Abstract
Introduction:
Methicillin resistant Staphylococcus aureus (MRSA) is a major human pathogen associated with nosocomial and community infections. mecA gene is considered one of the important virulence factors of S. aureus responsible for acquiring resistance against methicillin. The main objective of this study was to explore the prevalence, antibiotic susceptibility pattern, and mec A gene.
Methods:
A total of 39 isolates of S. aureus were isolated from 954 clinical specimens processed in Microbiology laboratory of Himal Hospital, Kathmandu. Antimicrobial susceptibility test (AST) was performed by Kirby-Bauer disc diffusion method using cefoxitin, and performed Polymerase Chain Reaction (PCR) for amplification of mecA gene in MRSA isolates.
Results:
Out of 954 clinical samples, (16.2%; 153/954) samples had bacterial growth. Among 153 culture positive isolates, 25.5% (39/153) were positive for S. aureus. Among 39 S. aureus (61.5%; 24/39) were multiple drug resistant (MDR). On AST, amoxicillin was detected as the least effective while vancomycin was the most effective. The prevalence of methicillin resistance was 46% (18/39) of which 72.2% (13/18) were positive for mecA gene in PCR assay.
Conclusion:
One in 4 culture positive isolates from the clinical specimens were S. aureus, of which almost two-thirds were MDR. Around half of the MDR showed MRSA and significant proportion of them were positive for mecA gene. This study concludes that the mecA gene is solely dependent for methicillin resistance in S. aureus but the presence of gene is not obligatory. PCR detection of the mecA gene is reliable, valid and can be suggested for the routine use in diagnostic laboratories.
Keywords: Staphylococcus aureus, MRSA, cefoxitin, mecA gene
Introduction
Methicillin Resistant S. aureus (MRSA) is a global threat causing serious infections in health facilities and the community, contributing 64% more deaths than the non-resistant form of the infections.1 It is estimated that one-third of the general population is colonized by S. aureus, and the global pooled prevalence of MRSA colonization is 1.3%.2 In 2014, the percentage of invasive MRSA isolates in Europe ranged from 0.9% in the Netherlands to 56% in Romania.3 The prevalence of MRSA infections in countries of South and East Asia, and the Western Pacific region ranged from 2.3% to 69.1%.4 MRSA led to adverse clinical outcomes compared with methicillin-sensitive S. aureus (MSSA).5 It often causes metastatic infections such as infective endocarditis (IE), septic arthritis, and osteomyelitis.3 More than 80 000 life-threatening infections are caused by MRSA and leads to 11 000 mortalities in a year.6,7
MRSA is generated when methicillin-susceptible S. aureus (MSSA) exogenously acquires a methicillin resistance gene, mecA, carried by a mobile genetic element known as staphylococcal cassette chromosome mec (SCCmec), and is considered to be transmissible across staphylococcal species.8 mecA gene is present in all MRSA strains that encodes penicillin binding protein 2a (PBP2a), which has a low tropism to all is β-lactam antibiotics, a cornerstone for producing MRSA phenomenon.9
MRSA are those strains of S. aureus that are resistant to the penicillinase-stable penicillin class of antibiotics such as methicillin, oxacillin, nafcillin, cloxacillin, and dicloxacillin by expression of mecA or other mechanisms, such as changes in affinity of penicillin binding proteins for oxacillin.10,11 Laboratory identification requires culture (on Mannitol Salt agar) that takes between 48 and 72 hours followed by susceptibility testing for MRSA which takes another 16 to 24 hours.12 The cefoxitin (30 µg) disk is used to detect MRSA by the disk diffusion method. S. aureus that are mecA positive should be considered as resistant to antibiotic oxacillin and other β-lactam group of antibiotics.13
Development of immunochromatographic test and molecular testing methods to detect MRSA have greatly reduced the time and labor required which can consequently help in improving infection control and reducing the cost.14 MRSA detection by real-time polymerase chain reaction (PCR) tests are capable of detecting genes specific to S. aureus. To distinguish MRSA strains from methicillin sensitive Staphylococcus aureus (MSSA), PCR methods target a portion of DNA where the MRSA-specific SCCmec gene of S. aureus from the samples within 1 to 3 hours.8 Molecular amplification of the mecA gene is recognized as a benchmark to diagnose MRSA in the community as these genes are highly conserved among staphylococcal species.15
Several studies in the past have reported the prevalence of MRSA infections in Nepal.16-22 Past studies conducted at different settings in Nepal have shown the prevalence of MRSA ranging from 26.1% to 57.1%.16-18,20,21,23 Increasingly in recent years, more studies have been carried out to explore the prevalence of MRSA infections in Nepal. Nevertheless, all these studies were mostly concentrated in phenotypic characterization with antimicrobial susceptibility test. This study explores the prevalence of MRSA infections, antimicrobial susceptibility patterns and detection of mecA gene by using molecular methods, PCR with objective to evaluate the usefulness of amplification of mecA gene and its reliability in the identification of MRSA strains.
Methods
Study area/collection of specimens
In the study period between September, 2017 and March, 2018, a total of 954 specimens such as Urine, Blood, and swabs from vagina, endo-cervix, intra-cervix, wound, ear, eye, semen; and other body fluids were collected from suspected patients in Himal Hospital, Kathmandu and were transferred to the laboratory for further processing.24 Himal Hospital is a tertiary care health center located at the heart of Kathmandu valley. It is a 100-bedded hospital that provides services to 300 to 500 out-patients in a day.
Specimen processing/identification tests/biochemical tests
The colonies grown were identified based on the morphology, Gram’s stain, and biochemical tests. S. aureus were confirmed using following tests: yellow colored colonies on mannitol salt agar, catalase positive, slide and tube coagulase positive, hydrolyzed gelatin, showed beta-hemolysis on blood agar, methyl red positive, Voges–Proskauer positive, nitrate reduction positive, fermentative, DNase producing, lactose, mannitol, maltose, mannose, sucrose and trehalose fermenting, and alkaline phosphatase positive.22,25
Antibiotic susceptibility testing (AST)
Antimicrobial susceptibility testing was performed by Kirby Bauer disc diffusion technique following clinical and laboratory standards institute (CLSI) guidelines, 2018.11 The concentration of suspension of the test organism was made equivalent to 0.5 McFarland standards. Lawn culture was performed on Mueller–Hinton agar plate. Antibiotic discs were placed over the lawn culture and the plate was incubated aerobically at 35°C for 24 hours. Finally, the plate was observed for zone of inhibition and interpreted according to CLSI guidelines, 2018 The antibiotic discs used were amoxicillin, amikacin (30 µg), ampicillin (10 µg), cefoxitin (30 µg), ciprofloxacin (5 µg), clindamycin (2 µg), chloramphenicol (30 µg), erythromycin (15 µg), gentamicin (10 µg), tetracycline (30 µg), cotrimoxazole (25 µg), vancomycin (30 µg), and meropenem (10 µg). Strains showing resistance to 3 or more than 3 different classes of antibiotics were considered multidrug resistant.26 All confirmed isolates were stored 2 sets: 1 set at +4°C (later sub-cultured to carry out phenotypic characterization) and another set frozen in tryptic soy broth containing 10% glycerol and stored at −70°C for molecular analysis.
Identification of methicillin resistant Staphylococcus aureus (MRSA) strains
Methicillin resistant S. aureus (MRSA) was identified using cefoxitin (30 µg) disks. Plates were incubated at 35°C. for a 24-hour incubation period. The diameter of the zone of inhibition (ZOI) of growth was recorded and interpreted as susceptible or resistant based on the CLSI guideline. S. aureus isolates were deemed methicillin resistant when the ZOI was ⩽21 mm with the cefoxitin disk.11
Detection of mecA by PCR technique
DNA was extracted from the MRSA isolates by chloroform: phenol extraction method as described.27 The primers used for mecA gene: forward primer of mecA gene (FP) 5′-ACT GCT ATC CAC CCT CAA AC-3′ and reverse primer (RP) 5′-CTG GTG AAG TTG TAA TCT GG-3′.28 Final 10 µl solution which included master mix of 5 µl, forward primer 1 µl, reverse primer 1 µl, DNA 1 µl, nuclease free water 2 µl were utilized for PCR. Thermal cycling was conducted for 120 seconds at 95°C (initial denaturation); 30 seconds at 95°C (denaturation) and 30 seconds at 56.2°C (annealing), 20 seconds at 72°C (extension), 29 amplification cycles using primer and 5 min at 72°C and preserved at 4°C. After completion of PCR thermal cycle, gel band was observed under UV light, yielded a 163-bp DNA fragment that was visualized by electrophoresis in 1.5% agarose gel stained with ethidium bromide staining and under UV light.28,29
Results
Distribution of bacterial genera, S. aureus, and MRSA in clinical samples
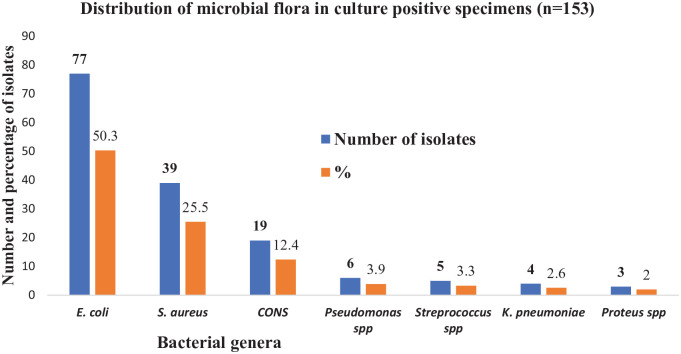
About 945 clinical specimens were examined for growth, among which 16.1% (153/945) samples had bacterial growth. Among 153 culture positive bacterial isolates (50.3%; 77/153) were E. coli followed by S. aureus (25.5%; 39/153), coagulase negative staphylococci (12.4%; 19/153), and Pseudomonas spp. (3.9%; 6/153) respectively (Figure 1).
Figure 1.
Distribution of bacterial genera among culture positive clinical specimens.
The prevalence of S. aureus 25.5% (39/153). Among 39 S. aureus, (61.5%; 24/39) were multiple drug resistant (MDR). Out of 39 S. aureus isolated, 46.1% (18/39) were MRSA (Table 1).
Table 1.
Distribution of S. aureus and MRSA in clinical specimens and according to age group, gender, and type of patients.
| Characters | Bacterial growth | S. aureus | P-value | MRSA | P-value |
|---|---|---|---|---|---|
| Clinical specimens | N (%) | N (%) | N (%) | ||
| Urine | 90 (58.9) | 9 (23. 1) | 4 (22.2) | ||
| Blood | 20 (13.1) | 4 (10.2) | 2 (11.1) | ||
| Wound swab | 25 (16.3) | 16 (41.1) | 7(38.8) | ||
| Pus | 15 (9.8) | 8 (20.5) | 4 (22.2) | ||
| Body fluid | 3 (1.9) | 2 (5.1) | 1(5.5) | ||
| Total | 153 | 39 | 18 | ||
| Age group in years | |||||
| <14 | 10 (25.6) | .9 | 2 (11.1) | .31 | |
| (15-45) | 21 (53.8) | 12 (66.7) | |||
| >45 | 8 (20.6) | 4 (22.2) | |||
| Gender | |||||
| Male | 22 (56.4) | .08 | 11 (61.1) | .11 | |
| Female | 17 (43.6) | 7 (38.9) | |||
| Type of patients | |||||
| Inpatients | 23 (59) | .001 | 13 (72.2) | .002 | |
| Outpatients | 16 (41) | 5 (27.8) | |||
Distribution of S. aureus and MRSA according to age, gender, and type of patients
Out of 39 S. aureus isolated, 53.8% (21/39) were isolated from age group: 15 to 45 years followed by age group <14 years (25.6%; 10/39) and age group >45 years 20.6%; 8/39) (Table 1).
Out of 18 MRSA, 66.7% (12/18) were isolated from the age group: 15 to 45 years followed by age group >45 (22.2%; 4/18) and age group <14 years (11.1%; 2/18) respectively. Male patients (61.1%; 11/18) had more MRSA than the female patients (38.9%; 7/18). Similarly, patients admitted in hospital (72.2%; 13/18) harbored more MRSA than outpatients (27.8%; 5/18) and found statistically significant (P < .05) (Table 1).
Antibiotic susceptibility patterns of S. aureus and MRSA isolates
All 39 S. aureus isolated were found 100% sensitive to vancomycin followed by amoxicillin (89.7%; 35/39) and cotrimoxazole (64.1%; 25/39) in decreasing order. All 18 MRSA isolates were found to be resistant to cefoxitin and amoxicillin followed by cotrimoxazole (83.3%; 15/18), and the prevalence of resistance was same across tetracycline, ciprofloxacin and erythromycin (72.2%; 13/18) (Table 2).
Table 2.
Antibiotic susceptibility pattern of S. aureus and MRSA.
| Antibiotics | S. aureus (n = 39) | MRSA (n = 18) | ||
|---|---|---|---|---|
| Sensitive N (%) | Resistant N (%) | Sensitive N (%) | Resistant N (%) | |
| Gentamycin | 28 (71.8) | 11(28.2) | 10 (55.5) | 8 (44.5) |
| Amikacin | 33 (84.6) | 6 (15.4) | 15 (83.3) | 3 (16.7) |
| Amoxicillin | 4 (10.3) | 35 (89.7) | 0 (0) | 18 (100) |
| Cefoxitin | 21 (53.9) | 18 (46.1) | 0 (0) | 18 (100) |
| Meropenem | 30 (76.9) | 9 (23.1) | 9 (50) | 9 (50) |
| Vancomycin | 39 (100) | 0 (0) | 18 (100) | 0 (0) |
| Erythromycin | 26 (66.7) | 13 (33.3) | 5 (27.8) | 13 (72.2) |
| Chloramphenicol | 31 (79.5) | 8 (20.5) | 8 (44.5) | 10 (55.5) |
| Ciprofloxacin | 25 (64.1) | 14 (35.9) | 5 (27.8) | 13 (72.2) |
| Cotrimoxazole | 14 (35.9) | 25 (64.1) | 3 (16.7) | 15 (83.3) |
| Tetracycline | 20 (51.3) | 19 (48.7) | 5 (27.8) | 13 (72.2) |
Gel electrophoresis and polymerase chain reaction (PCR)
Out of 18 MRSA isolates, 13 (72.2%) showed mecA gene on agarose gel containing extracted DNA, 5 µl EtBr, 1 kb marker, and on TAE buffer. Band was observed under UV light and the product size of mecA gene was 163 bp (Figure 2).
Figure 2.
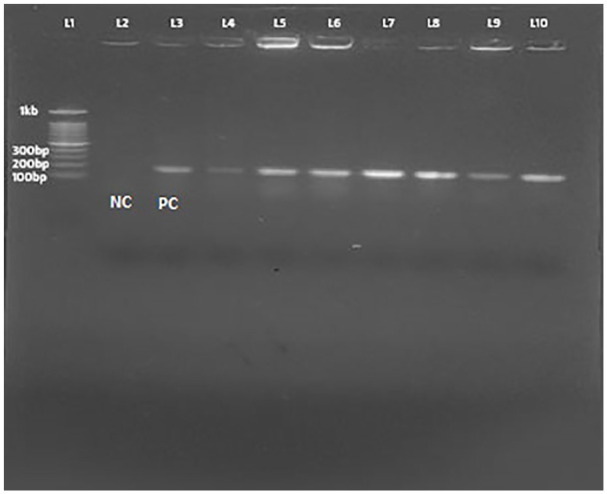
Visualization of amplified mecA gene under UV Trans-illuminator.
Abbreviations: L1, NEB 100 bp DNA marker; L2, Negative control S. aureus ATCC 29213; L3, Positive control S. aureus ATCC 49476; L4-L10, different clinical S. aureus isolates).
Discussion and Conclusion
Our study showed (16.2%) bacterial growth from different clinical specimens in culture. Among bacterial isolates, 1/4th of them were S. aureus. The findings of our study are consistent with other studies reported from School of Health & Allied Sciences, Pokhara University,22 Chitwan,16 B&B hospital, Lalitpur,30 Manipal Teaching Hospital, Pokhara,31 Lumbini Medical College and Teaching Hospital, Palpa,20 Alka Hospital, Lalitpur,23 and Birendra Military Hospital, Kathmandu.32
In our study, S. aureus were mostly sensitive to amikacin, meropenem, and gentamicin. The findings of our study resonate with previous studies reported from Nepal.33 Antimicrobial susceptibility pattern of MRSA showed most of the tested antibiotics to be resistant except vancomycin, and amikacin. These findings are in line with few previous studies,16,19,30,31 however, slightly contrasts with one past study, in which ciprofloxacin was sensitive to MRSA.34 Antimicrobial resistance is a global threat and MRSA has emerged as an important human pathogen with wide range of antibiotic resistance.6 Globally, the prevalence of MRSA is highly heterogeneous. Past reports of MRSA from Nepal reported prevalence of 15.4% to 26.0%17,35 and newer studies from various hospitals of Nepal reported higher prevalence: 26% to 69%.36,37 Most of the MRSA related studies conducted in Nepal, used only cefoxitin and/or oxacillin for screening MRSA.
Detection of mecA gene is the major evidence for the detection of MRSA isolate. This statement was approved by many studies for example in Sudan,38 Saudi Arabia,39 Iraq,40 Japan,41 India,42 Australia,43 and USA.44 However, our study showed low burden of the mecA gene (72.22%); this may raise questions to explore other intrinsic factors that may compete with mecA gene in producing resistance phenomenon in regions with high prevalence of MRSA. Nonetheless, the absence of mecA gene within resistant staphylococcal isolates have been reported globally.45 Additionally, moderate methicillin resistance was observed in isolates that lacked the mecA gene mutations.46
In our study, 13 (72.22%) out of 18 MRSA show mecA gene while 27.78% showed absence of mecA gene. A previous study in Nigeria reported the complete absence of 5 major SCCmec types and mecA genes as well as the gene product of PBP2a in isolates which were phenotypically MRSA suggesting a probability of hyper-production of β-lactamase as a cause of the phenomenon.47 Another study suggests that the specific alterations in different amino acids present in protein binding proteins cascade (PBPs 1, 2, and 3) could be the basis of resistance. These alterations were found to include 3 amino acid substitutions, which were identical and were present in PBPs 1, 2, and 3. Moreover, the same amino acid was found to have 2 other different substitutions in PBP1. Both the identical and different amino acid substitutions were observed in isolates from different multilocus types.48 These findings provided clear evidence that there are mechanisms other than the presence of mecA gene responsible for beta-lactam resistance of MRSA and the molecular methods alone are not enough to confirm characterization of MRSA isolates, a point that should be considered by regional and reference laboratories.
High prevalence of MRSA reported in this study warrants an attention for rational and regulated use of antibiotics based on the laboratory confirmation of the disease and antibiotic susceptibility tests. In addition to phenotypic identification of MRSA, PCR-based detection of MRSA is essential to monitor its spread and characterization. However, this is a pilot study in a single hospital. Future studies with larger sample size and from multiple settings are essential to generate evidence on the characteristics and role of mecA gene.
Strengths and Limitations
This study will be a useful reference for future studies to explore and expand on the wider prevalence of MRSA organisms in hospitals, patients, and community people. Since our study was based on a phenotypic detection method and detection of mecA gene by conventional PCR, genotypic characterization of new homologues of mecA (mecB, mecC, and mecD) and whole genome sequencing of MRSA strains are helpful for future studies.
Acknowledgments
We would like to express our sincere gratitude and admiration to all the patients for their involvement in the study.
Appendix
Abbreviations
AST Antimicrobial susceptibility test
CLSI Clinical and laboratory standard institute
MRSA Methicillin Resistant Staphylococcus aureus
MSSA Methicillin sensitive Staphylococcus aureus
PCR Polymerase chain reaction
SCCmec Staphylococcal cassette chromosome mec
ZOI Zone of inhibition
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by Departmental fund of Kantipur College of Medical Sciences, affiliated to Tribhuvan University, Kirtipur, Kathmandu, Nepal.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: All the authors made substantial contribution to the study. SNK and NA conceived and designed the study. SNK collected samples, investigated, and recorded the laboratory findings. NA, GK, KBA, and UTS supervised the laboratory work. KRR, MRB, and PG advised and formulated the methodology for the study. SNK, BD, and KRR drafted the original draft. KRR and BA are responsible for reviewing several versions of manuscript. Others helped to review and amend this manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval and Consent to Participate: The specimens and the data used in our study were derived from routine diagnostic database in Microbiology Laboratory of Himal Hospital, Naxal Kathmandu. Since the samples and data used in this study neither incurred direct involvement with the patients not interfered with the routine clinical care, formal ethics approval was deemed unnecessary complying with the guidelines of Nepal Health research Council.
ORCID iD: Komal Raj Rijal  https://orcid.org/0000-0001-6281-8236
https://orcid.org/0000-0001-6281-8236
References
- 1. World Health Organization. Fact sheet Antimicrobial Resistance. 2018. Accessed March 15, 2020 https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
- 2. Hassoun A, Linden PK, Friedman B. Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. Crit Care. 2017;21:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe in 2014. Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). 2017. Accessed March 15, 2020 http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-europe-2014.pdf
- 4. World Health Organization. Antimicrobial resistance: global report on surveillance. 2014. Accessed March 15, 2020 http://www.who.int/drugresistance/documents/surveillancereport/en/
- 5. van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. Predictors of mortality in Staphylococcus aureus Bacteremia. Clin Microbiol Rev. 2012;25:362-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pokharel S, Raut S, Adhikari B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob Health. 2019;4:e002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Center Diseases Control. Antibiotic/Antimicrobial resistance. 2019. Accessed July 25, 2019 https://www.cdc.gov/drugresistance/biggest_threats.html
- 8. Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010;54:4352-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aguayo-Reyes A, Quezada-Aguiluz M, Mella S, et al. Molecular basis of methicillin-resistance in Staphylococcus aureus. Rev Chilena Infectol. 2018;35:7-14. [DOI] [PubMed] [Google Scholar]
- 10. Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002;85:57-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute. M100: Performance standards for antimicrobial susceptibility testing: twenty fifth informational supplement edition. 28 ed CLSI; 2018. [Google Scholar]
- 12. Smyth RW, Kahlmeter G. Mannitol salt agar-cefoxitin combination as a screening medium for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:3797-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franklin RC, Matthew AW, Jeff A, Michael ND, George ME. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI Document M100-S22. Clinical and Laboratory Standards Institute; Vol. 32; 2012: 70-88. [Google Scholar]
- 14. Sturenburg E. Rapid detection of methicillin-resistant Staphylococcus aureus directly from clinical samples: methods, effectiveness and cost considerations. Ger Med Sci. 2009;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abd Al-Abbas MJ. Antimicrobial susceptibility of Enterococcus faecalis and a novel Planomicrobium isolate of bacterimia. Int J Med Med Sci. 2012;4:19-27. [Google Scholar]
- 16. Ansari S, Nepal HP, Gautam R, et al. Threat of drug resistant Staphylococcus aureus to health in Nepal. BMC Infect Dis. 2014;14:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumari N, Mohapatra TM, Singh YI. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in a tertiary-care hospital in Eastern Nepal. J Nepal Med Assoc. 2008;47:53-56. [PubMed] [Google Scholar]
- 18. Rijal KR, Shrestha N, Pahari N, et al. Methicillin resistance Staphylococcus aureus in patients visiting western regional hospital, Pokhara. JIOM. 2008;30:21-25. [Google Scholar]
- 19. Khanal LK, Jha BK. Prevalence of methicillin resistant Staphylococcus aureus (MRSA) among skin infection cases at a hospital in Chitwan, Nepal. Nepal Med Coll J. 2010;12:224-228. [PubMed] [Google Scholar]
- 20. Raut S, Bajracharya K, Adhikari J, Pant SS, Adhikari B. Prevalence of methicillin resistant Staphylococcus aureus in Lumbini Medical College and Teaching Hospital, Palpa, Western Nepal. BMC Res Notes. 2017;10:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhomi U, Rijal KR, Neupane B, et al. Status of inducible clindamycin resistance among macrolide resistant Staphylococcus aureus. Afr J Microbiol Res.2016;10:280-284. [Google Scholar]
- 22. Rijal KR, Pahari N, Shrestha BK, et al. Prevalence of methicillin resistant Staphylococcus aureus in school children of Pokhara. Nepal Med Coll J. 2008;10:192-195. [PubMed] [Google Scholar]
- 23. Shahi K, Rijal KR, Adhikari N, et al. Methicillin resistant Staphylococcus aureus: prevalence and antibiogram in various clinical specimens at Alka Hospital. TUJM. 2018;5:77-82. [Google Scholar]
- 24. Center for Disease Control and Prevention. Methicillian resistant Staphylococcus aureus (MRSA). 2017. Accessed March 14, 2020 https://www.cdc.gov/mrsa/healthcare/index.html
- 25. Kateete DP, Kimani CN, Katabazi FA, et al. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann Clin Microbiol Antimicrob. 2010;9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268-281. [DOI] [PubMed] [Google Scholar]
- 27. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Vol. 1, 2nd ed. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28. Vatansever L, Sezer C, Bilge N. Carriage rate and methicillin resistance of Staphylococcus aureus in food handlers in Kars City, Turkey. Springerplus. 2016;5:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oliveira DC, de Lencastre H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: a surprising observation. PLoS One. 2011;6:e23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belbase A, Pant ND, Nepal K, et al. Antibiotic resistance and biofilm production among the strains of Staphylococcus aureus isolated from pus/wound swab samples in a tertiary care hospital in Nepal. Ann Clin Microbiol Antimicrob. 2017;16:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhatta DR, Cavaco LM, Nath G, Gaur A, Gokhale S, Bhatta DR. Threat of multidrug resistant Staphylococcus aureus in Western Nepal. Asian Pac J of Trop Dis 2015;5:617-621. [Google Scholar]
- 32. Adhikari R, Pant ND, Neupane S, et al. Detection of methicillin resistant Staphylococcus aureus and determination of minimum inhibitory concentration of vancomycin for Staphylococcus aureus isolated from pus/wound swab samples of the patients attending a tertiary care hospital in Kathmandu, Nepal. Can J Infect Dis Med Microbiol. 2017;2017:2191532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sah P, Rijal KR, Shakya B, Tiwari BR, Ghimire P. Nasal carriage rate of Staphylococcus aureus in Hospital personnel of National Medical College and Teaching Hospital and their antibiotic susceptibility pattern. J Health Allied Sci. 2013;3:21-23. [Google Scholar]
- 34. Shrestha B. Comparative prevalence of MRSA in two Nepalese tertiary care hospitals. Open J Clin Diag. 2013;3:67. [Google Scholar]
- 35. Subedi S, Brahmadathan KN. Antimicrobial susceptibility patterns of clinical isolates of Staphylococcus aureus in Nepal. Clin Microbiol Infect. 2005;11:235-237. [DOI] [PubMed] [Google Scholar]
- 36. Bhatta DR, Cavaco LM, Nath G, et al. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC Infect Dis. 2016;16:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khanal LK, Adhikari RP, Guragain A. Prevalence of methicillin resistant Staphylococcus aureus and antibiotic susceptibility pattern in a tertiary hospital in Nepal. J Nepal Health Res Counc. 2018;16:172-174. [PubMed] [Google Scholar]
- 38. Maimona A, Eliman E, Suhair R, Miskelyemen AE, Mogahid ME. Emergence of vancomycin resistant and methcillin resistant Staphylococus aureus in patients with different clinical manifestations in Khartoum State. J Amer Sci. 2014;10:106-110. [Google Scholar]
- 39. Meshref AA, Omer MK. Detection of (mecA)gene in methicillin resistant Staphylococcus aureus (MRSA) at Prince A/Rhman Sidery. J Med Genet Genomic. 2011;3:41-45. [Google Scholar]
- 40. Al-Zu’bi E, Bdour S, Shehabi AA. Antibiotic resistance patterns of mecA-positive Staphylococcus aureus isolates from clinical specimens and nasal carriage. Microb Drug Resist. 2004;10:321-324. [DOI] [PubMed] [Google Scholar]
- 41. Hotta K, Ishikawa J, Ishii R, et al. Necessity and usefulness of detection by PCR of mecA and aac(6’)/aph(2”) genes for identification of arbekacin-resistant MRSA. Jpn J Antibiot. 1999;52:525-532. [PubMed] [Google Scholar]
- 42. Mehndiratta PL, Bhalla P, Ahmed A, Sharma YD. Molecular typing of methicillin-resistant Staphylococcus aureus strains by PCR-RFLP of SPA gene: a reference laboratory perspective. Indian J Med Microbiol. 2009;27:116-122. [DOI] [PubMed] [Google Scholar]
- 43. Cloney L, Marlowe C, Wong A, Chow R, Bryan R. Rapid detection of mecA in methicillin resistant Staphylococcus aureus using cycling probe technology. Mol Cell Probes. 1999;13:191-197. [DOI] [PubMed] [Google Scholar]
- 44. Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aziz HW, Al-Dulaimi TH, Al-Marzoqi AH, Ahmed NK. Phenotypic detection of resistance in Staphylococcus aureus isolates: detection of (mec A and fem A) gene in methicillin resistant Staphylococcus aureus (MRSA) by polymerase chain reaction. J Nat Sci Res. 2014;4:112-118. [Google Scholar]
- 46. Hiramatsu K, Ito T, Tsubakishita S, et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother. 2013;45:117-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olayinka BO, Olayinka AT, Obajuluwa AF, Onaolapo JA, Olurinola PF. Absence of mecA gene in methicillin-resistant Staphylococcus aureus isolates. Afr J Infect Dis. 2009;3:49-56. [Google Scholar]
- 48. Ba X, Harrison EM, Edwards GF, et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J Antimicrob Chemother. 2014;69:594-597. [DOI] [PMC free article] [PubMed] [Google Scholar]