Abstract
When nanoparticles are introduced into biological systems, host proteins tend to associate on the particle surface to form a protein layer termed the “protein corona” (PC). Identifying the proteins that constitute the PC can yield useful information about nanoparticle processing, bio-distribution, toxicity and clearance. Similarly, characterizing and identifying proteins within the PC from patient samples provides opportunities to probe disease proteomes and identify molecules that influence the disease process. Thus, nanoparticles represent unique probing tools for discovery of molecular targets for diseases. Here, we report a first review on target identification using nanoparticles in biological samples based on analysing physico chemical interactions. We also summarize the evolution of the PC surrounding various nano-systems, comment on PC signature, address PC complexity in fluids, and outline challenges associated with analysing the PC. In addition, the influence on PC formation of various nanoparticle parameters is summarized; nanoparticle characteristics considered include size, charge, temperature, and surface modifications for both organic and inorganic nanomaterials. We also discuss the advantages of nanotechnology, over other more invasive and laborious methods, for identifying potential diagnostic and therapeutic targets.
1. Introduction
The overall survival of cancer patients, excepting a few specific malignancies, has not improved dramatically over the last few decades[1]. To address the dismal outcome of most cancers it is essential that new molecular targets are identified in order to improve both diagnosis and treatment. To facilitate this goal of improving cancer outcomes, new technologies, methodologies and approaches are sorely needed. One such potential opportunity is provided by the adsorption of host-proteins to administered nanoparticles (NPs) to form the so-called protein corona (PC)[2]. Identifying the proteins that make up the PC could provide invaluable information about disease states in the specific patients to whom NPs are administered. For example, detection of biomarkers in patient samples at early stages of disease would be useful in selecting effective treatments. At this time, only a few diagnostic tests exist that have utility in early stage disease; early diagnostic tests include mammograms for breast cancer [3], PSA for prostate cancer[4], colonoscopy for colorectal cancer[5], CA-125 for ovarian cancer[6], oligomer species (AβOs) for Alzheimer’s disease (AD)[7], B-type natriuretic peptide (BNP) for cardiovascular disease (CVD) [8], and anti-myelin, interferon β-neutralizing antibodies for multiple sclerosis (MS) [9]. In these cases, diagnosis may be limited by specificity and sensitivity due to extremely low abundance of biomarkers and/or the swamping effect by non-specific high abundance proteins, as well as the use of invasive instrumentation and laborious procedures. Thus, there is a high need for the development of new targets to facilitate non-invasive and pain-free diagnostic tests that are effective in the early stages of disease.
Over the last few decades, nanotechnology has become increasingly important in the biomedical arena as the field of nanomedicine has exploded, particularly as applied to diagnosis, imaging and targeted drug delivery. Nanoparticles, including gold NPs (AuNPs), silver NPs (AgNPs), liposomes, super paramagnetic iron oxide (SPIONs), silica (SiO2), zinc oxide (ZnO), titanium dioxide (TiO2) and quantum dots (QDs) [10-13] are all widely used for the above applications. However, outcome is often limited by shear stress from biological fluids following administration [2, 14, 15]. Within 30 seconds of exposure to the biological milieu, the NP surface adsorbs many biomolecules, predominantly proteins, resulting in a new nano-bio interface which is called the protein corona (PC) [16, 17]. Although the term PC was not coined at the time, the concept of a PC has been known and evaluated since the 1980’s; for example, studies of adsorption of serum opsonins to liposomes were important in developing the surface PEGylation strategy for liposomes [18, 19] while other groups examined serum protein adsorption to NPs [20-22].Typically, the concentration of proteins that a NP is exposed to in vivo is about 60-80 g/L [23], and the host biological response to the NP critically depends on the nature of the proteins adsorbed i.e., its ‘corona signature’ [24]. The PC is dynamically regulated by hydrogen bonding, hydrophobic interactions, and Coulomb and Van der Waals forces [25], and depending on the nature of the unmodified NP core, proteins have a wide range of affinities for the NP’s surface[17].
Many factors affect the PC signature, these include characteristics of the NP (i.e. size, charge, and surface engineering), characteristics of the protein (i.e. molecular weight, isoelectric points, structure and folding) characteristics of the interaction (i.e. time, temperature, concentration) and the type of biological sample (i.e. plasma, urine, tissue lysate etc.) [2, 26-31]. Proteins in the PC vary with individual disease states (e.g. diabetes, rheumatism, cancer, obesity, haemodialysis, hyperfibrinogenaemia, hypercholesterolemia, haemophilia and pregnancy), as well as specific disease conditions (e.g. cancer stage or grade and tumor heterogeneity) [32-38].
To date, there are only a limited number of studies that have been performed to validate the clinical application of information derived from examining the constituent proteins of the PC [39, 40]. In one such study, Arvizo et al. examined the PC surrounding positively charged gold NPs and identified hepatoma derived growth factor (HDGF) as a biomarker for ovarian cancer [40]. Similarly, analysing the PC of 20nm self-therapeutic gold NPs, Giri et al, identified the proteins PPA1 and SMNDC1 as potential therapeutic targets for ovarian cancer [10]. Additionally, Schrittwieser et al. described a cobalt nanorods based bio-sensing system to detect the breast cancer biomarker, sHER2 in human serum and saliva [41]. Thus, NPs represent a valuable resource to interrogate biological systems for the identification of novel molecular players that could serve as diagnostic biomarkers and/or targets for therapeutic intervention.
In the present review we provide an update on the corona signature of various NPs with different surface properties, characterization of the PC signature, and how this information may be exploited to discover new molecular players in disease. The current study does not consider the use of NPs in the controlled release of cargo such as drugs or genes for various applications. How PC composition influences drug loading, release and uptake in cells and tissues of different origin is an area of emerging interest and the subject of a future review. The present review focuses on PC formation around various types of organic or inorganic nanomaterials and its importance for identifying new target proteins associated with disease progression that may be targeted to the benefit of patients. We also discuss the approaches used to date to understand and analyse PC formation and characterize the proteins present in the PC after incubation with biological fluids such as FBS, cell culture media or patient samples.
2. Protein corona ‘PC’ evolution
Despite the remarkable growth of nanomedicine, relatively little is known about the specific interaction of NPs with biological fluids. A decade ago, when the term PC was first used [15], NPs were already being applied for studies in various fields including biochemistry, toxicology, and colloidal science. However, prospective research on the PC is still required to drive applications of NPs in nanomedicine, including their use in diagnostics [42]. Nanoparticles, when exposed to biological fluids, for example through systemic administration or ex vivo incubation, interact with proteins and form a biological coating on their surface [2, 43, 44]. Subsequently this protein coat or corona around NPs dictates their biological responses including biodistribution, clearance and potential toxicity [45-52]. In addition to affecting NP behavior, the process of corona formation provides a tool for probing the local proteome [47]. Thus, characterization of the PC around NPs (unmodified and surface engineered) has the potential to provide molecular insight to characterize a disease and thereby help to identify new therapeutic targets.
Nanoparticles initially form a transient kinetically controlled “soft corona” consisting of the most abundant proteins. These highly abundant proteins are then slowly replaced by less abundant but higher affinity proteins to form the “hard corona” [43], a process determined by the Vroman effect [53]. In this process, protein binding is energetically favoured, enthalpy is reduced, the hydration layer around the NP is replaced, and finally an increase in entropy occurs [54]. Different proteins from biological samples will be adsorbed to the NP surface based on their affinity as shown in Figure 1. These changes in the PC with time can alter the bioavailability of NPs, trigger various biophysical processes, for example aggregation of NPs, and also may elicit immune responses which can eliminate NPs from the circulation. Cedervall et al. reported an isothermal titration colorimetric (ITC) method to study the affinity and stoichiometry of protein binding on NPs and used surface plasmon resonance (SPR) to study the association and disassociation of proteins on NPs [15]. They applied these techniques to determine how tailoring of copolymer NPs for size and hydrophobicity affects protein-NP interactions [15]. In addition to these physical studies, some theoretical and computational approaches have also been used to understand the interactions between proteins and NPs, including: 1) adoption of the Hill model to understand the dynamic equilibrium and kinetic coefficients for protein binding to NPs; 2) based on affinities of proteins, rate constants and stoichiometric analysis development of a dynamic model for evaluating the equilibrium composition of the PC [55] ; 3) development of a quantitative structure-activity relationship (QSAR) associated statistical modeling method using toxicity, blood circulation and NP biodistribution results; and 4) statistical analysis of biological surface adsorption index (BSAI) for binding coefficients of small molecules with NPs [56]. It is essential that these theoretical and experimental tools be used in combination to fully understand the protein NP interaction and the PC at various stages in its development.
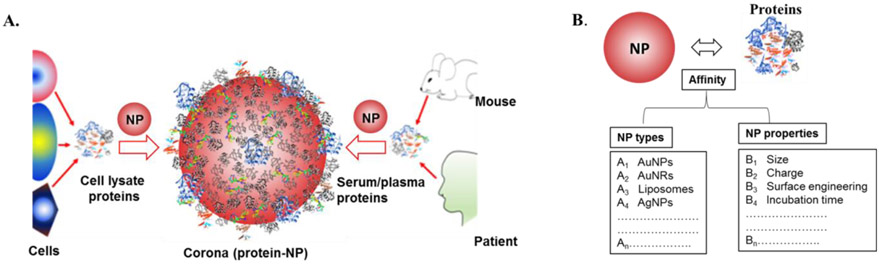
Figure 1.
The formation of nanoparticle protein corona and its interaction. (A) Proteins in either cellular lysates or serum/plasma when exposed to nanoparticle (NP), form a corona via adsorption of proteins on the nanoparticle surface. (B) The affinity-based interaction between proteins and nanoparticle depends on NP-related factors, including NP type (either organic or inorganic) and NP properties (size, charge, surface engineering and incubation time).
3. Nanoparticles in Proteomics
Human genome project (HGP) has identified approximately 20,300 protein-encoding genes in humans, although almost nothing is known about the knowledge or evidence of up to a third of them at protein level. [57]. Conventional approaches to study the proteome in biological samples include 2D gel electrophoresis and mass spectrometry techniques (e.g. MS, LCMS, and MALDI-TOF/TOF) [58, 59]. The major obstacles to successful proteome analyses are separation of the proteins of interest from a complex protein mixture, and low protein concentration making it difficult to obtain adequate sample for analysis [58, 60].
One strategy used to address the problems outlined above has been the introduction of nanomaterials into the field of proteomics to establish a new and rapidly evolving specialty research area termed nanoproteomics [61, 62]. Nanoproteomics provides the advantages of lower sample and reagent consumption, real-time multiplexed analysis, and a minimum number of high sensitivity assays in a short timespan [62, 63]. Thus, based on the unique properties of NPs, including tailorable surface properties and ease of separation from a mixture, nanoproteomics is emerging as an important technique to aid in diagnosis and identification of new molecular targets [64-66].
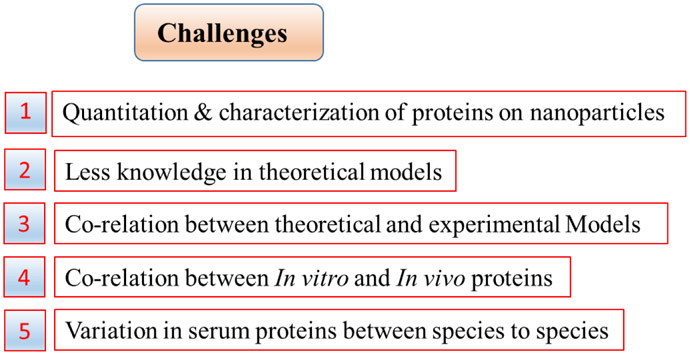
4. Challenges
Despite the existence of several models, as described above, to evaluate NP-protein interactions the need for improved quantitative studies is pressing and correlation between theoretical and empirical studies is essential to understanding PC formation. Figure 2 outlines the challenges to a full understanding of the NP-protein interaction, some of which are addressed in more detail below. Generally NPs are characterized and studied in aqueous solutions, however when introduced into a biological system they experience shear stress due to high protein and ion levels which in turn cause significant changes in their physicochemical properties. For example, when AgNPs, intended for use as antibacterial agents, were incubated in either media containing FBS or a buffer containing amino acids, the released toxic silver ions were trapped within the PC as non-toxic nano Ag2S; sulphidation of the AgNPs resulted in water insoluble Ag2S which decreased the available toxic Ag+ ions [67]. In another example, luminescent Cd-Se/ZnS quantum dots (QDs) are stable in mercaptopropionic acid (MPA), however in biologically equivalent fluids, within the physiological range of 150mM NaCl, the QDs were destabilized and precipitated resulting in colloidal instability. This colloidal instability can be further enhanced by divalent Ca+2 ions leading to aggregation of the particles[68]. Understanding the protein-protein interactions and protein displacement steps that occur when NPs are introduced to in vivo systems is also essential. It is known that when a NP is moved from one biological fluid to another the PC finger print can reflect the protein signature of the prior environment [69]. This suggests that the NP entry route, such as oral ingestion, inhalation, or intravenous injection, also play a key role in the PC composition and, thus, the ultimate fate of the NP. A further issue is that PC formation differs when NPs are subject to variations in protein exposure, this can result in variable behaviour in in vivo systems, including alterations depending on stage of particular disease. For example, albumin levels in plasma vary in diabetic and liver disorder patients [24], and a variation in PC formation is seen when NPs treated in plasma from normal and healthy patients are compared. This observation is further supported by a study in which bare polystyrene (100nm) NPs were incubated in the plasma of patients diagnosed with hypercholesteromia, diabetes or rheumatism and showed variable size increases of 18nm to 44nm [70]. Thus, in this regard, establishment of 3D-printed and virtual model systems are needed to unravel the complexities of in vivo PC formation
Figure 2.
Challenges in Nanoproteomics.
5. Methods used for identification of targets.
5.1. Genome analysis:
Genomic analysis (i.e. whole genome sequencing) is complete analysis of the genome in cells at a given time; it can detect genomic alterations, insertions, and duplications, as well as chromosome inversions and translocations that can be targeted for early detection of disease. For example, genome analysis identified BRCA1 and BRCA2 mutations in women with a family history of breast and ovarian cancers; BRCA testing now allows early detection of hereditary breast-ovarian cancer syndrome and thus appropriate preventive interventions. The Cologuard DNA test for aberrant NDRG4, BMP3 and KRAS mutations [71], and the Oncotype Dx Genomic score for early stage prostate cancer [72] are other examples of genome based tests for disease detection. However, application of complete genome analysis to new target identification is time and personnel intensive since it requires multiple techniques and specialties including bioinformatics and sequencing. For example, the gene expression profile of cervical cancer patients were combined with biomolecular network analysis and transcriptome data identified ephrin receptors, endothelin receptors, siRNAs and transcription factors as novel targets [73].
5.2. RNA sequencing:
RNA sequencing reveals the presence and level of RNA (transcriptome) in biological samples at a given time [74]; it can also reveal information about variations in the transcriptome i.e, post transcriptional modification, mutations and temporal changes. This can provide valuable information about the disease state and identify novel disease-associated targets [75]. For example, Kumar et al, used RNA sequencing to identify novel biomarkers for hematologic diseases [76]. In the first such study, Huang et al. sequenced RNA in exosomes recovered from human plasma and demonstrated the presence of abundant microRNAs (miRNA); they demonstrated roles for these miRNAs in various biological functions such as protein phosphorylation, angiogenesis, RNA splicing and chromosomal abnormality [77]. Thus, exosome miRNAs can yield valuable information on disease states. However, the RNA sequencing approach is also limited by the requirement for extensive technical support from multiple areas.
5.3. Proteomic Analysis:
Proteomic analysis (proteomics) is the identification and quantification of total proteins present in biological samples using mass spectrometry (MS) technique; it is a common and promising tool for identification of disease associated targets. Protein levels may vary during a disease process and could be used to monitor disease stage; in ovarian cancer proteomic analysis was used to demonstrate the presence of FK 506, Rho G protein disassociation inhibitor (RhoGDI) and glyoxalase-I in invasive cancer samples but not in the low malignant stage [78]. However, there are limitations to the identification of new targets by this methodology, including the presence of high abundance proteins such as albumin and immunoglobulins that can mask low abundant disease associated biomarkers [78]. The high dynamic range of total proteins and protein degradation during analysis are additional complexities in using this approach.
5.4. ELISA:
Enzyme linked immunosorbent assay (ELISA) is an accurate technique for detection and quantification of a target in biological samples [79, 80]; it is however limited to detection of known biomarkers since it requires antibodies to a specific antigen. ELISA also depends on multiple immune reaction steps which can itself lead to failure of the assay.
6. Advantages of the protein corona approach
Protein identification by tandem mass spectroscopy has emerged as a critical method to identify putative targets in a number of diseases including ovarian and pancreatic cancer [81-85].The proteomics approach can identify differentially expressed proteins in disease. However, differential expression of proteins in disease samples (tissues, cells, biological fluids) does not always correlate with clinical outcome. Functional characterization of the differentially expressed proteins to confirm their contribution to disease outcome is essential to define them as molecular targets. Unfortunately, functional validation of hundreds, if not thousands, of differentially expressed proteins to characterize a disease is not only cumbersome but such an endeavour may not ultimately lead to target identification. Protein-protein interaction (PPI) studies to create functional nodal networks may facilitate molecular target identification. Functional disruption of a hub node protein with higher network connectivity may exert enhanced therapeutic effects via disruption of other network proteins connected to this hub. Although this is a superior approach compared to pursuing individual differentially expressed proteins it suffers from the same limitations, since selected hub node proteins may not be relevant in disease pathology. Characterization of the PC around self-therapeutic NPs offers distinct advantages over these approaches and increases the success of identifying molecular targets. Because of the self-therapeutic nature of NPs, proteins present in the corona around these NPs may play pivotal role in disease progression. Identification of proteins in the PC of self-therapeutic NPs from healthy and disease samples along with comparative proteomics and bioinformatic analyses will likely identify novel molecular targets to characterize a disease.
7. Exploiting targets with nanoparticle - protein interactions ‘Sensing Targets with NPs’
In order to improve current treatment modalities, biological systems must be scrutinized to identify the biomolecules influencing the disease process. Figure 4 suggests a methodology for molecular target identification and analysis through PC study in biological systems. Using nanoproteomics to analyse the constituent proteins forming the PC around NPs is a viable approach to identification of such biomolecules and may generate information related directly to malignant sites. Thus, nanotechnology is a promising tool in the biomedical field to discover novel targets in disease sites or blood. For example, Hajipour et al., reported a gold nanotechnology sensing method for detection of Alzheimer’s disease and multiple sclerosis in patient plasma. In their methodology, spherical citrate, cysteine, cysteamine and polyethylene glycol coated AuNPs were incubated with 10% and 100% patient serum, and the PC responses were determined using UV-visible spectra, hierarchical cluster analysis (HCA), principal component analysis (PCA) and colorimetric differential profile (CDP) [86]. This method proved a simple, economic and quick process to detect neurodegenerative diseases. Others have used AuNP-GFP conjugates as sensors to discriminate between healthy and metastatic tissues, and this proved to be a robust and highly sensitive method to detect as little as 200ng of intracellular protein per nanoparticle [87]. You et al developed an array method using highly fluorescent AuNPs to detect seven specific proteins: namely, bovine serum albumin (BSA), cytochrome C, lipase, subtilisin A, β-galactosidase, acid phosphatase, and alkaline phosphatase. Six different cationic AuNPs were developed and each conjugated to a fluorescent polymer poly p-phenylene derivative, PPE-CO2. The polymer conjugated to the AuNP did not fluoresce, however when protein interacted with the AuNP-conjugate fluorescence was restored due to reverse quenching. Fluorescence patterns were highly characteristic for the individual protein even at nanomolar concentrations, establishing a highly versatile method to detect these particular proteins in biomedical applications [88]. In addition, several other groups have developed tailored AuNPs, by conjugation of specific ligands, for perceiving targets involved in signalling pathways. Recently, AuNP conjugates were developed with high affinity for His-tag containing proteins with the goal of detecting complex assemblies of the proteins [89]. In addition, aptamer-functionalized gold nanorods were successfully employed to detect thrombin in plasma of blood [89]. Other reports use NPs to improve antibody-antigen interactions, for example, alpha-fetoportin, a tumor marker, is detected in ranges from 100ng-400ng mL−1 and quantified by antibody-functionalized AuNPs [90].
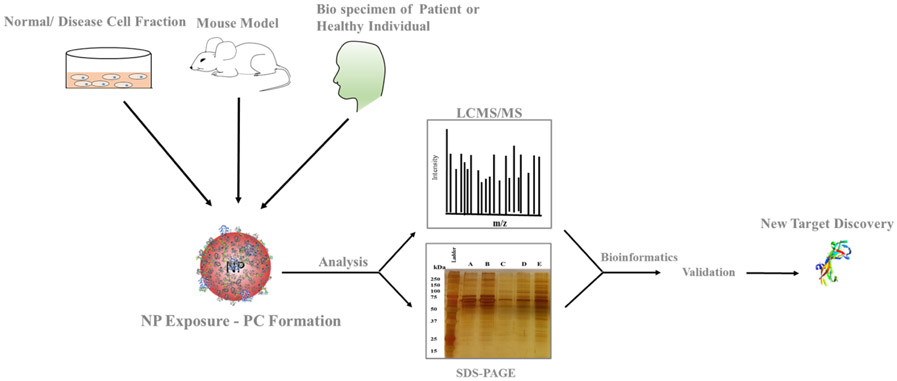
Figure 4.
Mode of study, NP-PC preparation to target discovery: Protein corona preparation can use disease cell fractions, mouse models of diseases or patient samples The chosen fraction is incubated with therapeutic nanoparticles. After isolation, a sample of the NP-PC is analyzed by gel electrophoresis to confirm protein adsorption NP. A second sample of the NP-PC is analyzed by MS/LCMS and compared to NP-PC from healthy samples to identify new targets. Bioinformatics analysis is done to find internal functional networks among the targets. Finally, identified targets will be analyzed in in vitro and in vivo studies for validation of its role in disease progression.
The surface engineering of NPs also influences interactions of the NP with proteins and modulates PC formation. Specific engineering features affecting these interactions include, tuning of the NP with different charges (i.e., positive, negative, zwitterion, or neutral) and the nature of the materials (i.e, hydrophilicity or hydrophobicity). Below we summarize how protein adsorption patterns vary between different types of NP when incubated in biological samples, and how surface modifications can help in identifying proteins from the samples.
7.1. Modulation of the Protein Corona: Influence of Size
It is well established that PC composition is impacted by NP size [91, 92]. This is especially true in the case of NPs in the range of 3-50 nm compared to those close to 100nm or greater. Lundqvist et al, demonstrated different protein compositions for the PCs of 9.5 nm and 76 nm sized silica NPs incubated in human serum [93]. The PC of the 9.5nm NPs contained complement factor H, plasminogen, gelsolin, serum paraoxonase/aryl esterase 1, fibrinogen alpha/gamma chain, complement factor H related protein1, complement C1q subcomponent subunit B, and ApoA1 while the 76nm particles contained coagulation factor XII, Histidine-rich glycoprotein, Histidine-rich glycoprotein, Apo E, Fibrinogen alpha chain, and ApoA1. In another study, the PC of iron oxide NPs of 30, 200, 400 nm sizes incubated in human plasma had only 20% of proteins shared across all particle sizes [94]. NP size clearly influences PC composition; different sized NPs could be exploited to gain different information about proteins in biological samples.
7.2. Modulation of the Protein Corona: Influence of Charge
A major advantages of NP systems is our ability to tune the surface properties in a precise manner. Surface functionalization of NPs to produce positive, negative and neutral surface charges facilitates different electrostatic interactions with proteins in biological systems and alters the PC accordingly. Elci et al. examined sub-organ distribution of intravenously injected 2nm-core AuNPs having variable surface charges (i.e. positive, negative and neutral). They demonstrated that the neutral NPs accumulated in Kupffer cells, the positively charged particles accumulated largely in hepatocytes and negatively charged were widely distributed in the liver. These results highlight the impact of surface charge on particle distribution to different cells of an organ [95]. Generally, positively charged NPs have a greater affinity for cells than do neutral and negatively charged particles [96, 97]. Although, in some cases, carboxyl group coated NPs show efficient cellular uptake due to pinocytosis or fast direct diffusion [98]. For enhanced drug delivery, positively charged NPs are the agents of choice, since they exhibit a preference for peripheral cells. In contrast to this, negatively charged particles are rapidly taken up deep into the tissues [99]. Additionally, neutrally charged NPs, for example zwitter ionic head groups on AuNPs, avoid non-specific biological interactions in biological fluids [100, 101]. Arvizo et al., recently reported a promising new approach by specifically tuning surface charge of NPs. They incubated differently charged gold nanoparticles i.e., +AuNPs and −AuNPs with lysates of a malignant ovarian cancer cell line (OV167), and found that the cancer-associated protein hepatoma derived growth factor (HDGF) exclusively bound to +AuNPs. In this way they identified HDGF as a possible target for ovarian cancer treatment. They also found that HSP-90 was detected only in −AuNPs [40]. These results clearly demonstrate that NP charge is crucial in determining the PC composition.
Overall, serum albumin, ApoA-II, alpha-fetoprotein, kinogen-1 complement C3, haemoglobin fetal subunit beta and prothrombonin are commonly associated with positively charged NPs; vitronectin, Fibrinogen-alpha-chain, Fibrinogen –gamma-chain and complement C3 with negatively charged particles; and serum albumin, α- 2- HS- glycoprotein and kinogen-1 with neutrally charged particles (Table 1 )[11, 12, 34, 93, 102-110]. NPs of different surface charge will capture different proteins to significantly impact the PC signature.
Table 1.
Proteins present PC of various NPs
| S No | Material | Name of Protein | Reference |
|---|---|---|---|
| 1 | Liposomes with DOPE, DOTAP, Cholesterol | Prothrombin, Fibrinogen, Vitamin K, Apolipoproteins (ApoA-1, II, IV, ApoC and Apo E) | (102) |
| 2 | PEGylated Liposomes | Coagulation factor XIII, Coagulation factor X, Complement protein-3, Prothrombin, Thrombospondin 1, Fibronectin, Kinogen-1, Apolipo protein A-IV | (103) |
| 3 | PLGA/PEG-PLGA Particles | Vitronectin, ApoE, Kininogen-1, Histidine-rich glycoproteins | (127) |
| 4. | Mesoporous lipid nanomaterials | Histidine rich glycoprotein, ApoA-II, ApoC-I, ApoC-III, Alpha-2 HS glycoprotein, ApoA-IV, serum albumin, ApoA-I, ApoE. | (135) |
| 5 | Iron oxide (SPIONS) with polyvinyl alcohol (PVA) | Alpha-2-Antiplasmin, ApoA-1, Cyt-P240 2C5, Alpha-2-HS-Glycoprotien, Complement Factor-B, H, Vitamine D binding protein, Seratotransferin, Prothrombonin, Serum Albumin, Alpha-fetoprotein and Kinogen-1 | (11) |
| 6 | Iron oxide (SPIONS) with Dextran | Charge factor alpha-1 anti-trypsin like protein, Heamoglobulin subunit -a I/II, Complement factor B, ApoA-II. Alpha-1 anti-proteinase, thyroxin binding globulin, endopin-1, fetuin B | (11) |
| 7 | Iron oxide with PEG/Glucose | Albumins, Complements, Coagulation, Transport proteins, Apolipoproteins, Fibrinogen | (12) |
| 8 | AgNPs (20nm) Citrate coated | Kininoogen-1, Fibrinogenin-a chain(FGA),Firbronegin beta chain, Fibrogenin gamma chain (FGG), Coagulation factor V(F5),Complement C4-B, Complement C3 (C3), Coagulation factor XI (F11), Ig-gamma −1 chain C region, IGHG1), Serum albumin (ALB), Apolipo proteins CI, CII, A-IV, Ig Kappa chain C regioin, APOA1,APOE, Vitronectin (VTN), Proteo glycon 4 (PRG-4) | (105) |
| 9 | AgNPs (20nm) Polyvenylpyrrolidine(PVP)coated | Plasma serine protease inhibitor (SERPINA 5), Plasma protease C1 inhibitor (SERPING1). | (105) |
| 10 | AuNPs (20nm) | SMNDC-1, PPA1, PI15 | (10) |
| 11 | AuNPs (nanorods, 50nm) | Serum albumin α-1-antiproteinase precursor, α-2-HS-glycoprotein, Apolipo protein A-1 precursor, hemoglobin fetal subunit beta, hemoglobin, Apolipoprotein A-II precursor, apolipo protein C-III | (107) |
| 12 | Quantum dots (DHLA-QDs) | Alpha 2-Macroglobulin chain A, Complement factor H, Complement C3b chain A, HSA, IgG Heavy chain, Apolipoprotein A1, IgG light chain | (140) |
| 13 | Quantum dots (DPA-QDs) | Alpha 2-Macroglobulin chain A, HSA, IgG Heavy chain, IgG light chain | (140) |
| 14 | Quantum dots (PEG-QDs) | Alpha 2-Macroglobulin chain A , HSA, IgG Heavy chain, Serum peroxonase, Apolipoprotein A1, IgG light chain | (140) |
| 15 | Silica nanoparticles (9.5 nm) | Complement factor H, Plasminogen, Gesolin,Serum paraxonase /arylestarase 1, Fibrinogen alpha/gamma chains,Complement factor H- related protein 1, Complement C1q subcomponent, subunit B, Apolipoprotein A1 | (93) |
| 16 | Silica nanoparticles (76 nm) | Coagulation factor XII, Histidine –rich glycoprotein, Fibrinogen alpha chain, ApoE, ApoA-1 | (93) |
| 17 | ZnO (30 nm) | Serum albumin precursor, Serum albumin, Fibronegin Alpha subunit, Fibronegin B beta chain, Fibrinogen gamma chain precursor, Vitronectin, Prealbumin, Alpha-1 inhibitor 3,Fibronectin isoform X2/X3, Alpha-1 macroglobulin, Serum protease inhibitor | (109) |
| 18 | TiO2 | Alpha-2 Macroglobulin, ALB protein, Alpha-2-HS protein, precursor, Alpha- 1-anti proteionase, Apolipo protein A-I precursor, Tetranectin, Haemoglobin subunit beta | (110) |
7.3. Centrifugation and surface area effect
In nanoproteomics, most protein analyses are performed on the pellet, i.e protein-particle complex isolated by centrifugation of the mixture. The problem with this approach is that data is limited to completely bound proteins and no information is generated regarding the partially bound proteins. In most cases, several parameters in the formation of the NP-protein complex are difficult to control and these dominate pellet formation [111]. The centrifugation method used to collect the NP-PC complex can influence adsorption of proteins to the NP surface, for example, when polystyrene NPs in PBS were incubated in 55% and 90% v/v FBS in PBS for 1h and the resulting NP-PCs were collected by conventional centrifugation or sucrose base gradient centrifugation method , the latter was more effective for NP-PC complex isolation than the former, illustrating the importance of NP-PC isolation method [111]. In another study, Nash and co-workers developed a magnetic separation method to effectively utilize stimuli responsive AuNPs for purification and rapid detection of biomarkers in human plasma while avoiding the co-precipitation of unbound protein with the AuNP-protein complex [112, 113].
Lundquist et. al., demonstrated the curvature of the silica NPs influences protein adsorption; 9.5 nm silica NPs adsorbed more protein than 76nm particles. In high protein concentrations, the PC signature of the 9.5 nm and 76 nm particles was similar and dominated by a single protein [93]. In another study, the PC of 200 nm silica NPs showed an increase in intensity of a 120kDa band with increasing plasma concentration where as it remained unchanged with 50 nm size particles possibly due to curvature affect leading to low surface coverage [114]. In summary, NP size and protein concentration are among the critical factors that influence PC signature. Smaller size NPs adsorb more protein than larger NPs of the same material signifying a role of surface area in PC formation.
7.4. Temperature
Mahmoudi et al., reported temperature dependent PC formation on superparamagnetic NPs grafted with different surface coatings [29, 115]. Polymer coated FePt NPs were incubated with either human serum albumin (HSA) or apo-transferin (apo-Tf) at temperatures ranging from 5- 41 °C. The highest protein affinity was found at the highest temperatures, however the hydrodynamic diameter of the PC remained the same at all temperatures. The authors also incubated super paramagnetic iron oxide FeOx NPs (SPIONS) having positive, negative or neutral surfaces in fetal bovine serum (FBS) at temperatures ranging from 5-41 °C. Between 37 41 °C, serotransferin [116], apolipoprotein A-1 [115], serum albumin (Mw = 66kDa)[115, 117] and α−2-HS glycoprotein (Mw = 49kDa) contributed to the temperature dependent PC formation [29]. Notably, the amount of adsorbed proteins in the PC was greater at 40°C than at lower temperatures, and particularly so when looking at the adsorption of α−2-HS glycoprotein on positively charged NPs. These data clearly demonstrate that corona signature is highly dependent on incubation temperature.
7.5. Effect of Nanoparticle concentration
PC signature is also affected by NP concentration [118]; Fedeli et al. examined effects of different concentrations of SiO2 NPs on the corona formation and underlying corona composition when incubated with human plasma or FBS. In 10% human plasma SiO2 NPs at 20μg /ml formed a PC consisting of anti-opsonin proteins like HRG and Kin-1 due to their higher affinity. In contrast, SiO2 NPs at 160 μg/ml evolved a PC with the same two proteins along with the pro-opsonins immunoglobulins, Fibronectin and LDL related Apo B100 and Apo E, HDL related Apo-I and Apo-II. Furthermore, SiO2 NPs incubated in 10% FBS had a PC containing Albumin, heamoglobulin, a-2 macrogolbulin, HDL associated Apo A-I, Apo A-II and complement factor were observed [118]. Interestingly, NPs incubated in human plasma at concentrations < 40 μg/ml had reduced cellular uptake by macrophages (about 90-95% inhibition); no such inhibition was seen following incubation in 10% FBS. Thus, NP dose, as well as specific biological sample, influence corona formation and its composition [118].
7.6. Effect of incubation time and concentration of biological fluids
It is well established that for NPs incubated in biological fluids, a PC is detectable within 30 seconds, and thereafter the PC varies with time [119]. Giri et al examined PC evolution over time around 20 nm AuNPs incubated in ovarian cancer cell lysates [10]. The bare AuNPs had hydrodynamic diameters (HDS) of 30 nm, but within 5 minutes HD increased to 283.5nm. Interestingly, HD decreased gradually thereafter; HD was 72.31nm after 1 h and 59.21 nm after 24 h. They also found that PC charge, assessed by zeta potential, was – 29.2 mV at 5 min, – 31.1mV at 15 min and –8.40 mV after 24h of incubation; bare AuNPs had a charge of – 43.3mV [10]. They concluded that the soft corona formed by low affinity high abundant proteins at initial time points was replaced by high affinity low abundant proteins leading to stabilization of HD and charge of NPs. Walczyk et al., assessed time dependent PC formation using 100 nm carboxylate surface coated polystyrene nanoparticles (PSCOOH) incubated in diluted plasma. They observed a relatively stable PC established within 1 h with subtle changes occurring thereafter up to 12h [120]. These results suggest that PC formation is highly dynamic, adding to the complexity in analysing the proteins making up the PC. In terms of the impact of protein concentration on PC composition, when 200 nm NPs of either SiO2 or PSOSO3 were incubated in 3% to 80% plasma, the PC evolution and composition varied significantly [120]. Using densitometry of SDS-PAGE bands the authors showed that the amount of protein in the PC of PSOSO3 NPs increased with plasma concentration, however, for SiO2 NPs the protein amount slightly decreased with increasing concentrations [114].
Saha et. al, reported a study on the PC of different cationic AuNPs with the same core size incubated in 10% and 50% human serum and studied their macrophage uptake [121]; about 60% of the PC proteins were either immunoglobulins or apolipoproteins at either serum concentration. However, the total number of proteins identified in 10% serum was greater than that in 50%, possibly due to the higher concentration of low affinity proteins in the 50% serum, which bind and mask the NP from other proteins in the sample; in 10% serum, the low abundance proteins would have greater access to the NP. Importantly, the percentage of apolipoproteins and immunoglobulins were similar with (50 %) high serum concentration and were identified low percent with increased hydrophobicity. Normally, NPs with associated apolipoproteins, coagulation factors and complement proteins have a greater tendency to be taken up by macrophages while those with immunoglobulins show the opposite. Here, among the PC proteins, C4BPA and IGLC2 had higher positive and negative correlations with macrophage uptake respectively [121]. Taken together the results demonstrated role NP of incubation time in biological samples that effects PC formation.
7.7. Static and dynamic fluidic Conditions
In biological systems, the blood flow rate varies among blood vessels in the circulatory system e.g., it is different in arteries, arterioles and capillaries for example; this may affect the PC signature. PC formation under different fluidic conditions has been studied using both liposomes and superparamagnetic NPs [122, 123]. Pozzi et al. examined the PC of liposomes under static and dynamic incubation in FBS; more low molecular weight proteins were adsorbed under dynamic incubation than static incubation [122].
8. Protein corona composition with different materials in detail
8.1. Organic materials
8.1.1. Liposomes:
Liposomes are the first choice among organic materials for delivery of cargo (e.g. drug or siRNA) in to cells. Recently, Hadjidemetriou et al., analysed the PC surrounding intravenously injected circulating PEGylated liposome (HSPC:Chol:DSPE-PEG-2000) in two different xenograft models, one a subcutaneous melanoma (B16F10) and the second a human lung carcinoma (A549) model with the goal of identifying cancer associated biomarkers[28]. Both low molecular weight and tumor-tissue leakage associated low abundant proteins were captured on the liposome surface, although neither were detected by direct plasma analysis. Furthermore, the total number of proteins detected in the PC was significantly higher than the number identified by direct plasma analysis [28]. Thus, analysis of the PC established in blood represents an important tool for identifying cancer biomarkers under in vivo conditions. In another study, Caputo et al., analysed the patient serum-derived PC of DOPG liposomes in pancreatic ductal adenocarcinoma (PDAC) patients with disease stages T1-T4, with the goal of correlating disease stage with PC composition [124]. PDAC tumor size and stage typically affected PC composition with stages T1, T2 and T3 being particularly associated with changes in proteins in the 25 to 50 kDa range. The proteins VEGF-2, VEGF-4 and CDK2 were identified in stage-1 whereas complement system proteins C3 and C5 which play major role in immune response [125]were identified in later stages, while between stages T1-T2 and T4 there was no difference in the PC [124]. Thus, analysis of the NP PC may be useful to predict disease stage and help in treatment selection. The most common lipids used in commercial liposomes are neutral dioleolyl phosphatidylethanolamine (DOPE), cholesterol and cationic 1, 2-dioleolyl-3-trimethyl ammonium- propane (DOTAP) and di methyl amino methane- carbamoyl (DC)-cholesterol. In general, liposomes of DOTAP and DC-Cholesterol interact with negatively charged plasma proteins including prothrombin, fibrinogen, vitamin K, and vitronectin (mentioned in Table-1). However, affinity for these proteins is reduced when DOTAP is replaced with neutral lipids. DOPE, in contrast, enhances affinity for serum albumin and apolipoproteins [102]. Cholesterol in liposomes induces affinity for IgG and complement proteins. Interestingly, DNA associated lipoplexes associate with basic immune globulin gamma (IgG-gamma) proteins more than similar liposomes without DNA [126].
8.1.2. PEGylated liposomes:
Palchetti et al. analysed the PC around circulating cationic PEGylated liposomes of multi-lipid composition under both static and dynamic incubation conditions [103]. The aim of the study was to determine the effect of shear stress on PC formation by comparing results from dynamic and static incubation [103]. The PC size under either static or dynamic incubation conditions was almost identical, however the corona of circulating liposomes had a higher negative charge than its counterpart. The authors identified a total of 217 proteins; 99 proteins were found only in the PC under dynamic conditions, i.e., analogous to circulating liposomes, 10 proteins were unique to the static corona and 108 proteins were common to both conditions. Adsorbed proteins were classified according to their biological functions. Analysis illustrated that less abundant proteins in the PC can affect the in vivo fate of liposomes but high abundance proteins have less effect [103]. Dynamic incubation exclusively resulted in adsorption of specific proteins such as coagulation factor XIII [(F13A), stabilization of fibrin clots], coagulation factor X [(FA10) conversion of prothrombin to thrombin] and protein Z [(PROZ) inhibition of activity of coagulation protease factor Za] which were absent in static incubation. Moreover, coagulation proteins, complement protein 3 (C3), prothrombin (THRB), thrombospondin 1 (TSP1), fibronectin (FINC), kinogen-1(KNG1), and apolipoprotein A-IV (APOA4) were found in higher abundance under dynamic incubation than static incubation (Table-1) [103]. Comparing common proteins in the PC of PEGylated liposomes to the PC of other types of PEGylated NP (Table 2), ApoA-II protein is present in the PEGylated AuNP corona; ApoA-IV is in the PC of PEGylated IONPs and silica NPs; proteins ApoE, seratotransferin, clusterin, vitamin D binding protein, haemoglobin subunit β−1, and prothrombin are common to IONPs; complement factor b is common to the PC of PEGylated silica NPS; complement factor H, firbrinogen γ- chain, fibrinogen β-chain proteins are in the PC of PEGylated gold and silica NPs; complement C3 and alpha-2 macrogloubulin proteins are common to the PC of with PEGylated gold NPs and IONPs.. Collectively, these results show that PC signature of circulating liposomes under dynamic flow is different from that in static fluid conditions. It is likely that under the conditions of flow only proteins with high affinity can stick to the particle surface resulting in the formation of an irreversible hard corona.
Table 2:
Common proteins in present in the corona of all PEGylated Liposomes, PLGA, gold, Iron oxide and silica nanoparticles. [12, 93, 127,129,130]
| Protein | Liposome | PLGA | Gold | IONPs | Silica |
|---|---|---|---|---|---|
| Alpha-2-HS-Glycoprotein | ∗ | ∗ | ∗ | ∗ | ∗ |
| Apolipoprotein | ∗ | ∗ | ∗ | ∗ | ∗ |
| Plasminogen | ∗ | ∗ | ∗ | ∗ | ∗ |
| Serum Albumin | ∗ | ∗ | ∗ | ∗ | ∗ |
| Alpha-2 macroglobulin | ∗ | ∗ | ∗ | ∗ | ∗ |
| Apolipoprotein A-1 | ∗ | ∗ | ∗ | ∗ | ∗ |
| Apolipo protein C –III | ∗ | ∗ | ∗ | – | ∗ |
| Anti-thrombonin –III | – | ∗ | – | – | ∗ |
| Apolipo protein C | – | – | ∗ | ∗ | ∗ |
| Apolipo protein C- IV | – | – | – | ∗ | ∗ |
| Apolipo protein A- II | ∗ | – | ∗ | – | – |
| Apolipo protein A- III | – | – | – | ∗ | ∗ |
| Apolipoprotein A- IV | ∗ | – | – | ∗ | ∗ |
| Coagulation Factor –X | – | – | – | ∗ | ∗ |
| Fibrinogen a chain | – | – | – | ∗ | ∗ |
| Seratotransferin | ∗ | – | – | ∗ | – |
| Apolipoprotein E | ∗ | – | – | ∗ | – |
| Vitamin D binding protein | ∗ | – | – | ∗ | – |
| Hemoglobin subunit beta-1 | ∗ | – | – | ∗ | – |
| Prothrombin | ∗ | – | – | ∗ | – |
| Clusterin | ∗ | – | – | ∗ | – |
| Alpha-2-macroglobulin | ∗ | – | ∗ | ∗ | – |
| Complement C3 | ∗ | – | ∗ | ∗ | – |
| Fibrinogen b chain | ∗ | – | ∗ | – | ∗ |
| Fibrinogen g chain | ∗ | – | ∗ | – | ∗ |
| Complement factor H | ∗ | − | ∗ | − | ∗ |
| Complement factor B | ∗ | − | − | − | ∗ |
| Fibronectin | − | − | ∗ | ∗ | − |
8.1.3. PLGA/PEG-PLGA particles:
It is known that the PC around injected NPs modulates the interactions of the NP with cells as well as its organ distribution in vivo. Sobczynski et al investigated how the PC affects the adhesion of vascular targeting PLGA particles to endothelial cells when intravenously injected, an important parameter for any vascular targeting material in humans [127]. There was a significant reduction in adhesion to endothelial cells in blood flow when compared to flow in buffer [127]. This was due to adsorption of certain high molecular weight proteins; following depletion of high molecular weight IgG the adhesion of NPs to endothelial cells was successfully restored. The type of targeting ligand (i.e. PLGA or Polystyrene) did not impact the negative adhesion effect to endothelial cells, rather it was entirely dependent on the targeting ligand density [127]. When incubated in human plasma the high density lipoproteins, vitronectin, ApoE, kininogen-1, histidine-rich glycoproteins were enriched in the PC (Table-1); a common characteristic of these adsorbed proteins was a pI below 7 [104]. In another study, Bertrand et al. examined the in vivo PC of a library of PEG-PLGA NPs 55nm, 90nm and 140 nm in size with different PEG (Mw 5K) density (10-50 PEG chains per 100 nm2). They found that PEG density played a key role in early clearance in of the NPs. Those NPs with 20 PEG chains per 100nm2 had prolonged circulation times, whereas those with PEG density below 20 PEG chains per 100nm2 area exhibited faster clearance. The results demonstrated that physiochemical characteristics of NPs significantly influence both circulation in blood and early distribution in vivo.[104] In general, ApoC-IV is found in the PC of PEGylated liposomes, PEGylated silica NPs and PEGylated IONPs NPs. Antithrombonin is also found in the PC of PEGylated PLGA particles and is common with PEGylated silica particles. Proteins which are common in the PCs of PEGylated PLGA and other PEGylated gold, liposome, IONPs and silica are listed in Table 2. [103, 128-130].
8.1.4. Mesoporous lipid nanomaterials:
As well as vesicles and liposomes, lipid containing multicomponent hydrated systems have been tested as delivery vehicles, with high protein entrapment, for controlled release applications in nanomedicine [131-134]. These nanosystems were self-assembled and mesoporous in nature with cubosome, spongosomes and hexososmes architecture. Recently Carla et al., studied the PC around mesoporous silica NPs with similar sizes (~120 nm) but different pore diameters following incubation in sera from 22 prostate cancer patients [135]. Mass/ LC-MS chromatography showed a PC enriched with low molecular weight proteins (Mwt < 50 kDa) predominantly adsorbed within the small pore (7.4nm) mesoporous NPs. The protein adsorption pattern differed between pore sizes of 7.4nm (Mwt < 50 kDa) and 14 nm (Mwt 50 to 70kDa) due to competition of proteins of different molecular weights for different pore sizes [135]. In the PC, histidine rich glycoprotein (59.6 kDa) and ApoA-II (11.2 kDa) increased from pore size 4.8nm to 14nm, whereas ApoC-I (9.3 kDa) , ApoC-III (10.9 kDa), Alpha-2 HS glycoprotein (39.3 kDa), ApoA-IV (45.4 kDa), and serum albumin (69.4 kDa) decreased. [135]. Protein lysozyme C (16.5 kDa) was enriched predominantly in 2.7nm pore size NPs whereas ApoA-I (30.8 kDa) and ApoE( 36.2 kDa) were enriched to similar levels at all pore sizes from 2.7 nm to 14nm. The authors concluded that due to their cubic structure and porous nature these mesoporous NPs can adsorb different types of proteins in nano-packets when exposed to serum or plasma, and that different pore sizes have discriminative adsorption properties. Thus, mesoporous NPs with tunable porous architecture may be useful to capture and identity different protein signatures dependent on pore size and may help to identity a protein-finger print to characterize a disease.
8.2. Inorganic nanomaterials:
8.2.1. Gold Nanoparticles:
Gold NPs (AuNPs) are widely used inorganic materials in biomedical applications due to their unique properties including high surface area and simple synthetic methods and characterization. Giri et al. analysed the PC of negatively charged self-therapeutic citrate coated 20nm AuNPs (with average zeta potential −43.3 mV) following incubation in lysates of either a human ovarian cancer cell line (A2780) or non-cancerous human ovarian surface epithelial (OSE) cells for various times i.e 5min, 15 min, 1 h, 6h and 24h [10]. The aim of the study was to identify new molecular targets involved in ovarian cancer progression. PC formation was assessed using UV-visible spectroscopy, DLS and zeta potential, and properties of the proteins in the corona were explored using tandem mass spectroscopy (MS/MS). Proteins in the PC had, on average, higher isoelectric points (pI) that all proteins in the cell lysates; mean pIs were 7.7 and 7.6 for the PC proteins from OSE and A2780 respectively both of which were significantly higher than the average for individual lysate proteins (6.5 and 6.3 for OSE and A2780 respectively). These data clearly demonstrated that protein binding to AuNPs was through electrostatic interactions and that protein charge played a fundamental role in PC formation. Interestingly, there was no difference in average molecular weights between PC proteins and the protein pool. Bioinformatics analysis of the PC proteins revealed that the RNA reorganization motif, RRM-1 domain was commonly found in PC proteins at both the 6 h and 24 h time points. Importantly, mRNA related proteins such as ELF1AX, PPA1, SMNDC1 and PARK7 were observed with a higher degree of nodal connectivity at the 6h time point. After 24h incubation, RPL12A, DEK, DDX46, GNA13 proteins were enriched in the PC. The translation related proteins ELF1AX and RPL10A might not represent specific targets for cancer cells, however the oncogenic related DEK protein might be an important target for ovarian cancer. Graph-theory based algorithms were also employed to identify cancer-specific hub nodes in biological networks, and three nodal hub proteins were identified: Pyrophosphate 1 (PPA1), Survival Motor Neuron Domain Containing 1 (SMNDC 1) and Peptidase inhibitor 15 (PI15). Using gene knockout studies, the oncogenic properties of PPA1 and SMNDC1 proteins in A2780 cells were confirmed [10]. This methodology was simple and convenient for the identification of novel oncogenic proteins from cancer cell line lysates in vitro. In another study, serum from prostate cancer patients was incubated with AuNPs and the resulting PC was probed in an immunoassay to detect serum protein biomarkers using an antibody mixture to identify adsorbed proteins[136]. The amount of VEGF protein adsorbed on NPs was decreased when compared with non-cancerous and less malignant cancer samples indicating that this methodology could be a novel and broad approach for prostate cancer biomarker detection. This study was performed to find the difference in PC composition between cancerous and non-cancer patient serum samples [136]. The PC on gold nanorods (AuNRs) has also been examined. Mahamoudi et al. identified several proteins in the corona when AuNRs were incubated in 10% FBS and 100% FBS under plasmonic heat induction by a laser, with the goal of determining the role of heat induction in PC formation. Proteins identified included serum albumin, α−1-antiproteinase precursor, α−2-HS-glycoprotein, apolipoprotein A-1 precursor, hemoglobin fetal subunit beta, hemoglobin, apolipoprotein A-II precursor, and apolipoprotein C-III; there were significant differences in the protein levels in the PC depending of treatment conditions [107]. In the PC of PEGylated AuNPs, ApoC-III protein were common to PEGylated silica NPs, ApoC was common with PEGylated IONPs and silica NPs. Alpha-2-macroglobulin and complement C3 were common with PEGylated liposome and IONPs; fibrinogen β chain, Fibrinogen γ chain, complement factor H were common with both PEGylated liposomes and silica NPs, and fibronectin was common with PEGylated IONPs (Table 2).
8.2.2. Iron oxide nanoparticles:
Currently, due to their magnetic properties, low toxicity and biodegradability super paramagnetic iron oxide NPs (SPIONs) are emerging as important materials for NP-based applications in the biomedical field[137]. Sakhulku et al. investigated the PC signature of SPIONS coated with two polymers: polyvinyl alcohol (PVA) and dextran [11]. The SPIONS, having different surface charges (positive, negative, and neutral), were injected in rats for in vivo corona formation analysis. Fifteen minutes after injection, 87% of PVA coated SPIONs, both neutral and negative were found in the bloodstream, which was significantly higher than the dextran coated SPIONs (50%) in the same compartment. A greater number of serum proteins were adsorbed to PVA coated SPIONs than to the dextran SPIONs; the proteins serotransferin, prothrombin, serum albumin, alpha-fetoprotein and kinogen-1 specifically bound to the PVA coated SPIONS but not in dextran coated particles. These results suggested that the adsorption of specific proteins to PVA-SPIONS decreased their uptake by the RES and thus increased their blood circulation time. This study further revealed that chemical composition and charge of the NPs were critical in PC formation and determining the fate of materials in vivo [11]. When charge was considered both neutral and negatively charged SPIONs had longer half-lives than the positively charged SPIONs. In the PC, proteins alpha-1 anti-trypsin like protein, haemoglobin subunit-α I/II, complement factor B, ApoA-II specifically bound to positively charged dextran SPIONS and alpha-1 anti-proteinase, thyroxin binding globulin, endopin-1, fetuinB, transthyretin, and haemoglobin subunit-α proteins were found exclusively on negatively charged dextran (mentioned in Table-1); none of these proteins bound to PVA SPIONs[11]. Interestingly, there were no specific proteins bound to neutral dextran coated SPIONS. These observations suggest that the PC is dependent on the surface engineering of NPs, i.e. corona composition is dependent on the nature of the surface modifications and overall charge of the NPs. Whether some proteins will present in the soft corona (low affinity) or hard corona (high affinity) will depend on the nature of the nanosystems (core nanoparticles) and their surface properties. Such information could be exploited to tune corona formation for personalized medicine in the future.
8.2.3. Iron oxide PEG/Glucose:
Stepein et al. evaluated and compared the in vitro and in vivo PC on 12 nm core iron oxide nanoparticles (IONPs) coated with either glucose (Glc) or polyethylene glycol PEG (5-KDa) polymer [12]. The PC analysis showed that PC composition affected in vivo bio distribution. The glucose functionalized nanoparticles i.e., IONPs-Glc were degraded more quickly in vitro whereas the opposite was true in vivo. PEG-coated NPs degraded and were cleared more quickly than glucose coated NPs in vivo. Furthermore, the distribution kinetics were also different in liver and spleen. This was due to a higher number of opsonins such as complement proteins and apolipoproteins adsorbed to the IONPs-Glc, as well as elevated levels of fibronectin on IONPs- Glc; fibronectin was known to be involved in Kupffer cell uptake. In additions, several other proteins such as coagulation factor XI, mannose binding protein C, C4b, and fibrinogen were only found on IONPs-Glc. These proteins were all known to be involved in NP clearance. In contrast, the PC of IONPs-PEG were enriched for albumin, acute phase protein α2-HS-glycoprotein, and kinogen which resulted in a longer circulation half-life [12]. Thus, it was evident that the PC around NPs influenced in vivo pharmacokinetic profile and ultimate fate of the NPs. In general, in the PC of PEGylated IONPs, gold and silica NPs protein ApoC was common to all. In addition, ApoC-IV was common with other PEGylated PLGA and silica NPs, ApoA-III was common with PEGylated liposome and silica NPs; coagulation Factor –X and Fibrinogen were common with PEGylated silica NPs; serotransferrin, ApoE, vitamin D binding protein, hemoglobin subunit b-1, prothrombin, and clusterin were common with PEGylated liposomes; and alpha-2-macroglobulim and complement C3 were common with PEGylated gold NPs and liposome (Table 2). These results show that the type of material used to modify the NP surface is critical to its affinity for proteins and dictates the NPs fate in vivo.
8.2.4. Silica Nanoparticles (SiO2 NPs):
Lundqvist et al. studied the PC of silica NPs and showed that corona formation was dependent on a number of factors, specifically: the biological sample from healthy individual with which the NP was incubated, such as whole blood, whole blood with EDTA, plasma, or serum; particle size; and ratio of particle surface area to protein concentration[93]. Proteins such as complement factor-H, plasminogen, gelsolin, serum paraoxonase, fibrinogen gamma chain, fibrinogen alpha chain, complement factor H-related protein 1, complement C1q subcomponent subunit B, apolipoprotein A-1, and fibrinogen alpha chain were found in 9.5nm silica particles, whereas, coagulation factor XII, histidine-rich glycoprotein, fibrinogen α chain, ApoE and apolipoprotein A1 were found on 76nm silica particles [93] (mentioned in Table-1). When PC of PEGylated silica NPs was considered fibrinogen β chain, fibrinogen γ chain and complement factor H were common with non PEGylated AuNPs and liposome; Complement factor B was common with PEGylated liposomes, apolipoprotein C –III was common with PEGylated liposomes, PLGA and AuNPs,; anti-thrombonin –III was common with PLGA; Apo C was common with AuNPs and IONPs; Apo C-IV was common with INOPs; ApoA-III was common PEGylated IONPs; ApoA-IV was common with both PEGylated IONPs and liposomes; and coagulation Factor–X, and fibrinogen α chain were common with PEGylated IONPs (Table 2). Common proteins found in the PC of PEGylated silica NPs and other PEGylated liposomes, PLGA, AuNPs, and IONPs were presented in Table 2. The corona obtained from various NP systems (surface modified and unmodified) from the same patient may collectively provide a unique protein-signature that may characterize the stage, grade and outcome of that particular individual leading to individualized medicine.
8.2.5. Silver nanoparticles:
Due to their strong antibacterial activity, silver nanoparticles (AgNPs) are widely used in consumer products and in biomedical applications. AgNPs can translocate into biological systems following dermal, inhalation or systematic administration. To date, there have been several studies reported on the PC of AgNPs. Huang et al. studied the plasma-derived PC of 20nm AgNPs coated with poly vinyl pyrimidine (AgNP-PVP-20) and citrate AgNP-CIT-20) [105]. Plasma proteins with molecular weight < 60 k Da were observed in the PC analysis using mass spectroscopy (MS) and label free quantitative techniques (LFQ). Proteomics analysis of the AgNPs PC showed acidic and low molecular weight plasma proteins selectively adsorbing to the NPs. Proteins related to platelet, coagulation, haemostasis, complement activation and immune responses (Table-1) were also adsorbed by the AgNPs [105], a possible explanation for the haemolysis and inhibition of proliferation and viability of lymphocytes seen on treatment with AgNPs at 10, 20, 30, 40 μg/ ml. These authors investigated the corona of AgNPs based on the previous report by Kwon et al., showing that AgNPs at specific sizes and doses induced human erythrocyte haemolysis by provoking pores formation [106]. Therefore, analysis of the PC around AgNPs may provide molecular insight on the anti-bacterial and hematological toxicities of the particles and thus provide information to further enhance anti-bacterial efficacy and alleviation of hematological toxicities.
8.2.6. Quantum dots (QDs):
Due to their excellent physical properties including optical and electrical properties, QDs are of increasing interest in the biomedical field [138, 139]. Stan et al., analysed the PC of Si/SiO2 QDs when incubated in bovine serum albumin (BSA), or cell culture medium with and without 10% FBS[108]. The results demonstrated that QDs altered the conformation of adsorbed proteins, i.e., the authors reported secondary structural changes in BSA in the PC. Under protein free condition QDs haemolysed red blood cells; this effect was attenuated in the presence of serum. These results showed that the formation of a PC in serum negated the haemolytic activity of QDs indicating that the interaction of QDs with serum proteins affect stability of cell membrane phospholipids and proteins [140]. In another study, Wang et al. examined the human serum derived PC around water solubilized CdSe/ZnS QDs functionalized with three different ligands: bidentate anionic dihydrolipoicacid (DHLA), zwitterion D-penicillamine (DPA) and poly (ethylene glycol) PEG [140]. They showed that corona size and binding efficiency of serum proteins was dependent on the type of surface ligand. For PEG-QDs, the PC attained only 2nm thick whereas for DPA-QD and DLHA-QDs the PC was 7.2 nm and 5.9nm thick respectively, suggesting that PEG molecules masked the QDs and reduced their interaction with serum proteins [141]. Results were similar to other observations on the human serum albumin (HSA) corona of PEGylated FePt [142]. Moreover, PC formation in serum was irreversible but when the same QDs were incubated with a single protein (HSA) the opposite was true. The most abundant proteins in the PC were immunoglobulins (IgG), HSA and to a lesser degree α2-macroglobulin in all QDs. ApoA1 was found in DHLA-QD and PEG-QDs, while complement proteins (factor H and C3) were adsorbed only on DHLA-QDs (Table-1). Characterization and identification of the PC around QDs may lead to a personalized disease-signature when QD-PC having different surface modifications interact with individual patient samples.
8.2.7. Zinc oxide (ZnO):
Zinc oxide NPs are used in sunscreen products, and the agricultural and food industries [143, 144]. Several studies described the interactions of ZnO NPs with proteins [109, 145]. Yu et al., reported a study with 289.6 nm and 28.3 nm sized ZnO particles in which they explored their interactions with biological fluids, specifically intestinal fluids, gastric fluids and plasma[109]. In rat plasma, the amount of protein (2152μg) adsorbed on 28.3 nm ZnO was greater than the amount (1544μg) on the larger ZnO particles. Fibronectin isoform CRA-β was detected only in 289.6 nm ZnO whereas, keratin K6, complement C1q subcomponent subunit B precursor, complement C3 precursor, SWItch/sucrose non-fermentable SWI/SNF–related matrix associated actin-dependent regulator of chromatin subfamily D member 3 were only in the 28.3 nm ZnO PC (mentioned in Table-1).The proteins serum albumin and fibrinogen were most abundant, and fibronectin was common to the PC of both ZnO NPs and played prominent roles in corona formation regardless of particle size [109]. These results demonstrated that the same nanoparticles materials with different sizes interact differently with biological samples and may be exploited to identify new molecular targets.
8.2.8. Titanium dioxide:
TiO2 nanoparticles (TiO2-NPs) have been used as nanomedicine including in medical implants, as well as in the food and cosmetics industries. [146]. However, TiO2-NPs are reported to have some associated toxicities [147, 148]. Borgognoni et al., examined the PC surrounding 120nm TiO2 NPs following incubation with primary human macrophage cells isolated from then peripheral blood of healthy volunteers [110]. The TiO2 NP surface mainly attracted secondary modified proteins that led to activation of cytokines. The PC signature of TiO2 particles below 1 μg/ml induced IL-6 cytokine while above 1 μg/ml IL-1β IL-10 and IL-6 were induced. IL-6 has a key role in autoimmune disease development and activation of acute phase proteins. [110]. Proteins alpha-2 macroglobulin, ALB, Alpha-2-HS protein precursor, Alpha-1-anti proteinase, Apolipoprotein A-I precursor, tetranectin, hemoglobin subunit beta were found in the PC of TiO2 NPs (mentioned in Table-1). In another case, TiO2 NPs in rats induced inflammation, oxidative stress and neutrophilia in lungs [148, 149].
Overall, proteins α−2-HS-glycoprotein, apolipoprotein, plasminogen, serum albumin, α−2-macroglobulin, ApoA-1, ApoC-III were common with all PEGylated PLGA, gold, IONPs and silica NPs (Table 2) [103, 128-130]. When considering charge: serum albumin, ApoA-II, Alpha-fetoprotein, Kinogen-1, complement-3, hemoglobulin subunit-beta and prothrombonin were common in the PC of all positively charged particles; Vitronectin, Fibrinogen-a-chain, Fibronogen- b-chain, Complement C3 were common to the PC of all negatively charged particles; and serum albumin, 2-HS-glycoprotein and kinogen-1 were common in PC of all neutral particles (Table-3).
Table 3.
Common Proteins adsorbed to all types of NPs with different charges[11,12,93,97,105,,107,109,110,127,140]
| Positive | Negative | Neutral |
|---|---|---|
| Serum albumin | Vitronectin | Serum albumin |
| Apolipoprotein A-II | Fibrinogen a- chain | a-2-HS-glycoprotein |
| Apolipoprotein A-II | Fibrinogen g- chain | Kinogen-1 |
| Alpha-fetoprotein | Complement C3 | |
| Kininogen-1 | ||
| Complement C3 | ||
| Hemoglobin fetal subunit beta | ||
| Prothrombin |
In conclusion, the above studies show that PC formation is highly complex in nature and its complete analysis require new methods. Each type of NP attracts particular proteins when incubated in a biological sample and the affinity of proteins towards NPs varies with the nature of NP surface. Each of NP surface properties such as size, charge and type of surface engineering play important roles in attracting various proteins. The PC signature is also dependent on other factors such as concentration of the NP and biological sample, type of biological sample, incubation time, temperature, and preparation methodology such as centrifugation conditions. The material coating a NP also plays an important role in adsorbing proteins from biospecimens that may result in a personalized corona signature. Personalized corona signature may aid in treatment modalities improving clinical outcome [150].
9. The Bio Corona: Need for evolving design
In order to fully exploit the interactions between NPs and proteins in biological species, there is a pressing need for improved surface engineered nanomaterials to bind specific targets and possibly self-direct to specific locations. In the human body, some proteins are expressed in one location, but are able to translocate and promote disease in a second location. For example, the protein human islet amyloid polypeptide (IAPP) is associated with type-2 diabetes and originates in the pancreas but is found in the brain where it promotes amyloidogenisis and Alzheimer’s disease [151]. Several oncogenic proteins associated with tumor progression circulate at all times in blood and other fluids. Scavenging of two such tumor markers, TIMP-1 and ICAM-1, in pancreatic cancer patient serum can determine their expression as a marker of disease stage [152]. The use of nanotechnology and the PC to analyse markers in this way is a simple non-invasive technique to monitor cancer-associated proteins. However, the choice of NP is critical since not all NPs adsorb proteins when exposed to biological fluids [101]. For example, Moyano et al., reported PC-free hydrophobic nanoparticles and demonstrated that 2nm gold core NPs fabricated with oligo ethylene glycol chain and zwitter ionic head groups associated with sulfo-betaine termini did not develop a PC and had extended stability in plasma [101]. Figure 5 is a straightforward outline for target identification using NP-PC methodology as well as other methods like genomic analysis, transcriptome analysis and proteomics. Careful design is critically important to ensure that NPs adsorb target proteins and/or avoid potentially confounding proteins. The PC on NPs can also stimulate uptake of NP into cells, and these uptake pathways can differ with cell type. For example, in the absence of serum, the internalization of polystyrene NPs by macrophages is via clathrin- and dynamin-dependent endocytosis whereas, in the presence of FBS alternative phagocytic processes predominate [153]. Recently, Oh et al. reported PC-free HER2 targeting silica NPs that effectively deliver cargo to tumors with minimal interaction with serum proteins and thus avoid phagocytosis by macrophages. This demonstrates that cloaking of NPs with suitable materials can preclude PC formation, prolong the NPs half-life, and ensure good therapeutic yields [154]. In another example of successful NP modification, BSA-coated gelatin-oleic NPs (GONs) had reduced uptake by human lung cancer cells A549, but enhanced uptake in HEK293 cells [155]. Thus, the PC composition is essential in determining a NPs interaction with proteins or membranes and its targeting to and uptake by both cancer and non-cancer cells.
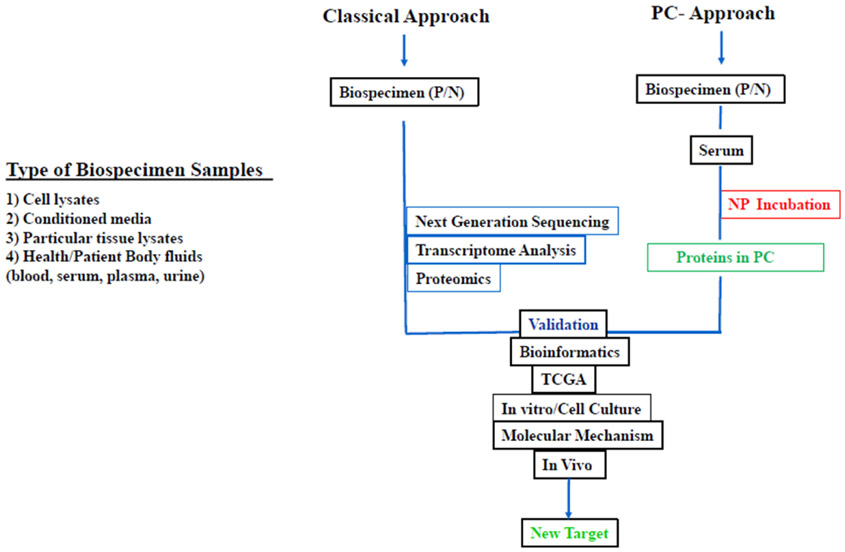
Figure 5.
Flow chart representing conventional techniques and the protein corona approach for discovery of new targets from patient biospecimen. In NP-PC approach, oncogenic and non-oncogenic proteins adsorb to therapeutic NPs surface upon incubation. Analysis of patient biospecimens using conventional approaches such as genomic sequencing, transcriptome, proteomics gives information about new targets but according to TCGA not all identified targets will have a role in disease progression.
The above described studies together all demonstrated that different NPs have affinity for different proteins present in the biological sample and NP surface nature such as size, charge and surface engineering play important role attracting various proteins from the given sample.
10. Future perspectives
Identifying new molecular targets involved in disease development and progression is essential for both early detection and devising novel therapeutic strategies to improve outcomes. Traditional approaches including gene microarray, RNA-seq, proteomics have been widely employed to identify disease specific or differentially regulated targets. Nanotechnology provides a complimentary approach; when introduced into biological systems, e.g. serum, plasma, urine, cell lysates or cellular secretome, nanoparticles capture biological molecules. The specific molecules captured depends on the surface properties of the nanoparticles as well as special affinity/avidity of biological molecules towards a particular surface type. Differential analysis of adsorbed molecules between normal healthy and malignant samples may lead to the identification of disease specific proteome components that are responsible for poor outcome. Personalized PCs may help in treatment decisions and improve patient prognosis [39, 150]. For example, this notion was recently verified using a cell culture-based assay that identified SMNDC1, PPA1 and PI15 as potential therapeutic targets in ovarian cancer. Figure 6 outlines how analysis of the NP-PC can be used to identify new molecular targets by comparing proteins captured on NPs after incubation with cancerous cells and non-cancerous cells. Comparative proteomics analysis followed by bioinformatics analysis will identify cancer and healthy control specific proteins. Further bioinformatics analysis may be used to create functional nodal networks with increasing network size based on network connectivity. Disrupting a nodal protein with higher connectivity is expected to exert stronger effects than those with lower connectivity, thus may identify effective regulators of tumor growth and metastasis. It is also possible to include specific targeting agents on the NP surface in order to trigger particular genes, proteins or pathways involved in disease progression. Lipophilic molecules, such as cationic triphenylphosphines (TPPs) selectively accumulate in mitochondria [156]. Thus, NP-TPP conjugate PC analysis could potentially provide valuable insight on novel mitochondrial proteins involved in signalling pathways for disease progression.
Figure 6.
I) Enrichment of proteins present on NP surface through Venn diagram after NP incubation in healthy and cancer cell lysates. II) Green color nodes represents proteins which are detected in NP-PC; different sizes of nodes indicate size of connected network among them derived from type of functional network. Different colored connecting lines indicate types of connectivity such as shared domain, co-expression, and biological pathways among proteins.
In conclusion, this review outlines several aspects related to improving PC formation including: 1) nanoparticle surface engineering, 2) analyzing the protein corona without altering its nature, and 3) prediction of new molecular regulator in the PC using proteomic analysis techniques and their validation for involvement in disease progression. Characterization and identification of PCs from patient samples before and after treatment has the potential to validate treatment outcome and therapeutic benefit. Protein-signatures (corona finger printing) obtained from the personalized PCs of specific patients and those patients outcomes could then be applied for treatment decisions in individuals with similar finger printing. Further development of nanotechnology in the biomedical field will lead to advanced and accurate methods that provide fundamental insights and expedite investigations utilizing nanoparticles in biomarker discovery as well as their future translation into clinical practice.
Figure 3.
Advantages of PC technique in identifying new targets in patient samples. Intrinsically, therapeutic nanoparticles attract disease associated proteins during their incubation with biological sample; adsorption of these proteins on the NP surface quenches their functional activity. Identification of these proteins constituting the NP-protein corona yield information about these unknown target proteins present in the patient sample. In contrast, while other techniques such as genomic sequencing, transcriptome analysis and proteomics also give information of new targets that are present in malignant cell lysates or patient sample, not all identified new targets will have a role in disease progression
Acknowledgements
This work was supported by CA213278 and CA136494 Team Science grants. Preparation of this publication was supported in part by the National Cancer Institute Cancer Center Support Grant P30CA225500 awarded to the University of Oklahoma Stephenson Cancer Center; services from the Office of Cancer Research were utilized.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019, CA Cancer J Clin, 69 (2019) 7–34. [DOI] [PubMed] [Google Scholar]
- [2].Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH, Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology, Nat Nanotechnol, 8 (2013) 772–781. [DOI] [PubMed] [Google Scholar]
- [3].Blackmore KM, Weerasinghe A, Holloway CMB, Majpruz V, Mirea L, O'Malley FP, Paroschy Harris C, Hendry A, Hey A, Kornecki A, Lougheed G, Maier BA, Marchand P, McCready D, Rand C, Raphael S, Segal-Nadler R, Sehgal N, Muradali D, Chiarelli AM, Comparison of wait times across the breast cancer treatment pathway among screened women undergoing organized breast assessment versus usual care, Can J Public Health, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haese A, Trooskens G, Steyaert S, Hessels D, Brawer M, Vlaeminck-Guillem V, Ruffion A, Tilki D, Schalken J, Groskopf J, Van Criekinge W, Multicenter optimization and validation of a 2-gene mRNA urine test for detection of clinically significant prostate cancer prior to initial prostate biopsy, J Urol, (2019) 101097JU0000000000000293. [DOI] [PubMed] [Google Scholar]
- [5].David Y, Ottaviano L, Park J, Iqbal S, Likhtshteyn M, Kumar S, Lyo H, Lewis AE, Lung BE, Frye JT, Huang L, Li E, Yang J, Martello L, Vignesh S, Miller JD, Follen M, Grossman EB, Confounders in Adenoma Detection at Initial Screening Colonoscopy: A Factor in the Assessment of Racial Disparities as a Risk for Colon Cancer, J Cancer Ther, 10 (2019) 269–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Colomban O, Tod M, Leary A, Ray-Coquard I, Lortholary A, Hardy-Bessard AC, Pfisterer J, Du Bois A, Kurzeder C, Burges A, Peron J, Freyer G, You B, Early Modeled Longitudinal CA-125 Kinetics and Survival of Ovarian Cancer Patients: A GINECO AGO MRC CTU Study, Clin Cancer Res, (2019). [DOI] [PubMed] [Google Scholar]
- [7].Benilova I, Karran E, De Strooper B, The toxic Abeta oligomer and Alzheimer's disease: an emperor in need of clothes, Nat Neurosci, 15 (2012) 349–357. [DOI] [PubMed] [Google Scholar]
- [8].Vasan RS, Biomarkers of cardiovascular disease: molecular basis and practical considerations, Circulation, 113 (2006) 2335–2362. [DOI] [PubMed] [Google Scholar]
- [9].Katsavos S, Anagnostouli M, Biomarkers in Multiple Sclerosis: An Up-to-Date Overview, Mult Scler Int, 2013 (2013) 340508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Giri K, Shameer K, Zimmermann MT, Saha S, Chakraborty PK, Sharma A, Arvizo RR, Madden BJ, McCormick DJ, Kocher JP, Bhattacharya R, Mukherjee P, Understanding protein-nanoparticle interaction: a new gateway to disease therapeutics, Bioconjug Chem, 25 (2014) 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H, Protein corona composition of superparamagnetic iron oxide nanoparticles with various physico-chemical properties and coatings, Sci Rep, 4 (2014) 5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stepien G, Moros M, Perez-Hernandez M, Monge M, Gutierrez L, Fratila RM, Las Heras M, Menao Guillen S, Puente Lanzarote JJ, Solans C, Pardo J, de la Fuente JM, Effect of Surface Chemistry and Associated Protein Corona on the Long-Term Biodegradation of Iron Oxide Nanoparticles In Vivo, ACS Appl Mater Interfaces, 10 (2018) 4548–4560. [DOI] [PubMed] [Google Scholar]
- [13].Ma Z, Bai J, Jiang X, Monitoring of the Enzymatic Degradation of Protein Corona and Evaluating the Accompanying Cytotoxicity of Nanoparticles, ACS Appl Mater Interfaces, 7 (2015) 17614–17622. [DOI] [PubMed] [Google Scholar]
- [14].Cedervall T, Lynch I, Foy M, Berggard T, Donnelly SC, Cagney G, Linse S, Dawson KA, Detailed identification of plasma proteins adsorbed on copolymer nanoparticles, Angew Chem Int Ed Engl, 46 (2007) 5754–5756. [DOI] [PubMed] [Google Scholar]
- [15].Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S, Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc Natl Acad Sci U S A, 104 (2007) 2050–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M, Understanding biophysicochemical interactions at the nano-bio interface, Nat Mater, 8 (2009) 543–557. [DOI] [PubMed] [Google Scholar]
- [17].Nguyen VH, Lee BJ, Protein corona: a new approach for nanomedicine design, Int J Nanomedicine, 12 (2017) 3137–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moghimi SM, Patel HM, Serum opsonins and phagocytosis of saturated and unsaturated phospholipid liposomes, Biochim Biophys Acta, 984 (1989) 384–387. [DOI] [PubMed] [Google Scholar]
- [19].Patel HM, Serum opsonins and liposomes: their interaction and opsonophagocytosis, Crit Rev Ther Drug Carrier Syst, 9 (1992) 39–90. [PubMed] [Google Scholar]
- [20].Leroux JC, Gravel P, Balant L, Volet B, Anner BM, Allemann E, Doelker E, Gurny R, Internalization of poly(D,L-lactic acid) nanoparticles by isolated human leukocytes and analysis of plasma proteins adsorbed onto the particles, J Biomed Mater Res, 28 (1994) 471–481. [DOI] [PubMed] [Google Scholar]
- [21].Gref R, Luck M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Muller RH, 'Stealth' corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption, Colloids Surf B Biointerfaces, 18 (2000) 301–313. [DOI] [PubMed] [Google Scholar]
- [22].Allemann E, Gravel P, Leroux JC, Balant L, Gurny R, Kinetics of blood component adsorption on poly(D,L-lactic acid) nanoparticles: evidence of complement C3 component involvement, J Biomed Mater Res, 37 (1997) 229–234. [DOI] [PubMed] [Google Scholar]
- [23].Caracciolo G, Farokhzad OC, Mahmoudi M, Biological Identity of Nanoparticles In Vivo: Clinical Implications of the Protein Corona, Trends Biotechnol, 35 (2017) 257–264. [DOI] [PubMed] [Google Scholar]
- [24].Nierenberg D, Khaled AR, Flores O, Formation of a protein corona influences the biological identity of nanomaterials, Rep Pract Oncol Radiother, 23 (2018) 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ke PC, Lin S, Parak WJ, Davis TP, Caruso F, A Decade of the Protein Corona, ACS Nano, 11 (2017) 11773–11776. [DOI] [PubMed] [Google Scholar]
- [26].Deng ZJ, Mortimer G, Schiller T, Musumeci A, Martin D, Minchin RF, Differential plasma protein binding to metal oxide nanoparticles, Nanotechnology, 20 (2009) 455101. [DOI] [PubMed] [Google Scholar]
- [27].Vogt C, Pernemalm M, Kohonen P, Laurent S, Hultenby K, Vahter M, Lehtio J, Toprak MS, Fadeel B, Proteomics Analysis Reveals Distinct Corona Composition on Magnetic Nanoparticles with Different Surface Coatings: Implications for Interactions with Primary Human Macrophages, PLoS One, 10 (2015) e0129008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hadjidemetriou M, Al-Ahmady Z, Kostarelos K, Time-evolution of in vivo protein corona onto blood-circulating PEGylated liposomal doxorubicin (DOXIL) nanoparticles, Nanoscale, 8 (2016) 6948–6957. [DOI] [PubMed] [Google Scholar]
- [29].Mahmoudi M, Abdelmonem AM, Behzadi S, Clement JH, Dutz S, Ejtehadi MR, Hartmann R, Kantner K, Linne U, Maffre P, Metzler S, Moghadam MK, Pfeiffer C, Rezaei M, Ruiz-Lozano P, Serpooshan V, Shokrgozar MA, Nienhaus GU, Parak WJ, Temperature: the “ignored” factor at the NanoBio interface, ACS Nano, 7 (2013) 6555–6562. [DOI] [PubMed] [Google Scholar]
- [30].Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE, Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy, Adv Drug Deliv Rev, 61 (2009) 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hadjidemetriou M, Al-Ahmady Z, Mazza M, Collins RF, Dawson K, Kostarelos K, In Vivo Biomolecule Corona around Blood-Circulating, Clinically Used and Antibody-Targeted Lipid Bilayer Nanoscale Vesicles, ACS Nano, 9 (2015) 8142–8156. [DOI] [PubMed] [Google Scholar]
- [32].Reiding KR, Vreeker GCM, Bondt A, Bladergroen MR, Hazes JMW, van der Burgt YEM, Wuhrer M, Dolhain R, Serum Protein N-Glycosylation Changes with Rheumatoid Arthritis Disease Activity during and after Pregnancy, Front Med (Lausanne), 4 (2017) 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sundsten T, Ostenson CG, Bergsten P, Serum protein patterns in newly diagnosed type 2 diabetes mellitus--influence of diabetic environment and family history of diabetes, Diabetes Metab Res Rev, 24 (2008) 148–154. [DOI] [PubMed] [Google Scholar]
- [34].Acharya SS, Dimichele DM, Rare inherited disorders of fibrinogen, Haemophilia, 14 (2008) 1151–1158. [DOI] [PubMed] [Google Scholar]
- [35].Rao G, Powell-Wiley TM, Ancheta I, Hairston K, Kirley K, Lear SA, North KE, Palaniappan L, Rosal MC, American L Heart Association Obesity Committee of the Council on, H. Cardiometabolic, Identification of Obesity and Cardiovascular Risk in Ethnically and Racially Diverse Populations: A Scientific Statement From the American Heart Association, Circulation, 132 (2015) 457–472. [DOI] [PubMed] [Google Scholar]
- [36].Riley CP, Zhang X, Nakshatri H, Schneider B, Regnier FE, Adamec J, Buck C, A large, consistent plasma proteomics data set from prospectively collected breast cancer patient and healthy volunteer samples, J Transl Med, 9 (2011) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Carboni D, Williams RK, Gasparri A, Ulivi G, Sukhatme GS, Rigidity-Preserving Team Partitions in Multiagent Networks, IEEE Trans Cybern, 45 (2015) 2640–2653. [DOI] [PubMed] [Google Scholar]
- [38].Pandey KB, Mishra N, Rizvi SI, Protein oxidation biomarkers in plasma of type 2 diabetic patients, Clin Biochem, 43 (2010) 508–511. [DOI] [PubMed] [Google Scholar]
- [39].Corbo C, Molinaro R, Tabatabaei M, Farokhzad OC, Mahmoudi M, Personalized protein corona on nanoparticles and its clinical implications, Biomater Sci, 5 (2017) 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Arvizo RR, Giri K, Moyano D, Miranda OR, Madden B, McCormick DJ, Bhattacharya R, Rotello VM, Kocher JP, Mukherjee P, Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles, PLoS One, 7 (2012) e33650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schrittwieser S, Pelaz B, Parak WJ, Lentijo-Mozo S, Soulantica K, Dieckhoff J, Ludwig F, Schotter J, Direct protein quantification in complex sample solutions by surface-engineered nanorod probes, Sci Rep, 7 (2017) 4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zheng T, Pierre-Pierre N, Yan X, Huo Q, Almodovar AJ, Valerio F, Rivera-Ramirez I, Griffith E, Decker DD, Chen S, Zhu N, Gold nanoparticle-enabled blood test for early stage cancer detection and risk assessment, ACS Appl Mater Interfaces, 7 (2015) 6819–6827. [DOI] [PubMed] [Google Scholar]
- [43].Monopoli MP, Aberg C, Salvati A, Dawson KA, Biomolecular coronas provide the biological identity of nanosized materials, Nature nanotechnology, 7 (2012) 779–786. [DOI] [PubMed] [Google Scholar]
- [44].Pareek V, Bhargava A, Bhanot V, Gupta R, Jain N, Panwar J, Formation and Characterization of Protein Corona Around Nanoparticles: A Review, J Nanosci Nanotechnol, 18 (2018) 6653–6670. [DOI] [PubMed] [Google Scholar]
- [45].Petros RA DJ, Strategies in the design of nanoparticles for therapeutic applications, Nat. Rev. Drug Discovery,9(2010) 615–627. [DOI] [PubMed] [Google Scholar]
- [46].Liang M, Lin IC, Whittaker MR, Minchin RF, Monteiro MJ, Toth I, Cellular Uptake of Densely Packed Polymer Coatings on Gold Nanoparticles, ACS Nano, 4 (2009) 403–413. [DOI] [PubMed] [Google Scholar]
- [47].Karmali PP, Simberg D, Interactions of nanoparticles with plasma proteins: implication on clearance and toxicity of drug delivery systems, Expert Opin Drug Delivery, 8 (2011) 343–357. [DOI] [PubMed] [Google Scholar]
- [48].Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE, Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy, Adv Drug Delivery Rev, 61 (2009) 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ehrenberg MS, Friedman AE, Finkelstein JN, Oberd√∂rster G.n., McGrath JL, The influence of protein adsorption on nanoparticle association with cultured endothelial cells, Biomaterials, 30 (2009) 603–610. [DOI] [PubMed] [Google Scholar]
- [50].Dobrovolskaia MA, McNeil SE, Immunological properties of engineered nanomaterials, Nat Nano, 2 (2007) 469–478. [DOI] [PubMed] [Google Scholar]
- [51].G.D. Dobrovolskaia MA, Weaver JL, Evaluation of nanoparticle immunotoxicity, Nat Nano, 4 (2009) 411–414. [DOI] [PubMed] [Google Scholar]
- [52].Chen D, Ganesh S, Wang W, Amiji M, Plasma protein adsorption and biological identity of systemically administered nanoparticles, Nanomedicine (Lond), 12 (2017) 2113–2135. [DOI] [PubMed] [Google Scholar]
- [53].Vroman L, Effect of absorbed proteins on the wettability of hydrophilic and hydrophobic solids, Nature, 196 (1962) 476–477. [DOI] [PubMed] [Google Scholar]
- [54].Pederzoli F, Tosi G, Vandelli MA, Belletti D, Forni F, Ruozi B, Protein corona and nanoparticles: how can we investigate on?, Wiley Interdiscip Rev Nanomed Nanobiotechnol, 9 (2017). [DOI] [PubMed] [Google Scholar]
- [55].Dell'Orco D, Lundqvist M, Oslakovic C, Cedervall T, Linse S, Modeling the time evolution of the nanoparticle-protein corona in a body fluid, PLoS One, 5 (2010) e10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Xia XR, Monteiro-Riviere NA, Riviere JE, An index for characterization of nanomaterials in biological systems, Nat Nanotechnol, 5 (2010) 671–675. [DOI] [PubMed] [Google Scholar]
- [57].Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, Deutsch EW, Domon B, Hancock W, He F, Hochstrasser D, Marko-Varga G, Salekdeh GH, Sechi S, Snyder M, Srivastava S, Uhlen M, Wu CH, Yamamoto T, Paik YK, Omenn GS, The human proteome project: current state and future direction, Mol Cell Proteomics, 10 (2011) M111 009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chandramouli K, Qian PY, Proteomics: challenges, techniques and possibilities to overcome biological sample complexity, Hum Genomics Proteomics, 2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH, Proteomics: Technologies and Their Applications, J Chromatogr Sci, 55 (2017) 182–196. [DOI] [PubMed] [Google Scholar]
- [60].Kearney P, Thibault P, Bioinformatics meets proteomics--bridging the gap between mass spectrometry data analysis and cell biology, J Bioinform Comput Biol, 1 (2003) 183–200. [DOI] [PubMed] [Google Scholar]
- [61].Jia L, Lu Y, Shao J, Liang XJ, Xu Y, Nanoproteomics: a new sprout from emerging links between nanotechnology and proteomics, Trends Biotechnol, 31 (2013) 99–107. [DOI] [PubMed] [Google Scholar]
- [62].Wilson SR, Vehus T, Berg HS, Lundanes E, Nano-LC in proteomics: recent advances and approaches, Bioanalysis, 7 (2015) 1799–1815. [DOI] [PubMed] [Google Scholar]
- [63].Ray S, Reddy PJ, Jain R, Gollapalli K, Moiyadi A, Srivastava S, Proteomic technologies for the identification of disease biomarkers in serum: advances and challenges ahead, Proteomics, 11 (2011) 2139–2161. [DOI] [PubMed] [Google Scholar]
- [64].Gonzalez-Gonzalez M, Jara-Acevedo R, Matarraz S, Jara-Acevedo M, Paradinas S, Sayagues JM, Orfao A, Fuentes M, Nanotechniques in proteomics: protein microarrays and novel detection platforms, Eur J Pharm Sci, 45 (2012) 499–506. [DOI] [PubMed] [Google Scholar]
- [65].Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy TM, Label-free biomarker detection from whole blood, Nat Nanotechnol, 5 (2010) 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Umehara S, Karhanek M, Davis RW, Pourmand N, Label-free biosensing with functionalized nanopipette probes, Proc Natl Acad Sci U S A, 106 (2009) 4611–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Miclaus T, Beer C, Chevallier J, Scavenius C, Bochenkov VE, Enghild JJ, Sutherland DS, Dynamic protein coronas revealed as a modulator of silver nanoparticle sulphidation in vitro, Nat Commun, 7 (2016) 11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Boldt K, Bruns OT, Gaponik N, Eychmuller A, Comparative examination of the stability of semiconductor quantum dots in various biochemical buffers, J Phys Chem B, 110 (2006) 1959–1963. [DOI] [PubMed] [Google Scholar]
- [69].Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K, The evolution of the protein corona around nanoparticles: a test study, ACS Nano, 5 (2011) 7503–7509. [DOI] [PubMed] [Google Scholar]
- [70].Jin H, Webb-Robertson BJ, Peterson ES, Tan R, Bigelow DJ, Scholand MB, Hoidal JR, Pounds JG, Zangar RC, Smoking, COPD, and 3-nitrotyrosine levels of plasma proteins, Environ Health Perspect, 119 (2011) 1314–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM, Multitarget stool DNA testing for colorectal-cancer screening, N Engl J Med, 370 (2014) 1287–1297. [DOI] [PubMed] [Google Scholar]
- [72].Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark-Langone KM, Snable J, Watson D, Falzarano SM, Magi-Galluzzi C, Klein EA, Quale C, Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies, BMC Genomics, 14 (2013) 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kori M, Yalcin Arga K, Potential biomarkers and therapeutic targets in cervical cancer: Insights from the meta-analysis of transcriptomics data within network biomedicine perspective, PLoS One, 13 (2018) e0200717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Casamassimi A, Federico A, Rienzo M, Esposito S, Ciccodicola A, Transcriptome Profiling in Human Diseases: New Advances and Perspectives, Int J Mol Sci, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shendure J, The beginning of the end for microarrays?, Nat Methods, 5 (2008) 585–587. [DOI] [PubMed] [Google Scholar]
- [76].Kumar A, Kankainen M, Parsons A, Kallioniemi O, Mattila P, Heckman CA, The impact of RNA sequence library construction protocols on transcriptomic profiling of leukemia, BMC Genomics, 18 (2017) 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L, Characterization of human plasma-derived exosomal RNAs by deep sequencing, BMC Genomics, 14 (2013) 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Jones MB, Krutzsch H, Shu H, Zhao Y, Liotta LA, Kohn EC, Petricoin EF 3rd, Proteomic analysis and identification of new biomarkers and therapeutic targets for invasive ovarian cancer, Proteomics, 2 (2002) 76–84. [PubMed] [Google Scholar]
- [79].Dou Y, Lv Y, Zhou X, He L, Liu L, Li P, Sun Y, Wang M, Gao M, Wang C, Antibody-sandwich ELISA analysis of a novel blood biomarker of CST4 in gastrointestinal cancers, Onco Targets Ther, 11 (2018) 1743–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Arya SK, Estrela P, Recent Advances in Enhancement Strategies for Electrochemical ELISA-Based Immunoassays for Cancer Biomarker Detection, Sensors (Basel), 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Paananen J, Fortino V, An omics perspective on drug target discovery platforms, Brief Bioinform, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Swiatly A, Plewa S, Matysiak J, Kokot ZJ, Mass spectrometry-based proteomics techniques and their application in ovarian cancer research, J Ovarian Res, 11 (2018) 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lee PY, Chin SF, Low TY, Jamal R, Probing the colorectal cancer proteome for biomarkers: Current status and perspectives, J Proteomics, 187 (2018) 93–105. [DOI] [PubMed] [Google Scholar]
- [84].Jones LH, Neubert H, Clinical chemoproteomics-Opportunities and obstacles, Sci Transl Med, 9 (2017). [DOI] [PubMed] [Google Scholar]
- [85].Nguyen C, West GM, Geoghegan KF, Emerging Methods in Chemoproteomics with Relevance to Drug Discovery, Methods Mol Biol, 1513 (2017) 11–22. [DOI] [PubMed] [Google Scholar]
- [86].Hajipour MJ, Ghasemi F, Aghaverdi H, Raoufi M, Linne U, Atyabi F, Nabipour I, Azhdarzadeh M, Derakhshankhah H, Lotfabadi A, Bargahi A, Alekhamis Z, Aghaie A, Hashemi E, Tafakhori A, Aghamollaii V, Mashhadi MM, Sheibani S, Vali H, Mahmoudi M, Sensing of Alzheimer's Disease and Multiple Sclerosis Using Nano-Bio Interfaces, J Alzheimers Dis, 59 (2017) 1187–1202. [DOI] [PubMed] [Google Scholar]
- [87].Rana S, Singla AK, Bajaj A, Elci SG, Miranda OR, Mout R, Yan B, Jirik FR, Rotello VM, Array-based sensing of metastatic cells and tissues using nanoparticle-fluorescent protein conjugates, ACS Nano, 6 (2012) 8233–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UH, Rotello VM, Detection and identification of proteins using nanoparticle-fluorescent polymer 'chemical nose' sensors, Nat Nanotechnol, 2 (2007) 318–323. [DOI] [PubMed] [Google Scholar]
- [89].Anthony KC, You C, Piehler J, Pomeranz Krummel DA, High-affinity gold nanoparticle pin to label and localize histidine-tagged protein in macromolecular assemblies, Structure, 22 (2014) 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [90].Nietzold C, Lisdat F, Fast protein detection using absorption properties of gold nanoparticles, Analyst, 137 (2012) 2821–2826. [DOI] [PubMed] [Google Scholar]
- [91].Nash MA, Yager P, Hoffman AS, Stayton PS, Mixed Stimuli-Responsive Magnetic and Gold Nanoparticle System for Rapid Purification, Enrichment, and Detection of Biomarkers, Bioconjugate Chemistry, 21 (2010) 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc Natl Acad Sci U S A, 105 (2008) 14265–14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lundqvist M, Augustsson C, Lilja M, Lundkvist K, Dahlback B, Linse S, Cedervall T, The nanoparticle protein corona formed in human blood or human blood fractions, PLoS One, 12 (2017) e0175871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hu Z, Zhang H, Zhang Y, Wu R, Zou H, Nanoparticle size matters in the formation of plasma protein coronas on Fe3O4 nanoparticles, Colloids Surf B Biointerfaces, 121 (2014) 354–361. [DOI] [PubMed] [Google Scholar]
- [95].Elci SG, Jiang Y, Yan B, Kim ST, Saha K, Moyano DF, Yesilbag Tonga G, Jackson LC, Rotello VM, Vachet RW, Surface Charge Controls the Suborgan Biodistributions of Gold Nanoparticles, ACS Nano, 10 (2016) 5536–5542. [DOI] [PubMed] [Google Scholar]
- [96].Cho EC, Xie J, Wurm PA, Xia Y, Understanding the role of surface charges in cellular adsorption versus internalization by selectively removing gold nanoparticles on the cell surface with a I2/KI etchant, Nano Lett, 9 (2009) 1080–1084. [DOI] [PubMed] [Google Scholar]
- [97].Huhn D, Kantner K, Geidel C, Brandholt S, De Cock I, Soenen SJ, Rivera Gil P, Montenegro JM, Braeckmans K, Mullen K, Nienhaus GU, Klapper M, Parak WJ, Polymer-coated nanoparticles interacting with proteins and cells: focusing on the sign of the net charge, ACS Nano, 7 (2013) 3253–3263. [DOI] [PubMed] [Google Scholar]
- [98].Shi X, Thomas TP, Myc LA, Kotlyar A, Baker JR Jr., Synthesis, characterization, and intracellular uptake of carboxyl-terminated poly(amidoamine) dendrimer-stabilized iron oxide nanoparticles, Phys Chem Chem Phys, 9 (2007) 5712–5720. [DOI] [PubMed] [Google Scholar]
- [99].Kim B, Han G, Toley BJ, Kim CK, Rotello VM, Forbes NS, Tuning payload delivery in tumour cylindroids using gold nanoparticles, Nat Nanotechnol, 5 (2010) 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Verma A, Stellacci F, Effect of surface properties on nanoparticle-cell interactions, Small, 6 (2010) 12–21. [DOI] [PubMed] [Google Scholar]
- [101].Moyano DF, Saha K, Prakash G, Yan B, Kong H, Yazdani M, Rotello VM, Fabrication of corona-free nanoparticles with tunable hydrophobicity, ACS Nano, 8 (2014) 6748–6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Caracciolo G, Liposome-protein corona in a physiological environment: challenges and opportunities for targeted delivery of nanomedicines, Nanomedicine : nanotechnology, biology, and medicine, 11 (2015) 543–557. [DOI] [PubMed] [Google Scholar]
- [103].Palchetti S, Colapicchioni V, Digiacomo L, Caracciolo G, Pozzi D, Capriotti AL, La Barbera G, Lagana A, The protein corona of circulating PEGylated liposomes, Biochim Biophys Acta, 1858 (2016) 189–196. [DOI] [PubMed] [Google Scholar]
- [104].Bertrand N, Grenier P, Mahmoudi M, Lima EM, Appel EA, Dormont F, Lim JM, Karnik R, Langer R, Farokhzad OC, Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics, Nat Commun, 8 (2017) 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Huang H, Lai W, Cui M, Liang L, Lin Y, Fang Q, Liu Y, Xie L, An Evaluation of Blood Compatibility of Silver Nanoparticles, Sci Rep, 6 (2016) 25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kwon T, Woo HJ, Kim YH, Lee HJ, Park KH, Park S, Youn B, Optimizing hemocompatibility of surfactant-coated silver nanoparticles in human erythrocytes, J Nanosci Nanotechnol, 12 (2012) 6168–6175. [DOI] [PubMed] [Google Scholar]
- [107].Mahmoudi M, Lohse SE, Murphy CJ, Fathizadeh A, Montazeri A, Suslick KS, Variation of protein corona composition of gold nanoparticles following plasmonic heating, Nano Lett, 14 (2014) 6–12. [DOI] [PubMed] [Google Scholar]
- [108].Wang H, Shang L, Maffre P, Hohmann S, Kirschhofer F, Brenner-Weiss G, Nienhaus GU, The Nature of a Hard Protein Corona Forming on Quantum Dots Exposed to Human Blood Serum, Small, 12 (2016) 5836–5844. [DOI] [PubMed] [Google Scholar]
- [109].Yu J, Kim HJ, Go MR, Bae SH, Choi SJ, ZnO Interactions with Biomatrices: Effect of Particle Size on ZnO-Protein Corona, Nanomaterials (Basel), 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Borgognoni CF, Mormann M, Qu Y, Schafer M, Langer K, Ozturk C, Wagner S, Chen C, Zhao Y, Fuchs H, Riehemann K, Reaction of human macrophages on protein corona covered TiO(2) nanoparticles, Nanomedicine : nanotechnology, biology, and medicine, 11 (2015) 275–282. [DOI] [PubMed] [Google Scholar]
- [111].Di Silvio D, Rigby N, Bajka B, Mayes A, Mackie A, Baldelli Bombelli F, Technical tip: high-resolution isolation of nanoparticle-protein corona complexes from physiological fluids, Nanoscale, 7 (2015) 11980–11990. [DOI] [PubMed] [Google Scholar]
- [112].Kavosi B, Salimi A, Hallaj R, Amani K, A highly sensitive prostate-specific antigen immunosensor based on gold nanoparticles/PAMAM dendrimer loaded on MWCNTS/chitosan/ionic liquid nanocomposite, Biosens Bioelectron, 52 (2014) 20–28. [DOI] [PubMed] [Google Scholar]
- [113].Nash MA, Yager P, Hoffman AS, Stayton PS, Mixed stimuli-responsive magnetic and gold nanoparticle system for rapid purification, enrichment, and detection of biomarkers, Bioconjug Chem, 21 (2010) 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA, Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles, J Am Chem Soc, 133 (2011) 2525–2534. [DOI] [PubMed] [Google Scholar]
- [115].Maffre P, Nienhaus K, Amin F, Parak WJ, Nienhaus GU, Characterization of protein adsorption onto FePt nanoparticles using dual-focus fluorescence correlation spectroscopy, Beilstein J Nanotechnol, 2 (2011) 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Jiang X, Weise S, Hafner M, Rocker C, Zhang F, Parak WJ, Nienhaus GU, Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding, J R Soc Interface, 7 Suppl 1 (2010) S5–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Rocker C, Potzl M, Zhang F, Parak WJ, Nienhaus GU, A quantitative fluorescence study of protein monolayer formation on colloidal nanoparticles, Nat Nanotechnol, 4 (2009) 577–580. [DOI] [PubMed] [Google Scholar]
- [118].Fedeli C, Segat D, Tavano R, Bubacco L, De Franceschi G, de Laureto PP, Lubian E, Selvestrel F, Mancin F, Papini E, The functional dissection of the plasma corona of SiO(2)-NPs spots histidine rich glycoprotein as a major player able to hamper nanoparticle capture by macrophages, Nanoscale, 7 (2015) 17710–17728. [DOI] [PubMed] [Google Scholar]
- [119].Docter D, Westmeier D, Markiewicz M, Stolte S, Knauer SK, Stauber RH, The nanoparticle biomolecule corona: lessons learned - challenge accepted?, Chem Soc Rev, 44 (2015) 6094–6121. [DOI] [PubMed] [Google Scholar]
- [120].Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA, What the cell “sees” in bionanoscience, J Am Chem Soc, 132 (2010) 5761–5768. [DOI] [PubMed] [Google Scholar]
- [121].Saha K, Rahimi M, Yazdani M, Kim ST, Moyano DF, Hou S, Das R, Mout R, Rezaee F, Mahmoudi M, Rotello VM, Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona, ACS Nano, 10 (2016) 4421–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Pozzi D, Caracciolo G, Digiacomo L, Colapicchioni V, Palchetti S, Capriotti AL, Cavaliere C, Zenezini Chiozzi R, Puglisi A, Lagana A, The biomolecular corona of nanoparticles in circulating biological media, Nanoscale, 7 (2015) 13958–13966. [DOI] [PubMed] [Google Scholar]
- [123].Sakulkhu U, Maurizi L, Mahmoudi M, Motazacker M, Vries M, Gramoun A, Ollivier Beuzelin MG, Vallee JP, Rezaee F, Hofmann H, Ex situ evaluation of the composition of protein corona of intravenously injected superparamagnetic nanoparticles in rats, Nanoscale, 6 (2014) 11439–11450. [DOI] [PubMed] [Google Scholar]
- [124].Caputo D, Cartillone M, Cascone C, Pozzi D, Digiacomo L, Palchetti S, Caracciolo G, Coppola R, Improving the accuracy of pancreatic cancer clinical staging by exploitation of nanoparticle-blood interactions: A pilot study, Pancreatology, 18 (2018) 661–665. [DOI] [PubMed] [Google Scholar]
- [125].Chen F, Wang G, Griffin JI, Brenneman B, Banda NK, Holers VM, Backos DS, Wu L, Moghimi SM, Simberg D, Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo, Nat Nanotechnol, 12 (2017) 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Capriotti AL, Caracciolo G, Caruso G, Foglia P, Pozzi D, Samperi R, Lagana A, DNA affects the composition of lipoplex protein corona: a proteomics approach, Proteomics, 11 (2011) 3349–3358. [DOI] [PubMed] [Google Scholar]
- [127].Sobczynski DJ, Charoenphol P, Heslinga MJ, Onyskiw PJ, Namdee K, Thompson AJ, Eniola-Adefeso O, Plasma protein corona modulates the vascular wall interaction of drug carriers in a material and donor specific manner, PLoS One, 9 (2014) e107408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ruiz A, Alpizar A, Beola L, Rubio C, Gavilan H, Marciello M, Rodriguez-Ramiro I, Ciordia S, Morris CJ, Morales MDP, Understanding the Influence of a Bifunctional Polyethylene Glycol Derivative in Protein Corona Formation around Iron Oxide Nanoparticles, Materials (Basel), 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Solorio-Rodriguez A, Escamilla-Rivera V, Uribe-Ramirez M, Chagolla A, Winkler R, Garcia-Cuellar CM, De Vizcaya-Ruiz A, A comparison of the human and mouse protein corona profiles of functionalized SiO2 nanocarriers, Nanoscale, 9 (2017) 13651–13660. [DOI] [PubMed] [Google Scholar]
- [130].Garcia-Alvarez R, Hadjidemetriou M, Sanchez-Iglesias A, Liz-Marzan LM, Kostarelos K, In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape, Nanoscale, 10 (2018) 1256–1264. [DOI] [PubMed] [Google Scholar]
- [131].Angelov B, Angelova A, Filippov SK, Drechsler M, Stepanek P, Lesieur S, Multicompartment lipid cubic nanoparticles with high protein upload: millisecond dynamics of formation, ACS Nano, 8 (2014) 5216–5226. [DOI] [PubMed] [Google Scholar]
- [132].Chen Y, Ma P, Gui S, Cubic and hexagonal liquid crystals as drug delivery systems, Biomed Res Int, 2014 (2014) 815981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Angelova A, Angelov B, Mutafchieva R, Lesieur S, Couvreur P, Self-assembled multicompartment liquid crystalline lipid carriers for protein, peptide, and nucleic acid drug delivery, Acc Chem Res, 44 (2011) 147–156. [DOI] [PubMed] [Google Scholar]
- [134].Angelova A, Drechsler M, Garamus VM, Angelov B, Liquid Crystalline Nanostructures as PEGylated Reservoirs of Omega-3 Polyunsaturated Fatty Acids: Structural Insights toward Delivery Formulations against Neurodegenerative Disorders, ACS Omega, 3 (2018) 3235–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Vidaurre-Agut C, Rivero-Buceta E, Romani-Cubells E, Clemments AM, Vera-Donoso CD, Landry CC, Botella P, Protein Corona over Mesoporous Silica Nanoparticles: Influence of the Pore Diameter on Competitive Adsorption and Application to Prostate Cancer Diagnostics, ACS Omega, 4 (2019) 8852–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Huo Q, Colon J, Cordero A, Bogdanovic J, Baker CH, Goodison S, Pensky MY, A facile nanoparticle immunoassay for cancer biomarker discovery, J Nanobiotechnology, 9 (2011) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Mahmoudi M, Sant S, Wang B, Laurent S, Sen T, Superparamagnetic iron oxide nanoparticles (SPIONs): development, surface modification and applications in chemotherapy, Adv Drug Deliv Rev, 63 (2011) 24–46. [DOI] [PubMed] [Google Scholar]
- [138].Utkin Y, Brain and Quantum Dots: Benefits of Nanotechnology for Healthy and Diseased Brain, Cent Nerv Syst Agents Med Chem, (2018). [DOI] [PubMed] [Google Scholar]
- [139].Quantum dots on your mind, Nat Nanotechnol, 13 (2018) 269. [DOI] [PubMed] [Google Scholar]
- [140].Stan MS, Cinteza LO, Petrescu L, Mernea MA, Calborean O, Mihailescu DF, Sima C, Dinischiotu A, Dynamic analysis of the interactions between Si/SiO2 quantum dots and biomolecules for improving applications based on nano-bio interfaces, Sci Rep, 8 (2018) 5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Wang H, Shang L, Maffre P, Hohmann S, Kirschhofer F, Brenner-Weiss G, Nienhaus GU, The Nature of a Hard Protein Corona Forming on Quantum Dots Exposed to Human Blood Serum, Small, (2016). [DOI] [PubMed] [Google Scholar]
- [142].Pelaz B, del Pino P, Maffre P, Hartmann R, Gallego M, Rivera-Fernandez S, de la Fuente JM, Nienhaus GU, Parak WJ, Surface Functionalization of Nanoparticles with Polyethylene Glycol: Effects on Protein Adsorption and Cellular Uptake, ACS Nano, 9 (2015) 6996–7008. [DOI] [PubMed] [Google Scholar]
- [143].Siddiqi KS, Ur Rahman A, Tajuddin A Husen, Properties of Zinc Oxide Nanoparticles and Their Activity Against Microbes, Nanoscale Res Lett, 13 (2018) 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Krol A, Pomastowski P, Rafinska K, Railean-Plugaru V, Buszewski B, Zinc oxide nanoparticles: Synthesis, antiseptic activity and toxicity mechanism, Adv Colloid Interface Sci, 249 (2017) 37–52. [DOI] [PubMed] [Google Scholar]
- [145].Choi SJ, Choy JH, Biokinetics of zinc oxide nanoparticles: toxicokinetics, biological fates, and protein interaction, Int J Nanomedicine, 9 Suppl 2 (2014) 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Bondarenko O, Juganson K, Ivask A, Kasemets K, Mortimer M, Kahru A, Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review, Arch Toxicol, 87 (2013) 1181–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Dhupal M, Oh JM, Tripathy DR, Kim SK, Koh SB, Park KS, Immunotoxicity of titanium dioxide nanoparticles via simultaneous induction of apoptosis and multiple toll-like receptors signaling through ROS-dependent SAPK/JNK and p38 MAPK activation, Int J Nanomedicine, 13 (2018) 6735–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Sager TM, Kommineni C, Castranova V, Pulmonary response to intratracheal instillation of ultrafine versus fine titanium dioxide: role of particle surface area, Part Fibre Toxicol, 5 (2008) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Rossi EM, Pylkkanen L, Koivisto AJ, Vippola M, Jensen KA, Miettinen M, Sirola K, Nykasenoja H, Karisola P, Stjernvall T, Vanhala E, Kiilunen M, Pasanen P, Makinen M, Hameri K, Joutsensaari J, Tuomi T, Jokiniemi J, Wolff H, Savolainen K, Matikainen S, Alenius H, Airway exposure to silica-coated TiO2 nanoparticles induces pulmonary neutrophilia in mice, Toxicol Sci, 113 (2010) 422–433. [DOI] [PubMed] [Google Scholar]
- [150].Garcia Vence M, Chantada-Vazquez MDP, Vazquez-Estevez S, Manuel Cameselle-Teijeiro J, Bravo SB, Nunez C, Potential clinical applications of the personalized, disease-specific protein corona on nanoparticles, Clin Chim Acta, 501 (2020) 102–111. [DOI] [PubMed] [Google Scholar]
- [151].Fawver JN, Ghiwot Y, Koola C, Carrera W, Rodriguez-Rivera J, Hernandez C, Dineley KT, Kong Y, Li J, Jhamandas J, Perry G, Murray IV, Islet amyloid polypeptide (IAPP): a second amyloid in Alzheimer's disease, Curr Alzheimer Res, 11 (2014) 928–940. [DOI] [PubMed] [Google Scholar]
- [152].Pan S, Brentnall TA, Chen R, Proteomics analysis of bodily fluids in pancreatic cancer, Proteomics, 15 (2015) 2705–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lunov O, Syrovets T, Loos C, Beil J, Delacher M, Tron K, Nienhaus GU, Musyanovych A, Mailander V, Landfester K, Simmet T, Differential uptake of functionalized polystyrene nanoparticles by human macrophages and a monocytic cell line, ACS Nano, 5 (2011) 1657–1669. [DOI] [PubMed] [Google Scholar]
- [154].Oh JY, Kim HS, Palanikumar L, Go EM, Jana B, Park SA, Kim HY, Kim K, Seo JK, Kwak SK, Kim C, Kang S, Ryu JH, Cloaking nanoparticles with protein corona shield for targeted drug delivery, Nat Commun, 9 (2018) 4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Nguyen VH, Meghani NM, Amin HH, Tran TTD, Tran PHL, Park C, Lee BJ, Modulation of serum albumin protein corona for exploring cellular behaviors of fattigation-platform nanoparticles, Colloids Surf B Biointerfaces, 170 (2018) 179–186. [DOI] [PubMed] [Google Scholar]
- [156].Han M, Vakili MR, Soleymani Abyaneh H, Molavi O, Lai R, Lavasanifar A, Mitochondrial delivery of doxorubicin via triphenylphosphine modification for overcoming drug resistance in MDA-MB-435/DOX cells, Mol Pharm, 11 (2014) 2640–2649. [DOI] [PubMed] [Google Scholar]