Abstract
Background:
We aimed to evaluate the prognostic significance of high expression of the miR-200 family of microRNAs in bladder cancer.
Methods:
Studies on the correlation between the miR-200 family and prognosis in patients with bladder cancer were searched in databases. Combined hazard ratios (HRs) were calculated based on HRs and 95% confidence intervals (CIs) for overall survival (OS), cancer-specific survival (CSS), and recurrence-free survival (RFS). Cochranes Q test and the I2 statistic were utilized to assess heterogeneity across the included studies. Potential publication bias was analyzed by Begg and Egger tests. The meta-analysis was conducted using RevMan 5.3 and Stata SE12.0.
Results:
Data from a total of 1150 patients from 8 studies were extracted. The meta-analysis revealed that high expression of the miR-200 family was correlated with better OS (pooled hazard ratio: 0.50, 95% confidence interval: 0.40–0.62), CSS (pooled hazard ratio: 0.36, 95% confidence interval: 0.22–0.59) and RFS (pooled hazard ratio: 0.48, 95% confidence interval: 0.36–0.65). Both Begg test and Egger test verified no publication bias within the included cohorts.
Conclusion:
The high expression of the miR-200 family is strongly associated with better prognosis in bladder cancer patients, which will improve bladder cancer management in clinical practice.
Keywords: bladder cancer, meta-analysis, miR-200 family, prognosis
1. Introduction
MicroRNAs (miRNAs) are a family of small noncoding RNAs of 18 to 22 nucleotides in length that regulate the expression of their target genes by affecting translation and reducing mRNA stability.[1] By affecting protein translation, miRNAs have been recognized as potent regulators of critical cellular processes, including proliferation, differentiation, apoptosis, stress response, and metabolism.[2,3] miRNAs are implicated in many diseases, including neurologic disorders, heart diseases, vascular diseases, and cancers.[4] miRNAs frequently reside in fragile sites and genomic regions involved in various cancers, suggesting that they play a potentially critical and complex role their pathogenesis.[5] Previous studies have confirmed that the aberrant expression of miRNAs is closely related to the prognosis of cancers.[6,7,8] Therefore, functional miRNAs could be promising prognostic biomarkers for various human cancers.
The tumor suppressor miR-200 family consists of miR-200a, miR-200b, miR-200c, miR-141, and miR-429. These 5 highly homologous members can be divided into 2 gene clusters based on the fact that they are expressed from 2 different polycistronic transcripts. The miR-200b/a/429 cluster is located on chromosome 1p36 and the miR-200c/141 cluster is located on chromosome 12p13.[9] The miR-200 family inhibits epithelial-mesenchymal transition (EMT) by regulating E-cadherin expression via suppression of zinc finger E-box-binding homeobox (ZEB)1 and ZEB2.[10] Previous studies have demonstrated that the miR-200 family is dysregulated in various human cancers and is closely related to the prognosis of cancers, such as colorectal cancer,[11] ovarian cancer,[12] and breast cancer.[13]
Bladder cancer is the second most common urological cancer. In 2018, the estimated numbers of new cases of and deaths from bladder cancer in the USA were 81,190 and 17,240, respectively.[14] Approximately 75% of newly diagnosed bladder cancer cases are non-muscle-invasive bladder cancer (NMIBC) while the remainder are muscle-invasive bladder cancer (MIBC). In NMIBC, patients suffer from a high rate of recurrence and progression. The 5-year recurrence rate of NMIBC ranges from 50% to 70%, and the rate of progression to MIBC in 5 years ranges from 10% to 30%.[15] In MIBC, the critical clinical concern is metastasis due to the high rate of mortality despite improved systemic therapy. There are currently prognostic biomarkers in bladder cancer that have prognostic significance including oncogenes, cell adhesion molecules, cell cycle regulatory proteins, and tumor-associated antigens.[16] However, more reliable prognostic biomarkers are still needed to better understand occurrence and progression of the disease.
Numerous previous studies have explored the prognostic value of the miR-200 family in patients with bladder cancer. However, their findings remain controversial. Thus, a meta-analysis based on the existing literature is needed to evaluate the prognostic significance of expression of the miR-200 family in bladder cancer.
2. Methods
2.1. Ethical statement
This systemic review and meta-analysis does not deal with original human or animal data and was performed according the Preferred Reporting Items for Systemic Reviews and Meta-Analysis (PRISMA) guidelines.[17]
2.2. Search strategy
All relevant studies were searched and identified in PubMed, Embase, Web of Science, and the Cochrane Library (updated on December 10, 2019). The databases were searched using the terms “bladder cancer OR carcinoma OR tumor” AND “miR-200 OR miR-200a OR miR-200b OR miR-200c OR miR-141 OR miR-429” AND “prognosis OR survival OR outcomes”.
2.3. Eligibility criteria
Searching was performed independently by 2 investigators. The titles and abstracts of all the articles were filtered according to the following eligibility criteria in this meta-analysis:
-
1.
the subject of the studies was limited to human beings;
-
2.
the publications were in English;
-
3.
the studies included evaluation of the effect of the miR-200 family on prognosis in bladder cancer; and the studies included HRs and 95% CIs or information to calculate them for prognosis-related outcomes.
2.4. Data extraction
All authors discussed disagreements until consensus was reached. A standardized form including the first authors name, year, country, number of patients enrolled, follow-up time, age, sex, and HRs, 95% CIs and P values was created to extract data from eligible publications. To minimize the risk of confusion for unmeasured values, the estimates were extracted from the largest adjusted model in the articles presenting multiple HRs.
2.5. Assessment of quality
The quality of all the included studies was assessed by 2 investigators independently. For meta-analyses, the Newcastle-Ottawa Scale (NOS) system was developed to assess nonrandomized studies.[18] The quality of the studies in this meta-analysis was evaluated by the NOS system in 3 categories: selection, comparability, and outcome, with 9 stars (Table 1). Those studies with a total score of ≤5 stars, 6–7 stars, and 8–9 stars were considered low quality, intermediate quality, and high quality, respectively. All included studies were of intermediate or high quality.
Table 1.
The Newcastle-Ottawa Scale scores for included studies.
| Items | Wszolek, 2011 | Yun, 2012 | Ratert, 2013 | Pignot, 2013 | Wang, 2015 | Martínez-Fernández, 2015 | Liu, 2018 | Wu, 2018 |
| Selection ☆☆☆☆ | ☆☆☆☆ | ☆☆☆ | ☆☆☆ | ☆☆☆ | ☆☆☆☆ | ☆☆☆ | ☆☆☆ | ☆☆☆☆ |
| Representativeness of the exposed cohort | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Selection of the non-exposed cohort | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Ascertainment of exposure | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Outcome of interest was not present at start of study | ☆ | NA | NA | NA | ☆ | NA | NA | ☆ |
| Comparability ☆☆ | ☆☆ | ☆☆ | ☆☆ | ☆☆ | ☆ | ☆☆ | ☆ | ☆ |
| Outcome ☆☆☆ | ☆☆☆ | ☆☆ | ☆☆☆ | ☆☆☆ | ☆☆☆ | ☆☆ | ☆☆ | ☆☆ |
| Assessment of outcome | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ |
| Follow-up time was long enough for outcomes to occur | ☆ | NA | ☆ | ☆ | ☆ | NA | ☆ | NA |
| Adequacy of follow-up of cohorts | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | NA | ☆ |
| Total scores | 9 | 7 | 8 | 8 | 8 | 7 | 6 | 7 |
NA = not available.
2.6. Statistical analysis
A meta-analysis of OS, CSS, and RFS was performed in this study. HRs with 95% CIs from each article were used to calculate combined HRs. Heterogeneity across the studies was evaluated with Cochranes Q test and Higgins I2 statistic. Those studies with P > .1 and I2 < 50% were considered to have significant heterogeneity. A pooled estimate was calculated with a fixed-effect model or a random-effect model if there was no significant heterogeneity. Potential publication bias was analyzed using Begg funnel plot and Egger linear regression tests. The stability of the results in the included studies was assessed by a sensitivity analysis. A P < .05 (two-tailed) was considered statistically significant. This meta-analysis was performed using RevMan 5.3 and Stata SE12.0 (Stata Corp LP, College Station, TX, USA) according to the PRISMA guidelines.[17]
3. Results
3.1. Characteristics of the studies retrieved
A total of 1152 articles were retrieved from databases, including PubMed, Embase, and the Cochrane Library, and 3 additional studies were identified from reference lists. Eighty one duplicate studies were excluded. A total of 978 publications, including reviews, letters, comments, nonrelevant studies, laboratory studies, and other urinary cancer studies, were removed. After carefully reviewing the remaining 96 articles, 88 articles that failed to evaluate the association between the miR-200 family and prognosis outcomes, had unavailable hazard ratios, were nonhuman studies, had duplicate data, or were published in a non-English language were excluded. Finally, eight cohort studies were analyzed in the meta-analysis. The screening procedure is shown in Figure 1, and the characteristics of the studies are shown in Table 2. These 8 articles were published from 2011 to 2018. Among them, 4 were published before 2015. Four, 3, and 1 of the studies were carried out in Asia, Europe, and America, respectively. The sample size ranged from 40 to 403 patients and a total of 1150 patients were included. The patients median age ranged from 60.1 to 73.0 years, and the percentage of male patients ranged from 66.7% to 83.1%. OS, CSS, and RFS were reported in 6, 2, and 5 articles, respectively.
Figure 1.
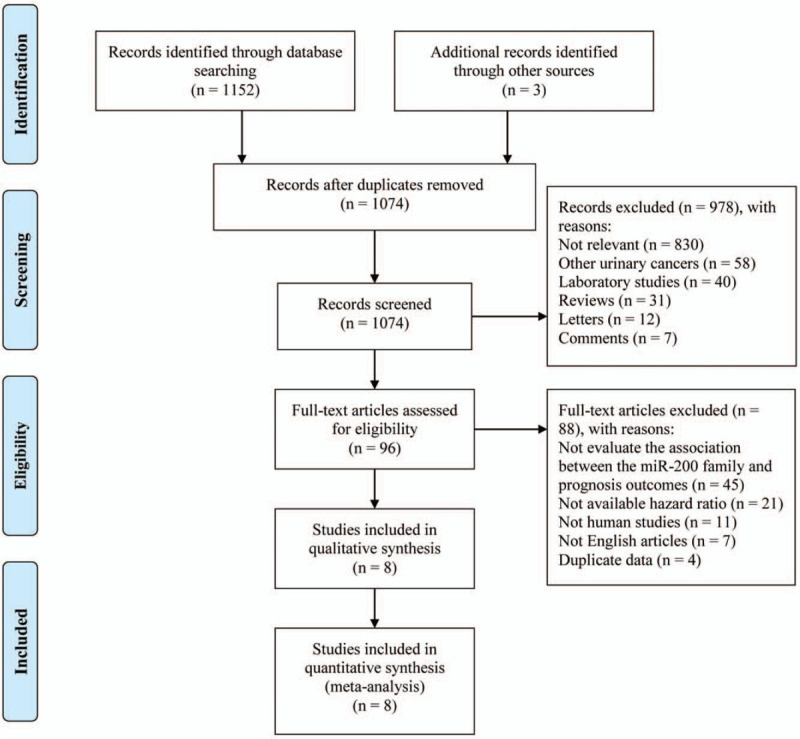
Flow diagram of PRISMA: processes of searching and identification for this review.
Table 2.
Characteristics of included studies.
| Study | Region | No. of patients | Study period | Follow-up (median, months) | Age (years) | Gender (male/female) | miRNA | Survival |
| Wszolek, 2011 | USA | 57 | 1990–2005 | 92 | 66.4 (34–90) | 38/19 | 141, 200 | CSS |
| Yun, 2012 | Korea | 207 | 2006–2012 | 41.3 | 63.5 ± 12.6 | 165/42 | 200 | RFS |
| Ratert, 2013 | Germany | 40 | 1998–2009 | 17 | 69 (50–92) | 32/8 | 141 | OS |
| Pignot, 2013 | France | 166 | 2001–2005 | 30.5 | 70 (31–91) | 138/28 | 200 | OS, RFS |
| Wang, 2015 | China | 114 | NA | 42.9 | 70.0 ± 10.1 | 86/28 | 141 | OS, CSS, RFS |
| Martínez-Fernández, 2015 | Spain | 87 | 2009–2012 | 28.8 | 73.0 (49–90) | 68/19 | 200 | OS, RFS |
| Liu, 2018 | China | 403 | NA | 41.6 | 60.1 (34–90) | 297/106 | 141, 200 | OS |
| Wu, 2018 | China | 76 | 2002–2006 | 36.5 | NA | 57/19 | 429 | OS, RFS |
CSS = cancer-specific survival, NA = not available, OS = overall survival, RFS = recurrence-free survival.
3.2. Survival outcomes
Prognostic outcomes, such as OS, CSS, and RFS, were quantified. The impact of the miR-200 family on OS was investigated in 6 studies with 886 bladder cancer patients. The forest plot showed that high expression of the miR-200 family was associated with better OS (pooled HR: 0.50, 95% CI: 0.40–0.62) and there was no significant heterogeneity in the Cochrane Q test (chi2 = 2.34, P = .89) and I2 test (I2 = 0%) (Fig. 2A). The impact of the miR-200 family on CSS was investigated in 2 studies including 171 patients with bladder cancer. The forest plot showed that high expression of the miR-200 family was associated with better CSS (pooled HR: 0.36, 95% CI: 0.22–0.59) and the Cochrane Q test (chi2 = 0.15, P = .93) and I2 test (I2 = 0%) did not show significant heterogeneity (Fig. 2B). The impact of the miR-200 family on RFS was investigated in 5 studies with 650 bladder cancer patients. The forest plot showed that high expression of the miR-200 family was associated with better RFS (pooled HR: 0.48, 95% CI: 0.36–0.65) and the Cochrane Q test (chi2 = 0.14, P = 1.00) and I2 test (I2 = 0%) did not show significant heterogeneity (Fig. 2C).
Figure 2.
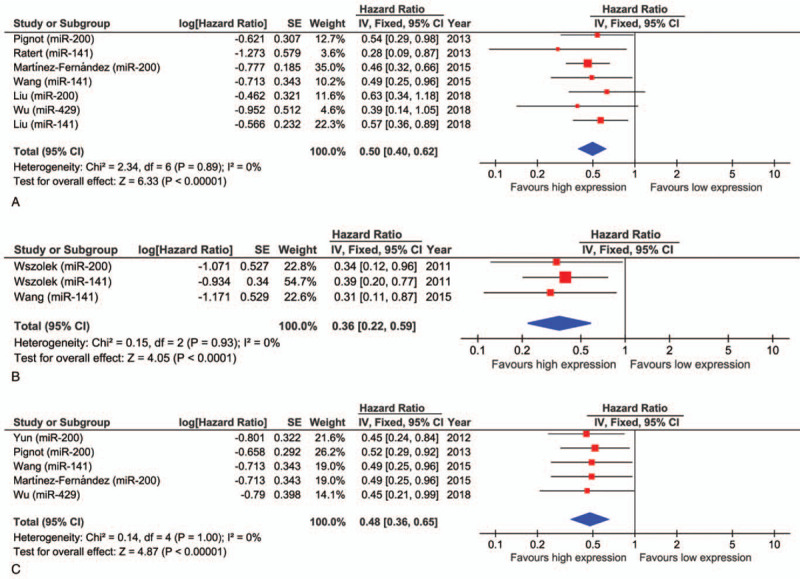
Forest plots demonstrating the effects of the miR-200 family on (A) OS, (B) CSS, and (C) RFS in patients with bladder cancer.
3.3. Subgroup data analysis
The subgroup analysis results based on year of publication, study setting, and NOS score are summarized in Table 3. The pooled HRs for survival outcome stratified by study setting showed that studies performed in Asia were associated with better RFS (pooled HR: 0.46, 95% CI: 0.31–0.69; P = .0001, I2 = 0%) but worse OS (pooled HR: 0.54, 95% CI: 0.40–0.74; P = .0001, I2 = 0%) than those studies in Europe. Pooled HRs for survival outcome revealed that better RFS (pooled HR: 0.46, 95% CI: 0.31–0.69; P = .0001, I2 = 0%) in studies with a NOS score of 7 than in studies with other NOS scores. Due to the limited number of publications, no further subgroup data analysis could be performed on the CSS studies.
Table 3.
Subgroup analysis for the association between the miR-200 family and the survivals.
| OS | RFS | |||||||||
| Heterogeneity | Heterogeneity | |||||||||
| Subgroup analysis | No. of studies | Pooled HR (95% CI) | P value | I2 (%) | P | No, of studies | Pooled HR (95% CI) | P value | I2 (%) | P |
| Overall | 6 | 0.50 (0.40–0.62) | <.01 | 0 | .89 | 5 | 0.48 (0.36–0.65) | <.01 | 0 | 1.00 |
| Study setting | ||||||||||
| Asia | 3 | 0.54 (0.40–0.74) | <.01 | 0 | .85 | 3 | 0.46 (0.31–0.69) | .01 | 0 | .98 |
| Europe | 3 | 0.46 (0.34–0.62) | <.01 | 0 | .61 | 2 | 0.51 (0.33–0.78) | <.01 | 0 | .90 |
| Year of publication | ||||||||||
| Before 2015 | 2 | 0.47 (0.27–0.79) | <.01 | 0 | .32 | 2 | 0.49 (0.32–0.74) | <.01 | 0 | .74 |
| After 2015 | 4 | 0.51 (0.40–0.64) | <.01 | 0 | .87 | 3 | 0.48 (0.32–0.72) | <.01 | 0 | .99 |
| NOS score | ||||||||||
| 6 | 1 | 0.59 (0.41–0.85) | <.01 | 0 | .79 | – | – | – | – | – |
| 7 | 2 | 0.45 (0.32–0.63) | <.01 | 0 | .75 | 3 | 0.46 (0.31–0.69) | <.01 | 0 | .98 |
| 8 | 3 | 0.48 (0.31–0.72) | <.01 | 0 | .61 | 2 | 0.51 (0.33–0.78) | <.01 | 0 | .90 |
CI = confidence interval, HR = hazard ratio, NOS = Newcastle-Ottawa Scale, OS = overall survival, RFS = recurrence-free survival.
3.4. Publication bias
Begg test and Egger test were performed to determine the publication bias in this meta-analysis. Funnel plots based on OS were determined by Begg test (Fig. 3A) and Egger test (Fig. 3B). Funnel plots based on CSS were determined by Begg test (Fig. 3C) and Egger test (Fig. 3D). The funnel plots of Begg test and Egger test based on RFS were shown in Figure 3E and F, respectively. The results of Begg and Egger tests are summarized in Table 4. Both tests verified that there was no publication bias within the included cohorts.
Figure 3.
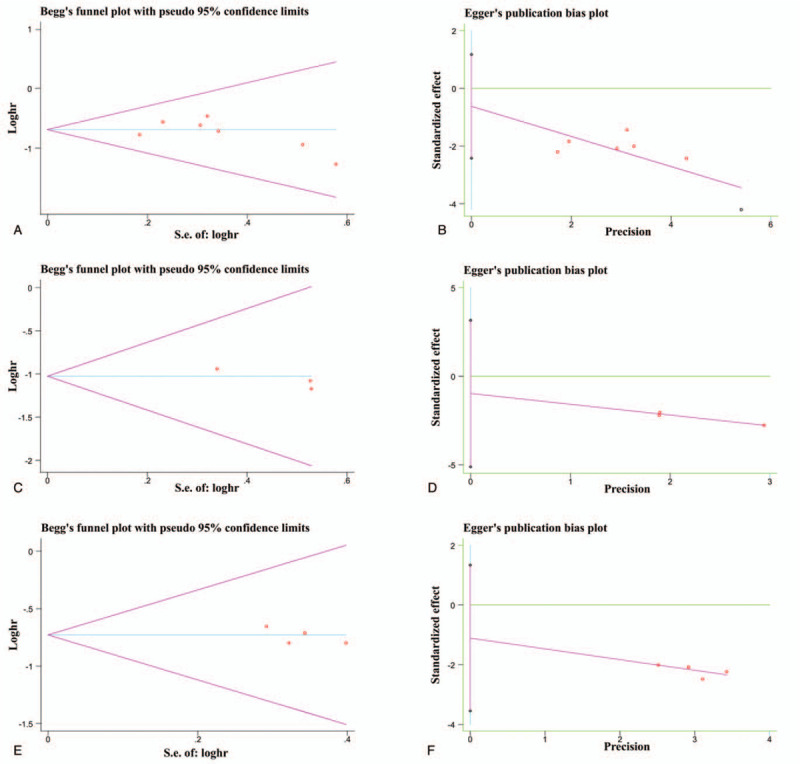
Funnel plots based on OS determined by (A) Begg test and (B) Egger test; based on CSS determined by (C) Begg test and (D) Egger test; and based on RFS determined by (E) Begg test and (F) Egger test.
Table 4.
Outcomes of Begg's and Egger's tests.
| Begg test | Egger test | |||||
| Test | OS | CSS | RFS | OS | CSS | RFS |
| p value | 0.368 | 0.296 | 0.624 | 0.413 | 0.205 | 0.244 |
CSS = cancer-specific survival, OS = overall survival, RFS = recurrence-free survival.
3.5. Sensitivity analysis
Sensitivity analysis was conducted to assess the stability of the studies and minimize the effect of individual research on conclusions. The included studies were sequentially omitted to assess whether any single study could have an impact on OS (Fig. 4A), CSS (Fig. 4B), or RFS (Fig. 4C). The sensitivity analysis suggested that the exclusion of any study did not alter the pooled results.
Figure 4.
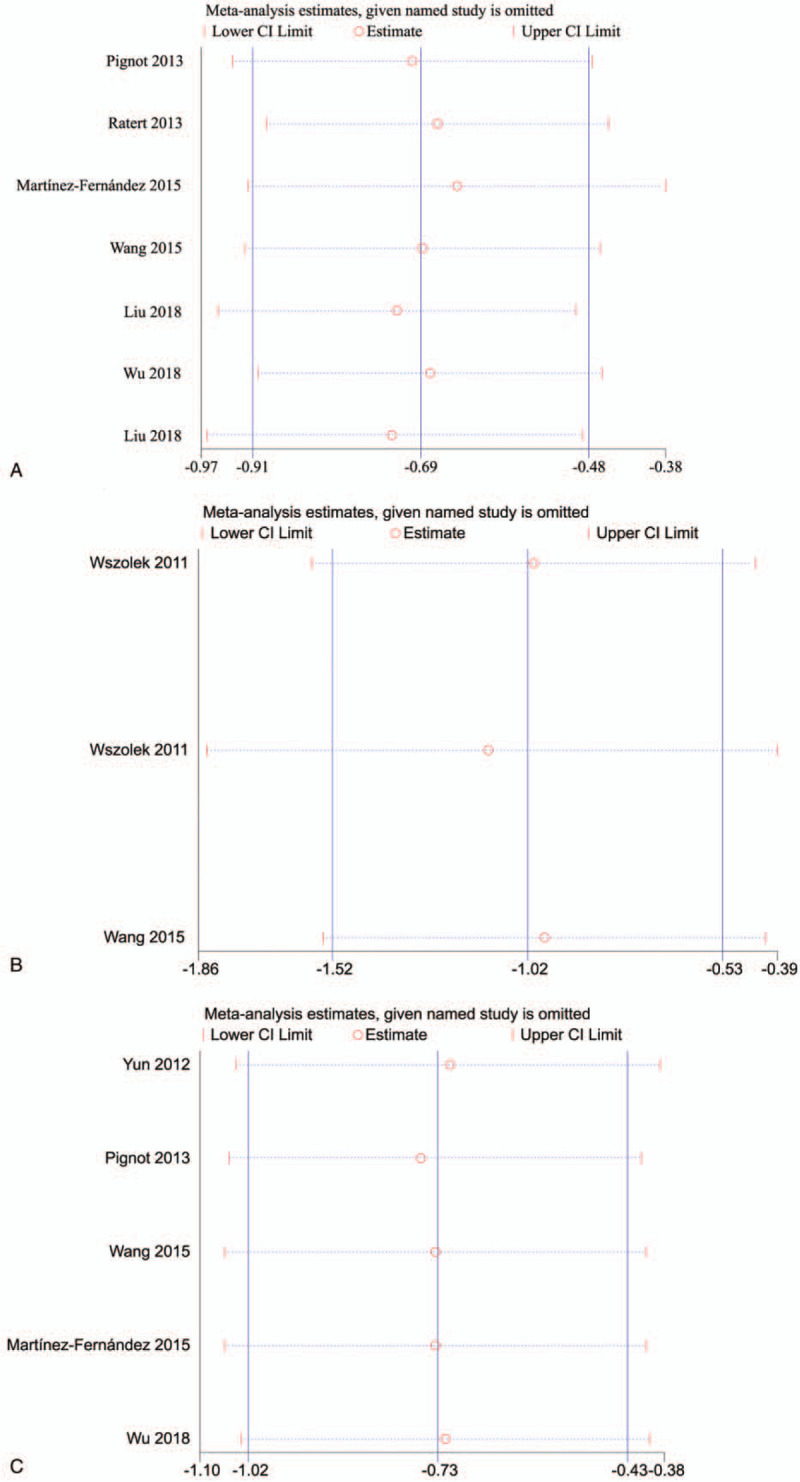
Sensitivity analysis for this meta-analysis. (A) Sensitivity analysis for OS; (B) sensitivity analysis for CSS; and (C) sensitivity analysis for RFS.
4. Discussion
Recently, many new prognostic biomarkers have been identified in bladder cancer, including the modified Glasgow Prognostic Score (mGPS),[19] circulating tumor cells (CTCs),[20] neutrophil/lymphocyte ratio (NLR)[21] and the combination of NLR, platelet/lymphocyte ratio (PLR), and lymphocyte/monocyte ratio (LMR),[22] and long noncoding RNAs.[23] The prognostic role of mGPS is strongly associated with RFC in urothelial bladder cancer patients.[19] CTCs are a predictor for progression and recurrence in NMIBC.[20] NLR and the combination of NLP, PLR, and LMR are prognostic markers for risk stratification in NMIBC.[21,22] The diagnostic and prognostic roles of long noncoding RNAs have been validated in bladder malignancies.[23] There is increasing evidence that miRNAs play important roles in tumorigenesis and cancer progression and are closely related to multiple cellular processes such as the cell cycle, angiogenesis, invasion, metastasis, and innate and adaptive immune responses.[24] To date, significant differences in miRNA expression have been observed in a variety of cancers analyzed by profiling and next-generation sequencing technologies.[25] Therefore, miRNAs have been considered novel potential biomarkers for cancer.
Recent studies have shown that members of the miR-200 family are involved in cancer progression and invasion. Cancer progression is closely related to the dynamic EMT process, during which epithelial cells lose their cell polarity and cell-cell adhesion and acquire the ability to migrate and invade by downregulating E-cadherin and upregulating vimentin expression.[24] It has been reported that members of the miR-200 family regulate EMT by targeting ZEB1 and ZEB2, members of the zinc-finger E-box binding homeobox family, resulting in dysregulation of E-cadherin.[26,27,28] The miR-200 members inhibit ZEB at the posttranscriptional level by binding to a highly conserved target site in the 3’-UTR. The functional relationship between ZEB factors and the miR-200 family in the double-negative feedback loop is called the ZEB/miR-200 feedback loop.[29] Several other tumor suppressor genes have also been reported to be potential targets for the miR-200 family, including BRD7, BAP1, GATA, CLOCK, and PTPN12.[30,31] Furthermore, miR-200 inhibits the self-renewal and differentiation of cancer stem cells, regulates cell division and apoptosis, and reverses chemoresistance.[32]
Previous studies have focused on the relationship between the miR-200 family and tumor prognosis, especially in ovarian cancer. A meta-analysis performed by Shi and Zhang that included 7 ovarian cancer-related studies,[12] and they found that high expression of the miR-200 family was associated with improved OS (HR = 0.34, 95% CI 0.20–0.58, P < .01) and progression-free survival (PFS) (HR = 0.64, 95% CI 0.50–0.82, P < .01). Another meta-analysis by Shi et al also found that high expression of the miR-200 family predicted better ovarian cancer prognosis than the low expression of the miR-200 family (OS: HR = 0.78, 95% CI 0.64–0.94, P = .01).[33]
Our study is the first meta-analysis to pool available data to evaluate the prognostic significance of the miR-200 family in bladder cancer. There was no significant heterogeneity in this meta-analysis. Our results suggest that high expression of the miR-200 family may predict improved survival of patients with bladder cancer. Subgroup analysis and sensitivity analysis showed that the results of this study were stable and reliable. The influence of the miR-200 family on the prognosis of bladder cancer may be explained by the potential inhibitory effect that these microRNAs have on EMT by targeting of ZEB1 and ZEB2.[26,34,35] Therefore, patients and clinicians may benefit from a better understanding of the protective role of the miR-200 family in bladder cancer. However, the included studies in this meta-analysis were retrospective, resulting in selection bias, and extensive population studies may be needed for further validation.
5. Conclusion
This meta-analysis demonstrates that high expression of the miR-200 family may predict better prognosis for patients with bladder cancer, which will improve bladder cancer management in clinical practice.
Author contributions
Conceptualization: Yanhui Mei, Jianbo Zheng, Yidong Fan.
Data curation: Yanhui Mei, Jianbo Zheng, Ping Xiang, Cheng Liu.
Formal analysis: Yanhui Mei, Jianbo Zheng, Ping Xiang, Yidong Fan.
Funding acquisition: Yidong Fan.
Investigation: Yanhui Mei, Jianbo Zheng, Ping Xiang, Cheng Liu, Yidong Fan.
Methodology: Yanhui Mei, Jianbo Zheng, Ping Xiang, Cheng Liu.
Resources: Jianbo Zheng.
Supervision: Yidong Fan.
Validation: Yanhui Mei, Yidong Fan.
Writing – original draft: Yidong Fan.
Writing – review & editing: Yanhui Mei, Jianbo Zheng, Ping Xiang, Cheng Liu, Yidong Fan.
Footnotes
Abbreviations: CIs = confidence intervals, CSS = cancer-specific survival, CTCs = circulating tumor cells, EMT = epithelial-mesenchymal transition, HRs = hazard ratios, LMR = lymphocyte/monocyte ratio, mGPS = modified Glasgow Prognostic Score, NLR = neutrophil/lymphocyte ratio, NMIBC = non-muscle-invasive bladder cancer, NOS = Newcastle-Ottawa Scale, OS = overall survival, PLR = platelet/lymphocyte ratio, RFS = recurrence-free survival, ZEB = zinc finger E-box-binding homeobox.
How to cite this article: Mei Y, Zheng J, Xiang P, Liu C, Fan Y. Prognostic value of the miR-200 family in bladder cancer: a systematic review and meta-analysis. Medicine. 2020;99:47(e22891).
YM and JZ contributed equally to this work.
This study was supported by the Medical and Health Science and Technology Development Project of Shandong Province, China (grant no.2017WS687).
The authors declare no conflicts of interest.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005;433:769–73. [DOI] [PubMed] [Google Scholar]
- [2].Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genes Dev 2005;15:200–5. [DOI] [PubMed] [Google Scholar]
- [3].Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature 2008;451:414–6. [DOI] [PubMed] [Google Scholar]
- [4].Hammond SM. An overview of microRNAs. Adv Drug Deliv rev 2015;87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yuan Y, Xu XY, Zheng HG, et al. Elevated miR-21 is associated with poor prognosis in non-small cell lung cancer: a systematic review and meta-analysis. Eur rev Med Pharmacol Sci 2018;22:4166–80. [DOI] [PubMed] [Google Scholar]
- [7].Huang Q, Song Q, Zhong W, et al. MicroRNA-10b and the clinical outcomes of various cancers: a systematic review and meta-analysis. Clin Chim Acta 2017;474:14–22. [DOI] [PubMed] [Google Scholar]
- [8].Zhou J, Dai W, Song J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun 2016;470:445–52. [DOI] [PubMed] [Google Scholar]
- [9].Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol 2008;5:115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi PW, Ng SW. The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. Intl J Mol Sci 2017;18:pii:E1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Peng Z, Zhu W, Dai J, et al. MicroRNA-200 as potential diagnostic markers for colorectal cancer: meta-analysis and experimental validation. Cell Mol Biol (Noisy-le-Grand, France) 2018;64:77–85. [PubMed] [Google Scholar]
- [12].Shi C, Zhang Z. The prognostic value of the miR-200 family in ovarian cancer: a meta-analysis. Acta Obestet Gynecol Scand 2016;95:505–12. [DOI] [PubMed] [Google Scholar]
- [13].Li X, Roslan S, Johnstone CN, et al. MiR-200 can repress breast cancer metastasis through ZEB1-independent but moesin-dependent pathways. Oncogene 2014;33:4077–88. [DOI] [PubMed] [Google Scholar]
- [14].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [15].Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet (London, England) 2016;388:2796–810. [DOI] [PubMed] [Google Scholar]
- [16].Ku JH, Kim WJ, Lerner SP, et al. Diagnostic and prognostic markers in bladder cancer. Dis Markers 2016;2016:2425091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Knobloch K, Yoon U, Vogt PM. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J Cranio Maxillofac Surg 2011;39:91–2. [DOI] [PubMed] [Google Scholar]
- [18].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [19].Ferro M, Cobelli O, Buonerba C, et al. Modified glasgow prognostic score is associated with risk of recurrence in bladder cancer patients after radical cystectomy: a multicenter experience. Medicine 2015;94:e1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Busetto GM, Ferro M, Giudice FD, et al. The prognostic role of circulating tumor cells (CTC) in high-risk non-muscle-invasive bladder cancer. Clin Genitourin Cancer 2017;15:e661–6. [DOI] [PubMed] [Google Scholar]
- [21].Vartolomei MD, Ferro M, Cantiello F, et al. Validation of Neutrophil-to-lymphocyte Ratio in a Multi-institutional Cohort of Patients With T1G3 Non-muscle-invasive Bladder Cancer. Clin Genitourin Cancer 2018;16:445–52. [DOI] [PubMed] [Google Scholar]
- [22].Cantiello F, Russo IG, Vartolomei MD, et al. Systemic inflammatory markers and oncologic outcomes in patients with high-risk non-muscle-invasive urothelial bladder cancer. Eur Urol Oncol 2018;1:403–10. [DOI] [PubMed] [Google Scholar]
- [23].Terracciano D, Ferro M, Terreri S, et al. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl Res 2017;184:108–17. [DOI] [PubMed] [Google Scholar]
- [24].Hong L, Han Y, Li S, et al. The malignant phenotype-associated microRNA in gastroenteric, hepatobiliary and pancreatic carcinomas. Expert Opin Biol Ther 2010;10:1693–701. [DOI] [PubMed] [Google Scholar]
- [25].Zhang L, Wei P, Shen X, et al. MicroRNA expression profile in penile cancer revealed by next-generation small RNA sequencing. PloS One 2015;10:e0131336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- [27].Vilming Elgaaen B, Olstad OK, Haug KB, et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer 2014;14:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Davalos V, Moutinho C, Villanueva A, et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2012;31:2062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009;11:1487–95. [DOI] [PubMed] [Google Scholar]
- [30].Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res 2007;67:8699–707. [DOI] [PubMed] [Google Scholar]
- [31].Park YA, Lee JW, Choi JJ, et al. The interactions between MicroRNA-200c and BRD7 in endometrial carcinoma. Gynecol Oncol 2012;124:125–33. [DOI] [PubMed] [Google Scholar]
- [32].Feng X, Wang Z, Fillmore R, et al. MiR-200, a new star miRNA in human cancer. Cancer Lett 2014;344:166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shi M, Mu Y, Zhang H, et al. MicroRNA-200 and microRNA-30 family as prognostic molecular signatures in ovarian cancer: a meta-analysis. Medicine 2018;97:e11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Korpal M, Lee ES, Hu G, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008;283:14910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]


