Abstract
During the last years a considerable therapeutic progress in melanoma patients with the RAF V600E mutation via RAF/MEK pathway inhibition and immuno-therapeutic modalities has been witnessed. However, the majority of patients relapse after therapy. Therefore, a deeper understanding of the pathways driving oncogenicity and metastasis of melanoma is of paramount importance. In this review, we summarize microRNAs modulating tumor growth, metastasis, or both, in preclinical melanoma-related in vivo models and possible clinical impact in melanoma patients as modalities and targets for treatment of melanoma. We have identified miR-199a (ApoE, DNAJ4), miR-7-5p (RelA), miR-98a (IL6), miR-219-5p (BCL2) and miR-365 (NRP1) as possible targets to be scrutinized in further target validation studies.
Keywords: Epithelial-mesenchymal transition, metastasis models, microRNA editing, microRNA inhibition and reconstitution of function, migration, invasion and proliferation, therapeutic targets, review
In 2018, 96,000 patients were diagnosed with melanoma in the US with a death toll of about 7,000 due to metastatic disease (1). Cutaneous-, mucosal- and ocular melanoma have been identified as major subtypes of melanoma (2). In this review, we focus on cutaneous melanoma with superficial spreading melanoma, nodular melanoma, lentigo maligna and acral melanoma as major subtypes (2). Oncogenic mutations have been identified in v-raf murine sarcoma viral oncogene homolog B1 (BRAF), cyclin-dependent kinase inhibitor 2A (CDKN2A), NRAS and p53 (3). Approximately 50% of melanomas harbor mutations in BRAF leading to aberrant activation of the mitogen-activated protein kinase (MAPK) signaling pathway (4). Clinical therapeutics for advanced-stage melanoma have improved dramatically with the development of V600E-BRAF and mitogen-activated protein kinase (MEK) inhibitors, immune checkpoint blocking antibodies and a modified oncolytic herpes virus, as approved therapeutic agents (5). Early clinical trials point to further clinical benefit by triple combinations of V600E-specific BRAF inhibitors, MEK inhibitors and immune checkpoint inhibitors (6-8). However, relapse due to drug resistance is observed in the vast majority of patients. Phenotype plasticity and interactions of transformed melanocytes with the tumor microenvironment are resistance-promoting factors (9,10). In order to identify new targets and modalities for therapeutic intervention in melanoma we reviewed the role of microRNAs (miRs) in the context of growth and metastasis of this disease. We focused on miRs for which efficacy in preclinical in vivo models has been shown, as mediators of tumor growth (TG), metastasis or both.
Metastasis of Melanoma
Melanoma preferentially metastasizes through the lymphatic route, but in a minority of cases dissemination through the haematogenous route has been observed (11). Before metastasis, melanoma cells extend to the adjacent epidermis and undergo epithelial-mesenchymal transition (EMT) (12,13). The following metastatic cascade includes steps such as intravasation, circulation as single cells or cell clusters coated with platelets, arrest in capillaries at a distant site, extravasation into the parenchyma of distant organs, formation of indolent micrometastases in supportive niches and finally activation of the dormant cells and colonisation of the respective target organs (14). Melanoma can colonize different organ sites either sequentially or synchronously (15-17). Major target organs of metastasis of subcutaneous melanoma are the lungs, brain, liver and bones (16,17). A total of 40-50% of patients with stage IV melanoma develop brain metastases (17). The lungs are the most frequent target organs of metastasis in patients with advanced subcutaneous melanoma (16). In contrast, uveal melanoma almost exclusively relapses to the liver, supporting the concept of compatibility of metastatic cells with host stroma (18). Once several metastases have formed, cells from each metastasis seed and reseed other tumors adding considerable complexity to the diversity of metastatic clones (14).
microRNAs and Their Role in Oncology
miRs are small RNAs with a length in the range of 22 nucleotides (nts) that direct post-transcriptional repression of target mRNA transcripts (19). Knock-out experiments have revealed important biological functions for many miRs (19). They are transcribed by polymerase II and approximately 1,900 miRs have been identified in humans (20,21). Corresponding miR genes can be localized in exons, introns or intergenic regions (22,23). In the canonical pathway, they are synthesized in the nucleus as pri-miRNAs containing a hairpin loop, processed by a micropocessor (DROSHA, DGCR8) to pre-miR hairpin structures and subsequently exported to the cytoplasm by exportin 5 (24,25). In the cytoplasm, pre-miRs are cleaved by DICER to produce a 20 nucleotides (nts) miR duplex with 5´ phosphate and a two nts overhang at each end. One strand of the miR duplex is loaded into the guide-strand channel of an Argonaute protein to form a silencing complex, whereas the passenger strand is discarded (24,25). Within the silencing complex miR seed sequence, nts 2-7 are poised to initially interact with target mRNAs. If pairing is very extensive, the target can be sliced, if not, translation is inhibited (24,25). One type of miR can target several distinct mRNAs and a single mRNA can be cleaved by several different types of miRs, thus enabling interference with several cancer-related networks with the potential of rewiring the transformed and metastatic state (26). The role of miRs in oncology was identified in the context of pathogenesis of B-cell chronic lymphatic leukemia (B-CLL). It was found that the genomic loci of miR-15a and miR-16-1 are deleted in the majority of patients with this disease (27,28). These miRs are involved in the cleavage of the anti-apoptotic protein B cell lymphoma 2 (BCL2) and deletion of these miRs recapitulates the human disease phenotype in mice (29). In addition to oncogenesis, miRs are involved in metastasis of different types of cancer. We recently summarized the role of miRs in metastasis of breast-, prostate, non-small cell lung carcinoma and ovarian cancer (30-33).
miRs Involved in Growth of Melanoma Cells With In Vivo Activity in Preclinical Models
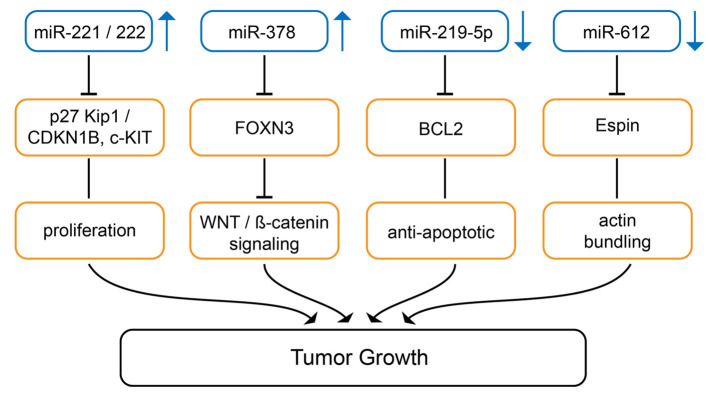
Up-regulated miRs promoting melanoma growth in preclinical in vivo models. The expression level of miRs-221 and -222 (Figure 1) correlates with melanoma progression (34). Antagomirs against miR-221 and -222 inhibit invasion and foci formation of Mel 665/1 melanoma cells and intratumoral bolus injection of antagomirs inhibits TG in nude mice (34). Tyrosine-protein kinase KIT (c-KIT receptor) and p27 Kip/cyclin-dependent kinase inhibitor 1B (CDKN1B) have been identified as targets of miR-221 and -222 (34). c-KIT receptor regulates melanogenesis, cell growth, migration and survival (35). During progression of melanoma, loss of c-KIT is a crucial event due to prevention of stem cell factor/c-KIT triggered apoptosis (36). CDKN1B inhibits cell cycle progression of melanoma cells by binding to cyclin-dependent kinase (CDK)/cyclin complexes (37).
Figure 1. miRs mediating tumor growth in melanoma-related preclinical in vivo models. miRs-221/-222 and miR-378 are up-regulated, miR-219-5p and miR-612 are down-regulated in melanoma in comparison to corresponding normal tissues. Arrows indicate up- or down-regulation. BCL2: Bcell lymphoma 2; c-KIT: tyrosine-protein kinase KIT; FOXN3: Forkheadbox N3; p27 Kip1/CDKN1B: cyclin-dependent kinase inhibitor 1B, WNT:wingless-type.
miR-378 (Figure 1) is up-regulated during progression of melanoma and potentiates migration, invasion and EMT in A375 and A875 melanoma cells (38). miR-378 promotes TG of A875 melanoma cells in vivo (38). It targets tumor suppressor forkhead box N3 (FOXN3), a member of the forkhead box (FOX) family which act as a tumor suppressor (TS) (39,40). FOXN3 also interacts with ß-catenin to promote down-regulation of wingless-type (WNT)/ß-catenin signaling proteins (38).
Down-regulated miRs inhibiting melanoma growth in preclinical in vivo models. miR-219-5p (Figure 1) is down-regulated in melanoma and correlates with unfavorable prognosis (39). Up-regulation of miR-219-5p in A375 melanoma cells with a transfected miR-mimetic inhibits proliferation, invasion and enhances apoptosis (39). A375 cells transfected with a miR-mimetic show decreased growth after subcutaneous implantation into the flanks of nude mice (39). BCL2 was identified as a direct target of miR-219-5p (39-42).
Low expression of miR-612 (Figure 1) was significantly associated with shorter overall and disease-free survival in melanoma patients (43). miR-612 suppresses growth, migration and invasion of SK-MEL-28 and A375 melanoma cells (43). A375 cells transfected with miR-612 grow significantly slower than control cells after subcutaneous implantation in nude mice. Espin, an actin binding protein, was identified as a direct target of miR-612. Espin is involved in actin cytoskeleton remodeling thus affecting cell growth, migration and invasion (44,45).
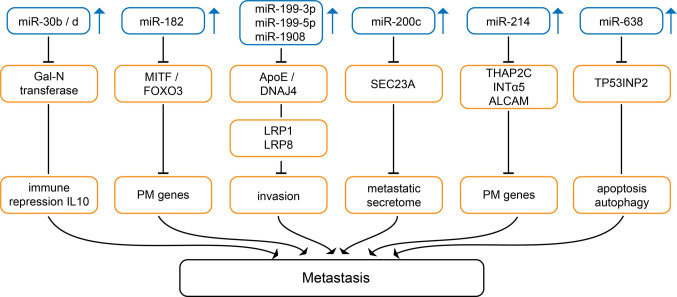
Up-regulated miRs mediating metastasis in preclinical in vivo melanoma-related models. miRs-30b and -30d (Figure 2) have redundant function and their expression marks the progression from primary to metastatic melanoma (46). Genomic amplification of miR-30b/d has been observed in a subset of melanoma patients (46). In melanoma cells 4L and 5B1, miRs-30b/d stimulate invasive capacity without affecting proliferation (46). Overexpression of miR-30b/d in B16F10 melanoma cells enhances lung metastases after tail vein injection (46). Inoculation of 5B1 cells transduced with miR-30d into the flanks of mice enhances local muscle invasion and metastasis to the lungs and the liver (46). miR-30d triggers infiltration of regulatory T-cells (Tregs) to the metastatic site mediated by interleukin 10 (IL10) (46). N-acetylgalactosaminyltransferase 7 (GALNT7) has been identified as a direct target of miR-30b/d (46). GALNT7 initiates mucin O-linked glycosylation in the Golgi apparatus by catalyzing transfer of N-acetylgalactosamine to Ser and Thr on target proteins (47). GALNT7 affects the structure of multiple transmembranes substrates determining their interaction with the extracellular microenvironment (47). Inactivation of GALNT7 has been shown to promote metastasis (46).
Figure 2. Up-regulated miRs mediating metastasis in melanoma-related in vivo models. Upward arrows indicate up-regulation of the corresponding miRs in melanoma versus corresponding normal tissues. ALCAM: Activated leukocyte cell adhesion molecule; Apo E: apolipoprotein E; DNAJ4:DNAJ heat shock protein family member A4; FOXO3: forkhead box O3; IL10: interleukin 10; INTα5: integrin α5; LRP1,8: low density lipoproteinrelated protein 1 or 8; PM: pro-metastatic; SEC23A: SEC23 homolog A; TFAP2C: transcription factor AP2γ; TP53INP2: tumor protein p53-inducible nuclear protein 2.
Expression of miR-182 (Figure 2) increases with progression from primary to metastatic melanoma (48). miR-182 stimulates cell migration and invasion of A375 and SK-MEL19 melanoma cells, whereas silencing of miR-182 triggers cell death (48). B16F10 cells transduced with miR-182 give rise to increased lung metastases after tail vein injection (48). Forkhead box 03 (FOXO3) and microphtalamia-associated transcription factor (MITF) have been identified as direct targets of miR-182 (48). FOXO3 functions as a tumor suppressor (TS) controling numerous regulators of the cell cycle and apoptosis such as proapoptotic BIM, a member of the BCL2 family (49-51). MITF is a basic helix-loop-helix leucine zipper transcription factor which controls lineage-specific pathway regulation in many cell types including melanocytes, osteoclasts and mast cells (52,53). MITF represses proliferation by activating cell cycle inhibitors such as p16INK4a which inhibits CDK4 and p21-CDK-interacting protein 1 (p21cip1) and controls cell migration by regulating the formin diaphanous1 (DIA1) which controls cell migration. DIA1 is a RAS homologue (RHO)-GTPases effector and is involved in polymerization of actin and coordination of cytoskeleton and microtubule networks at the cell periphery (54,55).
miRs-199-3p, -199-5p and -1908 (Figure 2) promote invasion in MeWo-LM2 and A375 melanoma cells as well as endothelial recruitment and angiogenesis (56). Their overexpression in the poorly metastatic MeWo cells causes colonization of the lungs more efficiently than their parental populations (56). Apolipoprotein E (ApoE) and DNA J heat shock factor protein family (Hsp40) member A4 (DNAJ4) have been identified as direct targets of miRs 199-3p, -199-5p and -1908 (56). ApoE targets low density receptor related protein 1 (LRP1) on melanoma cells and LRP8 on endothelial cells to suppress invasion of melanoma cells and migration of endothelial cells (56-60). Heat shock proteins, such as DNAJ4 have been shown to be involved in proliferation, differentiation and carcinogenesis (61).
miR-200c (Figure 2) was shown to target SEC 23 homolog A (SEC23A) (62). SEC23A interacts with components involved in anterograde versicle transport from the endoplasmic reticulum to the Golgi apparatus (63). Overexpression of miR-200c and SEC23A interference accelerate oligometastatic to polymetastatic progression of human melanoma cell line M14 after tail vein injection (63). The metastasis promoting activity of miR-200 is dependent on selective reprogramming of the secretome by depletion of SEC23A (62,64). As identifed by mass spectrometry, different proteins such as thrombospondin (65), transferrin (66), vitamin D binding protein (67), C-X-C chemokine receptor 4 (CXCR4) (68) and S100 calcium-binding protein A8 (S100A8) (69) may be involved in promoting metastasis by miR-200c.
miR-214 (Figure 2) expression in A375P melanoma cells enhances in vitro cell movement and improves cell adhesion to fibronectin, laminin and collagen (70). MA2 melanoma cells overexpressing miR-214 give rise to higher numbers of lung metastases in comparison to the control cell line after tail vein injection in severe combined immuno-deficicent (SCID) mice (70). Transcription factor AP2C (TFAP2C) has been identified as a direct target of miR-214 (70). Subsequent investigations have shown that depletion of mi-214 blocks dissemination of melanoma cells by repressing passage of melanoma cells through the blood endothelium by reducing the cell adhesion molecules integrin α5 (INTα5) and activated leukocyte adhesion molecule (ALCAM) (71-73). miR-214 affects TFAP2C directly and TFAP2A indirectly (74). These transcription factors play a role in regulating expression of genes in involved in invasion, extravasation and angiogenesis such as human epidermal growth factor receptor 2 (HER2) (proliferation), p21 (cell-cycle), c-KIT (TS), BCL2 (apoptosis), cluster of differentiation CD95 (CD95) (apoptosis), melanoma cell adhesion molecule (MCAM), mucin 18 (MUC18) and E-cadherin (adhesion), matrix metalloproteinase 2 (MMP2) (invasion), metastasis suppressor KAI-1 (invasion) and vascular endothelial growth factor (VEGF) as well as thrombin receptor (angiogenesis) (74,75). Especially, the role of ALCAM in melanoma progression is well documented (76,77). Therapeutic silencing of miR-214 has been shown to inhibit tumor progression in multiple mouse models (78).
miR-638 (Figure 2) is strongly up-regulated during melanoma progression (79). Transfection of miR-638 into SK-Mel-147 and SK-Mel-28 cells results in increased invasion (79). SK-Mel-147 overexpressing miR-638 show more metastatic nodules in the lungs after tail vein injection into NOD scid IL2R gamma chain knockout mice (NSG) in comparison to the control cell line (79). Knockdown of miR-638 induces p53-mediated apoptosis and autophagy in melanoma cells (79). p53 interacting protein 2 (TP53INP2) was identified as a direct target of miR-638 (79). TP53INP2 plays an important role as a TS through interaction with p53 (80,81).
Down-regulated miRs Inhibiting Metastasis in Preclinical Melanoma-related In Vivo Models
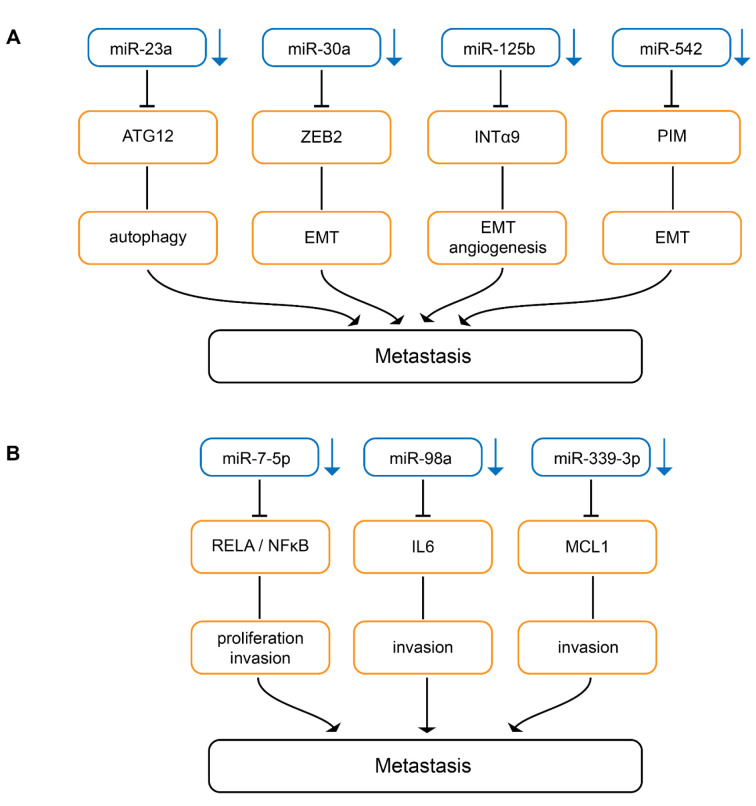
Down-regulated miRs inhibiting epithelial mesenchymal transition. Serum miR-23a (Figure 3A) has been identified as a potential biomarker for predicting melanoma metastasis and prognosis (82). Overexpression of miR-23a inhibits invasion, migration and epithelial mesenchymal transition (EMT) of A2058 and A375 melanoma cells (82). Subcutaneously implanted A2058 melanoma cells transduced with miR-23a give rise to decreased metastases in the lungs and the liver in nude mice without affecting TG (82). miR-23a down-regulation results from up-regulation of the runt-related transcription factor 2 (RUNX2) in melanoma (82,83). Autophagy-related 12 (ATG12) has been identifed as a direct target of miR-23a (82). ATG12 is essential for autophagy and promotes apoptosis through interaction with anti-apoptotic members of the BCL2 family (84,85). The miR-23-ATG12 axis suppresses melanoma invasion and migration through the AMP activated kinase (AMPK)/RHO A pathway (82). AMPK is a sensor for cellular energy and can be activated through depletion of ATP levels in cells with autophagy and exhibits anti-metastatic potential in melanoma (86). AMPK activates RHO GTPase family member A (RHO A) which mediates cytoskeletal organization by activation of RHO-associated kinases (ROCK)-myosin phosphatase target subunit 1(MYPT1) pathway during melanoma cell migration. MYPT1 is a regulatory subunit of protein phosphatase 1 (82,87,88).
Figure 3. Down-regulated miRs inhibiting metastasis in melanoma-related in vivo models. Downward arrows indicate down-regulation in melanoma compared to matching normal tissues. (A) miRs inhibiting metastases by attenuating autophagy, epithelial-mesenchymal transition and miR-editing. ATG 12: Autophagy related 12; EMT: epithelial mesenchymal transition; INTα9: integrin α9; PIM-1: serine/threonine-protein kinase PIM-1. (B) miRs inhibiting metastasis by interfering with signal-transduction pathways. IL6: Interleukin 6; MCL-1: induced myeloid leukemia cell differentiation protein; NFĸB: nuclear factor ĸB; Rel A: v-rel avian reticuloendotheliosis viral homolog A.
miR-30a (Figure 3A) suppresses EMT and lung metastasis of B16F10 transduced cells after tail vein injection (89). Zinc finger E-box binding homeobox 2 (ZEB2) has been identified as a direct target of miR-30a (89). ZEB2 binds to the E-cadherin promoter, resulting in down-regulation of promoter activity (90). Therefore, miR-30a increases E-cadherin expression and inhibits EMT (91). In addition to melanoma, miR-30a has been shown to be down-regulated and to inhibit EMT in non-small cell lung carcinoma (92).
miR-125b (Figure 3A) expression is decreased in melanoma tissues (93). miR-125b inhibits EMT in Mel Im and Mel Ju melanoma cells (93). Mel Im transfected with miR-125 expression constructs exhibt reduced lung metastasis after tail vein injection in nude mice (93). Integrin α9 (INTα9) was identified as a direct target of miR-125b (93). Knockdown of INTα9 expression phenocopies the effect of miR-125b overexpression in melanoma cells (93). It has been shown that INTα9 can promote melanoma metastasis by potentiating EMT (94).
miR-542 (Figure 3A) is down-regulated in melanoma cell lines and tissues (95). miR-542 inhibits EMT, cell division and migration of melanoma cells as well as lung metastasis after tail vein injection of miR-542-transfected melanoma cells (95). Proviral integration site for Moloney murine leukemia virus-1 (PIM-1), a serine-threonine kinase, has been identified as a direct target of miR-542 (93). PIM-1 controls EMT, cell motility, cell survival and is involved in cell metabolism (96). PIM-1 is an important target for oncology drug discovery (97).
Down-regulated miRs interfering with signaling. miR-7-5p (Figure 3B) inhibits migration and invasion of 1205 Lu melanoma cells and lung metastasis in Nod Scid Gamma (NSG) mice after tail vein injection (98). miR-7-5p inhibits migration of 1205 Lu and WM266-4 melanoma cells (98). Tail vein injection of 1205 Lu cells transduced with miR-7-5p results in decreased lung metastases in NSG mice (98). Nuclear factor ĸB (NFĸB) p65 subunit v-rel avian reticuloendotheliosis viral homolog A (RelA) has been identified as a direct target of miR-7-5p (98-100). miR-7-5p suppresses canonical nuclear factor ĸB (NFĸB) genes such as IL-1β, -6 and -8 and the effects of miR-7-5p on cell cycle, invasion and migration were recapitulated by RelA knockdown (98). RelA overexpression and low miR-7-5p levels are indicators of poor prognosis in melanoma (98).
Decreased expression of miR-98a (Figure 3B) is associated with lower overall survival in melanoma patients (101). B16F1 cells transduced with miR-98 exhibit reduced migration in vitro and decreased lung metastasis after tail vein injection (101). IL6 which promotes melanoma metastasis in vitro and in vivo has been identified as a direct target of miR-98 (101). IL6/IL6 receptor interaction via autocrine or paracrine loops plays an important role in tumor progression (102,103). In patients with melanoma, serum levels of IL6 have been correlated with worse prognosis (104).
miR-339-3p (Figure 3B) was identified in the context of a screening program for migration-accelerating or -inhibiting miRs (105). miR-339-3p inhibits invasion of A375 melanoma cells and five other melanoma cells derived from WM and MaMel cells (105). miR-339-3p transduced A375 cells exhibit reduced lung metastasis after tail vein injection in nude mice (105). Induced myeloid leukemia cell differentiation protein (MCL-1) was identified as a direct target of miR-339 (105). MCL-1 belongs to the BCL2 family of proteins and is required for the resistance of melanoma to anoikis (106). Overexpression of MCL-1 confers resistance to BRAF V600E inhibitors alone and in combination with mitogen-activated protein kinase 1/2 (MEK1/2) inhibitors in melanoma patients (107). Several programs for preclincial and clinical evaluation of MCL-1 inhibitors are presently ongoing (108,109).
Up-regulated miRs Promoting Growth and Metastasis in Preclinical Melanoma-related In Vivo Models
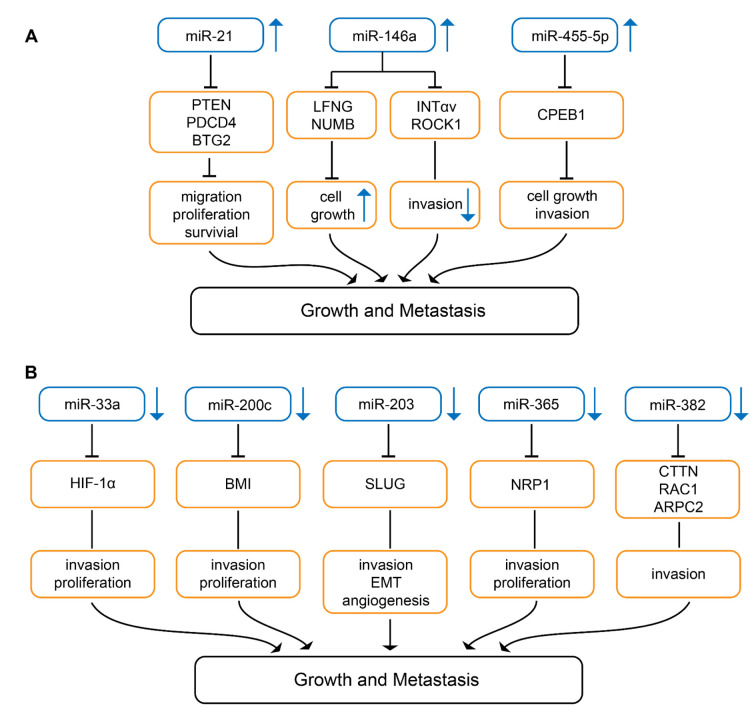
Knock-down of miR-21 (Figure 4A) in B16 melanoma cells inhibits cell proliferation and invasion, whereas apoptosis is promoted (110). Syngeneic mice injected with B16 cells into the tail vein exhibit large nodules in the lungs, whereas mice injected with miR-21 knock-down B16 cells give rise to only minor metastatic deposits (110). As direct targets of miR-21 TS genes phosphatase and tensin homolog (PTEN) and programmed cell death protein 4 (PDCD4) as well as anti-proliferative protein B-cell translocation gene 2 (BTG2) were identified (110). PTEN has phosphatase-dependent and phosphatase-independent (scaffold) activities regulating genome stability, migration and proliferation, cell survival and metabolism (111). PDCD4 inhibits proliferation, invasion and metastasis and induces apoptosis by binding to the translation initiation factor eIF4a, several transcription factors and other protein and modulates the function of the binding partners (112). BTG2 is down-regulated in many types of cancers and is involved in inhibition of proliferation, DNA repair and mRNA stability (113,114). In human melanoma, miR-21 is strongly expressed in comparison to naevi and nuclear miR-21 and may have an impact as a prognostic biomarker (115).
Figure 4. miRs involved in growth and metastasis in melanoma-related preclinical in vivo models. Upward arrows indicate up-regulation, downward arrows indicate down-regulation in melanoma compared to corresponding normal tissues. (A) miRs up-regulated in melanoma in comparison to corresponding normal tissues. BTG2: BTG2 family member 2; CPEB1: cytoplasmic polyadenylation element binding protein 1; INTαv: integrin αv; LFNG: lunatic fringe; NUMB: protein numb homolog; PDCD4: programmed cell death protein 4; PTEN: phosphatase and tensin homolog; ROCK 1: Rho-associated coiled-coil containing protein kinase 1. (B) miRs down-regulated in melanoma in comparison to corresponding normal tissues. ARPC2: Actin-related protein 2/3 complex subunit 2; BMI: B-lymphoma Mo-MLV insertion region 1 homolog; CTTN: cortactin; HIF-1α: hypoxia-inducible factor 1α; NRP-1: neuropilin 1; RAC1: ras-related C3 botulinum toxin substrate 1; SLUG: zinc finger protein SLUG.
miR-146a (Figure 4A) is a miR with a dual function: it can act as an onco-miR as well as a tumor-suppressive miR. It enhances TG, but inhibits metastasis (116). miR-146a is up-regulated during tumor progression, but is poorly expressed in circulating tumor cells (CTC) (116). In MA-2 and MC-1 melanoma cells down-modulation of miR-146a impairs cell proliferation as well as anchorage-independent growth in soft agar. miR-146a overexpressing MA-2 cells generate bigger tumors in comparison to control cells after subcutaneous implantation (116). miR-146a inhibits extravasation and lung metastasis of MA-2-miR-146a overexpressing melanoma cells after tail vein injection (116). miR-146a also affects dissemination from primary tumors. Down-modulation of miR-146a enhances dissemination of tumors to the lungs after subcutaneous implantation (116). miR-146a controls TG through its direct targets lunatic fringe (LNFG) and NUMB (116). Inhibition of NUMB and deletion of LNFG have been shown to activate the NOTCH1 pathway (117,118). LNFG is an O-fucose β1,3 N-acetyl-glucosminyl transferase which glycosylates epidermal growth factor (EGF)-like repeats in the extracellular domain of NOTCH, thus inhibiting JAGGED-mediated NOTCH activation (119). NUMB is a membrane-bound protein which is also involved in suppression of NOTCH signaling (120). miR-146a mediated down-regulation of direct targets INTαv and serine/threonine kinase RHO-associated, coiled-coil containing protein kinase 1 (ROCK1) affects motility, but not TG (116). ROCK1 mediates rounded cell migration, whereas INTαv is involved in mesenchymal-like invasion characterized by cell polarization and membrane protrusion (121). ROCK1 is a major effector of small GTPase RHO A and regulates the actomyosin cytoskeleton promoting contractile force generation and plays a role in mobility, metastasis and angiogenesis (122,123).
Wild-type miR-455-5p (Figure 4A) promotes melanoma growth and metastases, the A-to-I edited version inhibits these properties (124). miR-455-5p is edited by adenosine deaminase acting on RNA (ADAR1) (124). ADAR1-mediated RNA editing of miRs can affect their biogenesis or can change their binding sites (125,126). ADAR-1 expression and the edited version of miR-455-5p decrease during melanoma progression (124). SB2 melanoma cells transduced with wild-type miR-455-5p exhibit increased TG after subcutaneous implantation into nude mice and increased incidence of experimental lung metastasis (124). Cytoplasmic polyadenylation element binding protein 1 (CPEB1) has been identified as a direct target of wild-type miR-455-5p (124). CPEB1 binds to a specific RNA sequence in the 3´-UTR of the corresponding mRNA referred to as cytoplasmic polyadenylation element and is involved in translational control (127,128). CPEB1 has been identified as a TS in gastric and thyroid cancer (129,130). In contrast to the wild-type miR-455-5p, the edited version targets oncogenes such as integrin α2 (INTα2), mouse double minute 4 (MDM4) and ras homolog gene family member C (RHO-C) (124). The clinical relevance of these findings remains to be investigated.
Down-regulated miRs Inhibiting Growth and Metastasis in Melanoma-related Preclinical In Vivo Models
miR-33a (Figure 4B) is expressed at a low level in metastatic melanoma (131). miR-33a inhibits proliferation and invasion of human melanoma cells WM-35 and WM-451 (131). In vivo, A375 cells transduced with miR-33a showed reduced TG after subcutaneous implantation in comparison to control cells (131). Hyoxia-inducible factor-1α (HIF-1α) was identified as a direct target of miR-33a (131). HIF-1α exacerbates conversion of normal melanocytes to melanoma (132) and promotes metastasis by activation of tyrosine kinase SRC (133). Hypoxia controls metastasis of mouse and human melanoma cells (134). HIF-1α expression correlates with severity of disease and metastasis in melanoma patients (135).
miR-200c (Figure 4B) exhibits reduced expression in melanoma in comparison to naevi (136). miR-220c inhibits proliferation of melanoma cell lines WM115A, 1205 Lu, WM793, WM3523A and WM35 (136). In vivo growth in nude mice is inhibited after subcutaneous implantation into the flanks of miR-200c transduced WM115A cells in comparison to the non-transduced control cell line (136). Polycomb complex protein B lymphoma Mo-MLV insertion region 1 homolog (BMI-1) has been identified as a direct target of miR-200c (136). However, also other genes are down-regulated by miR-200c such as membrane transporters ATP-binding cassette superfamily G members 2 and 5 (ABCG2, ABCG5) and multi-drug resistance protein 1 (MDR1), whereas E-cadherin is up-regulated (136). BMI-1 knockdown phenocopies miR-200c overexpression (136). BMI-1 belongs to the polycomb group of proteins, a class of transcriptional repressors that orchestrate changes in chromatin structure to regulate gene activity (136). BMI-1 functions as a transcriptional repressor of the inhibitor of CDK4A/ARF tumor suppressor (INK4A/ARF) locus. INK4A inhibits cyclin-dependent kinase 4A (CDK4) activity and blocks entry into the cell cycle (137,138). The findings support the existence of an interconnected network of miR-200c, BMI-1, E-cadherin and MDR-1 (136-138). In addition to cancer cell proliferation, BMI-1 is involved in invasion, metastasis, self-renewal of cancer stem cells (CSC) and their differentiation and is a promising target for cancer therapy (139). Diminished expression of miR-200c correlates with the acquisition of a stem cell-like phenotype during melanoma progression (140-142). miR-200c is down-regulated in melanoma and acts as a TS (143).
miR-203 (Figure 4B) is decreased in metastatic melanoma and its expression level correlates with survival (144). miR-203 suppresses motility and sphere formation of melanoma cell lines BE, SK-Mel-28 and DFB (144). In vivo, BE cells transduced with miR-203 show decreased neo-vascularization of their xenografts (144). Reduced lung metastases was noted after tail vein injection of miR-203 transduced BE cells (144). SLUG (SNAI2), a zinc finger protein transcriptional repressor which down-regulates E-cadherin, was identified as a direct target of miR-203 (144). SLUG is involved in EMT and exerts anti-apoptotic activity (145). In melanoma, SLUG expression is essential for metastasis (146). Overexpression of SLUG is associated with lymph node metastasis, post-operative relapse and shorter patient survival in lung cancer and colorectal cancer patients (147,148).
miR-365 (Figure 4B) is down-regulated in human melanoma tissues relative to adjacent normal tissues (149). Ectopic expression of miR-365 in A375 and G361 melanoma cells attenuates cell proliferation and induces apoptosis in these cells (149). In vivo, miR-365 overexpression in A375 cells suppresses TG after subcutaneous implantation and reduces lung metastases after tail vein injection into nude mice (149). Neuropilin-1 (NRP-1) has been identified as a direct target of miR-365 (149). NRP-1 is a transmembrane glycoprotein that acts as a co-receptor for various members of the VEGF family and modulates the activity of other extracellular ligands such as transforming growth factor β (TGFβ), hepatocyte growth factor (HGF), fibroblast growth factor (FGF) and platelet-derived growth factor (PDGF), factors modulating angiogenesis and therefore is a potential target for treatment of melanoma (150-152).
miR-382 (Figure 4B) is decreased in more versus less aggressive primary melanomas (153). TG of melanoma cell line WM1361a is accelerated by expression of a miR-382 decoy (153). Lung metastasis is inhibited after subcutaneous implantation of 451Lu melanoma cells transfected with miR-383 into the flanks of nude mice (153). As direct targets of miR-382 cytoskeleton regulators cortactin (CTTN), actin-related protein 2 complex (ARPC2) and ras-related C3 botulinum substrate 1 (RAC1) have been identified (153). CTTN is a nodal point for regulation of the actin cytoskeleton, regulates lamellopodia, directs migration and matrix metalloproteinase activity (154,155). ARPC2 is a core component of the ARP2/3 complex which nucleates branched actin networks (156). RAC1 is an RHO-family GTPase that regulates a diverse set of cellular processes, particularly cell motility through regulation of actin polymerization (157,158).
Synopsis of Identified miRs
The identified miRs have been classified as miRs affecting TG only, metastasis only and TG as well as metastasis in preclinical in vivo efficacy models (Figures 1-4). These effects can be achieved by up-regulation or down-regegulation of the corresponding miRs in melanoma in comparison to naevi or adjacent normal tissues. A major subclass of the identified miRs targets transcription factors such as RelA (miR-7-5p), ZEB2 (miR-30a), HIF-1α (miR-33a), MITF and FOXN3 (miR-182), TFAP2C (miR-214), SLUG (miR-365), FOXN3 (miR-378) and CPEB1 (miR-455-5p). Another sub-category of the identified miRs targets mRNAs derived from cell-cycle and apoptosis-related genes such as BTG2, PDCD4 and PTEN (miR-21), BCL2 (miR-219-5p), BMI-1 (miR-200c), CDKN1B (miR-221, -222), MCL1 (miR-339-3p) and TP53INP2 (miR-638). Another group of the identified miRs targets components of the actin or actomyosin cytoskeleton, such as ROCK1 (miR-146a), ARPC2, CTTN and RAC1 (miR-382) as well as espin (miR-612). Autophagy is affected by miRs -23a and -638 and miRs -30a, -203 and -542 are involved in EMT-related pathways. An interesting observation is that an A-to-I edited miR (455-5p) can have an inhibitory impact on metastasis of melanoma cells in preclinical melanoma-related in vivo models due to change in target specificity induced by the editing process. Another observation is the finding that a component of the secretory pathway (SEC23A) can modulate the metastatic secretome. miR-146a is unique since it has a different impact on TG versus metastases. It promotes TG via targeting LFNG and NUMB resulting in activation of NOTCH signaling, whereas it inhibits metastases by targeting INTαv and ROCK1. LFNG as well as Gal-N-transferase (miR-30b/d) are sugar-modifying enzymes, which have an impact on metastasis and on growth as well as metastasis, respectively. Interestingly, one of the identified miRs promotes metastasis through targeting of the metabolic component ApoE. To our surprise, none of the identified miRs targets components of the rapidly accelerated fibrosarcoma (RAF)/MEK signaling pathway directly.
Clinical correlations are an important issue for prioritization of identified miRs and corresponding targets for therapeutic intervention. Up- or down-regulation of several miRs in melanoma tissues in comparison to corresponding control tissues holds true for miRs-7-5p, -23a, -30b/d, -98a, -125b, -182, -199a, -203, -214, -219-5p, -221/-222, -365, -378 and -638. In addition to deregulation, correlations with disease progression and corresponding survival data are available for an up-regulated and five down-regulated miRs. miR-199a (ApoE, DNAJ4) is up-regulated, miRs-7-5p (RelA), miR-98a (IL6), miR-203 (SLUG), miR-219-5p (BCL2) and miR-365 (NRP-1) are down-regulated. miR-219-5p regulates TG, miR-98a and -199a are drivers of metastasis, whereas miRs-203 and -365 have a combined impact on TG and metastasis.
Therapeutic aspects. Improved treatment options for advanced and metastatic melanoma are an important medical need. For miR-related targets, inhibition of up-regulated miRs or functional reconstitution of down-regulated miRs are the options for therapeutic intervention (more details in the following chapter). Alternatively, the identified targets can be modulated with small molecules or mAbs. However, one should keep in mind that miRs, in addition to the identified and validated targets, interfere with the messages of a several other genes. For the target of miR-219-5p, BCL2, a specific inhibitor, Venetoclax, has been approved by the FDA for treatment of CLL (159). Clinical studies with mAb MNRP1685A, which inhibits NRP-1, are currently ongoing (160). For IL6, the target of miR-98a, several IL6-inhibitory approved agents for treatment of inflammatory or myeloproliferative neoplasms are available (161). IL6-inhibitory agents are presently clinically evaluated for management of the side effects of chimeric antigen receptors (CARs)-based therapies (161).
As treatment options, prophylactic treatment of melanoma metastasis is problematic due to possible severe side-effects due to chronic treatment (162). With respect to treatment of established metastases, unprecedented responses have been achieved with programmed cell death protein 1 (PD1)- and PD1-ligand (PD-1L)-inhibitory mAbs (163-165). miRs inhibiting TG (miR-219-5p) and TG as well as metastases (miRs-203, -365) are possible candidates for treatment of metastatic melanoma as single agents or in combination with other agents such as checkpoint inhibitors. Expression of the corresponding target in metastases has to be assessed case by case. miRs affecting metastasis only (miR-7-5p, -98a, -199a) should be combined with a growth-inhibitory agent to optimize their therapeutic potential. However, it should be kept in mind, that treatment of established metastases is hampered by many critical issues such as their heterogeneous composition and disabled vasculature, resulting in poor drug penetration and resistance against drugs, which have a therapeutic impact on the corresponding primary tumor (162,166).
Technical aspects. As outlined, the therapeutic options of miR-related agents for treatment of advanced melanoma are inhibition of miRs or their functional reconstitution (167-169). miR inhibitors are single-stranded RNAs such as locked nucleic acids (LNA), 12-25 nts complementary to the corresponding mRNA (167). miR sponges contain multiple miR-binding sites which compete with the natural mRNA target for binding to the corresponding miR (169). Low-molecular weight drugs can affect the transcription of miRs or interfere with their secondary structure, but specificity issues associated with corresponding compounds remain to be resolved (167,169). Functional reconstitution can be performed with miR mimetics, double-stranded RNAs, designed to mimic endogenous mature miRs (168). The other option is to express the corresponding miR with plasmid or virus-based expression vectors in corresponding recipient cells (168).
However, several technical issues hampering the therapeutic application of miR-related agents remain to be resolved. Issues to be tackled are: design of efficient delivery systems, serum stability and removal of the complexed nucleic acids by the reticulo-endothelial system, renal excretion, immunogenicity and immuno-modulatory effects, hybridization-dependent and -independent off-target effects, haematological and hepatic toxicity, cytokine release syndrome (CRS), extravasation, entry into the target cell and endosomal escape (170-172). These issues have to be optimized case-by-case. Considerable progress has been achieved by improving the binding affinity, stability, pharmaco-kinetic and pharmaco-dynamic properties of miR-related agents for optimized target modulation (173-175). Progress has been achieved by stabilization of miR-targeting agents with polyethylene glycole, delivery as nanoparticles and combination with specific targeting packages and shielding to ameliorate immunogenicity and the potential to induce CRS (173-177). Complexation of miR-targeting agents to cell-penetrating viral proteins to accomplish endosomal escape is another important achievement (178,179).
Finally, the relationship between siRNA and miRs as therapeutic agents should be mentioned. siRNA and miRs share many similarities. Both are short duplex RNA molecules that exert gene-silencing effects at the post-transcriptional level by targeting mRNA. However, the MOA is different. siRNAs are highly specific for only one target, whereas miRs target several mRNAs with the capacity to interfere with several pathways (180).
Therapeutic Landscape
The field of miR-based therapeutics has witnessed several serious drawbacks in the past few years (181). Several clinical studies in patients with kidney disease, Hepatitis C infection and cancer were terminated due to serious side effects despite of promising efficacy data (181). Clinical evaluation of second generation of miRs with an improved side-effect profile is underway. MIRAGEN is presently running PhaseI/II clinical studies with COBOMARSEN (MRG-106), an LNA anti-miR-155, for treatment of several blood cancers such as cutaneous T-cell lymphoma, adult T-cell lymphoma and leukemia, diffuse large B-cell lymphoma and chronic lymphocytic leukemia with a very favourable side effect profile (181). A breakthrough in the field of RNA-related therapeutics is the FDA-approval of Onpattro (ALNYLAM Pharmaceuticals), an siRNA acting in the liver of patients with transerythyrin amyloidosis and neuropathy (182). The emerging importance of RNA-based therapeutics is emphasized by a recent deal between REGENERON and ALNYLAM covering 39 targets for diseases of the eye, central nervous system and liver (183).
Conflicts of Interest
SA and UB are and UHW was employed by Roche, Roche is interested in the diagnosis and therapy of cancer.
Authors’ Contributions
UHW devised the concept, coordinated and compiled the information for this review. UHW, SA and UB jointly prepared the review article and its Figures.
References
- 1.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin Oncol. 2019;69(5):363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 2.Das P, Deshmukh N, Badore N, Ghulaxe C, Patel P. A review article on melanoma. Pharm Sci and Res. 2016;8:112–117. [Google Scholar]
- 3.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm SJ, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, López-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 4.Liu W, Kelly JW, Trivett M, Murray WK, Dowling JP, Wolfe R, Mason G, Magee J, Angel C, Dobrovic A, McArthur GA. Distinct clinical and pathological features are associated with the BRAF T1799A(V600E)) mutation in primary melanoma. J Invest Dermatol. 2007;127:900–905. doi: 10.1038/sj.jid.5700632. [DOI] [PubMed] [Google Scholar]
- 5.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14:463–482. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan RJ, Hamid O, Gonzalez R, Infante JR, Patel MR, Hodi FS, Lewis KD, Tawbi HA, Hernandez G, Wongchenko MJ, Chang Y, Roberts L, Ballinger M, Yan Y, Cha E, Hwu P. Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat Med. 2019;25:929–935. doi: 10.1038/s41591-019-0474-7. [DOI] [PubMed] [Google Scholar]
- 7.Ribas A, Lawrence D, Atkinson V, Agarwal S, Miller WH Jr, Carlino MS, Fisher R, Long GV, Hodi FS, Tsoi J, Grasso CS, Mookerjee B, Zhao Q, Ghori R, Moreno BH, Ibrahim N, Hamid O. Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat Med. 2019;25:936–940. doi: 10.1038/s41591-019-0476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ascierto PA, Ferrucci PF, Fisher R, Del Vecchio M, Atkinson V, Schmidt H, Schachter J, Queirolo P, Long GV, Di Giacomo AM, Svane IM, Lotem M, Bar-Sela G, Couture F, Mookerjee B, Ghori R, Ibrahim N, Moreno BH, Ribas A. Dabrafenib, trametinib and pembrolizumab or placebo in BRAF-mutant melanoma. Nat Med. 2019;25:941–946. doi: 10.1038/s41591-019-0448-9. [DOI] [PubMed] [Google Scholar]
- 9.Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat Rev Cancer. 2019;19:377–391. doi: 10.1038/s41568-019-0154-4. [DOI] [PubMed] [Google Scholar]
- 10.Gurzu S, Beleaua MA, Jung I. The role of tumor microenvironment in development and progression of malignant melanomas - a systematic review. Rom J Morphol Embryol. 2018;59:23–28. [PubMed] [Google Scholar]
- 11.Kasumova GG, Haynes AB, Boland GM. Lymphatic versus hematogenous melanoma metastases: Support for biological heterogeneity without clear clinical application. J Invest Dermatol. 2017;137:2466–2468. doi: 10.1016/j.jid.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Zbytek B, Carlson JA, Granese J, Ross J, Mihm MC Jr, Slominski A. Current concepts of metastasis in melanoma. Expert Rev Dermatol. 2008;3:569–585. doi: 10.1586/17469872.3.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearlman RL, Montes de Oca MK, Pal HC, Afaq F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017;391:125–140. doi: 10.1016/j.canlet.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 15.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Disibio G, French SW. Metastatic patterns of cancers: results from a large autopsy study. Arch Pathol Lab Med. 2008;132:931–939. doi: 10.1043/1542165(2008)132[931:MPOCRF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Patel JK, Didolkar MS, Pickren JW, Moore RH. Metastatic pattern of malignant melanoma. A study of 216 autopsy cases. Am J Surg. 1978;135:807–810. doi: 10.1016/0002-9610(78)90171-x. [DOI] [PubMed] [Google Scholar]
- 18.Everett L, Copperman T. Metastatic uveal melanoma. N Engl J. 2019;380:1853. doi: 10.1056/NEJMicm1810596. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 22.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA. RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 24.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29:2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 27.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 28.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidle UH, Dickopf S, Hintermair C, Kollmorgen G, Birzele F, Brinkmann U. The role of micro RNAs in breast cancer metastasis: preclinical validation and potential therapeutic targets. Cancer Genomics Proteomics. 2018;15:17–39. doi: 10.21873/cgp.20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weidle UH, Epp A, Birzele F, Brinkmann U. The functional role of prostate cancer metastasis-related micro-RNAs. Cancer Genomics Proteomics. 2019;16:1–19. doi: 10.21873/cgp.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weidle UH, Birzele F, Nopora A. MicroRNAs as potential targets for therapeutic intervention with metastasis of non-small cell lung cancer. Cancer Genomics Proteomics. 2019;16:99–119. doi: 10.21873/cgp.20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidle UH, Birzele F, Kollmorgen G, Nopora A. Potential microRNA-related targets for therapeutic intervention with ovarian cancer metastasis. Cancer Genomics Proteomics. 2018;15:1–15. doi: 10.21873/cgp.20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felicetti F, Errico C, L Bottero L, Segnalini P, Stoppacciaro A, Mauro Biffoni, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C and Care A. The promyelocytic leukemia zinc finger–MicroRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;15:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 35.Alexeev V, Yoon K. Distinctive role of the cKit receptor tyrosine kinase signaling in mammalian melanocytes. J Invest Dermatol. 2006;126:1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- 36.Hussein MR, Haemel AK, Wood GS. Apoptosis and melanoma: molecular mechanisms. J Pathol. 2003;199:275–88. doi: 10.1002/path.1300. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Sanki A, Karim RZ, Thompson JF, Soon Lee C, Zhuang L, McCarthy SW, Scolyer RA. The role of cell cycle regulatory proteins in the pathogenesis of melanoma. Pathology. 2006;38:287–301. doi: 10.1080/00313020600817951. [DOI] [PubMed] [Google Scholar]
- 38.Sun M, Ma X, Tu C, Wang X, Qu J, Wang S, Xiao S. MicroRNA-378 regulates epithelial-mesenchymal transition and metastasis of melanoma by inhibiting FOXN3 expression through the Wnt/β-catenin pathway. Cell Biol Int. 2019;43:1113–1124. doi: 10.1002/cbin.11027. [DOI] [PubMed] [Google Scholar]
- 39.Kong X, Zhai J, Yan C, Song Y, Wang J, Bai X, Brown JAL, Fang Y. Recent advances in understanding FOXN3 in breast cancer, and other malignancies. Front Oncol. 2019;9:234. doi: 10.3389/fonc.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long J, Menggen Q, Wuren Q, Shi Q, Pi X. MiR-219-5p inhibits the growth and metastasis of malignant melanoma by targeting BCL-2. Biomed Res Int. 2017;2017:9032502. doi: 10.1155/2017/9032502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 42.Croce CM, Reed JC. Finally, an apoptosis-targeting therapeutic for cancer. Cancer Res. 2016;76:5914–5920. doi: 10.1158/0008-5472.CAN-16-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Zhang HL, Wang QY, Chen MJ, Liu LB. Overexpression of microRNA-612 restrains the growth, invasion, and tumorigenesis of melanoma cells by targeting espin. Mol Cells. 2018;41:119–126. doi: 10.14348/molcells.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taura A, Taura K, Koyama Y, Yamamoto N, Nakagawa T, Ito J, Ryan AF. Hair cell stereociliary bundle regeneration by espin gene transduction after aminoglycoside damage and hair cell induction by Notch inhibition. Gene Ther. 2016;23:415–423. doi: 10.1038/gt.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Zou J, Shen Z, Song E, Yang J. Whirlin interacts with espin and modulates its actin-regulatory function: an insight into the mechanism of Usher syndrome type II. Hum Mol Genet. 2012;1:692–710. doi: 10.1093/hmg/ddr503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, Bhardwaj N, Shao Y, Darvishian F, Zavadil J, Erlebacher A, Mahal LK, Osman I, Hernando E. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 48.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 50.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK, Brugge JS. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 52.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 53.Goding CR, Arnheiter H. MITF-the first 25 years. Genes and Devel. 2019;33:983–1007. doi: 10.1101/gad.324657.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 55.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing P, Liao R, Zhao J, Song F, Wang G, Chen K, Yang J. Roles of low-density lipoprotein receptor-related protein 1 in tumors. Chin J Cancer. 2016;35:6. doi: 10.1186/s40880-015-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roslan Z, Muhamad M, Selvaratnam L, Ab-Rahim S. The roles of Low-density lipoprotein receptor-related proteins 5, 6, and 8 in cancer: A review. J Oncol. 2019;2019:4536302. doi: 10.1155/2019/4536302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S. Heat shock proteins and cancer. Trends Pharmacol Sci. 2017;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 62.Sun Z, Zhou S, Tang J, Ye T, Li J, Liu D, Zhou J, Wang J, Rosie Xing H. Sec23a mediates miR-200c augmented oligometastatic to polymetastatic progression. EBioMedicine. 2018;37:47–55. doi: 10.1016/j.ebiom.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fromme JC, Orci L, Schekman R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008;18:330–336. doi: 10.1016/j.tcb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 64.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang T, Sun L, Yuan X, Qiu H. Thrombospondin-1 is a multifaceted player in tumor progression. Oncotarget. 2017;8:84546–8455. doi: 10.18632/oncotarget.19165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryschich E, Huszty G, Knaebel HP, Hartel M, Büchler MW, Schmid R. Transferrin receptor is a marker of malignant phenotype in human pancreatic cancer and in neuroendocrine carcinoma of the pancreas. Eur J Cancer. 2004;40:1418–1422. doi: 10.1016/j.ejca.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 67.Poynter JN, Jacobs ET, Figueiredo JC, Lee WH, Conti DV, Campbell PT, Levine AJ, Limburg P, Le Marchand L, Cotterchio M, Newcomb PA, Potter JD, Jenkins MA, Hopper JL, Duggan DJ, Baron JA, Haile RW. Genetic variation in the vitamin D receptor (VDR) and the vitamin D-binding protein (GC) and risk for colorectal cancer: results from the colon cancer family registry. Cancer Epidemiol Biomarkers Prev. 2010;19:525–536. doi: 10.1158/1055-9965.EPI-09-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mishan MA, Ahmadiankia N, Bahrami AR. CXCR4 and CCR7: Two eligible targets in targeted cancer therapy. Cell Biol Int. 2016;240:955–967. doi: 10.1002/cbin.10631. [DOI] [PubMed] [Google Scholar]
- 69.Xiong TF, Pan FQ, Li D. Expression and clinical significance of S100 family genes in patients with melanoma. Melanoma Res. 2019;29:23–29. doi: 10.1097/CMR.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pittà C, Pinatel E, Stadler MB, Provero P, Bernengo MG, Osman I, Taverna D. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30:1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Penna E, Orso F, Cimino D, Vercellino I, Grassi E, Quaglino E, Turco E, Taverna D. miR-214 coordinates melanoma progression by upregulating ALCAM through TFAP2 and miR-148b downmodulation. Cancer Res. 2013;73:4098–4111. doi: 10.1158/0008-5472.CAN-12-3686. [DOI] [PubMed] [Google Scholar]
- 72.Orso F, Quirico L, Virga F, Penna E, Dettori D, Cimino D, Coppo R, Grassi E, Elia AR, Brusa D, Deaglio S, Brizzi MF, Stadler MB, Provero P, Caselle M, Taverna D. miR-214 and miR-148b targeting inhibits dissemination of melanoma and breast cancer. Cancer Res. 2016;76:5151–5162. doi: 10.1158/0008-5472.CAN-15-1322. [DOI] [PubMed] [Google Scholar]
- 73.Huang R, Rofstad EK. Integrins as therapeutic targets in the organ-specific metastasis of human malignant melanoma. J Exp Clin Cancer Res. 2018;37:92. doi: 10.1186/s13046-018-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bar-Eli M. Searching for the ‘melano-miRs’: miR-214 drives melanoma metastasis. EMBO J. 2011;30:1880–1881. doi: 10.1038/emboj.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kołat D, Kałuzińska Ż, Bednarek AK, Płuciennik E. The biological characteristics of transcription factors AP-2α and AP-2γ and their importance in various types of cancers. Biosci Rep. 2019;39 :pii: BSR20181928. doi: 10.1042/BSR20181928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Kempen LC, van den Oord JJ, van Muijen GN, Weidle UH, Bloemers HP, Swart GW. Activated leukocyte cell adhesion molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. Am J Pathol. 2000;156:769–774. doi: 10.1016/S0002-9440(10)64943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genomics Proteomics. 2010;7:231–243. [PubMed] [Google Scholar]
- 78.Dettori D, Orso F, Penna E, Baruffaldi D, Brundu S, Maione F, Turco E, Giraudo E, Taverna D. Therapeutic silencing of miR-214 inhibits tumor progression in multiple mouse models. Mol Ther. 2018;26:2008–2018. doi: 10.1016/j.ymthe.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhattacharya A, Schmitz U, Raatz Y, Schönherr M, Kottek T, Schauer M, Franz S, Saalbach A, Anderegg U, Wolkenhauer O, Schadendorf D, Simon JC, Magin T, Vera J, Kunz M. miR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget. 2015;6:2966–2980. doi: 10.18632/oncotarget.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shahbazi J, Lock R, Liu T. Tumor protein 53-induced nuclear protein 1 enhances p53 function and represses tumorigenesis. Front Genet. 2013;4:80. doi: 10.3389/fgene.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seillier M, Peuget S, Gayet O, Gauthier C, N’Guessan P, Monte M, Carrier A, Iovanna JL, Dusetti NJ. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagydependent cell death. Cell Death Differ. 2012;19:1525–1535. doi: 10.1038/cdd.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y, Yi X, Ma J, Zhao T, Liu L, Jian Z, Liu L, Wang G, Gao T, Shi Q, Li C. Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics. 2017;7:2231–2249. doi: 10.7150/thno.18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deiana M, Dalle Carbonare L, Serena M, Cheri S, Parolini F, Gandini A, Marchetto G, Innamorati G, Manfredi M, Marengo E, Brandi J, Cecconi D, Mori A, Mina MM, Antoniazzi F, Mottes M, Tiso N, Malerba G, Zipeto D, Valenti MT. New insights into the runt domain of RUNX2 in melanoma cell proliferation and migration. Cells. 2018;7:pii; E220. doi: 10.3390/cells7110220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 85.Noda NN, Inagaki F. Mechanisms of autophagy. Annu Rev Biophys. 2015;44:101–22. doi: 10.1146/annurev-biophys-060414-034248. [DOI] [PubMed] [Google Scholar]
- 86.Cerezo M, Tichet M, Abbe P, Ohanna M, Lehraiki A, Rouaud F, Allegra M, Giacchero D, Bahadoran P, Bertolotto C, Tartare-Deckert S, Ballotti R, Rocchi S. Metformin blocks melanoma invasion and metastasis development in AMPK/p53-dependent manner. Mol Cancer Ther. 2013;12:1605–1615. doi: 10.1158/1535-7163.MCT-12-1226-T. [DOI] [PubMed] [Google Scholar]
- 87.Gayard M, Guilluy C, Rousselle A, Viollet B, Henrion D, Pacaud P, Loirand G, Rolli-Derkinderen M. AMPK alpha 1-induced RhoA phosphorylation mediates vasoprotective effect of estradiol. Arterioscler Thromb Vasc Biol. 2011;31:2634–2642. doi: 10.1161/ATVBAHA.111.228304. [DOI] [PubMed] [Google Scholar]
- 88.Sadok A, McCarthy A, Caldwell J, Collins I, Garrett MD, Yeo M, Hooper S, Sahai E, Kuemper S, Mardakheh FK, Marshall CJ. Rho kinase inhibitors block melanoma cell migration and inhibit metastasis. Cancer Res. 2015;75:2272–2284. doi: 10.1158/0008-5472.CAN-14-2156. [DOI] [PubMed] [Google Scholar]
- 89.Noori J, Sharifi M, Haghjooy Javanmard S. miR-30a inhibits melanoma tumor metastasis by targeting the E-cadherin and zinc finger E-box binding homeobox 2. Adv Biomed Res. 2018;7:143. doi: 10.4103/abr.abr_146_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1277. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J, Zhang H, Liu J, Tu X, Zang Y, Zhu J, Chen J, Dong L, Zhang J. miR-30 inhibits TGF-β1-induced epithelial-to-mesenchymal transition in hepatocyte by targeting Snail1. Biochem Biophys Res Commun. 2012;417:1100–1105. doi: 10.1016/j.bbrc.2011.12.121. [DOI] [PubMed] [Google Scholar]
- 92.Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044–2453. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 93.Zhang J, Na S, Liu C, Pan S, Cai J, Qiu J. MicroRNA-125b suppresses the epithelial-mesenchymal transition and cell invasion by targeting ITGA9 in melanoma. Tumour Biol. 2016;37:5941–5949. doi: 10.1007/s13277-015-4409-8. [DOI] [PubMed] [Google Scholar]
- 94.Gupta SK, Oommen S, Aubry MC, Williams BP, Vlahakis NE, Zhang J, Na S, Liu C, Pan S, Cai J, Qiu J. Integrin α9β1 promotes malignant tumor growth and metastasis by potentiating epithelial-mesenchymal transition. Oncogene. 2013;32:141–150. doi: 10.1038/onc.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rang Z, Yang G, Wang YW, Cui F. miR-542-3p suppresses invasion and metastasis by targeting the proto-oncogene serine/threonine protein kinase, PIM1, in melanoma. Biochem Biophys Res Commun. 2016;474:315–320. doi: 10.1016/j.bbrc.2016.04.093. [DOI] [PubMed] [Google Scholar]
- 96.Santio NM, Koskinen PJ. PIM kinases: From survival factors to regulators of cell motility. Int J Biochem Cell Biol. 2017;93:74–85. doi: 10.1016/j.biocel.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 97.Tursynbay Y, Zhang J, Li Z, Tokay T, Zhumadilov Z, Wu D, Xie Y. Pim-1 kinase as cancer drug target: An update. Biomed Rep. 2016;4:140–146. doi: 10.3892/br.2015.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giles KM, Brown RA, Ganda C, Podgorny MJ, Candy PA, Wintle LC, Richardson KL, Kalinowski FC, Stuart LM, Epis MR, Haass NK, Herlyn M, Leedman PJ. microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-ĸB. Oncotarget. 2016;7:31663–31680. doi: 10.18632/oncotarget.9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaltschmidt B, Greiner JFW, Kadhim HM, Kaltschmidt C. Subunit-Specific Role of NF-ĸB in Cancer. Biomedicines. 2018;6(2):44. doi: 10.3390/biomedicines6020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xia L, Tan S, Zhou Y, Lin J, Wang H, Oyang L, Tian Y, Liu L, Su M, Wang H, Cao D, Liao Q. Role of the NFĸB-signaling pathway in cancer. Onco Targets Ther. 2018;11:2063–2073. doi: 10.2147/OTT.S161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li F, Li XJ, Qiao L, Shi F, Liu W, Li Y, Dang YP, Gu WJ, Wang XG, Liu W. miR-98 suppresses melanoma metastasis through a negative feedback loop with its target gene IL-6. Exp Mol Med. 2014;46:e116. doi: 10.1038/emm.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Naugler WE, Karin M. The wolf in sheep’s clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med. 2008;14:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 103.Weidle UH, Klostermann S, Eggle D, Krüger A. Interleukin 6/interleukin 6 receptor interaction and its role as a therapeutic target for treatment of cachexia and cancer. Cancer Genomics Proteomics. 2010;7:287–302. [PubMed] [Google Scholar]
- 104.Hoejberg L, Bastholt L, Schmidt H. Interleukin-6 and melanoma. Melanoma Res. 2012;22:327–333. doi: 10.1097/CMR.0b013e3283543d72. [DOI] [PubMed] [Google Scholar]
- 105.Weber CE, Luo C, Hotz-Wagenblatt A, Gardyan A, Kordaß T, Holland-Letz T, Osen W, Eichmüller SB. miR-339-3p is a tumor suppressor in melanoma. Cancer Res. 2016;76:3562–3571. doi: 10.1158/0008-5472.CAN-15-2932. [DOI] [PubMed] [Google Scholar]
- 106.Boisvert-Adamo K, Longmate W, Abel EV, Aplin AE. Mcl-1 is required for melanoma cell resistance to anoikis. Mol Cancer Res. 2009;7:549–556. doi: 10.1158/1541-7786.MCR-08-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fofaria NM, Frederick DT, Sullivan RJ, Flaherty KT, Srivastava SK. Overexpression of Mcl-1 confers resistance to BRAFV600E inhibitors alone and in combination with MEK1/2 inhibitors in melanoma. Oncotarget. 2015;6:40535–40556. doi: 10.18632/oncotarget.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hird AW, Tron AE. Recent advances in the development of Mcl-1 inhibitors for cancer therapy. Pharmacol Ther. 2019;198:59–67. doi: 10.1016/j.pharmthera.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 109.Senichkin VV, Streletskaia AY, Zhivotovsky B, Kopeina GS. Molecular comprehension of Mcl-1: From gene structure to cancer therapy. Trends Cell Biol. 2019;29:549–562. doi: 10.1016/j.tcb.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 110.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286:39172–39178. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19:547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 112.Matsuhashi S, Manirujjaman M, Hamajima H, Ozaki I. Control mechanisms of the tumor suppressor PDCD4: Expression and functions. Int J Mol Sci. 2019;20:pii: E2304. doi: 10.3390/ijms20092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mao B, Zhang Z, Wang G. BTG2: a rising star of tumor suppressors (review) Int J Oncol. 2015;46:459–464. doi: 10.3892/ijo.2014.2765. [DOI] [PubMed] [Google Scholar]
- 114.Yuniati L, Scheijen B, van der Meer LT, van Leeuwen FN. Tumor suppressors BTG1 and BTG2: Beyond growth control. J Cell Physiol. 2019;234:5379–5389. doi: 10.1002/jcp.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saldanha G, Potter L, Lee YS, Watson S, Shendge P, Pringle JH. MicroRNA-21 expression and its pathogenetic significance in cutaneous melanoma. Melanoma Res. 2016;26:21–28. doi: 10.1097/CMR.0000000000000216. [DOI] [PubMed] [Google Scholar]
- 116.Raimo M, Orso F, Grassi E, Cimino D, Penna E, De Pittà C, Stadler MB, Primo L, Calautti E1, Quaglino P, Provero P, Taverna D. miR-146a exerts differential effects on melanoma growth and metastatization. Mol Cancer Res. 2016;14:548–562. doi: 10.1158/1541-7786.MCR-15-0425-T. [DOI] [PubMed] [Google Scholar]
- 117.Forloni M, Dogra SK, Dong Y, Conte D Jr, Ou J, Zhu LJ, Deng A, Mahalingam M, Green MR, Wajapeyee N. miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife. 2014;18:e01460. doi: 10.7554/eLife.01460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W, Cardiff RD, Yoon K, Gaiano N, Ling V, Beyene J, Zacksenhaus E, Gridley T, Leong WL, Guidos CJ, Perou CM, Egan SE. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer Cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, Vogt TF. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 120.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 121.Sanz-Moreno V. Tumour invasion: a new twist on Rac-driven mesenchymal migration. Curr Biol. 2012;22:R449–R451. doi: 10.1016/j.cub.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 122.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aspenström P. Activated Rho GTPases in cancer-the beginning of a new paradigm. Int J Mol Sci 19: 2018;pii:E3949. doi: 10.3390/ijms19123949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, Velazquez-Torres G, Nip KM, Zhu K, Brooks D, Jones SJ, Birol I, Mosqueda M, Wen YY, Eterovic AK, Sood AK, Hwu P, Gershenwald JE, Robertson AG, Calin GA, Markel G, Fidler IJ, Bar-Eli M. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 2015;17:311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maas S, Rich A, Nishikura K. A-to-I RNA editing: Recent news and residual mysteries. J Biol Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 126.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 128.Chen Y, Tsai YH, Tseng SH. Regulation of the expression of cytoplasmic polyadenylation element binding proteins for the treatment of cancer. Anticancer Res. 2016;36:5673–5680. doi: 10.21873/anticanres.11150. [DOI] [PubMed] [Google Scholar]
- 129.Caldeira J, Simões-Correia J, Paredes J, Pinto MT, Sousa S, Corso G, Marrelli D, Roviello F, Pereira PS, Weil D, Oliveira C, Casares F, Seruca R. CPEB1, a novel gene silenced in gastric cancer: a drosophila approach. Gut. 2012;61:1115–1123. doi: 10.1136/gutjnl-2011-300427. [DOI] [PubMed] [Google Scholar]
- 130.Zhang JH, Panicker LM, Seigneur EM, Lin L, House CD, Morgan W, Chen WC, Mehta H, Haj-Ali M, Yu ZX, Simonds WF. Cytoplasmic polyadenylation element binding protein is a conserved target of tumor suppressor HRPT2/CDC73. Cell Death Differ. 2010;17:1551–1565. doi: 10.1038/cdd.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou J, Xu D, Xie H, Tang J, Liu R, Li J, Wang S, Chen X, Su J, Zhou X, Xia K, He Q, Chen J, Xiong W, Cao P, Cao K. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1α. Cancer Biol Ther. 2015;16:846–855. doi: 10.1080/15384047.2015.1030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nys K, Maes H, Dudek AM, Agostinis P. Uncovering the role of hypoxia inducible factor-1α in skin carcinogenesis. Biochim Biophys Acta. 2011;1816:1–12. doi: 10.1016/j.bbcan.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 133.Hanna SC, Krishnan B, Bailey ST, Moschos SJ, Kuan PF, Shimamura T, Osborne LD, Siegel MB, Duncan LM, O’Brien ET 3rd, Superfine R, Miller CR, Simon MC, Wong KK, Kim WY. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. J Clin Invest. 2013;123:2078–2093. doi: 10.1172/JCI66715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, Bahadoran P, Lacour JP, Tartare-Deckert S, Bertolotto C, Ballotti R. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- 135.Giatromanolaki A, Sivridis E, Kouskoukis C, Gatter KC, Harris AL, Koukourakis MI. Hypoxia-inducible factors 1alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res. 2003;13:493–501. doi: 10.1097/01.cmr.0000056268.56735.4c. [DOI] [PubMed] [Google Scholar]
- 136.Liu S, Tetzlaff MT, Cui R, Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol. 2012;181:1823–1835. doi: 10.1016/j.ajpath.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 138.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 139.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen KG, Szakács G, Annereau JP, Rouzaud F, Liang XJ, Valencia JC, Nagineni CN, Hooks JJ, Hearing VJ, Gottesman MM. Principal expression of two mRNA isoforms (ABCB 5alpha and ABCB 5beta) of the ATP-binding cassette transporter gene ABCB 5 in melanoma cells and melanocytes. Pigment Cell Res. 2005;18:102–112. doi: 10.1111/j.1600-0749.2005.00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mihic-Probst D, Kuster A, Kilgus S, Bode-Lesniewska B, Ingold-Heppner B, Leung C, Storz M, Seifert B, Marino S, Schraml P, Dummer R, Moch H. Consistent expression of the stem cell renewal factor BMI-1 in primary and metastatic melanoma. Int J Cancer. 2007;121:1764–1770. doi: 10.1002/ijc.22891. [DOI] [PubMed] [Google Scholar]
- 142.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schüler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 143.Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lohcharoenkal W, Das Mahapatra K, Pasquali L, Crudden C, Kular L, Akkaya Ulum YZ, Zhang L, Xu Landén N, Girnita L, Jagodic M, Ståhle M, Sonkoly E, Pivarcsi A. Genome-wide screen for microRNAs reveals a role for miR-203 in melanoma metastasis. J Invest Dermatol. 2018;138:882–892. doi: 10.1016/j.jid.2017.09.049. [DOI] [PubMed] [Google Scholar]
- 145.Cho ES, Kang HE, Kim NH, Yook JI. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT) Arch Pharm Res. 2009;42:14–24. doi: 10.1007/s12272-018-01108-7. [DOI] [PubMed] [Google Scholar]
- 146.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shih JY, Tsai MF, Chang TH, Chang YL, Yuan A, Yu CJ, Lin SB, Liou GY, Lee ML, Chen JJ, Hong TM, Yang SC, Su JL, Lee YC, Yang PC. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res. 2005;11:8070–8078. doi: 10.1158/1078-0432.CCR-05-0687. [DOI] [PubMed] [Google Scholar]
- 148.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Int J Clin Exp Pathol. 2015;8:4913–4922. [PMC free article] [PubMed] [Google Scholar]
- 150.Graziani G, Lacal PM. Neuropilin-1 as therapeutic target for malignant melanoma. Front Oncol. 2015;5:125. doi: 10.3389/fonc.2015.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bagri A, Tessier-Lavigne M, Watts RJ. Neuropilins in tumor biology. Clin Cancer Res. 2009;15:1860–1864. doi: 10.1158/1078-0432.CCR-08-0563. [DOI] [PubMed] [Google Scholar]
- 152.Niland S, Eble JA. Neuropilins in the context of tumor vasculature. Int J Mol Sci. 2019;20:pii: E639. doi: 10.3390/ijms20030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hanniford D, Segura MF, Zhong J, Philips E, Jirau-Serrano X, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, Brown B, Osman I, Hernando E. Identification of metastasis-suppressive microRNAs in primary melanoma. J Natl Cancer Inst. 2015;107:dju494. doi: 10.1093/jnci/dju494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cosen-Binker LI, Kapus A. Cortactin: The gray eminence of the cytoskeleton. Physiology (Bethesda) 2006;21:352–361. doi: 10.1152/physiol.00012.2006. [DOI] [PubMed] [Google Scholar]
- 155.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Goley ED1, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 157.Smith LG, Li R. Actin polymerization: riding the wave. Curr Biol. 2004;14:R109–R111. [PubMed] [Google Scholar]
- 158.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 159.Pekarsky Y, Balatti V, Croce CM. BCL2 and miR-15/16: from gene discovery to treatment. Cell Death Differ. 2018;25:21–26. doi: 10.1038/cdd.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weekes CD, Beeram M, Tolcher AW, Papadopoulos KP, Gore L, Hegde P, Xin Y, Yu R, Shih LM, Xiang H, Brachmann RK, Patnaik A. A phase I study of the human monoclonal anti-NRP1 antibody MNRP1685A in patients with advanced solid tumors. Invest New Drugs. 2014;32:653–660. doi: 10.1007/s10637-014-0071-z. [DOI] [PubMed] [Google Scholar]
- 161.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, Tian T, Wei Z, Madan S, Sullivan RJ, Boland G, Flaherty K, Herlyn M, Ruppin E. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24:1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Karivedu V, Eldessouki I, Taftaf A, Zhu Z, Makramalla A, Karim NA. Nivolumab and Ipilimumab in the treatment of metastatic uveal melanoma: A single-center experience. Case Rep Oncol Med. 2019;2019:3560640. doi: 10.1155/2019/3560640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kilickap S, Guven DC, Aktepe OH, Aktas BY, Dizdar O. Complete responses to two different anti-PD1 agents in a metastatic melanoma patient. J Oncol Pharm Pract. 2019;29:1078155219858657. doi: 10.1177/1078155219858657. [DOI] [PubMed] [Google Scholar]
- 166.Anderson RL, Balasas T, Callaghan J, Coombes RC, Evans J, Hall JA, Kinrade S, Jones D, Jones PS, Jones R, Marshall JF, Panico MB, Shaw JA, Steeg PS, Sullivan M, Tong W, Westwell AD, Ritchie JWA, Cancer Research UK and Cancer Therapeutics CRC Australia Metastasis Working Group A framework for the development of effective anti-metastatic agents. Nat Rev Clin Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review) Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104–1110. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.MacLeod AR, Crooke ST. RNA therapeutics in oncology: Advances, challenges, and future directions. J Clin Pharmacol. 2017;57(Suppl 10):S43–S59. doi: 10.1038/gt.2011.50. [DOI] [PubMed] [Google Scholar]
- 171.Li Z, Rana TM. Therapeutic targeting of microRNAs: Current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 172.Garofalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Curr Pharm Des. 2014;20:5328–5335. doi: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- 173.Malek A, Merkel O, Fink L, Czubayko F, Kissel T, Aigner A. In vivo pharmacokinetics, tissue distribution and underlying mechanisms of various PEI(-PEG)/siRNA complexes. Toxicol Appl Pharmacol. 2009;236:97–108. doi: 10.1016/j.taap.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 174.Wang AZ, Langer R, Farokhzad OC. Nanoparticle delivery of cancer drugs. Annu Rev Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 175.Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, Bertin SL, Reppen TW, Chu Q, Blokhin AV, Hagstrom JE, Wolff JA. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 177.Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q, Hoekstra M, Kandasamy P, Kel’in AV, Milstein S, Taneja N, O’Shea J, Shaikh S, Zhang L, van der Sluis RJ, Jung ME, Akinc A, Hutabarat R, Kuchimanchi S, Fitzgerald K, Zimmermann T, van Berkel TJ, Maier MA, Rajeev KG, Manoharan M. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 178.Copolovici DM, Langel K, Eriste E, Langel U. Cell-penetrating peptides: Design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 179.Thorén PE, Persson D, Isakson P, Goksör M, Onfelt A, Nordén B. Uptake of analogs of penetratin, Tat(48-60) and oligoarginine in live cells. Biochem Biophys Res Commun. 2003;307:100–107. doi: 10.1016/s0006-291x(03)01135-5. [DOI] [PubMed] [Google Scholar]
- 180.Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jones D. Setbacks shadow microRNA therapies in the clinic. Nat Biotechnol. 2018;36:909–910. doi: 10.1038/nbt1018-909. [DOI] [PubMed] [Google Scholar]
- 182.Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Disc. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 183.Cormac S. Billion-dollar deal propels RNAi to CNS frontier. Nat Biotechnol. 2019;37:699–706. doi: 10.1038/d41587-019-00014-7. [DOI] [PubMed] [Google Scholar]