Abstract
We have developed a deep learning-based approach to improve image quality of single-shot turbo spin-echo (SSTSE) images of female pelvis. We aimed to compare the deep learning-based single-shot turbo spin-echo (DL-SSTSE) images of female pelvis with turbo spin-echo (TSE) and conventional SSTSE images in terms of image quality.
One hundred five and 21 subjects were used as training and test sets, respectively. We performed 6-fold cross validation. In the training process, low-quality images were generated from TSE images as input. TSE images were used as ground truth images. In the test process, the trained convolutional neural network was applied to SSTSE images. The output images were denoted as DL-SSTSE images. Apart from DL-SSTSE images, classical filtering methods were adopted to SSTSE images. Generated images were denoted as F-SSTSE images. Contrast ratio (CR) of gluteal fat and myometrium and signal-to-noise ratio (SNR) of gluteal fat were measured for all images. Two radiologists graded these images using a 5-point scale and evaluated the image quality with regard to overall image quality, contrast, noise, motion artifact, boundary sharpness of layers in the uterus, and the conspicuity of the ovaries. CRs, SNRs, and image quality scores were compared using the Steel-Dwass multiple comparison tests.
CRs and SNRs were significantly higher in DL-SSTSE, F-SSTSE, and TSE images than in SSTSE images. Scores with regard to overall image quality, contrast, noise, and boundary sharpness of layers in the uterus were significantly higher on DL-SSTSE and TSE images than on SSTSE images. There were no significant differences in the CRs, SNRs, and respective scores between DL-SSTSE and TSE images. The score with regard to motion artifacts was significantly higher on DL-SSTSE, F-SSTSE, and SSTSE images than on TSE images. The score with regard to the conspicuity of ovaries was significantly higher on DL-SSTSE images than on F-SSTSE, SSTSE, and TSE images (P < .001).
DL-SSTSE images showed higher image quality as compared with SSTSE images. In comparison with conventional TSE images, DL-SSTSE images had acceptable image quality while keeping the advantage of the motion artifact-robustness and acquisition time efficiency in SSTSE imaging.
Keywords: artificial intelligence, convolutional neural network, deep learning, magnetic resonance imaging, single-shot
1. Introduction
Magnetic resonance imaging (MRI) is an established noninvasive method of diagnostic imaging. Compared with computed tomography (CT), MRI provides superior soft tissue contrast and is free of ionizing radiation. In gynecological MRI, turbo spin-echo (TSE) imaging has been mainly used for pelvic MRI protocols.[1–5] However, TSE requires a long imaging time. Along with the TSE imaging, single-shot TSE (SSTSE) imaging has been developed to reduce acquisition time and motion artifacts.[6] SSTSE images have a relatively poor signal-to-noise ratio (SNR) and blurring compared with TSE images, because all of the signal data in k-space is obtained after a single radiofrequency pulse and the echo train length is extended.[7] SSTSE imaging techniques are routinely used in clinical MRI of the abdomen.[7,8] Because a trade-off exists between spatial resolution and SNR in MRI, long acquisition time is required for magnetic resonance (MR) images with high spatial resolution and SNR. To solve this issue, the obtainment of MR images keeping the advantages of TSE in image quality and of SSTSE in time efficiency has been investigated for years.
Recently, convolutional neural networks (CNN) have been widely applied for image segmentation, classification, recognition, and image super-resolution.[9–14] Noise reduction techniques using CNN have also been proposed in the field of medical imaging.[15] Several studies have shown that deep learning-based super-resolution or denoise approaches were successfully applied to low-quality MR images to shorten imaging time.[16,17] However, few studies have shown the impact of the deep learning-based approaches that improve the image quality of SSTSE image. High-quality SSTSE images have the potential to be a good substitute to TSE images. In addition, although several deep learning-based approaches have already been proposed for brain MR images and the preliminary results were encouraging,[13] applying the approaches to pelvic or abdominal MR images is still challenging due to the respiratory or peristalsis motion.
In the present study, we have developed a CNN-based approach to improve image quality of SSTSE images of the female pelvis obtained in a shorter imaging time. Thus, we aimed to compare deep learning-based single-shot turbo spin-echo (DL-SSTSE) images of the female pelvis with conventional TSE images and SSTSE images in terms of image quality including contrast, noise, motion artifact, and the conspicuity of the uterus and ovary.
2. Materials and methods
2.1. Patients
This retrospective study was approved by the institutional review board and the need to obtain informed consent was waived. A total of 182 women with suspected uterus or ovarian diseases who underwent MRI of pelvis between April 2018 and March 2019 were included in this study. If patients underwent MRI several times during the period, only the images obtained in the first time were used. Patients with no T2-weighted SSTSE sagittal images (n = 53), or TSE sagittal images (n = 3) acquired were excluded. Among the remaining 126 patients (mean age 50.9 ± 15.4 years), 105 subjects were used as a training set, and 21 subjects were randomly used as a test set. We performed 6-fold cross-validation to confirm the performance consistency. Clinical characteristics of the patients are described in Table 1.
Table 1.
Clinical characteristics for patient groups.
| Characteristics | Patients (n = 126) |
| Age (y)∗ | 50.9 ± 15.4 |
| Disease | |
| Ovarian tumor | 42 |
| Leiomyoma | 39 |
| Leiomyosarcoma | 1 |
| Endometrial cancer | 8 |
| Endometriosis | 7 |
| Cervical intraepithelial neoplasia | 5 |
| Endometrial polyp | 4 |
| Cervical cancer | 6 |
| Adenomyosis | 2 |
| Ectopic pregnancy | 2 |
| Gestational trophoblastic disease | 2 |
| Endometrial hyperplasia | 2 |
| Uterine prolapse | 1 |
| Vulvar tumor | 1 |
| Inguinal tumor | 1 |
| Ovarian hemorrhage | 1 |
| Precocious puberty | 1 |
| Placental Remnant | 1 |
Data are represented as means ± standard deviation.
2.2. MRI protocol
A 1.5-T MRI system (Prodiva 1.5T; Philips Healthcare, Best, the Netherlands) with dS-Torso Cardiac coil and dS-NVS Spine coil were used. After acquisition of the scout images, 2-dimensional T2-weighted SSTSE sagittal images were acquired as the detailed scout images, and 2-dimensional T2-weighted TSE sagittal images were acquired. The acquisition parameters for SSTSE images were as follows: TR, 2000 ms; TE, 100 ms; FOV, 250 − 320 × 250 − 320 mm; acquisition matrix, 320 × 160; reconstruction matrix, 512 × 512; acceleration factor for parallel imaging, 2.0; slices, 28–38; slice thickness/gap, 4.0/0.8 mm; fold-over direction, AP; echo-train length, 57; bandwidth, 277.0–289.4 Hz/pixel; and imaging time, 56 s−1 minutes 16 seconds. Half-Fourier imaging technique was used. The acquisition parameters for TSE images were as follows: TR, 3809−5166 ms; TE, 100 ms; FOV, 250−320 × 250−320 mm; acquisition matrix, 320 × 320; reconstruction matrix, 512 × 512; acceleration factor for parallel imaging, 2.0; slices, 28−38; slice thickness/gap, 4.0/0.8 mm; echo-train length, 20; bandwidth, 273.2 Hz/pixel; and imaging time, 3 minutes 11 s−4 min 18 seconds. MultiVane technique, which has been developed as periodically rotated overlapping parallel lines with enhanced reconstruction technique, was used for robust motion correction.[18]
2.3. Data preparation
TSE images were used as ground truth images. As input, low-quality images generated from TSE, instead of SSTSE images, were used in the dataset. Although TSE images were obtained with a motion artifact suppression technique, the images included peristalsis artifacts.[19] Misregistration is always randomly caused by peristalsis between SSTSE and TSE images even if registration techniques are applied.[20,21] Hence, it is difficult to optimize the training dataset of SSTSE and TSE images and accuracy cannot be gained. Instead, input images were generated from TSE images to simulate SSTSE images by contrast-adjustment, downsampling, adding noise, and blurring images with image processing software (ImageJ version 1.52a; National Institutes of Health, Bethesda, MD) as performed in several studies.[13,22]
In the contrast-adjustment process, the intensity of each pixel on the TSE image was transformed corresponding to the following equation:
where Snew is the post-transformed intensity of a pixel, γ is a slope which controls linear-transformation of a pixel value, Sold is the pretransformed intensity of a pixel, and Smax is the maximum intensity of all pixels. Specifically, 0.85 for γ was applied to equalize the image contrasts of postprocessed images to those of SSTSE images. Then, images were downsampled to half resolution in the phase encode direction by the nearest neighbor method, and a Gaussian noise with a standard deviation of 100 was added as an image noise. Finally, a Gaussian filter with a kernel of a standard deviation of 1 voxel was applied for image blurring.
2.4. Convolutional neural network algorithm and training process
The network algorithm based on U-Net is shown in Figure 1. U-Net architecture has been successfully applied for various biomedical imaging applications including synthetic image generation.[23,24] It includes a contraction part (left side) and an expansion part (right side). The contraction part is composed of 4 sequential stacks. Each stack includes 2 convolutional layers with a size of 3 × 3, where a batch normalization (BN) layer and a parametric rectified linear unit (PReLU) layer were followed by each convolutional layer. In addition, a 2 × 2 max pooling layer with stride 2 was followed for downsampling. The number of feature channels is doubled after each downsampling step. The expansion part is composed of a deconvolutional layer with a size of 2 × 2 and stride 2 for up-sampling, which halves the number of feature maps, a concatenation layer with the corresponding cropped feature maps from the contraction part, and 2 convolutional layers with a size of 3 × 3, where a BN layer and a PReLU layer were followed by each convolutional layer. At the final layer a convolutional layer with a size of 3 × 3 is used to convolute 64 feature maps to 1 feature map. To optimize the parameters of the network, we used mean squared error as the loss function to minimize the loss value between the generated high-quality image and the ground truth TSE image. Adam with α = 0.001, β1 = 0.9, β2 = 0.999 was used as optimizer. Total epochs, batch size, and computation time were 100, 2, and about 6 hours, respectively. All the CNN algorithms were implemented with the deep learning platform (Neural Network Console; Sony Network Communications, Tokyo, Japan) and NVIDIA Tesla V100 GPU.
Figure 1.

Architecture of the U-Net-based CNN scheme. In the contraction part, the low-quality images generated from TSE images were used as input images. Each blue box includes a convolutional layer with a size of 3 × 3, a BN layer, and a PReLU layer. Each purple box represents a max pooling layer for downsampling. Each white box represents a concatenate layer. Each green box represents a deconvolution layer. A yellow box represents a final convolutional layer. The TSE images were used as ground truth images. The image size and the number of feature channels from each convolutional layer are listed near each blue box. In the test process, the trained CNN model was applied to the datasets of SSTSE images, which were used as input images. The output images were denoted as DL-SSTSE images. BN = batch normalization layer, CNN = convolutional neural network, Conv = convolutional layer, DL-SSTSE = deep learning-based single-shot turbo spin-echo, PReLU = parametric rectified linear unit, SSTSE = single-shot turbo spin-echo, TSE = turbo spin-echo.
2.5. Test process
The trained CNN model was applied to the dataset of input SSTSE images. The output images generated by the trained CNN model were denoted as DL-SSTSE images.
2.6. Image enhancement by filtering algorithms
Apart from the CNN algorithm, classical filtering methods were adopted to SSTSE images to enhance the image quality. For smoothing and sharpening, the median filter of radius 2.0 pixel and unsharp masking of radius 1.5 pixel were adopted to the images. Generated images were denoted as F-SSTSE images. For contrast-adjustment, the intensity of each pixel on the image was transformed corresponding to the equation described as section 2.3. Specifically, 1.18 for γ was applied to equalize the image contrasts of postprocessed images to those of TSE images.
2.7. Image analysis
2.7.1. Quantitative analysis
The contrast ratio (CR) and SNR were measured to assess the qualities of SSTSE, DL-SSTSE, F-SSTSE, and TSE images. CRs of gluteal fat and myometrium were measured using the following equation:
where SIglutealfat is the mean signal intensity (SI) of the region of interest (ROI) in gluteal fat (mean area 2.17 cm2 ± 1.47, range 0.44–6.30 cm2) and SImyometrium is the SI of the ROI in the myometrium (mean area 0.54 cm2 ± 0.20, range 0.20–1.03 cm2). When placing the ROIs, care was taken to avoid areas of possible pathological findings.
The usual SNR measurements could not be performed because the parallel imaging technique was used.[25] Thus, estimated SNRs of gluteal fat were measured using the following equation:
where SIglutealfat is the mean SI of ROI in gluteal fat and SDglutealfat is the standard deviation (SD) of the SI in the same ROI.
2.7.2. Qualitative analysis
Two radiologists (OY and NO, with 28 years and 12 years of experience in genitourinary MRI, respectively) reviewed T2-weighted SSTSE, DL-SSTSE, F-SSTSE, and TSE images in a random order. Because MultiVane technique was used in TSE imaging, wrap-around artifacts occurred and the FOV of TSE images became round shaped.[18] Therefore, parts of 320 × 320 pixel size were extracted from the center of all images before reviewing.
First, the radiologists independently graded images using a 5-point scale to assess image quality based on 6 separate categories: overall image quality, contrast, noise, motion artifacts, boundary sharpness of the zonal layers in the uterine corpus, and conspicuity of ovaries and follicles. Overall image quality, contrast, and noise were rated as follows: 1 = unacceptable, 2 = poor, 3 = mild, 4 = good, and 5 = excellent. Motion artifact was rated as follows: 1 = marked, 2 = moderate, 3 = mild, 4 = minimal, and 5 = absent. Boundary sharpness of the zonal layers in the uterine corpus and conspicuity of ovaries and follicles were rated as follows: 1 = unable to see, 2 = blurry but visualized, 3 = acceptable, 4 = good, and 5 = excellent. After the independent evaluation, discrepant scores were reconciled by consensus and scores were determined. In the evaluation of ovaries and follicles, 7 patients were excluded because of unclear detection in any sagittal images.
2.8. Statistical analysis
The interobserver agreement for independent image quality scores was evaluated with weighted κ statistics.[26] A weighted kappa value was considered to indicate as follows: poor agreement; 0.01 to 0.20, slight; 0.21 to 0.40, fair; 0.41 to 0.60, moderate; 0.61 to 0.80, good; and 0.81 to 1.00, excellent. CRs, SNRs, and image quality scores were compared using the Steel-Dwass multiple comparison tests,[27] because the samples did not have equal variances by Bartlett test. Statistical analysis was performed with statistical software (R version 3.3.2; R Foundation for Statistical Computing, Vienna, Austria). A P value < .05 was considered statistically significant.
3. Results
3.1. Quantitative analysis
Median CRs of gluteal fat and myometrium in SSTSE, DL-SSTSE, F-SSTSE, and TSE images are shown in Figure 2. CRs were significantly higher in DL-SSTSE (0.39 vs 0.30; P < .001), F-SSTSE (0.39 vs 0.30; P < .001), and TSE images (0.39 vs 0.30; P < .001) than in SSTSE images. There were no significant differences in the CRs between DL-SSTSE and TSE images (P = .98). There were also no significant differences in the CRs between F-SSTSE and TSE images (P = .99).
Figure 2.
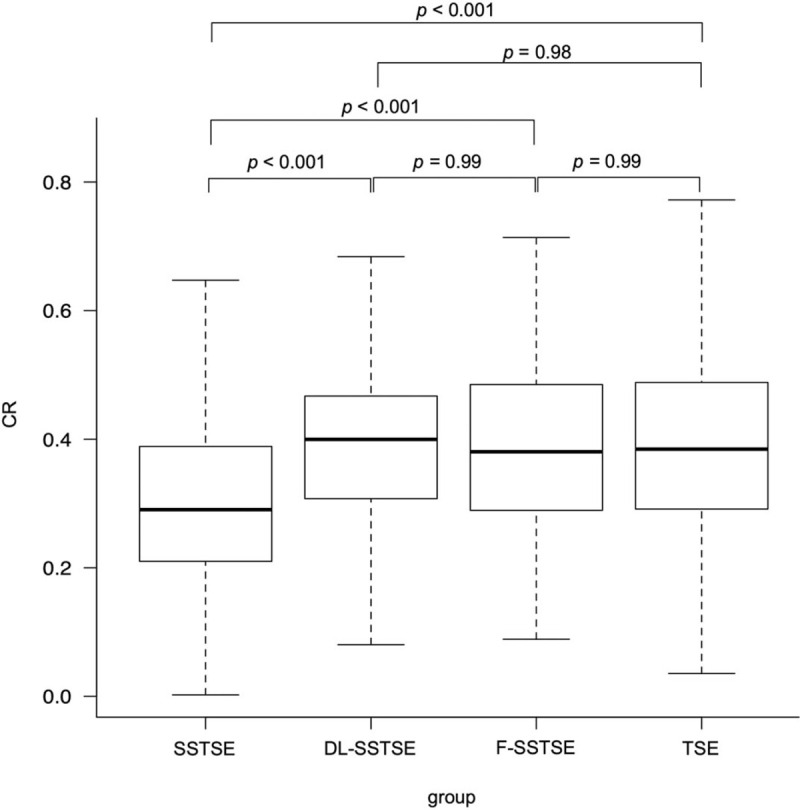
CRs of gluteal fat and myometrium. The median CRs for SSTSE, DL-SSTSE, F-SSTSE, and TSE images were 0.30 (0.21–0.39), 0.39 (0.31–0.47), 0.39 (0.29–0.48), and 0.39 (0.29–0.48), respectively. CRs of gluteal fat and myometrium were significantly higher in DL-SSTSE (P < .001), F-SSTSE (P < .001), and TSE images (P < .001) than in SSTSE images. CR = contrast ratio, DL-SSTSE = deep learning-based single-shot turbo spin-echo, F-SSTSE = filtering method-based single-shot turbo spin-echo, SSTSE = single-shot turbo spin-echo, TSE = turbo spin-echo.
Median SNRs of gluteal fat in SSTSE, DL-SSTSE, F-SSTSE, and TSE images are shown in Figure 3. SNRs were significantly higher in DL-SSTSE (31.91 vs 22.54; P < .001), F-SSTSE (29.32 vs 22.54; P < .001), and TSE images (29.04 vs 22.54; P < .001) than in SSTSE images. There were no significant differences in the SNRs between DL-SSTSE and TSE images (P = .11). There were also no significant differences in the SNRs between F-SSTSE and TSE images (P = .99).
Figure 3.
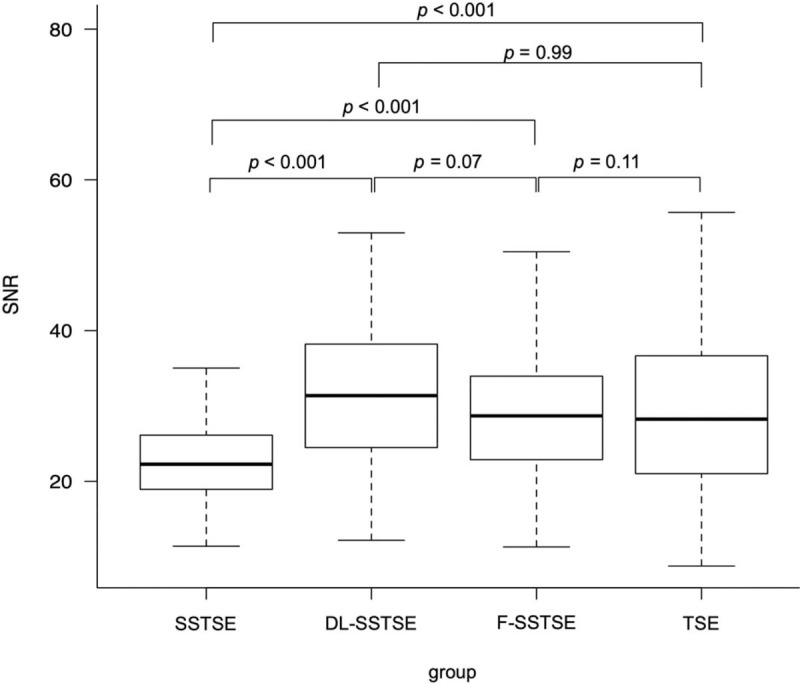
SNRs of gluteal fat. The median SNRs for SSTSE, DL-SSTSE, F-SSTSE, and TSE images were 22.54 (18.93–26.12), 31.91 (24.49–38.19), 29.32 (22.89–33.67), and 29.04 (21.03–36.61), respectively. SNRs of gluteal fat were significantly higher in DL-SSTSE (P < .001), F-SSTSE (P < .001), and TSE images (P < .001) than in SSTSE images. DL-SSTSE = deep learning-based single-shot turbo spin-echo, F-SSTSE = filtering method-based single-shot turbo spin-echo, SNR = signal-to-noise ratio, SSTSE = single-shot turbo spin-echo, TSE = turbo spin-echo.
3.2. Qualitative analysis
The representative DL-SSTSE images, along with the ground truth TSE, F-SSTSE, and SSTSE images are shown in Figure 4. Our proposed CNN algorithm improved the image quality of uterus and ovary in SSTSE images.
Figure 4.
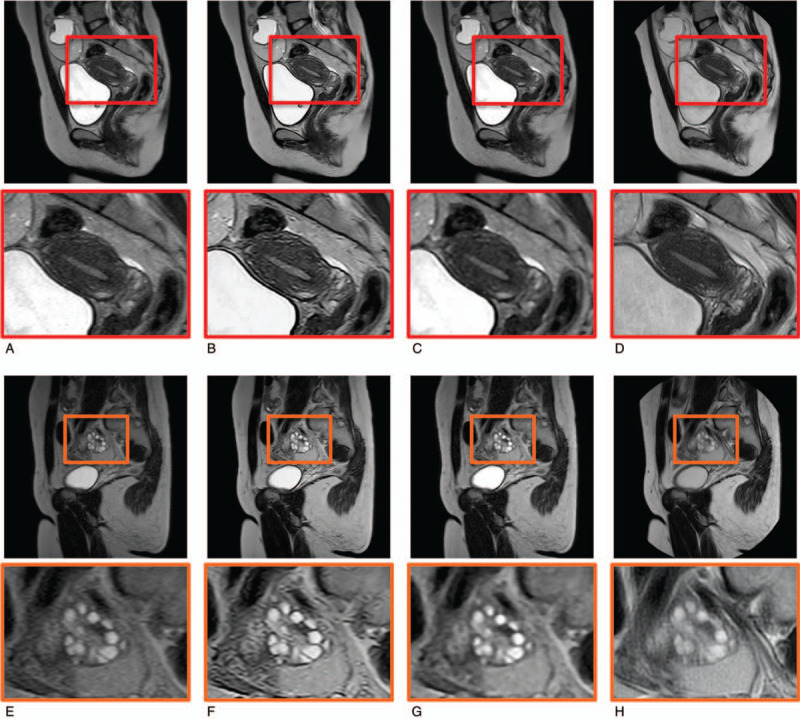
Representative T2-weighted MR images and magnified images of uterus and ovary. The image quality of contrast, noise, and sharpness of uterus on DL-SSTSE images (B) were improved compared with the SSTSE images (A), and were comparable to those of TSE images (D). However, motion artifact was sometimes greater on TSE images (H) compared with the DL-SSTSE images (F) or SSTSE images (E). The conspicuity of the ovaries on DL-SSTSE images was improved compared with the SSTSE images. On the other hand, image qualities of F-SSTSE images (C and G) were not improved compared with SSTSE images because classical filtering methods cannot improve the spatial resolution and image noise at the same time. Dl-SSTSE = deep learning-based single-shot turbo spin-echo, F-SSTSE = filtering method-based single-shot turbo spin-echo, SSTSE = single-shot turbo spin-echo, TSE = turbo spin-echo.
The image quality scores for SSTSE, DL-SSTSE, F-SSTSE, and TSE images are shown in Table 2. The interobserver agreement between the 2 reviewers for evaluating the image quality score was excellent (K = .94). Scores with regard to contrast and noise were significantly higher on DL-SSTSE, F-SSTSE, and TSE images than on SSTSE images. There were no significant differences between DL-SSTSE, F-SSTSE, and TSE images. Scores with regard to overall image quality and boundary sharpness of the zonal layers in the uterine corpus were significantly higher on DL-SSTSE and TSE images than on SSTSE and F-SSTSE images. There were no significant differences between DL-SSTSE and TSE images. The score with regard to motion artifact was significantly higher on DL-SSTSE, F-SSTSE, and SSTSE images than on TSE images. There were no significant differences between DL-SSTSE, F-SSTSE, and SSTSE images. The score with regard to the conspicuity of ovaries and follicles was significantly higher on DL-SSTSE images than on SSTSE, F-SSTSE, and TSE images. There were no significant differences between SSTSE, F-SSTSE, and TSE images.
Table 2.
Image quality scores for SSTSE, DL-SSTSE, F-SSTSE, and TSE images.
| P values | ||||||||||
| SSTSE images | DL-SSTSE images | F-SSTSE images | TSE images | SSTSE vs DL-SSTSE | SSTSE vs F-SSTSE | DL-SSTSE vs F-SSTSE | DL-SSTSE vs TSE | TSE vs SSTSE | TSE vs F-SSTSE | |
| Over all image quality | 4 (3–4) | 4 (4–5) | 4 (3–4) | 4 (4–5) | < 0.001 | 0.57 | <0.001 | 0.98 | < 0.001 | < 0.001 |
| Contrast | 3 (3–4) | 4 (4–5) | 4 (4–5) | 4 (4–5) | < 0.001 | < 0.001 | 0.97 | 0.64 | < 0.001 | 0.88 |
| Noise | 4 (3–4) | 5 (4–5) | 5 (4–5) | 5 (4–5) | < 0.001 | < 0.001 | 0.99 | 0.83 | < 0.001 | 0.93 |
| Motion artifact | 5 (4–5) | 5 (5–5) | 5 (5–5) | 4 (3–4) | 0.64 | 0.97 | 0.87 | < 0.001 | < 0.001 | < 0.001 |
| Boundary sharpness of the zonal layers in the uterus | 4 (3–4) | 4 (4–5) | 4 (3–4) | 4 (3–5) | < 0.001 | 0.99 | < 0.001 | 0.99 | < 0.001 | < 0.001 |
| Conspicuity of ovaries and follicles | 4 (4–4) | 5 (4–5) | 4 (3.25–4) | 4 (3–4) | < 0.001 | 0.99 | < 0.001 | < 0.001 | 0.99 | 0.99 |
DL-SSTSE = deep learning-based single-shot turbo spin-echo, F-SSTSE = filtering method-based single-shot turbo spin-echo, SSTSE = single-shot turbo spin-echo, TSE = turbo spin-echo.
4. Discussion
In the present study, we showed that DL-SSTSE images outperformed the SSTSE images for CR, SNR, and image qualities except for motion artifact. Moreover, DL-SSTSE images were comparable to TSE images for CR, SNR, and image quality with regard to contrast, noise, and boundary sharpness of the 3 zonal layers in the uterine corpus. In addition, DL-SSTSE images had significantly higher image qualities with regard to motion artifacts and the conspicuity of the ovary than TSE images.
Contrasts of DL-SSTSE images were significantly higher than those of SSTSE images, while no significant differences from those of TSE images. In our acquisition protocols, echo-train length of SSTSE imaging was 57, while that of TSE imaging was 20. CNN can improve image contrast regardless of the acquisition parameters if appropriate dataset and algorithms are designed. SNRs of DL-SSTSE images were higher than those of both SSTSE and TSE images. In clinical situations, TSE imaging is routinely performed to ensure adequate image quality because SSTSE imaging results in a poor SNR. The CNN is an effective denoise approach without producing image blurring, as Park et al[28] reported.
The spatial resolution in MRI is mainly influenced by matrix size, field-of-view, and slice thickness. Although increasing the matrix size improves the spatial resolution, it results in a longer imaging time or lower SNR. In our acquisition protocols, the matrix number of phase encoding steps in SSTSE images was half of that in TSE images. In addition, SSTSE images are blurred compared with TSE images, theoretically because of the features of single-shot imaging.[7] The echo train length of 57 in SSTSE imaging was longer than that in TSE imaging. From our results, it is evident that DL-SSTSE images had better boundary sharpness in the uterine corpus and conspicuity of ovaries and follicles than SSTSE images, thus the CNN can CNN can improve the spatial resolution and produce less-blurred images. On the other hand, classical filtering methods cannot improve the spatial resolution and image noise at the same time.
T2-weighted TSE imaging has been acquired in most pelvic MR imaging protocols. However, it requires long imaging times and causes motion artifacts. Recently, compressed sensing (CS) approach has been developed and widely used in clinical situation for acceleration of MR acquisition speed.[29] CS exploits image sparsity to reconstruct high-quality images from the undersampled k-space data, reducing the imaging time of MRI by up to 50%, depending on the sequence.[30,31] CS is a promising technique for accelerating the acquisition speed of MRI. However, it does not suppress motion artifacts. In our study, default acquisition times were approximately 1 min for T2-weighted SSTSE imaging and approximately 4 min for T2-weighted TSE imaging. In addition, grading scores with regard to motion artifacts of DL-SSTSE images were significantly higher than those of TSE. Motion artifacts degrade the image quality of the uterus and ovaries and can potentially impair the radiologist's ability to make an accurate diagnosis. CNN-based DL-SSTSE images can keep the advantage of the motion artifact robustness and the acquisition speed acceleration of SSTSE images.
Our study had several limitations. First, in the process of input image generation, a Gaussian noise was added to the images. A noise in MRI has been traditionally modeled by a Rician distribution with constant noise power at each voxel,[32] while noise reduction, coil uniformity correction, or parallel imaging techniques dramatically change spatial noise characteristics into more complex patterns in modern MRI systems.[33,34] If the complex noise distribution in MRI can be simulated more precisely, a further improvement of the image quality could be achieved. Second, the proposed CNN emphasized image artifacts, such as Gibbs ringing artifacts; nevertheless, they did not influence the grading scores. Third, we did not evaluate the image quality of pelvic benign or malignant tumors. Further study is necessary to investigate whether DL-SSTSE images can serve as substitutes for TSE images to diagnose female pelvic diseases. Fourth, we did not compare the performance of the proposed network algorithm with other networks. Further investigation should be performed to optimize the network algorithm for improving the SSTSE images.
In conclusion, DL-SSTSE images of the female pelvis showed higher image quality as compared with SSTSE images. In comparison with conventional TSE images, DL-SSTSE images had acceptable image quality while keeping the advantage of the motion artifacts-robustness and acquisition time efficiency of SSTSE imaging. Thus, it has the potential to be a good substitute to TSE images for evaluation of the female pelvis.
Author contributions
Conceptualization: Tomofumi Misaka.
Data curation: Yukino Ota.
Formal analysis: Masanobu Uemura.
Methodology: Tomofumi Misaka.
Software: Takuma Kobayashi.
Visualization: Nobuyuki Asato, Yukihiko Ono.
Writing – original draft: Tomofumi Misaka.
Writing – review & editing: Kensuke Umehara, Junko Ota.
Footnotes
Abbreviations: CNN = convolutional neural network, CR = contrast ratio, CT = computed tomography, DL-SSTSE = deep learning-based single-shot turbo spin-echo, MR = magnetic resonance, MRI = magnetic resonance imaging, ROI = region of interest, SD = standard deviation, SI = signal intensity, SNR = signal-to-noise ratio, SSTSE = single-shot turbo spin-echo, TSE = turbo spin-echo.
How to cite this article: Misaka T, Asato N, Ono Y, Ota Y, Kobayashi T, Umehara K, Ota J, Uemura M, Ashikaga R, Ishida T. Image quality improvement of single-shot turbo spin-echo magnetic resonance imaging of female pelvis using a convolutional neural network. Medicine. 2020;99:47(e23138).
The authors have no funding information to disclose.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Siddiqui N, Nikolaidis P, Hammond N, et al. Uterine artery embolization: pre- and post-procedural evaluation using magnetic resonance imaging. Abdom Imaging 2013;38:1161–77. [DOI] [PubMed] [Google Scholar]
- [2].Bazot M, Stivalet A, Daraï E, et al. Comparison of 3D and 2D FSE T2-weighted MRI in the diagnosis of deep pelvic endometriosis: preliminary results. Clin Radiol 2013;68:47–54. [DOI] [PubMed] [Google Scholar]
- [3].Sala E, Wakely S, Senior E, et al. MRI of malignant neoplasms of the uterine corpus and cervix. Am J Roentgenol 2007;188:1577–87. [DOI] [PubMed] [Google Scholar]
- [4].Hori M, Kim T, Onishi H, et al. Uterine tumors: comparison of 3D versus 2D T2-weighted Turbo Spin-Echo MR Imaging at 3.0 T—initial experience. Radiology 2011;258:154–63. [DOI] [PubMed] [Google Scholar]
- [5].Pui MH, Yan Q, Xu B, et al. MRI of gynecological neoplasm 2004;28:143–52. [DOI] [PubMed] [Google Scholar]
- [6].Semelka RC, Kelekis NL, Thomasson D, et al. HASTE MR imaging: description of technique and preliminary results in the abdomen. J Magn Reson Imaging 1996;6:698–9. [DOI] [PubMed] [Google Scholar]
- [7].Bhosale P, Ma J, Choi H. Utility of the FIESTA pulse sequence in body oncologic imaging: review. Am J Roentgenol 2009;192: 6_supplement: S83–93. [DOI] [PubMed] [Google Scholar]
- [8].Bosmans H, Van Hoe L, Gryspeerdt S, et al. Single-shot T2-weighted MR imaging of the upper abdomen: preliminary experience with double-echo HASTE technique. AJR Am J Roentgenol 1997;169:1291–3. [DOI] [PubMed] [Google Scholar]
- [9].Lindgren Belal S, Sadik M, Kaboteh R, et al. Deep learning for segmentation of 49 selected bones in CT scans: first step in automated PET/CT-based 3D quantification of skeletal metastases. Eur J Radiol 2019;113:89–95. [DOI] [PubMed] [Google Scholar]
- [10].Ciritsis A, Rossi C, Eberhard M, et al. Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making. Eur Radiol 2019;29:5458–68. [DOI] [PubMed] [Google Scholar]
- [11].McCoy DB, Dupont SM, Gros C, et al. Convolutional neural network-based automated segmentation of the spinal cord and contusion injury: deep learning biomarker correlates of motor impairment in acute spinal cord injury. AJNR Am J Neuroradiol 2019;40:737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chaudhari AS, Fang Z, Kogan F, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med 2018;80:2139–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shi J, Liu Q, Wang C, et al. Super-resolution reconstruction of MR image with a novel residual learning network algorithm. Phys Med Biol 2018;63:085011. [DOI] [PubMed] [Google Scholar]
- [14].Umehara K, Ota J, Ishida T. Application of super-resolution convolutional neural network for enhancing image resolution in chest CT. J Digit Imaging 2018;31:441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gholizadeh-Ansari M, Alirezaie J, Babyn P. Deep learning for low-dose CT denoising using perceptual loss and edge detection layer. J Digit Imaging 2020;33:504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kidoh M, Shinoda K, Kitajima M, et al. Deep learning based noise reduction for brain MR imaging: tests on phantoms and healthy volunteers. Magn Reson Med Sci 2019;19:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jiang D, Dou W, Vosters L, et al. Denoising of 3D magnetic resonance images with multi-channel residual learning of convolutional neural network. Jpn J Radiol 2018;36:566–74. [DOI] [PubMed] [Google Scholar]
- [18].Pipe JG, Gibbs WN, Li Z, et al. Revised motion estimation algorithm for PROPELLER MRI. Magn Reson Med 2014;72:430–7. [DOI] [PubMed] [Google Scholar]
- [19].Lane BF, Vandermeer FQ, Oz RC, et al. Comparison of sagittal T2-weighted BLADE and fast spin-echo MRI of the female pelvis for motion artifact and lesion detection. Am J Roentgenol 2011;197:W307–13. [DOI] [PubMed] [Google Scholar]
- [20].Peterlík I, Courtecuisse H, Rohling R, et al. Fast elastic registration of soft tissues under large deformations. Med Image Anal 2018;45:24–40. [DOI] [PubMed] [Google Scholar]
- [21].Nasr B, Le Ven F, Savean J, et al. Characterization of the physiological displacement of the aortic arch using non-rigid registration and MR imaging. Eur J Vasc Endovasc Surg 2017;53:282–9. [DOI] [PubMed] [Google Scholar]
- [22].Shi F, Cheng J, Wang L, et al. LRTV: MR image super-resolution with low-rank and total variation regularizations. IEEE Trans Med Imaging 2015;34:2459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ryu K, Shin N-Y, Kim D-H, et al. Synthesizing T1 weighted MPRAGE image from multi echo GRE images via deep neural network. Magn Reson Imaging 2019;64:13–20. [DOI] [PubMed] [Google Scholar]
- [24].Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017;44:1408–19. [DOI] [PubMed] [Google Scholar]
- [25].Heverhagen JT. Noise measurement and estimation in MR imaging experiments. Radiology 2007;245:638–9. [DOI] [PubMed] [Google Scholar]
- [26].Kundel HL, Polansky M. Measurement of observer agreement. Radiology 2003;228:303–8. [DOI] [PubMed] [Google Scholar]
- [27].Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 1952;47:583. [Google Scholar]
- [28].Park J, Hwang D, Kim KY, et al. Computed tomography super-resolution using deep convolutional neural network computed tomography super-resolution using deep convolutional neural network. Phys Med Biol 2018;63:145011. [DOI] [PubMed] [Google Scholar]
- [29].Donoho DL. Compressed sensing. IEEE Trans Inf Theory 2006;52:1289–306. [Google Scholar]
- [30].Feng L, Benkert T, Block KT, et al. Compressed sensing for body MRI. J Magn Reson Imaging 2017;45:966–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kijowski R, Rosas H, Samsonov A, et al. Knee imaging: rapid three-dimensional fast spin-echo using compressed sensing. J Magn Reson Imaging 2017;45:1712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magn Reson Med 1995;34:910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rabanillo I, Aja-Fernandez S, Alberola-Lopez C, et al. Exact calculation of noise maps and ${g}$ -Factor in GRAPPA using a ${k}$ -space analysis. IEEE Trans Med Imaging 2018;37:480–90. [DOI] [PubMed] [Google Scholar]
- [34].Baselice F, Ferraioli G, Pascazio V, et al. Bayesian MRI denoising in complex domain. Magn Reson Imaging 2017;38:112–22. [DOI] [PubMed] [Google Scholar]


