Graphical abstract
Keywords: SARS-CoV-2, Coronavirus, Therapy, Small molecules, Organic synthesis
Highlights
-
•
The repositioning of approved drugs is a promising approach for treating COVID-19.
-
•
Several approved drugs have shown anti-SARS-CoV-2 activity and are discussed herein.
-
•
In this review, the synthetic approaches toward such molecules was highlighted.
-
•
The mechanisms of action and pharmacological properties of such drugs is discussed.
Abstract
In December 2019, a new variant of SARS-CoV emerged, the so-called acute severe respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus causes the new coronavirus disease (COVID-19) and has been plaguing the world owing to its unprecedented spread efficiency, which has resulted in a huge death toll. In this sense, the repositioning of approved drugs is the fastest way to an effective response to a pandemic outbreak of this scale. Considering these facts, in this review we provide a comprehensive and critical discussion on the chemical aspects surrounding the drugs currently being studied as candidates for COVID-19 therapy. We intend to provide the general chemical community with an overview on the synthetic/biosynthetic pathways related to such molecules, as well as their mechanisms of action against the evaluated viruses and some insights on the pharmacological interactions involved in each case. Overall, the review aims to present the chemical aspects of the main bioactive molecules being considered to be repositioned for effective treatment of COVID-19 in all phases, from the mildest to the most severe.
1. Introduction
In the last two decades, the human vulnerability to emerging viral diseases has become undeniably clear, especially since three zoonotic coronaviruses have emerged: SARS-CoV-1, MERS-CoV and SARS-CoV-2 [1]. SARS-CoV-1 (Severe Acute Respiratory Syndrome) initially surfaced in 2003 in China, while MERS-CoV (Middle East respiratory syndrome) was first identified in the Kingdom of Saudi Arabia in 2012, and SARS-CoV-2, the etiologic agent of COVID-19, can be traced back to December 2019 in China [2], [3].
The COVID-19 outbreak started in Wuhan, Hubei province, China and has quickly spread worldwide. Indeed, the World Health Organization (WHO) declared, on January 30th 2020, that the rapid eruption of the disease caused by the new coronavirus (COVID-19) constitutes a public health emergency of international importance, the organization's highest level of alert, as defined by international health regulations. On March 11th 2020, WHO characterized COVID-19 as a pandemic. Currently, over 200 countries have reported cases of COVID-19, and by the end of June 2020, almost 10 million cases had already been confirmed, with nearly 500,000 deaths worldwide [4], [5]. Still, according to WHO, most COVID-19 patients (about 80%) may be asymptomatic, while approximately 20% require specialized health care due to symptoms like difficulty in breathing, and among those, 5% may require respiratory support through the use of mechanical ventilators.
In this context, coronaviruses (CoVs) are a large group of viruses that cause mild to severe diseases in humans and are currently divided into four genera (alpha, beta, gamma and deltacoronavirus). Alphacoronaviruses affect swines, felines, dogs, bats, and two types that affect humans have been described: HCoV-NL63 and HCoV-229E. Betacoronaviruses comprise a large number of mammal-infecting viruses, as well as 5 human pathogens - HCoV-OC43, HCoV-HKU1, and the three viruses that have caused recent human epidemics: SARS-CoV-1, MERS-CoV and SARS-CoV-2. Gammacoronaviruses include viruses specific to whales and birds, and Deltacoronaviruses are isolated from various mammalian and avian species [6], [7]. At this point, it is important to highlight that SARS-CoV-2 is already responsible for 10 times more deaths than the total sum of SARS-CoV-1 and MERS-CoV, and its death toll is increasing daily at a global scale [4]. Like other CoVs, SARS-CoV-2 is characterized by a plus single-stranded RNA with large enveloped nucleocapsids that are capable of infecting a variety of hosts, including animals and humans [8], [9]. Whilst MERS-CoV and SARS-CoV-1 share some common virological and clinical features there are several important differences [10].
Currently, the most effective response to the SARS-CoV-2 pandemic has been quarantine and social distancing measures, which avoid the contact between infected and uninfected individuals. Such approaches have the aim of flattening the virus’s spread curve, consequently reducing the burden over the health care systems. In this sense, the development of vaccines that may prevent the contamination by SARS-CoV-2 is highly desirable, and some initiatives are already on the way - those consist of an mRNA strand containing the information for a SARS-CoV-2 coronavirus protein, called Spike (S), or based on a modified adenovirus expressing the Spike protein, called a viral chimera (hybrid of two viruses). This attenuated adenovirus can be applied to humans for the development of antibodies [11].
On the other hand, strategies for the treatment COVID-19 are also being currently investigated, namely (i) the use of convalescent plasma therapy that involves removing blood plasma from patients recovered from the coronavirus, with the potential of decreasing the severity and/or duration of the disease, as these individuals would already have developed immunity to the disease, ((ii) the repositioning of clinically available drugs, which consists on the use of drugs already approved for the treatment of other diseases, in order be redirected toward COVID-19 patients, bypassing the clinical and regulatory trial period associated to the development of new drugs. This latter strategy may be faster than the development a new antiviral agent from scratch, or even of a new vaccine. As a consequence, according to the International Clinical Trials Registry Platform search portal, 927 clinical trials for COVID-19 have been registered [12]. Taking into consideration that the repositioning of clinically approved drugs is the fastest way to an effective response to a pandemic outbreak [13], various classes of drugs such as antidiarrheal agents, antimalarial agents, cyclophilin inhibitors, interferons, kinase inhibitors, neurotransmitter inhibitors, anticholinergics, nucleic acid synthesis inhibitors, protease inhibitors, protein synthesis inhibitors, selective estrogen receptor modulators, and sterol metabolism inhibitors have demonstrated in vitro and in vivo antiviral efficacy against MERS-CoV and SARS-CoV-1, and are currently being evaluated against SARS-CoV-2 [14].
In light of the aforementioned concepts, in this review we aim to provide a comprehensive and critical discussion on the chemical aspects surrounding the drugs currently being studied as candidates in the COVID-19 therapy. We intend to provide the general chemical community with an overview on the synthetic pathways related to such molecules, as well as their mechanisms of action against the evaluated coronaviruses and insights on the pharmacological interactions related to the antiviral effect of these drugs. It is worth highlighting that the drugs discussed in this manuscript have showed the potential to treat COVID-19 itself by inhibiting SARS-CoV-2, and not only its symptoms, which implies that drugs such as azithromycin and dexamethasone, among others, are not in the scope of this review. Overall, the review intends to present the chemical aspects of the main bioactive molecules being considered to be repositioned for effective treatment of COVID-19 in all phases, from the mildest to the most severe.
2. Chemical aspects of the main bioactive molecules with potential to treat COVID-19
2.1. Antiviral compounds
In the beginning of 2020, SARS-CoV-2, a coronavirus that shares 79.5% genetic homology to SARS-CoV-1, both being descendants of bat coronaviruses within the Betacoronavirus genus, was identified as the causal agent of COVID-19 [15]. With basis on this information, antiviral compounds with reported activity against SARS-CoV-1 or other coronaviruses were among the first ones evaluated as possible treatments for COVID-19. In addition, drugs with a broad-spectrum antiviral activity against other types of virus e.g. filoviruses and paramyxoviruses have also been considered. As a consequence, antiviral compounds widely used to treat HIV, influenza and hepatitis C, among others, have been showing the best results in the treatment of the SARS-CoV-2 infection.
2.1.1. Peptideomimetics: Lopinavir, ritonavir and atazanavir
Proteases are enzymes that cleave peptide bonds between protein aminoacids, which, in turn, are important for viral replication and production of new infectious virions. Hence, the inhibition of such enzymes has become an important target for the development of antiviral agents, specially antiretrovirals. In the rational design of protease inhibitors, these substances are required to mimic a peptide substrate, and therefore are classified as peptideomimetics [16].
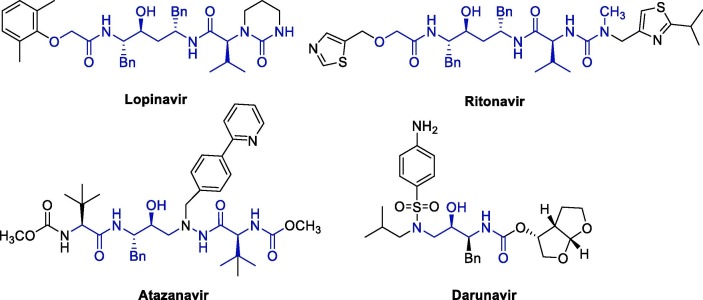
Originally, protease inhibitors are drugs used to treat HIV infections, being lopinavir (LPV), ritonavir (RTV), atazanavir (ATV) and darunavir (DRV) the main representatives of this class (Fig. 1 ) [16].
Fig. 1.
Structure of protease inhibitors used in the treatment of HIV infections.
In this context, CoV-type viruses encode proteases that are involved in the proteolytic processing of polyproteins, playing an important role in viral replication. Thus, due to their main role in viral maturation, which are essential in the viral life cycle, CoV proteases emerge as a promising antiviral target [17]. Considering this, the inhibitory activity of this class of peptideomimetics against SARS-CoV-1 and MERS-CoV has been investigated through in vivo and in vitro assays in recent years [18]. Most notably, it was discovered that 3CLpro, an important protease for the maturation of coronavirus, would be vital to its life cycle, constituting an important target in the search for anti-SARS-CoV-2 agents [19]. Molecular modeling studies performed in 2008 indicated that the SARS-CoV-1 3CLpro enzyme could be inhibited by the combination of LPV and RTV [20].
In 2003, Chan and coworkers undertook a retrospective cohort study in 75 patients with severe acute respiratory syndrome, and pointed out that their treatment with LPV/RTV was associated with reduced death and intubation rates when compared to the ones who received the standard treatment protocol [21]. This information paved the path for the studies involving the use of LPV/RTV in patients with COVID-19.
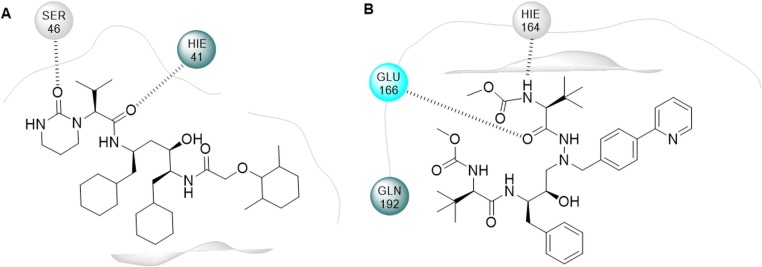
Importantly, molecular docking studies have been carried out to assess the profile of affinity of these drugs with SARS-CoV-2 and SARS-CoV-1 3CLpro, and DRV showed the best results, which indicates its potential to become a potential anti-COVID-19 drug.[19] ATV was also able to fit into SARS-CoV-2 Mpro protease active site, having displayed a greater affinity than LPV (Fig. 2 ). In this sense, enzymatic assays confirmed that ATV inactivates Mpro, and in vitro assays on cells infected with SARS-CoV-2 indicated that ATV was able to inhibit viral replication alone or in combination with ritonavir (ATV/RTV EC50 = 0.5 ± 0.08 µM, ATV EC50 = 2.0 ± 0.12 µM). It is important highlighting that the use of ATV would highly advantageous due to its bioavailability in the respiratory tract. On that account, the aforementioned data suggests that ATV is a strong candidate for clinical evaluation in the treatment of COVID-19 [22].
Fig. 2.
2D representation of the interactions of (a) LPV and (b) ATV in the Mpro active site [300], [22].
Recently, it was reported that a 54-year-old patient, the third SARS-CoV-2 infection case diagnosed in Korea, was treated with LPV/RTV(Kaletra® - LPV 200 mg / RTV 50 mg, orally). The treatment was started on the tenth day after the symptoms appearance, and the viral β-coronavirus load started to decrease in the second day of treatment, with no detectable or little coronavirus titers being observed since then [23].
In a similar path, epidemiological information and clinical characteristics were collected from patients admitted between January 22 and February 11 to the Xixi Hospital in Hangzhou, China. LPV was administered to ten patients in the initial stage of the disease (3–7 days after the appearance of the symptoms), and among these, three had to interrupt the treatment because of adverse effects such as liver problems. In addition, hypokalemia and eosinophilia were also observed in some patients. However, the small number of patients accompanied in this study makes it inconclusive, and a higher sampling is required to confirm the effectiveness of the LPV treatment [24].
LPV is produced in combination with RTV (Kaletra®) by Abbot pharmaceuticals [25], [26]. The LPV/RTV association is administered orally and is presented as coated tablets, in two dosages: LPV/RTV 200 mg + 50 mg, and LPV/RTV 100 mg + 25 mg. This association is important because RTV substantially increases LPV’s exposure through the inhibition of the cytochrome P450 3A4 enzyme [26]. The absolute bioavailability of the LPV/RTV in humans has not been established, but the oral bioavailability of LPV in rats was estimated to be 25%. At therapeutic concentrations, LPV is 98–99% bound to albumin [26], while LPV has a low penetrability in the Central Nervous System (0.02% brain plasma ratio) [27]. LPV is rapidly metabolized by CYP3A into three metabolites: M1, M2 and M3, each less active. As RTV acts by inhibiting this enzyme, the rate of LPV metabolism is reduced. The half-life of lopinavir is 5–6 h, with a clearance of 6–7 L/h [26].
Although LPV is very effective, some adverse reactions are evident: diarrhea, nausea, asthenia, stomatitis, and fever, as well as high levels of total bilirubin, triglycerides, and liver enzymes. Additionally, anemia and leukopenia are also reported adverse effects [26]. A clinical research conducted in January 2020 with 148 patients who were admitted to the Shanghai Public Health Clinical Center hospital with COVID-19 identified that LPV/RTV should be administered with caution, as abnormal liver function was observed in 37.2% of these patients [28]. Recently, the results of a randomized, controlled, open-label, platform trial, involving over 5000 hospitalized patients were reported [29]. The patients were administered a combination of LPV/RTV (400 mg and 100 mg, respectively), and the same 28-day mortality, duration of hospital stay, and risk of progressing to invasive mechanical ventilation or death were observed for both the patients in the LPV/RTV and control groups, which led the researchers to conclude that the treatment of hospitalized COVID-19 patients with lopinavir-ritonavir is not effective.
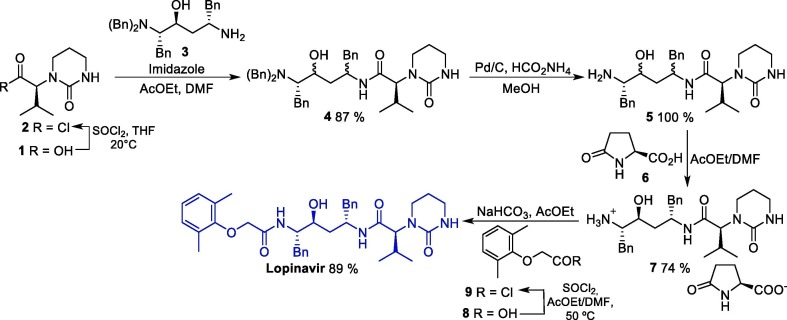
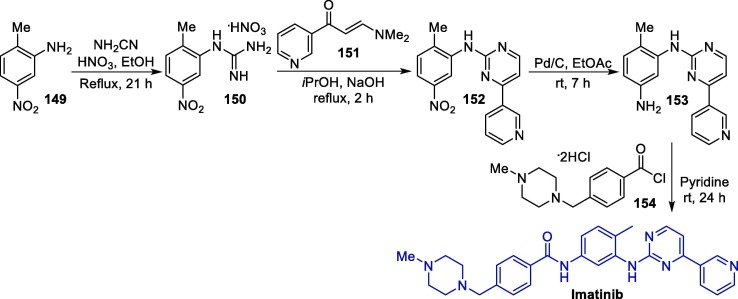
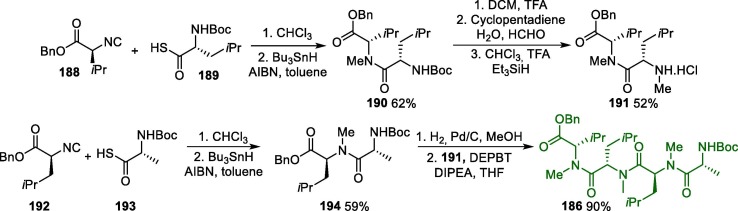
The synthetic route towards LPV was described in 2000, and initiates with the conversion of protected diamino alcohol 1 into acyl chloride 2, which in turn reacts with compound 3 to furnish protected intermediate 4. Next, 4 undergoes a debenzylation reaction to produce amino intermediate 5, which is treated with L-pyroglutamic acid 6 to giving salt 7 as a single diastereoisomer in high yield (74%). Finally, the condensation of 7 with acyl chloride 9 leads to lopinavir in a 58% overall yield (Scheme 1 ) [30].
Scheme 1.
Synthetic route of lopinavir.
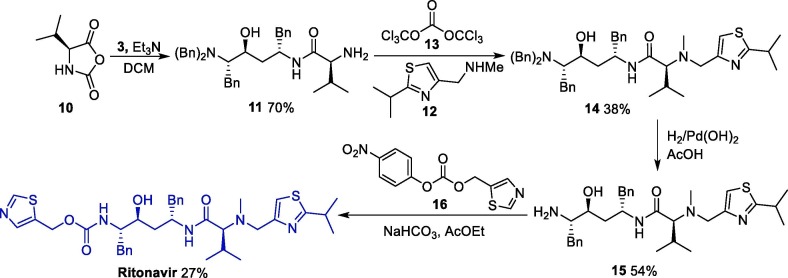
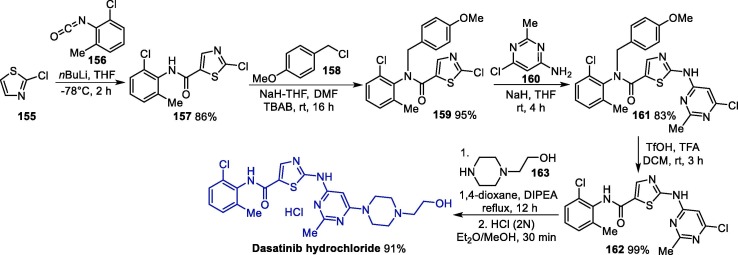
As for RTV, a patent describes its synthesis from the same diamino alcohol 3 in four steps (Scheme 2 ). First, the diamino alcohol 5 is reacted with N-carboxivalin anhydride 10, forming the intermediate 11, which reacts with 1-(2-isopropylthiazol-4-yl)-N-methylmethanamine 12 in the presence of bis-trichloromethyl carbonate 13 as a catalyst, affording intermediate 14. An amine deprotection step leads to the primary amine 15, which gives ritonavir after reacting with thiazole derivative 16 [31].
Scheme 2.
Synthetic route towards ritonavir.
Atazanavir (Reyataz®) is produced by Bristol-Myers Squibb Pharmaceuticals. ATV is administered orally in 150, 200 or 300 mg tablets. After ingestion, it is rapidly absorbed, reaching maximum concentration after 2.5 h. ATV binds to human serum proteins in an 86% rate, and its absorption is highly dependent on gastric pH, so the ingestion of tablets with food increases this effect. Still, ATV can interact with drugs that modify gastric pH, changing its bioavailability [32].
The ATV biotransformation generates monooxygenated and deoxygenated metabolites. Since ATV also inhibits the CYP3A function, it can be administered in combination with other antiretrovirals with the aim of increasing its plasma concentrations, which can improve the therapeutic effect. Similarly to LPV, the association with RTV increases the plasma level of ATV, and this association may increase plasma levels tenfold in lung tissue [33], [33].
The main adverse effects related to the use of ATV are skin rash as well as increased serum cholesterol, amylase and serum bilirubin. There are also some concerns related to other ATV adverse effects, especially in cases of elevated bilirubin: immune reconstitution syndrome (for example, Graves' disease, polymyositis, Guillain-Barré syndrome), nephrolithiasis/cholelithiasis, angioedema, chronic renal failure, complete atrioventricular block, diabetes mellitus and erythema multiforme [34]. Due to its low selectivity index, ATV may increase the concern about the adverse effects resulting from its administration to patients with COVID-19, and so far its use in the treatment of this disease has not been evaluated [35].
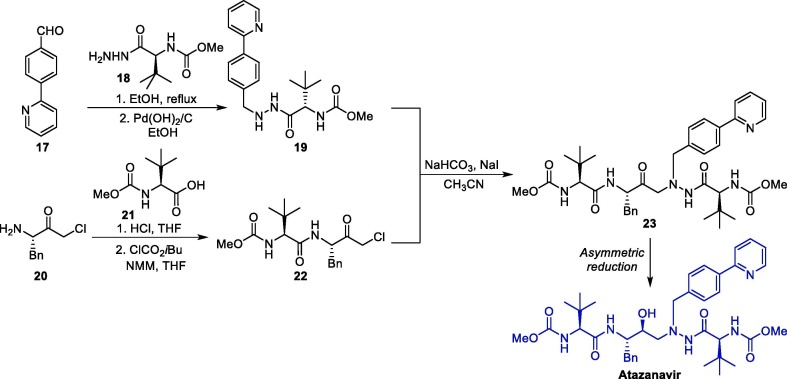
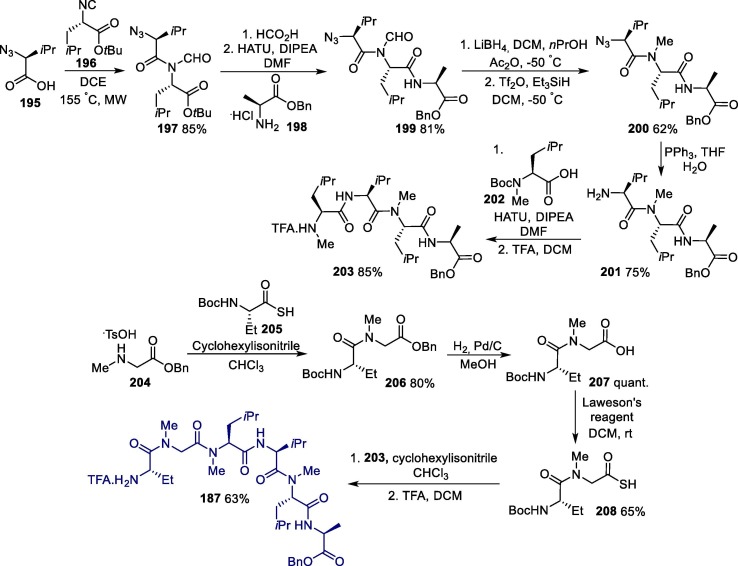
The synthesis of ATV can be accomplished efficiently via a convergent synthesis protocol followed by a stereoselective reduction step (Scheme 3 ) [36]. In this convergent synthesis, the two key intermediates 19 and 22 were prepared, respectively, from the reaction between pyridinyl benzaldehyde 17 and carbamate 18, and between (S)-3-amino-1-chloro-4-phenylbutan-2-one 20 and methoxycarbonyl-L-tert-leucine 21. Then, the aliphatic nucleophilic substitution reaction between these two intermediates leads to amino ketone 23, which furnishes atazanavir after a stereoselective reduction reaction. The diastereoselectivity of the reduction step can be achieved through the use of lithium tri-tert-butoxyaluminum hydride in diethyl ether as a reducing agent, and under these conditions, the desired aminoalcohol is obtained through a Felkin-Anh control with a 28:1 diastereoisomeric ratio [37].
Scheme 3.
Synthetic route towards of atazanavir. NMM = N-Methylmorpholine.
2.1.2. Nucleoside/nucleotide analogues: Remdesivir, tenofovir, favipiravir and sofosbuvir
The nucleosides and nucleotide analogues discussed in this topic are effective antiviral drugs that act by preventing viral replication in infected cells. In order to be incorporated into the viral RNA replication process, those substances have to be intracellularly converted to their triphosphate derivatives (NTP) [37], and then interfere with viral RNA synthesis at the level of the RNA-dependent RNA polymerase (RdRp). In this sense, the repositioning of nucleoside and nucleotide analogues for the treatment of COVID-19 is a promising strategy, since those could inhibit the viral replication of SARS-CoV-2 though the same pathway.
Remdesivir (GS-5734) emerged from a collaboration between Gilead Sciences, the U.S. Centers for Disease Control and Prevention (CDC) and the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) as part of Gilead's research into developing an Ebola virus (EBOV) disease treatment program. Currently, this drug is still undergoing clinical trials, but is the leading drug candidate for the treatment of EBOV infections.
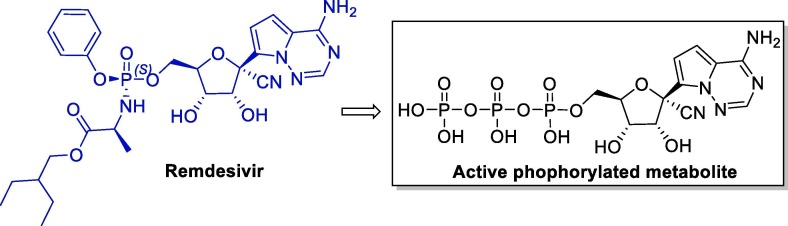
Remdesivir stood out among several other drugs for its sub-micromolar activity against EBOV in studies carried out in human microvascular endothelial cells (HMVEC-TERT). The 1:1 isomeric mixture of phosphoramidate Sp (Remdesivir) and its Rp isomer has also proven to be very potent against EBOV in HeLa and HMVEC cells (Fig. 3 ) [38], [39].
Fig. 3.
Structure of remdesivir and its active metabolite.
Remdesivir is a prodrug, being converted to its active phosphorylated form under physiological conditions. Studies carried out with several nucleoside analogs have shown that the nitrile group bound to the anomeric carbon is fundamental to the selectivity against viral polymerases, and also confers low toxicity (CC50 < 0.01–0.15 μM) [40].
The antiviral activity of remdesivir against several coronaviruses (SARS, MERS, contemporary human CoV and bat-CoVs) has already been proven [40], [41] and biochemical data from recombinant respiratory syncytial virus (RSV) RdRp suggested that the primary mechanism of action was through delayed chain termination [38], [42], [43].
With the rapid increase in the number of COVID-19 cases and no therapeutic alternatives available, two clinical tests using remdesivir were initiated in China, one with mild and moderate cases and other with severe ones [44], [45]. Even though both studies were suspended after the epidemic was well-controlled in China, the results of the randomized, double-blind, placebo-controlled, multicenter clinical trial conducted with adults with severe COVID-19 were published without reaching the predetermined sample size. The trial was conducted at ten hospitals in Wuhan, Hubei, China, with 237 patients and no statistically significant clinical benefits could be associated with remdesivir within these limited sample size [46].
In light of the promising results showed by remdesivir against SARS-CoV-2 in vitro, other research groups set out to conduct large-scale clinical trials. Recently, the final results of a double-blind, randomized, placebo-controlled trial of intravenous remdesivir clinical trial conducted with 1062 adults who were hospitalized with COVID-19 were disclosed [47]. The authors concluded that remdesivir reduces the recovery time in adults hospitalized with COVID-19 with lower respiratory tract infection (10 days vs 15 in the placebo group) and also led to a lower mortality (6.7% at day 15 vs 11.9% in the placebo group). However, considering that the mortality of patients is still considered high, it became clear that the treatment with an antiviral drug alone may not be enough, and future strategies should evaluate those in combination with other therapeutic approaches. Another randomized clinical trial involving 538 hospitalized patients with moderate COVID-19 has showed that patients who received a 5-day course of remdesivir had a significantly clinical improvement at day 11 than those who were given standard care [48].
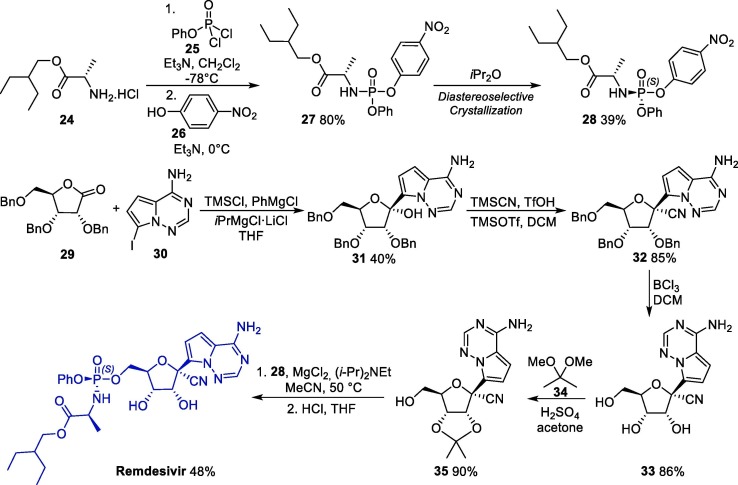
Considering the amount of remdesivir required for biological in vitro and in vivo evaluations, a robust synthetic method towards this molecule is highly desirable. An interesting method for the synthesis of remdesivir involves the diastereoselective crystallization of intermediate 28 from 27, which was obtained from the reaction between 2-ethylbuthyl-L-alanine 24 with 25, followed by a substitution step with p-nitropenol 26 (Scheme 4 ). In parallel, iodinated derivative 30 is reacted with ribolactone 29 in the presence of PhMgCl and TMSCl, giving C-glycosylation product 31 [49]. Next, the cyanation of 31 with TMSCN affords 32 in >95:5 anomeric ratio, and its subsequent treatment with boron trichloride leads to ribonucleoside 33. Next, a protection reaction with 2,2-dimethoxypropane 34 furnishes the corresponding acetonide 35, which is reacted with fragment 34 and gives remdesivir after a deprotection step [40].
Scheme 4.
Synthetic route towards remdesivir.
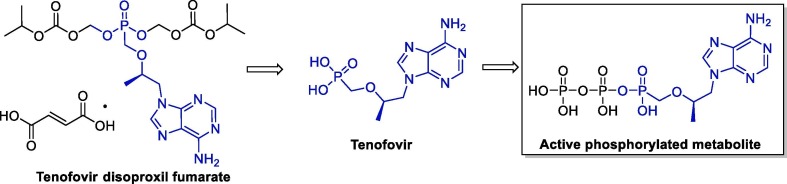
Tenofovir disoproxil fumarate (TDF, Viread® - Gilead Sciences) is an oral prodrug of tenofovir, a nucleotide analogue that has activity against retroviruses such as HIV-1, HIV-2 and hepadnaviruses. TDF is promptly converted to tenofovir after absorption, and is metabolized intracellularly to the active phosphorylated metabolite, a competitive inhibitor of HIV-1 reverse transcriptase that terminates the growth of the DNA chain (Fig. 4 ). Interestingly, tenofovir has longer serum and intracellular half-lives (17 h and ≥60 h, respectively) than most nucleoside analogues, which allows daily administration. It is also important mentioning that the tenofovir pharmacokinetics are dose-dependent and comparable in healthy and HIV-infected individuals; the recommended oral dosage of in adults is 300 mg/day [50].
Fig. 4.
Structure of TDF, tenofovir and their active metabolite.
TDF is approved for the treatment of HIV-1 infection in adults as combination therapy with other antiretroviral drugs. Tenofovir is also commercialized in combination with emtricitabine (Truvada®) as an FDA-approved prescription drug for pre-exposure prophylaxis to reduce the risk of HIV infection [51], [52], [53].
In 2020, a docking study showed that tenofovir and other four approved drugs (galidesivir, remdesivir, sofosbuvir, and ribavirin) can bind to SARS-CoV-2 RdRp with binding energies similar to those of native nucleotides, inhibiting the function of the proteins and eventually leading to viral eradication [54].
Another important study in this field evaluated the antiviral efficacies of three FDA-approved formulations containing, respectively, hydroxychloroquine sulfate, a lopinavir-ritonavir combination, and an emtricitabine-tenofovir combination, against SARS-CoV-2 using a highly susceptible ferret infection model. The ferrets were treated with the antivirals, and of the three drugs, the emtricitabine-tenofovir was only one that caused a noticeable reduction in the overall clinical scores as well as a shorter duration of the clinical symptoms. Gastrointestinal symptoms were also evaluated, but none of the tested antiviral drugs appreciably diminished the gastrointestinal SARS-CoV-2 replication in infected ferrets [55]. Recently, polymerase extension experiments have confirmed that the active metabolites of tenofovir are permanent terminators for the SARS-CoV-2 RdRp [56]. In addition, in vitro studies with infected Vero CCL-81 cells have shown that the treatment with TDF reduced the released viral genome amount by 15-fold, while no detectable cytotoxicity was observed [57].
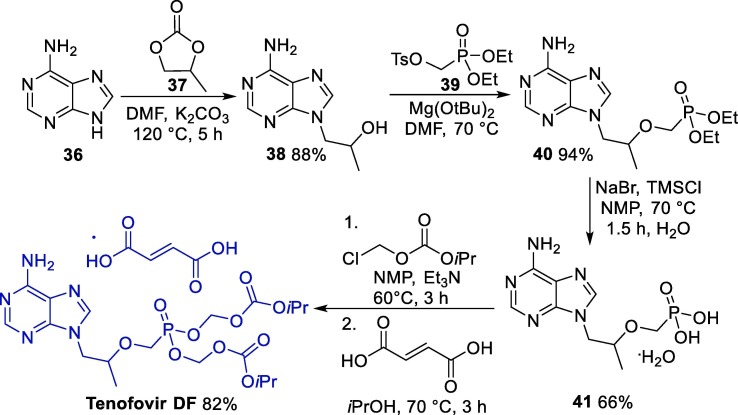
A versatile approach for the synthesis of tenovir DF starts with the reaction between adenine 36 and R-propylene carbonate 37 in the presence of sodium hydroxide, giving alkylated intermediate 38, which is reacted with tosylate 39, furnishing 40, which was further hydrolyzed to 41 (Scheme 5 ) [58]. Next, the alkylation of 41 with chloromethyl isopropyl carbonate followed by treatment with fumaric acid gives rise to tenofovir DF.
Scheme 5.
Synthetic route towards TDF. NMP = N-Methyl-2-pyrrolidone.
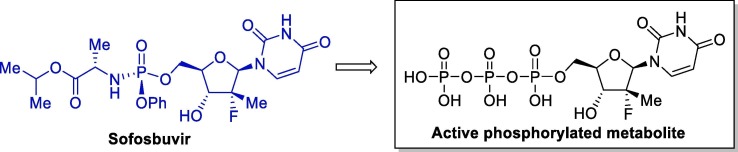
Sofosbuvir (Solvadi®, Gilead Sciences) is an antiviral agent approved by the FDA in 2013 for the treatment of chronic hepatitis C in combination with other antiviral compounds. This drug is a pyrimidine nucleotide analogue that acts by inhibiting hepatitis C virus (HCV) NS5B polymerase, and is administered orally once-daily in the treatment of chronic hepatitis C. Sofosbuvir is a pro-drug that undergoes intracellular metabolism to form its pharmacologically active form, which is incorporated into HCV RNA by NS5B polymerase and acts as a chain terminator (Fig. 5 ) [59].
Fig. 5.
Chemical structure of sofosbuvir and its active metabolite.
In 2017, in vitro studies with multiple human tumor cell lines and isolated human fetal- derived neuronal stem cells infected with Zika virus (ZIKV) revealed that sufosbuvir inhibits the replication and infection of several ZIKV strains with EC50 values ranging from 1 to 5 μM [60]. Importantly, the oral treatment of mice with sofosbuvir protected those against ZIKV-induced death.
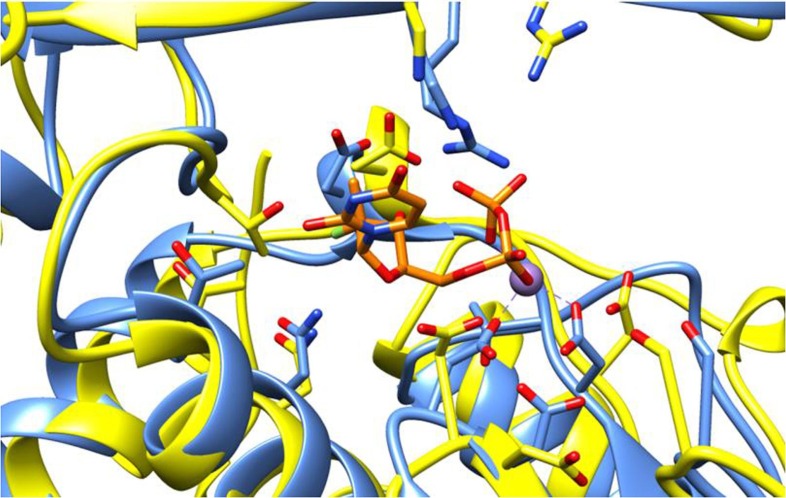
In a recent study, available at a preprint platform, it was reported that sofosbuvir was able to inhibit SARS-CoV-2 in HuH-7 and Calu-3 cells with EC50 values of 6.2 and 9.5 μM, respectively, but was inactive in Vero cells. The activity of this compound against SARS-CoV-2 was attributed to the homology in the nucleotide uptake channel shared by the RNA polymerases of HCV NS5B and SARS-CoV-2 nsp12. Indeed, a recent in silico has performed the structural superposition of the SARS-CoV-2 polymerase domain (nsp12) with HCV NS5B bound to sofosbuvir and observed that the inhibitor can be fitted into the nsp12′s active site with no steric hindrances, and that the residues that participated in the sofosbuvir binding are well-conserved in the SARS-CoV-2 active site (Fig. 6 ) [61]. Similar results were observed by other research groups [55].
Fig. 6.
Structural superposition of the HCV NS5B and SARS-CoV-2 polymerases. HCV NS5B is depicted in blue, SARS-CoV-2 nsp12 in yellow, sofosbuvir in orange [62]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
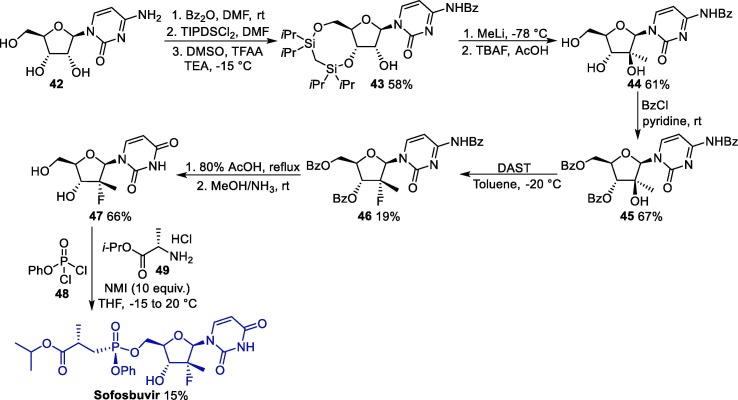
The synthesis of sofosbuvir can be accomplished starting from cytidine 42, which undergoes protection of the NH and hydroxyl groups of the furan moiety to furnish 43, which is submitted to a methylation reaction followed by deprotection conditions, giving 44 (Scheme 6 ) [62]. Next, 44 reacts with benzyl chloride, leading to 45, and undergoes a fluorination step to form 46, which undergoes a deprotection step to give 47. Finally, compound 47 is used in an NMI-promoted multicomponent reaction in the presence of 48 and 49, giving rise to sofosbuvir.
Scheme 6.
Synthetic route towards sofosbuvir. TIPDSCl2 = 1,3-Dichloro-1,1,3,3-tetraisopropyldisiloxane, DAST = Diethylaminosulfur trifluoride, NMI = N-Methylimidazole.
Favipiravir (Avigan®) is a small pyrazine compound which was developed by Fujifilm Toyama Chemical and approved in Japan in 2014 for the treatment of infections caused by avian influenza or novel influenza resistant to neuraminidase inhibitors [63]. Favipiravir is marketed as 200 mg tablets. According to its pharmacokinetic profile, after oral administration, Favipiravir reaches its maximum concentration two hours after ingestion, with a half-life equivalent to 2–5.5 h. It has a plasma protein binding rate of 54% and is metabolized in the liver mainly by aldehyde oxidase (AO), and partially by xanthine oxide, producing inactive metabolites that are eliminated by the liver [64]. It was observed that favipiravir presents a risk of teratogenicity and embryotoxicity [65].
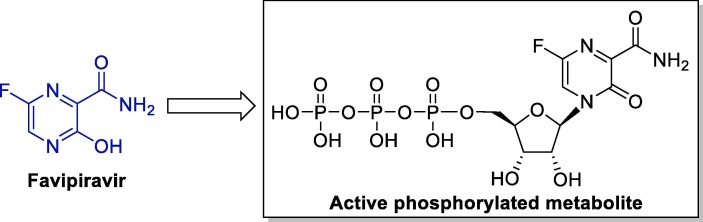
Mechanistic studies of its activity against influenza viruses have showed that favipiravir acts as a prodrug: when entering the cell it undergoes intracellular phosphoribosylation to its active form, favipiravir ribofuranosyl-5B-triphosphate (Fig. 7 ). This active triphosphate compound is recognized as a substrate by RdRp, and potently and selectively inhibits the RNA polymerase activity by interrupting the nucleotide incorporation process during viral RNA replication [64], [66], [67]. Additionally, favipiravir has a carboxamide group that allows it to make hydrogen bonds like the ones made by guanine, and thus, it can be considered a guanine analogue [39].
Fig. 7.
Chemical structure of favipiravir and its active metabolite.
Due to its marked antiviral properties and the urgency in finding therapeutic options for COVID-19, favipiravir is currently the target of several in vitro and in vivo studies [64]. A recent work conducted with Vero E6 cells has shown that favipiravir is active in vitro against SARS-CoV-2, although it presented a high EC50 value when compared to chloroquine or remdesivir (61.88 μM vs 6.90 and 1.76 μM for chloroquine and remdesivir, respectively) [68].
A non-randomized study in China conducted with 80 COVID-19 patients has revealed that patients treated with favipiravir had a significant reduction in the time of SARS-CoV-2 viral clearance when compared to that of patients treated with lopinavir/ritonavir. Patients in both groups were treated concomitantly with inhaled IFN-alfa until viral release for a maximum of 14 days [69]. The average viral clearance time for patients treated with favipiravir was 4 days, while in patients treated with lopinavir/ritonavir, this time was 11 days. It was found that on day 14 of treatment, approximately 91% of the people treated with favipiravir showed radiographic improvement, and at the same time, for those treated with lopinavir/ritonavir this value was around 62%. As for adverse effects, there was a much lower percentage in the group treated with favipiravir (11.4% vs 55.6%) [70].
In another clinical trial, baloxavir marboxil and favipiravir were used in patients with COVID-19 randomized in a 1:1:1 ratio in three groups: baloxavir marboxil, favipiravir and control groups. The study aimed to evaluate both the efficacy and safety of these drugs to patients being treated with the antivirals lopinavir/ritonavir or darunavir/cobicistat or arbidol confirmed with COVID-19 and who tested positive for SARS-CoV- 2 [70]. Unfortunately, the addition of one or the two drugs in the studied dosages led to no improvements in the results when compared to the current treatments. However, the obtained data is useful for carrying out additional studies in order to reach the best form of administration of these antivirals as anti-COVID-19 drugs [71].
An additional randomized trial concluded that patients treated with favipiravir have superior recovery rate (71.4%) and shorter duration of fever and cough relief time than the ones in the umifenovir group (55.8% recovery rate) [71].
In this sense, the results of such preliminary clinical tests show that favipiravir is a promising candidate for the treatment of COVID-19. Indeed, several clinical tests examining the anti-SARS-CoV-2 potential of favipiravir (alone or combined with other drugs) are underway worldwide.
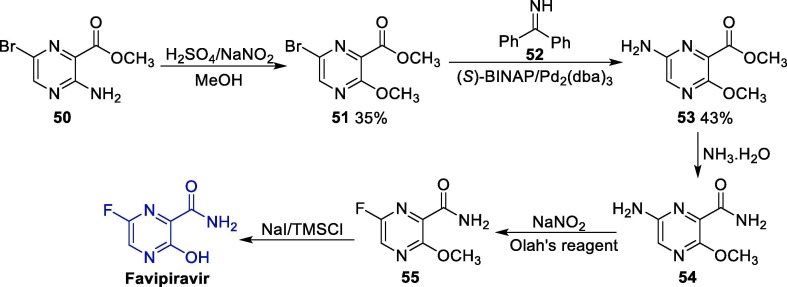
Favipiravir can be obtained via several different synthetic routes, and the original one was disclosed in 2000, having methyl 3-amino-6-bromopyrazine-2-carboxylate 50 as starting material (Scheme 7 ) [72]. In this case, bromopyrazine 1 initially undergoes a diazotization/alcoholysis reaction to furnish intermediate 51, which is reacted with imine 52 in the presence of a palladium catalyst to produce amine pyrazine 53. Next, the ester group in 53 is converted to an amide one after reaction with ammonia, leading to intermediate 54, which is submitted to a Schiemann fluorination using Olah’s reagent (70% hydrofluoric acid/30% pyridine) to give 55. Finally, the demethylation of 55 affords favipiravir [73].
Scheme 7.
Synthesis of favipiravir from methyl 3-amino-6-bromopyrazine-2-carboxylate.
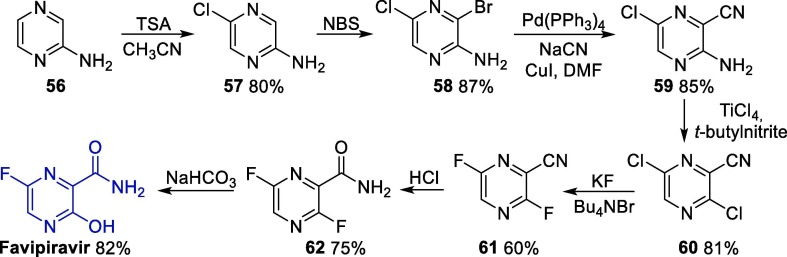
Although this synthetic route involves only five steps, it comprises the use of the highly toxic Olah’s reagent. However, other routes are available for the synthesis of favipiravir, involving several steps and the use of different starting materials such as 3-hydroxypyrazine-2-carboxamide and 3-aminopyrazine-2-carboxylic acid [74]. On the other hand, a more recent approach was published in 2019 and comprises the use of the commercially available reagent 2-aminopyrazine 56 as starting material, enabling the obtainment of favipiravir in seven steps (Scheme 8 ). Here, 2-aminopyrazine 56 is initially converted to chloro-substituted pyrazine 57, which next undergoes a bromination step, giving intermediate 58. Next, a nitrile group is inserted into the pyrazine ring under palladium catalysis, leading to 59, which is transformed into dichloro intermediate 60 upon reaction with titanium tetrachloride in the presence of tert-butylnitrile. Then, 60 undergoes a double fluorine substitution to furnish 61, and the hydration of the nitrile group in this intermediate leads to amide 62, which furnishes favipiravir after a nucleophilic substitution reaction of the fluorine bound to C3 [74].
Scheme 8.
Synthesis of favipiravir starting from 2-aminopyrazine. TSA = N-chloro-N-methoxy-4-methylbenzenesulfonamide.
2.1.3. Indole derivatives: Umifenovir
Umifenovir (Arbidol®) is an indole derivative first marketed in 1993 for the prophylactic treatment of infections caused by influenza A and B viruses [74]. Produced by Pharmstandard, it is still currently used in Russia and China to treat influenza infections [75]. Umifenovir is marketed in 50 and 100 mg capsules, being administered orally. The pharmacokinetics is limited, presenting rapid absorption and reaching the maximum concentration in 1.6–1.8 h. It is a slow elimination drug, with a half-life of 16 to 21 h, and may be administered twice a day [76].
The drug’s anti-influenza mechanism of action is related to arbidol's ability to bind to the haemagglutinin (HA) protein [77]. The haemagglutinin (HA) protein is a homotrimeric glycoprotein found on the surface of the influenza virus, and it is essential for its infectivity. This protein is responsible for allowing the influenza virus binding to the sialic acid present on the surface of the target cells (respiratory tract cells or erythrocytes). As a result of this interaction, the virus is internalized in the host cell. Once umifenovir binds to the HA protein, this glycoprotein is prevented from binding to sialic acid, so the virus is no longer able to penetrate the host cell [78].
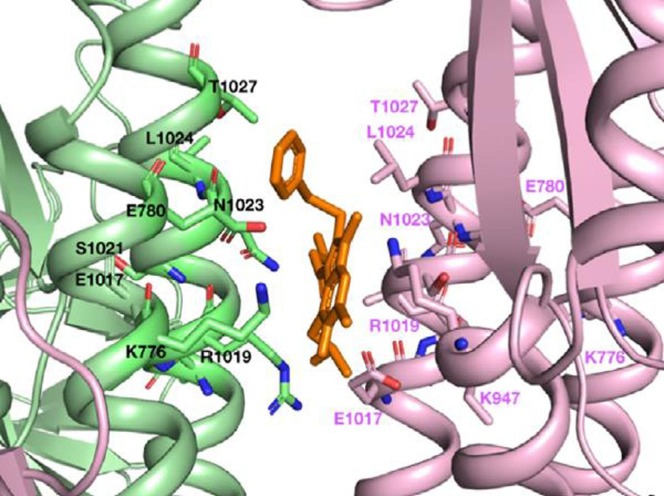
The structural similarity between the SARS-CoV-2 peak and the influenza virus (H3N2) HA glycoproteins justifies the fact that drugs that are capable of binding to HA can also do so to the SARS-CoV-2 spike protein. This fact was evidenced by molecular modeling studies, wherein was demonstrated that umifenovir is able to bind to the protein peak, preventing its trimerization, which would be a determining factor for the mechanism of cell adhesion (Fig. 8 ) [78].
Fig. 8.
Umifenovir (in orange) binding region in SARS-CoV-2 spike glycoprotein. Reprinted from International Journal of Antimicrobial Agents, 56, N. Vankadari, “Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein”, Page 2, with permission of Elsevier. Copyright 2020. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Recently in 2020, in vitro studies performed with Vero cells confirmed that arbidol efficiently inhibits SARS-CoV-2 infection with an EC50 of 4.11 μM. The author also determined that arbidol was able to efficiently block both viral entry and post-entry stages, and also concluded that the drug prevented the viral attachment and release of SARS-CoV-2 from the intracellular vesicles. Importantly, the EC50 of arbidol against SARS-CoV-2 led the authors to suggest that the dose of arbidol currently recommended by the Chinese Guidelines (200 mg, 3 times/day) should be elevated in order to achieve ideal therapeutic efficacy to inhibit the SARS-CoV-2 infection [79].
A clinical trial was conducted at Wuhan Jinyintan Hospital, in 2020, from February 2 to March 20 conducted to evaluate the effectiveness and safety of umifenovir in the treatment of COVID-19 patients. In this study, 81 patients were evaluated: 45 received 200 mg of umifenovir three times a day, and 36 were in the control group. The authors concluded that baseline clinical and laboratory characteristics were similar in the two groups, and patients in the umifenovir group had a longer hospital stay than those in the control [80]. Although such results may seem discouraging, further clinical trials with higher doses of umifenovir may be required in order to verify its clinical efficiency against the SARS-CoV-2 infection.
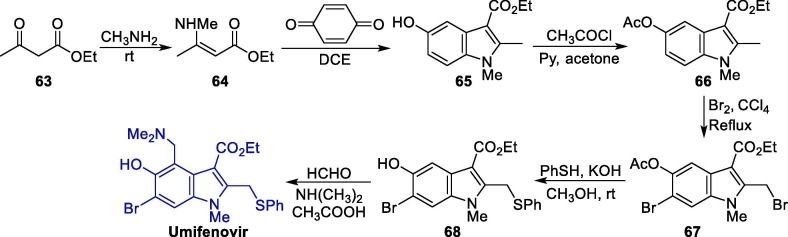
The synthesis of umifenovir was described in 2006 starting from the reaction between ethyl acetoacetate 63 and methylamine, giving enaminone 64, which next undergoes a Nentizescu condensation reaction with 1,4-benzoquinone to produce indole derivative 65 (Scheme 9 ). Then, an acetylation reaction is carried out to protect the hydroxyl group in 65, producing 66, which is converted to 67 after a bromination step. The reaction of intermediate 67 with thiophenol in basic medium leads to the formation of 68, which finally affords umifenovir after a Mannich reaction [81].
Scheme 9.
Synthetic route towards umifenovir.
2.2. Antiparasitic drugs
Based on the hypothesis that some antimalarial medications present immunomodulatory, anti-inflammatory and weakly antithrombotic effects, those have been tested against SARS-CoV-2, expectedly acting as a prophylactic, or even preventing/limiting the disease’s symptoms [82]. In that way, aminoquinoline-type molecules such as chloroquine, hydroxychloroquine and amodiaquine were among the first tested compounds against SARS-CoV-2. However, several controversial results on these drugs have arisen, causing important discussions within the scientific community. In this context, other antiparasitic drugs, such as mefloquine, nitazoxanide, ivermectin and emetine, have also been studied. In this section we provide a discussion on the pharmacological aspects that led these compounds to be considered as therapeutic options for COVID-19, as well as the synthetic routes towards them.
2.2.1. 4-Aminoquinolines: Chloroquine, hydroxichloroquine and amodiaquine
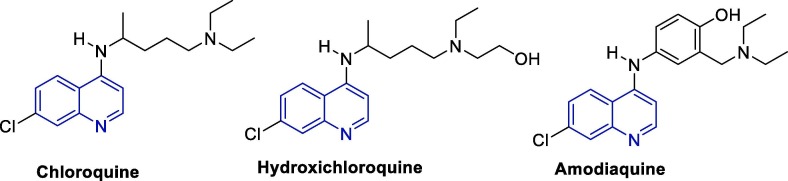
Due to their pronounced antiparasitic effects, quinoline-based antimalarials are the most used drugs in the initial treatment and prevention of malaria. In the last decades, the activity of such drugs has also been evaluated against several type of viruses, including coronaviruses [83], [84], [85], [86], [87], [88], which caused this class of compounds to be pointed as a therapeutic option for COVID-19 [69], [89], [90], Fig. 9 shows the structures of chloroquine (CQ), hydroxychloroquine (HCQ) and amodiaquine (ADQ), all members of the aminoquinoline class of antimalarial drugs that have also been pointed as promising candidates for the treatment of COVID-19.
Fig. 9.
Chemical structure chloroquine, hydroxichloroquine and amodiaquine.
Chloroquine was the first clinically used antimalarial drug and served as a prototype for the development of the following ones. This drug is usually marketed as a diphosphate salt, and can be found in 250 mg tablets (equivalent to 150 mg of the drug). CQ is also used as an immunomodulator for rheumatic diseases such as systemic lupus erythematosus rheumatoid arthritis, juvenile idiopathic arthritis and Sjogren's syndrome (chronic autoimmune rheumatic disorder) [91], [92], [93]. Additionally, it can also be used by patients with liver abscess caused by Entamoeba histolytica [94]. Despite their therapeutic indications, several side effects are reported for chloroquine, hydroxychloroquine and other antimalarials, ranging from mild, e.g. nausea and headache, to severe ones, e.g. cardiotoxicity, hypotension, vasodilation, suppressed myocardial function, cardiac arrhythmia and arrest [95], [96], [97], [98].
Hydroxychloroquine (Plaquenil®) and amodiaquine (Amodaquine®) are less toxic than chloroquine. Generally, HCQ is found as a sulfate salt in the form of tablets containing 200 mg of the drug, and is a common medication in regions where the malaria incidence is still sensitive to QC. Just as QC, HCQ is quickly absorbed by the gastrointestinal tract and has a high volume of distribution through the blood plasma [99]. As for ADQ, it is marketed as a dihydrate hydrochloride salt in combination with artesunate (270 mg of ADQ and 100 mg of artesunate), and used for the treatment of patients affected by QC-resistant malaria. Due to hepatic biotransformation, ADQ is converted to its main desethylated metabolite, which is also active, albeit to a lesser extent [100], [100].
In the last decades, different research groups have evaluated the in vitro anti-coronavirus activity of CQ, HCQ and ADQ [101], [102], [103]. Since the beginning of 2020, efforts have been directed to the evaluation of the anti-SARS-CoV-2 activity of such compounds [69], [104], [105]. Table 1 summarizes the main results reported in this context. The different values regarding the in vitro activity observed for the same virus, in some cases, can be attributed to methodological differences, such as the specific multiplicity of infection (MOI) applied to each test.
Table 1.
Reported anti-coronavirus activity for CQ, HCQ and ADQ.
| Drug | MERS-CoV (µM) | SARS-CoV-1 (µM) | SARS-CoV-2 (µM) |
|---|---|---|---|
| CQ diphosphate | EC50 = 6.275e[103] | IC50 = 8.8a[102] | IC50 = 46.80c[106] |
| EC50 = 3.0b[104] | EC50 = 6.538f[103] | EC50 = 1.13d[69] | |
| EC50 = 4.1b[104] | EC50 = 5.47c[105] | ||
| HCQ sulfate | EC50 = 8.279e[103] | EC50 = 7.966f[103] | IC50 = 11.17c[106] |
| EC50 = 0.72c[105] | |||
| ADQ dihydrochoride dihydrate | EC50 = 6.212e[103] | EC50 = 1.274f[103] | IC50 = 4.94c[106] |
MOI not reported.
MOI = 0.005.
MOI = 0.01.
MOI = 0.05.
MOI = 0.1.
MOI = 1.0.
The broad antiviral spectrum associated with 4-aminoquinoline derivatives can be explained with basis on different modes of action, depending on the virus. One of these mechanisms involves the inhibition of quinone reductase 2, an important enzyme for the biosynthesis of sialic acids; such acids are the structural components of the sugar chains present in the cell's transmembrane proteins and are important for cell recognition by the virus. In the case of SARS-CoV-1, and probably SARS-CoV-2, CQ and its analogs appear to affect different stages of viral replication [90], [106].
Furthermore, these aminoquinoline drugs cause the alkalization of several cellular components, including endosomal contents, affecting the mechanisms of viral entry into the host cell by the pH-dependent endocytic pathway in a complementary way [90]. An adequate acidic medium on a cellular level is important for virus successful replication, since a low pH is required for the functional activity of cathepsins, which are proteins necessary for the proper cleavage of the viral S protein and, consequently, for the formation of the autophagosome in the virus internalization process. Also, lysosome acidification leads to the disintegration of the viral capsule and the release of its genomic material and viral enzymes in the cytosol. Maintaining a low local pH is similarly important for other processes in the viral replication cycle, such as proper viral proteins maturation and virion assembly [100]. As a matter of fact, structure–activity relationship (SAR) studies on the antimalarial activity of these substances, the weakly basic groups present in their structure, namely the heterocyclic nitrogen and the tertiary amine group present in the side chain, are highly important for their activity [107], [108], [109], [110].
CQ and HCQ also appear to reduce the terminal glycosylation levels on the angiotensin-converting enzyme 2 (ACE2) receptor, thus being active in the initial stage of viral internalization in the cell. Recently, in silico studies demonstrated that CQ is capable of binding to sialic acids and gangliosides with high affinity; a new type of interaction between gangliosides and the 111–162 domain of the N-terminal portion of the SARS-CoV-2 S protein has also been identified, highlighting that this peptide sequence is fully conserved in clinical isolates worldwide. Considering that this protein domain is possibly responsible for facilitating the contact of the virus with the ACE2 receptors, it is a possible pharmacological target for anti-COVID-19 drugs [107]. In this study, the interaction mode between CQ/HCQ and 9-O-acetyl-N-acetylneuraminic acid (9-O-Ac-SIA, the sialic acid subtype most easily recognized by SARS-CoV-2) was initially simulated, and most of the overall favorable energy variation has arisen from ionic interactions between the drug’s positively charged quinoline nitrogen and the negatively charged carboxylate group present in the sialic acid (Fig. 10 ). Additionally, other weaker Van der Waals and OH-π interactions involving the CQ heteroaromatic system and N-aliphatic side chain help in the complex stabilization [107].
Fig. 10.
(a) Mills projection of the 9-O-acetyl-sialic acid subtype. (b) Simplified representation of the interactions between CQ/HCQ e o 9-O-Ac-SIA.
The interaction between these drugs and the GM1 membrane ganglioside was also simulated, and it was demonstrated that, in this system, two molecules of CQ are capable of interacting with the sugar moieties on the GM1 ganglioside, mimicking the way in which the viral S protein interacts with that membrane component. In this case, HCQ showed an even greater interaction than CQ, which was attributed to a complementary hydrogen bond from the hydroxyl group present in the side chain of this substance. Other weaker non-covalent interactions were also observed. In that way, CQ and HCQ have the potential of preventing the interaction of viral S protein with membrane gangliosides, preventing cell recognition. The greater binding affinity observed for HCQ is in line with its higher anti-SARS-CoV-2 in vitro activity when compared to CQ [107].
Although the in vitro studies with CQ and HCQ have been showing promising results, there is still little evidence about the safety and efficacy of CQ and HCQ to treat patients infected with SARS-CoV-2. On the other hand, some preliminary studies of several clinical trials have been published in the last months, and conflicting conclusions were drawn.
A clinical trial conducted at Tongji Hospital, Wuhan, China with 550 COVID-19 patients in critical condition and under mechanical ventilation showed that HCQ dramatically decreased the fatality rate on critically ill patients, in addition to decreasing the inflammatory cytokine storm [111]. A second example is the open-label non-randomized clinical trial performed with a combination of HCQ and azithromycin involving a restricted number of 36 patients, from which it was concluded that the drugs caused a decrease in their viral load [112].
Conversely, a third clinical trial conducted with 1376 patients hospitalized with COVID-19 concluded that there was no decrease or increase in the risk of intubation or death on patients whom received HCQ when compared to those who did not receive this drug [113].
Other recent publications, however, have highlighted the negative results on the clinical use of these drugs on COVID-19 patients, some of which have appointed a high risk of intubation or even death due to their side effects [114], [115]. Indeed, a clinical trial conducted with 1438 patients suggested that there is a higher incidence of heart failure in patients who received HCQ/azithromycin, with no observed improvement in hospital mortality [116]. In light of these observations, another clinical trial involving 81 patients with severe cases of COVID-19 led to the conclusion that a high dosage of HQC should not be recommended for patients with severe COVID-19 due to of its potential safety risks, especially when taken concomitantly with azithromycin and oseltamivir. However, such results cannot be extrapolated to patients with non-severe COVID-19 [117].
A study that had reported severe adverse events in COVID-19 patients treated with hydroxychloroquine or chloroquine was retracted from the Lancet due to issues related to the veracity of the data, and in sequence, another article by the same authors using data from the same Surgisphere database was also retracted from the New England Journal of Medicine [118]. However, several recent randomized clinical trials have pointed out that HCQ does not reduce mortality in hospitalized patients with COVID-19 [119]. The HCQ arm of the WHO RECOVERY trial, for instance, was shutdown with preliminary results having exposed that HQC does not lead to any appreciable reduction in mortality (25.7% with HCQ vs 23.5% with usual care) [120]. Indeed, after the disclosure of the WHO RECOVERY report, the FDA has revoked the use of chloroquine and hydroxychloroquine in the EUA in June 5, 2020, and the WHO and the National Institutes of Health have stopped all trials and the use of such drugs in hospitalized patients owing to the nonexistence of benefits [121]. The final results of the RECOVERY clinical trials were recently disclosed, confirming its preliminary findings that HCQ is not an effective treatment for patients hospitalized with COVID-19 independently of age, sex, race, time since illness onset, level of respiratory support, and baseline-predicted risk [122]. Moreover, the results suggested that the patients who received HCQ underwent hospitalization for a longer duration, and a higher risk of invasive mechanical ventilation or death was observed for the HCQ patients when compared to the ones who received usual care.
Overall, at this point there is robust data available to indicate that CQ and its analogs do not benefit hospitalized patients with COVID-19, and may even cause severe adverse effects such as cardiac complications when high doses are administered. It should be noted that for severe cases, there is no indicative that its use is responsible for reversing the infection. However, there is still a gap to be explored regarding the use of CQ/HCQ as prophylactic agents or in patients with less severe SARS-CoV-2 infection by means of comprehensive, randomized and double-blind studies.
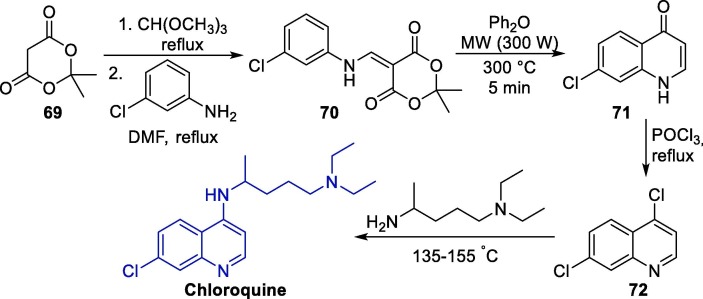
The synthesis of these three aminoquinoline derivatives usually involves a nucleophilic aromatic substitution (SNAr) step with a 4-haloquinoline precursor and a suitable amine [123], [124], [125], [126], [127]. An example of this method directed toward QC synthesis was described in 2005 [125]. Initially, Meldrum's acid 69 is converted into the intermediate 70 through a condensation step with trimethyl ortho-formate followed by the reaction with 3-chloroaniline. Intermediate 70 is then subjected to a decarboxylative pyrolytic cyclization under microwave irradiation to form oxoquinoline 71, which is treated with phosphoryl chloride to furnish 4,7-dichloroquinoline 72. Finally, 72 is converted to CQ via a regioselective SNAr reaction with N1,N1-diethylpentane-1,4-diamine (Scheme 10 ).
Scheme 10.
Seminal synthetic approach towards chloroquine.
Over the last decades, several interesting methods were reported for the synthesis of such compounds, for instance using cross-coupling as an alternative to the classic SNAr methods [126].
As for amodiaquine, few methods are available for its preparation. A particularly interesting approach involves initial conversion of 4,7-dichloroquine 73 into 7-chloro-4-(4-hydroxyanilino)quinoline 74 through a regioselective SNAr step followed by the one-pot addition of formaldehyde and diethylamine (Scheme 11 ) [127]. This tandem procedure allowed the preparation of the amodiaquine dihydrochloride salt in a 92% yield after the addition of concentrated hydrochloric acid and crystallization.
Scheme 11.
One-pot procedure for the synthesis of amodiaquine.
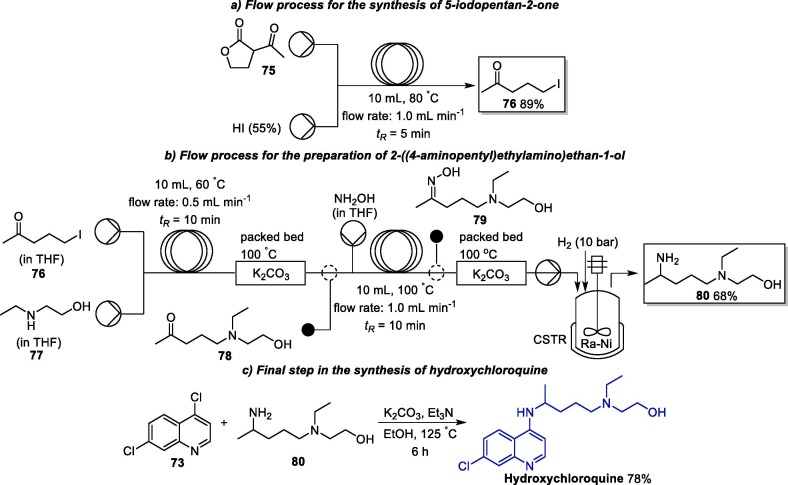
An interesting process for the multigram synthesis of HCQ involving a continuous flow (CF) process was published in 2018 (Scheme 12 ) [128]. Initially, 5-iodopentan-2-one 76 is produced with 89% yield from the decarboxylative ring-opening of lactone 75 in the presence of a hydroiodic acid solution under CF conditions (1.0 mL min−1 flow rate and 5 min of residence time) [128]. Next, the flow conditions for the conversion of 76 into 80 were individually optimized and, subsequently, telescoped in a CF reaction process, which allowed the isolation of 80 with an overall yield of 68%. In this process, initially, 5-iodopentan-2-one 76 reacted with 2-(ethylamino)ethan-1-ol 77, giving intermediate 78, which undergoes reductive amination using hydroxylamine in the presence of Raney-Nickel (Ra-Ni) as catalyst and molecular hydrogen. In final step, 80 was reacted with 4,7-dichloroquinoline 73 under batch conditions in the presence of potassium carbonate and triethylamine, giving rise to HCQ in a 78% yield.
Scheme 12.
Flow process for the synthesis of hydroxychloroquine.
2.2.2. Other quinoline derivatives: Mefloquine
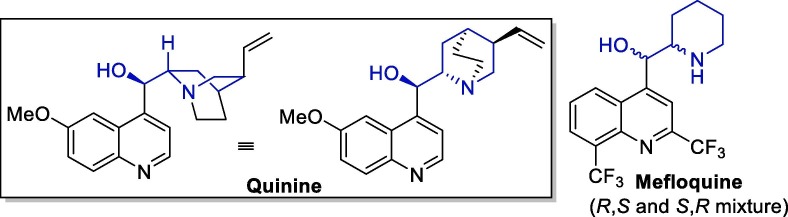
Mefloquine is a 4-alkyl substituted quinoline derivative functionalized with a (piperidin-2-yl)methanolyl side chain that can be viewed as a simplified version of the (5-vinylquinuclidin-2-yl)methanolyl side chain present in quinine, a natural antimalarial agent found in the bark of Cinchona and Quina trees (Fig. 11 ) [128]. This drug was developed in the 1970s by the United States Army, and is sold as a racemic mixture by Hoffmann-La Roche [129].
Fig. 11.
Quinine and mefloquine structural representations.
MQ is easily permeable through the blood–brain barrier, and its liver metabolization occurs via the cytochrome P450 system [130]. MQ is distributed extensively in the tissues and is eliminated slowly in the feces and bile, mainly as a carboxylic acid metabolite [131].
In addition to the 4-aminoquinoline derivatives discussed in the previous topic, it has already been demonstrated that MQ blocks the cytopathic effect of coronavirus in cell cultures and prevents its replication, which infers that it can be considered for repositioning in COVID-19 therapies. Its effectiveness as an inhibitory agent against MERS-CoV and SARS-CoV-1 is already known (EC50 values of 7.416 μM and 15.553 μM, respectively) [103]. More recently, it has also been demonstrated that MQ, in the form of its hydrochloride salt, has an inhibitory effect against SARS-CoV-2 with an IC50 value of 7.11 and 8.06 μM (MOI of 0.004 and 0.01, respectively). Additionally, its immunosuppressive effect prevents the activation of an inflammatory response caused by the virus [106].
It is important to note that this drug is not widely used because of its toxicity to the central nervous system [132]. In fact, in 2013, its potential neuropsychiatric effects started to be mandatorily highlighted in the USA labels [131]. However, MQ can be regarded as a model in the development of new bioactive agents for COVID-19 treatment [34].
The antimalarial mode of action associated with MQ involves pH increasing on the cellular vesicular structures important for the parasite's replication cycle, being similar to that described for CQ, HCQ and ADQ. Due to the similarities already discussed, it is coherent to assume that, in the same way, MQ inhibits viral replication by acting as an alkalinizing agent toward the content of endosomal vesicles related to the virus internalization, protein maturation and virion assembly processes. This effect can be associated to the two basic nitrogen atoms present in its structure [108], [109], [110], [111].
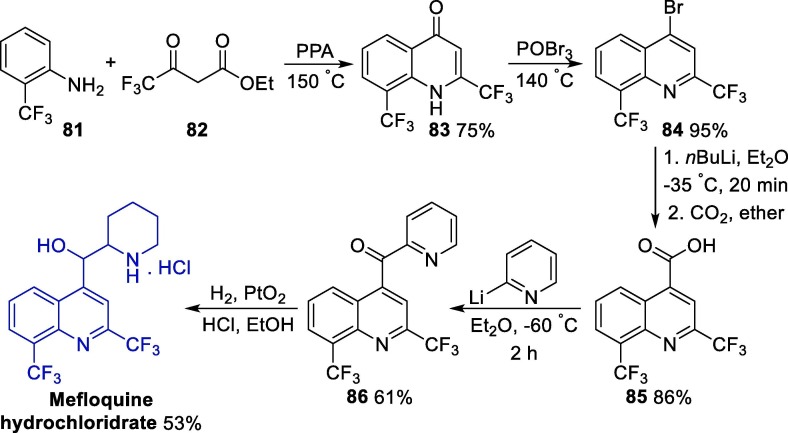
One of the first synthetic methods for the preparation of mefloquine was published in 1971 and involved a linear five-step route (Scheme 13 ) [129]. Initially, a polyphosphoric acid-mediated cyclization of o-trifluoromethylaniline 81 with ethyl trifluoromethylacetoacetate 82 leads to the oxyquinoline intermediate 83, which was then converted to the 4-bromoquinoline 84 after treatment with phosphorus(V) oxybromide. Next, the carboxylation of intermediate 84 via a transmetallization step with n-butyllithium followed by treatment with carbon dioxide gives intermediate 85. The reaction of cinclioninic acid 85 with a 2-pyridyllithium excess led to the formation of ketone 86, which was then reduced using a platinum(IV) catalyst in the presence of hydrochloric acid, affording mefloquine hydrochloride salt. This classic method furnishes the desired product with a 20% overall yield.
Scheme 13.
Classical method for the synthesis of mefloquine. PPA = polyphosphoric acid.
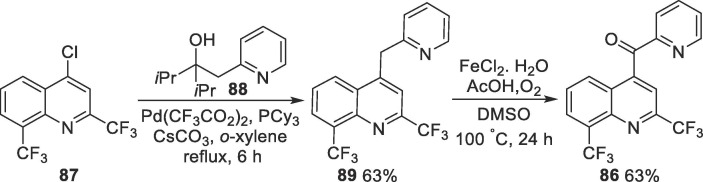
More recently, a new synthetic method was described for the benzylic oxidation of diaryl methylenes, and this strategy was applied to the synthesis of intermediate 86 (Scheme 14 ) [133]. In this case, the authors used commercially available 4-chloro-2,8-di(trifluoromethyl)quinoline 87 as a starting material in a palladium-catalyzed cross-coupling reaction with alcohol 88, leading to intermediate 89. Then, 89 is oxidized to ketone 86 using molecular oxygen in the presence of ferrous chloride. The main advantage of this alternative is the application of more easily handled reagents, in comparison to the highly reactive organolithium species used in the classical method.
Scheme 14.
Alternative method for the synthesis of intermediate 86.
2.2.3. Thiazolides derivatives: Nitazoxanide
Nitazoxanide (NTZ, Alinia®) is an FDA-approved synthetic nitrothiazolyl-salicylamide derivative that has been used for treating gastrointestinal infections caused by Cryptosporidium parvum and Giardia lamblia in millions of adults and children since 2004. This drug causes minimal side effects, with an excellent safety record for a variety of indications [134].
NTZ is available in the form of tablets (for 12-year-old and older patients) and oral suspension (1-yeard-old and older patients), and is marketed under different brand names in different countries, such as Heliton® (Argentina), Annita® (Brazil), Celectan® (Colombia, Ecuador, Venezuela), Repinox® (Costa Rica, Dominican Republic, Guatemala, Honduras, Nicaragua, Panama, El Salvador), Nodik® (Guatemala), Daxon® (Mexico), Dexidex® (Mexico), Kidonax® (Mexico), Pacovanton® (Mexico), Paramix® (Mexico), Anelmin® (Paraguay) and Colufase® (Peru) [135].
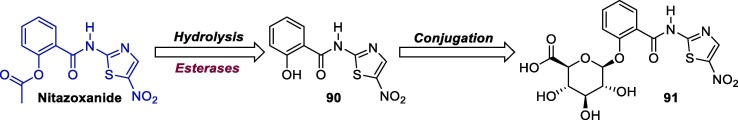
Once ingested, NTZ is rapidly hydrolyzed by plasma esterases into its active metabolite, tizoxanide (desacetyl-nitazoxanide, 90), which undergoes a conjugation reaction and is transformed into tizoxanide glucuronide 91 (Scheme 15 ). Its active metabolite is excreted in the urine, bile and feces, while tizoxanide glucuronide 91 is excreted in urine and bile. These metabolites are free of mutagenic effects [135].
Scheme 15.
NTZ structure and its main metabolites.
Both NTZ and its active metabolite, tizoxanide (desacetyl-nitazoxanide), have shown inhibitory effect against a broad range of both DNA and RNA viruses in cell culture assays [64], [136], [137], [138]. In special, NTZ has action against influenza A, dengue, yellow fever, Japanese encephalitis virus, rotavirus, norovirus, hepatitis B and C and human immunodeficiency virus [139]. In addition, NTZ has also been shown to inhibit the replication of the pathogenic human coronaviruses MERS-CoV and SARS-CoV-2. Importantly, the compound exhibits antiviral effect against SARS-CoV-2 at a low-micromolar concentration (EC50 = 2.12 μM, CC50 > 35.53 μM, SI > 16.76) [69]. It has been pointed out that NTZ’s broad antiviral spectrum is a consequence of its interference with host-regulated pathways involved in viral replication; the compound acts by upregulating the innate antiviral mechanisms by broadly amplifying cytoplasmic RNA sensing and type I interferon (IFN) pathways.
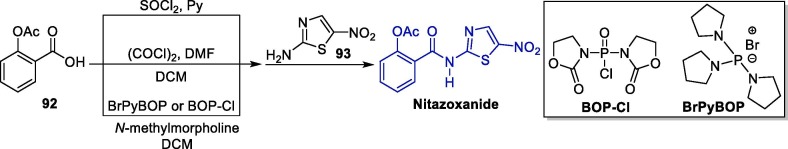
The synthesis of NTZ can be achieved in a few steps initiating with the conversion of 2-acetoxybenzoic acid 92 to the corresponding acid chloride or anhydride, which can be achieved using different approaches, followed by the reaction with 2-amino-5-nitrothiazole 93 (Scheme 16 ) [139], [140].
Scheme 16.
Synthetic route towards nitazoxanide.
2.2.4. Avermectins: Ivermectin
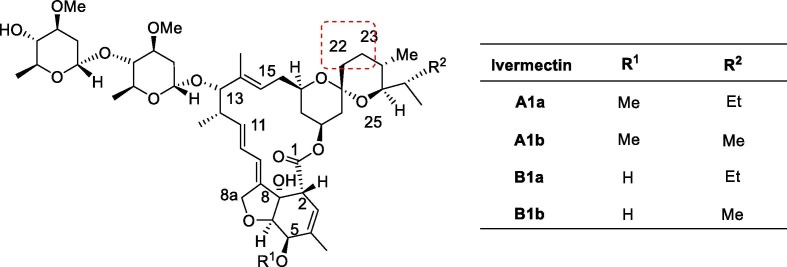
Avermectins (AVMS, Fig. 12 ) are naturally occurring 16-membered macrocyclic polyketides isolated from the fermentation of the fungus Streptomyces avermitilis. AVMS were discovered in 1967 by the Nobel Prize winner Satoshi Omura of the Japanese Kitasato Institute and ever since have found applications in humans and animals as antiparasitic agents, and in agriculture, as insecticides [141]. Ivermectin, a semi-synthetic derivative of the avermectins introduced by Merck, is a mixture of two homologous compounds 5-O-dimethyl- 22,23-dihydroavermectin B1a (not less than 80%) and B1b (not more than 20%), each of which being prepared from the avermectin family of fermentation products by selective reduction at the cis-22–23-double bond of avermectin B1. These two components, B1a and B1b have very similar biological and toxicological properties [142].
Fig. 12.
Structures of some ivermectins.
Ivermectin is one of the most important drugs against parasitic infections in veterinary and human medicine, and in humans its metabolism occurs in the liver by cytochrome P450 enzymes [143]. Its pharmacokinetics are characterized by slow absorption, wide tissue distribution because of its high lipid solubility, low metabolism, and slow excretion phase, with most of the dose eliminated unchanged in feces [144]. Ivermectin does not easily cross the blood–brain barrier in humans, as it is excluded by a P-glycoprotein drug pump [145].
Ivermectin has been shown to have in vitro antiviral activity against a broad range of viruses, such as human immunodeficiency virus (HIV-1), dengue fever, influenza, Zika and Ebola viruses [146]. As an anti-HIV agent, it acts by inhibiting the interaction between the HIV-integrase protein (IN) and the importin (IMP) α/ß1 heterodimer, which is responsible for IN nuclear import of integrase protein. However, this compound was also able to inhibit nuclear import of viral proteins, e.g. simian virus SV40 large tumor antigen and dengue virus (DENV) non-structural protein 5 [147], [148]. Still, it has been shown that it is active against RNA viruses such as DENV 1–4, West Nile Virus, and influenza [149], [150], [151]. Its wide activity is attributed to the fact that many RNA viruses rely by on IMPα/β1 during infection. Considering that SARS-CoV-1 proteins are important for IMPα/β1 during infection, it was inferred that ivermectińs nuclear transport inhibitory activity could also work well against SARS-CoV-2. At 24 h, there was a 93% reduction in viral RNA present in the supernatant (indicative of released virions) of samples treated with ivermectin compared to the vehicle DMSO. Indeed, in vitro studies conducted with Vero/hSLAM showed that 24 h after the addition of ivermectin, a 93% reduction in viral RNA was observed, and no more viral material was present in the medium after 72 h [147].
A recent observational study conducted with 280 patients with confirmed SARS-CoV-2 infection has shown that the use of ivermectin (single dose of 200 μg kg−1) led to a lower mortality rate (15% vs 25.2% in patients who were not treated with ivermectin), especially in patients who required higher inspired oxygen or ventilatory support (38.8 vs 80.7%) [152]. Another pilot clinical trial conduced in Iraq evaluated the administration of ivermectin (single dose of 200 μg kg−1 at the admission day) to patients taking HCQ (400 mg twice a day in the first day and 200 mg twice a day for 5 days plus a single dose of 500 mg single dose of azithromycin in the first day and 250 mg a day for 5 days), and observed that the patients in the ivermectin group had shorter hospital stays than the other group (7.6 vs 13.2 days, respectively) [153]. Although several clinical trials involving ivermectin are still ongoing, the results of a large-scale randomized study are not available yet.
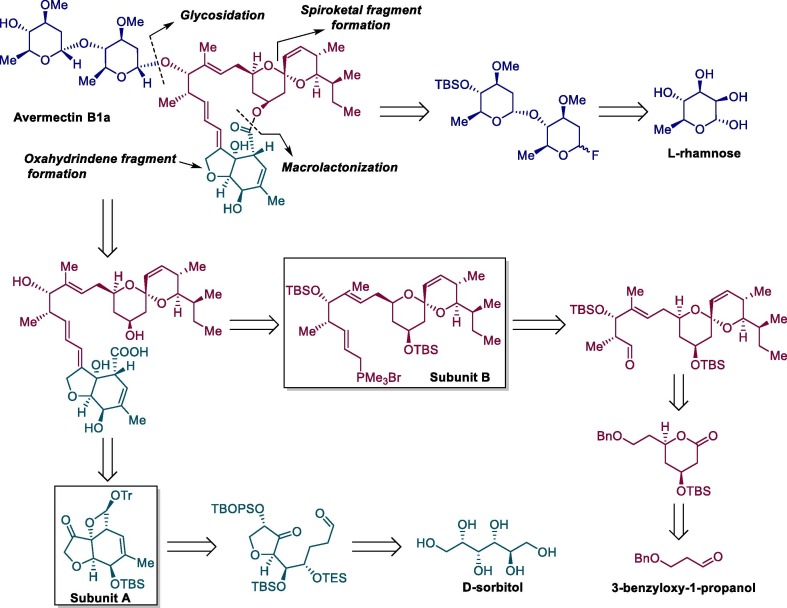
Because of the importance of avermectins, their complex biosynthetic pathway [154], [155], [156], [157] and total synthesis studies [158], [159], [160], [161], [162] have been extensively studied by different research groups. Scheme 17 shows the retrosynthetic analysis for ivermectin B1a from commercially available reagents, D-sorbitol, 3-benzyloxy-1-propanol and L-rhamnose [163].
Scheme 17.
Retrosynthetic design for ivermectin B1a.
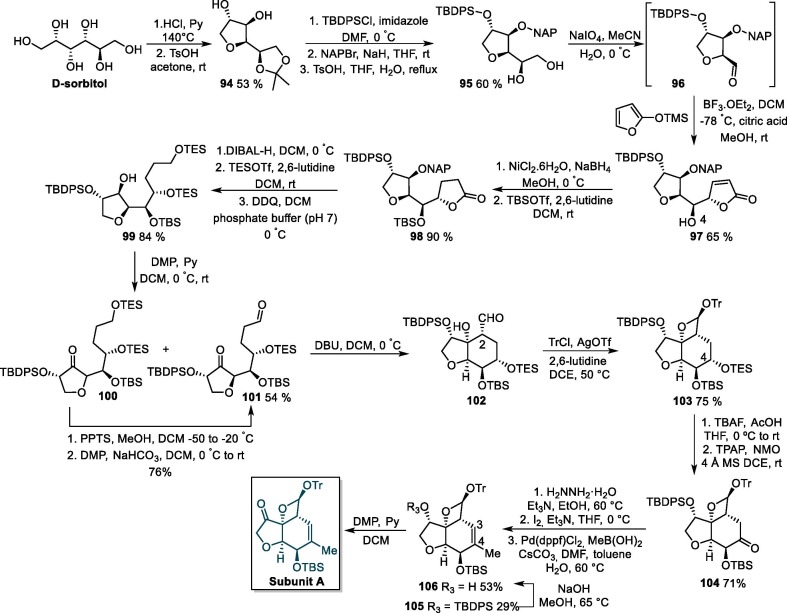
The synthesis of the subunit A was initiated via the acid-promoted cyclization of D-sorbitol followed by an acetonization reaction, giving acetonide derivative 94, which next undergoes a protection step with t-butyldiphenylchlorosilane (TBDPSCl) and 2-naphthylmethyl (NAP) bromide to produce 95 (Scheme 18 ). The selective hydrolysis and oxidative cleavage of 95 with sodium metaperiodate furnishes aldehyde 96, which is subsequently transformed into substituted lactone 97 by reacting with 2-(trimethylsiloxy)furan in the presence of BF3.OEt2. Then, the double bond in the furan-2(5H)-one ring of 97 is hydrogenated, and lactone derivative 98 is formed after a protection step. Next, 98 is submitted to a reduction/protection/NAP removal sequence to furnish 99, which is oxidized to bis-carbonyl compound 101 using the Dess-Martin periodinane in the presence of pyridine. Besides the major product 101, analog compound 100 is also formed in the aforementioned oxidation step, but can be promptly converted to 101 using conventional hydrolysis and oxidation reactions. The treatment of 101 with DBU generates cyclic compound 102, which is used in the next step without purification due to its instability on silica gel [164].
Scheme 18.
Synthetic route towards ivermectin B1a subunit A. TBDPSCl = t-butyldiphenylchlorosilane, NAPBr = 2-naphthylmethyl bromide, DMP = Dess-Martin periodinane, PPTS = pyridinium p-toluenesulfonate.
The reaction of 102 with trityl triflate (prepared in situ from trityl chloride and silver triflate) produces trityl oxetane acetal 103 as a single isomer, and the selective cleavage of the C-4 TES ether of 103 followed the ruthenium-catalyzed oxidation of the resulting alcohol affords ketone derivative 104. Next, a hydrazone formation/iodination/Suzuki-Miyaura coupling sequence leads to 105, which undergoes a selective removal of the TBDPS group to give compound 106. Finally, a Dess-Martin oxidation step gives rise to ivermectińs subunit A [164].
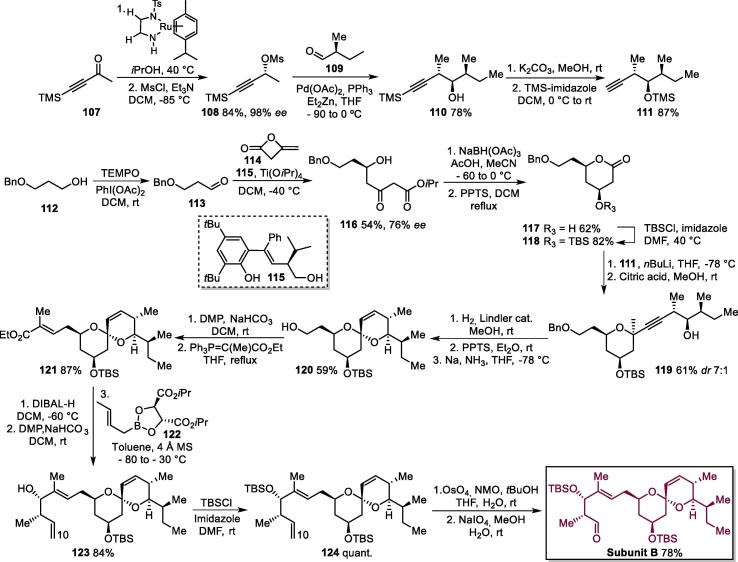
The next stage involved the preparation of subunit B (Scheme 19 ). Initially, the Noyori asymmetric hydrogenation of commercially available 4-(trimethylsilyl)-3-butyn-2-one 107 followed by mesylation gives 108, which is reacted with 109 in a Pd-catalyzed Marshall propargylation reaction to produce homopropargylic alcohol 110. Next, the removal of the trimethylsilyl group of terminal alkyne 110 in basic medium followed by the protection of the hydroxyl group affords intermediate 111 [164].
Scheme 19.
Synthetic route towards ivermectin B1a subunit B. PPTS = pyridinium p-toluenesulfonate. DMP = Dess-Martin Periodinane, NMO = N-methylmorpholine N-oxide.
In parallel, alcohol 112 is submitted to a TEMPO oxidation, leading to aldehyde 113, which is treated with diketene 114 in the presence of Ti(OiPr)4 and a catalytic amount of chiral Schiff base 115 to furnish δ-hydroxy-β-ketoester 116. The anti-selective reduction of 116 and acid-catalyzed lactonization furnishes 117, which undergoes a t-butyldimethylsilyl (TBS) protection step to produce 118. Next, alkyne 111 is treated with BuLi to produce the corresponding Li acetylide, which is reacted with lactone 118 to give methyl acetal derivative 119 by selective cleavage of TMS ether group with citric acid in methanol. Acetal 119 was transformed into spiroacetal compound 120 via a Lindler reduction/acid treatment/Birch reduction sequence, and the oxidation of 120 with the Dess-Martin periodinane followed by a reaction with ethyl 2-(triphenylphosphoranylidene) propionate gives 121. The compound 121 was converted into homoallyl alcohol 123 via DIBAL-H reduction/DMP oxidation/condensation with (E)-crotyl boronate 122. Next, alcohol 123 is converted to tert-butyldimethylsilyl ether 124, which undergoes dihydroxylation of its alkene moiety with osmium tetroxide (OsO4) followed by oxidative cleavage with sodium periodate NaIO4 to form subunit B [164].
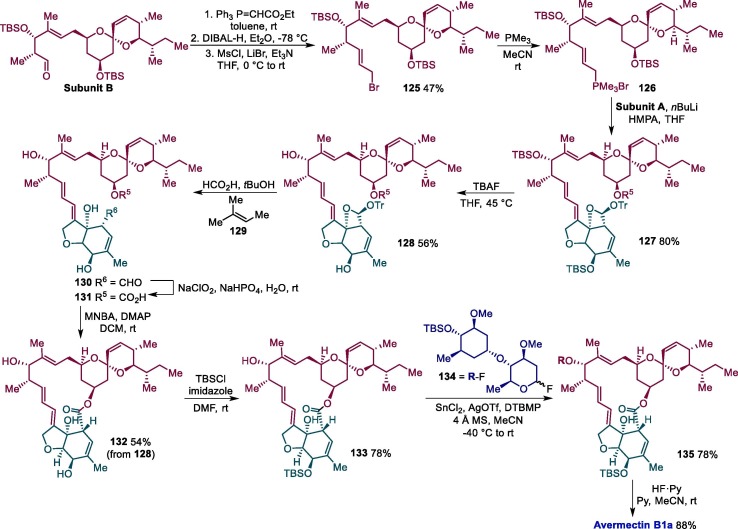
Subunit B is next submitted to a Horner-Wadsworth-Emmons condensation with (carbethoxymethylene)triphenylphosphorane followed by DIBAL-H reduction and conversion of the alcohol group to mesylate accompanied by the nucleophilic displacement of the mesylate group by LiBr, resulting in intermediate 125 (Scheme 20 ). The reaction of 125 with trimethylphosphine then leads to the formation of trimethyl phosphonium salt 126, with undergoes an E-selective Wittig reaction with subunit A, furnishing compound 127. Tetra-n-butylammonium fluoride (TBAF) is then used to deprotect silyl ether group in 127, leading to 128, which has the trityl group removed by formic acid to produce 130. A subsequent Krause-Pinnick oxidation gives compound 131, which undergoes an intramolecular cyclization promoted by 2-methyl-6-nitrobenzoic anhydride (MNBA), affording 132. The C5-hydroxy group of avermectin B1a aglycon 132 is next selectively protected with as a tert-butyldimethylsilyl ether, producing 133, and submitted to a stereoselective glycosylation reaction with 134 mediated by SnCl2-AgOTf to give the desired α-glycoside 135, which gives rise to avermectin B1a after the deprotection of silyl ether protecting group [164].
Scheme 20.
Final step in the total synthesis of avermectin B1a. HMPA = Hexamethylphosphoramide, TBAF = tetra-n-butylammonium fluoride, DTBMP = 2,6-di-tert-butyl-4-methylpyridine.
2.2.5. Isoquinoline alkaloids: Emetine
Emetine is an isoquinoline alkaloid isolated from the root of the Psychotria ipecacuanha, a medicinal plant of the Rubiaceae family. Emetine is a drug used both as an anti-protozoal approved for treatment of ameobiasis, and as an emetic agent. However, its potential cardiotoxicity has restricted its clinical use in recent years. The drug is administered intramuscularly or subcutaneously, but not intravenously, due to the potentialized toxicity resulting from the increased bioavailability.
Emetine inhibits protein synthesis in eukaryotic cells by irreversibly blocking ribosome movement along the mRNA strand, and inhibits DNA replication in the early S phase of the cell cycle. This is accomplished by binding to the 40S subunit of the ribosome. Also, emetine was identified as a specific inhibitor of HIF-2α protein stability and transcriptional activity [164], [165], [166]. The in vitro antiviral activity of emetine has been evaluated against SARS-CoV-2, and it was observed that this compound may effectively inhibit virus replication in Vero E6 cells with EC50 values under 100 μM. The therapeutic plasma concentration of emetine alone may reach 0.156 μM, which is below its EC50 against SARS-CoV-2, and the toxic plasma concentration is 0.5 mg/mL (1.04 μM). In order to reduce emetine effective concentration below the maximal therapeutic plasma concentration, the combination of remdesivir and emetine was explored, and a synergy effect was observed. Remarkably, remdesivir at 6.25 μM in combination with emetine at 0.195 μM may achieve a 64.9% inhibition in viral yield [167].
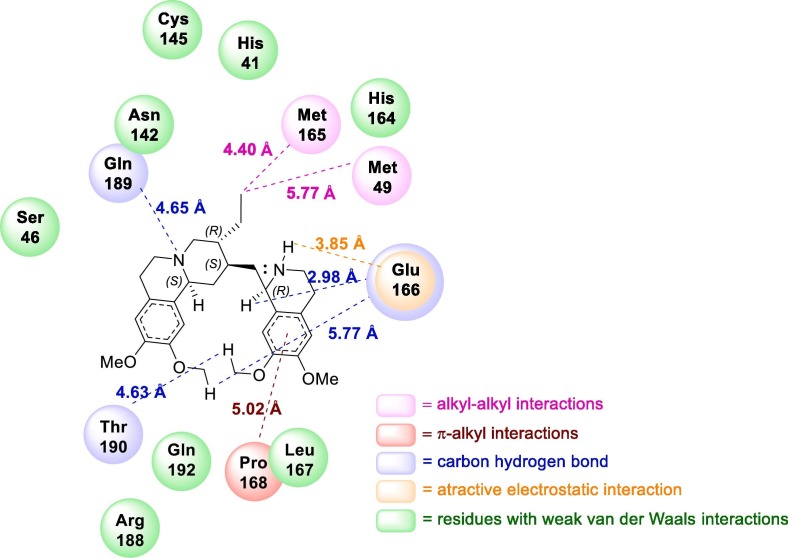
Docking studies led to the conclusion that emetine binds to the active site of Mpro, the SARS-CoV-2 main protease, through attractive electrostatic interaction with Glu166, alkyl hydrophobic interaction with Met49, and Met165, π-alkyl interaction with Pro168 (Fig. 13 ). The inhibition of this enzyme prevents the protein mutarotation of the virus, blocking the viral replication, and the further spread of the infection [167].
Fig. 13.
2D Representation of the interaction plots within the active site of SARS-CoV-2 MPro.
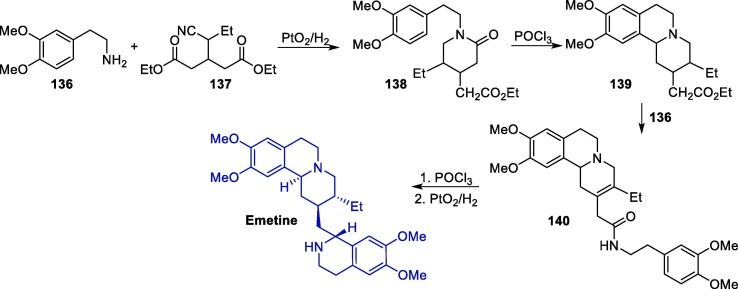
The synthesis of emetine has been reported in the literature since 1950 [168], [169], [170], [171], [172], [173], [174], [175], [176]. Various synthetic routes towards emetine have been reported, several of which using 2-(3,4-dimethoxyphenyl)ethylamine 136 as a starting material. An interesting route starts with the reaction between 136 and compound 137, resulting in piperidone intermediate 138, which undergoes an intramolecular cyclization in the presence of phosphorus oxychloride (POCl3), leading to isoquinoline intermediate 139 (Scheme 21 ) [177]. The subsequent reaction of 139 with 136 furnished 140, which is submitted to a second intramolecular cyclization step followed by hydrogenation to give rise to emetine.
Scheme 21.
Synthetic route towards emetine.
2.3. Anticancer drugs
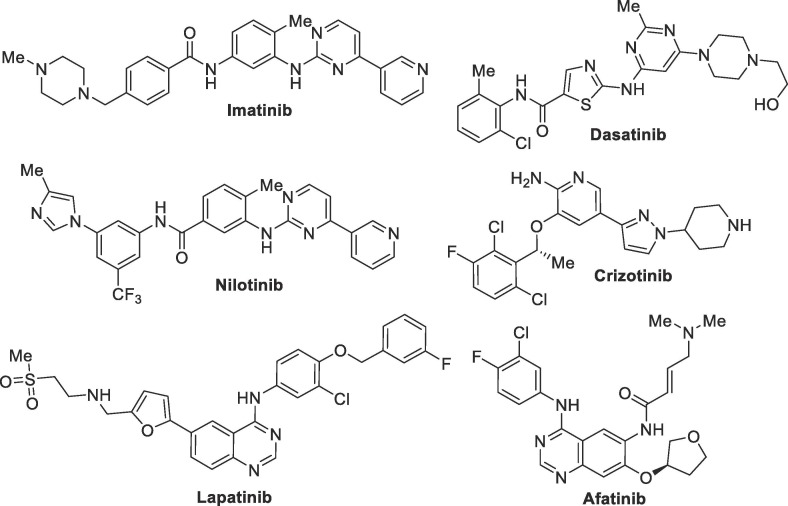
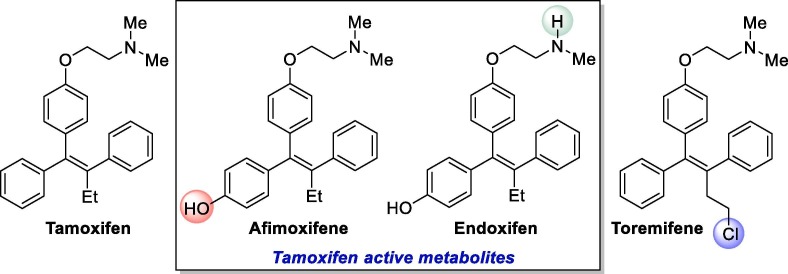
Besides antiviral and antiparasitic substances, some clinically developed anticancer drugs have also shown potential of being repurposed for the treatment of COVID-19. In this group are the nucleoside analogue gemcitabine, tyrosine kinase inhibitors imatinib and dasatinib, as well as estrogen inhibitor tamoxifen. Such drugs were evaluated in the past against several types of viruses, including SARS-CoV-1, which prompted the interest in their evaluation against SARS-CoV-2 as possible COVID-19 treatments.
2.3.1. Nucleoside analogues: Gemcitabine
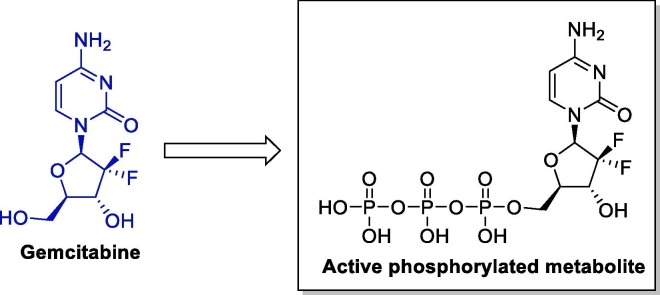
Gemcitabine (Gemzar®) is an anticancer drug, originally developed by the Eli Lilly Company back in the 1980 s. This compound is a fluorinated nucleoside analogue and is commercially available as an HCl salt. Interestingly, Gemcitabine is a pro-drug (Fig. 14 ) that undergoes intracellular phosphorylation to produce its active triphosphate form (Gemcitabine-RTP), which acts by inhibiting DNA synthesis by impairing ribonucleotide reductase, essential for de novo pyrimidine biosynthesis, leading to cell apoptosis. As a nucleotide analog, the incorporation of gemcitabine into DNA blocks the processing and chain elongation by the DNA polymerase complex, producing a cytostatic effect in the cell cycle. The gemcitabine triphosphate metabolite is also incorporated into RNA, thus inhibiting RNA synthesis [177], [178], [179].
Fig. 14.
Structure of gemcitabine and its active phosphorylated metabolite.
On the other hand, this drug has a short plasma half-life, which in some ways restricts its clinical success. Gemcitabine hydrochloride is intravenously administered, and its dosage ranges from 1000 to 1250 mg.m−2, depending on the type of cancer [180]. This drug has proven to be effective for several types or cancer, and its administration is adjusted depending on the type of cancer; in pancreatic cancer cases, gemcitabine is administered as a sole agent, while in non-small cell lung and bladder cancer, it is combined with cisplatin. Other examples include ovarian cancer, in which the drug it is given earlier than carboplatin, but in breast cancer, after paclitaxel.
In 2010, it was reported that the combination of decitabine with gemcitabine reduced HIV infectivity by 73% at low concentrations, with the antiviral activity of the combination being attributed to a rise in the HIV mutation rate [181]. In 2011, the in vivo antiretroviral activity of this drug was described using the mouse model for murine leukemia, targeting ribonucleotide reductase and causing suppression of the influenza virus RNA transcription and replication [182], [183]. Later in 2014, the evaluation of the in vitro activity of this chemotherapeutic drug, among others, against MERS-CoV and SARS-CoV-1 was reported [103]. Gemcitabine showed activity against both viruses, with EC50 values of 1.216 and 4.957 µM for MERS-CoV and SARS-CoV-1, respectively. On the other hand, a preliminary recent study has revealed that gemtabine showed no in vitro activity against SARS-CoV-2 [106]. Recently, studies have shown that gemcitabine exhibited in vitro activity against SARS-CoV-2, with EC50 values of 1.24 µM [184]. Additionally, the combination of gemcitabine with oxysophoridine showed a synergic effect against SARS-CoV-2.
The vast applications of gemcitabine as a potent chemotherapy and antiviral agent have been attracting growing attention in the last decades, and therefore, several synthetic pathways towards this compound have been developed [185]. The first synthetic procedure towards gemcitabine was established by the Lilly research laboratories back in the 1980 s and was published in 1988 (Scheme 22 ) [186].
Scheme 22.
Synthetic route towards chemotherapic drug gemcitabine. TBDMSOTf = tert-butyldimethylsilyl trifluoromethanesulfonate.
The initial step in this route is the Reformatsky reaction between (R)-glyceraldehyde 141 and ethyl bromodifluoroacetate 142 under typical conditions (zinc in THF/diethyl ether medium), giving intermediate 143 as a diastereoisomeric mixture which, after chromatographic separation, leads to the anti-adduct in a 65% yield (Scheme 22). Then, a deprotection/intramolecular cyclization sequence promoted by Dowex 50 furnishes lactone 144, which next has the alcohol groups protected as TBDMS ethers, giving 145, and is subsequently reduced with DIBAL-H to furnish difluororibose intermediate 146. Next, the hydroxyl group at the anomeric carbon of 146 is converted to a better leaving group upon mesylation, which results in intermediate 147. Finally, the reaction of 147 with 148 in the presence of TMSOTf followed by a deprotection step and chromatographic separation gives rise to gemcitabine.
The low yields and diastereoisomeric ratio observed in the original gemcitabine synthesis led to the search for more efficient steps, not only in what regards purification methods, but also synthetic routes. In order to improve this transformation, the most explored approach is the development of new synthetic methods for the construction of the difluorinated ribose sugar, especially for the introduction of the fluorine groups [186].
2.3.2. Pyrimidine derivatives: Imatinib and dasatinib
The protein tyrosine kinase (PTK) is a group of proteins that promotes the phosphorylation of proteins by catalyzing the transfer of ATP phosphate groups to their tyrosine residues, thereby transferring signals to regulate cell growth, differentiation and death, as well as a series of other important biochemical processes. In this sense, PTK disorders may lead to a series of diseases, including tumor invasion, metastasis and neovascularization, and also tumor chemotherapy resistance [187]. Therefore, in the last decades PTK has been regarded as an important target for the research and development of antitumor drugs, even being included in the so-called targeted cancer therapy.
The first tyrosine kinase inhibitor (TKI), imatinib, was approved by the FDA in 2001, and paved the path for the development of highly selective and effective drugs for the treatment of chronic myeloid leukemia (CML), non-small cell lung cancer and renal cell carcinoma, presenting relatively low side effects [188]. Indeed, some of these drugs have become the first-line for the treatment of several types of cancer, and to date more than 20 kinds of TKI have been approved by the FDA [189]. Fig. 15 shows the structure of some FDA-approved TKI drugs.
Fig. 15.
Chemical structure of FDA-approved tyrosine kinase inhibitors.
Imatinib (Gleevec® or Glivec®) is administered as a mesylate salt, and is currently used for the treatment of newly diagnosed chronic myeloid leukemia patients, and also those have failed interferon-α therapy, inducing major cytogenetic responses in 80–90% of the former, and 65–90% of the latter [189], [190], [191].
The first synthetic procedure towards imatinib was reported back in 1996 (Scheme 23 ) [192], [193], [194]. In this approach, 2-amino-4-nitrotoluene 149 is reacted with cyanamide in the presence of nitric acid to form guanidine nitrate intermediate 150, which is next condensed with α,β-unsaturated carbonyl compound 151 in basic medium, leading to nitro pyrimidine intermediate 152. Next, 152 undergoes reduction using palladium on carbon as catalyst, yielding 153, which is finally reacted with 4-(4-methyl-piperazinomethyl)-benzoyl chloride 154 in the presence of pyridine to give imatinib. Unfortunately, the yields of the several steps were not mentioned in neither patents nor the subsequent paper, but some authors have later reported low yields for this process, which led to the development of new and more efficient processes [195], [196], [197], [198].
Scheme 23.
Synthetic route towards the tyrosine kinase inhibitor imatinib.
Although imatinib is effective for most chronic patients, some become resistant or intolerant to this drug in the chronic or advanced phases of the disease, which led to the development of more potent TKIs such as dasatinib [199], [200]. The crystalline monohydrate form of dasatinib was approved by the FDA in 2006, and it is currently commercialized under the brand name Sprycel®, being the first-line drug for the treatment of chronic, accelerated, myeloid or lymphoid blast phase of Philadelphia chromosomal translocation chronical myeloid leukemia in cases of resistance or intolerance to imatinib [201].
The first synthetic route towards dasatinib was published in 2006 (Scheme 24 ) [202]. Initially, 2-chlorothiazole 155 undergoes a sulphur-directed ortho-lithiation and subsequent nucleophilic reaction with 1-chloro-2-isocyanato-3-methylbenzene 156, yielding thiazole-functionalized carboxamide 157. Next, the NH group of the amide moiety in 157 is protected after reacting with 4-methoxybenzyl chloride 158, giving 159, which then undergoes a sodium hydride-promoted coupling reaction with 6-chloro-2-methylpyrimidin-4-amine 160, leading to intermediate 161. The deprotection of the NH group in 161 followed by the reaction with 2-(piperazin-1-yl)ethan-1-ol 163 and reaction with hydrochloric acid furnishes dasatinib hydrochloride salt with an overall yield of 61% after six steps.
Scheme 24.
Synthetic route towards the tyrosine kinase inhibitor dasatinib.
In a work published in 2014, which evaluated the activity of a series of clinically developed drugs against MERS-CoV and SARS-CoV in vivo and in vitro, not only imatinib but also dasatinib showed activity against both viruses [103]. Imatinib mesylate inhibited the activity of MERS-CoV and SARS-CoV with an EC50 of 17.689 and 9.823 µM, respectively, while these values for dasatinib were significantly lower (5.468 and 2.100 µM, respectively). Although much more research is still required, such results indicate the potential of these TKI drugs for treating coronavirus infections.
Later, further studies were conducted in order to determine when in the life cycle imatinib was inhibiting the replication of SARS-CoV [203]. For that, imatinib was administered in the first 4 h and also after 5 h to Vero E6 cells and Calu-3 cells infected with the virus. The authors found that when imatinib was added in the first 4 h of infection, the virus production was significantly inhibited in a dose-dependent manner, but when added 5 h after infection, the SARS-CoV-1 levels were significantly higher when compared to the drug added before infection. In this sense, it was inferred that imatinib inhibits a step in the replication cycle before RNA production.
In a study that was conducted by the same group involving the evaluation of the activity of several FDA-approved drugs against SARS-CoV-2, it was shown that imatinib mesylate was able to inhibit this virus with an IC50 value of 3.24 μM. Additionally, a recent in silico study has shown that the use of losartan and imatinib, in combination, may be effective for the treatment of acute respiratory distress syndrome in patients infected with SARS-CoV-2 [204]. It is proposed that losartan acts by blocking AT1 receptors, antagonizing the effect of angiotensin II, while imatinib may regulate the local immunological responses.
Also, a recently published case report on a CML patient suggested that TKI therapy may protect patients who are infected with SARS-CoV-2 [205]. Currently, a clinical trial is ongoing in the Netherlands to evaluate imatinib in the prevention of pulmonary vascular leak in COVID-19 patients [206].
2.3.3. Triphenyl olefins: Tamoxifen and toremifene
Selective Estrogen Receptor Modulators (SERMs) are estrogen inhibitors that display pronounced biological activities regarding the prevention and/or treatment of disorders like breast cancer and osteoporosis [207]. SERMs act selectively on estrogen target tissues through the ER system, and are expected to act as agonist in some tissues such as bone, liver and the cardiovascular system, an antagonist in breast and brain, and a mixed agonist/antagonist on uterus. In this group, an important drug is tamoxifen, which usually is commercialized in the citrate form and can be found under a variety of commercial names (Nolvadex®, Genox®, Tamifen® and others). Interestingly, tamoxifen itself presents a relatively low affinity for the ERs, but acts as a pro-drug, being afimoxifene (4-hydroxytamoxifen) and endoxifen (N-desmethyl-4-hydroxytamoxifen) the active metabolites (Fig. 16 ). Importantly, the E/Z tamoxifen isomers present divergent activity, with the Z-isomer being antiestrogenic, while the E-isomer is a potent estrogen agonist; a similar trend is observed for the tamoxifen metabolites [208].
Fig. 16.
Structure of the prodrug tamoxifen and its main active metabolites, afimoxifene and endoxifen.
Another widely used antiestrogen compound is toremifene, a chlorinated derivative of tamoxifen that has been extensively used in the treatment of breast cancer. Importantly, several clinical trials have investigated the effectiveness of toremifene as a first-line endocrine therapy for postmenopausal women with advanced breast cancer, and concluded that this medication has a comparable antitumor activity to tamoxifen. However, contrary to tamoxifen, toremifene has not presented any carcinogenic potential to the liver of female rats even when administered at high dosages, what is related to a lower potential of forming DNA reactive intermediates [209].
As reported in 2014, tamoxifen citrate presents activity against both MERS-CoV and SARS-CoV-1, with EC50 values of 10.117 and 92.886 µM, respectively [103]. A recent study has shown that tamoxifen is effective in the inhibition of SARS-CoV-2, albeit with a significantly higher IC50 value than that of imatinib mesylate (34.12 μM vs 3.24 μM for imatinib) [106].
Back in 2013, an in vitro study that screened several FDA-approved compounds found that toremifene acts a potent inhibitor of EBOV infection [210]. Both Vero E6 and Hep G2 cells were used in this study, and toremifene citrate was able to inhibit the activity of EBOV in both cell lines with very promising IC50 values (0.162 and 0.0255 µM for Vero E6 and Hep G2, respectively). In vivo studies were also conducted in this work, and have shown that the anti-EBOV activity took place even in the absence of detectable estrogen receptor expression. It was concluded that toremifene inhibits virus entry after internalization, which seems to be an off-target effect where there is an interference in a late step in viral entry, likely affecting the triggering of fusion. The antiviral properties of toremifene against EBOV were further studied, and it was found that this compound interacts with and destabilizes the Ebola virus glycoprotein (GP) [211]. In this study, the binding site of toremifene was determined to be on the surface of the GP, which generates a decrease in the stability of the viral GP, preventing viral fusion. Toremifene has also shown in vitro activity against MERS-CoV and SARS-CoV-1, having presented EC50 values of 12.915 and 11.969 µM, respectively [103].
In this sense, the pronounced antiviral activity of toremifene has led this compound to be included in several studies that aimed to evaluate its possible effect against SARS-CoV-2. A recent in vitro study conducted with Vero cells infected with SARS-CoV-2 showed that this compound is a promising inhibitor for the new coronavirus, having presented an IC50 of 3.58 µM (CC50 = 10.11 µM, SI = 2.82) [130]. Importantly, similar results were observed by other research groups, which have determined a IC50 of 4.77 µM (CC50 = 20.51 µM, SI = 4.3) for toremifene against SARS-CoV-2 [106].
Recently, in silico studies have indicated that toremifene may act by inhibiting the SARS-CoV-2 spike glycoprotein, which is responsible for facilitating the fusion of the viral and cell membranes. It was proposed that an interaction between the dimethylamine moiety of toremifene and residues Q954 and N955 in heptad repeat 1 (HR1) takes place, perturbing the structure. Besides that, the authors also found that toremifene and the methyltransferase non-structural protein (NSP) 14 interact strongly, and that could inhibit the viral replication [212].
Due to its marked anticancer activity, several synthetic routes towards tamoxifen have been developed in the last decades. One of the seminal reports was made in 1966, which used a triarylethylene as intermediate to obtain the target compound in the form of a mixture of diastereoisomers [213]. Later in the same year, another group reported an approach for the synthesis of tamoxifen from the reaction between phenylmagnesium bromide 165 and compound 164, giving carbinol 166 as intermediate, which is then dehydrated to furnish a mixture of E/Z diastereoisomers as products, and those can be separated using recrystallization techniques (Scheme 25 a) [214]. Other authors have also given their contributions to this field, having developed another viable method to produce carbinol 166, in this case using organomagnesium reagent 168 and 1,2-diphenyl-1-butanone 167 (Scheme 25 b) [215].
Scheme 25.
Synthetic approaches towards tamoxifen, an estrogen inhibitor.
Besides with the route involving a carbinol intermediate showed in Scheme 4, several other approaches have been developed for the synthesis of tamoxifen, e.g. the use of other organometallic nucleophiles, McMurry reductive coupling, as well as carbometallation, multicomponent, aldol and Wittig-Horner reactions [216].
As for the synthesis of toremifene, the first approach was reported back in 1983 (Scheme 26 ) [217]. Initially, substituted benzophenone 169 is reacted with cinnamaldehyde 170 in the presence of lithium aluminum hydride, leading to triphenyl compound 171. Finally, the dehydration of 171 gives rise to alkene 172, which affords toremifene after a chlorination step.
Scheme 26.
Seminal synthetic approach towards toremifene.
Later in 2014, an alternative method employing milder reaction conditions for the synthesis of toremifene was reported (Scheme 27 ) [218]. In this case, 4-hydroxybenzaldehyde 173 is initially reacted with compound 174 to produce 175 in two steps. Then compound 175 is alkylated with 2-chloro-N,N-dimethylethanamine 176, giving 172, which undergoes chlorination to give toremifene in a Z:E ratio greater than 5:1.
Scheme 27.
Alternative synthetic route towards toremifene.
2.4. Antihypertensive drugs: Losartan, an imidazole derivative
Losartan was developed in 1986 by Merck & Co. and approved by the FDA in 1995 as an angiotensin II receptor blocker (ARB) antihypertensive drug. It consists of a non-peptide derivative capable of interacting with AT1 receptors in order to antagonize the action of angiotensin II. Its structure was rationally planned based on that of compounds S8307 and S8308, both previously discovered sartan analogues which presented selective and competitive antagonistic action for AT1 receptors but low potency, a short action time and low oral bioavailability (Fig. 17 ). Subsequent structural comparison studies with angiotensin II, the natural substrate for AT1, led to important rationalizations regarding their pharmacodynamic interactions. An important observation was that angiotensin II has two carboxylate groups positioned around the N-terminal portion, both directly involved in the interaction with the receptor. Thus, the presence of a second acidic group in the inhibitor structure could provide increased activity. Derivative EXP-6155 was developed later and presented an activity about 10 times higher, but still with little absorption by the gastrointestinal tract. Subsequent optimizations led to the biphenyl derivative EXP-7711, which displays a higher lipophilicity and consequent good oral bioavailability. The bioisosteric replacement of the carboxyl group with the tetrazole moiety led to losartan. Even though this compound is not actually a prodrug since it displays a potent antihypertensive effect, it is also partially converted into metabolite EXP-3174, also an AT1 antagonist even more potent than the original molecule, which contributes to increase the potency observed for this drug [219], [220], [221].
Fig. 17.
First non-peptide AT1 antagonists, including losartan and its active metabolite.
The acidic group position in the biphenyl system is crucial for biological activity. In the case of losartan, the ortho position of the tetrazole cycle is essential for pharmacodynamic interactions and for the best oral potency of the medication. Although the carboxylic isomers are active intravenously, the tetrazole system leads to a higher binding affinity with the receptor, probably by inducing the bioactive conformation on the biphenyl so that it can fit into the receptor binding site. At the same time, this group establishes an ionic interaction with a positively charged amino acid residue. In the imidazole cycle, the saturated 4-carbon side chain at C-2 is necessary for the spatially favorable hydrophobic interaction with the non-polar aminoacid residues in the receptor. Bulky, electronegative and lipophilic substituents, e.g. chlorine, at C-4 favor interactions, and the presence of hydrogen-bond a acceptor is required at C-5 (Fig. 18 ) [220].
Fig. 18.
Model representation of losartan interaction with the AT1 receptor.
Back in 2005, in vivo studies identified that there is a molecular connection between SARS pathogenesis and the renin-angiotensin system in lung failure, and that the modulation of this system could be used to protect individuals with SARS and other related viruses [222]. It is also speculated that treatment with ARB such as losartan and valsartan may modify the physiological levels of angiotensin II, leading to the production of Ang1-7, with maintenance of vasodilation and hypertensive regulation, improving pulmonary function, thus being a more favorable strategy than the use of ACE inhibitors, such as captopril or enalapril [223].
Due to concerns about the continued use of ACE/ACE2 inhibitors and ARB drugs, a study that aimed to assess whether the use of these drugs may be associated with an increased severity of the symptoms in patients with COVID-19 was conducted [224]. The study, which evaluated 511 COVID-19 patients with previous known hypertension, showed that in elderly patients (above 65 years of age) those who used ARB before hospitalization were associated with positive effects related to pneumonia-related morbidity and mortality, being less likely to develop severe lung disease [225]. These data were also corroborated by other published studies.
A recent in silico study has concluded that losartan and the anticancer drug imatinib could be efficient drugs to treat ARDS (acute respiratory distress syndrome) in COVID-19 patients. The drugs would act by occupying and distorting the binding of the RBD (receptor-binding domain) of the viral S protein with ACE2 by changing the conformational shape of the N-terminal α-helix where the RBD binds. In this sense, it was suggested that low doses of losartan and imatinib could be clinically studied to treat ARDS in patients with COVID-19 [205].
Since the discover of losartan, several research groups have explored alternative synthetic routes for its preparation. In 1991, when losartan had already been discovered but not yet been approved for clinical use in the USA, the synthesis of this molecule along with a series of imidazole analogues with antihypertensive activity and good oral bioavailability was described (Scheme 28 ). The method, however, requires a number of sequential reaction steps and afforded a 27% overall yield. Initially, oxazolidine 177 reacts with 4-toluylmagnesium bromide 178 for the construction of the biphenyl system, giving intermediate 179, which has its oxazolidine moiety converted into a cyano group through treatment with phosphoryl chloride in the presence of pyridine, leading to 180. Next, the cyano group in 180 is converted into a tetrazole ring via reaction with sodium azide in the presence of tributyltin chloride and is sequentially protected with a triphenylmethyl group, which leads to the formation of 181. This intermediate, in turn, suffers benzylic bromination in the presence of N-bromosuccinimide (NBS) and dibenzoylperoxide as initiator, affording 182, which is next submitted to a nucleophilic displacement reaction with imidazole 183 as a nucleophile and reduction of the aldehyde carbonyl with sodium borohydride to produce alcohol 184. A final of tetrazole deprotection step via treatment with diluted hydrochloric acid furnishes losartan [220].
Scheme 28.
Synthetic approach towards losartan.
Later, more simple and efficient method for the synthesis of this compound were described, for instance involving C—C coupling reactions, C—H activation, denitrogenative ortho-arylation reaction of benzotriazonones [226], [227], [228], [229].
2.5. Immunosuppressants: Cyclosporin, a macrocyclic polypeptide
Discussions on the role of an exacerbated innate immune response in the development of respiratory symptoms in patients with SARS-CoV-1 and, as recently observed, with SARS-CoV-2, raised the hypothesis that the use of immunosuppressants could be strategic for the treatment of these patients. This effect probably stems from the ability of these viruses to invade specific cells of the immune system, such as macrophages, modulating the host immune responses [230].
In order to better understand this process, some features related to the interactions between the virus and the host cell, such as is the central role that mitochondrial metabolism plays in the antiviral response at the cellular level, must be considered. Once the viral RNA is detected in the cytosol of the infected cell, mitochondrial antiviral proteins (MAVS) are activated and trigger the phosphorylation of transcription factors and cytokine expression genes, essential for the immune response [231]. In addition, the increased cell’s energy demand can lead to the failure of mitochondrial functions, which, in turn, triggers the activation of caspases, leading to irreversible cellular damage, being possibly one of the main pathological mechanisms for more severe forms of COVID-19 [231].
In this context, calcineurin inhibitors such as cyclosporine A may provide important protection against these processes. Neoral® and Gengraf® are some of the brand names for cyclosporin A (CysA), which acts by reversing the protein denaturation triggered in the cytosol by the MAVS, rehabilitating the cell at the same time that they act on cyclophilins in other cellular processes, also preventing mitochondrial collapse. These same mechanisms also justify the cardioprotective effect associated with the use of this medication in patients with myocardial infarction [231].
CysA also has shown significant antiviral activities, especially against RNA viruses such as betacoronaviruses [231]. The antiviral activity observed in these cases results from its interactions with cellular cyclophilins, inhibiting their isomerase function, which the virus modulates to produce nucleocapsid proteins necessary for genomic encapsulation and viral replication [231].
Cyclophyllin is one of the identified cellular components capable of interacting with the viral Nsp1 protein in the step sequence that leads to viral genome replication. It is an enzyme capable of catalyzing s-cis/s-trans isomerization in prolyl-peptide bonds so that the proteins cleavage is possible. CypA is the human cyclophyllin of greatest biological importance, being associated with these isomerization processes necessary for the biosynthesis of cellular proteins [232]. This enzyme is selectively inhibited by cyclosporine A, affecting the host cell protein biosynthesis itself, but also of the various viral proteins necessary for its replication. At the same time, it is precisely the formation of the CysA/CypA complex that leads to immunosuppression, preventing the activation of the transcriptional nuclear regulatory factor of active T cells (NFAT). Thus, it is estimated that CypA is an important pharmacological target to be considered in the treatment of the so-called coronavirus infections, possibly including COVID-19 [232], [232]. Indeed, a recent study evaluated the in vitro activity of a series of FDA-approved compounds against SARS-CoV-2 and it was observed that cyclosporine A showed potential antivirus activity with an IC50 of 5.82 µM (CC50 > 50 and SI 8.59) [130].
Cyclosporin A is a cyclic polypeptide isolated from the fungus Tolypocladium inflatum and comprises eleven aminoacid residues [233]. Its good oral bioavailability and metabolic resistance towards physiological hydrolases are directly associated with the presence of the N-methyl groups present in seven of the eleven amide nitrogen atoms present in the structure. Its pharmacodynamic effect is directly associated to the presence of the unusual (4R)-4-((E)-2-butenyl)-4-methyl-L-threonine residue (MeBmt) and with the primary sequence MeBmt-Abu-Sar-MeLeu, both highlighted in red and in bold, respectively, in Fig. 19 [234].
Fig. 19.
Different representations for the structure of cycloporin A. (a) Angular chemical representation. (b) Amino acid residues sequence associated with the drug's pharmacodynamic interaction. (c) Peptide primary structure representation. d) Linear representation highlighting the intramolecular hydrogen bonds. MeBmt = (4R)-4-((E)-2-butenyl)-4-methyl-L-threonine residue; Abu = α-aminobutyric acid residue; Sar = sarcosine residue; MeLeu = N-methylleucine residue; MeVal = N-methylvaline residue; Ala = alanine residue; Val = valine residue; - - - - = Hydrogen bonds.
Cyclosporin A’s total synthesis was first published in 1984 after a series of scientists devoted efforts towards the synthesis of the unusual aminoacids present in its structure [235]. This allowed later structural changes to be explored to establish relationships between the structure and the bioactivity [236]. Since the seminal work published in 1984 [236], a few research groups have been able to describe complete synthetic alternatives for the construction of this molecule. In this context, it is worth highlighting the efforts of some scientists in view of the partial synthesis of specific structural fragments and the unusual MeBmt amino acid, an important feature in this structure [237], [238].
The most recent example of a Ciclosporin A total synthesis dates back to 2010. The authors explored the unique reactivity of organic isonitriles, which were used as precursors for the tertiary amide groups present in CysA structure. In the synthesis design, the target structure was fragmented into three structural units that were explored separately and condensed at the end (Scheme 29 ) [235].
Scheme 29.
Partial retrosynthetic rationalization toward cyclosporin A total synthesis.
Fragment 185, corresponding to the acetonide MeBmt residue, had already had its synthesis reported by other researchers [238], [239]. The preparation of the tetrapeptide fragment 186 was conducted with the convergent synthesis of the two dipeptide units, 191 and 194. Both units could be obtained through the reaction between isonitriles and appropriate thioacids, followed by selective functional groups deprotection and peptide condensation steps (Scheme 30 ).
Scheme 30.
Synthetic route towards tetrapeptide fragment 186. AIBN = azobisisobutyronitrile, DETBT = 3-(diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one.
The hexapeptide fragment 183 had its synthesis started through reaction between azide 195 and isonitrile 196 under microwave irradiation, leading to the N-formylamide intermediate 197; the N-formyl group was inserted with the intention of avoiding unwanted side epimerization reactions in the following steps (Scheme 31 ).
Scheme 31.
Synthetic strategy towards the hexapeptide fragment 187. HATU = 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate.
The C-terminal portion was then expanded by deprotecting the carboxylic acid in 197 followed by condensation with the aminoacid 198, giving 199. Subsequently, the mentioned N-formyl group had to be chemoselectively reduced with the aim of maintaining the integrity of the peptide structure, leading to 200, which underwent a reduction of the azide group with triphenylphosphine, giving 201, and subsequent condensation with the protected amino acid 202 led to derivative 203. In the next stage, thioacid 205 was condensed with aminoacid 204 through a cyclohexylisonitrile-mediated reaction, giving 206. After the C-terminal portion debenzylation, which furnished 207, Lawesson's reagent was applied under microwave irradiation in order to provide dipeptide thioacid 208. Fragment 187 was prepared by coupling thioacid 208 with the previously synthesized tetrapeptide 203, followed by a protonation step on the N-terminal portion with trifluoroacetic acid.
Next, fragments 187 and 185 were coupled using dicyclohexylcarbodiimide (DCC) and hydroxybenzotriazole (HOBt) in N-methylmorpholine (NMM) as solvent, leading to the heptapeptide 209 in 75% yield (Scheme 32 ). Finally, the tetrapeptide fragment 209 was subjected to hydrogenolysis to deprotect its C-terminal portion and, subsequently, condensation with the heptapeptide 209 in the presence of benzotriazol-1-yl-oxitripyrrolidinophosphonium hexafluorophosphate (PyBOP), giving 210.
Scheme 32.
Final step in the total synthesis of cyclosporin A.
After sequential N- and C-terminal deprotections, the linear undecapeptide 210 was then subjected to macrolactamization under microwave irradiation in the presence of HOBt and cyclohexylisonitrile. Curiously, no linear polymerization products were observed, leading to the formation of the cyclosporine macrocycle with 54% yield.
In this example, the distinct reactivity of isocyanides and thioacids was explored for the construction of a macrocyclic polypeptide of complex structure and great medicinal interest. All the performed reactions afforded good yields and, although being a long synthetic route, it certainly represents an interesting alternative to the preceding works.
2.6. Antipsychotics and antihistamines
2.6.1. Phenothiazine derivatives: Chlorpromazine, fluphenazine and promethazine
Phenothiazines are pioneering medications developed in the 1950s for the treatment of behavioral disorders (Fig. 20 ) [239]. Although phenothiazines act primarily by blocking dopamine receptors, they also have anticholinergic, antihistamine, and antiemetic effects. Chlorpromazine and fluphenazine are commonly used antipsychotic drugs, while promethazine, even though presenting a weak antipsychotic activity, is usually prescribed as an antihistamine and a sedative in many countries.
Fig. 20.
Structure of promethazine, fluphenazine and chlorpromazine.
Chlorpromazine (CPZ) (Largactil®, Thorazine® and others) was developed in the early 1950s, being a pioneer antipsychotic drug prescribed for patients with psychiatric disorders such as schizophrenia. When compared to the treatments available until its discovery, this substance revolutionized what would be called neuropharmacology due to its remarkable effectiveness in controlling arousal and agitation, as well as in relieving psychotic symptoms. Its clinical success has led to new studies for the development of new antipsychotics [240]. An example of an antipsychotic developed from CPZ is fluphenazine (FPZ) (Prolixin®), which features a piperazine-derived substituent and is used in the treatment of chronic psychoses, appearing to be virtually equal in effectiveness to chlorpromazine [241], [242], [243].
Promethazine (PMZ), widely known by its brand name Phenergan®, was also designed as an antipsychotic drug from structural modifications using CPZ as a prototype. The difference in its N-alkyl chain, however, led to a structure with less affinity for dopaminergic receptors and greater affinity for histamine receptors, with a H1-antagonist effect, making its clinical use completely redirected from the originally intended application [244], [245], [246].
Recently, it was observed that there was a lower prevalence of more severe forms of COVID-19 in psychiatric patients using antipsychotic medication. From this observation, researchers raised the hypothesis that the psychotropic drugs already used by these patients could lead to protective effects with regard to symptom evolution. In a recent publication, the authors presented findings that suggest that CPZ can be applied in the treatment of COVID-19 patients after a phase II single blind randomized controlled therapeutic trial evaluation [247].
CPZ, FPZ and PMZ have already shown to present antiviral activity, including against MERS-CoV and SARS-CoV-1, although their antiviral clinical application has not yet been explored [103], [248], [248]. Recently, a study evaluating the in vitro inhibitory ability of CPZ against SARS-CoV-2 replication in human cells proved the anti-SARS-CoV-2 activity of this substance with an IC50 of approximately 10 μM, further corroborating the proposal to reposition this drug for the treatment of COVID-19 [249].
The antiviral effect associated with these drugs is a consequence of the inhibition of clathrin-mediated endocytosis [250], [251]. In summary, endocytosis is a cellular process through which extracellular components are internalized in the cell via the invagination of the membrane phospholipid bilayer followed by the formation of specific vesicles called endosomes. Such structures can be directed to excretory pathways to externalize their content, or they can, at a given moment, fuse with lysosomes containing enzymes and specific conditions for their degradation for cellular use. The main function of the endocytic mechanism is the recycling or removal of protein components, but it plays an important role in the establishment of several diseases of viral origin [251]. To understand the importance of endocytosis in viral infection, it is necessary to understand some of the morphological characteristics associated with coronaviruses. These are plus-stranded RNA viruses surrounded by four specific proteins with the function of encapsulating the viral genome, namely, the nucleocapsid protein (N), the membrane glycoprotein (M), the envelope protein (E) and the spike glycoprotein (S). The latter is directly involved in the internalization of the virus by the cell, for which it is divided into two fragments conventionally called S1 and S2, the first responsible for the interaction and binding with the receptor and the second for membrane fusion. Studies indicate that, after cell infection, the virus tends to accumulate in lysosomal structures, typically associated with the endocytic process. In this context, clathrin-dependent endocytosis and cathepsin-mediated protein S cleavage are two steps directly involved in the establishment of viral infection [251].
In this sense, the endocytic pathway has been pointed as an important pharmacological target to be considered in the identification of new drugs for the treatment of COVID-19. Indeed, both PMZ and FPZ presented a moderate MERS-CoV S-protein mediated cell–cell fusion inhibiting activity, with EC50 values of about 17 and 15 μM, respectively [252]. CPZ also inhibited MERS-CoV replication at both early and post-entry stages with an EC50 of about 5 μM and a CC50 of 21 μM [253]. It is important to note, however, that, to date, no study has been published directly relating SARS-CoV-2 infection to the endocytic pathway. Nevertheless, due to the similarities already identified with SARS-CoV-1, it is coherent to suggest that this process is also present in the mechanism of SARS-CoV-2 infection [251], [252].
Complementarily, CPZ's biodistribution profile may represent a major advantage in the treatment of acute respiratory syndrome associated with SARS-CoV-2 infection, since its concentration in the lungs and saliva tends to be higher than in blood plasma, increasing its potential for both treatment of COVID-19 and its prevention, respectively. Furthermore, the fact that it is easily able to cross the blood–brain barrier could, in theory, prevent the development of neuropathological forms of COVID-19 [131], [248].
Regarding the pharmacodynamic aspects of the mechanism of phenothiazine drugs clathrin-mediated endocytosis inhibition, some observations can be highlighted. An interesting study described the use of molecular dynamics simulations to explore the interactions between CPZ and phospholipid monolayers, focusing on the effect of drug ionization in this interaction. The study focused on the Langmuir monolayer models, simulating dipalmotoylphosphatidylcholine (DPPC) and dipalmitoylphosphatidylglycerol (DPPG) as zwitterionic and anionic components, respectively [254]. The authors identified a distribution in the electrically neutral form of CPZ within the non-polar phospholipid part, while the cationized form, prevalent in physiological pH, is located at the lipid-water interface. The ammonium group present in protonated CPZ, in this case, seems to be important for the establishment of hydrogen bonds with the phosphate groups of the phospholipids polar heads, while the hydrocarbon chain and the phenothiazine group seems to continue interacting through hydrophobic forces inside the monolayer, as represented in Fig. 21 [255].
Fig. 21.
CPZ/phospholipid monolayer interactions suggested by molecular dynamics simulation studies.
Considering that the antiviral activity described by CPZ involves precisely the inhibition of the virus internalization step via the endocytic pathway, this study may provide valuable clues on the pharmacodynamic interactions between the drug and the cell membrane of the host cell.
The classic method for the synthesis of these N-alkylated phenothiazine drugs consists of a N-alkylation reaction using a suitable base and alkylating agent. An important aspect of the reactivity of this heterocycle, however, is that the N—H bond is weakly acidic, and usually the use of strong bases such as alkali metal amides or even organometallic bases is required [255], [256]. An example of this approach is the PMZ synthesis protocol developed in the early 1940s, in which promethazine is obtained as a mixture of enantiomers via the alkylation of phenothiazine 211 with 1-dimethylamino-2-propylchloride 212 (Scheme 33 ) [257], [258].
Scheme 33.
Early protocol for the synthesis of promethazine as a mixture of enantiomers.
A modification of this method employing Aliquat 336 as a phase transfer agent and potassium hydroxide as a suitable base in the absence of solvents was described in 1985 [256].
For over four decades, both enantiomers of promethazine were considered to display the same therapeutic activities, hence it has been administered as racemate. However, some findings have shown that the two enantiomers exhibit different physiological and pharmacological effects, which has prompted the development of stereoselective approaches for the synthesis of PMZ [259], [260]. The first chemoenzymatic stereodivergent synthesis of PMZ was reported in 2014 (Scheme 34 ) [247]. In the first step, racemic 1-(10H-phenothiazin-10-yl)propan-2-ol (±)-214 was synthesized with a 77% yield via the regioselective ring-opening of propylene oxide 213 with phenothiazine 211 in the presence of butyllithium. The central chiral scaffold was obtained via a lipase-mediated kinetic resolution protocol employing Novozym 435 and Lipozyme TL IM as biocatalysts, which furnished both enantiomeric forms, (R)-(−)-214 and (S)-(+)-214, with high enantioselectivity (up to >99% ee) from the racemate (±)-214. The enantiomers (S)-(+)-214 and (R)-(−)-214 were further transformed into bromide derivatives employing phosphorous tribromide, giving (R)-(+)-216 and (S)-(−)-216, respectively. Finally, the reaction of (R)-(+)-216 and (S)-(−)-216 with methylamine gave compounds (R)-(+)-217 and (S)-(−)-217 as products, respectively.
Scheme 34.
Chemoenzymatic synthesis of enantioenriched enantiomers of promethazine.
Recently, the preparation of a library of psychotropic drug analogs with neuroprotective potential was published [261]. In this work, an alternative method for the construction of CPZ was achieved via a reductive N-alkylation strategy using of a different alkylating compound in the presence of triethylsilane (TES) as a reducing agent was presented (Scheme 35 ). For this, initially nitrile 218 is treated with excess sodium borohydride and nickel sulfate as catalyst, giving an amine intermediate that undergoes acetylation in situ with acetic anhydride to form acetamide 219. Next, compound 219 is reacted with 2-chlorophenothiazine 220 in the presence of TES and TFA, furnishing intermediate 221, which undergoes an amide alkaline hydrolysis step under microwave irradiation to produce 22. In the last step, an Eschweiler-Clarke reaction is conducted to accomplish the dimethylation of the primary amine 222, finally giving rise to CPZ with a 25% overall yield.
Scheme 35.
Synthetic approach towards chlorpromazine.
As for FPZ, its synthesis can be accomplished via different approaches, which usually focus in the construction of the distinguished N-alkyl group in its structure. In one of these strategies, 2-trifluoromethylphenothiazine 223 is condensed with tetrahydropyranyl ether 224 in the presence sodamide followed by treatment in acidic medium to give intermediate 225 (Scheme 36 ) [262]. The resulting alcohol 225 is then reacted with tosyl chloride to produce tosylate 226, which is next reacted with piperazine to give intermediate 227. Finally, the reaction of intermediate 227 with ethylene oxide leads to FPZ [263].
Scheme 36.
Synthetic approach towards fluphenazine.
2.7. Antietilism drugs: Disulfiram, a carbothioamide derivative
Disulfiram (DSF, Antabuse®) was synthesized in 1881 and first used as a compounding agent to accelerate the manufacturing process of rubber. In 1948 it was proposed to be used in the treatment of chronic alcoholism, being approved by the FDA in 1951. It is a non-toxic and low-cost drug with high bioavailability (greater than 80%) and a half-life of 60–120 h, and is usually prescribed in a single-day dose of 250 mg, with at least 12 h of abstinence, but some patients may require higher doses. Symptoms of DSF overdose include irritation, slight drowsiness, unpleasant taste, mild gastrointestinal disturbances and orthostatic hypotension. DSF must be administered orally or through sub-dermal implants, and the recommended duration of treatment is one year [264], [265], [266].
DSF is an organic disulfide that can be easily obtained from the oxidation of dithiocarbamate 229 by using an oxidizing agent such as hydrogen peroxide and carbon disulphide; sodium hypochlorite can also be employed in this reaction, affording high yields and high purity products (Scheme 37 ) [267], [268].
Scheme 37.
Synthetic approach towards disulfiram.
DSF and its metabolites are known inhibitors of enzymes recognized as pharmacological targets in the treatment of microbial and viral diseases, which is a consequence of the reactivity of DSF toward specific cysteine residues directly involved in the enzymés stability and activity.
In this context, a promising pharmacological target for the identification of substances with anti-SARS-CoV-2 activity is the virus 3C-like protease (3CLpro). This enzyme is responsible for processing the first replicase-translated polypeptides into a series of native proteins that are essential for viral replication in the host cell. Considering this, the identification of a 3CLpro inhibitor can represent an advance in drug repositioning for COVID-19 treatment.
Considering these concepts, a research group conducted in silico studies that have revealed DSF́s binding interactions with the main active site cavity of the SARS-CoV-2 main protease, 3CLpro [269]. DSF contains chemical groups that can react with the cysteinés thiols exposed on the 3CLpro surface. A Cys145 residue in the active site is located at 3.5 Å from a histidine-residue (His41) and therefore, His41 may participate in the drug’s sulfhydryl activation (Fig. 22 a). Molecular docking studies have shown that DSF binds to the active site of the enzyme with relative proximity to the thiol group of Cys145 (Fig. 22b). The model infers that DSF obstructs the function of the catalytic dyad Cys145 and His41, and consequently predict that this moleculés binding, as well as of its metabolites, will result in competitive inhibition of the viral protease [270].
Fig. 22.
Binding models of DSF into SARS-CoV-2 main protease, 3CLpro. (a) Depiction of the overall structure showing the Cys145 and His41 catalytic residues located in the active site cavity and (b) SARS-CoV-2 3CLpro-DSF. [270]. Reprinted from Journal of Biomolecular Structure and Dynamics, N. Lobo-Galo, M. Terrazas-López, A. Martínez-Martínez, Á.G. Díaz-Sánchez. “FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication” with permission of Taylor & Francis Ltd. Copyright 2020.
The use of DSF against COVID-19 can be advantageous for many reasons, but specially because it is a long-lasting drug, and upon ingestion, one fifth of the administered dose is active even after one week, avoiding long administration periods. The pharmacokinetics studies of DSF indicate that a relatively small fraction of the compound is excreted in urine after 6 h of administration, maintaining a half-life of 6 h in the plasma, while the rest is transformed into other metabolites such as thiol-reactive species that can also be reactive towards Cys thiol groups [270].
The inhibitory potential of DSF and its metabolites on 3CLpro is proposed to occur through a mechanism involving initial non-covalent binding to the protein followed by a nucleophilic attack by the thiol group in Cys145. This would result in a more stable covalently bound enzyme-inhibitor complex, leading ultimately to the enzyme irreversible inactivation (Scheme 38 ) [270].
Scheme 38.
Possible mechanism of reaction between DSF and 3CLpro, SARS-CoV-2́s main protease.
In this sense, DSF could potentially be tested for clinical therapeutic use against COVID-19. The FDA-approved drug is used to facilitate the treatment of chronic alcoholism, as it targets the hepatocyte mitochondrial acetaldehyde dehydrogenase (ALDH-2), an enzyme to which the drug and its metabolites interacts by blocking its catalytic cysteine site. Interestingly, other cysteine-dependent enzymes that can be inhibited by DSF are the SARS-CoV-1 and MERS-CoV betaine aldehyde dehydrogenase, carboxikinase, urease and papain-like protease. Additionally, DSF and its metabolites can penetrate human cells and consequently have a high potential to block the proteolytic function of the 3CLpro, essential for SARS-CoV-2 replication [270].
2.8. Antifungal agents: Terconazole, a triazole derivative
Terconazole (TCZ, Terazol®) is an antifungal drug approved by the FDA against vulvovaginitis in 1987 that is primarily used for fungal and yeast vaginal infections (or vaginal candidiasis) [270], [271]. TCZ inhibits ergosterol synthesis by inhibiting the 14-alpha-demethylase (lanosterol 14-alpha-demethylase); the depletion of ergosterol in the fungal membrane disrupts its structure and many of its functions, leading to the inhibition of the fungal growth [272].
TCZ is administered in the form of a vaginal cream or suppository. It is used in the local treatment of vulvovaginal candidiasis, and the recommended dosage is 40 mg (as 0.8% vaginal cream) or 80 mg (as a pessary) at bedtime for 3 nights or 20 mg (as 0.4% cream) at bedtime for 7 nights. The oral LD50 values were found to be 1741 and 849 mg/kg for the male and female in rat. After application, 5–16% of TCZ is absorbed, and the systemically absorbed drug is then metabolized in the liver and excreted in urine and feces.
In 2014 it was reported that TCZ was able to inhibit both MERS-CoV and SARS-CoV-1 infections with EC50 of 12.203 µM and 15.327 µM, respectively [103]. Recently, the same group reported that TCZ presented good results regarding SARS-CoV-2 inhibition, presenting an IC50 value of 11.92 µM (MOI of 0.004) at non-cytotoxic concentrations (CC50 41.46 and SI 3.48) [106].
One of the early synthetic routes towards TCZ starts with a ketalization reaction of 2,4-dichloroacetophenone 231 with glycerol 232, giving intermediate 233, which undergoes a subsequent bromination reaction to furnish intermediate 234 (Scheme 39 ) [273], [274]. Next, the reaction of alcohol 234 with benzoyl chloride in dry pyridine afforded ether derivative 235, which is then coupled triazole 236 in the presence of sodium hydride, affording 237. Intermediate 237 was converted into the respective mesylate 238 with 87% yield. In parallel, the reductive alkylation of 10 with acetone leads to 11, and its subsequent demethylation with hydrobromic acid solution gives piperazine intermediate 12 in 85%. Finally, TCZ is obtained through the reaction of intermediates 12 with 9 in the presence of sodium hydride [275].
Scheme 39.
Synthetic route towards terconazole.
2.9. Antibacterial agents: Teicoplanin, a glicopeptide
Teicoplanin (Targocid®), a semisynthetic glycopeptide antibiotic isolated from Actinoplanes teichomyceticus, is used in the treatment of serious infections caused by aerobic and anaerobic Gram-positive bacteria such as staphylococcus aureus and streptococcus [275]. This compound was first isolated in 1978, and its structure was described in 1984 simultaneously by two research groups [276], [277]. Teicoplanin’s antimicrobial activity depends essentially on the duration of time the substance level is higher than the minimum inhibitory concentration (MIC) of the pathogen. Teicoplanin diffuses rapidly into the skin, subcutaneous tissue, myocardium, lung, pleural fluid, bone, synovial fluid, skin vesicle fluid, and slowly into the cerebrospinal fluid. This drug can be administered intravenously or intramuscularly once daily following an initial loading dose [278]. Bioavailability of a single intramuscular injection of 3–6 mg/kg is more than 90%. Teicoplanin is poorly absorbed by the normal gastrointestinal tract after oral administration; 40% of the oral dose is found in the stool as an active drug. After six daily intramuscular administrations of 200 mg, the mean maximum teicoplanin concentration amounts to 12.1 mg/L and occurs 2 h after administration. According to most recent studies, the elimination half-life of teicoplanin varies from 100 to 170 h. Teicoplanin exhibited linear pharmacokinetics at dose range of 2–25 mg/kg [279].
Teicoplanin inhibits the growth of susceptible organisms by interfering with cell-wall biosynthesis; the biosynthesis of peptidoglycan is blocked by specific binding to terminal amino acyl-D-alanyl-D-alanine residues in the cell wall, preventing any further peptidoglycan synthesis and bacterial growing [276], [280]. The drug is composed of five major components that can form complexes with the C-terminal L-Lys-D-Ala-D-Ala subunits of lipid II peptidoglycan precursors, and this binding inhibits transglycosylation and transpeptidation, leading to disturbed cell wall synthesis in Gram-positive bacteria [281], [282].
In 2016, it was reported that this antibiotic has inhibitory effects against MERS-CoV and SARS-CoV-1 with IC50 values of 0.63 and 3.76 µM, respectively. The proteolysis of glycoprotein by cathepsin L is required for the membrane fusion of SARS-CoV-1, and teicoplanin potently inhibits the activity of this enzyme in a dose-dependent manner. The IC50 value of teicoplanin against cathepsin L (208.0 μM) indicates that high doses are required for the inhibition to be effective, which could be a consequence of the relatively low sensitivity of cathepsin L. This fact demonstrates the consistency between the inhibition of virus entry and the inhibition of cathepsin L activity, and shows that the target molecule of teicoplanin is cathepsin L [282]. It is noteworthy that the hydrophobic groups of the glycopeptide antibiotics might play an important role in the interactions with cathepsin L. Drugs like vancomycin do not have hydrophobic groups, which may be the reason why they do not inhibit such viral infections. The inhibitory effects of teicoplanin and its derivatives on HIV-1, HCV, influenza viruses, flaviviruses, FIPV, and SARS-CoV-1 have also been reported, further supporting that the antiviral target for glycopeptide antibiotics is a common host factor [282].
Recently, teicoplanin was described as a potent SARS-CoV-2 inhibitor in a dose-dependent manner, with IC50 value of 1.66 μM [283]. It was found that during the invasion phase, SARS-CoV-2 binds to the ACE2 on the surface of host cells. The interaction between the S protein and ACE2 triggers conformational changes within the S protein, making it susceptible to activation by host cell protease. SARS-CoV-2 enters the endosome of the cell, and cathepsin L can further cleave the S protein and activate the membrane fusion. Teicoplanin potently prevents the SARS-CoV-2 S protein activation by directly inhibiting the enzymatic activity of cathepsin L. These results corroborate the potential use of teicoplanin against COVID-19 [284], [285]. The recommended plasma concentration of teicoplanin for clinical use to inhibit Gram-positive bacteria is 15 mg/L (8.78 μM), which corresponds to a dose of 400 mg/day. Considering that the IC50 of this molecule against SARS-CoV-2 was only 1.66 uΜ, much lower than the routine use, a dose of 400 mg/day could be considered for patients with SARS-CoV-2 infection. Additionally, in order to prevent virus infection and amplification at a stage as early as possible, the use of teicoplanin for SARS-CoV-2 would be recommended in the early stages. In that way, teicoplanin could function as a dual inhibitor for the SARS-CoV-2 infection and co-infection with Gram-positive bacteria [284].
Structurally, teicoplanin is a complex of five major molecules with a common aglycopeptide core which differ in the length and branching of the aliphatic chain (A2-1, A2-2, A2-3, A2-4 and A2-5, Fig. 23 ). Such chemical complexity makes its synthesis and purification challenging [286].
Fig. 23.
Chemical structure of the teicoplanin complex.
In spite of the considerable challenge, a few research groups have endeavored into developing routes for the synthesis of teicoplanin. For instance, efforts were made in order to establish the synthesis of the teicoplanin aglycopeptide core (R1 = R2 = R3 = H) [287], [288]. Initially, modified phenylalanine 244 was obtained via a Schöllkopf alkylation of bromide 242 using Schöllkopf reagent 243, followed by N-protection with Teoc-OBt. Aminoacid precursor 248 was prepared employing the Sharpless asymmetric aminohydroxylation reaction of precursor 246, giving 247, which underwent protection with methoxyethoxymethyl chloride (MEMCl) and further hydrogenation, while 250 was prepared in a single step from 249 (Scheme 40 ).
Scheme 40.
Synthesis of the precursor residues 245, 248 and 250 of the teicoplanin aglycone. Teoc-OBt = 1-[2-(trimethylsilyl)ethoxycarbonyloxy]benzotriazole. MEMCl = 2-methoxyethoxymethyl chloride.
Next, the FG dipeptide with a biaryl ether bridge was synthetized using an aromatic nucleophilic substitution reaction (Scheme 41 ). First, the coupling of 248 and 250 was conducted in basic medium in the presence of 18-crown-6 and 4 Å molecular sieves, giving intermediate 251, which underwent a hydrogenation reaction followed by diazonium salt formation and treatment with cuprous oxide/nitrate to furnish the corresponding phenol 252. Subsequently, a four-step O-methylation/protection of the primary alcohol/MEM and N-Boc deprotection/N-Boc reprotection and trifluoroacetamide hydrolysis sequence afforded the amine 253, which is coupled with 245 using EDC to produce 254. Then, 254 was converted into 255 via two-step oxidation process – Dess-Martin periodinane oxidation followed by treatment with sodium chlorite [288], [289].
Scheme 41.
Synthesis of EFG tripeptide precursor 255 of teicoplanin aglycone. EDC = 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide.
In parallel, the ABCD ring system was obtained starting with the protection of (2S,3R)-β-hydroxy-β-(4-fluoro-3-nitrophenyl)alaninate 256 as TBDMSO ether 257. The O-methylation of 258 with iodomethane affords the ester 259, which is coupled with 257 using EDC, providing 260, which was O-protected with TBS to give 261. A second coupling reaction of 261 with (R)-N-Boc-(3,5-dihydroxy-4-methoxyphenyl)glycine 262 using EDC leads to 263, and a subsequent TBDMS-deprotection with TBAF produces 264. Then, intermediate 264 undergoes a cyclization reaction in the presence of potassium carbonate and calcium carbonate to afford 265, which is submitted to an ester deprotection step followed by a nitro reduction to afford 267. Next, 267 was subjected to a diazotization reaction followed by the Sandmeyer substitution of the corresponding diazonium salt 268 with chloride to give 269 (Scheme 42 ) [288], [289], [290], [291].
Scheme 42.
Synthesis of the ABCD ring system (part A) of teicoplanin aglycone. EDC = 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide, HOBt = hydroxybenzotriazole, TBAF = tetra-n-butylammonium fluoride.
Next, the Suzuki coupling reaction of 269 with aminoacid 270 provided a 1:1.3 mixture of R:S isomers, which after separation, gave (S)-271. A subsequent silyl ether deprotection with TBAF leads to 272, which underwent a methyl ester hydrolysis and N-Cbz deprotection to produce the intermediate 274. A macrolactamization of 274 using EDC furnishes 275, which was converted into the amine 276 via a N-Boc deprotection step (Scheme 43 ) [288], [289], [290], [291].
Scheme 43.
Synthesis of the ABCD ring system (part B) of teicoplanin aglycone. EDC = 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide, HOBt = hydroxybenzotriazole.
Next, DEPBT 277 was used in the coupling reaction between fragments 255 and 276, furnishing intermediate 278, which underwent a macrocyclization reaction using cesium fluoride to give intermediate 279 with a 3:1 preference for the natural stereochemistry. The alcohol protection with OTBS ether, followed by a N-protection with Troc group gives the intermediate 281, and subsequently, a nitro reduction affords 282. The diazotization and Sandmeyer substitution of 282 provided intermediate 283 (Scheme 44 ) [289], [290].
Scheme 44.
Synthesis of the key intermediate 283 of teicoplanin aglycone. DEPBT = 3-(Diethoxyphosphoryloxy)-1,2,3-benzotriazin-4(3H)-one.
Subsequently, primary alcohol 283 underwent oxidation with Dess-Martin periodinane (DMP) followed by a N-Troc deprotection reaction with Zn-Pb to afford carboxylic acid 284. Next, a macrolactonization reaction of 284 was carried out to provide 285, which undergoes a MEM deprotection with B-bromocatecholborane 286, followed by a two-step alcohol oxidation to form intermediate 287. Finally, an O-TBS and N-Boc deprotection reaction using aluminum tribromide in ethanethiol (AlBr3-EtSH) furnished the teicoplanin aglycon (288) (Scheme 45 ) [290].
Scheme 45.
Final step in the teicoplanin aglycone synthesis.
2.10. Therapeutic antibodies
Since the beginning of the pandemic, several FDA-approved drugs have had their in vitro activity tested against SARS-CoV-2 replication, and hopes were raised that those could be used as therapeutic options for COVID-19. However, the administration of a single antiviral drug so far does not seem to be an effective option against this disease [291].
On the other hand, the treatment with hyperimmune plasma and intravenous immunoglobulin infusions has already proven to be safe and effective for several diseases, including the 2014 Ebola outbreak in west Africa [292]. However, the broad variety of antibodies in convalescent plasma may possibly lead to tissue damage, as some of these antibodies are virus-neutralizing, while others might have off-target effects. Taking into consideration that an extensive immune dysregulation, including the cytokine storm syndrome, is a chief contributor to the aggravation of COVID-19, the time frame of this type of treatment has to be carefully planned, as suggested by early studies conducted in Wuhan, China [293].
In this scenario, neutralizing human/humanized monoclonal antibodies appear as a viable alternative passive immune therapy, since those have the ability of precisely acting on specific neutralizing sites on the SARS-CoV-2 spike protein. These antibodies can be produced through cloning from the B cells of recovered patients or even be genetically engineered.
Back in February 2020, a research group reported that a SARS-CoV-1-specific human monoclonal antibody (CR3022) could bind potently to SARS-CoV-2 RBD without overlapping with its ACE2 binding site [294]. Conversely, it was also observed that known potent SARS-CoV-1-specific neutralizing antibodies were not able of binding to the SARS-CoV-2 spike protein, which suggests that understanding the differences between the RBD of these viruses is crucial for the development of effective passive immune therapies. Later, the human antibody 47D11 was discovered using ELISA techniques and it was able to potently inhibit SARS-CoV-1 and SARS-CoV-2 infections in VeroE6 cells. Although a precise inhibition mechanism was not described, the authors found that 47D11 binds to the S1B RBD [295].
In June 2020, another research group disclosed their results on the isolation of four monoclonal antibodies from a convalescent patient and the evaluation of their neutralization abilities [296]. It was found that two of those antibodies (B38 and H4) act by blocking the binding between the spike glycoprotein RBD of SARS-CoV-2 and ACE2, displaying an additive inhibitive effect. Importantly, in vivo studies conducted in mice have proven that these antibodies can significantly reduce virus titers in infected lungs. Additionally, detailed studies of the SARS-CoV-2 RBD-B38 complex structure have shown that most residues on the epitope overlap with the RBD-ACE2 binding interface, which justifies the antibody’s blocking effect and neutralizing capacity (Fig. 24 ).
Fig. 24.
(a) Superposition of SARS-CoV-2 RBD, B38 and RBD-hACE2 [Protein Data Bank (PDB) ID 6LZG]. The B38 heavy chain is colored in cyan, the light chain in green, SARS-CoV RBD in magenta and hACE2 in light pink. [Protein Data Bank (PDB) ID 6LZG]. (b) The residues on RBD involved in both B38 and hACE2 binding (labeled in red) and involved in hACE2-RBD binding (highlighted in light pink) [297]. Reprinted with permission from AAAS. Copyright 2020. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Another interesting work in this field described the use of high-throughput single-cell RNA sequencing from 60 convalescent patients’ B cells to identify SARS-CoV-2 neutralizing antibodies [297]. Among the 14 identified neutralized antibodies, the most potent was BD-368-2, which exhibited high anti-SARS-CoV-2 activity in vitro and therapeutic efficacy in SARS-CoV-2-infected hACE2-transgenic mice. The group was also able to solve the structure of one neutralizing monoclonal antibody-SARS-CoV-2 spike ectodomain trimer complex by using cryo-electron microscopy, having confirmed that its epitope overlaps with the RBD/ACE2 binding moiety.
Another promising approach in this field involves the use of hyperimmune equine plasma produced from the injection of a recombinant SARS-CoV-2 spike protein RBD [298]. In this case, the authors verified that the RBD was highly effective in triggering high-titer neutralizing antibody response in horses. Then, immunoglobulin F(ab′)2 fragments were obtained from the equine antisera through the removal of the Fc region from the immunoglobulins. Interestingly, the neutralization tests conducted with the live virus have demonstrated that the RBD-specific F(ab′)2 fragments were able to efficiently inhibit SARS-CoV-2 in vitro. In a similar path, another research group produced an anti-COVID-19 hyperimmune F(ab′)2 equine serum, but in this case the prefusion trimeric spike glycoprotein, which has a higher immunogenicity when compared to smaller protein fragments such as the RBD, was used as antigen [299]. Remarkably, the authors found that the neutralization titers present in the equine F(ab′)2 sera were considerably higher than those of human convalescent plasma.
3. Final remarks
Undoubtedly, the COVID-19 pandemic has surged as the greatest scientific challenge of modern times. The virus efficient transmission and pronounced virulence, in addition to its relatively high lethality, has urged satisfactory results not only regarded to vaccines but also efficient therapeutic options. Furthermore, the fast spread of information and the access of the lay population to it, as well as the governments’ struggle for a solution to the pandemic, often focused on political and commercial interests, further increase the pressure over the scientific community in the search for positive results against COVID-19.
To date, at least 125 vaccines against COVID-19 are under development, with about a dozen under advanced phases 2 and 3 clinical trials in humans. However, vaccine development is a slow process and only solves part of the problem. Therefore, the repositioning of clinically approved drugs seems to be the fastest response to this pandemic outbreak.
In view of all these concepts, this review aimed to consolidate the main pharmacological and synthetic aspects surrounding the clinically developed small molecules with promising activity against SARS-CoV-2. We hope this paper eases the understanding of the rationale involved in the repositioning of drugs for emerging diseases and inspires the scientific community to plan new molecular designs to fight COVID-19.
The results discussed herein show that important advances have already been achieved in the past few months, and considering the short period of time since the first COVID-19 documented case and the great volume of significant reports as a result of the joint work of the scientific community worldwide, remain impressive.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Fellowships granted by CNPq, CAPES and FAPERJ are gratefully acknowledged. This work was partially supported by FAPERJ grant numbers E-26/010.101106/2018, E-26/202.800/2017 and CNPq 301873/2019-4. The authors also acknowledge Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001.
References
- 1.Solomon T., Baylis M., Brown D. Zika virus and neurological disease-approaches to the unknown. Lancet. Infect. Dis. 2016;16:402–404. doi: 10.1016/S1473-3099(16)00125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Eng. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Eng. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 Coronavirus pandemic. Available at https://www.worldometers.info/coronavirus/ (accessed in June 27th, 2020).
- 6.Arsene A.L., Dumitrescu I., Drăgoi C.M., Udeanu D.Y., Lupuliasa D., Jinga V., Drăgănescu D., Dinu-Pîrvu C.E., Dragomiroiu G.T.A.B., Blejan I.E., Moisi R.E., Nicolae A.C., Moldovan H., Popa D.E., Velescu B.S., Ruță S. A new era for the therapeutic management of the ongoing covid-19 pandemic. Farmacia. 2020;68:185–196. [Google Scholar]
- 7.https://www.who.int/health-topics/coronavirus#tab=tab_1.
- 8.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Tawfiqd J.A., Zumla A., Memish Z.A. Coronaviruses: severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus in travelers. Curr. Opin. Infect. Dis. 2014;27:411–417. doi: 10.1097/QCO.0000000000000089. [DOI] [PubMed] [Google Scholar]
- 11.Lee N., McGeer A. The starting line for COVID-19 vaccine development. Lancet. 2020;395:1815–1816. doi: 10.1016/S0140-6736(20)31239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organisation, COVID-19 Trials – International Clinical Trials Registry Platform (ICTRP), 2020.
- 13.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 14.V.K. Maurya, S. Kumar, M.L.B. Bhatt, S.K. Saxena, Therapeutic development and drugs for the treatment of COVID-19, Medical Virology: From Pathogenesis to Disease Control, Coronavirus Disease 2019 (COVID-19), Springer, Singapore, 2020, pp. 109–126.
- 15.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behja W., Jemal M. Anti-HIV drug discovery, development and synthesis of delavirdine: review article. Int. Res. J. Pure Appl. Chem. 2019;20:1–16. [Google Scholar]
- 17.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1752802. (in press) [DOI] [PubMed] [Google Scholar]
- 18.Yao T., Qian J., Zhu W., Wang Y., Wang G. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J. Med. Virol. 2020;92:556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sang P., Tian S., Meng Z., Yang L. Anti-HIV drug repurposing against SARS-CoV-2. RSC Adv. 2020;10:15775–15783. doi: 10.1039/d0ra01899f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nukoolkarn V., Lee V.S., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J. Theor. Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan K.S., Lai S.T., Chu C.M., Tsui E., Tam C.Y., Wong M.M.L., Tse M.W., Que T.L., Peiris J.S.M., Sung J., Wong V.C.W., Yuen K.Y. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med. J. 2003;9:399–406. [PubMed] [Google Scholar]
- 22.Fintelman-Rodrigues N., Sacramento C.Q., Lima C.R., Silva F.S., Ferreira A.C., Mattos M., Freitas C.S., Soares V.C., Dias S.S.G., Temerozo J.R., Miranda M., Matos A.R., Bozza F.A., Carels N., Alves C.R., Siqueira M.M., Bozza P.T., Souza T.M.L. Atazanavir inhibits SARS-CoV-2 replication and pro-inflammatory cytokine production. BioRXiv. 2020:1–28. doi: 10.1101/2020.04.04.020925. [DOI] [Google Scholar]
- 23.Adapted from reference 33.
- 24.Lim J., Jeon S., Shin H.-Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.-W., Kang Y.M., Lee B., Park S.-J. Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F., Yu W., Xu A., Zhang Y., Xuan W., Yan T., Pan K., Zhang J. Patients of COVID-19 may benefit from sustained Lopinavir-combined regimen and the increase of Eosinophil may predict the outcome of COVID-19 progression. Int. J. Infect. Dis. 2020;95:183–191. doi: 10.1016/j.ijid.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangum E.M., Graham K.K. Lopinavir-ritonavir: A new protease inhibitor. Pharmacotherapy. 2011;21:1352–1363. doi: 10.1592/phco.21.17.1352.34419. [DOI] [PubMed] [Google Scholar]
- 27.Reddy A.V.R., Garaga S., Akshinamoorthy C.T., Naidu A. Synthesis and characterization of impurities in the production process of lopinavir. Sci. Pharm. 2015;83:49–63. doi: 10.3797/scipharm.1407-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson P.J., Corbellino M., Ottaviani S., Prelle A., Stebbing J., Casalini G. CNS penetration of potential anti–COVID–19 drugs. J. Neurol. 2020;267:1880–1882. doi: 10.1007/s00415-020-09866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Z., Chen L., Li J., Cheng X., Yang J., Tian C., Zhang Y., Huang S., Liu Z., Cheng J. Clinical features of COVID-19-related liver damage. Clin. Gastroenterol. Hepatol. 2020;18:1561–1566. doi: 10.1016/j.cgh.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horby P.W., Mafham M., Bell J.L., Linsell L., Staplin N., Emberson J.R., Palfreeman A., Raw J., Elmahi E., Prudon B., Green C., Carley S., Chadwick D., Davies M., Wise M.P., Baillie J.K., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Lim W.S., Montgomery A., Rowan K., Juszczak E., Haynes R., Landray M.J. Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)32013-4. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoner E.J., Cooper A.J., Dickman D.A., Kolaczkowski L., Lallaman J.E., Liu J., Oliver-Shaffer P.A., Patel K.M., Paterson J.B., Plata D.J., Riley D.A., Sham H.L., Stengel P.J., Tien J.J. Synthesis of HIV Protease Inhibitor ABT-378 (Lopinavir) Org. Proc. Res. Dev. 2000;4:264–269. [Google Scholar]
- 32.P. Bellani, M. Frigerio, P. Castoldi, A process for the synthesis of ritonavir, PCT Int. Appl. (2001), WO 2001021603 A1 20010329.
- 33.Gaspar G., Monereo A., Garcia-Reyne A., de Guzman M. Fanconi syndrome and acute renal failure in a patient treated with tenofovir: a call for caution. AIDS. 2004;18:351–352. doi: 10.1097/00002030-200401230-00035. [DOI] [PubMed] [Google Scholar]
- 34.Arshad U., Neary M., Pertinez H., Box H., Tatham L., Rajoli R.K.R., Curley P., Sharp J., Liptrott N.J., Valentijn A., David C., O’Neill P., Rannard S.P., Bray P.G., Aljayyoussi G., Pennington S., Ward S.A., Back D.J., Khoo S.H., Biagini G., Owen A. Prioritisation of potential anti-SARS-CoV-2 drug repurposing opportunities based on ability to achieve adequate plasma and target site concentrations derived from their established human pharmacokinetics. Clin. Pharmacol. Ther. 2020;108:775–790. doi: 10.1002/cpt.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.UpToDate®, Atazanavir: Drug information [Internet]. UpToDate®, 2020 [cited 2020 Apr 13]. Available from: https://www.uptodate.com/contents/atazanavirdruginformation?search=atazanavir&source=panel_search_result&selectedTitle=1~118&usage_type=panel&kp_tab=drug_general&display_rank=1#F25707939.
- 36.Yamamoto N., Matsuyama S., Hoshino T., Yamamoto N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. BioRxiv. 2020 doi: 10.1101/2020.04.06.026476. [DOI] [Google Scholar]
- 37.Fan X., Song Y.-L., Long Y.-Q. An efficient and practical synthesis of the HIV protease inhibitor atazanavir via a highly diastereoselective reduction approach. Org. Proc. Res. Dev. 2008;12:69–75. [Google Scholar]
- 38.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R.R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Clercq E. New nucleoside analogues for treatment of hemorrhagic fever virus infections. Chem. Asian J. 2019;14:3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f ][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 41.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Green N., Ott R.D., Isaacs R.J., Fang H. Cell-based assaysto identify inhibitors of viral disease. Expert Opin. Drug Discov. 2008;3:671–676. doi: 10.1517/17460441.3.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ClinicalTrials.gov, A trial of remdesivir in adults with mild and moderate COVID-19. https://clinicaltrials.gov/ct2/show/NCT04252664 (Accessed October 13, 2020).
- 46.ClinicalTrials.gov, A trial of remdesivir in adults with severe COVID-19, https://clinicaltrials.gov/ct2/show/NCT04257656 (Accessed October 13, 2020).
- 47.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., de Castilla D.L., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the Treatment of Covid-19 – Final Report. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2007764. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spinner C.D., Gottlieb R.L., Criner G.J., López J.R.A., Cattelan A.M., Viladomiu A.S., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A., Chai L.Y.A., Roestenberg M., Tsang O.T.Y., Bernasconi E., Le Turnier P., Chang S.-C., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wang H., Gaggar A., Brainard D.M., McPhail M.J., Bhagani S., Ahn M.Y., Sanyal A.J., Huhn G., Marty F.M. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19. A randomized clinical trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krasovskiy A., Knochel P. A LiCl-mediated Br/Mg exchange reaction for the preparation of functionalized aryl- and heteroarylmagnesium compounds from organic bromides. Angew. Chem. Int. Ed. 2004;43:3333–3336. doi: 10.1002/anie.200454084. [DOI] [PubMed] [Google Scholar]
- 51.Kearney B.P., Flaherty J.F., Shah J. Tenofovir disoproxil fumarate clinical pharmacology and pharmacokinetics. Clin. Pharmacokinet. 2004;43:595–612. doi: 10.2165/00003088-200443090-00003. [DOI] [PubMed] [Google Scholar]
- 52.Schooley R.T., Ruane P., Myers R.A., Beall G., Lampiris H., Berger D., Chen S.-S., Miller M.D., Isaacson E., Cheng A.K. Study 902 Team, Tenofovir DF in antiretroviral-experienced patients: results from a 48-week, randomized, double-blind study. AIDS. 2002;16:1257–1263. doi: 10.1097/00002030-200206140-00008. [DOI] [PubMed] [Google Scholar]
- 53.Squires K., Pozniak A.L., Pierone G., Jr., Steinhart C.R., Berger D., Bellos N.C., Becker S.L., Wulfsohn M., Miller M.D., Toole J.J., Coakley D.F., Cheng A. Study 907 Team, Tenofovir disoproxil fumarate in nucleoside-resistant HIV-1 infection: a randomized trial. Ann. Int. Med. 2003;139:313–321. doi: 10.7326/0003-4819-139-5_part_1-200309020-00006. [DOI] [PubMed] [Google Scholar]
- 54.Louie M., Hogan C., Hurley A., Simon V., Chung C., Padte N., Lamy P., Flaherty J., Coakley D., Di Mascio M., Perelson A.S., Markowitz M. Determining the antiviral activity of tenofovir disoproxil fumarate in treatment-naive chronically HIV-1-infected individuals. AIDS. 2003;17:1151–1156. doi: 10.1097/00002030-200305230-00006. [DOI] [PubMed] [Google Scholar]
- 55.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park S.-J., Yu K.-M., Kim Y.-I., Kim S.-M., Kim E.-H., Kim S.-G., Kim E.J., Casel M.A.B., Rollon R., Jang S.-G., Lee M.-H., Chang J.-H., Song M.-S., Jeong H.W., Choi Y., Chen W., Shin W.-J., Jung J.U., Choia Y.K. Antiviral Efficacies of FDA-Approved Drugs against SARS-CoV-2 Infection in Ferrets. mBio. 2020;11:e01114–e01120. doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Chien, T.K. Anderson, S. Jockusch, C. Tao, S. Kumar, X. Li, J.J. Russo, R.N. Kirchdoerfer, J. Ju, Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, bioRxiv preprint (2020). 10.1101/2020.03.18.997585. [DOI] [PMC free article] [PubMed]
- 58.Clososki G.C., Soldi R.A., da Silva R.M., Guaratini T., Lopes J.N.C., Pereira P.R.R., Lopes J.L.C., dos Santos T., Martins R.B., Costa C.S., de Carvalho A.N., da Silva L.L.P., Arruda E., Lopes N.P. Tenofovir disoproxil fumarate: New chemical developments and encouraging in vitro biological results for SARS-CoV-2. J. Braz. Chem. Soc. 2020;31:1552–1556. [Google Scholar]
- 59.Ripin D.H.B., Teager D.S., Fortunak J., Basha S.M., Bivins N., Boddy C.N., Byrn S., Catlin K.K., Houghton S.R., Jagadeesh S.T., Kumar K.A., Melton J., Muneer S., Rao L.N., Rao R.V., Ray P.C., Reddy N.G., Reddy R.M., Shekar K.C., Silverton T., Smith D.T., Stringham R.W., Subbaraju G.V., Talley F., Williams A. Process improvements for the manufacture of tenofovir disoproxil fumarate at commercial scale. Org. Proc. Res. Dev. 2010;14:1194–1201. [Google Scholar]
- 60.Keating G.M., Vaidya A. Sofosbuvir: First global approval. Drugs. 2014;74:273–282. doi: 10.1007/s40265-014-0179-7. [DOI] [PubMed] [Google Scholar]
- 61.Bullard-Feibelman K.M., Govero J., Zhu Z., Salazar V., Veselinovic M., Diamon M.S., Geiss B.J. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res. 2017;137:134–140. doi: 10.1016/j.antiviral.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jácome R., Campillo-Balderas J.A., de León S.P., Becerra A., Lazcano A. Sofosbuvir as a potential alternative to treat the SARS-CoV-2 epidemic. Sci. Rep. 2020;10:9294. doi: 10.1038/s41598-020-66440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barth R., Rose C.A., Schöne O. Synthetic routes to sofosbuvir. In: Časar Z., editor. Synthesis of Heterocycles Contemporary Medicinal Chemistry. Springer International Publishing; Switzerland: 2016. pp. 51–88. [Google Scholar]
- 64.Wu R., Wang L., Kuo H.-C.D., Shannar A., Peter R., Chou P.J., Li S., Hudlikar R., Lu X., Liu Z., Poiani G.J., Amorosa L., Brunetti L., Kong A.-N. An update on current therapeutic drugs treating COVID-19. Curr. Pharmacol. Rep. 2020;6:56–70. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du Y.-X., Chen X.-P. Favipiravir: Pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108:242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 66.Nagata T., Lefor A.K., Hasegawa M., Ishii M. Favipiravir: a new medication for the Ebola vírus disease pandemic. Disaster Med. Public Health Prep. 2015;9:79–81. doi: 10.1017/dmp.2014.151. [DOI] [PubMed] [Google Scholar]
- 67.Furuta Y., Gowen B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favirapir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.(a) Furuta Y., Takahashi K., Kuno-Maekawa M., Sangwa H., Uehara S., Kosaki K., Nomura N., Egwa H., Shiraki K. Mechanism of Action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn Acad. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-NCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lou Y., Liu L., Yao H., Hu X., Su J., Xu K., Luo R., Yang X., He L., Lu X., Zhao Q., Liang T., Qiu Y. Clinical outcomes and plasma concentrations of baloxavir marboxil and favipiravir in COVID-19 patients: An exploratory randomized, controlled trial. MedRxiv. 2020 doi: 10.1101/2020.04.29.20085761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.C. Chen, Y. Zhang, J. Huang, P. Yin, Z. Cheng, J. Wu, S. Chen, Y. Zhang, B. Chen, M. Lu, Y. Luo, L. Ju, J. Zhang, Favipiravir versus Arbidol for COVID-19: A randomized clinical trial, MedRxiv (2020). 10.1101/2020.03.17.20037432. This trial is registered with Chictr.org.cn (ChiCTR2000030254). [DOI]
- 73.Y. Furuta, H. Egawa, Nitrogenous heterocyclic carboxamide deriva- tives or salts thereof and antiviral agents containing both, European Patent Office (2020) WO, 00/10569.
- 74.Guo Q., Xu M., Guo S., Zhu F., Xie Y., Shen J. The complete synthesis of favipiravir from 2–aminopyrazine. Chem. Pap. 2019;73:1043–1051. [Google Scholar]
- 75.Boriskin Y.S., Leneva I.A., Pecheur E.I., Polyak S.J. Arbidol: a broadspectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 2008;15:997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- 76.Leneva I.A., Russell R.J., Boriskin Y.S., Hay A.J. Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antiviral Res. 2009;81:132–140. doi: 10.1016/j.antiviral.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 77.Deng P., Zhong D., Yu K., Zhang Y., Wang T., Chena X. Pharmacokinetics, Metabolism, and Excretion of the Antiviral Drug Arbidol in Humans. Antimicrob. Agent. Chemother. 2013;57:1743–1755. doi: 10.1128/AAC.02282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein? Int. J. Antimicrob. Agent. 2020;56:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edinger T.O., Pohl M.O., Stertz S. Entry of influenza A virus: host factors and antiviral targets. J. Gen. Virol. 2014;95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 80.Wang X., Cao R., Zhang H., Liu J., Xu M., Hu H., Li Y., Zhao L., Li W., Sun X., Yang X., Shi Z., Deng F., Hu Z., Zhong W., Wang M. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lian N., Xie H., Lin S., Huang J., Zhao J., Lin Q. Umifenovir treatment is not associated with improved outcomes inpatients with coronavirus disease 2019: a retrospective study. Clin. Microbiol. Infect. 2020;26:917–921. doi: 10.1016/j.cmi.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chai H., Zhao Y., Zhao C., Gong P. Synthesis and in vitro anti-hepatitis B virus activities of some ethyl 6-bromo-5-hydroxy-1H-indole-3-carboxylates. Bioorg. Med. Chem. 2006;14:911–917. doi: 10.1016/j.bmc.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 83.(a) Maisch B. SARS-CoV-2 as potential cause of cardiac inflammation and heart failure. Is it the virus, hyperinflammation, or MODS? Hertz. 2020;45:321–322. doi: 10.1007/s00059-020-04925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertens. Res. 2020;43:648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coombs K., Mann E., Edwards J., Brown D.T. Effects of chloroquine and cytochalasin B on the infection of cells by Sindbis virus and vesicular stomatitis virus. J. Virol. 1981;37:1060–1065. doi: 10.1128/jvi.37.3.1060-1065.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Savarino A., Di Trani L., Donatelli I., Cauda R., Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savarino A. Use of chloroquine in viral diseases. Lancet. 2011;11:653–654. doi: 10.1016/S1473-3099(11)70092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferraris O., Moroso M., Pernet O., Emonet S., Ferrier Rembert A., Paranhos-Baccalà G., Peyrefitte C.N. Evaluation of Crimean-Congo hemorrhagic fever virus in vitro inhibition by chloroquine and chlorpromazine, two FDA approved molecules. Antiviral Res. 2015;118:75–81. doi: 10.1016/j.antiviral.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delogu I., de Lamballerie X. Chikungunya disease and chloroquine treatment. J. Med. Virol. 2011;83:1058–1059. doi: 10.1002/jmv.22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Farias K.J.S., Machado P.R.L., Muniz J.A.P.C., Imbeloni A.A., da Fonseca B.A.L. Antiviral activity of chloroquine against dengue virus type 2 replication in aotus monkeys. Viral Immunol. 2015;28:161–169. doi: 10.1089/vim.2014.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antiviral Res. 2020;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meinão I., Sato E., Andrade L., Ferraz M., Atra E. Controlled trial with chloroquine diphosphate in systemic lupus erythematosus. Lupus. 1996;5:237–241. doi: 10.1177/096120339600500313. [DOI] [PubMed] [Google Scholar]
- 93.Wozniacka A., Lesiak A., Narbutt J., McCauliffe D.P., Sysa-Jedrzejowska A. Chloroquine treatment influences proinflammatory cytokine levels in systemic lupus erythematosus patients. Lupus. 2006;15:268–275. doi: 10.1191/0961203306lu2299oa. [DOI] [PubMed] [Google Scholar]
- 94.Mota L.M.H., da Cruz B.A., Brenol C.V., Pereira I.A., Rezende-Fronza L.S., Bertolo M.B., Pinheiro G.R.C. Consenso 2012 da Sociedade Brasileira de Reumatologia para o tratamento da artrite reumatoide. Rev. Bras. Reumat. 2012;52:152–174. [Google Scholar]
- 95.Wilmot A.J., Powell S.J., Adams E.B. Chloroquine compared with chloroquine and emetine combined in amebic liver abscess. Am. J. Trop. Med. Hyg. 1959;8:623–624. doi: 10.4269/ajtmh.1959.8.623. [DOI] [PubMed] [Google Scholar]
- 96.Alkadi H.O. Antimalarial drug toxicity: A review. Chemotherapy. 2007;53:385–391. doi: 10.1159/000109767. [DOI] [PubMed] [Google Scholar]
- 97.Taylor W.R.J., White N.J. Antimalarial drug toxicity: A review. Drug. Saf. 2004;27:25–61. doi: 10.2165/00002018-200427010-00003. [DOI] [PubMed] [Google Scholar]
- 98.Pavsic N., Mraz J., Strazar Z.D., Gabrijelcic J., Toplisek J., Kozelj M., Prokselj K. Shunt reversal due to deterioration of chloroquine-induced cardiomyopathy in a patient with primary Sjögren syndrome. Int. J. Cardiol. 2016;215:145–146. doi: 10.1016/j.ijcard.2016.04.112. [DOI] [PubMed] [Google Scholar]
- 99.Tonnesmann E., Kandolf R., Lewalter T. Chloroquine cardiomyopathy – a review of the literature. Immunopharmacol. Immunotoxicol. 2013;35:434–442. doi: 10.3109/08923973.2013.780078. [DOI] [PubMed] [Google Scholar]
- 100.Lawrenson A.S., Cooper D.L., O’Neill P.M., Berry N.G. Study of the antimalarial activity of 4-aminoquinoline compounds against chloroquine-sensitive and chloroquine-resistant parasite strains. J. Mol. Model. 2018;24:237. doi: 10.1007/s00894-018-3755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cairns M., Cisse B., Sokhna C., Cames C., Simondon K., Ba E.H., Trape J.F., Gaye O., Greenwood B.M., Milligan P.J.M. Amodiaquine dosage and tolerability for intermittent preventive treatment to prevent malaria in children. Antimicrob. Agent. Chemother. 2010;54:1265–1274. doi: 10.1128/AAC.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Keyaerts E., Vijgen L., Maes P., Neyts J., Ranst M.V. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Comm. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of middle east respiratory syndrome coronavirus infection. Antimicrob. Agent. Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., Van Nieuwkoop S., Bestebroer T.M., Van Den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agent. Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weston S., Haupt R., Logue J., Matthews K., Frieman M. FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro. BioRxiv. 2020 doi: 10.1101/2020.03.25.008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agent. 2020;55:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Foley M., Tilley L. Quinoline antimalarials: mechanisms of action and resistance and prospects for new agents. Pharmacol. Ther. 1998;79:55–87. doi: 10.1016/s0163-7258(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 109.Egan T.J. Quinoline antimalarials. Exp. Opin. Therapeut. Pat. 2001;11:185–209. [Google Scholar]
- 110.O’Neill P.M., Ward S.A., Berry N.G., Jeyadevan J.P., Biagini G.A., Asadollaly E., Park B.K., Bray P.G. A medicinal chemistry perspective on 4-aminoquinoline antimalarial drugs. Curr. Top. Med. Chem. 2006;6:479–507. doi: 10.2174/156802606776743147. [DOI] [PubMed] [Google Scholar]
- 111.Heinink A. Expert opinion on therapeutic patents: Foreword. Exp. Opin. Therapeut. Pat. 2010;20:185–209. doi: 10.1517/13543776.2010.518674. [DOI] [PubMed] [Google Scholar]
- 112.Yu B., Li C., Chen P., Zhou N., Wang L., Li J., Jiang H., Wang D.-W. Low dose of hydroxychloroquine reduces fatality of critically ill patients with COVID-19. Sci. China Life Sci. 2020;63:1617–1618. doi: 10.1007/s11427-020-1751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;382:2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tang W., Cao Z., Han M., Wang Z., Chen J., Sun W., Wu Y., Xiao W., Liu S., Chen E., Chen W., Wang X., Yang J., Lin J., Zhao Q., Yan Y., Xie Z., Li D., Yang Y., Liu L., Qu J., Ning G., Shi G., Xie Q. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369:m1849. doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mercuro N.J., Yen C.F., Shim D.J., Maher T.R., McCoy C.M., Zimetbaum P.J., Gold H.S. Risk of QT interval prolongation associated with use of hydroxychloroquine with or without concomitant azithromycin among hospitalized patients testing positive for coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:1036–1041. doi: 10.1001/jamacardio.2020.1834. https://dx.doi.org/10.1001%2Fjamacardio.2020.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosenberg E.S., Dufort E.M., Udo T., Wilberschied L.A., Kumar J., Tesoriero J., Weinberg P., Kirkwood J., Muse A., DeHovitz J., Blog D.S., Hutton B., Holtgrave D.R., Zucker H.A. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York State. JAMA. 2020;323:2493–2502. doi: 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., Mourão M.P.G., Brito-Sousa J.D., Baía-da-Silva D., Guerra M.V.F., Hajjar L.A., Pinto R.C., Balieiro A.A.S., Pacheco A.G.F., Santos J.D.O., Jr, Naveca F.G., Xavier M.S., Siqueira A.M., Schwarzbold A., Croda J., Nogueira M.L., Romero G.A.S., Bassat Q., Fontes C.J., Albuquerque B.C., Daniel-Ribeiro C.-T., Monteiro W.M., Lacerda M.V.G. Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: A randomized clinical trial. JAMA Netw. Open. 2020;3:e208857. doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mehra M.R., Desai S.S., Ruschitzka F., Patel A.N. RETRACTED: Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020 doi: 10.1016/S0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Funck-Brentano C., Nguyen L.S., Salem J.-E. Retraction and republication: cardiac toxicity of hydroxychloroquine in COVID-19. Lancet. 2020;396:e2–e3. doi: 10.1016/S0140-6736(20)31528-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.P. Horby, M. Landray, Statement from the chief investigators of the randomised evaluation of COVID-19 therapy (RECOVERY) trial on hydroxychloroquine: no clinical benefit from use of hydroxychloroquine in hospitalised patients with COVID-19. June 5, 2020. https://www.recoverytrial.net/files/hcq-recovery-statement-050620-final-002.pdf (accessed October 13, 2020).
- 122.WHO, “Solidarity” clinical trial for COVID-19 treatments. Posted 6 July 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/ global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial- for-covid-19-treatments (accessed October 13, 2020).
- 123.Horby P., Mafham M., Linsell L., Bell J.L., Staplin N., Emberson J.R., Wiselka M., Ustianowski A., Elmahi E., Prudon B., Whitehouse T., Felton T., Williams J., Faccenda J., Underwood J., Baillie J.K., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Lim W.S., Montgomery A., Rowan K., Tarning J., Watson J.A., White N.J., Juszczak E., Haynes R., Landray M.J. N. Eng. J. Med. 2020 doi: 10.1056/NEJMoa2022926. (in press) [DOI] [Google Scholar]
- 124.Johnson W.S., Buell B.G. A new synthesis of chloroquine. J. Am. Chem. Soc. 1952;74:4513–4516. [Google Scholar]
- 125.Madrid P.B., Sherrill J., Liou A.P., Weisman J.L., DeRisi J.L., Guy R.K. Synthesis of ring-substituted 4-aminoquinolines and evaluation of their antimalarial activities. Bioorg. Med. Chem. Lett. 2005;15:1015–1018. doi: 10.1016/j.bmcl.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 126.Margolis B.J., Long K.A., Laird D.L.T., Ruble J.C., Pulley S.R. Assembly of 4-aminoquinolines via palladium catalysis: a mild and convenient alternative to SNAr methodology. J. Org. Chem. 2007;72:2232–2235. doi: 10.1021/jo062168u. [DOI] [PubMed] [Google Scholar]
- 127.J.M. Fortunak, A.A. Kulkarni, C. King, Green chemistry synthesis of the malaria drug amodiaquine and analogs thereof, WO2013138200A1, 2013.
- 128.Yu E., Mangunuru H.P.R., Telang N.S., Kong C.J., Verghese J., Gilliland S.E., Ahmad S., Dominey R.N., Gupton B.F. High-yielding continuous-flow synthesis of antimalarial drug hydroxychloroquine. Beilstein J. Org. Chem. 2018;14:583–592. doi: 10.3762/bjoc.14.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumar M.S., Nageshwar Y.V.D., Meshram H.M. A facile and alternative method for the synthesis of mefloquine. Synth. Comm. 1996;26:1913–1919. [Google Scholar]
- 130.Jeon S., Ko K., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020;64(2020):e00819–20. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dyall J., Gross R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Hensley L.E., Frieman M.B., Jahrling P.B. Middle east respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77:1935–1966. doi: 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Piedade R., Traub S., Bitter A., Nüssler A.K., Gil J.P., Schwab M., Burk O. Carboxymefloquine, the major metabolite of the antimalarial drug mefloquine, induces drug-metabolizing enzyme and transporter expression by activation of pregnane X receptor. Antimicrob. Agents Chemother. 2015;59:96–104. doi: 10.1128/AAC.04140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tse E.G., Korsik M., Todd M.H. The past, present and future of anti-malarial medicines. Malar. J. 2019;18:93. doi: 10.1186/s12936-019-2724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sterckx H., De Houwer J., Mensch C., Herrebout W., Tehrani K.A., Maes B.U.W. Base metal-catalyzed benzylic oxidation of (aryl)(heteroaryl)methanes with molecular oxygen. Beilstein J. Org. Chem. 2016;12:144–153. doi: 10.3762/bjoc.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fox L.M., Saravolatz L.D. Nitazoxanide: A new thiazolide antiparasitic agent. Clin. Infect. Dis. 2005;40:1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- 136.Magill A., Ryan E., Hill D., Solomon T. ninth ed. Elsevier; 2013. Nitoxanide. Book: Hunter’s Tropical Medicine and Emerging Infectious Disease; pp. 1106–1107. [Google Scholar]
- 137.Rossignol J.-F., El-Gohary Y.M. Nitazoxanide in the treatment of viral gastroenteritis: a randomized double-blind placebo-controlled clinical trial. Aliment. Pharmacol. Ther. 2006;24:1423–1430. doi: 10.1111/j.1365-2036.2006.03128.x. [DOI] [PubMed] [Google Scholar]
- 138.Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S., Nambu A., Bavari S., Soloveva V., Sadukhan S., Cassell G.H., Geisbert T.W., Hur S., Goldfeld A.E. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits ebola virus. iScience. 2019;19:1265–1276. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rossignol J.-F. Nitazoxanide: A first-in-class broad-spectrum antiviral agent. Antiviral Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stachulski A.V., Pidathala C., Row E.C., Sharma R., Berry N.G., Iqbal M., Bentley J., Allman S.A., Edwards G., Helm A., Hellier J., Korba B.E., Semple J.E., Rossignol J.-F. Thiazolides as novel antiviral agents. 1. Inhibition of Hepatitis B, virus replication. J. Med. Chem. 2011;54:4119–4132. doi: 10.1021/jm200153p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cabré-Castellví J., Palomo-Coll A., Palomo-Coll A.L. Convenient synthesis of carboxylic acid anhydrides using N,N-bis[2-oxo-3-oxazolidinyl]phosphorodiamidic chloride. Synthesis. 1981:616–620. [Google Scholar]
- 142.Voorhis, W. C. V.; Huijsduijnen, R. H. V.; Wells, T. N. C., Profile of William C. Campbell, Satoshi Ōmura, and Youyou Tu, 2015 nobel laureates in physiology or medicine. Proc. Natl. Acad. Sci. U S A. 112(52) (2015) 15773–15776. [DOI] [PMC free article] [PubMed]
- 143.Soyuncu S., Oktay C., Berk Y., Ekekena C. Abamectin intoxication with coma and hypotension. Clin. Toxicol. 2007;45:299–300. doi: 10.1080/15563650601072225. [DOI] [PubMed] [Google Scholar]
- 144.Abokwidir M., Fleischer A.B. An emerging treatment: Topical ivermectin for papulopustular rosácea. J. Dermatolog. Treat. 2015;26:379–380. doi: 10.3109/09546634.2014.991672. [DOI] [PubMed] [Google Scholar]
- 145.Canga A.G., Prieto A.M.S., Liébana M.J.D., Martínez N.F., Vega M.S., Vieitez J.J.G. The pharmacokinetics and interactions of ivermectin in humans-A mini-review. AAPS J. 2008;10:42–46. doi: 10.1208/s12248-007-9000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chandler R.E. Serious neurological adverse events after ivermectin-do they occur beyond the indication of onchocerciasis? Am. J. Trop. Med. Hyg. 2018;98:382–388. doi: 10.4269/ajtmh.17-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. Antiviral Res. 2020;18:104767–104784. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wagstaff K.M., Rawlinson S.M., Hearps A.C., Jans D.A. An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biol. Screen. 2011;16:192–200. doi: 10.1177/1087057110390360. [DOI] [PubMed] [Google Scholar]
- 149.Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tay M.Y.F., Fraser J.E., Chan W.K.K., Moreland N.J., Rathore A.P., Wang C., Vasudevan S.G., Jans D.A. Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013;99:301–306. doi: 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 151.Yanga S.N.Y., Atkinson S.C., Wang C., Lee A., Bogoyevitch M.A., Borg N.A., Jans D.A. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Res. 2020;177:104760. doi: 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- 152.Götz V., Magar L., Dornfeld D., Giese S., Pohlmann A., Höper D., Kong B.-W., Jans D.A., Beer M., Haller O., Schwemmle M. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 2016;6:23138. doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.J.C. Rajter, M. Sherman, N. Fatteh, F. Vogel, J. Sacks, J.-J. Rajter, ICON (Ivermectin in COvid Nineteen) study: Use of Ivermectin is Associated with Lower Mortality in Hospitalized Patients with COVID19, medRxiv pre-print (2020). 10.1101/2020.06.06.20124461. [DOI] [PMC free article] [PubMed]
- 154.F. I Gorial, S. Mashhadani, H.M. Sayaly, B.D. Dakhil, M.M. AlMashhadani, A.M. Aljabory, H.M. Abbas, M. Ghanim, J.I. Rasheed, Effectiveness of ivermectin as add-on therapy in COVID-19 management (Pilot trial), medRxiv pre-print (2020). 10.1101/2020.07.07.20145979. [DOI]
- 155.Yoon Y.J., Kim E.-S., Hwang Y.-S., Choi C.-Y. Avermectin: biochemical and molecular basis of its biosynthesis and regulation. Appl. Microbiol. Biotechnol. 2004;63:626–634. doi: 10.1007/s00253-003-1491-4. [DOI] [PubMed] [Google Scholar]
- 156.Zhuo Y., Zhang T., Wang Q., Cruz-Morales P., Zhang B., Liu M., Barona-Gómez F., Zhang L. Synthetic biology of avermectin for production improvement and structure diversification. Biotechnol. J. 2014;9:316–325. doi: 10.1002/biot.201200383. [DOI] [PubMed] [Google Scholar]
- 157.Ikeda H., Ohmura S. Avermectin biosynthesis. Chem. Rev. 1997;97:2591–2609. doi: 10.1021/cr960023p. [DOI] [PubMed] [Google Scholar]
- 158.Zhang J., Yan Y.-J., An J., Huang S.-X., Wang X.-J., Xiang W.-S. Designed biosynthesis of 25-methyl and 25-ethyl ivermectin with enhanced insecticidal activity by domain swap of avermectin polyketide synthase. Microb. Cell. Fact. 2015;14:152. doi: 10.1186/s12934-015-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ley S.V., Armstrong A., Diez-Martin D., Ford M.J., Grice P., Knight J.G., Kolb H.C., Madin A., Marby C.A., Mukherjee S., Shaw A.N., Slawin A.M.Z., Vile S., White A.D., Williams D.J., Woods M. Total synthesis of the anthelmintic macrolide avermectin B1a. J. Chem. Soc. Perkin Trans. 1991;1:667. [Google Scholar]
- 160.Díez-Martín D., Grice P., Kolb H.C., Ley S.V., Madin A. Total synthesis of avermectin B1a: Synthesis of the C11–C25 spiroacetal fragment. Synlett. 1990:326–328. [Google Scholar]
- 161.Armstrong A., Ley S.V. Total synthesis of avermectin B1a: Planning of the synthesis and preparation of the C1–C10 “Southern” hydrobenzofuran fragment. Synlett. 1990:323–325. [Google Scholar]
- 162.(a) Hanessian S., Ugolini A., Dub D., Hodges P.J., Andre C. Synthesis of (+)-Avermectin B1. J. Am. Chem. Soc. 1986;108:2777–2778. [Google Scholar]; (b) White J.D., Bolton G.L., Dantanarayana A.P., Fox C.M.J., Hiner R.N., Jackson R.W., Sakuma K., Warrier U.S. Total Synthesis of the Antiparasitic Agent Avermectin B la. J. Am. Chem. Soc. 1995;117:1908–1939. [Google Scholar]
- 163.Hanessian S., Ugolini A., Hodges P.J., Beaulieu P., Dubd D., André C. Progress in natural product chemistry by the chiron and related approaches-synthesis of avermectin B1a. Pure Appl. Chem. 1987;59:299–316. [Google Scholar]
- 164.Yamashita S., Hayashi D., Nakano A., Hayashi Y., Hirama M. Total synthesis of avermectin B1a revisited. J. Antibiot. 2015;69:31–50. doi: 10.1038/ja.2015.47. [DOI] [PubMed] [Google Scholar]
- 165.Assis M.C., Giulietti A.M. Diferenciação morfológica e anatômica em populações de “ipecacuanha” – Psychotria ipecacuanha (Brot.) Stokes (Rubiaceae) Rev. Bras. Bot. 1999;22:205–216. [Google Scholar]
- 166.The National Center for Advancing Translational Sciences (NCATS), Emetine, 2020. https://drugs.ncats.io/substances?q=%22Emetine%22 (accessed 18 July 2020).
- 167.Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.-F., Chen D., Yan Hui K.P., Chu D.K.W., Chan M.C.W., Cheung P.P.-H., Huang X., Peiris M., Yen H.-L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Das S., Sarmah S., Lyndem S., Roy A.S. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1763201. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Evstigneeva R.P., Livshits R.S., Zakharkin L.L., Bainova M.S., Preobrazhenskii N.A. Dokl. Akad. Nauk SSSR. 1950;75:539. [Google Scholar]
- 170.Barash M., Osbond J.M., Wickens J.C. Chemical constitution and amœbicidal action. Part IV. Synthesis of emetine and stereoisomers of emetine. J. Chem. Soc. 1959:3530–3543. [Google Scholar]
- 171.Openshaw H.T., Whittaker N. The synthesis of emetine and related compounds. Part IV. A new synthesis of 3-substituted 1,2,3,4,6,7-hexahydro-9,10-dimethoxy-2-oxo-11bH-benzo[a]quinolizines. J. Chem. Soc. 1963;1449:1461. [Google Scholar]
- 172.Szántay C., Töke L., Kolonits P. Synthesis of protoemetine. A new total synthesis of emetine. J. Org. Chem. 1966;31:1447–1451. doi: 10.1021/jo01343a030. [DOI] [PubMed] [Google Scholar]
- 173.Van Tamelen E.E., Placeway C., Schiemenz G.P., Wright I.G. Total syntheses of dl-ajmalicine and emetine. J. Am. Chem. Soc. 1969;91:7359–7371. [Google Scholar]
- 174.Kametani T., Suzuki Y., Terasawa H., Ihara M. Studies on the syntheses of heterocyclic compounds. Part 766. A total stereoselective synthesis of emetine and (±)-dihydroprotoemetine. J. Chem. Soc., Perkin Trans. 1979;1:1211–1217. [Google Scholar]
- 175.Feyns L.V., Grady L.T. Emetine Hydrochloride. Anal. Profiles Drug Substances. 1981;10:289–335. [Google Scholar]
- 176.S. Sharma, N. Anand, Natural products, in: Approaches to Design and Synthesis of Antiparasitic Drugs, first ed., Elsevier Science, vol. 25, 1997, pp. 347–383.
- 177.R.S. Vardanyan, V.J. Hruby, Drugs for treating protozoan infections, in: Synthesis of Essential Drugs, Elsevier Science, first ed., 2006, pp. 559–582.
- 178.Silvestris N., Cinieri S., La Torre I., Pezzella G., Numico G., Orlando L., Lorusso V. Role of gemcitabine in metastatic breast cancer patients: a short review. Breast. 2008;17:220–226. doi: 10.1016/j.breast.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 179.Vulfovich M., Rocha-Lima C. Novel advances in pancreatic cancer treatment. Expert Rev. Anticancer Ther. 2008;8:993–1002. doi: 10.1586/14737140.8.6.993. [DOI] [PubMed] [Google Scholar]
- 180.Lorusso D., Di Stefano A., Fanfani F., Scambia G. Role of gemcitabine in ovarian cancer treatment. Ann. Oncol. 2006;17:v188–V194. doi: 10.1093/annonc/mdj979. [DOI] [PubMed] [Google Scholar]
- 181.Sweetman S.C. 37th ed. Royal Pharmaceutical Society; London: 2011. Martindale: The Complete Drug Reference. [Google Scholar]
- 182.Clouser C.L., Patterson S.E., Mansky L.M. Exploiting drug repositioning for discovery of a novel HIV combination therapy. J. Virol. 2010;84:9301–9309. doi: 10.1128/JVI.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Clouser C.L., Holtz C.M., Mullett M., Crankshaw D.L., Briggs J.E., Chauhan J., VanHoutan I.M., Patterson S.E., Mansky L.M. Analysis of the ex vivo and in vivo antiretroviral activity of gemcitabine. PLoS One. 2011;6:e15840. doi: 10.1371/journal.pone.0015840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Denisova O.V., Kakkola L., Feng L., Stenman J., Nagaraj A., Lampe J., Yadava B., Aittokallioa T., Kaukinenc P., Aholac T., Kuivanend S., Vapalahtid O., Kantelee A., Tynellf J., Julkunenf I., Kallio-Kokkog H., Paavilainenh H., Hukkanenh V., Elliotti R.M., De Brabanderj J.K., Saelensk X., Kainova D.E. Obatoclax, saliphenylhalamide, and gemcitabine inhibit influenza a virus infection. J. Biol. Chem. 2012;287:35324–35332. doi: 10.1074/jbc.M112.392142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang Y.-N., Zhang Q.-Y., Li X.-D., Xiong J., Xiao S.-Q., Wang Z., Zhang Z.-R., Deng C.-L., Yang X.-L., Wei H.-P., Yuan Z.-M., Ye H.-Q., Zhang B. Gemcitabine, lycorine and oxysophoridine inhibit novel coronavirus (SARSCoV-2) in cell culture. Emerg. Microb. Infect. 2020;9:1170–1173. doi: 10.1080/22221751.2020.1772676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brown K., Dixey M., Weymouth-Wilson A., Linclau B. The synthesis of gemcitabine. Carbohyd. Res. 2014;387:59–73. doi: 10.1016/j.carres.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 187.Hertel L.W., Kroin J.S., Misner J.W., Tustin J.M. Synthesis of 2-deoxy-2,2-difluoro-D-ribose and 2-deoxy-2,2'-difluoro-D-ribofuranosyl nucleosides. J. Org. Chem. 1988;53:2406–2409. [Google Scholar]
- 188.Arora A., Scholar E.M. Role of tyrosine kinase inhibitors in cancer therapy. J. Pharmacol. Exp. Ther. 2005;315:971–979. doi: 10.1124/jpet.105.084145. [DOI] [PubMed] [Google Scholar]
- 189.M.D. Moen, K. McKeage, G.L. Plosker, M.A.A. Siddiqui, Imatinib: a review of its use in chronic myeloid leukaemia, Drugs 67 (200) 299–320. [DOI] [PubMed]
- 190.Solassol I., Pinguet F., Quantin X. FDA- and EMA-approved tyrosine kinase inhibitors in advanced EGFR-mutated non-small cell lung cancer: safety, tolerability, plasma concentration monitoring, and management. Biomolecules. 2019;9:668. doi: 10.3390/biom9110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kantarjian H.M., Talpaz M., O'Brien S., Jones D., Giles F., Garcia-Manero G., Faderl S., Ravandi F., Rios M.B., Shan J., Cortes J. Survival benefit with imatinib mesylate versus interferon-α-based regimens in newly diagnosed chronic-phase chronic myelogenous leukemia. Blood. 2006;108:1835–1840. doi: 10.1182/blood-2006-02-004325. [DOI] [PubMed] [Google Scholar]
- 192.Savage D.G., Antman K.H. Imatinib mesylate – a new oral targeted therapy. N. Engl. J. Med. 2002;346:683–693. doi: 10.1056/NEJMra013339. [DOI] [PubMed] [Google Scholar]
- 193.J. Zimmermann, Pyrimidin derivatives and process for their preparation, EP 0564409 A1 19931006, 1993.
- 194.J. Zimmermann, Pyrimidine derivatives and processes for the preparation thereof, US5521184A, 1996.
- 195.Zimmermann J., Buchdunger E., Mett H., Meyer T., Lydon N.B., Traxler P. Phenylamino-pyrimidine (PAP) - derivatives: a new class of potente and highly selective PDGF-receptor autophosphorylation inhibitors. Bioorg. Med. Chem. Lett. 1996;6:1221–1226. [Google Scholar]
- 196.Kompella A., Adibhatla B.R.K., Muddasani P.R., Rachakonda S., Gampa V.K., Dubey P.K. A Facile total synthesis for large-scale production of imatinib base. Org. Process Res. Dev. 2012;16:1794–1804. [Google Scholar]
- 197.Ivanov A.S., Shishkov S.V. Synthesis of imatinib: a convergent approach revisited. Monatsh. Chem. 2009;140:619–623. [Google Scholar]
- 198.Hopkin M.D., Baxendale I.R., Ley S.V. A flow-based synthesis of imatinib: the API of Gleevec. Chem. Commun. 2010;46:2450–2452. doi: 10.1039/c001550d. [DOI] [PubMed] [Google Scholar]
- 199.Kinigopoulou M., Filippidou M., Gogou M., Giannousi A., Fouka P., Ntemou N., Alivertis D., Georgis C., Brentas A., Polychronidou V., Voulgari P., Theodorou V., Skobridis K. An optimized approach in the synthesis of imatinib intermediates and analogues. RSC Adv. 2016;6:61458–61467. [Google Scholar]
- 200.Kantarjian H., Shah N.P., Hochhaus A., Cortes J., Shah S., Ayala M., Moiraghi B., Shen Z., Mayer J., Pasquini R., Nakamae H., Huguet F., Boqué C., Chuah C., Bleickardt E., Bradley-Garelik M.B., Zhu C., Szatrowski T., Shapiro D., Baccarani M. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 2010;362:2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 201.Keam S.J. Dasatinib in chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. Biodrugs. 2008;22:59–69. doi: 10.2165/00063030-200822010-00007. [DOI] [PubMed] [Google Scholar]
- 202.Tokarski J.S., Newitt J.A., Chang C.Y.J., Cheng J.D., Wittekind M., Kiefer S.E., Kish K., Lee F.Y.F., Borzillerri R., Lombardo L.J., Xie D.L., Zhang Y.Q., Klei H.E. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 203.Das J., Chen P., Norris D., Padmanabha R., Lin J., Moquin R.V., Shen Z., Cook L.S., Doweyko A.M., Pitt S., Pang S., Shen D.R., Fang Q., de Fex H.F., McIntyre K.W., Shuster D.J., Gillooly K.M., Behnia K., Schieven G.L., Wityak J., Barrish J.C. 2-aminothiazole as a novel kinase inhibitor template. Structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J. Med. Chem. 2006;49:6819–6832. doi: 10.1021/jm060727j. [DOI] [PubMed] [Google Scholar]
- 204.Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Friemank M.B. Abelson Kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.R. Nejat, A.S. Sadr, Are losartan and imatinib effective against SARS-CoV2 pathogenesis? A Pathophysiologic-based in silico study, ChemRxiv preprint (2020). [DOI] [PMC free article] [PubMed]
- 206.Abruzzese E., Luciano L., D’Agostino F., Trawinska M.M., Pane F., De Fabritiis P. SARS-CoV-2 (COVID-19) and Chronic Myeloid Leukemia (CML): a case report and review of ABL kinase involvement in infection. Mediterr. J. Hematol. Infect. Dis. 2020;12:e2020031. doi: 10.4084/MJHID.2020.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.European Clinical Trials Register. Availabe at https://www.clinicaltrialsregister.eu/ctr-search/trial/2020-001236-10/N, 2020 (Accessed 21 May 2020).
- 208.Patel H.K., Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol. Therapeut. 2018;186:1–24. doi: 10.1016/j.pharmthera.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 209.Jordan V.C., Murphy C.S. Endocrine pharmacology of antiestrogens as antitumor agents. Endocrine Rev. 1990;11:578–610. doi: 10.1210/edrv-11-4-578. [DOI] [PubMed] [Google Scholar]
- 210.Wiseman L.R., Goa K.L. Toremifene,A review of its pharmacological properties and clinical efficacy in the management of advanced breast cancer. Drugs. 1997;54:141–160. doi: 10.2165/00003495-199754010-00014. [DOI] [PubMed] [Google Scholar]
- 211.Johansen L.M., Brannan J.M., Delos S.E., Shoemaker C.J., Stossel A., Lear C., Hoffstrom B.G., DeWald L.E., Schornberg K.L., Scully C., Lehár J., Hensley L.E., White J.M., Olinger G.G. FDA-approved selective estrogen receptor modulators inhibit ebola virus infection. Sci. Transl. Med. 2013;5:190ra79. doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.W.R. Martin, F. Cheng, Repurposing of FDA-approved toremifene to treat COVID-19 by blocking the spike glycoprotein and NSP14 of SARS-CoV-2, ChemRxiv preprint (2020). 10.26434/chemrxiv.12431966.v1. [DOI] [PMC free article] [PubMed]
- 214.Robson J.M., Schönberg A. Oestrous reactions including mating produced by triphenylethylene. Nature. 1937;140:196. [Google Scholar]
- 215.Bedford G.R., Richardson D.N. Preparation and identification of cis and trans isomers of a substituted triarylethylene. Nature. 1966;212:733–734. [Google Scholar]
- 216.D.J. Collins, J.J. Hobbs, C.W. Emmens, Antiestrogenic and antifertility compounds. 4. 1,1,2 Triarylalkan-1-ols and 1,1,2-triarylalk-1-enes containing basic ether groups, J. Med. Chem. 14 (1971) 952–957. [DOI] [PubMed]
- 217.Kasiotisa K.M., Haroutounian S.A. Tamoxifen: a synthetic overview. Curr. Org. Chem. 2012;16:335–352. [Google Scholar]
- 218.R.J. Toivola, A.J. Karjalainen, K.O.A. Kurkela, M.L. Soderwall, L.V.M. Kangas, G.L. Blanco, H.K. Sunduiqst, Triphenylalkane and -alkene derivatives and their use, EP 95875 A2 19831207, 1983.
- 219.H. Hao, J. Gage, L. Jiuyuan, Z. Enxuan, Synthesis of toremifene, CN104230723A, 2014.
- 220.Carini D.J., Duncia J.V., Aldrich P.E., Chiu A.T., Johnson A.L., Pierce M.E., Price W.A., Santella J.B., Wells G.J., Wexler R.R., Wong P.C., Yoo S.E., Timmermans P.B.M.W.M. Nonpeptide Angiotensin II Receptor Antagonists: The Discovery of a Series of N-(Biphenylylmethyl)Imidazoles as Potent, Orally Active Antihypertensives. J. Med. Chem. 1991;34:2525–2547. doi: 10.1021/jm00112a031. [DOI] [PubMed] [Google Scholar]
- 221.Chiu A.T., Carini D.J., Johnson A.L., McCall D.E., Price W.A., Thoolen M.J.M.C., Wong P.C., Taber R.I., Timmermans P.B.M.W.M. Non-Peptide Angiotensin II Receptor Antagonists. II. Pharmacology of S-8308. Eur. J. Pharmacol. 1988;157:13–21. doi: 10.1016/0014-2999(88)90465-7. [DOI] [PubMed] [Google Scholar]
- 222.P.C. Wong, W.A. Price, A.T. Chiu, N.Y. Wong, J.V. Duncia, D.J. Carini, A.L. Johnson, P.B.M.W.M. Timmermans, EXP 6803, A Nonpeptide Angiotensin II Receptor Antagonist. Cardiovasc. Drug Rev. 7(4) (1989) 285–300.
- 223.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.G. Froldi, What could be the better choice between ACE inhibitors and AT1R antagonists in coronavirus disease 2019 (COVID-19) patients? J. Med. Virol. 92 (2020) 2302–2303. 10.1002/jmv.25974. [DOI] [PubMed]
- 225.Sriram K., Insel P.A. Risks of ACE inhibitor and ARB usage in COVID-19: Evaluating the evidence. Clin. Pharmacol. Ther. 2020;108:236–241. doi: 10.1002/cpt.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Y. Liu, F. Huang, J. Xu, P. Yang, Y. Qin, M. Cao, Z. Wang, X. Li, S. Zhang, L. Ye, J. Lv, J. Wei, T. Xie, H. Gao, K.-F. Xu, F. Wang, L. Liu, C. Jiang, Anti-hypertensive angiotensin II receptor blockers associated to mitigation of disease severity in elderly COVID-19 patients, medRxiv preprint (2020). 10.1101/2020.03.20.20039586. [DOI]
- 227.Shuangxia F., Zheng G., Yelv T., Hui L., Guofang J. An efficient and green synthetic route to losartan. J. Chem. Res. 2015;39:451–454. [Google Scholar]
- 228.Ding Y.J., Li Y., Dai S.Y., Lan Q., Wang X.S. Pd(II)-catalyzed, controllable C-H mono-/diarylation of aryl tetrazoles: concise synthesis of losartan. Org. Biomol. Chem. 2015;13:3198–3201. doi: 10.1039/c4ob02453b. [DOI] [PubMed] [Google Scholar]
- 229.Seki M., Nagahama M. Synthesis of angiotensin II receptor blockers by means of a catalytic system for C-H activation. J. Org. Chem. 2011;76:10198–10206. doi: 10.1021/jo202041e. [DOI] [PubMed] [Google Scholar]
- 230.Thorat V.H., Upadhyay N.S., Cheng C.H. Nickel-catalyzed denitrogenative ortho-arylation of benzotriazinones with organic boronic acids: an efficient route to losartan and irbesartan drug molecules. Adv. Synth. Catal. 2018;360:4784–4789. [Google Scholar]
- 231.Sanchez-Pernaute O., Romero-Bueno F.I., O’Callaghan A.S. Why choose cyclosporin A as first-line therapy in COVID-19 pneumonia. Reumat. Clín. 2020 doi: 10.1016/j.reuma.2020.03.001. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ma-Lauer Y., Zheng Y., Malešević M., von Brunn B., Fischer G., von Brunn A. Influences of cyclosporin A and non-immunosuppressive derivatives on cellular cyclophilins and viral nucleocapsid protein during human coronavirus 229E replication. Antiviral Res. 2020;173:104620. doi: 10.1016/j.antiviral.2019.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92:2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lawen A. Biosynthesis of cyclosporins and other natural peptidyl prolyl cis/trans isomerase inhibitors. Biochim. Biophys. Acta. 1850;2015:2111–2120. doi: 10.1016/j.bbagen.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 235.Wu X., Stockdill J.L., Wang P., Danishefsky S.J. Total synthesis of cyclosporine: access to N-methylated peptides via lsonitrile coupling reactions. J. Am. Chem. Soc. 2010;132:4098–4100. doi: 10.1021/ja100517v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wenger R.M. Synthesis of cyclosporine. Total syntheses of ‘Cyclosporin A’ and ‘Cyclosporin H’, two fungal metabolites isolated from the species tolypocladium inflatum GAMS. Helv. Chim. Acta. 1984;67:502–525. [Google Scholar]
- 237.Aebi J.D., Dhaon M.K., Rich D.H. A short synthesis of enantiomerically pure (2S, 3R, 4R, 6E)-3-hydroxy-4-methyl-2-(methylamino)-6-octenoic acid, the unusual C-9 amino acid found in the immunosuppressive peptide cyclosporine. J. Org. Chem. 1987;52:2881–2886. [Google Scholar]
- 238.Evans D.A., Weber A.E. Asymmetric glycine enolate aldol reaction. J. Am. Chem. Soc. 1986;108:6757–6761. [Google Scholar]
- 239.Tung R.D., Rich D.H. Total synthesis of the unusual cyclosporin amino acid MeBMT. Tetrahedron Lett. 1987;28:1139–1142. [Google Scholar]
- 240.Ohlow M.J., Moosmann B. Phenothiazine: The seven lives of pharmacology’s first lead structure. Drug Discov. Today. 2011;16:119–131. doi: 10.1016/j.drudis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 241.Ban T.A. Fifty years chlorpromazine: A historical perspective. Neuropsychiatr. Dis. Treat. 2007;3:495–500. [PMC free article] [PubMed] [Google Scholar]
- 242.Darlíng H.F. Fluphenazine: A prelímínary study. Dis. Nerv. Syst. 1959;20:167–170. [PubMed] [Google Scholar]
- 243.Tardy M., Huhn M., Engel R.R., Leucht S. Fluphenazine versus low-potency first-generation antipsychotic drugs for schizophrenia. Cochrane Database Syst. Rev. 2014;8:CD009230. doi: 10.1002/14651858.CD009230.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.A. Appert-Colin, N. Callizot, Use of piperazine phenothiazine derivatives in the manufacture of a medicament with neuroprotector and/or neurotrophic effects on cns and/or pns, EP1470818 A1, 2014.
- 245.Yanai K. Anticholinergic activity of antihistamines. Clin. Neurophysiol. 2012;123:633–634. doi: 10.1016/j.clinph.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 246.Biel J.H., Martin Y.C. Organic synthesis as a source of new drugs. Adv. Chem. 1971;108:81–122. [Google Scholar]
- 247.Borowiecki P., Paprocki D., Dranka M. First chemoenzymatic stereodivergent synthesis of both enantiomers of promethazine and ethopropazine. Beilstein J. Org. Chem. 2014;10:3038–3055. doi: 10.3762/bjoc.10.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Plaze M., Attali D., Petit A.-C., Blatzer M., Simon-Loriere E., Vinckier F., Cachia A., Chrétien F., Gaillard R. Repurposing of chlorpromazine in COVID-19 treatment: The ReCoVery study. L’Encephale. 2020;46:S35–S39. doi: 10.1016/j.encep.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cong Y., Hart B.J., Gross R., Zhou H., Frieman M., Bollinger L., Wada J., Hensley L.E., Jahrling P.B., Dyall J., Holbrook M.R. MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS One. 2018;13:e0194868. doi: 10.1371/journal.pone.0194868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.M. Plaze, D. Attali, M. Prot, A. Petit, M. Blatzer, F. Vinckier, L. Levillayer, F. Perin-dureau, A. Cachia, G. Friedlander, F. Chrétien, E. Simon-Loriere, R. Gaillard, Inhibition of the replication of SARS-CoV-2 in human cells by the FDA-approved drug chlorpromazine, bioRxiv preprint (2020). 10.1101/2020.05.05.079608. [DOI] [PMC free article] [PubMed]
- 251.Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pawar A.Y. Combating devastating COVID -19 by drug repurposing. Int. J. Antimicrob. Agent. 2020;56:105984. doi: 10.1016/j.ijantimicag.2020.105984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Liang R., Wang L., Zhang N., Deng X., Su M., Su Y., Hu L., He C., Ying T., Jiang S., Yu F. Development of small-molecule MERS-CoV inhibitors. Viruses. 2018;10:721. doi: 10.3390/v10120721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.McFadden G. Gleevec casts a pox on poxviruses. Nat. Med. 2005;11:711–712. doi: 10.1038/nm0705-711. [DOI] [PubMed] [Google Scholar]
- 255.Pickholz M., Oliveira O.N., Skaf M.S. Interactions of chlorpromazine with phospholipid monolayers: Effects of the ionization state of the drug. Biophys. Chem. 2007;125:425–434. doi: 10.1016/j.bpc.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 256.Murakami A. NII-electronic library service. Chem. Pharm. Bull. 1970;2091 [Google Scholar]
- 257.M. Sorokina, Application Publication (10) Pub. No.: US 2019/2019, 1.
- 258.Ban T.A. The role of serendipity in drug discovery. Dialogues Clin. Neurosci. 2006;8:335–344. doi: 10.31887/DCNS.2006.8.3/tban. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.P. Charpentier, Phenotazines, US2530451, 1950.
- 260.E.J. Boland, J. McDonough, Phenothiazine enantiomers as agents for the prevention of bone loss, WO110458A1, 2004.
- 261.E.J. Boland, Phenothiazine enantiomers as agents for the prevention of bone loss, US0258650 A1, 2006.
- 262.Uliassi E., Pena-Altamira L.E., Morales A.V., Massenzio F., Petralla S., Rossi M., Roberti M., Gonzalez L.M., Martinez A., Monti B., Bolognesi M.L. A focused library of psychotropic analogues with neuroprotective and neuroregenerative potential. ACS Chem. Neurosci. 2019;10:279–294. doi: 10.1021/acschemneuro.8b00242. [DOI] [PubMed] [Google Scholar]
- 263.R.S. Vardanyan, V.J. Hruby, Antipsychotics (Neuroleptics), in: Synthesis of Essential Drugs, Elsevier Science, 1st Ed., 2006, chapter 6, pp. 83–101.
- 264.G.E. Ullyot, S. Kline, New perfluoroalkylphenothiazine derivatives, US 3058979, 1962.
- 265.Suh J.J., Pettinati H.M., Kampman K.M., O’Brien C.P. The status of disulfiram a half of a century later. J. Clin. Psychopharmacol. 2006;26:290–302. doi: 10.1097/01.jcp.0000222512.25649.08. [DOI] [PubMed] [Google Scholar]
- 266.Franck J., Jayaram-Lindstrom N. Pharmacotherapy for alcohol dependence: status of current treatments. Curr. Opin. Neurobiol. 2013;23:692–699. doi: 10.1016/j.conb.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 267.Gaval-Cruz M., Weinshenker D. Mechanisms of disulfiram-induced cocaine abstinence: antabuse and cocaine relapse. Mol. Intervent. 2009;9:175–187. doi: 10.1124/mi.9.4.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.H.S. Adams, L. Meuser, Method of Manufacturing Tetra-Alkylated Thiuramdisulphides, US 1782111, 1930.
- 269.G.C. Bailey, Manufacture of Organic Disulphides, US1796977, 1931.
- 270.Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., Díaz-Sánchez A.G. FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1764393. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Chavan B.B., Radhakrishnanand L.P.G.P., Kosuri E.R., Kalariya P.D., Talluri M.V.N.K. Development of stability indicating UPLC method for terconazole and characterization of acidic and oxidative degradation products by UPLC-Q-TOF/MS/MS and NMR. New J. Chem. 2018;42:10761–10773. [Google Scholar]
- 272.Van Cutsem J., Van Gerven F., Zaman R., Janssen P.A.J. Terconazole – A new broad-spectrum antifungal. Chemotherapy. 1983;29:322–331. doi: 10.1159/000238215. [DOI] [PubMed] [Google Scholar]
- 273.Isaacson D.M., Tolman E.L., Tobia A.J., Rosenthale M.E., McGuire J.L., Bossche H.V., Janssen P.A.J. Selective inhibition of 14α-desmethyl sterol synthesis in Candida albicans by terconazole, a new triazole antimycotic. J. Antimicrob. Chemother. 1988;21:333–343. doi: 10.1093/jac/21.3.333. [DOI] [PubMed] [Google Scholar]
- 274.Heeres J., Backx L.J.J., Mostmans J.H., Van Cutsem J. Antimycotic imidazoles. Part 4. Synthesis and antifungal activity of ketoconazole, a new potent orally active broad-spectrum antifungal agent. J. Med. Chem. 1979;22:1003–1005. doi: 10.1021/jm00194a023. [DOI] [PubMed] [Google Scholar]
- 275.Heeres J., Hendrickx R., Van Cutsem J. Antimycotic Azoles 6. Synthesis and antifungal properties of terconazole, a novel triazole ketal. J. Med. Chem. 1983;26:611–613. doi: 10.1021/jm00358a032. [DOI] [PubMed] [Google Scholar]
- 276.Paretti F. Structure and mechanism of action of teicoplanin. J. Hosp. Infect. 1986;7:79–83. doi: 10.1016/0195-6701(86)90011-3. [DOI] [PubMed] [Google Scholar]
- 277.Parenti F., Beretta G., Berti M., Arioli V. Teichomycins, new antibiotics from actinoplanes teichomyceticus nov. Sp. I. description of the producer strain, fermentation studies and biological properties. J. Antibiot. 1978;31:276–283. doi: 10.7164/antibiotics.31.276. [DOI] [PubMed] [Google Scholar]
- 278.Hunt A.H., Molloy R.M., Occolowitz J.L., Marconi G.G., Debono M. Structure of the major glycopeptide of the teicoplanin complex. J. Am. Chem. Soc. 1984;106:4891–4895. [Google Scholar]
- 279.Brogden R.N., Peters D.H. Teicoplanin. A reappraisal of its antimicrobial activity, pharmacokinetic properties and therapeutic efficacy. Drugs. 1994;47:823–854. doi: 10.2165/00003495-199447050-00008. [DOI] [PubMed] [Google Scholar]
- 280.EMC, Targocid 400mg powder for solution for injection/infusion or oral solution, 2020. https://www.medicines.org.uk/emc/product/2927/smpc (accessed 18 July 2020).
- 281.Parenti F., Schito G.C., Courvalin P. Teicoplanin chemistry and microbiology. J. Chemother. 2000;12:5–14. doi: 10.1080/1120009x.2000.11782312. [DOI] [PubMed] [Google Scholar]
- 282.Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., Zhang H. Glycopeptide antibiotics potently inhibit cathepsin L in the Late endosome/lysosome and block the entry of ebola virus, middle east respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291:9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Andrault P.M., Samsonov S.A., Weber G., Coquet L., Nazmi K., Bolscher J.G., Lalmanach A.C., Jouenne T., Brömme D., Pisabarro M.T., Lalmanach G., Lecaille F. Antimicrobial peptide LL-37is both a substrate of cathepsins S and K and a selective inhibitor of cathepsin L. Biochemistry. 2015;54:2785–2798. doi: 10.1021/acs.biochem.5b00231. [DOI] [PubMed] [Google Scholar]
- 284.J. Zhang, X. Ma, F. Yu, J. Liu, F. Zou, T. Pan, H. Zhang, Teicoplanin potently blocks the cell entry of 2019-nCoV, bioRxiv preprint (2020). 10.1101/2020.02.05.935387. [DOI]
- 285.Baron S.A., Devaux C., Colson P., Raoult D., Rolain M.J. Teicoplanin: an alternative drug for the treatment of coronavirus COVID-19? Int. J. Antimicrob. Agent. 2020;56:105944. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Serafin M.B., Bottega A., Foletto V.S., da Rosa T.F., Hörner A., Hörner R. Drug repositioning is an alternative for the treatment of coronavirus COVID-19. Int. J. Antimicrob. Agent. 2020;55:105969. doi: 10.1016/j.ijantimicag.2020.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yushchuk O., Ostash B., Truman A.W., Marinelli F., Fedorenko V. Teicoplanin biosynthesis: unraveling the interplay of structural, regulatory, and resistance genes. Appl. Microbiol. Biotechnol. 2020;104:3279–3291. doi: 10.1007/s00253-020-10436-y. [DOI] [PubMed] [Google Scholar]
- 288.Boger D.L., Kim S.H., Miyazaki S., Strittmatter H., Weng J.-H., Mori Y., Rogel O., Castle S.L., McAtee J.J. Total synthesis of the teicoplanin aglycon. J. Am. Chem. Soc. 2000;122:7416–7417. doi: 10.1021/ja003835i. [DOI] [PubMed] [Google Scholar]
- 289.Boger D.L., Kim S.H., Mori Y., Weng J.-H., Rogel O., Castle S.L., McAtee J.J. First and second generation total synthesis of the teicoplanin aglycon. J. Am. Chem. Soc. 2001;123:1862–1871. doi: 10.1021/ja003835i. [DOI] [PubMed] [Google Scholar]
- 290.Boger D.L., Miyazaki S., Kim S.H., Wu J.H., Loiseleur O., Castle S.L. Diastereoselective total synthesis of the vancomycin aglycon with ordered atropisomer equilibrations. J. Am. Chem. Soc. 1999;121:3226–3227. [Google Scholar]
- 291.Boger D.L., Miyazaki S., Kim S.H., Wu J.H., Castle S.L., Loiseleur O., Jin Q. Total Synthesis Of The Vancomycin Aglycon. J. Am. Chem. Soc. 1999;121:10004–10011. [Google Scholar]
- 292.Cao B., Hayden F.G. Antiviral monotherapy for hospitalised patients with COVID-19 is not enough. Lancet. 2020 doi: 10.1016/S0140-6736(20)32078-X. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Sahr F., Ansumana R., Massaquoi T.A., Idriss B.R., Sesay F.R., Lamin J.M., Baker S., Nicol S., Conton B., Johnson W., Abiri O.T., Kargbo O., Kamara P., Goba A., Russell J.B.W., Gevao S.M. Evaluation of convalescent whole blood for treating Ebola virus disease in Freetown, Sierra Leone. J. Infect. 2017;74:302–309. doi: 10.1016/j.jinf.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Tian X., Lia C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infec. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wang C., Li W., Drabek D., Okba N.M.A., Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:225. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Y. Wu, F. Wang, C. Shen, W. Peng, D. Li, C. Zhao, Z. Li, S. Li, Y. Bi, Y. Yang, Y. Gong, H. Xiao, Z. Fan, S. Tan, G. Wu, W. Tan, X. Lu, C. Fan, Q. Wang, Y. Liu, C. Zhang, J. Qi, G.F. Gao, F. Gao, L. Liu. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding. [DOI] [PMC free article] [PubMed]
- 298.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Pan X., Zhou P., Fan T., Wu Y., Zhang J., Shi X., Shang W., Fang L., Jiang X., Shi J., Sun Y., Zhao S., Gong R., Chen Z., Xiao G. Immunoglobulin fragment F(ab’)2 against RBD potently neutralizes SARS-CoV-2 in vitro. Antiviral Res. 2020;182:104868. doi: 10.1016/j.antiviral.2020.104868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.L.E.R. Cunha, A.A. Stolet, M.A. Strauch, V.A.R. Pereira, C.H. Dumard, P.N. C. Souza, J.G. Fonseca, F.E. Pontes, L.G.R. Meirelles, J.W.M. Albuquerque, C.Q. Sacramento, N. Fintelman-Rodrigues, T.M. Lima, R.G.F. Alvim, R.B. Zingali, G.A.P. Oliveira, TM.L. Souza, A. Tanuri, A.M.O. Gomes, A.C. Oliveira, H.L.M. Guedes, L.R. Castilho, J.L. Silva. Equine hyperimmune globulin raised against the SARS-CoV-2 spike glycoprotein has extremely high neutralizing titers. bioRxiv preprint. DOI: 10.1101/2020.08.17.254375. [DOI]