Abstract
Osteosarcoma (OS) is a cancerous tumor in a bone. We aimed to identify the critical genes involved in OS progression, and then try to elucidate the molecular mechanisms of this disease. The microarray data of GSE32395 was used for the present study. We analyzed differentially expressed genes (DEGs) in OS cells compared with control group by Student’s t-test. The significant enriched gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathways were analyzed for upregulated genes and downregulated genes, respectively. In addition, a protein-protein interaction (PPI) network was constructed. GO and KEGG enrichment analyses were conducted for genes in the PPI network. In total, 183 DEGs, including 100 upregulated DEGs and 83 downregulated DEGs were screened. The upregulated DEGs were significantly enriched in 2 KEGG pathways, such as “Glycosaminoglycan biosynthesis-chondroitin sulfate” and the downregulated DEGs were significantly enriched in 12 pathways, including “cell adhesion molecules,” “pentose phosphate pathway” and “allograft rejection.” GO enrichment analysis indicated that the upregulated DEGs were significantly involved in biological process, such as “multicellular organismal metabolic process” and “limb morphogenesis,” while the downregulated DEGs were significantly enriched in biological process, such as “Positive regulation of pathway-restricted SMAD protein phosphorylation.” The PPI network included 84 interactions and 51 nodes. The “glycosaminoglycan biosynthesis-chondroitin sulfate pathway,” “microtubule motor activityfunction,” and “regulation of mitosis process” were significantly enriched by genes in PPI network. In particular, CENPE, PRC1, TTK, and PLK4 had higher degrees in the PPI network. The interactions between TTK and PLK4 as well as CENPE and PRC1 may involve in the OS development. These 4 genes might be possible biomarkers for the treatment and diagnosis of OS.
Keywords: osteosarcoma, molecular mechanisms, differentially expressed genes
Introduction
Osteosarcoma (OS), originating from osteoblasts, is the most common bone malignant tumor, which mainly afflicts young adults or children.1 Currently, the main therapy for OS is conventional chemotherapy, but metastatic OS exhibits resistance to conventional chemotherapy.2 Early surgical resection combined with multi-agent chemotherapy could lead to a long-term survivor, while patients with metastatic OS had a poor prognosis with survival rate of less than 20%.3 Consequently, researches on the molecular mechanisms of OS may helpful for the development of novel target therapies for OS patients.
Considerable researches have been conducted to investigate the molecular mechanism of OS and several genes that play important roles have been identified. For example, glycogen synthase kinase-3β (GSK-3β) was found to play oncogenic effect on OS cells, and that GSK-3β repression can restrain the pathway of nuclear factor-κB (NF-κB), which can lead to the apoptosis of OS cells.4 Pi et al. found that Aurora-B might promote the progression of OS activating the PTK2/PI3K/Akt-NF-κB pathway.5 Besides, cyclin-dependent kinases (CDKs) are important for cell division and cell cycle regulation, and play important roles in OS development through affecting numerous pathways, such as those of cell cycle control.6 Moreover, it has been shown that CDK4 and CDK9 were found have potential to be prognostic marker and therapeutic target in OS.7,8 Overall, the pathogenesis of OS is multifactorial and thus more attentions upon it are needed.
In this current study, in order to have a better understanding of OS, differentially expressed genes (DEGs) in OS cells were identified. Meanwhile, we carried out of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses for the DEGs. In addition, we built a protein-protein interaction (PPI) network to investigate the pivotal DEGs related with the progression of OS in-depth. Finally, the key genes in PPI network were validated in 4 OS cell lines. These key DEGs could be beneficial for uncovering the molecular mechanisms of OS and might be potential targets for the therapy of OS.
Materials and Methods
Sources of Data
The gene expression profile of the GSE32395 dataset including 7 OS cell lines (HOS osteosarcoma cell line CRL-1543, HOS-58 osteosarcoma cell line, U2-OS osteosarcoma cell line HTB-96, Saos osteosarcoma cell line HTB-85, MNNG/HOS osteosarcoma cell line CRL-1547, SJSA osteosarcoma cell line CRL-2098, and MG-63 osteosarcoma cell line CRL-1427, OS group) and 2 control cells (L87/4 human stem cells and hFOB 1.19 human osteoblasts, control group) was downloaded from Gene Expression Omnibus (GEO) database, the platform of which was GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version].
Data Preprocessing and Analysis of DEGs
The gene expression matrix was obtained by the preprocessing of the raw data as performed in Bioconductor AFFY package.9 The data were preprocessed by the Robust Multichip Averaging (RMA) algorithm.10 Next, identification of DEGs between the OS and the control group was carried out by 2-tailed Student’s t-tests (P < 0.05, |log2 fold change (FC)| ≥ 2).
Gene Functional Annotation
Transcription factor (TF) was identified by the functional annotation of DEGs using the TRANSFAC database (http://gene-regulation.com/pub/databases.html).11 Furthermore, tumor suppressor genes (TSGs) and oncogenes were also screened through the database of TSG12 and Tumor Associated Genes (TAG).13
KEGG Pathway and GO Functional Enrichment Analyses of DEGs
KEGG pathway14 and GO functional15 analyses were carried out to screen important pathways and the functional terms of the upregulated genes and downregulated genes, respectively, using Database for Annotation, Visualization and Integrated Discovery16 with a P value of < 0.05.
PPI Network Construction
PPI network was built using search tool for the retrieval of interacting genes (STRING, version 9.1),17 and visualized in Cytoscape. Only interactions with a combined score > 0.4 were hold. The proteins with higher degree of interactions were regarded as the hub nodes.
Real-Time PCR Verification of Key Genes
Four kinds of human OS cell lines, including MG63, HOS, Saos and U2OS cells, and the human mesenchymal stem cells were purchased from iCell Bioscience Inc. (Shanghai, China). Cells were cultured in minimum essential medium supplemented with 10% fetal bovine serum, 1% streptomycin (100 µg/ml) and penicillin (100 U/ml) at 37°C and 5% CO2.
Total RNA from cells were extracted from cells using Trizol reagent (Thermo Fisher Scientific Inc.). cDNA was synthesized from total RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc.). qPCR was then performed on ABI7500 with SYBR Green qPCR Master Mix (Thermo Fisher Scientific Inc.). GAPDH served as internal control. Primers are given in Table 1. The expression of gene was calculated using the 2−ΔΔCt method.
Table 1.
Primers for Quantitative Real-Time PCR.
| Primer | Direction | Sequence (5’-3’) |
|---|---|---|
| CENPE | Forward | AAGACCGAGCTTTCTTACAAGA |
| Reverse | CTACAGTTTGCAGCGTAGAATC’ | |
| PRC1 | Forward | CCTATTCTGAGTTTGCGAAGGA |
| Reverse | TGATCAGGGCTTCTCAGGAC | |
| TTK | Forward | TGGCCAACCTGCCTGTTT |
| Reverse | AATGCATTCATTTGCTGAAGAAGA | |
| PLK4 | Forward | CCTTATCACCTCCTCCTTC |
| Reverse | CCAAGTCCTTCATTTGTAACC | |
| GAPDH | Forward | TGACAACTTTGGTATCGTGGAAGG |
| Reverse | AGGCAGGGATGATGTTCTGGAGAG |
Statistical Analysis
Experimental data were represented as mean ± standard deviation. Comparisons were performed using 1-way analysis of variance with post-hoc test of Bonferroni test in Graphpad Prism. P < 0.05 was regarded as significant level.
Results
DEGs in OS Cell Lines Compared With Control Cell
After analysis, 183 DEGs were identified, including 100 upregulated genes and 83 downregulated genes (Table 2). Among these DEGs, 5 upregulated TFs (ZHX1, PBX1, NR1D1, MAFB and LEF1) and 2 downregulated TFs (SIM2 and HMGB2), as well as 8 upregulated TAGs (6 TSGs and 2 uncertain genes for tumor development) and 7 downregulated TSGs were further identified. The upregulated TSGs were UNC5B, PARK2, MTSS1, MIR181B1, FBXO32 and ADAMTS9; the other upregulated genes were MAFB and CDR. The downregulated TSGs were TPM1, RB1, PTPRJ, PPP2CB, PCDH10, IGFBP7 and DAB2.
Table 2.
The List of 188 Differentially Expressed Genes.
| Category | Genes |
|---|---|
| The upregulated genes | ABCA1, ABHD17A, ADAMTS9, APLP1, ARL4C, ARRDC3, BRINP1, CACNB4, CC2D2A, CD163L1, CDR1, CHMP4C, CHST11, COL11A2, DAPK2, DCLK1, DDIT4, DOK6, DYNC1I1, EPHX1, EYA2, FAM27C, FAM49A, FBXO32, FOLR1, FOXRED2, FRMD4B, GATS, GHR, GSE1, HSPB8, IFNL3, INSR, IRX3, KAZALD1, KIAA1161, KLRC2, LAMA4, LARGE, LDLRAD4, LEF1, LOC100506127, LRRC15, MAFB, MAN1A1, MGMT, MIR181B1, MIR21, MTRF1 L, MTSS1, NBPF4, NCALD, NCAM1, NHSL1, NR1D1, NUPR1, PARK2, PBX1, PCDHGA4, PCDHGB1, PCDHGB4, PCDHGB6, PCDHGC3, PCOLCE2, PCSK5, PPFIBP2, PRB1, PSG1, RABGAP1 L, RDH10, RFTN2, RGCC, RGS3, SCUBE1, SEPT5, SERINC5, SESN3, SGCD, SLC4A8, SLC7A8, SNORA42, SNORD104, ST3GAL1, STEAP1B, SYBU, SYNPO, SYTL2, TENC1, TNFRSF19, TOX, TRPS1, TUBA4A, UNC5B, UST, UTY, WISP1, WLS, ZDHHC23, ZHX1, ZNF608 |
| The downregulated genes | AKAP2, ALCAM, ANLN, ANXA3, BNC1, BTBD10, CD248, CENPE, CEP170, COL4A1, DAB2, DBF4, DIAPH3, DSE, FAM216A, FAM64A, FARP1, FSTL1, FZD2, GPR126, GPR176, HIST1H2BB, HIST1H4D, HLA-A, HLA-B, HMGB2, IGFBP7, KDM6A, KIF20B, KIF23, KLHL7, KNSTRN, KRT8, LPCAT2, MARCH4, MB21D2, MEST, METAP1, MICB, MTMR10, MYL9, NDFIP2, NMI, NRIP3, NRXN3, OSMR, PCDH10, PDLIM5, PFKP, PLK4, PPP2CB, PRC1, PRPS1, PTPLA, PTPN9, PTPRJ, PVRL3, RAB3IP, RAP1GDS1, RB1, RBPMS, RCN1, RHOJ, RNY5, RP2, SCRN1, SH3BGRL2, SHOC2, SIM2, SLC9A7, SNORD25, SNX25, SPTLC2, STARD4, STK10, STK32B, TAGLN, TMOD3, TPM1, TTK, TUBB6, UEVLD, UFSP2 |
KEGG Pathway Enrichment Analysis for DEGs
Based on the criterion of P < 0.05, the upregulated DEGs were significantly enriched in “glycosaminoglycan biosynthesis-chondroitin sulfate” and “arrhythmogenic right ventricular cardiomyopathy.” Meanwhile, the downregulated DEGs were significantly enriched in 12 pathways, such as “cell adhesion molecules” (CAMs), “pentose phosphate pathway,” “allograft rejection,” “type I diabetes mellitus” and “cell cycle.” The results of significantly enriched pathways are presented in Table 3.
Table 3.
KEGG Pathway Enrichment Analysis of Differentially-Expressed Genes.
| KEGG pathway | Gene counts | P-value |
|---|---|---|
| Upregulated | ||
| Glycosaminoglycan biosynthesis– chondroitin sulfate | 2 | 6.21 × 10-3 |
| Arrhythmogenic right ventricular cardiomyopathy | 3 | 7.34 × 10-3 |
| Downregulated | ||
| Cell adhesion molecules | 5 | 2.90 × 10-4 |
| Pentose phosphate pathway | 2 | 6.66 × 10-3 |
| Allograft rejection | 2 | 1.20 × 10-2 |
| Graft vs. host disease | 2 | 1.50 × 10-2 |
| Type I diabetes mellitus | 2 | 1.64 × 10-2 |
| Cell cycle | 3 | 1.86 × 10-2 |
| Autoimmune thyroid disease | 2 | 2.35 × 10-2 |
| Natural killer cell mediated cytotoxicity | 3 | 2.37 × 10-2 |
| Phagosome | 3 | 3.21 × 10-2 |
| Viral myocarditis | 2 | 4.06 × 10-2 |
| Adherens junction | 2 | 4.38 × 10-2 |
| Antigen processing and presentation | 2 | 4.71 × 10-2 |
KEGG: Kyoto Encyclopedia of Genes and Genomes; Gene Counts: number of genes.
GO Term Results for DEGs
Table 4 shows the top 5 GO terms. The upregulated DEGs were significantly enriched in 92 biological process (BP) terms, 8 cellular component (CC) terms and 28 molecular function (MF) terms. The upregulated DEGs were significantly involved in “multicellular organismal metabolic process,” “limb morphogenesis” and “hemophilic cell adhesion.” These genes were mostly located on “golgi apparatus,” “growth hormone receptor complex” and “collagen type XI.” Most of these genes possess the molecular function of “insulin-like growth factor binding,” “integrase activity” and “α-tubulin binding.” Meanwhile, the downregulated DEGs were obviously enriched in 58 BP terms, 18 CC terms and 21 MF terms. The suppressed processes in OS included “Positive regulation of pathway-restricted SMAD protein phosphorylation,” “Peptidyl-tyrosine dephosphorylation” and “Regulation of mitosis.” The downregulated genes were mostly located on “Spindle,” “major histocompatibility complex (MHC) class I protein complex” and “striated muscle thin filament.” The primary functions of the downregulated DEGs were “WW domain binding,” “polo kinase kinase activity” and “chondroitin-glucuronate 5-epimerase activity.”
Table 4.
TOP 5 Significantly Enriched BP, CC and MF for Differentially-Expressed Genes.
| GO ID | Term | Gene Counts | P-value |
|---|---|---|---|
| Upregulated | |||
| GO:0044236_BP | Multicellular organismal metabolic process | 5 | 5.24 × 10-4 |
| GO:0035108_BP | Limb morphogenesis | 5 | 7.66 × 10-4 |
| GO:0007156_BP | Homophilic cell adhesion | 5 | 8.18 × 10-4 |
| GO:0048703_BP | Embryonic viscerocranium morphogenesis | 2 | 1.19 × 10-3 |
| GO:0009636_BP | Response to toxic substance | 4 | 4.60 × 10-3 |
| GO:0005794_CC | Golgi apparatus | 16 | 3.90 × 10-4 |
| GO:0070195_CC | Growth hormone receptor complex | 1 | 5.16 × 10-3 |
| GO:0005592_CC | Collagen type XI | 1 | 1.03 × 10-2 |
| GO:0005899_CC | Insulin receptor complex | 1 | 1.54 × 10-2 |
| GO:0031982_CC | Vesicles | 11 | 2.13 × 10-2 |
| GO:0005520_MF | Insulin-like growth factor binding | 3 | 2.58 × 10-4 |
| GO:0008907_MF | Integrase activity | 1 | 5.03 × 10-3 |
| GO:0005509_MF | Calcium ion binding | 9 | 6.23 × 10-3 |
| GO:0043014_MF | α-tubulin binding | 2 | 6.39 × 10-3 |
| GO:0042277_MF | Peptide binding | 4 | 7.32 × 10-3 |
| Downregulated | |||
| GO:0010862_BP | Positive regulation of pathway-restricted SMAD protein phosphorylation | 3 | 2.42 × 10-4 |
| GO:0060391_BP | Positive regulation of SMAD protein import into nucleus | 2 | 9.63 × 10-4 |
| GO:0035335_BP | Peptidyl-tyrosine dephosphorylation | 3 | 1.32 × 10-3 |
| GO:0007088_BP | Regulation of mitosis | 4 | 1.52 × 10-3 |
| GO:0051301_BP | Cell division | 8 | 2.20 × 10-3 |
| GO:0005819_CC | Spindle | 9 | 9.35 × 10-7 |
| GO:0042612_CC | MHC class I protein complex | 2 | 9.44 × 10-4 |
| GO:0005865_CC | Striated muscle thin filament | 2 | 2.03 × 10-3 |
| GO:0005900_CC | Oncostatin-M receptor complex | 1 | 1.26 × 10-2 |
| GO:0005945_CC | 6-phosphofructokinase complex | 1 | 1.26 × 10-2 |
| GO:0050699_MF | WW domain binding | 2 | 4.91 × 10-3 |
| GO:0042801_MF | Polo kinase kinase activity | 1 | 5.02 × 10-3 |
| GO:0047757_MF | Chondroitin-glucuronate 5-epimerase activity | 1 | 5.02 × 10-3 |
| GO:0008017_MF | Microtubule binding | 4 | 6.70 × 10-3 |
| GO:0042605_MF | Peptide antigen binding | 2 | 8.64 × 10-3 |
GO: Gene Ontology; BP: biological process; CC: cellular component; MF: molecular function; Gene Counts: number of genes.
PPI Network of DEGs
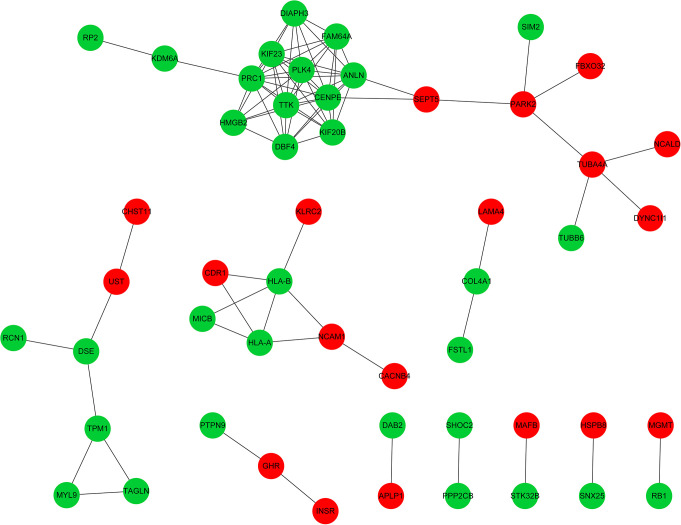
Figure 1 shows the PPI network, which included 84 interactions and 51 nodes. For this network, the connectivity degree of each node was calculated and Table 5 shows the nodes with a degree of ≥ 2. According to the degrees of the nodes, centromere-associated protein E (CENPE), protein regulator of cytokinesis 1 (PRC1), phosphotyrosine picked threonine-protein kinase (TTK) and polo-like kinase 4 (PLK4) were selected as the hub nodes, as they interacted with more than10 nodes, indicating their crucial roles in the PPI network.
Figure 1.
Protein-Protein Interaction (PPI) Network of Differentially Expressed Genes (DEGs) in Osteosarcoma. Red nodes represent upregulated DEGs; green nodes represent downregulated DEGs; gray lines stand for the interaction between 2 proteins. DEGs, differentially-expressed genes.
Table 5.
Differentially-Expressed Genes With >2 Degrees of Connectivity in the Protein-Protein Interaction Network.
| Gene | Degrees |
|---|---|
| CENPE | 11 |
| PRC1 | 11 |
| TTK | 10 |
| KIF23 | 10 |
| ANLN | 10 |
| PLK4 | 10 |
| FAM64A | 8 |
| DBF4 | 8 |
| KIF20B | 8 |
| DIAPH3 | 7 |
| HMGB2 | 6 |
| HLA-B | 5 |
| PARK2 | 4 |
| HLA-A | 4 |
| TUBA4A | 4 |
| SEPT5 | 3 |
| DSE | 3 |
| TPM1 | 3 |
| NCAM1 | 3 |
| CDR1 | 2 |
| UST | 2 |
| KDM6A | 2 |
| COL4A1 | 2 |
| MYL9 | 2 |
| TAGLN | 2 |
| GHR | 2 |
| MICB | 2 |
A total of 14 pathways were enriched by DEGs in PPI network, such as “glycosaminoglycan biosynthesis-chondroitin sulfate” and “natural killer cell mediated cytotoxicity” (Table 6). Furthermore, DEGs in the PPI network were also significantly enriched in 81 BP terms, 22 CC terms and 30 MF terms. The top 5 GO terms for each were listed in Table 7. For the BP terms, the dramatically enriched processes were mainly related with the “regulation of mitosis” and “cell division.” For the CC terms, the significantly enriched components were mainly correlated with the “spindle” and “MHC class I protein complex.” For the MF terms, the significantly enriched functions were mainly associated with “microtubule motor activity” and “chondroitin-glucuronate 5-epimerase activity.”
Table 6.
KEGG Pathway Enrichment Analysis of Differentially-Expressed Genes in the Protein-Protein Interaction Network.
| KEGG Pathway | Gene counts | P-value |
|---|---|---|
| Glycosaminoglycan biosynthesis-chondroitin sulfate | 3 | 1.12 × 10-4 |
| Phagosome | 5 | 4.76 × 10-4 |
| Natural killer cell mediated cytotoxicity | 4 | 2.77 × 10-3 |
| Antigen processing and presentation | 3 | 4.38 × 10-3 |
| Small cell lung cancer | 3 | 5.99 × 10-3 |
| Allograft rejection | 2 | 1.14 × 10-2 |
| Graft-vs.-host disease | 2 | 1.39 × 10-2 |
| Type I diabetes mellitus | 2 | 1.52 × 10-2 |
| Cell cycle | 3 | 1.67 × 10-2 |
| Cell adhesion molecules | 3 | 2.02 × 10-2 |
| Autoimmune thyroid disease | 2 | 2.18 × 10-2 |
| Pathogenic Escherichia coli infection | 2 | 2.51 × 10-2 |
| Viral myocarditis | 2 | 3.79 × 10-2 |
| Cardiac muscle contraction | 2 | 4.51 × 10-2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; Gene Counts, number of genes.
Table 7.
TOP 5 Significantly Enriched BP, CC and MF Terms for Differentially-Expressed Genes in the Protein-Protein Interaction Network.
| GO ID | Term | Gene Counts | P-value |
|---|---|---|---|
| GO:0007088_BP | Regulation of mitosis | 5 | 2.40 × 10-5 |
| GO:0051301_BP | Cell division | 8 | 1.93 × 10-4 |
| GO:0002480_BP | Antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent | 2 | 3.79 × 10-4 |
| GO:0006266_BP | DNA ligation | 2 | 5.76 × 10-4 |
| GO:0030208_BP | Dermatan sulfate biosynthetic process | 2 | 5.76 × 10-4 |
| GO:0007080_BP | Mitotic metaphase plate congression | 2 | 8.14 × 10-4 |
| GO:0005819_CC | Spindle | 8 | 4.68 × 10-7 |
| GO:0042612_CC | MHC class I protein complex | 2 | 4.38 × 10-4 |
| GO:0000139_CC | Golgi membrane | 7 | 1.00 × 10-3 |
| GO:0070195_CC | Growth hormone receptor complex | 1 | 2.87 × 10-3 |
| GO:0001725_CC | Stress fiber | 2 | 6.78 × 10-3 |
| GO:0003777_MF | Microtubule motor activity | 4 | 9.65 × 10-5 |
| GO:0047757_MF | Chondroitin-glucuronate 5-epimerase activity | 1 | 3.13 × 10-3 |
| GO:0005200_MF | Structural constituent of cytoskeleton | 3 | 3.16 × 10-3 |
| GO:0042605_MF | Peptide antigen binding | 2 | 3.44 × 10-3 |
| GO:0042169_MF | SH2 domain binding | 2 | 4.48 × 10-3 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; MHC, major histocompatibility complex; TAP, transporter associated with antigen processing; Gene counts, number of genes.
Real-Time PCR Verification of the mRNA Expression of Key Genes
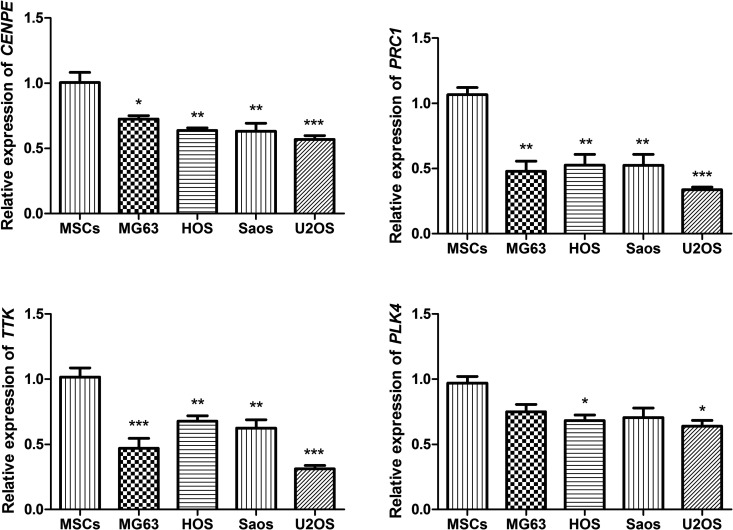
The CENPE, PRC1, TTK and PLK4 were selected as the hub nodes from PPI network. Based on the microarray results, these 4 genes were all significantly downregulated in OS cells. We performed real-time PCR to validate their expression in 4 kinds of human OS cell lines, including MG63, HOS, Saos and U2OS cells. As shown in Figure 2, the expression of CENPE, PRC1 and TTK were all significantly decreased in all 4 OS cells compared with the human mesenchymal stem cells (P < 0.05). The expression of PLK4 was significantly decreased in HOS and U2OS cells (P < 0.05), and its expression levels in MG63 and Saos were also decreased in MG63 and Saos cells, though not reach the significant level (P > 0.05).
Figure 2.
Real-time PCR analysis of CENPE, PRC1, TTK and PLK4 in 4 kinds of osteosarcoma cells, including MG63, HOS, Saos and U2OS cells and human mesenchymal stem cells (MSCs). *P < 0.05, ** P < 0.01, *** P < 0.001 compared with the human mesenchymal stem cells.
Discussion
In the current study, 100 upregulated and 83 downregulated DEGs in OS compared with control groups were obtained. Pathway enrichment analysis revealed that the upregulated genes were significantly enriched in “Glycosaminoglycan biosynthesis-chondroitin sulfate” and “arrhythmogenic right ventricular cardiomyopathy” and the downregulated genes were significantly enriched in 12 pathways, such as “CAMs,” and “pentose phosphate pathway.” GO-BP analysis suggested the upregulated genes were significantly enriched in 92 BP terms, such as “multicellular organismal metabolic process,” and the downregulated genes were significantly enriched “positive regulation of pathway-restricted SMAD protein phosphorylation process.” Furthermore, CENPE, PRC1, TTK and PLK4 were selected as the hub nodes in PPI network. DEGs in this network were significantly enriched in the “glycosaminoglycan biosynthesis-chondroitin sulfate” pathway, “regulation of mitosis process” and “microtubule motor activity” functions.
Our results showed that the upregulated DEGs and the DEGs in PPI network were obviously enriched in the “glycosaminoglycan biosynthesis-chondroitin sulfate” pathway and GO-BP of “multicellular organismal metabolic process.” The downregulated DEGs were significantly enriched in “CAMs” pathway and GO-BP of “positive regulation of pathway-restricted SMAD protein phosphorylation process.” Glycosaminoglycans are part of proteoglycans and defects in glycosaminoglycan synthesis could lead bone diseases.18 Besides, alteration of glycosaminoglycan in extracellular matrix was reported to be involved in cell proliferation and differentiation.19 Previous evidence has shown that glycosaminoglycan biosynthesis-chondroitin sulfate has relevance with growth and proliferation in human OS cells of MG63 and Saos cells.19,20 A previous study indicated that CAMs involved in the process of the metastasis of OS21 and CAM1 was found to be a diagnostic marker for OS.22 The SMAD signaling pathway plays role in some cancer cellular processes, e.g. proliferation and apoptosis.23 In addition, the present results showed that the regulation of mitosis biology process and microtubule motor activity molecular function was significantly enriched in the PPI network. Studies indicated that microtubule motor activity involved in the mitosis of OS cells and could promote OS cell proliferation.24,25 Therefore, our present study is in line with the previous study, suggesting that glycosaminoglycan biosynthesis-chondroitin sulfate pathway, regulation of mitosis biology process and microtubule motor activity may play critical roles in the progression of OS.
In the PPI network, CENPE was the top node and interacted with 11 proteins, including PRC1. These 2 genes were closely associated with the biological process of cell division. CENPE is a mitotic motor whose inactivation disrupts spindle checkpoint function.26,27 Furthermore, the incidences of spontaneous lymphomas and lung tumors are elevated in mice with reduced levels of CENPE, which agrees with its decreased level in OS group, suggesting that low CENPE level may facilitate the progression of OS.28 In another bioinformatics-based study, CENPE was also identified as downregulated gene and a high-degree node in PPI network of osteosarcoma.29 However, CENPE was reported to be upregulated in several types of cancer, including lung cancer, esophageal cancer and breast cancer.30-32 It is reported that PRC1 plays key roles in the central spindle and cytokinesis formation.33 A previous study testified that PRC1 may involve in growth of tumor cell and may be a possible target for anti-breast cancer drugs.34 However, it has not been reported whether PRC1 could be a target for OS treatment. Therefore, we speculate that the interaction between PRC1 and CENPE may be critical for the progression of OS.
The PPI network in the current study also showed that TTK was a hub node with a higher degree that directly interacted with PLK4. TTK plays significant roles in centrosome duplication, the proper execution of mitosis and tumor cell proliferation. In addition, significant overexpression of TTK is found in some human tumors.35 Caldarelli et al showed that selective TTK inhibitors can inhibit U2OS cells proliferation by promoting mitotic override.36 Moreover, TTK has been observed to affect the NF-κB signaling pathway, and NF-κB activation is common in carcinomas, mainly promoted by inflammatory cytokines within the tumor microenvironment.37 PLK4, a member of the polo-like kinase family, is a critical regulator of centriolar duplication.38 In vitro, evidence has demonstrated that the depletion of PLK4 with small interfering RNA results in the decrease of U2OS cells proliferation.37 In addition, the overexpression of PLK4 contributes to amplification of centrosomes, while the deletion of it decreases the number of centriole in U2OS cells. Furthermore, PLK4 is directly targeted by NF-κB, which is absolutely critical for cell proliferation in U2OS cells. For OS, Tang et al. indicated that the combined use of NF-κB inhibitors and chemotherapy drugs can improve the in vitro and in vivo effects of the chemotherapy drugs.4 Therefore, we considered that the interaction between TTK and PLK4 may involve in the centrosome duplication and the growth of OS cells through the activation of NF-κB pathway in OS.
However, this study has some limitations. First, because of lacking OS patients, the expression levels of key genes including CENPE, PRC1, TTK, and PLK4 were not validated in OS tissue or serum samples. Second, the interactions among proteins as well as the molecular mechanism of these key proteins in regulating OS development warrant further investigation.
In conclusion, “glycosaminoglycan biosynthesis-chondroitin sulfate” pathway, “microtubule motor activity,” and the “regulation of mitosis biology process” may be critical for OS progression. The interaction between PRC1 and CENPE may be important for OS progression. The interaction between PLK4 and TTK may be involved in the centrosome duplication and the growth of OS cells through NF-κB pathway activation in OS. CENPE, PRC1, TTK, and PLK4 may be biomarkers for the diagnosis and treatment of OS. However, further studies with a larger sample size are warranted to confirm these results.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Statement: Not applicable. Our study did not contain human or animal trials.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Daliang Kong  https://orcid.org/0000-0003-2591-8398
https://orcid.org/0000-0003-2591-8398
References
- 1. Thanapprapasr K, Nartthanarung A, Thanapprapasr D, Jinawath A. pFAK-Y397 overexpression as both a prognostic and a predictive biomarker for patients with metastatic osteosarcoma. Plos One. 2017;12(8):e0182989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. [DOI] [PubMed] [Google Scholar]
- 3. Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13(4):357–368. [DOI] [PubMed] [Google Scholar]
- 4. Tang QL, Xie XB, Wang J, et al. Glycogen synthase kinase-3beta, NF-kappaB signaling, and tumorigenesis of human osteosarcoma. J Natl Cancer Inst. 2012;104(10):749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pi WS, Cao ZY, Liu JM, et al. Potential molecular mechanisms of AURKB in the oncogenesis and progression of osteosarcoma cells: a label-free quantitative proteomics analysis. Technol Cancer Res Treat. 2018;18:1533033819853262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Cheng L, Wang C, Jing J. Cell cycle kinases in osteosarcoma: potential for therapeutic intervention. Curr Pharm Des. 2016;22(31):4830–4834. [DOI] [PubMed] [Google Scholar]
- 7. Ma H, Seebacher NA, Hornicek FJ, Duan Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in osteosarcoma. EBioMed. 2019;39:182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Y, Shen JK, Yu Z, Hornicek FJ, Kan Q, Duan Z. Expression and therapeutic implications of cyclin-dependent kinase 4 (CDK4) in osteosarcoma. Biochim Biophy Acta Mol Basis Dis. 2018;1864(5 Pt A):1573–1582. [DOI] [PubMed] [Google Scholar]
- 9. Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. [DOI] [PubMed] [Google Scholar]
- 10. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- 11. Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9(4):326–332. [DOI] [PubMed] [Google Scholar]
- 12. Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41(database issue):D970–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen JS, Hung WS, Chan HH, Tsai SJ, Sun HS. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics. 2013;29(4):420–427. [DOI] [PubMed] [Google Scholar]
- 14. Arakawa K, Kono N, Yamada Y, Mori H, Tomita M. KEGG-based pathway visualization tool for complex omics data. In Silico Biol. 2005;5(4):419–423. [PubMed] [Google Scholar]
- 15. Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(Suppl 4):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 17. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41(database issue):D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sasarman F, Maftei C, Campeau PM, Brunel-Guitton C, Mitchell GA, Allard P. Biosynthesis of glycosaminoglycans: associated disorders and biochemical tests. J Inherit Metab Dis. 2016;39(2):173–188. [DOI] [PubMed] [Google Scholar]
- 19. Nikitovic D, Zafiropoulos A, Tzanakakis GN, Karamanos NK, Tsatsakis AM. Effects of glycosaminoglycans on cell proliferation of normal osteoblasts and human osteosarcoma cells depend on their type and fine chemical compositions. Anticancer Res. 2005;25(4):2851–2856. [PubMed] [Google Scholar]
- 20. Kumarasuriyar A, Murali S, Nurcombe V, Cool SM. Glycosaminoglycan composition changes with MG-63 osteosarcoma osteogenesis in vitro and induces human mesenchymal stem cell aggregation. J Cell Physiol. 2009;218(3):501–511. [DOI] [PubMed] [Google Scholar]
- 21. Schiano C, Grimaldi V, Casamassimi A, et al. Different expression of CD146 in human normal and osteosarcoma cell lines. Med Oncol. 2012;29(4):2998–3002. [DOI] [PubMed] [Google Scholar]
- 22. Inoue T, Hagiyama M, Enoki E, et al. Cell adhesion molecule 1 is a new osteoblastic cell adhesion molecule and a diagnostic marker for osteosarcoma. Life Sci. 2013;92(1):91–99. [DOI] [PubMed] [Google Scholar]
- 23. Su E, Han X, Jiang G. The transforming growth factor beta 1/SMAD signaling pathway involved in human chronic myeloid leukemia. Tumori. 2010;96(5):659–660. [DOI] [PubMed] [Google Scholar]
- 24. Wang R, Dong K, Lin F, et al. Inhibiting proliferation and enhancing chemosensitivity to taxanes in osteosarcoma cells by RNA interference-mediated downregulation of stathmin expression. Mol Med. 2007;13(11-12):567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. [DOI] [PubMed] [Google Scholar]
- 26. Rao CV, Yamada HY, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis. 2009;30(9):1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mirzaa G, Vitre B, Carpenter G, et al. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum Genet. 2014;133(8):1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11(1):25–36. Epub December 28, 2006. [DOI] [PubMed] [Google Scholar]
- 29. Yang XR, Xiong Y, Duan H, Gong RR. Identification of genes associated with methotrexate resistance in methotrexate-resistant osteosarcoma cell lines. J Orthop Surg Res. 2015;10:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shan L, Zhao M, Lu Y, et al. CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. Int J Oncol. 2019;55(1):257–266. [DOI] [PubMed] [Google Scholar]
- 31. Hao X, Qu T. Expression of CENPE and its prognostic role in non-small cell lung cancer. Open Med (Wars). 2019;14:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu X, Luo X, Feng G, et al. CENPE expression is associated with its DNA methylation status in esophageal adenocarcinoma and independently predicts unfavorable overall survival. PloS One. 2019;14(2):e0207341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15(7):371–377. [DOI] [PubMed] [Google Scholar]
- 34. Shimo A, Nishidate T, Ohta T, Fukuda M, Nakamura Y, Katagiri T. Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells. Cancer Sci. 2007;98(2):174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Colombo R, Caldarelli M, Mennecozzi M, et al. Targeting the mitotic checkpoint for cancer therapy with NMS-P715, an inhibitor of MPS1 kinase. Cancer Res. 2010;70(24):10255–10264. [DOI] [PubMed] [Google Scholar]
- 36. Caldarelli M, Angiolini M, Disingrini T, et al. Synthesis and SAR of new pyrazolo [4, 3-< i> h</i>] quinazoline-3-carboxamide derivatives as potent and selective MPS1 kinase inhibitors. Bioorg Med Chem Letts. 2011;21(15):4507–4511. [DOI] [PubMed] [Google Scholar]
- 37. Ledoux AC, Sellier H, Gillies K, Iannetti A, James J, Perkins ND. NFkappaB regulates expression of Polo-like kinase 4. Cell Cycle. 2013;12(18):3052–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van de Weerdt BC, Medema RH. Polo-like kinases: a team in control of the division. Cell Cycle. 2006;5(8):853–864. [DOI] [PubMed] [Google Scholar]