is essential to identify the molecular and cellular mechanisms by which the immune complexes formed during therapy induce protective immunity without adverse effects such as exacerbated release of cytokines, activation of the complement system and autoimmune reactions. Evaluation of the important aspects of mAb administration, such as dosage, route and potential drug interactions, is required. In addition to the possibility of its combination with other therapies to achieve sterilizing immunity and, then, determine the best means to explore them therapeutically. Although resistance to the therapeutic effects of mAbs is rare, it is possible in the case of development of neutralizing antibodies by the patient's immune system. Thus, future studies are needed to complement this proposed new technology.
CRISPR‐Cas against viral infections
The system formed by Clustered Regularly Interspaced Short Palindromic Repeats, denominated CRISPR, enables genome editing through DNA cleavage by an endonuclease Cas9, guided by an RNA sequence that is capable of pairing with the bases of a target sequence. CRISPR was developed from the molecular mechanisms of the bacterial immune system against bacteriophages. This technique has been used to quickly, easily and efficiently modify endogenous genes in a wide variety of clinically important cell types and in organisms that have traditionally been challenging to genetically manipulate [89]. CRISPR was quickly adopted for efficient gene knockout and insertion of genetic material of interest at specific gene sites in the most diverse of species [90].
In classical therapies, antiviral treatments block viral replication and decrease the symptoms of infection; however, they do not have the ability to destroy the invading virus due to evasion by mutations or by establishing a latent infection [91]. However, the CRISPR‐Cas system reaches the viral genome or host genes that enable persistence of the virus [90]. The principal mechanisms of action suggested for CRISPR‐Cas are reported below:
Modification of receptors for viral entry: Entry of the virus into the host cell is mediated by interactions between the viral proteins and cell membrane receptors. CRISPR‐Cas‐induced editing of receptor genes can prevent virus‐receptor binding and restrict the virus from entering and spreading, in addition to interfering with viral tropism. Since these receptors also aid in the replication and packaging of the viral genome, modulating these receptors can impede viral multiplication. Both approaches would result in a reduction in the number of viral particles.
Segmentation of the host viral factors: The virus essentially depends on the host proteins for its replication and propagation. Using the CRISPR‐Cas technique, some of the genes that encode proteins fundamental to viruses can be silenced (by knock‐down method), thereby preventing viral replication and making the virus more susceptible to the host immune response.
Induction of host transcriptional restriction factors: Restriction of these factors occurs by the coupling between inactive Cas9 (dCas9) and viral RNA, blocking replication and/or leading to a reduction in the transcription of viral RNA and the number of virions.
Excision and deletion of the integrated viral genome: In cases of viruses which integrate their DNA into the host genome, some viral genes may be excised using CRISPR‐Cas, with subsequent deletion and inactivation of these genes and reintegration of the host genome.
All four of these strategies will lead to a reduction in viral replication (Figure 4 ).
Figure 4.
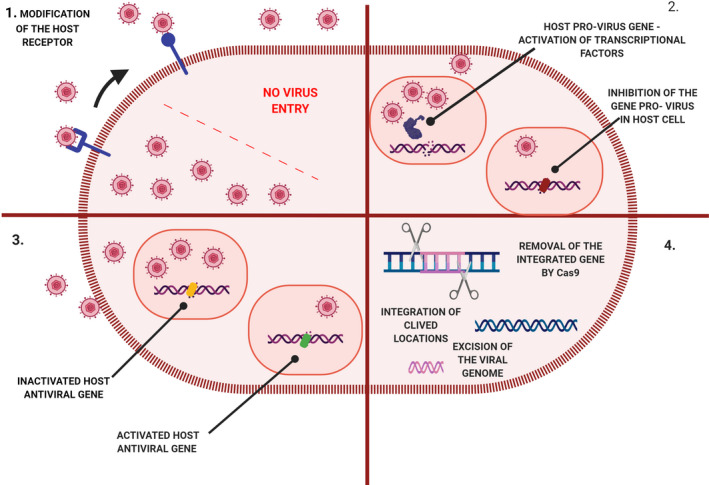
Schematic representation of different CRISPR‐Cas strategies with antiviral action aimed at host and viral genomes: 1. Modification of receptors for viral entry, preventing the entry of virions. 2. Segmentation of host viral factors by silencing provirus genes. 3. Induction of the expression of host transcriptional restriction factors ‐ activation of the antiviral gene. 4. Excision and deletion of the integrated viral genome.
Some viruses with DNA genomes can survive in host cells in the form of episomes, such as HSV and HBV [92, 93] or get integrated into host chromosomes, like in the case of HPV [94]. Others, like HIV, have a single‐stranded RNA genome and a reverse transcriptase to transcribe that RNA into double‐stranded DNA to be integrated into the host DNA [95]. Consequently, CRISPR‐Cas‐directed cleavage of the viral genome inactivates replication and controls subsequent infection [96].
The antiviral activity of the CRISPR‐Cas system has already been demonstrated in vivo and was successfully validated in a leukaemia patient who did not have a detectable viral load of HIV‐1 after receiving bone marrow with a homozygous mutation of delta type 5 CC chemokine receptor 32 (CCR5Δ32) [97], which is essential for the infection of immune cells. Due to the low prevalence of CCR5Δ32 genes, scientists used genome editing techniques to generate homozygous induced pluripotent stem cells (IPSCs), mimicking the naturally occurring mutation [8]. These genomic alterations in IPSCs can be differentiated into monocytes and macrophages and have been shown to be resistant to challenge tests for infection in vitro [98].
HBV contains circular DNA that is converted to covalently closed circular DNA (cccDNA) at the start of replication in infected hepatocytes [99]. The first instance of the use of CRISPR‐Cas that limited HBV infection in vitro and in vivo was published by Lin et al. [100], who reported a significant reduction in viral protein expression. Additionally, in animal models with persistent HBV infections, the CRISPR system could cleave the intrahepatocyte plasmid containing the viral genome, resulting in a reduction in serum antigen levels. Several studies have shown that DNA editing, guided by CRISPR‐Cas, was able to eliminate HBV in cell cultures [99]. Other studies have searched for conserved sequences in the HBV genome in databases and have shown that CRISPR‐Cas can cleave these regions, suggesting a broader action against HBV of different genotypes. Furthermore, it can also be advantageous in multiplex systems [101] for simultaneous cleavage of multiple genes, increasing their efficiency of inactivation and depletion of the viral genome [99]. However, further studies are needed to guarantee the therapeutic potential of this technique and ensure the elimination of copies of the viral cccDNA genes present in hepatocytes.
Human papillomavirus is a double‐stranded DNA virus that infects cells in the stratified epithelium and is widely associated with cervical cancer. It infects the basement membrane of the tissue, expressing oncoproteins such as E6 and E7 and interrupts the normal cell cycle by activating telomerases and cell immortalization by degrading suppression factors such as the p53 protein (via E6) and inactivating the retinoblastoma protein (Rb) (via E7). Despite the existence of a vaccine as a prophylactic measure for HPV infection [102], effective therapies are still needed, and CRISPR‐Cas has been suggested to impede cancer progression [91]. Zhen et al. [103] were able to inactivate the viral oncogenes E6 and E7 to restore p53/Rb levels and induce apoptosis in HPV16‐infected SiHa cells. Furthermore, they managed to significantly reduce the growth of tumours by expressing anti‐E6 and anti‐E7 gRNA. Thus, the CRISPR‐Cas system shows promise for treatments, and even cures, of many viral diseases in humans [91]. Additionally, many in vitro and in vivo strategies are being devised to prevent infections with the help of the CRISPR‐Cas technique [104].
The discovery of the CRISPR‐Cas9 system has revolutionized all fields of science, including virology. Direct manipulation of viral genomes using CRISPR‐Cas9 has enabled systematic studies on cis‐ and trans‐elements encoded in the genomes. Viral genome‐specific mutagenesis using CRISPR‐Cas9 led to the development of a new antiviral technology, especially against chronic viral infections, in the hope of eliminating the need for continuous medication in the near future. However, as most published studies have been carried out on cell culture‐based systems, there is a great need to evaluate the therapeutic potentials in vivo. The development of efficient multiplexed CRISPR systems that can be delivered in vivo using single‐vector vehicles will be important for allowing efficient virus targeting without the risk of selection of escape mutants. Further studies are required to investigate immune responses to exogenously expressed CRISPR‐Cas9 and devise strategies to mask this system and thus reduce their immunogenicity. High‐fidelity Cas9 variants have shown their efficacy in the field of genome editing by reducing off‐target effects [105]. The application of these variants in eradicating viral genome sequences from the host genome may offer a new antiviral perspective. Despite numerous challenges that need to be resolved, their full healing potential should be able to motivate the development of new antiviral therapies.
Conclusion
Due to the epidemiological importance of viral diseases and the difficulties faced in controlling them, the search for new drugs and antiviral therapeutic options is essential and urgent. This review presented an update on the antiviral drugs approved in the recent years and new treatment perspectives, such as the use of nanoparticles, monoclonal antibodies, and the CRISPR‐Cas system. Due to the structural characteristics and genetic variability of many viruses, this search is endless, but imperative in the control of infections, especially in cases of strains resistant to conventional antivirals.
Conflict of Interest
All the authors declare that there are no editorial and financial conflicts of interest (e.g. consultancy, stock ownership, equity interests, patent or licensing agreements) regarding this manuscript.
References
- 1. Jain S. Epidemiology of viral pneumonia. Clin. Chest Med. (2017) 38 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang H., Yang Z., Zhu H., Mo Q., Tua H. High HIV‐1 prevalence and viral diversity among entry‐exit populations at frontier ports of China, 2012–2016: a cross‐sectional molecular epidemiology study. Infect. Gent. Evol. (2018) 65 231–237. [DOI] [PubMed] [Google Scholar]
- 3. Bellei N., Melchior T.B. H1N1: pandemia e perspectiva atual. J. Bras. Patol. Med. Lab. (2011) 47 611–617. [Google Scholar]
- 4. World Health Organization . Coronavirus disease (COVID‐19) situation report. WHO, Congo, 2020. p. 141. [Google Scholar]
- 5. Caini S., Mora D., Olmedo M. et al. The epidemiology and severity of respiratory viral infections in a tropical country: Ecuador, 2009–2016. J. Infect. Public. Heal. (2019) 12 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abid F.B., Abukhattab M., Ghazouani H. et al. Epidemiology and clinical outcomes of viral central nervous system infections. Int. J. Infect. Dis. (2018) 73 85–90. [DOI] [PubMed] [Google Scholar]
- 7. Nunes M.L., Carlini C.R., Marinowic D. et al. Microcephaly and Zika virus: a clinical and epidemiological analysis of the current outbreak in Brazil. J. Pediatr. (2016) 92 230–240. [DOI] [PubMed] [Google Scholar]
- 8. Chen S., Yu X., Guo D. CRISPR‐Cas targeting of host genes as an antiviral strategy. Viruses (2018) 10 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bryan‐Marrugo O.L., Ramos‐Jiménez J., Barrera‐Saldañ H., Rojas‐Martínez A., Vidaltamayo R., Rivas‐Estilla A.M. History and progress of antiviral drugs: from acyclovir to direct‐acting antiviral agents (DAAs) for hepatitis C. Medic. Univer. (2015) 17 165–174. [Google Scholar]
- 10. Organização das Nações Unidas Brasil . Vírus ebola deixa mais de 150 crianças órfãs ou desacompanhadas na República Democrática do Congo. São Paulo: ONU [Updating on 2018 Sep 24, Cited 2020 Jan 19]. Available in: https://nacoesunidas.org/virus‐ebola‐deixa‐mais‐de‐150‐criancas‐orfas‐oudesacompanhadas‐na‐republica‐democratica‐do‐congo/
- 11. Britto M.A. Fármacos recentes usados para o tratamento da infecção pelo HIV‐1: enfuvirtida, maraviroc, raltegravir e etravirina. Rev. Ciênc. Farm. Básica Apl. (2011) 32 159–168. [Google Scholar]
- 12. García‐Serradilla M., Risco C., Pacheco B. Drug repurposing for new, efficient, broad spectrum antivirals. Virus Res. (2019) 264 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Field H.J., De Clerk E. Antiviral drugs ‐ a short history of their discovery and development. Microbiol. Today. (2004) 31 60–62. [Google Scholar]
- 14. De Clercq E. Milestones in the discovery of antiviral agents: nucleosides and nucleotides. Acta Pharm. Sin. B. (2012) 2 535–548. [Google Scholar]
- 15. Littler E., Oberg B. Achievements and challenges in antiviral drug discovery. Antivir. Chem. Chemother. (2005) 16 155–168. [DOI] [PubMed] [Google Scholar]
- 16. Vigant F., Santos N.C., Lee B. Broad‐spectrum antivirals against viral fusion. Nat. Rev. Microbiol. (2015) 13 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaudhuri J.A., Symons J.A., Deval J. Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antivir. Res. (2018) 155 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitra B., Thapa R.J., Guo H., Block T.M. Host functions used by hepatitis B virus to complete its life cycle: Implications for developing host‐targeting agents to treat chronic hepatitis B. Antivir. Res. (2018) 158 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Villa T.G., Feijoa‐Siota L., Rama J.L.R., Ageitos J.M. Antivirals against animal viruses. Biochem. Pharmacol. (2017) 133 97–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dewdney T.G., Wang Y., Kovari I.A., Reiter S.J., Kovari L.C. Reduced HIV‐1 integrase flexibility as a mechanism for raltegravir resistance. J. Struct. Biol. (2013) 184 245–250. [DOI] [PubMed] [Google Scholar]
- 21. Beigel J.H., Bao Y., Beeler J. et al. Oseltamivir, amantadine, and ribavirin combination antiviral therapy versus oseltamivir monotherapy for the treatment of influenza: a multicentre, double‐blind, randomised phase 2 trial. Lancet Infect. Dis. (2017) 17 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sonnenfeld G., Merigan T.C. The role of interferon in viral infections. Semin. Immunopathol. (1979) 2 311–338. [Google Scholar]
- 23. Fensterl V., Sen G.C. Interferons and viral infections. BioFactors (2009) 35 14–20. [DOI] [PubMed] [Google Scholar]
- 24. Kawai T., Akira S. The role of pattern‐recognition receptors in innate immunity: update on Toll‐like receptors. Nat. Immunol. (2010) 11 373–384. [DOI] [PubMed] [Google Scholar]
- 25. Vincent M.J., Bergeron E., Benjannet S. et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. (2005) 2 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Savarino A., Boelaert J., Cassone A., Majori G., Gauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect Dis. (2003) 3 722–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosenberg E.S., Dufort E.M., Udo T. et al. Association of treatment with hydroxychloroquine or azithromycin with in‐hospital mortality in patients with COVID‐19 in New York State. JAMA Netw. (2020) 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang W., Cao Z., Han M. et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ (2020) 369 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mercuro N.J., Yen C.F., Shim D.J. et al. COVID‐19. JAMA Cardiol. (2019) 2020 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borba M.G.S., Val F.F.A., Sampaio V.S.S. et al. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection: A randomized clinical trial. JAMA Netw. (2020) 4 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boulware D.R., Pullen M.F., Bangdiwala A.S. et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid‐19. N. Engl. J. Med. (2020) 383 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitià O., Ubals M., Corbacho M. et al. A cluster‐randomized trial of hydroxychloroquine as prevention of Covid‐19 transmission and disease. medRxiv. (2020) 1–23. [Google Scholar]
- 33. Horby P., Mafham M., Linsell L. et al. Effect of hydroxychloroquine in hospitalized patients with COVID‐19: preliminary results from a multi‐centre, randomized, controlled trial. medRxiv. (2020) 1–25. [Google Scholar]
- 34. Skipper C.P., Pastick K.A., Enger N.W. et al. Hydroxychloroquine in nonhospitalized adults with early COVID‐19. Ann. Intern. Med. (2020) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cao B.I., Wang Y., Wen D.et al. A Trial of Lopinavir‐Ritonavir in Adults Hospitalized with Severe Covid‐19. N. Engl. J. Med. (2020) 382 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. National Institutes of Health . COVID‐19 treatment guidelines. Updating on 2020 July 30, Cited on 2020 Aug 20. Available in: https://www.covid19treatmentguidelines.nih.gov/whats‐new/.
- 37. UpToDate . Coronavirus disease 2019 (COVID‐19): management in hospitalized adults. [Updating on 2020 Aug 18, Cited on 2020 Aug 20. Available in: https://www.uptodate.com/contents/coronavirus‐disease‐2019‐covid‐19‐management‐in‐hospitalized‐adults.
- 38. Beigel J.H., Tomashed K.M., Dodd L.E. et al. Remdesivir for the treatment of Covid‐19—preliminary report. N. Engl. J. Med. (2020) 1–12. [DOI] [PubMed] [Google Scholar]
- 39. Wang Y., Zhang D., Du G. et al. Remdesivir in adults with severe COVID‐19: a randomised, double‐blind, placebo‐controlled, multicentre trial. Lancet (2020) 395 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grein J., Ohmagari N., Sin D. et al. Compassionate use of remdesivir for patients with severe Covid‐19. N. Engl. J. Med. (2020) 382 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong R.W., Lingwood C.A., Ostrowski M.A., Cabral T., Cochrane A. Cardiac glycoside/aglycones inhibit HIV‐1 gene expression by a mechanism requiring MEK1/2‐ ERK1/2 signaling. Sci. Rep. (2018) 8 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szymańska E., Orłowski P., Winnicka K. et al. Multifunctional tannic acid/silver nanoparticle‐based mucoadhesive hydrogel for improved local treatment of HSV infection: In vitro and in vivo studies. Int. J. Mol. Sci. (2018) 19 387–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sabet S., George M.A., El‐Shorbagy H.M. et al. Gelatin nanoparticles enhance delivery of hepatitis C virus recombinant NS2 gene. PLoS One (2017) 12 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jamali A., Mottaghitalab F., Abdoli A. et al. Inhibiting influenza virus replication and inducing protection against lethal influenza virus challenge through chitosan nanoparticles loaded by siRNA. Drug Deliv. Transl. Res. (2018) 8 12–20. [DOI] [PubMed] [Google Scholar]
- 45. Marasco W.A., Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. (2007) 25 1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Glass Z., Huang M., Chen Z.Y., Xu Q. Ex vivo cell based CRISPR/Cas9 genome editing for therapeutic applications. Biomaterials (2020) 234 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Das A.T., Binda C.S., Berkhout B. Elimination of infectious HIV DNA by CRISPR–Cas9. Curr. Opin. Virol. (2019) 38 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Food and Drug Administration (FDA) . FDA approved drug products. [Cited 2019 Dec 15]. Available in: https://www.centerwatch.com/directories/1067
- 49. Weber I.T., Harrison R.W. Decoding HIV resistance: from genotype to therapy. Future Med. Chem. (2017) 13 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arca‐Lafuente S., Martínez‐Román P., Mate‐Cano I., Madrid R., Briz V. Nanotechnology: a reality for diagnosis of HCV infectious disease. J. Infect. (2019) 80 8–15. [DOI] [PubMed] [Google Scholar]
- 51. Kumar R., Nayak M., Sahoo G.C. et al. Iron oxide nanoparticles based antiviral activity of H1N1 influenza A virus. J. Infect. Chemother. (2019) 25 325–329. [DOI] [PubMed] [Google Scholar]
- 52. Cagno V., Andreozzi P., D’Alicarnasso M. et al. Broad‐spectrum non‐toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat. Mater. (2018) 17 195–203. [DOI] [PubMed] [Google Scholar]
- 53. Sur S., Rathore A., Dave V., Reddy K.R., Chouhan R.S., Sadhu V. Recent developments in functionalized polymer nanoparticles for efficient drug delivery system. Nano‐Structures and Nano‐Objects. (2019) 20 100397. [Google Scholar]
- 54. Dey P., Bergmann T., Cuellar‐Camacho J.L. et al. Multivalent flexible nanogels exhibit broad‐spectrum antiviral activity by blocking virus entry. ACS Nano (2018) 12 6429–6442. [DOI] [PubMed] [Google Scholar]
- 55. Hu C.M.J., Chen Y.T., Fang Z.S., Chang W.S., Chen H.W. Antiviral efficacy of nanoparticulate vacuolar ATPase inhibitors against influenza virus infection. Int. J. Nanomed. (2018) 13 8579–8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Javan F., Vatanara A., Azadmanesh K., Nabi‐Meibodi M., Shakouri M. Encapsulation of ritonavir in solid lipid nanoparticles: in vitro anti‐HIV‐1 activity using lentiviral particles. J. Pharm. Pharmacol. (2017) 69 1002–1009. [DOI] [PubMed] [Google Scholar]
- 57. Yadavalli T., Sukla D. Could zinc oxide tetrapod nanoparticles be used as an effective immunotherapy against HSV‐2? Nanomedicine. (2016) 11 2239–2242. [DOI] [PubMed] [Google Scholar]
- 58. Ogata M., Umemura S., Sugiyama N. et al. Synthesis of multivalent sialyllactosamine‐carrying glyco‐nanoparticles with high affinity to the human influenza virus hemagglutinin. Carbohydr. Polym. (2016) 153 96–104. [DOI] [PubMed] [Google Scholar]
- 59. Park H.J., Jeon E.J., Lee J.S. et al. Galactosylated lipidoid nanoparticles for delivery of small interfering RNA to inhibit hepatitis C viral replication in vivo . Adv. Healthc. Mater. (2016) 5 2931–2941. [DOI] [PubMed] [Google Scholar]
- 60. Das Neves J., Nunes R., Rodrigues F., Sarmento B. Nanomedicine in the development of anti‐HIV microbicides. Adv. Drug. Deliv. Rev. (2016) 103 57–75. [DOI] [PubMed] [Google Scholar]
- 61. Murugan K., Dinesh D., Paulpandi M. et al. Nanoparticles in the fight against mosquito‐borne diseases: bioactivity of Bruguiera cylindrica‐synthesized nanoparticles against dengue virus DEN‐2 (in vitro) and its mosquito vector Aedes aegypti (Diptera: Culicidae). Parasitol. Res. (2015) 114 4349–4361. [DOI] [PubMed] [Google Scholar]
- 62. Yang X., Li C., Huang C. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale (2015) 5 1–9. [DOI] [PubMed] [Google Scholar]
- 63. Hang X., Peng H., Song H., Qi Z., Miao X., Xu W. Antiviral activity of cuprous oxide nanoparticles against hepatitis C virus in vitro . J. Virol. Methods. (2015) 222 150–157. [DOI] [PubMed] [Google Scholar]
- 64. McReynolds S., Jiang S., Guo Y. et al. Characterization of the prefusion and transition states of severe acute respiratory syndrome coronavirus S2‐HR2. Biochemistry (2008) 47 6802–6808. [DOI] [PubMed] [Google Scholar]
- 65. Sivasankarapillai V.S., Pillai A.M., Rahdar A. et al. On facing the SARS‐CoV‐2 (COVID‐19) with combination of nanomaterials and medicine: possible strategies and first challenges. Nanomaterials. (2020) 10 852–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schmitt V., Kesch C., Jackson J.K. et al. Design and characterization of injectable poly (lactic‐co‐glycolic acid) pastes for sustained and local drug release. Pharm. Res. (2020) 37 36–48. [DOI] [PubMed] [Google Scholar]
- 67. Dibo M., Battocchio E.C., Souza L.M.S. et al. Antibody therapy for the control of viral diseases: an update. Curr. Pharm. Biotechnol. (2019) 20 1108–1121. [DOI] [PubMed] [Google Scholar]
- 68. Salazar G., Zhang N., Fu T.M., Na Z. Antibody therapies for the prevention and treatment of viral infections. NPJ Vaccines. (2017) 2 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pelegrin M., Naranjo‐Gomez M., Piechaczyk M. Antiviral monoclonal antibodies: can they be more than simple neutralizing agents? Trends Microbiol. (2015) 23 653–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Food and Drug Administration (U.S.) . Palivizumab product approval information. [Updating on 1998, Cited 2020 Jan 18]. Available in: https://www.accessdata.fda.gov/drugsatfda_docs/appletter/1998/palimed061998L.htm
- 71. Emu B., Fessel J., Schrader S. et al. Phase 3 study of ibalizumab for multidrug‐resistant HIV‐1. N. Engl. J. Med. (2018) 379 645–654. [DOI] [PubMed] [Google Scholar]
- 72. Iacob S.A., Iacob D.G. Ibalizumab targeting CD4 receptors, an emerging molecule in HIV therapy. Front. Microbiol. (2017) 8 2323–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. AIDSinfo (USA) . AIDSinfo. [Updating on 27 Apr 2020, Cited 2020 Jan 30] Available in: https://aidsinfo.nih.gov/drugs/584/3bnc117/0/professional
- 74. Gaudinski M.R., Coates E.E., Houser K.V. et al. Safety and pharmacokinetics of the Fc‐modified HIV‐1 human monoclonal antibody VRC01LS: a phase open‐label clinical trial in healthy adults. PLoS Med. (2018) 15 1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sapparapu G., Fernandez E., Kose N. et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature (2016) 540 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. The New York Times (New York) . Inovio's DNA‐Encoded Monoclonal Antibody (dMAb™) Platform Leaps Forward with First‐in‐Human Trial. [Updating on 7 Jan 2019, Cited 2020 Jan 30] Available in: https://markets.on.nytimes.com/research/stocks/news/press_release.asp?docTag=201901070800PR_NWS_USPRXPH15873&feedID=600&press_symbol=136927
- 77. Davey R.T., Jr‐Dodd L., Proschan M.A. et al. A randomized, controlled trial of ZMapp for Ebola virus infection. N. Engl. J. Med. (2016) 375 1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Qiu X., Wong G., Audet J. et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature (2014) 514 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moekotte A.L., Huson M.A.M., Van der Ende A.J. et al. Monoclonal antibodies for the treatment of Ebola virus disease. Expert Opin. Investig. Drugs. (2016) 25 1325–1335. [DOI] [PubMed] [Google Scholar]
- 80. Corti D., Misasi J., Mulangu S. et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science (2016) 351 1339–1342. [DOI] [PubMed] [Google Scholar]
- 81. Wilson J.A., Hevey M., Bakken R. et al. Epitopes involved in antibody mediated protection from Ebola virus. Science (2000) 287 1664–1666. [DOI] [PubMed] [Google Scholar]
- 82. Qiu X., Audet J., Lv M. et al. Two mAb cocktail protects macaques against the Makona variant of Ebola virus. Sci. Transl. Med. (2016) 8 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease‐19 (COVID‐19). Asian Pac. J. Allergy. (2020) 38 10–18. [DOI] [PubMed] [Google Scholar]
- 84. Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS‐CoV‐2 and other human coronaviruses. Trends Immunol. (2020) 41 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tian X., Li C., Huang A. et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerg. Microbes Infect. (2020) 9 382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hoffmann M., Kleine‐Weber H., Schroeder S. et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell (2020) 181 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hwang W.C., Lin Y., Santelli E. et al. Structural basis of neutralization by a human anti‐severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. (2006) 281 34610–34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhou P., Yang X.L., Wang X.G. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature (2020) 579 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sander J.D., Joung J.K. CRISPR‐Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. (2014) 32 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Soppe J.A., Lebbink R.J. Antiviral goes viral: harnessing CRISPR/Cas9 to combat viruses in humans. Trends. Microbiol. (2017) 25 833–850. [DOI] [PubMed] [Google Scholar]
- 91. Koujah L., Shukla D., Naqvi A.R. CRISPR‐Cas based targeting of host and viral genes as an antiviral strategy. Semin. Cell Dev. Biol. (2019) 18 30108–30113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cong L., Ann‐Ran F., Cox D. et al. Multiplex genome engineering using CRISPR/Cas systems. Science (2013) 339 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science (2012) 337 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mali P., Yang L., Esvelt K.M. et al. RNA‐guided human genome engineering via Cas9. Science (2013) 339 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bhaya D., Davison M., Barrangou R. CRISPR‐Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. (2011) 45 273–297. [DOI] [PubMed] [Google Scholar]
- 96. Wiedenheft B., Sternberg S.H., Doudna J.A. RNA‐guided genetic silencing systems in Bacteria and Archaea. Nature (2012) 482 331–338. [DOI] [PubMed] [Google Scholar]
- 97. Allers K., Hütter G., Hofmann J. et al. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood (2011) 117 2791–2799. [DOI] [PubMed] [Google Scholar]
- 98. Ye L., Wang J., Beyer A.I. et al. Seamless modification of wild type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. (2014) 111 9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Seeger C., Sohn J.A. Targeting hepatitis b virus with CRISPR/Cas9. Mol. Ther. Nucleic Acids. (2014) 3 216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lin S.R., Yang H.C., Kuo Y.T. et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo . Mol. Ther. Nucleic Acids. (2014) 3 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shlomai A., Rice C.M. Getting rid of a persistent troublemaker to cure hepatitis. Science (2014) 343 1212–1213. [DOI] [PubMed] [Google Scholar]
- 102. Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. (2010) 10 550–560. [DOI] [PubMed] [Google Scholar]
- 103. Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16‐positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. (2014) 450 1422–1426. [DOI] [PubMed] [Google Scholar]
- 104. Sanches‐da‐Silva G.N., Medeiros L.F.S., Lima F.M. The potential use of the CRISPR‐Cas system for HIV‐1 gene therapy. Int. J. Genomics. (2019) 2019 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bayat H., Omidi M., Rajabibazl M., Sabri S., Rahimpour A. The CRISPR growth spurt: from bench to clinic on versatile small RNAs. J. Microbiol. Biotechnol. (2017) 2 207–218. [DOI] [PubMed] [Google Scholar]


