Abstract
The coronavirus disease 2019 (COVID‐19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has affected millions of people worldwide. Characterization of the immunological mechanisms involved in disease symptomatology and protective response is important to progress in disease control and prevention. Humans evolved by losing the capacity to synthesize the glycan Galα1‐3Galβ1‐(3)4GlcNAc‐R (α‐Gal), which resulted in the development of a protective response against pathogenic viruses and other microorganisms containing this modification on membrane proteins mediated by anti‐α‐Gal immunoglobulin M (IgM)/IgG antibodies produced in response to bacterial microbiota. In addition to anti‐α‐Gal antibody‐mediated pathogen opsonization, this glycan induces various immune mechanisms that have shown protection in animal models against infectious diseases without inflammatory responses. In this study, we hypothesized that the immune response to α‐Gal may contribute to the control of COVID‐19. To address this hypothesis, we characterized the antibody response to α‐Gal in patients at different stages of COVID‐19 and in comparison with healthy control individuals. The results showed that while the inflammatory response and the anti‐SARS‐CoV‐2 (Spike) IgG antibody titers increased, reduction in anti‐α‐Gal IgE, IgM, and IgG antibody titers and alteration of anti‐α‐Gal antibody isotype composition correlated with COVID‐19 severity. The results suggested that the inhibition of the α‐Gal‐induced immune response may translate into more aggressive viremia and severe disease inflammatory symptoms. These results support the proposal of developing interventions such as probiotics based on commensal bacteria with α‐Gal epitopes to modify the microbiota and increase α‐Gal‐induced protective immune response and reduce severity of COVID‐19.
Keywords: antibody, coronavirus, COVID‐19, immunology, microbiota, α‐Gal
1. INTRODUCTION
The coronavirus disease 19 (COVID‐19), a pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has rapidly evolved from an epidemic outbreak to a disease affecting the global population. SARS‐CoV‐2 infects human host cells by binding to the angiotensin‐converting enzyme 2 (ACE2) receptor. 1 It has been established that COVID‐19 mainly affects the respiratory tract, but as a systemic disease, it affects multiple processes including the gastrointestinal, cardiovascular, neurological, hematopoietic, and immune systems. 2 Several days after the onset of symptoms, the SARS‐CoV‐2 infection becomes more systemic and affects various organs with inflammatory responses and lymphocytopenia. 2 Lymphocytopenia is likely caused by the direct lethal effect of SARS‐CoV‐2 on lymphocytes with the ACE2 receptor 3 and the release of pro‐inflammatory cytokines such as tumor necrosis factor‐α (TNF‐α), interleukin 1 (IL‐1) and IL‐6 that induce apoptosis in lymphocytes. 4 The “cytokine storm syndrome (CSS)” has been associated with COVID‐19 through the activation of the nuclear factor‐kB (NF‐kB) innate immune pathway resulting in the upregulation of pro‐inflammatory cytokines. 5 Lymphocytopenia in patients with COVID‐19 along with the rise in neutrophils has been associated with worse disease prognosis. Consequently, patients with respiratory distress syndrome in intensive care unit (ICU) show lower lymphocyte counts and higher mortality when compared to other COVID‐19 patients. 6 , 7 Additionally, COVID‐19 patients suffer dysbacteriosis in the gut and lung microbiota due to enrichment of opportunistic pathogens and depletion of beneficial commensals, which recommends the development of interventions such as probiotics to reduce the severity of COVID‐19 through modification of the microbiota composition. 1 , 8 , 9
Humans evolved by losing the capacity to synthesize the glycan Galα1‐3Galβ1‐(3)4GlcNAc‐R (α‐Gal), which resulted in the development of a protective response of anti‐α‐Gal IgM/IgG antibodies against pathogenic viruses (e.g., HIV), bacteria (e.g., Mycobacterium) and parasites (e.g., Plasmodium) containing this modification on membrane proteins. 10 , 11 , 12 , 13 , 14 The natural IgM/IgG antibodies against α‐Gal are produced in response to bacteria with this modification in the microbiota. 10 In addition to anti‐α‐Gal antibody‐mediated pathogen opsonization, this glycan induces various immune mechanisms such as B‐cell maturation, macrophage response, activation of the complement system, upregulation of pro‐inflammatory cytokines through the Toll‐like receptor 2 (TLR2)/NF‐kB innate immune pathway, and TLR‐mediated induction of the anti‐inflammatory nuclear factor‐erythroid 2‐related factor 2 signalling pathway. 14 , 15 , 16 In conjunction, the immune response to α‐Gal in animal models has shown protection against infectious diseases without inflammatory responses. 10 , 12 , 13 , 14 , 17
Based on these results, we have hypothesized that the immune response to α‐Gal may play a role in the person‐to‐person variability in COVID‐19 disease symptoms with a putative protective capacity. 18 First, if the virus contains α‐Gal, it would be possible to limit the zoonotic transmission of SARS‐CoV‐2 by antibody‐mediated opsonization. 18 Secondly, boosting α‐Gal‐mediated protective immune and anti‐inflammatory responses may contribute to the control of COVID‐19 while increasing protection to pathogens with α‐Gal on their surface that negatively affect the individual response to SARS‐CoV‐2. 14 , 18
To address this hypothesis, herein we characterized the antibody response to α‐Gal in patients at different stages of COVID‐19 and in comparison with healthy control individuals. The results showed that while the inflammatory response and the anti‐SARS‐CoV‐2 (Spike) IgG antibody titers increased, reduction in anti‐α‐Gal antibody titers and alteration of anti‐α‐Gal antibody isotype composition correlated with COVID‐19 severity. These results suggested that the inhibition of the α‐Gal‐induced immune response translates into more aggressive viremia and severe disease symptoms.
2. MATERIALS AND METHODS
2.1. COVID‐19 patients and healthy control individuals
A retrospective case‐control study was conducted in patients suffering from COVID‐19 admitted to the University General Hospital of Ciudad Real (HGUCR), Spain from March 1 to April 15, 2020. The infection by SARS‐CoV‐2 was confirmed in all patients included in the study by the real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) assay from Abbott Laboratories (Abbott RealTime SARS‐COV‐2 assay, Abbott Park, IL) from upper respiratory tract samples after hospital admission. Clinical features, as well as laboratory determinations, were obtained from patients' medical records. The patients were grouped into (a) hospital discharge (n = 27), (b) hospitalized (n = 29) and (c) ICU (n = 25) (Table 1). Patients were hospitalized for developing a moderate‐severe clinical condition with radiologically demonstrated pneumonia and failure in blood oxygen saturation. Patients with acute respiratory failure who needed mechanical ventilation support were admitted to a hospital ICU. Patients were discharged from the hospital due to the clinical and radiological improvement of pneumonia caused by the SARS‐CoV‐2, along with the normalization of analytical parameters indicative of inflammation, such as C‐reactive protein (CRP), d‐dimer, and blood cell count (Table 1). Samples from asymptomatic COVID‐19 cases with positive anti‐SARS‐CoV‐2 IgG antibody titers but negative by RT‐PCR (n = 10) were collected on May 22‐29, 2020 and included in the analysis. Samples from healthy control individuals (individuals without a record of tick bites and allergic reactions; n = 37, sex ratio F/M = 1.1, 41.4 ± 11.1 years old) were collected before the COVID‐19 pandemic in April 2019. The use of samples and individual′s data was approved by the Ethical and Scientific Committee (University Hospital of Ciudad Real, C‐352, and SESCAM C‐73).
Table 1.
Clinical parameters and laboratory tests of COVID‐19 symptomatic cohort
| Parameters | Hospital discharge | Hospitalized | ICU | f ratio | p |
|---|---|---|---|---|---|
| Age (year) | 61.0 ± 18.0 | 73.7 ± 12.6 | 57.2 ± 14.6 | 9.196 | <.001 |
| Female/male sex ratio | 0.9 | 0.8 | 0.7 | 0.039 | .962 |
| Neutrophils (103 cells/µl) | 7.0 ± 4.0 | 7.7 ± 4.4 | 14.2 ± 9.6 | 12.116 | <.001 |
| Neutrophils (%) | 68.9 ± 14.1 | 76.8 ± 10.8 | 85.1 ± 10.9 | 13.771 | <.001 |
| Lymphocytes (103 cells/µl) | 1.5 ± 0.5 | 1.2 ± 0.6 | 1.1 ± 0.7 | 4.223 | .018 |
| Lymphocytes (%) | 19.0 ± 10.1 | 13.9 ± 8.4 | 8.4 ± 7.4 | 11.521 | <.001 |
| Neutrophil–lymphocyte count ratio (NLR) | 5.4 ± 4.2 | 10.1 ± 10.0 | 18.8 ± 14.8 | 14.231 | <.001 |
| D‐dimer (ng/ml) | 712 ± 623 | 1514 ± 1528 | 6528 ± 9436 | 7.066 | .002 |
| C‐reactive protein (CRP) (mg/dl) | 1.0 ± 1.4 | 4.4 ± 5.7 | 10.4 ± 9.7 | 17.558 | <.001 |
Note: The patients were grouped into hospital discharge (n = 27), hospitalized (n = 29) and ICU (n = 25). The results (average ± SD) were compared between different groups by one‐way ANOVA test.
Abbreviations: ANOVA, analysis of variance; COVID‐19, coronavirus disease 2019; ICU, intensive care unit.
2.2. Serum and saliva samples
Serum samples were collected for confirmed COVID‐19 patients and healthy control individuals. Nursing personnel at the HGUCR extracted blood samples. Blood samples were drawn in a vacutainer tube without anticoagulant. The tube remained at rest for 15–30 min at room temperature (RT) for clotting. Subsequently, the tube was centrifuged at 1500g for 10 min at RT to remove the clot and obtain the serum sample. Serum samples were heat‐inactivated for 30 min at 56°C and conserved at ‐20°C until used for analysis. 19 Saliva samples from asymptomatic COVID‐19 cases were collected and stored at ‐20°C until used for analysis.
2.3. Determination of antibody titers against SARS‐CoV‐2
Antibody titers specific for the recognition of virus infection based on IgG against SARS‐CoV‐2 Spike (EI 2606‐9601G) and Nucleocapsid (EI 2606‐9601‐2G) proteins and IgA (EI 2606‐9601A) were determined by ELISA (Euroimmun) following the manufacturer′s indications. 19 , 20 Briefly, 100 µl of the calibrator, positive and negative controls and serum samples at 1:100 dilution was added to the 96‐microwell plate coated with SARS‐CoV‐2 proteins and incubated for 1 h at 37°C. After washing three times with 300 µl/well of wash buffer, 100 µl/well of enzyme conjugate (peroxidase‐labelled anti‐human IgG or IgA) were added and incubated for 30 min at RT. Then, after 3 washes with 300 µl/well of wash buffer, 100 µl/well of chromogen substrate solution were added and incubated for 15 min (EI 2606‐9601‐2 G) or 30 min (EI 2606‐9601 G; EI 2606‐9601 A) at RT. Finally, the colorimetric reaction was stopped with 100 µl/well of stop solution and the absorbance was measured in a spectrophotometer (Thermo Fisher Scientific) at O.D. of 450 nm. Results were evaluated semiquantitatively by calculating the ratio between O.D. of the sample and the O.D of the calibrator, those under 0.8 considered as negative and those over 1.1 as positive.
2.4. Determination of antibody titers against α‐Gal
High absorption capacity polystyrene microtiter plates were coated with 50 ng of BSA coated with α‐Gal (BSA‐α‐Gal, thereafter named α‐Gal; Dextra) per well in carbonate‐bicarbonate buffer (Sigma‐Aldrich) and used for ELISA. After an overnight incubation at 4°C, coated plates were washed once with 100 µl/well PBS with 0.05% Tween 20 (PBST; Sigma‐Aldrich), blocked with 100 µl/well of 1% human serum albumin (HAS) in PBST (Sigma‐Aldrich) for 1 h at RT and then washed four times with 100 µl/well of PBST. Human serum and saliva samples were diluted 1:100 and 1:2, respectively, in PBST with 1% HAS and 100 µl/well were added into the wells of the antigen‐coated plates and incubated for 1 h at 37°C. Plates were washed four times with PBST and 100 µl/well of goat anti‐human immunoglobulins‐peroxidase IgG (FC specific; Sigma‐Aldrich), IgM (µ‐chain specific; Sigma‐Aldrich), IgE (ɛ‐chain specific; Sigma‐Aldrich), and IgA (heavy chain specific; Bio‐Rad) secondary antibodies diluted 1:1000, v/v in blocking solution were added and incubated for 1 h at RT. Plates were washed four times with 100 µl/well of PBST and 100 µl/well of 3,3,′5,5‐tetramethylbenzidine TMB (Promega) were added and incubated for 20 min at RT. Finally, the reaction was stopped with 50 µl/well of 2 N H2SO4 and the O.D. was measured in a spectrophotometer at 450 nm. The average of two technical replicates per sample was used for analysis after background (coated wells incubated with PBS and secondary antibodies) subtraction. Reference values for serum immunoglobulin levels 21 were considered in the analysis of the profile of anti‐α‐Gal antibody isotypes. Although this methodology has been previously validated, 22 , 23 the anti‐α‐Gal immunoglobulin E (IgE) antibody titers were determined in sera from selected individuals (n = 8) with α‐Gal syndrome, anaphylaxis, and skin local reactions to tick bites and healthy individuals without record of tick bites and allergic reactions using the ImmunoCAP Phadia 250 automated platform (Thermo Fisher Scientific) with the commercial ImmunoCap α‐Gal bovine Thyroglobulin kit according to the manufacturer′s instructions. 22 These values were used to draw a trendline (R 2 = 0.83) to calculate the corresponding kU/l values for the ELISA O.D. at 450 nm values using the formula IgE titers (kU/l) = 100 × EXP ([O.D.450 nm – 2.9]/0.2). Positive anti‐α‐Gal IgE levels were considered at a cut‐off value of 0.35 kU/l. 23
2.5. Determination of IL‐1 and IL‐4 serum levels
Serum levels of IL‐1 and IL‐4 were determined by ELISA (Invitrogen) following the manufacturer′s instructions. Briefly, 96‐microwell plates coated in duplicate with anti‐human IL‐1β or IL‐4 were washed twice with 400 µl/well of wash buffer and 100 µl of human IL‐1β or IL‐4 standard (20.00 pg/ml) at serial dilutions (1:2, 1:4, 1:8, 1:16, 1:32), 100 µl/well of sera at 1:2 dilution, and 100 µl/well of sample diluent as negative control. Then, 50 µl/well of biotin‐conjugate were added to all wells. After incubation for 2 h at RT and three washes with 400 µl/well of wash buffer, 100 µl/well of streptavidin‐HRP were added to all wells. After incubation for 1 h at RT and 3 washes with 400 µl/well of wash buffer, 100 µl/well of 3,3′,5,5′‐tetramethylbenzidine or TMB substrate solution were added to all wells. As soon as the Standard 1 well reached an O.D. of 0.9 at 620 nm, the colorimetric reaction was stopped with 100 µl/well of stop solution and the absorbance was measured in a spectrophotometer (Thermo Fisher Scientific) at O.D. of 450 nm. Human IL‐1β or IL‐4 concentration (pg/ml) in each sample was calculated from the obtained standard curve.
2.6. Statistical analysis
The ELISA O.D. at 450 nm values were compared between different groups by one‐way analysis of variance (ANOVA) test (p = .05; https://www.socscistatistics.com/tests/ANOVA/default2.aspx). Clinical parameters and laboratory tests of COVID‐19 symptomatic cohort (Table 1) were compared between different groups by one‐way ANOVA test (p = .05). Pairwise comparisons between groups were conducted by Student′s t test (p = .05). A Spearman Rho (r s) correlation analysis (p = .05; https://www.socscistatistics.com/tests/spearman/default2.aspx) was conducted between anti‐SARS‐CoV‐2 Spike IgG titers and COVID‐19 disease severity (2 = asymptomatic, 3 = hospital discharge, 4 = hospitalized, 5 = ICU), anti‐α‐Gal IgA, IgE, IgM and IgG antibody titers and disease severity (1 = healthy, 2 = asymptomatic, 3 = hospital discharge, 4 = hospitalized, 5 = ICU), and for anti‐α‐Gal IgA and IgG antibody titers between serum and saliva samples.
3. RESULTS
3.1. Inflammatory biomarkers are associated with severity in COVID‐19 patients
In the blood cell analysis, the ICU patients showed a higher lymphocytopenia, percentage and neutrophil counts when compared to hospital discharge and hospitalized individuals (p < .001; Figure 1A and Table 1). The cellular and biochemical indicators of systemic inflammation, neutrophil‐lymphocyte count ratio (NLR), C‐reactive protein (CRP), and D‐dimer levels were higher in ICU patients when compared to other patients (p < .002; Figure 1A and Table 1).
Figure 1.
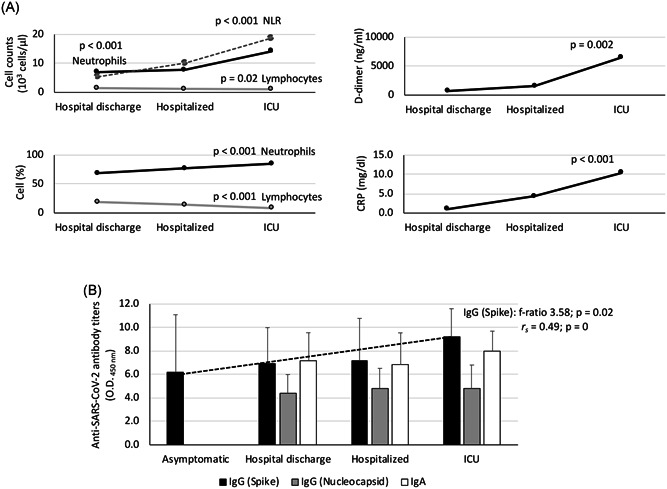
Laboratory tests in COVID‐19 patients. A, Cellular and biochemical indicators of systemic inflammation included neutrophils (cell counts and percent), lymphocytes (cell counts and percent), NLR, D‐dimer and CRP levels (Table 1). B, Serum anti‐SARS‐CoV‐2 IgA, IgG (spike) and IgG (nucleocapsid) antibody levels were determined by ELISA. The patients were grouped as asymptomatic (n = 10), hospital discharge (n = 27), hospitalized (n = 29) and ICU (n = 25). The results were compared between different groups by one‐way ANOVA test (p < .05). A Spearman rho (r s) correlation analysis (p < .05) was conducted between anti‐Spike IgG antibody titers and disease severity (2 = asymptomatic, 3 = hospital discharge, 4 = hospitalized, 5 = ICU). ANOVA, analysis of variance; COVID‐19, coronavirus disease 2019; CRP, C‐reactive protein; ELISA, enzyme‐linked immunosorbent assay; ICU, intensive care unit; IgA, immunoglobulin A; NLR, neutrophil‐lymphocyte count ratio; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
All patients had pneumonia associated with COVID‐19. Hospitalized patients in the plant were treated with oxygen therapy with a fraction of inspired oxygen (FiO2) above 50% and with the objective of reaching an O2 saturation above 95%. When, despite the oxygen therapy, patients presented an O2 saturation below 90%, a sustainable rate of breaths per minute (bpm) > 30 and an increase in respiratory work and assisted mechanical ventilation was required, and consequently they were admitted to ICU. Although more severe symptoms have been associated with elderly patients, herein older patients were recorded in the hospitalized and not the ICU group (p < .001; Table 1). The healthy and asymptomatic individuals did not show symptoms of inflammation. These results corroborated a higher inflammation rate in the most critical COVID‐19 patients independently of the age factor.
3.2. Immune response to SARS‐CoV‐2 increased with severity in COVID‐19 patients
All COVID‐19 symptomatic patients showed both IgA and IgG antibody titers against SARS‐CoV‐2 (Figure 1B). In asymptomatic cases, only IgG antibody titers were determined, and all tested positive (Figure 1B). However, only the IgG titers against the SARS‐CoV‐2 Spike protein significantly increased in accordance with disease symptoms (p = .02; Figure 1B) with a positive correlation (r s > 0; p = 0; Figure 1B). These results showed that COVID‐19 patients were immunocompetent despite the inflammatory response.
3.3. Immune response to α‐Gal varied in COVID‐19 patients
The serum IgA, IgE, IgM and IgG antibody response to α‐Gal was characterized in healthy individuals and COVID‐19 patients at different disease stages (Figures 2 and 3a). The calculated anti‐α‐Gal IgE levels were below (8.3E − 5 to 3.4E − 02 kU/l) the cut‐off value of 0.35 kU/l used for the diagnosis of the α‐Gal syndrome. A negative correlation was observed for IgE, IgM, and IgG between anti‐α‐Gal antibody titers and disease severity (r s < 0; p = 0; Figure 3A). The anti‐α‐Gal IgA antibody titers did not vary between the different groups (p = .21136; Figure 3A) nor correlate with disease severity (r s = 0.02; p = .91; Figure 3A). For anti‐α‐Gal IgM and IgG antibodies, the titers decreased from healthy to ICU individuals (p < .00001; Figures 2 and 3a). However, in asymptomatic cases, the anti‐α‐Gal IgE titers were higher than in healthy individuals and symptomatic COVID‐19 patients (p < .000001; Figure 3A). In COVID‐19 patients, the IgE but not IgM and IgG antibody titers were higher in hospitalized patients than in hospital discharge and ICU cases (p < .05; Figure 2).
Figure 2.
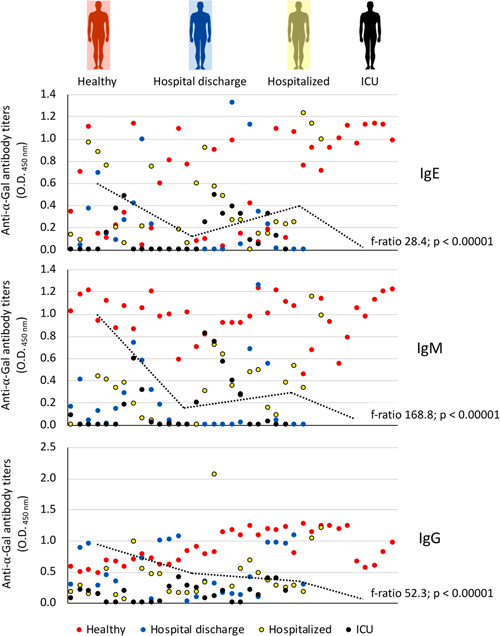
Anti‐α‐Gal antibody response in COVID‐19 symptomatic patients and healthy controls. The IgE, IgM, and IgG anti‐α‐Gal antibody titers were determined by ELISA. Individuals were grouped as healthy controls (n = 37), hospital discharge (n = 27), hospitalized (n = 29) and ICU (n = 25). Values for individuals from all groups are shown in different colors and distributed across the entire X‐axis. The results were compared between different groups by one‐way ANOVA test (p < .05). ANOVA, analysis of variance; ELISA, enzyme‐linked immunosorbent assay; ICU, intensive care unit; IgE, immunoglobulin E; α‐Gal, Galα1‐3Galβ1‐(3)4GlcNAc‐R
Figure 3.
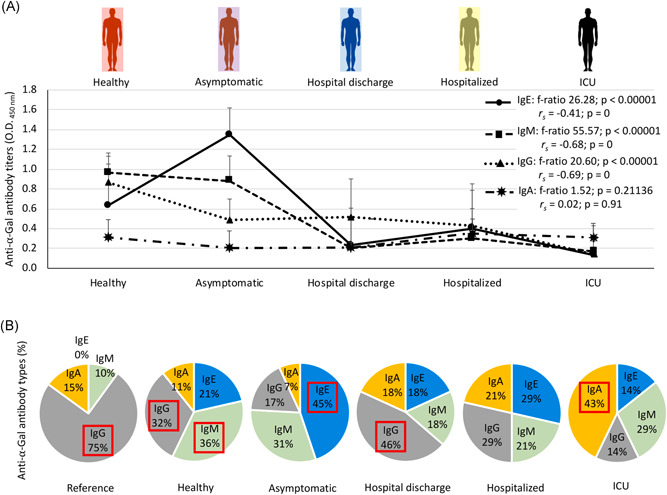
Serum anti‐α‐Gal antibody response in COVID‐19 asymptomatic and symptomatic cases and healthy controls. A, The IgA, IgE, IgM and IgG anti‐α‐Gal antibody titers were determined by ELISA. Individuals were grouped as healthy controls (n = 37), asymptomatic (n = 10), hospital discharge (n = 27), hospitalized (n = 29) and ICU (n = 25). The results were compared between different groups by one‐way ANOVA test (p < .05). A Spearman rho (r s) correlation analysis (p < .05) was conducted between anti‐α‐Gal IgA, IgE, IgM and IgG antibody titers and disease severity (1 = healthy, 2 = asymptomatic, 3 = hospital discharge, 4 = hospitalized, 5 = ICU). B, Profile of anti‐α‐Gal antibody isotype (shown as percentage of antibody titers) for each group. Reference values for serum immunoglobulin levels were included. Antibody isotypes with highest representation on each group are highlighted in red. ANOVA, analysis of variance; COVID‐19, coronavirus disease 2019; ELISA, enzyme‐linked immunosorbent assay; ICU, intensive care unit; IgA, immunoglobulin A; α‐Gal, Galα1‐3Galβ1‐(3)4GlcNAc‐R
The profile of anti‐α‐Gal antibody isotypes was qualitatively compared between groups including reference values for serum immunoglobulin levels (Figure 3B). The results evidenced that anti‐α‐Gal IgE and IgM antibodies are more abundant than reference values even in healthy individuals. However, the most abundant anti‐α‐Gal antibodies varied from IgM/IgG in healthy individuals to IgE (asymptomatic), IgG (hospital discharge), none (hospitalized) and IgA (ICU) in COVID‐19 cases (Figure 3B). These results suggested a role for anti‐α‐Gal IgA, which increased in relative representation in ICU patients by four‐fold (vs. healthy individuals), six‐fold (vs. asymptomatic cases) and two‐fold (vs. hospital discharge and hospitalized cases).
Despite differences in absolute values due to dilutions of the samples used for ELISA (1:100 for serum vs. 1:2 for saliva), as expected, anti‐α‐Gal IgA but not IgG antibody titers were higher in saliva than in serum samples (p = .0002; Figure 4A) but without a significant correlation (p > .05). The saliva anti‐α‐Gal IgA antibody titers were similar between asymptomatic COVID‐19 cases and healthy individuals (p = .3049; Figure 4B).
Figure 4.
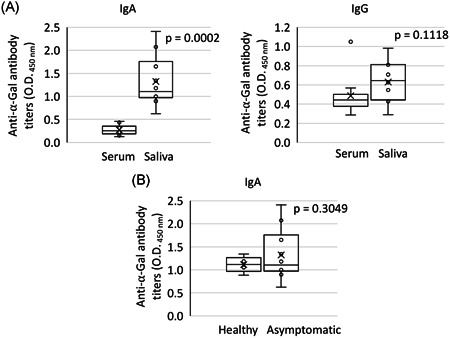
Salivary anti‐α‐Gal antibody response in COVID‐19 asymptomatic cases and healthy controls. A, The anti‐α‐Gal IgA and IgG antibody titers were determined by ELISA and compared in asymptomatic cases between serum and saliva samples by Student′s t‐test (p < .05; n = 10). B, The anti‐α‐Gal IgA antibody titers in saliva were determined by ELISA and compared between asymptomatic COVID‐19 cases and healthy individuals by Student′s t‐test (p < .05; n = 10). COVID‐19, coronavirus disease 2019; ELISA, enzyme‐linked immunosorbent assay; ICU, intensive care unit; IgA, immunoglobulin A; α‐Gal, Galα1‐3Galβ1‐(3)4GlcNAc‐R
The analysis of the cytokine response was focused on anti‐inflammatory IL‐4 and pro‐inflammatory IL‐1 serum levels. For both IL‐4 and IL‐1, serum levels did not vary significantly with disease symptoms and were similar in COVID‐19 patients than in healthy individuals (p > .4). In hospitalized patients only, three and two different cases showed cytokine levels above 20 pg/ml (IL‐4) and 0.2 pg/ml (IL‐1), respectively. However, these results may be affected by different interventions and treatments to which patients were subjected that alter cytokine serum levels.
4. DISCUSSION
Systemic inflammation is associated with changes in the quantity and composition of circulating blood cells and has been identified as the primary basic mechanism resulting in disability and increased mortality in COVID‐19. 24 As previously reported, 25 , 26 in the blood cell analysis of cellular and biochemical indicators of systemic inflammation, ICU patients showed a higher lymphocytopenia independent of the age factor associated with more severe COVID‐19 symptoms. 27
The results of our study showed a negative correlation between anti‐α‐Gal antibody titers and COVID‐19 disease severity. However, these results raised the question of whether the observed reduction in the anti‐α‐Gal antibody response at the population level is a consequence or a cause of COVID‐19 symptomatology. In our study, COVID‐19 patients were immunocompetent with a positive correlation between anti‐SARS‐CoV‐2 Spike antibody titers and disease severity. 28 Therefore, our results suggested that the decrease in the anti‐α‐Gal antibody response occurred by mechanisms different from humoral immunosuppression and as a consequence of SARS‐CoV‐2 infection.
In addition to the observed negative correlation between anti‐α‐Gal IgE, IgM, and IgG antibody titers and COVID‐19 disease severity, our results showed differences in the profile of anti‐α‐Gal antibody isotypes in COVID‐19 cases that may be associated with different disease stages (Figure 5). These results suggested that higher anti‐α‐Gal IgE levels in asymptomatic cases may reflect an allergic response mediated by this glycan, which reflects the trade‐off associated with the immune response to α‐Gal that benefits humans by providing immunity to pathogen infection while increasing the risk of developing allergic reactions to this molecule. 12 , 13 , 17 In healthy individuals as in hospital discharge cases, the higher representation of anti‐α‐Gal IgM and/or IgG antibodies may be associated with a protective response to COVID‐19. However, in hospitalized patients, the representation of anti‐α‐Gal antibody isotypes did not vary, which could reflect the absence of protection. Finally, the higher representation of anti‐α‐Gal IgA antibodies in ICU patients may be associated with the inflammatory response observed in these cases. In accordance with these results, it was recently shown in endogenous α‐Gal‐negative turkeys that treatment with probiotic bacteria with high α‐Gal content results in protection against aspergillosis through reduction by still unknown mechanisms in the pro‐inflammatory anti‐α‐Gal IgA response in the lungs. 29
Figure 5.
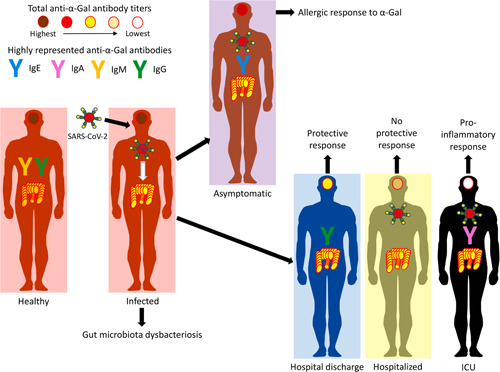
A negative correlation between anti‐α‐Gal antibody titers and COVID‐19 disease severity and differences in the profile of anti‐α‐Gal antibody isotypes may be associated with different disease stages. Our hypothesis is that the dysbacteriosis observed in COVID‐19 patients translates into a reduction in total anti‐α‐Gal antibody titers and alteration of anti‐α‐Gal antibody isotype composition due to the reduction in the microbiota of α‐Gal‐containing commensal bacteria. COVID‐19, coronavirus disease 2019; IgA, immunoglobulin A; α‐Gal, Galα1‐3Galβ1‐(3)4GlcNAc‐R; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
In Spain, differences have been observed in the number of reported cases per 100 000 people by age and sex, with more females at age 20–59 and males at age 60–89 with a higher mortality in males. 30 In this study, differences in age, but not sex, were observed in symptomatic COVID‐19 cases (Table 1). However, the youngest cases corresponded to ICU patients and healthy control individuals, thus reducing the possible effect of age and sex on the observed disease symptoms. Furthermore, currently a clear correlation has not been found between age, sex, and the antibody response to α‐Gal. 31 , 32 , 33
The protective response of anti‐α‐Gal IgM/IgG antibodies against pathogenic organisms containing this modification on membrane proteins has been well documented. 10 , 11 , 12 , 13 , 14 , 17 , 23 In contrast, IgE antibody response against α‐Gal has been associated with the allergy to mammalian meat or α‐Gal syndrome and other diseases such as atopy, coronary artery disease and atherosclerosis. 33 , 35 , 36 , 37 In this study and based on anti‐α‐Gal IgE levels, all individuals were negative for α‐Gal syndrome. 38
The anti‐α‐Gal IgM/IgG antibodies can protect from infection by opsonizing pathogens with α‐Gal on their surface. 10 , 12 , 13 In preliminary analyses, it was suggested that blood type O individuals are less susceptible to COVID‐19 than other blood type groups, 39 , 40 a finding that was recently confirmed by genetic analyses. 41 ABO blood groups contain highly fucosylated antigens, 42 , 43 a property shared with the glycans present in SARS‐CoV‐2. 44 The glycans on envelope glycoproteins are linked to host‐cell glycosylation machinery and thus the viral surface envelope is dominantly covered by host‐derived glycans. 17 , 44 For example, glycosylation in spike asparagine (N343) is highly fucosylated with 98% of detected glycans bearing fucose residues. 44 Accordingly, the monoclonal antibody S309 that neutralizes SARS‐CoV‐2 binds core fucose moieties in N343 and N‐acetylglucosamine (GlcNAc), a structural glycan found in both SARS‐CoV‐2 44 and ABO blood groups. 42 , 43 Human cells do not express α‐Gal, but SARS‐CoV‐2 can carry structurally similar blood group B antigen when it replicates in cells expressing the blood group enzyme. 45 , 46 Therefore, the blood type B antigen can be then targeted by pre‐existing and cross‐reactive anti‐α‐Gal and anti‐B antibodies produced by blood type B‐negative individuals, 45 , 46 thus preventing infection by blocking the virus cell attachment and entry. Other immune‐mediated mechanisms may be also activated in response to α‐Gal, 14 , 15 , 16 which can be activated by SARS‐CoV‐2 expressing blood type B antigen on their envelope. Although not addressed in this study, these findings prompted us to consider that blood type O individuals could produce antibodies against A and B antigens that in addition to IgM/IgG antibodies against α‐Gal, which cross‐react with the structurally similar blood B antigen, 46 could be involved in a polyvalent recognition of the SARS‐CoV‐2 Spike that may be implicated in the human protection to COVID‐19.
In our study, we did not collect information on ABO blood type in COVID‐19 patients. However, in a related study with a similar group of patients (n = 73; 9, 40 and 24 hospital discharge, hospitalized and ICU patients, respectively), the results did not show significant differences in ABO blood group distribution. Nevertheless, the results suggest that the ABO blood factor should be considered when evaluating the antibody response to α‐Gal. Blood type O individuals, who produce anti‐A and anti‐B antibodies, can be protected against SARS‐CoV‐2 particles carrying blood antigens A or B. However, blood type A and B individuals, who produce either anti‐A or anti‐B antibodies, would be protected only against SARS‐CoV‐2 particles carrying antigen A or B, respectively. Therefore, both blood type A and B individuals will be highly susceptible to SARS‐CoV‐2 particles coming from blood type O individuals because these viral particles do not carry either blood antigen A or B on the envelope. 45 Assuming equal replicative fitness for viruses replicating in cells expressing any of the ABO blood group enzymes, an epidemiological dynamics would be created in which, after a large proportion of the population being exposed and infected by SARS‐CoV‐2, the frequency of individuals with blood types B and A would be equally represented among COVID‐19 patients. In contrast, blood type O individuals would be underrepresented, relative to the frequency of these blood groups in the general population. These predictions were corroborated in a large study (n = 750 000) even after adjusting for age, sex, body mass index, race, ethnicity, and co‐morbidities. 47 Therefore, the blood type's effects are not explained by other risk factors including age, sex, race, ethnicity, hypertension, diabetes mellitus, obesity, and cardiovascular and respiratory diseases, 40 which support the immunological considerations discussed above.
Based on the fact that natural antibodies against α‐Gal are produced in response to bacteria with this modification in the microbiota, 10 our hypothesis is that the dysbacteriosis observed in COVID‐19 patients 34 translates into a reduction in total anti‐α‐Gal antibody titers and alteration of anti‐α‐Gal antibody isotype composition due to the reduction in the microbiota of α‐Gal‐containing commensal bacteria and other still uncharacterized mechanisms (Figure 5). Alternatively, and hypothetically, individuals with higher α‐Gal content in the microbiota may be less susceptible to COVID‐19. Additionally, the pulmonary microbiota can be affected with the presence of gut bacteria in the lungs. 9
In conclusion, according to these results and previous findings in retrovirus, 48 , 49 the inhibition of the α‐Gal‐induced immune response may translate into more aggressive viremia and severe disease inflammatory symptoms. 50 These results further encourage addressing the proposal of developing interventions such as probiotics based on commensal bacteria with α‐Gal epitopes to modify the microbiota and increase the α‐Gal‐induced protective immune response and reduce the severity of COVID‐19. 8 , 18 Furthermore, as recently proposed, the production of coronavirus or virus‐like particles (VLPs) in non‐catarrhine mammalian cells may be used to produce vaccines with α‐Gal‐containing antigens to induce anti‐α‐Gal protective response and increase vaccine efficacy for the control of COVID‐19. 51 , 52 , 53
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
José Miguel Urra, Christian Gortázar, and José de la Fuente designed the study; Elisa Ferreras‐Colino, Marinela Contreras, Carmen M. Cabrera, Isabel G. Fernández de Mera, José Miguel Urra, Christian Gortázar, and José de la Fuente performed the experiments; José de la Fuente, José Miguel Urra, Elisa Ferreras‐Colino, Marinela Contreras, Carmen M. Cabrera, Isabel G. Fernández de Mera, and Alejandro Cabezas‐Cruz analyzed data; José Miguel Urra, Christian Gortázar, A.M., Margarita Villar, and José de la Fuente supervised the project. All authors wrote the manuscript.
ACKNOWLEDGMENTS
This study was partially supported by the Consejería de Educación, Cultura y Deportes, JCCM, Spain, project CCM17‐PIC‐036 (SBPLY/17/180501/000185). We thank Antonio Mas (University of Castilla La Mancha, UCLM, Spain) for the critical reading of the manuscript. We acknowledge UCLM, Spain support to Grupo SaBio. MC was funded by the Ministerio de Ciencia, Innovación y Universidades, Spain (grant FJC‐2018‐038277‐I). IGFM was supported by the UCLM. MV was supported by the UCLM and the Fondo Europeo de Desarrollo Regional, FEDER, EU.
Urra JM, Ferreras‐Colino E, Contreras M, et al. The antibody response to the glycan α‐Gal correlates with COVID‐19 disease symptoms. J Med Virol. 2021;93:2065‐2075. 10.1002/jmv.26575
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID‐19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS‐CoV‐2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aggarwal S, Gollapudi S, Gupta S. Increased TNF‐alpha‐induced apoptosis in lymphocytes from aged humans: changes in TNF‐alpha receptor expression and activation of caspases. J Immunol. 1999;162(4):2154‐2161. [PubMed] [Google Scholar]
- 5. Horowitz RI, Freeman PR, Bruzzese J. Efficacy of glutathione therapy in relieving dyspnea associated with COVID‐19 pneumonia: a report of 2 cases. Respir Med Case Rep. 2020;30:101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China [published online ahead of print, 2020 Feb 7]. JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borges do Nascimento IJ, Cacic N, Abdulazeem HM, et al. Novel coronavirus infection (COVID‐19) in humans: a scoping review and meta‐analysis. J Clin Med. 2020;9(4):941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Pierro F. A possible probiotic (S. salivarius K12) approach to improve oral and lung microbiotas and raise defenses against SARS‐CoV‐2. Minerva Med. 2020;111(3):281‐283. [DOI] [PubMed] [Google Scholar]
- 9. Fanos V, Pintus MC, Pintus R, Marcialis MA. Lung microbiota in the acute respiratory disease: from coronavirus to metabolomics. J Pediatr Neonat Individual Med 2020;9:e090139. [Google Scholar]
- 10. Yilmaz B, Portugal S, Tran TM, et al. Gut microbiota elicits a protective immune response against malaria transmission. Cell. 2014;159(6):1277‐1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galili U. Evolution in primates by "Catastrophic‐selection" interplay between enveloped virus epidemics, mutated genes of enzymes synthesizing carbohydrate antigens, and natural anti‐carbohydrate antibodies. Am J Phys Anthropol. 2019;168(2):352‐363. [DOI] [PubMed] [Google Scholar]
- 12. Cabezas‐Cruz A, Hodžić A, Román‐Carrasco P, et al. Environmental and molecular drivers of the α‐Gal syndrome. Front Immunol. 2019;10:1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de la Fuente J, Pacheco I, Villar M, Cabezas‐Cruz A. The alpha‐Gal syndrome: new insights into the tick‐host conflict and cooperation. Parasit Vectors. 2019;12(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pacheco I, Contreras M, Villar M, et al. Vaccination with alpha‐Gal protects against mycobacterial infection in the zebrafish model of tuberculosis. Vaccines. 2020;8(2):195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cabezas‐Cruz A, Mateos‐Hernández L, Pérez‐Cruz M, et al. Regulation of the immune response to α‐Gal and vector‐borne diseases. Trends Parasitol. 2015;31(10):470‐476. [DOI] [PubMed] [Google Scholar]
- 16. Yin S, Cao W. Toll‐like receptor signaling induces Nrf2 pathway activation through p62‐triggered Keap1 degradation. Mol Cell Biol. 2015;35(15):2673‐2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galili U Human natural antibodies to mammalian carbohydrate antigens as unsung heroes protecting against past, present, and future viral infections. Antibodies. 2020;9(2):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de la Fuente J. The exquisite corpse for the advance of science. Art Sci. 2020;4:43. [Google Scholar]
- 19. Burbelo PD, Riedo FX, Morishima C, et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):206‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang MS, Lu YT, Ho ST, et al. Antibody detection of SARS‐CoV spike and nucleocapsid protein. Biochem Biophys Res Commun. 2004;314(4):931‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schroeder HW, Jr , Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125(2 suppl 2):S41‐S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mateos‐Hernández L, Villar M, Moral A, et al. Tick‐host conflict: immunoglobulin E antibodies to tick proteins in patients with anaphylaxis to tick bite. Oncotarget. 2017;8(13):20630‐20644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cabezas‐Cruz A, Mateos‐Hernández L, Alberdi P, et al. Effect of blood type on anti‐α‐Gal immunity and the incidence of infectious diseases. Exp Mol Med. 2017;49(3):e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Z, Fu Z, Huang W, Huang K. Prognostic value of neutrophil‐to‐lymphocyte ratio in sepsis: a meta‐analysis. Am J Emerg Med. 2020;38(3):641‐647. [DOI] [PubMed] [Google Scholar]
- 26. Urra JM, Cabrera CM, Porras L, Ródenas I. Selective CD8 cell reduction by SARS‐CoV‐2 is associated with a worse prognosis and systemic inflammation in COVID‐19 patients. Clin Immunol. 2020;217:108486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du N, Chen H, Zhang Q, et al. A case series describing the epidemiology and clinical characteristics of COVID‐19 infection in Jilin Province. Virulence. 2020;11(1):482‐485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salazar E, Kuchipudi SV, Christensen PA, et al. Relationship between anti‐spike protein antibody titers and SARS‐CoV‐2 in vitro virus neutralization in convalescent plasma. Preprint. bioRxiv. 2020;2020.06.08.138990.
- 29. Mateos‐Hernández L, Risco‐Castillo V, Torres‐Maravilla E, et al. Gut microbiota abrogates Anti‐α‐Gal IgA response in lungs and protects against experimental Aspergillus infection in poultry. Vaccines. 8, 2020:285. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Global Health 5050 . COVID‐19: Data disaggregated by age and sex. Available at: https://globalhealth5050.org/covid19/age-and-sex-data/#1589893682295-1abaac66-2013. Accessed August 26, 2020.
- 31. Fischer J, Lupberger E, Hebsaker J, et al. Prevalence of type I sensitization to alpha‐gal in forest service employees and hunters. Allergy. 2017;72(10):1540‐1547. [DOI] [PubMed] [Google Scholar]
- 32. Mateo Borrega M, Garcia B, Larramendi C, et al. IgE‐mediated sensitization to galactose‐α‐1,3‐ galactose (α‐Gal) in urticaria and anaphylaxis in Spain: geographical variations and risk factors. J Investig Allergol Clin Immunol. 2019;29(6):436‐443. [DOI] [PubMed] [Google Scholar]
- 33. Gonzalez‐Quintela A, Dam Laursen AS, Vidal C, Skaaby T, Gude F, Linneberg A. IgE antibodies to alpha‐gal in the general adult population: relationship with tick bites, atopy, and cat ownership. Clin Exp Allergy. 2014;44(8):1061‐1068. [DOI] [PubMed] [Google Scholar]
- 34. Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID‐19 during time of hospitalization. Gastroenterology. 2020;S0016‐5085(20):34701‐34706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Nunen S, O'Connor KS, Clarke LR, Boyle RX, Fernando SL. The association between Ixodes holocyclus tick bite reactions and red meat allergy. Intern Med J. 2007;39:A132. [DOI] [PubMed] [Google Scholar]
- 36. Wilson JM, Schuyler AJ, Schroeder N, Platts‐Mills TA. Galactose‐α‐1,3‐galactose: atypical food allergen or model IgE hypersensitivity? Curr Allergy Asthma Rep. 2017;17(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson JM, McNamara CA, Platts‐Mills TAE. IgE, α‐Gal and atherosclerosis. Aging. 2019;11(7):1900‐1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de la Fuente J, Cabezas‐Cruz A, Pacheco I. Alpha‐gal syndrome: challenges to understanding sensitization and clinical reactions to alpha‐gal. Expert Rev Mol Diagn. 2020:1‐7. [DOI] [PubMed] [Google Scholar]
- 39. Wu Y, Feng Z, Li P, Yu Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID‐19 [published online ahead of print, 2020 Jun 17]. Clin Chim Acta. 2020;509:220‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zietz M, Tatonetti NP Testing the association between blood type and COVID‐19 infection, intubation, and death. Preprint. medRxiv. 2020;2020.04.08.20058073. [DOI] [PMC free article] [PubMed]
- 41. Ellinghaus D, Degenhardt F, Bujanda L, et al. Genomewide association study of severe Covid‐19 with respiratory failure. N Engl J Med. 2020. 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cooling L. Blood groups in infection and host susceptibility. Clin Microbiol Rev. 2015;28(3):801‐870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider M, Al‐Shareffi E, Haltiwanger RS. Biological functions of fucose in mammals. Glycobiology. 2017;27(7):601‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science. 2020;369(6501):330‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Breiman A, Ruvën‐Clouet N, Le, Pendu J. Harnessing the natural anti‐glycan immune response to limit the transmission of enveloped viruses such as SARS‐CoV‐2. PLOS Pathog. 2020;16(5):e1008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rispens T, Derksen NI, Commins SP, Platts‐Mills TA, Aalberse RC. IgE production to α‐gal is accompanied by elevated levels of specific IgG1 antibodies and low amounts of IgE to blood group B. PLOS One. 2013;8(2):e55566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. 23andMe . 23andMe finds evidence that blood type plays a role in COVID‐19. https://blog.23andme.com/23andme-research/blood-type-and-covid-19/. Accessed September 4, 2020.
- 48. Rother RP, Fodor WL, Springhorn JP, et al. A novel mechanism of retrovirus inactivation in human serum mediated by anti‐alpha‐galactosyl natural antibody. J Exp Med. 1995;182(5):1345‐1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rother RP, Squinto SP. The alpha‐galactosyl epitope: a sugar coating that makes viruses and cells unpalatable. Cell. 1996;86(2):185‐188. [DOI] [PubMed] [Google Scholar]
- 50. Ritchie AI, Singanayagam A. Immunosuppression for hyperinflammation in COVID‐19: a double‐edged sword? Lancet. 2020;395(10230):1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de la Fuente J, Gortázar C, Cabezas‐Cruz A, et al. Boosting anti‐alpha‐Gal immune response to control COVID‐19. Royal Society Open Science Stage1 Registered Report. 2020:020. 10.17605/OSF.IO/XHDPU. date of in‐principle acceptance: 15/4/2. [DOI] [Google Scholar]
- 52. Chen JM. SARS‐CoV‐2 replicating in nonprimate mammalian cells probably have critical advantages for COVID‐19 vaccines due to anti‐Gal antibodies: a minireview and proposals. J Med Virol. 2020. 10.1002/jmv.26312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bogani G, Raspagliesi F, Ditto A, de la Fuente J. The adoption of viral capsid‐derived virus‐like particles (VLPs) for disease prevention and treatments. Vaccines. 2020;8(3):E432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


