Abstract
Polyphenols based bioactive compounds from vegetables and fruits are known for impressive antioxidant activity. Ingestion of these antioxidants may promote human health against cardiovascular diseases and cancer. Mango is a popular tropical fruit with special taste, high nutritional value and health-enhancing metabolites. The aim was to investigate the diversity of phytochemicals between two mango cultivars of china at three stages of fruit maturity. We used ESI-QTRAP-MS/MS approach to characterize comprehensively the metabolome of two mango cultivars named Hongguifei (HGF) and Tainong (TN). HPLC was used to quantify selected catechin based phenolic compounds. Moreover, real-time qPCR was used to study the expression profiles of two key genes (ANR and LAR) involved in proanthocyanidin biosynthesis from catechins and derivatives. A total of 651 metabolites were identified, which include at least 257 phenolic compounds. Higher number of metabolites were differentially modulated in peel as compared to pulp. Overall, the relative quantities of amino acids, carbohydrates, organic acids, and other metabolites were increased in the pulp of TN cultivar. While the contents of phenolic compounds were relatively higher in HGF cultivar. Moreover, HPLC based quantification of catechin and derivatives exhibited cultivar specific variations. The ANR and LAR genes exhibited an opposite expression profile in both cultivars. Current study is the first report of numerous metabolites including catechin-based derivatives in mango fruit. These findings open novel possibilities for the use of mango as a source of bioactive compounds.
Subject terms: Plant sciences, Plant molecular biology
Introduction
Fruits of tropical and subtropical regions are appreciated as energy suppliers, as well as for the presence of health-enhancing metabolites. Plant origin secondary metabolites are the focus of research for numerous health-beneficial properties and antioxidant activities. Mango (Mangifera indica (L.) Lam.) is an important tropical fruit and rank fifth in global production (55.6 million tonnes/year) after banana, apple, grapes, and oranges1. Numerous cultivars of mango are found worldwide, which show variations in fruit peel color, size, shape, and composition2,3. Apart from being consumed fresh, mangoes are also used to make desserts, juices, pickles, marmalades and jam4. During processing, a significant portion of the fruit is removed, which generates millions of tons of mango waste every year. However, the peel of mango fruit may be interesting for the presence of high levels of health-promoting compounds2,4. It seems in line with the efforts to explore the cost-effective potential of agri-waste for industrial use or to reduce its negative effects on the environment. The identification and reclamation of important metabolites from mango or its byproducts is a difficult task and its completion would promote the revaluation of mango as a natural source of antioxidants/bioactive compounds. In this regard, large scale metabolite profiling seems a promising way to explore metabolite potential of mango fruit.
Proanthocyanidins (PAs) are oligo/polymeric flavonoids that are naturally present in many vegetables, nuts, seeds and fruits5. Flavan-3-ols constitute the structural units of PAs and consist of C6-C3-C6 based flavonoid skeleton. There are two most common forms of these metabolites as 2,3-cis-(–)-epicatechin and 2,3-trans-(+)-catechin6. Flavan-3-ols (as monomers or as PAs) promote plant resistance against various biotic and abiotic stresses7. These metabolites possess numerous pharmacological properties, hence involved in scavenging free radicals, antimicrobial, antioxidant, anti-nutritional, anti-cancer and cardiac protection activities8,9. Quantitative and qualitative differences in the phytochemical profiles of mango cultivars may contribute to distinguishing their health-promoting properties.
The biochemical composition of fruits (sugars, organic acids, flavonoids, etc.) predominantly influences the consumer preference for visual features and taste. Several researchers have investigated the nutritional composition of mangoes10–12. However, very limited information is available about the identities of metabolites, which govern important properties of this fruit. In addition, there is a lack of studies for comprehensive identification, documentation, and quantification of flavonoids and other secondary metabolites. Instead of thoroughly evaluating all of the phytochemicals, researchers have attempted to study only particular metabolites of mango10,13,14. Recent advancements in widely-targeted metabolomics (supported by techniques like LC–MS/MS) have made possible a prompt and ultra-sensitive detection of a huge number of metabolites15,16. The liquid chromatography-tandem mass spectrometry is employed in the current study to identify and detect relative quantities of metabolites from two mango cultivars exhibiting contrasting features in terms of shape, size, taste and peel color2. This study aims at the revelation of the metabolic variations between two mango cultivars (from China) with distinct features2,17 and offers valuable data for appraising its nutritional importance in industrial utilization and breeding strategies.
Results and discussion
Metabolic profiling
Previous studies have reported the quantification of individual metabolite classes in mango pulp and peel10,18–24. In most of these studies, a standard metabolite was used to identify an exact compound or a relevant group of metabolites. However, based on these studies it was difficult to envisage comprehensive metabolic dynamics in mango. In current study, a total of 651 metabolites were annotated/ identified in three growth stages (Table S1), which include 54 nucleotides and their derivatives; 21 carbohydrates and their derivatives; 99 amino acids and their derivatives; 67 lipids and their derivatives; 6 indole derivatives; 8 alcohols and polyols; 2 terpenoids; 5 alkaloids; 21 vitamins and their derivatives; 72 organic acids; 257 phenolic compounds and 38 other metabolites.
Principal component analysis (PCA) for metabolite profiles
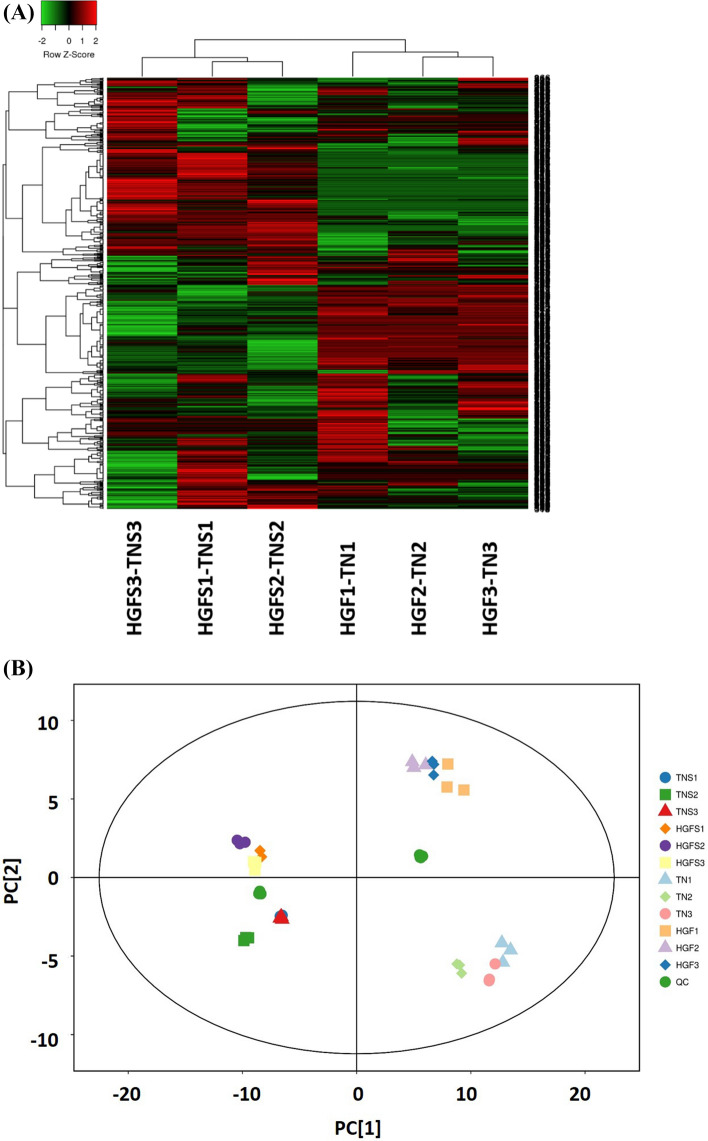
To assess relative variations in metabolic profiles (for 651 metabolites), we used multivariate statistics. Hierarchical cluster analysis (HCA), for relative differences in accumulation patterns at three growth stages, arranged metabolites from both cultivars into three groups (Fig. 1A). Peel and pulp samples exhibit distinct metabolite profiles in terms of upregulated or downregulated compounds in a cultivar and growth stage-specific manner (Fig. 1A, Table S2). The metabolites in peel at stages 1 and 2 are clustered in the same column as compared to third stage metabolites. On the other hand, metabolites at stages 2 and 3 are clustered in a similar column for pulp samples as compared to the first stage. It suggests the diversity of metabolites in peel samples at the third stage of fruit growth.
Figure 1.
HCA and PCA analysis of relative metabolite variations in peel and pulp samples of both cultivars. Both the HCA and PCA analysis was performed using all the metabolites annotated in current study. (A) Heat map for HCA. An online tool (heatmapper)25 was used to visualize the metabolite variations. The complete linkage hierarchical clustering was used for normalization. Each column represents a pairwise comparison of metabolites from both cultivars at a particular stage, while each row represents a metabolite. The red color is an indication of a higher concentration in HGF and green color represents a higher concentration of the metabolite in TN. (B) PC1 and PC2 score plots for pulp and peel between both cultivars. TN1, TN2, TN3, HGF1, HGF2, and HGF3 represent pulp samples of TN and HGF cultivars at first, second and third stage respectively. Similarly, TNS1, TNS2, TNS3, HGFS1, HGFS2, and HGFS3 represent peel samples of TN and HGF cultivars at first, second and third stage respectively. QC, mix represents quality control samples.
PCA is extensively applied in chemometric experiments to extract and rationalize important facts from biological systems with multivariate descriptions. By using this analysis, we can determine the core arrangement of variables in terms of principal components. According to PCA plots (Fig. 1B), the QC samples (mix) formed a close cluster, which indicates the similarity of metabolic profiles and stability/repeatability of analysis. Consistently, based on the PC1, a clear separation could be observed between the peel and pulp metabolites. Additionally, both cultivars are clearly distinguished by PC2 (Fig. 1B). This finding suggests the existence of distinct metabolic programs in peels and pulps. Moreover, both cultivars exhibit discrete metabolites in respective tissues that could be the basis of their contrasting features.
Partial least-squares discriminant analysis (PLS-DA) for differential metabolites
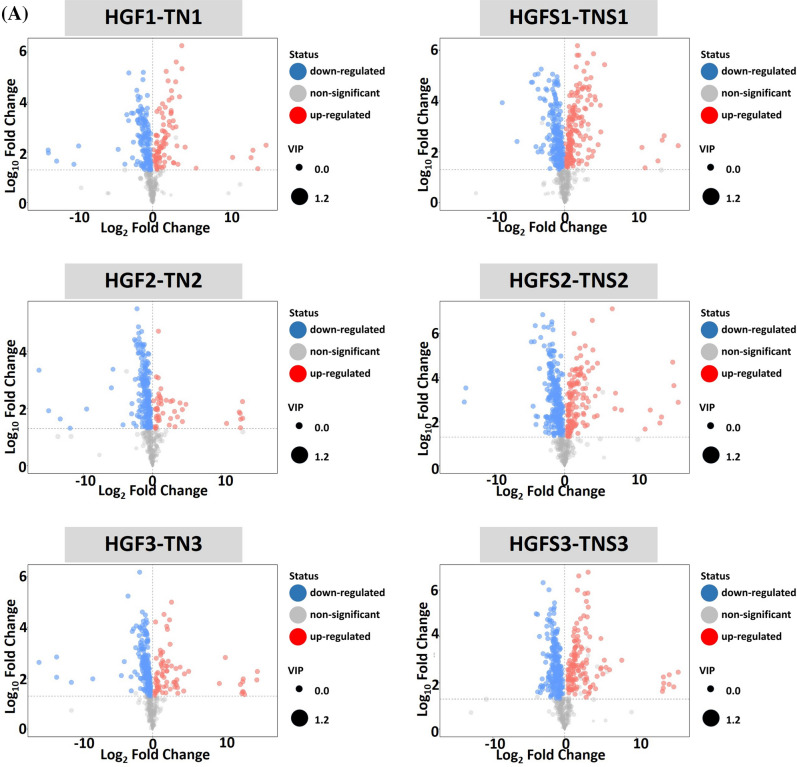
For the estimation of potential contributions of metabolites in cultivar-specific features, pairwise comparisons were performed using OPLS-DA models among three stages of mango fruit for pulp and peel (as inter-cultivar pairs). As a result, higher predictability (Q2) and strong goodness of fit (R2X, R2Y) were observed for these models. (Table S3; Fig. S1). All the values of R2X, R2Y and Q2 in OPLS-DA are above 0.7 and even many of them are close to 1 (Table S3), indicating the models is very good. Besides, R2Y in the permutation test of OPLS-DA of each group is very close to 1 (Fig. S1), suggesting the established model conforms to the real situation of sample data. The Q2 in each model is very close to 1, which shows that the model can well explain the difference between the two groups of samples. The model has no over fitting phenomenon and is very stable. For further improving our understanding of metabolic variations, we performed a differential metabolite screening among all detected metabolites for fold-change and the projection scores or VIP values. The metabolites were considered differentially expressed if the p value was less than 0.05 and the VIP value was greater than 1. The results of this screening are presented using Volcano plots in Fig. 2A and summarized in Table S2.
Figure 2.
Volcano plots and Ven diagrams for differentially expressed metabolites in peel and pulp samples. (A) volcano plots; (B) Ven diagram of pulp samples; (C) Ven diagram of peel samples. TN1, TN2, TN3, HGF1, HGF2, and HGF3 represent pulp samples of TN and HGF cultivars at first, second and third stage respectively. Similarly, TNS1, TNS2, TNS3, HGFS1, HGFS2, and HGFS3 represent peel samples of TN and HGF cultivars at first, second and third stage respectively.
To study the cultivar-specific relative quantification of metabolites in the pulp, a comparison was drawn between pulp metabolites of HGF and TN cultivars. At 1st stage of fruit growth, there were 231 differential metabolites (87 and 144 up-regulated in HGF and TN, respectively), 252 metabolites (47 and 205 up-regulated in HGF and TN, respectively) at 2nd stage, and 261 metabolites (76 and 185 up-regulated in HGF and TN, respectively) at 3rd stage (Fig. 2B, HGF1-TN1, HGF2-TN2, HGF3-TN3). In other words, a higher number of metabolites were up-regulated in TN cultivar as compared to HGF.
Metabolites in pulps
To address the prospective importance of metabolite concentration in fruit maturity features, it was observed that sebacate and ρ-hydroxyphenyl acetic acid (organic acids); indole and epigallocatechin (catechin derivative) were present in very high amounts (> 10 log_fold) in TN pulp (Table S2, represented by blue color in HGF3-TN3). On the other hand in HGF cultivar, 6-hydroxynicotinic acid (vitamins); rosinidin o-hexoside (Anthocyanins); protocatechuic aldehyde (catechin derivatives); syringic acid, 1-o-beta-d-glucopyranosyl sinapate (hydroxycinnamoyl derivatives); chrysin o-hexoside, chrysin 5-o-glucoside (toringin) (flavone); o-feruloyl quinic acid (quinate and its derivatives); eriodictyol c-hexoside (flavone c-glycosides); and aromadedrin (flavonol) were present in very high amounts (> 10 log_fold). Overall, the relative quantities of amino acids, carbohydrates, organic acids, and other metabolites were higher in the pulp of TN cultivars. While the contents of phenolic compounds were relatively higher in HGF cultivar. It explains better sweetness/taste of TN and better quality of HGF pulp17. Among others, syringetin (a flavonol) was detected only at the 2nd stage as ~ 10.71 log_fold higher contents in HGF as compared to TN. This compound has never been reported in mango. Besides, it was just reported in grape and wine26. This metabolite can induce human osteoblast differentiation through bone morphogenetic protein‐2/extracellular signal‐regulated kinase 1/2 pathway26. It can also enhance radio-sensitivity more effectively in cancer cells than in normal cells through enhancement of the caspase-3-mediated apoptosis pathway27. Moreover, growth stage or cultivar-specific variations in metabolite contents may provide important information in identifying respective molecular markers for mango fruit selection at different growth stages (Fig. 2B).
Metabolites in peels
In a comparison of peel samples between HGF and TN cultivars, the upregulated metabolites were as follows: 357 at 1st stage (165 HGF vs 192 TN), 379 at 2nd (138 HGF vs 241 TN) and 375 at 3rd (136 HGF vs 239 TN) (Fig. 2C). However, in the comparison of differential metabolites between growth stages, a higher number of metabolites were upregulated in lateral growth stages. Likewise pulp metabolites, TN peel exhibited up-regulation of a higher number of metabolites as compared to peel of HGF (Fig. 2C).
With a view to exploit the potential of mango peel as a source of valuable metabolites that vary between these two cultivars at very high levels (> 13 log_fold), following metabolites were detected in HGF peel (as compared to TN): DIMBOA glucoside (cyclic hydroxamic acid); spinacetin (flavone); sissotrin (isoflavone); C-hexosyl-chrysoeriol o-hexoside (flavone C-glycosides); cucurbitacin D (terpenoids); gentisic acid (benzoic acid derivatives); Vanillic acid (hydroxycinnamoyl derivatives); N-sinapoyl hydroxycoumarin (coumarins) (Table S2, HGFS3-TNS3). These metabolites mainly constituted flavonoids and may be considered as representative differential metabolites for the different peel colors in both cultivars (Table S2). DIMBOA glucoside represented the most upregulated compound in the peel of HGF cultivar (> 15 log_fold). It belongs to benzoxazinoids (a group of cyclic hydroxamic acids), which are found prevalently in the members of family Poaceae. This metabolite has been reported from Secale cereale L., Triticum aestivum L. and Zea mays L.28. The benzoxazinoid derivatives were discovered in nature in the 1950s and have been attracting significant scientific interest in nutrition and pharmaceutics during the past decade29. Benzoxazinoid hydroxamic acids have been reported that exhibit phytotoxic activities, playing a significant role in plant defense against fungi, bacteria, insects, and participating in allelopathy mechanisms28,30. In this context, it is the first report of DIMBOA-glucoside outside the grass family. Similarly, cucurbitacins are identified as tetracyclic triterpenoids and belong to the Cucurbitaceae family. They are known to have diverse pharmacological activities including antimicrobial activities, anti-inflammatory, antitumor and cardiovascular properties31. Cucurbitacin D (Table S2, HGFS3-TNS3; > 13 log_fold) effectively inhibits glucose uptake and lactate production in metastatic prostate cancer cells via modulating glucose metabolism32. These findings open novel possibilities for the use of mango peel as a source of plant bioactive compounds.
Differential metabolic pathways
To obtain detailed pathway information, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/) was used to map the differential metabolites between both cultivars (Fig. S2, Table S4). The identified metabolic pathways with differential metabolites compared between pulps or peels of both cultivars are shown in Fig. S2A and S2B. These pathways were mainly involved in aminoacyl-tRNA biosynthesis, purine metabolism, glucosinolate biosynthesis, phenylpropanoid biosynthesis (PP), flavonoid biosynthesis and pathways involved in amino acid metabolism (Table S4). These pathways are also involved in the metabolism of plant secondary metabolites. Flavonoids are considered one of the major contributors to crucial features of fruits. The PP pathway in pulp and peel samples differed between both cultivars for differentially expressed metabolites (DEMs) (Fig. S2A and S2B). It is possible that the expression of flavonoid biosynthesis-related genes could be related to genotypic differences.
Phenolic compounds
As the phenolic compounds constituted the largest group of metabolites identified in this analysis, so we decided to analyze this group in detail (Table 1). Previously, a variable but limited number of phenolic compounds were reported in mango fruit10,19,33. Mango fruit generally contains two groups of phenolic acids i.e., hydroxybenzoic and hydroxycinnamic acid derivatives. In literature, hydroxybenzoic acids like protocatechuic acid, vanillic acid, ρ-hydroxybenzoic acid, gallic acid, syringic acid and hydroxycinnamic acids like caffeic acid, chlorogenic acid, ferulic acid, and ρ-coumaric acid have been reported in mango. Although, the metabolic content and type vary with geographical location, plant age and variety2. In current study, the majority of metabolites are reported for the first time in mango (Table 1).
Table 1.
All the phenolic compounds identified in mango fruit (pulp and peel) of both cultivars during three growth stages.
| Sr. no | Class | Metabolite | Precursor ions Q1 (Da) | Product ions Q3 (Da) | Rt. (min) | Molecular weight (Da) | Ionization model | KEGG.ID |
|---|---|---|---|---|---|---|---|---|
| 1 | Anthocyanins | Cyanidin 3-O-glucosyl-malonylglucoside | 697.1 | 696.9 | 2.24 | 697.1 | [M+H]+ | – |
| 2 | Anthocyanins | Delphinidin 3-O-glucoside (Mirtillin) | 465.1 | 303.1 | 2.26 | 465.1 | [M+H]+ | C12138 |
| 3 | Anthocyanins | Pelargonin | 595 | 271.8 | 2.38 | 595 | [M+H]+ | C08725 |
| 4 | Anthocyanins | Petunidin 3-O-glucoside | 479 | 317 | 2.56 | 479 | [M+H]+ | C12139 |
| 5 | Anthocyanins | Cyanidin 3-O-glucoside (Kuromanin) | 449.1 | 287.3 | 2.59 | 449.1 | [M+H]+ | C08604 |
| 6 | Anthocyanins | Cyanidin O-syringic acid | 465.1 | 285.3 | 2.59 | 466.1 | [M−H]− | – |
| 7 | Anthocyanins | Pelargonidin 3-O-β-D-glucoside (Callistephin chloride) | 433.1 | 271 | 2.83 | 433.1 | [M+H]+ | – |
| 8 | Anthocyanins | Malvidin 3-O-glucoside (Oenin) | 493.2 | 331.6 | 2.92 | 493.2 | [M+H]+ | C12140 |
| 9 | Anthocyanins | Delphinidin | 303 | 149.3 | 2.98 | 303.24 | [M+H]+ | C05908 |
| 10 | Anthocyanins | Peonidin O-malonylhexoside | 547.1 | 503.4 | 3 | 548.1 | [M−H]− | – |
| 11 | Anthocyanins | Cyanidin O-diacetyl-hexoside-O-glyceric acid | 619.1 | 531.3 | 3.26 | 620.1 | [M−H]− | – |
| 12 | Anthocyanins | Rosinidin O-hexoside | 477.1 | 315.6 | 3.32 | 477.1 | [M+H]+ | – |
| 13 | Anthocyanins | Cyanidin | 287 | 231.6 | 3.54 | 287.24 | [M+H]+ | C05905 |
| 14 | Anthocyanins | Pelargonidin | 271 | 215.1 | 3.85 | 271.24 | [M+H]+ | C05904 |
| 15 | Anthocyanins | Peonidin | 301.1 | 273.6 | 3.95 | 301.1 | [M+H]+ | C08726 |
| 16 | Benzoic acid derivatives | Anthranilate O-hexosyl-O-hexoside | 460.1 | 118.2 | 0.78 | 461.1 | [M−H]− | – |
| 17 | Benzoic acid derivatives | Gallic acid | 169 | 122.8 | 1.74 | 170.022 | [M−H]− | C01424 |
| 18 | Benzoic acid derivatives | 2,5-dihydroxy benzoic acid O-hexside | 315.1 | 152.1 | 1.84 | 316.1 | [M−H]− | – |
| 19 | Benzoic acid derivatives | Gallic acid O-Hexoside | 331 | 313.7 | 2.02 | 332 | [M−H]− | – |
| 20 | Benzoic acid derivatives | Syringic acid O-glucoside | 359.1 | 197.1 | 2.26 | 360.1 | [M−H]− | – |
| 21 | Benzoic acid derivatives | 2,5-dihydroxybenzoic acid (Gentisic acid) | 153 | 108.1 | 2.5 | 154.027 | [M−H]− | C00628 |
| 22 | Benzoic acid derivatives | 2,4-Dihydroxybenzoic acid | 153 | 109 | 2.75 | 154.027 | [M−H]− | – |
| 23 | Benzoic acid derivatives | Methyl gallate | 183 | 124.1 | 3.14 | 184.0372 | [M−H]− | – |
| 24 | Benzoic acid derivatives | p-Aminobenzoate | 137.3 | 94.3 | 3.14 | 136.3 | [M+H]+ | C00568 |
| 25 | Benzoic acid derivatives | 4-Hydroxybenzaldehyde | 121 | 91.9 | 3.72 | 122.037 | [M−H]− | C00633 |
| 26 | Benzoic acid derivatives | Ethyl gallate | 197.1 | 124.1 | 3.87 | 198.0528 | [M−H]− | – |
| 27 | Benzoic acid derivatives | Vanillin | 151 | 136.1 | 4 | 152.0473 | [M−H]− | C00755 |
| 28 | Benzoic acid derivatives | 8-Methyl-2-oxo-4-phenyl-2H-chromen-7-yl 4-(hexyloxy)benzoate | 457.2 | 191.5 | 4.48 | 456.2 | [M+H]+ | – |
| 29 | Benzoic acid derivatives | Benzoic acid | 121 | 77 | 4.57 | 122.0368 | [M−H]− | C00180 |
| 30 | Catechin derivatives | ( +)-Gallocatechin (GC) | 307 | 248.1 | 2.27 | 306.074 | [M+H]+ | C12127 |
| 31 | Catechin derivatives | Protocatechuic acid O-glucoside | 315.1 | 153.2 | 2.44 | 316.1 | [M−H]− | – |
| 32 | Catechin derivatives | Protocatechuic acid | 153.1 | 109.1 | 2.48 | 154.027 | [M−H]− | C00230 |
| 33 | Catechin derivatives | Epigallocatechin (EGC) | 307 | 139.1 | 2.73 | 306 | [M+H]+ | C12136 |
| 34 | Catechin derivatives | Epigallocatechin (EGC) | 305 | 125 | 2.76 | 306 | [M−H]− | C12136 |
| 35 | Catechin derivatives | Catechin | 291.1 | 139.1 | 2.99 | 290.079 | [M+H]+ | C06562 |
| 36 | Catechin derivatives | Protocatechuic aldehyde | 137.1 | 137 | 3.06 | 138.032 | [M−H]− | C16700 |
| 37 | Catechin derivatives | L-Epicatechin | 289 | 78.8 | 3.18 | 290.3 | [M−H]− | C09727 |
| 38 | Catechin derivatives | Epigallate catechin gallate (EGCG) | 459 | 139.1 | 3.32 | 458.085 | [M+H]+ | C09731 |
| 39 | Catechin derivatives | Catechin–catechin–catechin | 865.1 | 407.2 | 3.44 | 866.1 | [M−H]− | – |
| 40 | Catechin derivatives | Epicatechin-epiafzelechin | 561.1 | 271.3 | 3.61 | 562.1 | [M−H]− | – |
| 41 | Catechin derivatives | Epicatechin gallate (ECG) | 441.3 | 289.1 | 3.89 | 442.3 | [M−H]− | – |
| 42 | Cholines | Choline | 104.1 | 60.2 | 0.76 | 103.1 | [M+H]+ | C00114 |
| 43 | Cholines | O-Phosphocholine | 184 | 83.2 | 0.78 | 183 | [M+H]+ | C00588 |
| 44 | Cholines | sn-Glycero-3-phosphocholine | 258.2 | 125.2 | 0.78 | 258.2 | [M+H]+ | C00670 |
| 45 | Cholines | Acetylcholine | 147.1 | 88 | 0.85 | 146.1181 | [M+H]+ | C08201 |
| 46 | Coumarins | Esculetin (6,7-dihydroxycoumarin) | 177 | 133.1 | 3.24 | 178.027 | [M−H]− | C09263 |
| 47 | Coumarins | Daphnetin | 179 | 179 | 3.31 | 178.027 | [M+H]+ | C03093 |
| 48 | Coumarins | O-Feruloyl 2-hydroxylcoumarin | 339.1 | 177.5 | 3.32 | 338.1 | [M+H]+ | – |
| 49 | Coumarins | O-Feruloyl 3-hydroxylcoumarin | 339.1 | 177.5 | 3.34 | 338.1 | [M+H]+ | – |
| 50 | Coumarins | N-sinapoyl hydroxycoumarin | 369.1 | 207.5 | 3.82 | 368.1 | [M+H]+ | – |
| 51 | Coumarins | O-Feruloyl 4-hydroxylcoumarin | 339.1 | 177.5 | 3.88 | 338.1 | [M+H]+ | – |
| 52 | Coumarins | Scopoletin (7-Hydroxy-5-methoxycoumarin) | 193.1 | 178.1 | 4 | 192.042 | [M+H]+ | C01752 |
| 53 | Coumarins | Scoparone | 207.1 | 207.1 | 4.73 | 206.058 | [M+H]+ | C09311 |
| 54 | Coumarins | 3,4-Dihydrocoumarin | 149.2 | 107 | 5.63 | 148.052 | [M+H]+ | C02274 |
| 55 | Coumarins | 6-MethylCoumarin | 161 | 105.1 | 5.96 | 160.052 | [M+H]+ | – |
| 56 | Flavanone | Afzelechin (3,5,7,4′-Tetrahydroxyflavan) | 275 | 139.1 | 3.4 | 274.084 | [M+H]+ | C09320 |
| 57 | Flavanone | Hesperetin 5-O-glucoside | 463.1 | 301.1 | 3.85 | 464.132 | [M−H]− | – |
| 58 | Flavanone | Hesperetin O-malonylhexoside | 549.2 | 387.3 | 3.99 | 550.2 | [M−H]− | – |
| 59 | Flavanone | Naringenin 7-O-glucoside (Prunin) | 433.1 | 122.9 | 4.22 | 434.1213 | [M−H]− | C09099 |
| 60 | Flavanone | Naringenin O-malonylhexoside | 521 | 317.6 | 4.5 | 520 | [M+H]+ | – |
| 61 | Flavanone | Liquiritigenin | 255 | 119 | 5.16 | 256.074 | [M−H]− | C09762 |
| 62 | Flavanone | Butein | 271.1 | 135.1 | 5.49 | 272.069 | [M−H]− | C08578 |
| 63 | Flavanone | Phloretin | 273.1 | 167.1 | 5.56 | 274.084 | [M−H]− | C00774 |
| 64 | Flavanone | Naringenin chalcone | 273.1 | 153.1 | 5.57 | 272.069 | [M+H]+ | C06561 |
| 65 | Flavanone | Naringenin | 273.1 | 153.1 | 5.59 | 272.0685 | [M+H]+ | C00509 |
| 66 | Flavanone | Isoliquiritigenin | 255 | 119.1 | 6.09 | 256.074 | [M−H]− | C08650 |
| 67 | Flavanone | 7-O-Methyleriodictyol | 301.1 | 135.1 | 6.28 | 302.079 | [M−H]− | – |
| 68 | Flavanone | 4′-Hydroxy-5,7-dimethoxyflavanone | 299.1 | 74.8 | 6.78 | 300.1 | [M−H]− | – |
| 69 | Flavanone | Isosakuranetin (4′-Methylnaringenin) | 287.1 | 161.1 | 6.81 | 286.084 | [M+H]+ | C05334 |
| 70 | Flavanone | Pinocembrin (Dihydrochrysin) | 257.1 | 153 | 7.05 | 256.074 | [M+H]+ | C09827 |
| 71 | Flavanone | Xanthohumol | 355.2 | 178.9 | 8.4 | 354.147 | [M+H]+ | C16417 |
| 72 | Flavone | Chrysoeriol O-hexosyl-O-malonylhexoside | 709.1 | 547.3 | 2.49 | 710.1 | [M−H]− | – |
| 73 | Flavone | Acacetin O-acetyl hexoside | 487.1 | 283.2 | 2.63 | 488.1 | [M−H]− | – |
| 74 | Flavone | Luteolin O-hexosyl-O-hexosyl-O-hexoside | 771.1 | 609.5 | 2.83 | 772.1 | [M−H]− | – |
| 75 | Flavone | Selgin 5-O-hexoside | 479.1 | 302.8 | 3.52 | 478.1 | [M+H]+ | – |
| 76 | Flavone | Tricin O-sinapic acid | 535 | 329.4 | 3.75 | 536 | [M−H]− | – |
| 77 | Flavone | Tricin O-saccharic acid | 521.1 | 329.3 | 3.81 | 522.1 | [M−H]− | – |
| 78 | Flavone | Luteolin 7-O-glucoside (Cynaroside) | 449.1 | 287.1 | 3.87 | 448.101 | [M+H]+ | C03951 |
| 79 | Flavone | Chrysoeriol O-acetylhexoside | 503.1 | 341.3 | 3.94 | 504.1 | [M−H]− | – |
| 80 | Flavone | Apigenin 7-O-neohesperidoside (Rhoifolin) | 579.2 | 271.1 | 4.01 | 578.1636 | [M+H]+ | C12627 |
| 81 | Flavone | Apigenin 7-rutinoside (Isorhoifolin) | 579.2 | 271.1 | 4.01 | 578.1636 | [M+H]+ | – |
| 82 | Flavone | Chrysoeriol O-rhamnosyl-O-glucuronic acid | 621.1 | 299.4 | 4.07 | 622.1 | [M−H]− | – |
| 83 | Flavone | Tricin di-O-hexoside | 655.2 | 331.7 | 4.16 | 654.2 | [M+H]+ | – |
| 84 | Flavone | Syringetin 5-O-hexoside | 509.2 | 347.6 | 4.17 | 508.2 | [M+H]+ | – |
| 85 | Flavone | Syringetin 7-O-hexoside | 509.3 | 283.6 | 4.17 | 508.3 | [M+H]+ | – |
| 86 | Flavone | Selgin O-hexosyl-O-hexoside | 641.1 | 479.5 | 4.39 | 640.1 | [M+H]+ | – |
| 87 | Flavone | Spinacetin | 347 | 288 | 4.41 | 346 | [M+H]+ | – |
| 88 | Flavone | Chrysoeriol O-malonylhexoside | 549.1 | 301.7 | 4.55 | 548.1 | [M+H]+ | – |
| 89 | Flavone | Tricin O-malonylhexoside | 579.1 | 331.7 | 4.56 | 578.1 | [M+H]+ | – |
| 90 | Flavone | 7,4′-Dihydroxyflavone | 255.1 | 137.1 | 4.57 | 254.058 | [M+H]+ | C12123 |
| 91 | Flavone | Apigenin 4-O-rhamnoside | 417.1 | 270.9 | 4.9 | 416.111 | [M+H]+ | – |
| 92 | Flavone | Chrysoeriol O-sinapoylhexoside | 669 | 301.6 | 4.92 | 668 | [M+H]+ | – |
| 93 | Flavone | Chrysin 5-O-glucoside (Toringin) | 417.1 | 255.7 | 5 | 416.1 | [M+H]+ | – |
| 94 | Flavone | Luteolin | 287.1 | 287.1 | 5 | 286.1 | [M+H]+ | C01514 |
| 95 | Flavone | Acetyl-eriodictyol O-hexoside | 491.1 | 287.3 | 5.16 | 492.1 | [M−H]− | – |
| 96 | Flavone | Chrysin O-malonylhexoside | 503 | 255.6 | 5.24 | 502 | [M+H]+ | – |
| 97 | Flavone | Butin | 273.1 | 153.1 | 5.59 | 272.069 | [M+H]+ | C09614 |
| 98 | Flavone | Apigenin | 271.1 | 215.1 | 5.63 | 270.0528 | [M+H]+ | C01477 |
| 99 | Flavone | Tricin | 331.1 | 315.8 | 5.74 | 330.1 | [M+H]+ | – |
| 100 | Flavone | Chrysoeriol | 301.1 | 286.1 | 5.77 | 300.0634 | [M+H]+ | C04293 |
| 101 | Flavone | Tricin 7-O-acetylglucoside | 535.3 | 487.2 | 5.78 | 534.3 | [M+H]+ | – |
| 102 | Flavone | Amentoflavone | 539.1 | 403 | 5.88 | 538.09 | [M+H]+ | C10018 |
| 103 | Flavone | Baicalein (5,6,7-Trihydroxyflavone) | 269.1 | 251.1 | 5.94 | 270.053 | [M−H]− | C10023 |
| 104 | Flavone | Chrysin | 255.1 | 69.7 | 6.95 | 254.0579 | [M+H]+ | C10028 |
| 105 | Flavone | sakuranetin | 287.1 | 287.1 | 6.96 | 286.084 | [M+H]+ | C09833 |
| 106 | Flavone | Acacetin | 283.1 | 268 | 7.06 | 284.069 | [M−H]− | C01470 |
| 107 | Flavone | Nobiletin | 403.1 | 373.1 | 7.06 | 402.132 | [M+H]+ | C10112 |
| 108 | Flavone | Velutin | 313.1 | 298.3 | 7.22 | 314.1 | [M−H]− | – |
| 109 | Flavone | Tangeretin | 373.1 | 373.1 | 7.54 | 372.121 | [M+H]+ | C10190 |
| 110 | Flavone C-glycosides | 8-C-hexosyl-hesperetin O-hexoside | 627.1 | 430 | 2.78 | 626.1 | [M+H]+ | – |
| 111 | Flavone C-glycosides | 6-C-hexosyl-luteolin O-hexoside | 611.1 | 329 | 3.09 | 610.1 | [M+H]+ | – |
| 112 | Flavone C-glycosides | Eriodictiol C-hexosyl-O-hexoside | 613.1 | 300.3 | 3.09 | 612.1 | [M+H]+ | – |
| 113 | Flavone C-glycosides | C-hexosyl-chrysoeriol O-hexoside | 625.2 | 463.6 | 3.37 | 624.2 | [M+H]+ | – |
| 114 | Flavone C-glycosides | Eriodictyol C-hexoside | 449.1 | 329.3 | 3.37 | 450.1 | [M−H]− | – |
| 115 | Flavone C-glycosides | 6-C-hexosyl-hesperetin O-hexoside | 627.1 | 447.3 | 3.41 | 626.1 | [M+H]+ | – |
| 116 | Flavone C-glycosides | Luteolin 6-C-glucoside | 449.1 | 300 | 3.45 | 448.1 | [M+H]+ | – |
| 117 | Flavone C-glycosides | Luteolin C-hexoside | 449.1 | 329.6 | 3.45 | 448.1 | [M+H]+ | – |
| 118 | Flavone C-glycosides | C-hexosyl-luteolin O-hexosyl-O-salicylic acid | 731.1 | 431.5 | 3.55 | 730.1 | [M+H]+ | – |
| 119 | Flavone C-glycosides | C-hexosyl-apigenin O-pentoside | 565.1 | 397.8 | 3.6 | 564.1 | [M+H]+ | – |
| 120 | Flavone C-glycosides | di-C,C-hexosyl-apigenin | 595.1 | 415.4 | 3.68 | 594.1 | [M+H]+ | – |
| 121 | Flavone C-glycosides | Vitexin 2″-O-β-L-rhamnoside | 579 | 433.1 | 3.68 | 578.164 | [M+H]+ | C12628 |
| 122 | Flavone C-glycosides | C-hexosyl-luteolin O-p-coumaroylhexoside | 757.2 | 757 | 3.73 | 756.2 | [M+H]+ | – |
| 123 | Flavone C-glycosides | 8-C-hexosyl-luteolin O-hexoside | 611.1 | 299.8 | 3.75 | 610.1 | [M+H]+ | – |
| 124 | Flavone C-glycosides | Apigenin C-glucoside | 433.1 | 271.7 | 3.78 | 432.1 | [M+H]+ | – |
| 125 | Flavone C-glycosides | Isovitexin | 431.1 | 431.1 | 3.79 | 432.1056 | [M−H]− | C01714 |
| 126 | Flavone C-glycosides | Naringenin C-hexoside | 435.1 | 339.7 | 3.79 | 434.1 | [M+H]+ | – |
| 127 | Flavone C-glycosides | Acacetin C-hexoside | 447.1 | 298 | 3.84 | 446.1 | [M+H]+ | – |
| 128 | Flavone C-glycosides | O-methylnaringenin C-pentoside | 419.1 | 383.9 | 3.84 | 418.1 | [M+H]+ | – |
| 129 | Flavone C-glycosides | C-hexosyl-luteolin O-feruloylpentoside | 757.1 | 739 | 3.87 | 756.1 | [M+H]+ | – |
| 130 | Flavone C-glycosides | Chrysoeriol 8-C-hexoside | 463.1 | 367.8 | 3.89 | 462.1 | [M+H]+ | – |
| 131 | Flavone C-glycosides | Chrysin C-hexoside | 417.2 | 381.9 | 4 | 416.2 | [M+H]+ | – |
| 132 | Flavone C-glycosides | C-pentosyl-apigenin O-p-coumaroylhexoside | 711.2 | 693.1 | 4.1 | 710.2 | [M+H]+ | – |
| 133 | Flavone C-glycosides | C-pentosyl-chrysoeriol 7-O-feruloylhexoside | 771.2 | 177.5 | 4.25 | 770.2 | [M+H]+ | – |
| 134 | Flavone C-glycosides | 8-C-hexosyl-apigenin O-feruloylhexoside | 771.2 | 753 | 4.26 | 770.2 | [M+H]+ | – |
| 135 | Flavone C-glycosides | Apigenin 6-C-pentoside | 403.1 | 367.7 | 4.3 | 402.1 | [M+H]+ | – |
| 136 | Flavone C-glycosides | Apigenin 8-C-pentoside | 403.1 | 367.6 | 4.32 | 402.1 | [M+H]+ | – |
| 137 | Flavonol | Quercetin 5-O-malonylhexosyl-hexoside | 713.1 | 713.1 | 2.96 | 712.1 | [M+H]+ | – |
| 138 | Flavonol | Dihydromyricetin | 321.1 | 153.1 | 3.52 | 320.053 | [M+H]+ | C02906 |
| 139 | Flavonol | Myricetin 3-O-galactoside | 479.1 | 317.3 | 3.53 | 480.09 | [M−H]− | – |
| 140 | Flavonol | Fustin | 289 | 215.1 | 3.65 | 288.063 | [M+H]+ | C01378 |
| 141 | Flavonol | Quercetin 7-O-rutinoside | 611.2 | 303.7 | 3.7 | 610.2 | [M+H]+ | – |
| 142 | Flavonol | Kaempferol 3,7-dirhamnoside (Kaempferitrin) | 579.2 | 433.1 | 3.74 | 578.164 | [M+H]+ | C16981 |
| 143 | Flavonol | Kaempferol 3-O-robinobioside (Biorobin) | 595.2 | 287 | 3.76 | 594.159 | [M+H]+ | – |
| 144 | Flavonol | Quercetin O-acetylhexoside | 505.1 | 301.2 | 3.8 | 506.1 | [M−H]− | – |
| 145 | Flavonol | Quercetin 4′-O-glucoside (Spiraeoside) | 465 | 302.9 | 3.86 | 464.096 | [M+H]+ | – |
| 146 | Flavonol | Kaempferol 3-O-galactoside (Trifolin) | 449 | 286.9 | 3.87 | 448.101 | [M+H]+ | C12626 |
| 147 | Flavonol | Quercetin 3-O-glucoside (Isotrifoliin) | 465 | 302.8 | 3.87 | 464.096 | [M+H]+ | C05623 |
| 148 | Flavonol | Quercetin 3-α-L-arabinofuranoside (Avicularin) | 435.1 | 303 | 3.98 | 434.0849 | [M+H]+ | – |
| 149 | Flavonol | Kaempferide | 301.1 | 259.1 | 4.13 | 300.1 | [M+H]+ | C10098 |
| 150 | Flavonol | Dihydroquercetin (Taxifolin) | 303.1 | 125.1 | 4.15 | 304.058 | [M−H]− | C01617 |
| 151 | Flavonol | Isorhamnetin 5-O-hexoside | 479.2 | 317.8 | 4.17 | 478.2 | [M+H]+ | – |
| 152 | Flavonol | Kaempferol 3-O-rhamnoside (Kaempferin) | 431 | 285.1 | 4.48 | 432.106 | [M−H]− | C16911 |
| 153 | Flavonol | Aromadedrin (Dihydrokaempferol) | 287.1 | 125.1 | 4.62 | 288.063 | [M−H]− | C00974 |
| 154 | Flavonol | Myricetin | 319 | 319 | 4.7 | 318.038 | [M+H]+ | C10107 |
| 155 | Flavonol | Quercetin-3,4′-O-di-β-glucopyranoside | 627.1 | 465.2 | 4.83 | 626.1 | [M+H]+ | – |
| 156 | Flavonol | Kaempferol 7-O-rhamnoside | 433.1 | 287 | 4.94 | 432.106 | [M+H]+ | – |
| 157 | Flavonol | Morin | 301 | 151.1 | 5.11 | 302.04265 | [M−H]− | C10105 |
| 158 | Flavonol | Quercetin | 303 | 303 | 5.13 | 302.043 | [M+H]+ | C00389 |
| 159 | Flavonol | Laricitrin | 333 | 305.2 | 5.15 | 332.053 | [M+H]+ | C12633 |
| 160 | Flavonol | Kaempferol | 285 | 214 | 5.73 | 286.048 | [M−H]− | C05903 |
| 161 | Flavonol | Syringetin | 347 | 287.1 | 5.8 | 346.069 | [M+H]+ | C11620 |
| 162 | Flavonol | Isorhamnetin | 315.1 | 300.1 | 5.85 | 316.058 | [M−H]− | C10084 |
| 163 | Flavonol | Di-O-methylquercetin | 329.1 | 314.3 | 5.91 | 330.1 | [M−H]− | – |
| 164 | Flavonol | Ayanin | 345.2 | 177.5 | 6.33 | 344.2 | [M+H]+ | C04444 |
| 165 | Flavonol | Rhamnetin (7-O-methxyl quercetin) | 317 | 317 | 6.43 | 316.058 | [M+H]+ | C10176 |
| 166 | Flavonol | 3,7-Di-O-methylquercetin | 329 | 314 | 6.63 | 330.074 | [M−H]− | C01265 |
| 167 | Flavonol | Troxerutin (Trihydroxyethyl rutin) | 347.3 | 285.4 | 6.66 | 346.251 | [M+H]+ | – |
| 168 | Flavonol | Kumatakenin | 315.1 | 300 | 7.23 | 314.079 | [M+H]+ | – |
| 169 | Flavonolignan | Tricin 4′-O-(syringyl glyceryl)ether | 557.2 | 331.9 | 4.44 | 556.2 | [M+H]+ | – |
| 170 | Flavonolignan | Tricin 7-O-β-guaiacylglycerol | 527.1 | 331.7 | 5.44 | 526.1 | [M+H]+ | – |
| 171 | Flavonolignan | Tricin 4′-O-syringyl alcohol | 497.1 | 331.8 | 5.75 | 496.1 | [M+H]+ | – |
| 172 | Flavonolignan | Tricin 4′-O-β-guaiacylglycerol | 527.1 | 331.7 | 5.84 | 526.1 | [M+H]+ | – |
| 173 | Hydroxycinnamoyl derivatives | Caftaric acid | 311.1 | 149.2 | 2.16 | 312.1 | [M−H]− | – |
| 174 | Hydroxycinnamoyl derivatives | O-Caffeoyl maltotriose | 665.1 | 323.4 | 2.23 | 666.1 | [M+H]+ | – |
| 175 | Hydroxycinnamoyl derivatives | Syringin | 371.1 | 209.2 | 2.58 | 372.142 | [M+H]+ | C01533 |
| 176 | Hydroxycinnamoyl derivatives | Coniferin | 341 | 179.1 | 2.72 | 342.132 | [M−H]− | C00761 |
| 177 | Hydroxycinnamoyl derivatives | Caffeic acid O-glucoside | 341 | 179.2 | 2.86 | 342 | [M−H]− | – |
| 178 | Hydroxycinnamoyl derivatives | Homovanillic acid | 181.1 | 137.1 | 3.04 | 182.0579 | [M+H]+ | C05582 |
| 179 | Hydroxycinnamoyl derivatives | Hydroxy-methoxycinnamate | 195.1 | 177.5 | 3.22 | 194.1 | [M+H]+ | – |
| 180 | Hydroxycinnamoyl derivatives | 1-O-β-d-Glucopyranosyl sinapate | 385.1 | 223.2 | 3.26 | 386.1 | [M−H]− | – |
| 181 | Hydroxycinnamoyl derivatives | 6-Hydroxymethylherniarin | 207.1 | 147.4 | 3.31 | 206.1 | [M+H]+ | – |
| 182 | Hydroxycinnamoyl derivatives | Vanillic acid | 169 | 111 | 3.35 | 168.042 | [M+H]+ | C06672 |
| 183 | Hydroxycinnamoyl derivatives | Syringic acid | 197.1 | 122.9 | 3.38 | 198.0528 | [M−H]− | C10833 |
| 184 | Hydroxycinnamoyl derivatives | Feruloyl syringic acid | 375.2 | 137.6 | 3.47 | 374.2 | [M−H]− | – |
| 185 | Hydroxycinnamoyl derivatives | p-Coumaryl alcohol | 149 | 130.1 | 3.67 | 150.068 | [M−H]− | C02646 |
| 186 | Hydroxycinnamoyl derivatives | 3-(4-Hydroxyphenyl)propionic acid | 165.1 | 92.9 | 3.84 | 166.063 | [M−H]− | C01744 |
| 187 | Hydroxycinnamoyl derivatives | p-Coumaric acid | 163 | 119 | 3.86 | 164.047 | [M−H]− | C00811 |
| 188 | Hydroxycinnamoyl derivatives | Coniferyl alcohol | 179.1 | 146.1 | 3.88 | 180.079 | [M−H]− | C00590 |
| 189 | Hydroxycinnamoyl derivatives | Sinapyl alcohol | 209 | 179.1 | 3.88 | 210.089 | [M+H]+ | C02325 |
| 190 | Hydroxycinnamoyl derivatives | Ferulic acid | 193.1 | 134.1 | 4.07 | 194.0579 | [M+H]+ | C01494 |
| 191 | Hydroxycinnamoyl derivatives | 3-Hydroxy-4-methoxycinnamic acid | 193.1 | 134.1 | 4.08 | 194.0579 | [M−H]− | – |
| 192 | Hydroxycinnamoyl derivatives | 2-Methoxybenzoic acid | 151 | 136.1 | 4.16 | 152.0473 | [M−H]− | – |
| 193 | Hydroxycinnamoyl derivatives | p-Coumaraldehyde | 149.1 | 131 | 4.42 | 148 | [M−H]− | – |
| 194 | Hydroxycinnamoyl derivatives | Resveratrol | 229.1 | 135 | 4.59 | 228.079 | [M−H]− | C03582 |
| 195 | Hydroxycinnamoyl derivatives | Sinapinaldehyde | 207.1 | 177.1 | 4.61 | 208 | [M−H]− | – |
| 196 | Hydroxycinnamoyl derivatives | Coniferylaldehyde | 179.1 | 123 | 4.64 | 178.063 | [M−H]− | C02666 |
| 197 | Hydroxycinnamoyl derivatives | Pinoresinol | 357.1 | 136.1 | 5.41 | 358.142 | [M+H]+ | – |
| 198 | Hydroxycinnamoyl derivatives | 4-Methoxycinnamic acid | 177 | 145.2 | 5.5 | 178 | [M−H]− | – |
| 199 | Hydroxycinnamoyl derivatives | 3,4-Dimethoxycinnamic acid | 207.1 | 192.1 | 5.52 | 208.1 | [M−H]− | – |
| 200 | Hydroxycinnamoyl derivatives | trans-cinnamaldehyde | 133.1 | 115 | 5.97 | 132.0575 | [M+H]+ | C00903 |
| 201 | Hydroxycinnamoyl derivatives | Caffeic aldehyde | 165.1 | 95.5 | 6.04 | 164.1 | [M+H]+ | C10945 |
| 202 | Hydroxycinnamoyl derivatives | Syringaldehyde | 183.1 | 165.5 | 6.58 | 182.1 | [M−H]− | – |
| 203 | Hydroxycinnamoyl derivatives | Methyleugenol | 179 | 138 | 7.3 | 178.099 | [M−H]− | C10454 |
| 204 | Hydroxycinnamoyl derivatives | Caffeyl alcohol | 317.2 | 281.3 | 7.47 | 316 | [M+H]+ | C09066 |
| 205 | Isoflavone | Daidzein 7-O-glucoside (Daidzin) | 417.1 | 255.1 | 3.36 | 416.111 | [M+H]+ | C10216 |
| 206 | Isoflavone | Glycitin | 445 | 282.1 | 3.54 | 446.121 | [M+H]+ | C16195 |
| 207 | Isoflavone | Genistein 7-O-Glucoside (Genistin) | 433 | 270.9 | 4.01 | 432.106 | [M−H]− | C09126 |
| 208 | Isoflavone | Formononetin 7-O-glucoside (Ononin) | 429.1 | 267.1 | 4.59 | 430.126 | [M+H]+ | C10509 |
| 209 | Isoflavone | 2′-Hydroxygenistein | 287 | 217.1 | 4.89 | 286.048 | [M−H]− | C12134 |
| 210 | Isoflavone | Daidzein | 255.1 | 199.1 | 4.89 | 254.0579 | [M+H]+ | C10208 |
| 211 | Isoflavone | Orobol (5,7,3′,4′-tetrahydroxyisoflavone) | 285 | 257.1 | 5.08 | 286.048 | [M−H]− | C10510 |
| 212 | Isoflavone | Sissotrin | 447.1 | 285.1 | 5.16 | 446.121 | [M−H]− | C05376 |
| 213 | Isoflavone | Formononetin (4′-O-methyldaidzein) | 269.1 | 269.1 | 6.33 | 268.074 | [M+H]+ | C00858 |
| 214 | Isoflavone | Prunetin | 283 | 268.1 | 6.97 | 284.069 | [M−H]− | C10521 |
| 215 | Phenolamides | Spermidine | 146.2 | 72.1 | 0.56 | 145.2 | [M+H]+ | C00315 |
| 216 | Phenolamides | Spermine | 203 | 112 | 0.62 | 202 | [M+H]+ | C00750 |
| 217 | Phenolamides | Putrescine | 89 | 71.9 | 0.64 | 88.1 | [M+H]+ | C00134 |
| 218 | Phenolamides | Agmatine | 131.1 | 72.1 | 0.75 | 130.1 | [M+H]+ | C00179 |
| 219 | Phenolamides | 1,5-Diaminopentane | 103 | 86.1 | 0.76 | 102.116 | [M+H]+ | C01672 |
| 220 | Phenolamides | N-Acetylputrescine | 131 | 71.9 | 0.8 | 130.11061 | [M+H]+ | C02714 |
| 221 | Phenolamides | N-hexosyl-p-coumaroyl putrescine | 397.1 | 147.4 | 1.73 | 396.1 | [M+H]+ | – |
| 222 | Phenolamides | N-Caffeoyl putrescine | 251.1 | 233.5 | 1.97 | 250.1 | [M+H]+ | C03002 |
| 223 | Phenolamides | N-(4′-O-glycosyl)-p-coumaroyl agmatine | 439.1 | 147.4 | 2.11 | 438.1 | [M+H]+ | – |
| 224 | Phenolamides | N′,N″,N″′-p-coumaroyl-cinnamoyl-caffeoyl spermidine | 584.2 | 325.8 | 2.42 | 583.2 | [M+H]+ | – |
| 225 | Phenolamides | N′, N″-di-p-coumaroylspermine | 495.3 | 478.4 | 2.43 | 494.3 | [M+H]+ | – |
| 226 | Phenolamides | N′-Feruloyl putrescine | 265.1 | 177.5 | 2.44 | 264.1 | [M+H]+ | – |
| 227 | Phenolamides | N-Caffeoyl agmatine | 293.2 | 234.5 | 2.45 | 292.2 | [M+H]+ | – |
| 228 | Phenolamides | N-Sinapoyl putrescine | 295 | 207.6 | 2.77 | 294 | [M+H]+ | – |
| 229 | Phenolamides | N′, N″-disinapoylspermidine | 558.3 | 264.8 | 3.75 | 557.3 | [M+H]+ | – |
| 230 | Proanthocyanidins | Procyanidin B3 | 577.1 | 407.3 | 2.79 | 578.1424 | [M−H]− | – |
| 231 | Proanthocyanidins | Procyanidin A3 | 577.1 | 425.4 | 2.92 | 576.1 | [M+H]+ | – |
| 232 | Proanthocyanidins | Procyanidin B2 | 579.1 | 127.3 | 3.03 | 578.1424 | [M+H]+ | – |
| 233 | Proanthocyanidins | Procyanidin A1 | 575 | 285.3 | 3.68 | 576.1268 | [M−H]− | – |
| 234 | Proanthocyanidins | Procyanidin A2 | 577 | 425.9 | 4.06 | 576.1268 | [M+H]+ | C10237 |
| 235 | Quinate and its derivatives | Quinic acid | 191 | 85 | 0.92 | 192.063 | [M−H]− | C00296 |
| 236 | Quinate and its derivatives | p-Coumaroyl quinic acid O-glucuronic acid | 513.1 | 191.2 | 1.82 | 514.1 | [M−H]− | – |
| 237 | Quinate and its derivatives | Quinacyl syringic acid | 371.1 | 179.2 | 1.85 | 372.1 | [M−H]− | – |
| 238 | Quinate and its derivatives | Homovanilloyl quinic acid | 355.1 | 181.2 | 1.89 | 356.1 | [M−H]− | – |
| 239 | Quinate and its derivatives | 5-O-p-coumaroyl quinic acid O-hexoside | 499.1 | 163.2 | 2.1 | 500.1 | [M−H]− | – |
| 240 | Quinate and its derivatives | O-Feruloyl quinic acid | 369.1 | 177.5 | 2.12 | 368.1 | [M+H]+ | – |
| 241 | Quinate and its derivatives | Quinic acid O-di-glucuronic acid | 543.1 | 191.2 | 2.13 | 544.1 | [M−H]− | – |
| 242 | Quinate and its derivatives | Neochlorogenic acid (5-O-Caffeoylquinic acid) | 353 | 191.1 | 2.35 | 354.095 | [M−H]− | C17147 |
| 243 | Quinate and its derivatives | 1-O-Caffeoyl quinic acid | 353.1 | 191.1 | 2.38 | 354.095 | [M−H]− | – |
| 244 | Quinate and its derivatives | 3-O-p-coumaroyl quinic acid O-hexoside | 499.2 | 173.2 | 2.45 | 500.2 | [M−H]− | – |
| 245 | Quinate and its derivatives | 5-O-p-coumaroyl shikimic acid O-hexoside | 481.1 | 445.4 | 2.62 | 482.1 | [M−H]− | – |
| 246 | Quinate and its derivatives | Chlorogenic acid (3-O-Caffeoylquinic acid) | 353.1 | 191.1 | 2.72 | 354.0951 | [M−H]− | C00852 |
| 247 | Quinate and its derivatives | 1-O-p-Coumaroyl quinic acid | 337.1 | 155.8 | 2.85 | 338.1 | [M−H]− | – |
| 248 | Quinate and its derivatives | 4-O-Caffeoyl quinic acid (criptochlorogenic acid) | 353.1 | 191.2 | 3 | 354.1 | [M−H]− | – |
| 249 | Quinate and its derivatives | 3-O-Feruloyl quinic acid | 369.1 | 177.5 | 3.01 | 368.1 | [M+H]+ | C02572 |
| 250 | Quinate and its derivatives | O-Sinapoyl quinic acid | 399 | 207.5 | 3.17 | 398 | [M+H]+ | – |
| 251 | Quinate and its derivatives | 3-O-p-coumaroyl shikimic acid O-hexoside | 481.1 | 319.3 | 3.23 | 482.1 | [M−H]− | – |
| 252 | Quinate and its derivatives | 5-O-p-Coumaroylquinic acid | 337 | 275.8 | 3.26 | 338 | [M−H]− | – |
| 253 | Quinate and its derivatives | 3-O-p-Coumaroyl quinic acid | 337.1 | 190.9 | 3.3 | 338.1 | [M−H]− | – |
| 254 | Quinate and its derivatives | 1-O-Feruloyl quinic acid | 369.1 | 207.5 | 3.42 | 368.1 | [M+H]+ | – |
| 255 | Quinate and its derivatives | Chlorogenic acid methyl ester | 367 | 179.1 | 3.64 | 368.111 | [M−H]− | – |
| 256 | Quinate and its derivatives | 5-O-p-Coumaroyl shikimic acid | 321.1 | 147.5 | 3.81 | 320.1 | [M+H]+ | – |
| 257 | Quinate and its derivatives | 3-O-p-Coumaroyl shikimic acid | 319 | 145.3 | 3.93 | 320 | [M−H]− | – |
Absolute quantification of catechin derivatives and total proanthocyanidins (PAs) contents
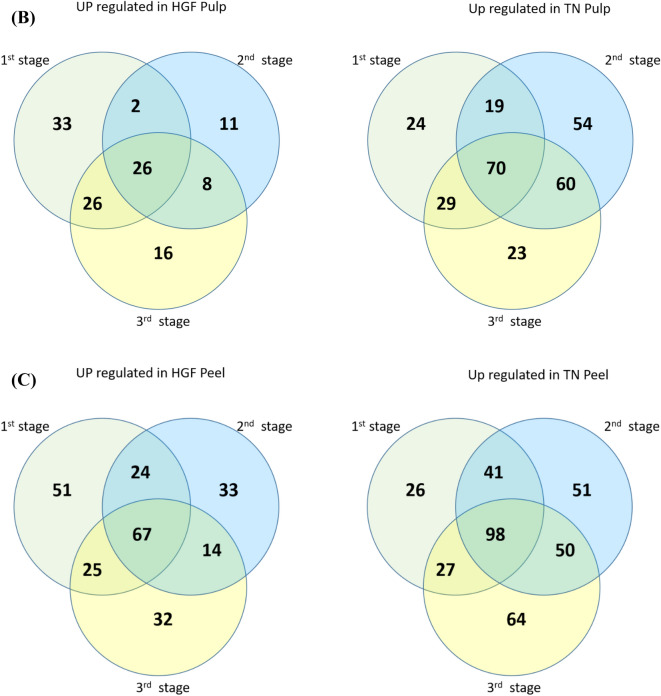
Current study reports developmental stage dependent variation (Fig. 3A) in total proanthocyanidins (PAs) contents using a method described by Dong et al.34. Pulp samples of both cultivars contain similar quantities of PAs. However, there is an increasing trend for PAs contents towards fruit maturity in both cultivars (Fig. 3B). Fruit peels of both cultivars contain higher amount of PAs as compared to pulps. For TN cultivar, the concentration of peel PAs remained in close range across three growth stages. However, there was a massive increase in PAs content of HGF cultivar towards maturity (Fig. 3B). Very few studies have documented the concentration of PAs in mango and it is often influenced by several factors including tissue type, geographical area and method of extraction33,35–37. Pulp of mangoes from USA were reported to contain 12.8 mg proanthocyanidins/100 g fresh weight. Two different extraction methods yielded 0.18 and 0.48 mg PAs per 100 g DW of mangoes from Spain36. Similarly, procyanidins A2 (14–78 µg /ml), B1 (29–88 µg /ml) and B2 (0–10 µg /ml) were reported in peel liqueurs of mangoes from Brazil37.
Figure 3.
Fruit color and total proanthocyanidin (PA) contents of Hongguifei (HGF) and Tainong (TN) mango.
The phenolic compounds are among the major contributors that are accountable for antioxidant properties in fruits, vegetables, whole grains and other plant-based materials38. The TN mango fruit contains higher levels of total phenolics (TPs) and total flavonoids as compared to HGF2. However, total anthocyanin contents (TAs) were reported to be higher in HGF as compared to TN2. In phenylpropanoid biosynthesis pathway, both anthocyanins and proanthocyanins belong to the terminal steps39. Therefore, the increase in PAs (Fig. 3) potentially correlates with anthocyanins2 in HGF mango.
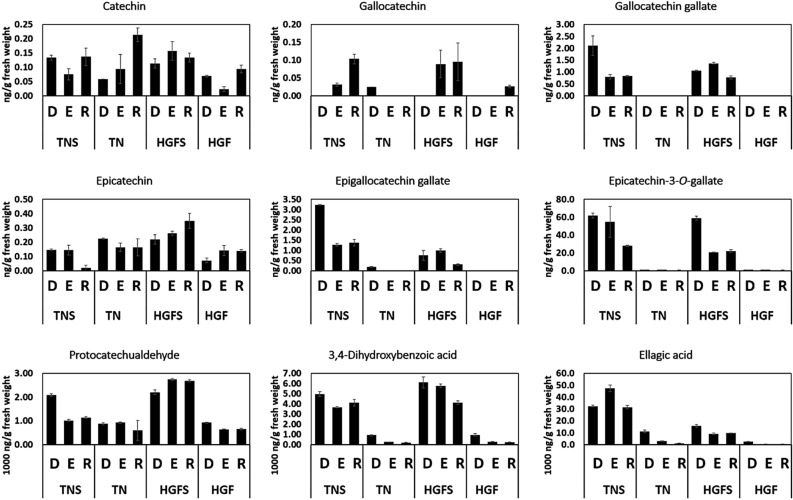
In order to validate the results of metabolome based estimation of relative quantities of catechin and derivatives, absolute quantities of catechin and its derivatives were calculated in peel and pulp samples of both cultivars using HPLC. It was observed that the concentrations of catechin, gallocatechin, gallocatechin gallate, epicatechin, epicatechin gallate, epicatechin-3-O-gallate, protocatechuic aldehyde, 3,4-Dihydroxybenzoic acid and ellagic acid were differentially present in both cultivars for pulp and peel samples (Fig. 4). Overall, peel samples contained higher amounts of these metabolites that decreased with the age of fruit40. Moreover, fold change modifications of these metabolites follow similar trend (with minor variations) as discussed above (Table S2). In previous studies, these compounds were either individually reported as catechine41, epicatechin42, protocatechuic acid2 and ellagic acid or in terms of total proanthocyanidin and tannins10. The composition of phenolic compounds in peels has attracted a crucial importance for mango in calculating functional food mixtures43.
Figure 4.
HPLC based quantification of Catechin, derivatives and phenolic acids in the pulp and peel of both cultivars. TNS and HGFS represent quantification in peel of respective cultivars. TN and HGF represent quantification in pulp of respective cultivars. The letters D, E and R represent development, enlargement and ripening of mango fruit. The use of commercial standards for the quantification of catechin derivatives and phenolic acids along with linear equations, correlation coefficients, LOD’s, and LOQ’s values are presented in Table S5.
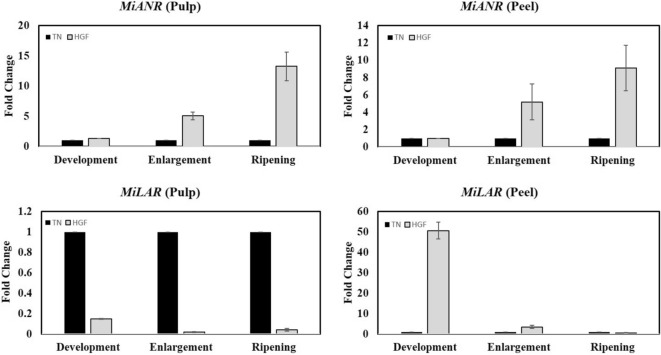
Differential expression of MiANR and MiLAR genes
The genes encoding anthocyanidin reductase (ANR) and leucoanthocyanidin reductase (LAR) enzyme have been cloned and characterized in plants including poplar, buckwheat, lotus and fruits such as grapevine, strawberry, persimmon, apple, and mango (for references see44). The expression patterns of these genes are highly correlated with PA accumulation in many plants. To determine whether the differential expression of catechin/derivatives correlated with the transcript abundance of MiANR and MiLAR, the expression levels of these genes were analyzed in fruits of both cultivars using relative qRT-PCR (Fig. 5). In pulp samples, both genes exhibited an opposite expression profile i.e., the expression of MiANR increased with fruit maturity in HGF and MiLAR followed similar pattern in TN cultivar. In peel samples of both cultivars, the relative difference of expression increased (with fruit maturity) for MiANR and decreased (with fruit maturity) for MiLAR (Fig. 5). These results suggested that transcription of LAR and ANR seems controlled by a feedback mechanism45. It means a higher concentration of catechin may stimulate the expression of LAR and higher levels of epicatechin can promote the expression of anthocyanidin synthase (ANS) and ANR. In addition, there could be a potential competition between LAR and ANR enzymes. The activity of both of these reductases (ANR and LAR) is dependent on NAPDH. Therefore, if one of them is overexpressed, it will decrease the availability of NAPDH for the other enzyme. It is expected that such competition may govern the mutual inhibition of LAR and ANR expression observed in this study. Altogether, the PA biosynthesis is likely co-regulated by structural genes such as LAR and ANR, and the mutual inhibition between the ANR and LAR expression may affect PA accumulation16. A lack of clear association between the transcripts of LAR/ANR and the catechin/derivatives has already been reported in other plants as well40,46. Earlier studies indicate a potential importance of ANR and LAR genes in the biosynthesis of galloylated catechins47. Therefore, it could be explained in terms of variable amounts of catechin/derivatives and the fact that different forms of catechin are potentially interconvertible.
Figure 5.
Relative RT-qPCR analysis of MiANR and MiLAR gene.
In each comparison, the default expression value of both genes was adjusted to one in samples obtained from TN cultivar.
Materials and methods
Plant material, sample preparation and extraction
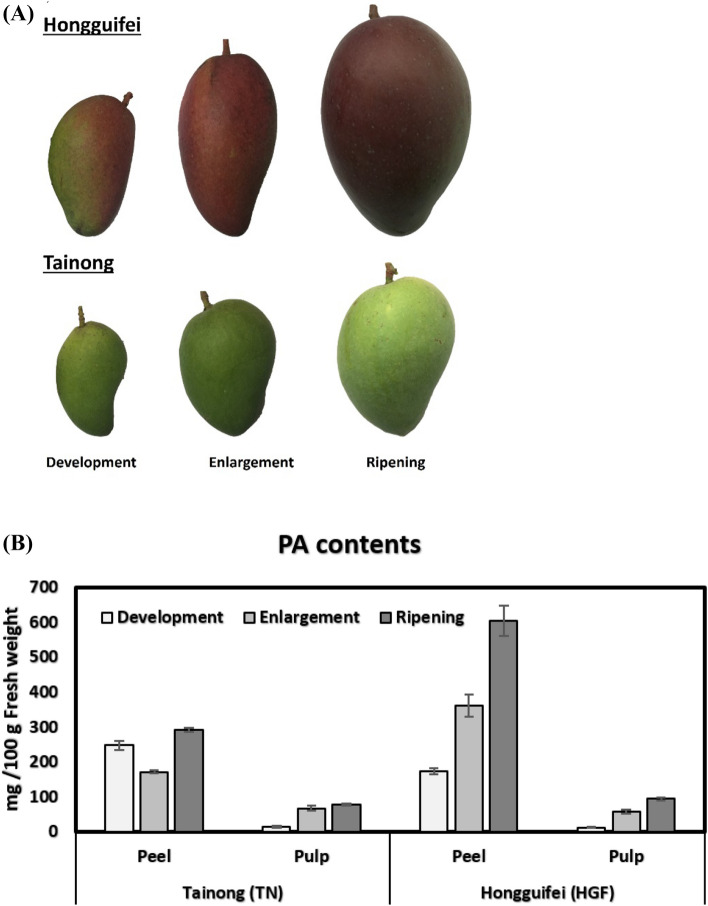
Current study compares metabolites of fruit peel and pulp of two mango varieties named Tainong (TN) and Hongguifei (HGF), during three different developmental stages of fruit (Development or 1st, Enlargement or 2nd, and Ripening or 3rd). Both varieties were maintained at a farm located at Basuo town, Dongfang County, Hainan province. The TN mango was developed by the Fengshan tropical horticulture branch institute of the Taiwan Agricultural Test Institute and introduced to southern provinces of China including Hainan and Guangdong in 1994. The TN fruit is oval-shaped and relatively small in size that weighs up to 300 g. Mature fruit peel is dark green to greenish-yellow. The peel is slightly thicker and ductile, which is good for storage and transportation The HGF mango, also known as Hongjinglong, is native to Taiwan. It is a hybrid of 'Irwin' and 'Kate' and was introduced to Hainan in the 1990s48,49. The HGF mango fruit is oblong, the top of the fruit is small and the fruit is relatively large (300–500 g). Mature fruit peel is purple-red and green (Fig. 3A).
Fruit samples from both varieties were collected at 40 (Developmental or 1st), 65 (Enlargement or 2nd), and 90 (Ripening or 3rd) days after full bloom (DAFB) respectively. Samples for each stage consisted of 10 fruits from 10 mango trees. Fruits of each variety were peeled and cored, and the flesh was cut into small sections. Fruit samples at each stage were mixed and immediately frozen in liquid nitrogen, and then stored at − 80 °C until used.
A mixer mill with zirconia bead (MM 400, RETSCH) was used to grind the freeze-dried samples for 90 s at 30 Hz. Then, an overnight extraction (at 4 °C using 70% aqueous methanol) was performed for 100 mg powder. Before LC–MS analysis, the samples were centrifuged at 10,000×g for 10 min followed by filtration (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China, https://www.anpel.com.cn/).
HPLC analysis
An LC–ESI–MS/MS system (HPLC, Shim-pack UFLC SHIMADZU CBM30A system, www.shimadzu.com.cn/; MS, Applied Biosystems4500 Q TRAP, www.appliedbiosystems.com.cn/) was used to analyze the sample extracts. The analytical parameters were as follow: HPLC column, waters ACQUITY UPLC HSS T3 C18 (1.8 µm, 2.1 mm * 100 mm); solvent system, water (0.04% acetic acid): acetonitrile (0.04% acetic acid); gradient program, 100:0 V/V at 0 min, 5:95 V/V at 11.0 min, 5:95 V/V at 12.0 min, 95:5 V/V at 12.1 min, 95:5 V/V at 15.0 min; flow rate, 0.40 mL/min; temperature, 40 °C; injection volume: 5 μl. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS.
ESI-Q TRAP-MS/MS
Triple quadrupole-linear ion trap mass spectrometer (QTRAP; API 4500 Q TRAP LC/MS/MS System) was used for Linear Ion Trap (LIT) and triple quadrupole (QQQ) scans. The equipment contained an ESI Turbo Ion-Spray interface, which was operated in a positive and negative ion-mode and the data was analyzed using analyst 1.6 software (AB SCIEX). The chromatographic method (e.g., mobile phase composition, pH, elution gradient) is the same in both ESI acquisition modes. Following conditions were used for the source of ESI operation: turbo spray (ion-source); 550 °C (source temperature); 5500 V (ion spray voltage or IS); GSI (ion source gas I), GSII (gas II), CUR (curtain gas) were set at 55, 60, and 25.0 psi, respectively; CAD (the collision gas) was set at high. Polypropylene glycol (10 and 100 μmol/L) was used to tune the instrument and for calibration of mass in QQQ and LIT modes, respectively. The collision gas (N2) was set to 5 psi during QQQ scans based MRM analysis. A specific set of MRM transitions were monitored for each period according to the metabolites eluted within this period.
Quantitative and qualitative principles of metabolites
Based on the public database and the self-built database MWDB (metware) of metabolite information, the first-order spectrum and two-level spectral data of spectral detection were qualitatively analyzed. The structural analysis of metabolites is referenced by knapsack (https://kanaya.naist.jp/KNApSAcK/), Massbank (https://www.massbank.jp/), Metlin (https://metlin.scripps.edu/index.php), MoTo DB (https://www.ab.wur.nl/moto/), hmdb (https://www.hmdb.ca/), and other existing mass spectrometry public databases. The quantification of metabolites was accomplished using the multi-reaction monitoring model of the triple four-stage rod mass spectrometry (multiple reaction monitoring, MRM). The detection standard of MRM is based on the parameters including Q1, Q3, RT, DP, CE from the database, which was built using the standards. The relative content of the compounds was determined by the signal intensity of Characteristic fragment ion Q3. The range of DP (declustering potential) was − 80 to 80 V and CE (collision energy) was − 50 to 50 V. In MRM mode, the four levers first filter the precursor ions of the target, the ions matching substances with different molecular weights are excluded for initial removal of disturbance, and the precursor ions are induced by the collision chamber to form many fragments of ions. The fragment ions are then filtered through the triple four-pole filter to select characteristic fragment ions, eliminate non-target ion interference, make the quantification more accurate, and improve repeatability. After obtaining the data of the different samples of the metabolite spectra, the peak area integral of all the material mass spectra was obtained, and the mass spectra of the same metabolites in different samples were corrected by integral correction.
Statistical analysis
The SIMCA14.1 software package (V14.1, Sartorius Stedim Data Analytics AB, Umea, Sweden) was used for principal component analysis (PCA) and orthogonal projections to latent structures-discriminate analysis (OPLS-DA). PCA showed the distribution of the original data. In order to obtain a higher level of group separation and to get a better understanding of variables responsible for classification, supervised OPLS-DA were applied. Based on OPLS-DA, a loading plot was constructed, which showed the contribution of variables to differentiate between two groups. The first principal component of variable importance in the projection (VIP) was calculated to refine this analysis. The VIP values above one were designated as differential metabolites. In the second step, Student’s t-test was used to assess the remaining variables and variables with p value > 0.05 were discarded between two comparison groups. In addition, commercial databases including KEGG50 https://www.genome.jp/kegg/ and MetaboAnalyst https://www.metaboanalyst.ca/ were used to search for the pathways of metabolites.
Determination of total proanthocyanidins (PAs) content
The proanthocaynidins content of the extracts were determined using the method described by DongRuixia[12]. Calibration curve was prepared by mixing methanol solution of standard proanthocyanidin (1 mL; 0.2–1 mg/mL) with 6 mL of 4% (g/v) vanillic aldehyde and 3 mL of concentrated HCl. After capping and shaking the tube, it was incubated in the dark for 15 m at 30℃ ± 1. The absorbance was measured at 500 nm (UVmini-1240, Shimadzu Corporation, Kyoto, Japan) with methanol as blank control and the standard curve was plotted. 1 mL of each of the extract solution in methanol (0.1 g mL−1) was also mixed with the above mentioned reagents, After incubation for 30 min, the absorbance was measured to determine proanthocyanidins content. The concentration of proanthocyanidin of samples was calculated using the following equation based on a PAs standard curve: y = 1.038x + 0.046 Where X is the absorbance and y is the proanthocyanidin equivalent. For the precentage of PAs content in tested samples, the equation is as follow: D = (v Cn/1000 W) 100%. All tests were conducted in triplicate D: the percentage of PAs content of samples; V: The constant volumn of sample; C: the concentration of proanthocyanidin of samples (mg/mL); n: dilution times; W: weight of sample(dry weight). Then the percentage of PAs conent of samples was converted to mg/ 100 g FW based on the ratio of fresh and dry weight of mango.
RNA extraction and relative RT-qPCR analysis
Peel and pulp samples were used for RNA extraction with the help of RNAprep Pur Plant Kit for polysaccharides and polyphenolics-rich samples (TIANGEN Biotech, Beijing) following the instructions of the manufacturer. The concentration of RNA was estimated from each sample through NanoDrop spectrophotometer (BERTHOLD, Germany). All-in-one First-Strand Synthesis Mastermix, with DNaseI (NOVA BIOMED, China) was used to reverse transcribe 1.0 μg of total RNA. The qRT-PCR analysis was performed using an APPLIED BIOSYSTEMS StepOnePlus Real-Time PCR System and TB GREEN Premix Ex Taq II, Tli RNaseH plus kit (TAKARA). The Oligo Calculator (https://mcb.berkeley.edu/labs/krantz/tools/oligocalc.html) was used for designing the gene-specific primers and NCBI Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to verify primer specificity. The expression data were normalized using MiActin gene (GenBank accession number HQ830244) as an internal control. Following primers were used for RT-qPCR: MiANR (Fow-TCCAAGACCCTGGCTGAAAG; Rev-CTGGCGTAAGAGAAGGACCA), MiLAR (Fow-ATTAAACCAGCTCCCTCTCG; Rev-CACATCATGCCCAAACTCAG), and MiActin (Fow-GCTTGCCTATGTTGCCCTTGACTA; Rev-GCATCGGAATCTCTCAGCTCCAAT). An equal amount of cDNA template was used for each sample including the internal control. The qPCR analysis was repeated in three independent experiments.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conclusions
The current study provides a global picture of metabolite dynamics between two mango cultivars by conducting an analysis of widely-targeted metabolomics based on LC–MS/MS data. The differential accumulation or absence of particular metabolites from either of cultivars indicates underlying differential metabolism. Important metabolites including catechin, its derivatives and procyanidins only up-regulated in HGF pulp and peel samples. The current study revealed that the expression of MiANR (a key gene of the PP pathway) was significantly higher in both pulp and peel samples of HGF cultivar at all three stages of fruit development. Moreover, total proanthocyanidin contents and relative flavan-3-ols/procyanidins were also higher in HGF cultivar. It seems obvious that such variations are directly responsible for the detected differences in relative quantities of flavonoid. This study documented changes in absolute contents of important catechin/ derivatives and expression profile of key genes involved in their biosynthesis for three growth stages. Such knowledge of mango fruit will be helpful for producers in adding value to the fruit and increasing antioxidant components. Moreover, different derivatives show distinct contents towards maturity and higher concentrations in peels as compared to pulp. It advocates the need for further research to improve contents of these metabolites in edible portion of mango.
Supplementary information
Acknowledgements
The authors acknowledge the China Science and Technology Exchange Center (CSTEC) for the postdoc fellowship of Farrukh Azeem through Talented Young Scientist Program (TYSP).
Author contributions
Conceptualization, L.T. and F.A.; methodology, F.A. and L.T.; software, C.Z.; validation, F.A., H.N. and G.Y.; formal analysis, F.A. and J.Z.; resources, L.T.; data curation, F.A.; writing—original draft preparation, F.A. and L.T.; writing—review and editing, H.N. and R.Z.; supervision, L.T.; project administration, L.T. and R.Z.; funding acquisition, L.T.
Funding
This research was supported by central public-interest scientific institution basal research fund for the Chinese Academy of Tropical Agricultural Sciences (project no.1630092019006 and 630092018007), and the earmarked fund for the Belt and Road Tropical Project (BARTP-07).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Farrukh Azeem, Email: azeuaf@hotmail.com.
Rulin Zhan, Email: zhanrulin555@163.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-75636-y.
References
- 1.FAOSTAT. Food and Agriculture Organization of the United Nations (FAOSTAT). https://www.fao.org/faostat/en/#data (accessed 19 March 2019) (2019).
- 2.Abbasi AM, et al. Comparative assessment of phenolic content and in vitro antioxidant capacity in the pulp and peel of mango cultivars. Int. J. Mol. Sci. 2015;16:13507–13527. doi: 10.3390/ijms160613507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbasi AM, et al. Phytochemical composition, cellular antioxidant capacity and antiproliferative activity in mango (Mangifera indica L.) pulp and peel. Int. J. Food Sci. Technol. 2017;52:817–826. doi: 10.1111/ijfs.13341. [DOI] [Google Scholar]
- 4.Jahurul MHA, et al. Mango (Mangifera indica L.) by-products and their valuable components: a review. Food Chem. 2015;183:173–180. doi: 10.1016/j.foodchem.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 5.Terahara N. Flavonoids in foods: a review. Nat. Prod. Commun. 2015;10:1934578X1501000. doi: 10.1177/1934578X1501000334. [DOI] [PubMed] [Google Scholar]
- 6.Pfeiffer J, et al. Biosynthesis of flavan 3-ols by leucoanthocyanidin 4-reductases and anthocyanidin reductases in leaves of grape (Vitis vinifera L.), apple (Malus × domestica Borkh.) and other crops. Plant Physiol. Biochem PPB. 2006;44:323–334. doi: 10.1016/j.plaphy.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Harding SA. Condensed tannins: arbiters of abiotic stress tolerance? Tree Physiol. 2019;39:341–344. doi: 10.1093/treephys/tpz005. [DOI] [PubMed] [Google Scholar]
- 8.Burton-Freeman BM, Sandhu AK, Edirisinghe I. Mangos and their bioactive components: adding variety to the fruit plate for health. Food Funct. 2017;8:3010–3032. doi: 10.1039/C7FO00190H. [DOI] [PubMed] [Google Scholar]
- 9.Smeriglio A, Barreca D, Bellocco E, Trombetta D. Proanthocyanidins and hydrolysable tannins: occurrence, dietary intake and pharmacological effects. Br. J. Pharmacol. 2017;174:1244–1262. doi: 10.1111/bph.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira BG, et al. Chemical profile of mango (Mangifera indica L.) using electrospray ionisation mass spectrometry (ESI-MS) Food Chem. 2016;204:37–45. doi: 10.1016/j.foodchem.2016.02.117. [DOI] [PubMed] [Google Scholar]
- 11.Pierson JT, et al. Phytochemical extraction, characterisation and comparative distribution across four mango (Mangifera indica L.) fruit varieties. Food Chem. 2014;149:253–263. doi: 10.1016/j.foodchem.2013.10.108. [DOI] [PubMed] [Google Scholar]
- 12.Sogi DS, Siddiq M, Greiby I, Dolan KD. Total phenolics, antioxidant activity, and functional properties of ‘Tommy Atkins’ mango peel and kernel as affected by drying methods. Food Chem. 2013;141:2649–2655. doi: 10.1016/j.foodchem.2013.05.053. [DOI] [PubMed] [Google Scholar]
- 13.Nawab A, Alam F, Haq MA, Lutfi Z, Hasnain A. Mango kernel starch-gum composite films: physical, mechanical and barrier properties. Int. J. Biol. Macromol. 2017;98:869–876. doi: 10.1016/j.ijbiomac.2017.02.054. [DOI] [PubMed] [Google Scholar]
- 14.Ajila CM, Bhat SG, Prasada Rao UJS. Valuable components of raw and ripe peels from two Indian mango varieties. Food Chem. 2007;102:1006–1011. doi: 10.1016/j.foodchem.2006.06.036. [DOI] [Google Scholar]
- 15.Wang A, et al. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam) Food Chem. 2018;260:124–134. doi: 10.1016/j.foodchem.2018.03.125. [DOI] [PubMed] [Google Scholar]
- 16.Yan N, et al. A comparative UHPLC-QqQ-MS-based metabolomics approach for evaluating Chinese and North American wild rice. Food Chem. 2019;275:618–627. doi: 10.1016/j.foodchem.2018.09.153. [DOI] [PubMed] [Google Scholar]
- 17.RuoNan X, Chen M, Qun Z, ChunHua L, XinFeng Y. Characteristic analysis on fruit quality of major varieties in main mango producing regions of Hainan. J. South. Agric. 2018;49:2511–2517. [Google Scholar]
- 18.Engels C, Gänzle MG, Schieber A. Fractionation of Gallotannins from mango (Mangifera indica L.) kernels by high-speed counter-current chromatography and determination of their antibacterial activity. J. Agric. Food Chem. 2010;58:775–780. doi: 10.1021/jf903252t. [DOI] [PubMed] [Google Scholar]
- 19.Hu K, Dars AG, Liu Q, Xie B, Sun Z. Phytochemical profiling of the ripening of Chinese mango (Mangifera indica L.) cultivars by real-time monitoring using UPLC-ESI-QTOF-MS and its potential benefits as prebiotic ingredients. Food Chem. 2018;256:171–180. doi: 10.1016/j.foodchem.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Khakimov B, et al. A comprehensive and comparative GC–MS metabolomics study of non-volatiles in Tanzanian grown mango, pineapple, jackfruit, baobab and tamarind fruits. Food Chem. 2016;213:691–699. doi: 10.1016/j.foodchem.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Pino JA, Mesa J, Muñoz Y, Martí MP, Marbot R. Volatile Components from mango (Mangifera indica L.) cultivars. J. Agric. Food Chem. 2005;53:2213–2223. doi: 10.1021/jf0402633. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro SMR, Barbosa LCA, Queiroz JH, Knödler M, Schieber A. Phenolic compounds and antioxidant capacity of Brazilian mango (Mangifera indica L.) varieties. Food Chem. 2008;110:620–626. doi: 10.1016/j.foodchem.2008.02.067. [DOI] [Google Scholar]
- 23.Singh UP, et al. Characterization of phenolic compounds in some Indian mango cultivars. Int. J. Food Sci. Nutr. 2004;55:163–169. doi: 10.1080/09637480410001666441. [DOI] [PubMed] [Google Scholar]
- 24.Sung J, Suh JH, Chambers AH, Crane J, Wang Y. Relationship between sensory attributes and chemical composition of different mango cultivars. J. Agric. Food Chem. 2019;67:5177–5188. doi: 10.1021/acs.jafc.9b01018. [DOI] [PubMed] [Google Scholar]
- 25.Babicki S, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu YL, Liang HL, Hung CH, Kuo PL. Syringetin, a flavonoid derivative in grape and wine, induces human osteoblast differentiation through bone morphogenetic protein-2/extracellular signal-regulated kinase 1/2 pathway. Mol. Nutr. Food Res. 2009;53:1452–1461. doi: 10.1002/mnfr.200800483. [DOI] [PubMed] [Google Scholar]
- 27.Bando SI, Hatano O, Takemori H, Kubota N, Ohnishi K. Potentiality of syringetin for preferential radiosensitization to cancer cells. Int. J. Radiat. Biol. 2017;93:286–294. doi: 10.1080/09553002.2017.1242815. [DOI] [PubMed] [Google Scholar]
- 28.Niemeyer HM. Hydroxamic acids derived from 2-hydroxy-2 H-1,4-benzoxazin-3(4 H )-one: key defense chemicals of cereals. J. Agric. Food Chem. 2009;57:1677–1696. doi: 10.1021/jf8034034. [DOI] [PubMed] [Google Scholar]
- 29.Adhikari KB, et al. Absorption and metabolic fate of bioactive dietary benzoxazinoids in humans. Mol. Nutr. Food Res. 2013;57:1847–1858. doi: 10.1002/mnfr.201300107. [DOI] [PubMed] [Google Scholar]
- 30.Guo B, et al. Extract from maize (Zea mays L.): antibacterial activity of DIMBOA and its derivatives against Ralstonia solanacearum. Molecules. 2016;21:1397. doi: 10.3390/molecules21101397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasanthi HR, ShriShriMal N, Das DK. Phytochemicals from plants to combat cardiovascular disease. Curr. Med. Chem. 2012;19:2242–2251. doi: 10.2174/092986712800229078. [DOI] [PubMed] [Google Scholar]
- 32.Sikander M, et al. Cucurbitacin D reprograms glucose metabolic network in prostate cancer. Cancers (Basel) 2019;11:364. doi: 10.3390/cancers11030364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruales J, et al. Biological active ecuadorian mango ‘Tommy Atkins’ ingredients-an opportunity to reduce agrowaste. Nutrients. 2018;10:1–14. doi: 10.3390/nu10091138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong RX, Li LX, Wang XI, Xu RR. Determination of Proanthocyanidin content in plants. J. Tea Bus. 2008;30:67–69. [Google Scholar]
- 35.Gu L, et al. Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J. Nutr. 2004;134:613–617. doi: 10.1093/jn/134.3.613. [DOI] [PubMed] [Google Scholar]
- 36.Dorta E, González M, Lobo MG, Laich F. Antifungal activity of mango peel and seed extracts against clinically pathogenic and food spoilage yeasts. Nat. Prod. Res. 2016 doi: 10.1080/14786419.2015.1115995. [DOI] [PubMed] [Google Scholar]
- 37.Coelho EM, et al. Bioactive compounds and antioxidant activity of mango peel liqueurs (Mangifera indica L.) produced by different methods of maceration. Antioxidants. 2019;8:102. doi: 10.3390/antiox8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almeida MMB, et al. Bioactive compounds and antioxidant activity of fresh exotic fruits from northeastern Brazil. Food Res. Int. 2011;44:2155–2159. doi: 10.1016/j.foodres.2011.03.051. [DOI] [Google Scholar]
- 39.Freixas Coutin JA, et al. Proanthocyanidin accumulation and transcriptional responses in the seed coat of cranberry beans (Phaseolus vulgaris L.) with different susceptibility to postharvest darkening. BMC Plant Biol. 2017;17:89. doi: 10.1186/s12870-017-1037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, et al. Changes in catechin contents and expression of catechin biosynthesis-associated genes during early cucumber fruit development. Acta Physiol. Plant. 2019;41:130. doi: 10.1007/s11738-019-2925-7. [DOI] [Google Scholar]
- 41.Velderrain-Rodríguez G, et al. Gallic acid content and an antioxidant mechanism are responsible for the antiproliferative activity of ‘Ataulfo’ mango peel on LS180 Cells. Molecules. 2018;23:695. doi: 10.3390/molecules23030695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arts ICW, van de Putte B, Hollman PCH. Catechin contents of foods commonly consumed in The Netherlands. 1. Fruits, vegetables, staple foods, and processed foods. J. Agric. Food Chem. 2000;48:1746–1751. doi: 10.1021/jf000025h. [DOI] [PubMed] [Google Scholar]
- 43.Can-Cauich CA, et al. Tropical fruit peel powders as functional ingredients: evaluation of their bioactive compounds and antioxidant activity. J. Funct. Foods. 2017;37:501–506. doi: 10.1016/j.jff.2017.08.028. [DOI] [Google Scholar]
- 44.Tan L, et al. Biochemical and functional characterization of anthocyanidin reductase (ANR) from Mangifera indica L. Molecules. 2018;23:2876. doi: 10.3390/molecules23112876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Shi Z, Maximova S, Payne MJ, Guiltinan MJ. Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 2013;13:202. doi: 10.1186/1471-2229-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao L, et al. Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 2015;6:243. doi: 10.3389/fpls.2015.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L-Q, Wei K, Cheng H, Wang L-Y, Zhang C-C. Accumulation of catechins and expression of catechin synthetic genes in Camellia sinensis at different developmental stages. Bot. Stud. 2016;57:31. doi: 10.1186/s40529-016-0143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang J, Liu Y, Zhou B, Ke K, Lin T. Characteristics and properties of Tainong No. 1 mango and cultivation techniques for its early-bearing and high yield. Guangdong Agric. Sci. 2001;3:33–36. [Google Scholar]
- 49.Wang C. Tainong No. 1 mango, a mango variety suitable for cultivation in Southern China. China Fruits. 1999;1999:58. [Google Scholar]
- 50.Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.