Abstract
Transcriptional regulatory networks control context-specific gene expression patterns and play important roles in normal and disease processes. Advances in genomics are rapidly increasing our ability to measure different components of the regulation machinery at the single-cell and bulk population level. An important challenge is to combine different types of regulatory genomic measurements to construct a more complete picture of gene regulatory networks across different disease, environmental, and developmental contexts. In this review, we focus on recent computational methods that integrate regulatory genomic data sets to infer context specificity and dynamics in regulatory networks.
Keywords: Gene regulatory networksGene regulation, Enhancer, Promoter, Single cell, Data
Introduction
Transcriptional regulatory networks specify the connections between regulatory proteins and target genes and control the context-specific expression of genes. These networks play important roles in diverse disease, developmental, environmental, and evolutionary contexts [1,2]. The structure of regulatory networks involves many components including but not limited to cis-regulatory elements (CREs), transcription factors, and chromatin remodelers [3] that interact through processes such as three-dimensional organization of the genome [2,4], chromatin modifications [5], and genome accessibility [6]. Advances in next-generation sequencing tools have greatly expanded our ability to measure different aspects of the regulatory machinery. For instance, transcription factor (TF) binding and histone modifications (ChIP-seq [7]), transcription (RNA-seq [8]), chromatin accessibility (DNase-seq, ATAC-seq [6]), and genome-wide three-dimensional organization (chromosome conformation capture [9]) have been measured across individuals [10], diseases [11], developmental stages [12], and environments [13] in both bulk populations and single cells [14].
Using these diverse readouts of the cellular state and regulatory landscape (Figure 1A), there are two major approaches to model gene regulatory networks [15]. The first approach predicts regulatory proteins, for example, TFs, chromatin remodelers, and signaling proteins that determine a gene’s expression level (Figure 1B). The second approach identifies CREs affecting gene expression levels (Figure 1C), that is, TF binding sites in or near the promoter, or distal enhancers interacting with the promoter through chromatin looping [2]. In this review, we discuss advances from the last two years in these two types of computational approaches addressing three main challenges: inferring genome-scale context-specific transcriptional regulatory networks to predict regulatory proteins of individual or groups of genes, predicting target genes of CREs, and integrating single-cell multiomics data sets for gene regulatory network inference.
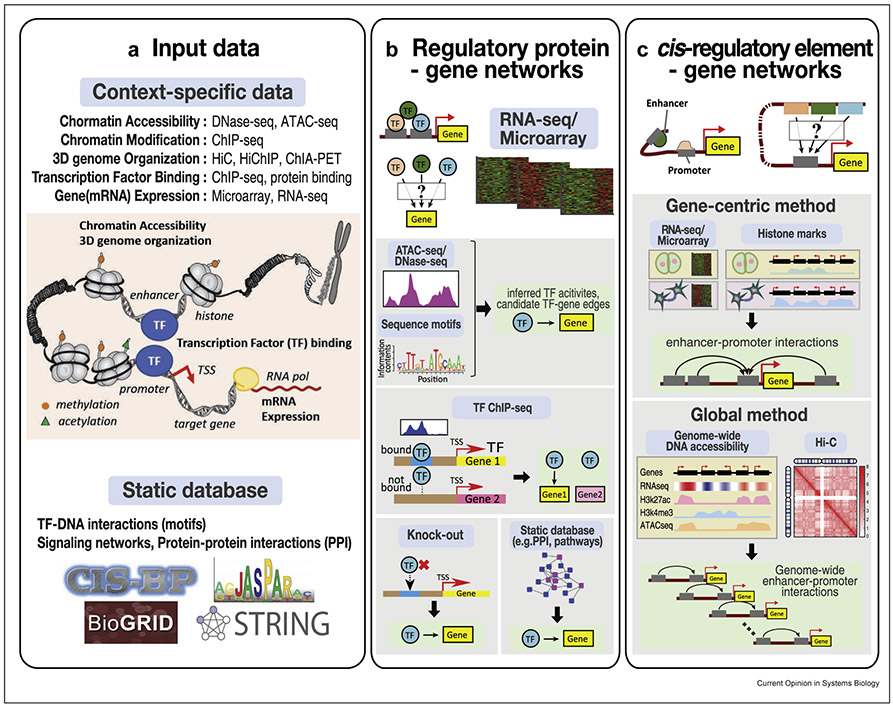
Figure 1. Data types used in integrative approaches for two main types of regulatory networks:
(a) Input data for inferring regulatory networks can be categorized into context-specific and static data sets. Context-specific gene regulatory events are measured across different layers of gene regulation using various genomic technologies. DNase-seq or ATAC-seq measure open regions of the chromatin and can be leveraged to identify regulatory elements. Chromosome conformation capture (3C) techniques such as Hi-C, HiChIP, and ChIA-PET allow global mapping of 3D genome organization. ChIP-seq and protein binding assays are used for detecting transcription factor (TF) binding to DNA. Transcriptomic assays measure expression levels of regulators and target genes under different conditions. Static databases often used in integrative inference of regulatory networks are DNA-binding motif collections such as JASPAR and CIS-BP, predefined signaling networks, and protein–protein interactions (PPIs). Databases of functional gene annotations and pathways are used for evaluating gene regulatory networks but can also be used as another source of data for integration. (b) Regulatory networks that capture the relationship between regulatory proteins, such as transcription factors and signaling proteins, and target genes. Gene expression measured with RNA-seq or microarrays is the primary data type for inferring these networks. Integrative inference approaches integrate other data types with gene expression such as sequence motifs, filtered by available accessibility (DNAse-seq, ATAC-seq) measurements, TF ChIP-seq, and gene perturbation data. (c) Cis-regulatory networks represent the noncoding regulatory sequences such as enhancers that regulate genes. These networks can be inferred either by learning a gene-centric model that predicts gene expression based on the chromatin state and/or accessibility of candidate enhancers across multiple conditions or a global model that learns predictive rules of enhancer–promoter interactions based on chromatin marks, architectural proteins, RNA-seq, and chromosome conformation (3C) data.
Integrating data to infer genome-scale context-specific transcriptional regulatory networks
A context-specific regulatory network is defined as a network that captures interactions between regulatory proteins and target genes in a particular context, such as a developmental stage, tissue, disease, or environment (Figure 1B and 2A). While there are a large number of mathematical formalisms to represent regulatory networks [16], we focus on statistical models, which include information-theoretic methods [17], dependency networks [18,19], Bayesian networks [20], and Gaussian graphical models [21]. Given a particular modeling formalism, a simple approach to infer a context-specific regulatory network is to use sequence-specific motifs on gene promoters to first define a comprehensive set of possible regulatory edges and then prune out edges with low statistical support using context-specific expression as evidence. Statistical support is assessed by Pearson’s correlation or by regressing candidate regulators to the expression of a target gene. The motif-derived edges can be filtered by requiring the presence of context-specific ATAC-seq or DNase-seq peak overlapping the motif instance [22-26]. This approach identified the regulatory network for Huntington disease [22] and early brain development [26] and predicted key regulators in neurodegenerative diseases which were experimentally validated, for example, SMAD3 [22] and POU3F2 [26]. Although straightforward to implement, this approach can only add edges between TFs and targets with known motifs and may miss regulators with unknown specificity, and accessible motifs are not sufficient to comprehensively capture true regulatory interactions.
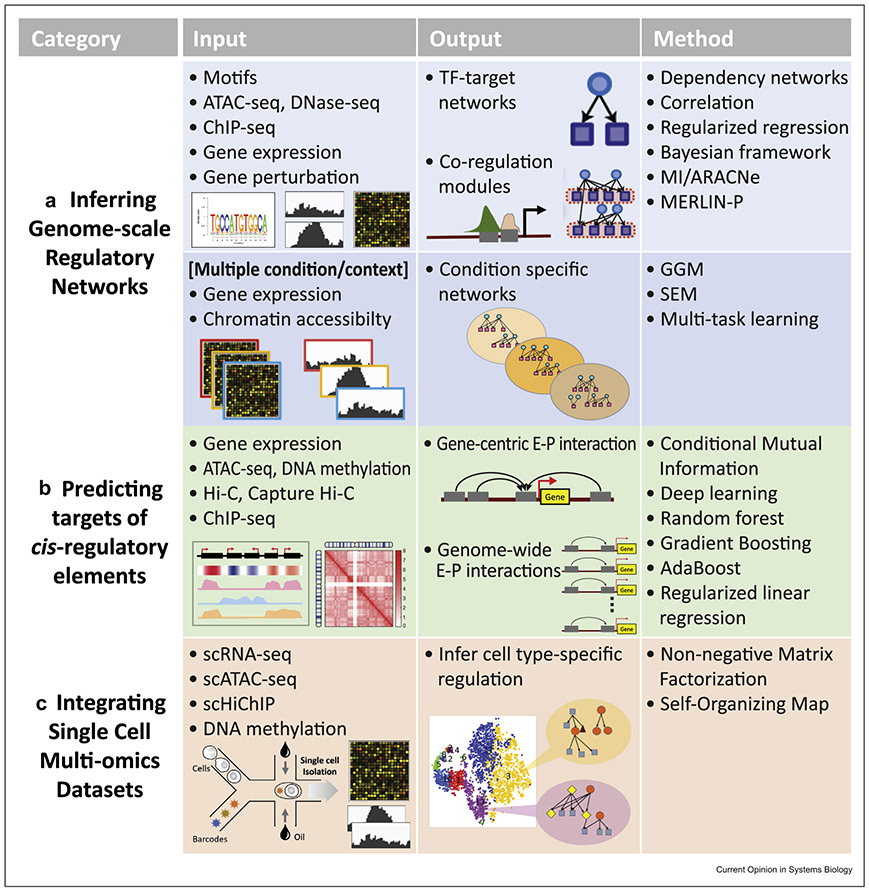
Figure 2. Classes of problems in gene regulation that integrate diverse data types and computational approaches to address these problems.
For each class of problems, the input data, outputs, and the algorithmic components used are shown. (a) Inference of genome-scale regulatory networks in a specific condition or across multiple conditions. These methods learn the structure of the network by predicting relationships between TFs and target genes and integrate gene expression with static data from public databases and context-specific data from ChIP-seq and ATAC-seq experiments. Methods use a variety of approaches to infer networks, including Gaussian graphical models (GGMs), dependency networks, mutual information (MI). Context-specific networks are learned by joint inference of networks across multiple conditions. Multi-task learning is a popular framework to enable information sharing across different contexts. (b) Predicting target genes of regulatory sequence elements. These methods use regulatory genomic data sets such as ChIP-seq, ATAC-seq, and 3D genome organization assays to identify targets of cis-regulatory elements. Regression and classification approaches have been developed to either predict counts or discriminate between an interacting and noninteracting pair. (c) Integration of different types of single-cell omic measurements to identify cell type–specific gene regulatory networks. These methods leverage single-cell RNA-seq (scRNA-seq) and scATAC-seq and occasionally bulk data sets. Methods typically use clustering and dimensionality reduction, for example, via matrix factorization, to define data set–specific clusters followed by linking the clusters. Some methods focus on genic regions, while others can incorporate auxiliary data sets such as HiChIP to link to genes. GGM: Gaussian graphical model; SEM: structural equation model; TF: transcription factor.
Incorporating prior to constrain network structure and estimate regulator activity
To enable more comprehensive network inference, sequence-specific motif-derived edges can be incorporated as prior knowledge to encourage the network inference algorithm to include edges from the prior. Prior-based regulatory network inference can use either a structure prior approach [18,20] or parameter prior approach [24]. With the structure-based prior, one puts a prior probability distribution on the graph structure, such that an edge with motif support is more likely to be included in the regulatory network model. For example, Chasman et al. [23] used DNase-seq filtered motifs as a structure prior for a network inference algorithm [18] and context-specific gene expression data of hindbrain and spinal cord development to infer a regulatory network in neuroepithelial stem cells. The parameter prior framework has been used within a regularized regression setting [24], which learns a sparse network by penalizing edge addition while optimizing prediction of a target’s expression level. The prior can be used to reduce the penalty for addition of an edge in the network [24].
Newer methods have used the prior network to additionally estimate hidden TF activity (TFA) to overcome the assumption that mRNA levels must correlate with the regulator’s protein activity on a gene’s promoter [24,27]. TFA estimation takes in an initial, noisy regulatory network and expression and models the expression as a product of the hidden TFAs and a refined network structure [27]. The TFA can be estimated independently followed by network inference, for example, mLASSO-StARS [24], or in a single iterative algorithm, for example, NetRex [27]. mLASSO-StARS [24] learns a dependency network by solving a set of independent LASSO regression problems, one per gene. Both mRNA and TFA are used as potential predictors of a target gene’s expression. NetRex [27] jointly estimates both the TFA and the network to explain the expression data by using regularization to penalize the mismatch in the number of edges of the inferred and prior networks. NetRex updates the prior network until the rewired network and the estimated TFA optimally explains the expression data. Both NetRex and mLASSO-StARS need to specify the hyperparameters to obtain a final network structure. mLASSO-StARS uses StARS to select the hyperparameters by controlling the average instability of edges. NetRex uses a grid search over hyperparameters and obtains the final result from the consensus of multiple nearly optimal settings. NetRex has additional constraints that can better capture coregulatory relationships in gene modules but can make network inference computationally expensive. These approaches were shown to improve the quality of inferred networks and were useful for identifying key regulators and targets in mammalian [23,24] and insect [27] systems.
Network inference across multiple contexts
Often data from multiple contexts are available and a key question is to define regulatory networks for each context and identify similarities and differences. Although one approach would be to infer networks for each context independently [28], examining multiple contexts simultaneously could improve the quality of the inferred networks [21,29], make comparisons easier [30,31], and help in scenarios with low sample sizes [32]. To enable simultaneous network inference across multiple conditions, several approaches have used multi-task learning (MTL) (Figure 2A) [33]. In MTL, related tasks are jointly solved while sharing the information across tasks. In one class of methods, a regulatory network is modeled as a Gaussian graphical model (GGM), which represents the genome-wide expression levels of genes as a multivariate Gaussian and the network structure by a precision matrix. One GGM approach, JRmGRN [21], models each condition’s network as sum of two sparse precision matrices, one for condition-specific components and the second for shared component across all conditions. JRmGRN additionally regularized the shared network to favor more hub genes and was more effective at identifying shared hubs and context-specific network components than existing approaches. While JRmGRN models each condition with one precision matrix, another GGM-based approach, NETI2 [31], additionally models the intrasample heterogeneity, which is common in tumor samples because they represent a mix of cancerous and noncancerous cells. NETI2 infers cancer subtype–specific networks and a network for noncancerous cells shared across the subtypes. The proportion of cancerous and noncancerous cells for each sample is known; however, the expression of each cell type is not known and is estimated using an expectation–maximization algorithm. On TCGA data [34] for different breast cancer subtypes, NETI2 captured subtype-specific gene hubs that exhibited reduced connections in the noncancerous network.
Regulatory networks across multiple conditions have also been modeled using dependency networks [35] and structural equation models (SEMs) [29]. Compared with GGMs, these models can capture directed edges and are therefore more suitable for representing regulatory networks. In addition, dependency networks are computationally faster and often outperform GGMs [19]. One approach, FSSEM [29], uses SEMs to infer a regulatory network for two conditions from gene expression and genetic variation, available in expression quantitative trait locus data sets to better identify the effect of TFs on target genes. SEMs are a class of statistical models that represent observed measurements as a linear combination of other observed or latent variables. To enable sharing, FSSEM uses a penalized framework to minimize the difference in the network between the two conditions. Inferelator-AMuSR [35] is a dependency network-based approach that uses MTL with a block-sparse penalty to encourage similarity between networks while learning overall sparse networks. The regression weight of a regulator across conditions is considered a block, and the sparsity criteria select a small number of conserved blocks, while allowing some regulators to be condition specific. Both FSSEM and Inferelator-AMuSR inferred more accurate networks than when inferring networks independently, suggesting a benefit of joint inference across multiple conditions.
Often the number of samples for a given condition may be too few to predict the regulators of individual genes. To overcome the small sample size, it is better to group genes based on expression levels and model regulatory relationships at the group level at the expense of losing gene-level regulatory information [32]. PSIONIC [32] is an example of this framework that learns patient-specific TF regression weights in multiple cancers using an approach called grouping and overlapping in MTL [36]. In PSIONIC, ATAC-seq–filtered TF motifs from enhancers and promoters associated with a gene are used to predict gene expression. Instead of learning a regression vector for each patient profile independently, PSIONIC’s grouping and overlapping in MTL approach decomposes the matrix of regression vectors into lower dimensional latent regulatory programs and tumor-specific weights representing the contribution of each regulatory program to the expression of each sample. PSIONIC was significantly better at predicting expression in test samples than a single task model, suggesting that sharing information across tumors while learning regulatory programs is advantageous.
To summarize, current methods for genome-scale regulatory network inference have used prior knowledge to constrain the network structure and estimate hidden regulator activity [24,37]. Across multiple contexts, MTL-based approaches have improved network inference, especially for the shared components [21,29]. The type of network inference algorithm depends upon the number of gene expression samples and the availability of sequence-specific motifs and matched ATAC-seq measurements for each context of interest.
Data integration for predicting CREs controlling expression of target genes.
Identification of CREs that regulate a gene’s context-specific expression, proximally or distally, is a grand challenge in gene regulation [3-5]. A number of approaches exist (refer the study by Xu et al. [38] for a comprehensive review); here, we review recent methods that integrate multiple types of regulatory genomic data to predict such interactions. These methods can be grouped into (Figure 1C) (a) gene-centric approaches, which predict the regulatory elements by learning a model for each gene [39-41], and (b) global approaches, which learn a single model for all gene–element pairs [42,43].
Gene-centric methods to predict cis-regulatory interactions
Gene-centric methods leverage gene expression and a small number of regulatory signals from multiple conditions to explain the expression level of a gene as a function of the regulatory activity of CREs. Current methods have used regularized regression [40,44] or mutual information [41] to link elements to genes (Figure 2B, [38]). These methods also vary in the size of the neighborhood around a gene to search for regulatory elements, the specific regulatory signal used to correlate or predict expression, and the number of cell lines examined. In particular, FOCS and Vijayabaskar et al. [40,44] used regression to select CREs for a gene within the 100 Kb and 500 Kb region, respectively, around the transcription start site (TSS). Instead, MICMIC [41] used conditional mutual information to predict which regulatory elements, defined by CpG methylation levels, within the 300 Kb window of a gene’s TSS explain the expression of the gene. MICMIC is suitable for large-scale studies that have matched RNA and DNA methylation measurements, for example, in cancer [45]. The activity-by-contact (ABC) model [39] predicted enhancer–gene connections assuming that an en-hancer’s quantitative effect on a gene’s expression de-pends on its strength (activity), weighted by the contact count of the enhancer. The activity is estimated from H3K27ac and DNaseI accessibility for the element and count from a Hi-C or related experiment. The ABC score is the relative effect of an element on a gene’s expression and accurately captured the measurements of CRISPRi-FlowFish experiments, which perturb en-hancers and quantify the effects on gene expression. The ABC model is conceptually the simplest model and does not have any parameters; however, it might need more careful calibration compared with regression-based or information-theoretic approaches.
Global methods to infer cis-regulatory interactions
Global methods in contrast learn a single predictive model of interaction presence or strength by leveraging cell type–specific measurements such as gene expression, histone modifications, and TF binding. These approaches vary depending upon the input data types and whether they use a classification or regression framework (Figure 2B). Recent classification-based approaches have relied on sequence as the input features to predict interactions (e.g, SPEID [46]) and accessible sequences (e.g., DeepTACT [47]), while others have used sequence conservation and chromatin marks (e.g, EPIP [48]). A variety of classification algorithms have been used including AdaBoost (EPIP [48]) and deep neural networks (DeepTACT [47], SPEID [46]). Methods that rely on sequence are broadly applicable across many contexts but might be limited in capturing cell line–specific information compared with methods that incorporate additional data sets such as accessibility and chromatin marks.
Classification approaches rely on a training set of true enhancer–promoter interactions and could be susceptible to the definition of the positive label set. Recent approaches have attempted to predict the contact count directly using regression instead of discriminating between interactions and noninteractions. For example, HiC-Reg [42] trains a random forest regression model to predict Hi-C contact counts by integrating published high-resolution Hi-C data sets with epigenomic marks and architectural proteins. Another approach, 3DPredictor, predicted Hi-C contact counts with gradient boosting and features derived from RNA-seq and CTCF ChIP-seq data [43]. While HiC-Reg predicts counts for all pairs of genomic loci, including pairs with nonpromoter regions, 3DPredictor focuses on enhancer–promoter pairs. Regression-based approaches have the advantage that they do not need positive labels of interactions, and their outputs can be used to identify topologically associating domains.
To summarize, the advantage of gene-centric approaches is that they are specific to each gene and can model multiple enhancer elements per gene. However, most of these approaches learn a single model for all contexts and require sufficient samples for model learning. Global approaches can predict interactions in a single biological context but typically model only one enhancer per gene and require multiple measurements for the same context. Selection of a specific tool for predicting CREs of a gene should be guided by (a) the number of available samples with gene expression and regulatory signals, (b) availability of Hi-C or similar measurements, and (c) need to identify interaction pairs or examine higher-order organizational units.
Methods to learn regulatory networks from single-cell omics data types
Single-cell techniques are rapidly increasing our ability to measure multiple types of omic profiles, such as transcriptomes [49], epigenomes [50], methylomes [51], and 3D genome organization [52] for individual cells. Several methods have been developed [53] and benchmarked for inferring gene regulatory networks from scRNA-seq data [54]. More recently, the availability of scRNA-seq and scATAC-seq data from the same underlying subpopulations (but not necessarily from the same cells) offers an exceptional opportunity to infer cell type–specific gene regulatory networks (Figure 2C).
A key challenge with integrating scRNA-seq and scATAC-seq data is the lack of correspondence between cells as they come from different populations. To address this issue, one strategy is to cluster or project the scRNA-seq and scATAC-seq data separately and then integrate the clusters. Bravo González et al. [55] leveraged the spatial information of cell types in the Drosophila eye-antennal disc tissue to integrate scRNA-seq and scATAC-seq data. Briefly, cell types are defined using the scRNA-seq and scATAC-seq measurements independently, followed by assigning each cell a pseudotime based on their position on the distal–proximal (antennal cells) or anterior–posterior (eye cells) axis. Each cell was also mapped onto a spatial map of virtual cells based on the cell type annotations and their relative position in pseudotime. Once mapped, transcriptomic and epigenomic profiles are available for the same virtual cell, which was used to derive links between enhancers and genes using random forest regression to predict expression based on accessibility at candidate regions in a gene-centric manner.
In contrast, SOMatic [56] integrated scATAC-seq and scRNA-seq data with self-organizing maps (SOMs), which are based on neural networks and used for dimensionality reduction and visualization of high-dimensional data on a 2D map. SOMs were first trained on scATAC-seq and scRNA-seq data separately; then, SOM units, each representing a cluster of genes or regulatory regions, were grouped into metaclusters so that SOM units nearby on the 2D map remain close in the cluster. A linking function was used to link genes and genomic regions into Linked SOM metaclusters of regulatory regions around the TSS that also overlap with ATAC-seq peaks. Linked SOM metaclusters with enriched motifs were used to construct a regulatory network. Compared with Gonzalez-Blas et al., SOMatic does not require additional spatial information to integrate the scRNA-seq and scATAC-seq data. However, it is dependent on the ability to reliably link a scATAC-seq cluster with a scRNA-seq cluster. SOMatic also does not predict enhancer–promoter interactions.
While SOMatic and González-Blas et al. both first define clusters and then link them, DC3 [57] uses an alternative approach by jointly factoring the scRNA-seq and scATAC-seq data by integrating single-cell or bulk HiChIP data available for the same condition. The HiChIP data enable DC3 to incorporate long-range interactions between enhancers and genes. DC3 uses a joint non-negative matrix factorization (NMF) approach where the scRNA-seq and scATAC-seq data sets are each represented by an NMF decomposition term but the NMF factors are coupled to maximize the concordance with HiChIP. Importantly, DC3 is flexible to incorporate single-cell or bulk RNA-seq, ATAC-seq, and HiChIP. The output of DC3 provides the gene expression, accessibility, and long-range interaction profile for each subpopulation, which can be used to identify important regulators based on differential expression, accessibility, and regulatory networks based on the enhancer–gene interaction profile.
To summarize, recent integrative methods for regulatory network inference from single-cell data sets use dimensionality reduction or clustering to define cell populations independently [55,56] or jointly [57] from scRNA-seq and scATAC-seq data. Methods that independently define clusters use additional postprocessing to link the clusters but do not require additional long-range data sets (e.g., HiChIP or Hi-C). On the other hand, methods such as DC3 align cell type–specific gene expression patterns to accessibility patterns by incorporating long-range interaction profiles. Regulatory networks are defined by identification of CREs associated with scATAC-seq clusters linked to scRNA-seq clusters.
Conclusion
In this review, we covered recent approaches to integrate regulatory genomic data sets to predict sequence or protein regulators of context-specific gene expression from bulk and single-cell data sets (Supplementary Table 1). Despite the availability of several promising methods, network inference remains challenging and we envision development in several areas going forward. One area is to develop methods that can combine both cis and trans regulatory information to define context-specific regulatory networks, while accounting for both distal and proximal interactions of CREs. In addition to being more comprehensive, such integrated regulatory networks would help determine the role of noncoding variation in gene expression modules and pathways in complex traits [58]. Another emerging area is to integrate single-cell and bulk data sets to gain insights into cell type–specific regulatory networks [57] and to extract cell type–specific signatures from complex patient samples [59], where single-cell assays might be difficult or expensive. Such approaches could significantly advance our understanding of cell type–specific regulatory networks derived from disease samples. A third challenge is to develop systematic benchmarks for evaluation of regulatory networks in mammalian systems. This would require a concerted effort from experimental and computational researchers to test model-driven predictions in a high-throughput manner. Recent technological advances such as Perturb-Seq [60] and Perturb-ATAC [61] offer powerful tools to perturb and monitor transcriptional state and enable high-throughput validation of network predictions. Such benchmarks would be important for the development and application of regulatory network-based methods for examining normal and disease processes.
Supplementary Material
Acknowledgements
SR acknowledges the support of an NIH grant (R01GM117339), the James McDonell Foundation Grant (#3194-133-349500-4-AAB5159), and UW Data Science Foundation grant. BB is supported by the Genomic Sciences Training Program (NHGRI 5T32HG002760) and an NIH grant (R01HG010045). JS was additionally supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A3A12033291).
Footnotes
Conflict of interest statement
Nothing declared.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.coisb.2020.09.005.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.van der Lee R, Correard S, Wasserman WW: Deregulated regulators: disease-causing cis variants in transcription factor genes. Trends Genet 2020, 36:523–539. [DOI] [PubMed] [Google Scholar]
- 2.Spitz F, Furlong E: Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012, 13: 613–626. [DOI] [PubMed] [Google Scholar]
- 3.The ENCODE Project Consortium: An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Steensel B, Furlong EEM: The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 2019, 20:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzarides T: Chromatin modifications and their function. Cell 2007, 128:693–705. [DOI] [PubMed] [Google Scholar]
- 6.Klemm SL, Shipony Z, Greenleaf WJ: Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 2019, 20: 207–220. [DOI] [PubMed] [Google Scholar]
- 7.Furey TS: ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat Rev Genet 2012, 13:840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M: RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009, 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempfer R, Pombo A: Methods for mapping 3D chromosome architecture. Nat Rev Genet 2020, 21:207–226. [DOI] [PubMed] [Google Scholar]
- 10.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, Mohammadi P, Park Y, Parsana P, Segrè AV, et al. : Genetic effects on gene expression across human tissues. Nature 2017, 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Santpere G, Kawasawa YI, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. : Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 2018:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K, et al. : An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Genome Res 2020, 30: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yus E, Lloréns-Rico V, Martínez S, Gallo C, Eilers H, Blötz C, Stülke J, Lluch-Senar M, Serrano L: Determination of the gene regulatory network of a genome-reduced bacterium highlights alternative regulation independent of transcription factors. Cells 2019, 9:143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chappell L, Russell AJC, Voet T: Single-cell (Multi)omics technologies. Annu Rev Genom Hum Genet 2018, 19:15–41. [DOI] [PubMed] [Google Scholar]
- 15.Thompson DA, Regev A: Fungal regulatory evolution: cis and trans in the balance. FEBS Lett 2009, 583:3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HD, Shay T, O’Shea EK, Regev A: Transcriptional regulatory circuits: predicting numbers from alphabets. Science 2009, 325:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Favera RD, Califano A: ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinf 2006, 7:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siahpirani AF, Roy S: A prior-based integrative framework for functional transcriptional regulatory network inference. Nucleic Acids Res 2017, 45 2221–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P: Inferring regulatory networks from expression data using tree-based methods. PloS One 2010, 5:e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werhli AV, Husmeier D: Reconstructing gene regulatory networks with bayesian networks by combining expression data with multiple sources of prior knowledge. Stat Appl Genet Mol Biol 2007, 6, 15. [DOI] [PubMed] [Google Scholar]
- 21.Deng W, Zhang K, Liu S, Zhao PX, Xu S, Wei H: JRmGRN: joint reconstruction of multiple gene regulatory networks with common hub genes using data from multiple tissues or conditions. Bioinformatics 2018, 34:3470–3478. [DOI] [PubMed] [Google Scholar]
- 22.Ament SA, Pearl JR, Cantle JP, Bragg RM, Skene PJ, Coffey SR, Bergey DE, Wheeler VC, MacDonald ME, Baliga NS, et al. : Transcriptional regulatory networks underlying gene expression changes in Huntington’s disease. Mol Syst Biol 2018, 14:e7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chasman D, Iyer N, Fotuhi Siahpirani A, Estevez Silva M, Lippmann E, McIntosh B, Probasco MD, Jiang P, Stewart R, Thomson JA, et al. : Inferring regulatory programs governing region specificity of neuroepithelial stem cells during early hindbrain and spinal cord development. Cell Syst 2019, 9: 167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.•.Miraldi ER, Pokrovskii M, Watters A, Castro DM, De Veaux N, Hall JA, Lee J-Y, Ciofani M, Madar A, Carriero N, et al. : Leveraging chromatin accessibility for transcriptional regulatory network inference in T Helper 17 Cells. Genome Res 2019, 29:449–463.This approach uses a dependency network learning framework to combine gene expression with ATAC-seq data to reconstruct genome-wide regulatory networks. In addition to using ATAC-seq data to define a structure prior, this approach estimates TF activity that enables an improved regulatory network prediction.
- 25.Mallm J-P, Iskar M, Ishaque N, Klett LC, Kugler SJ, Muino JM, Teif VB, Poos AM, Großmann S, Erdel F, et al. : Linking aberrant chromatin features in chronic lymphocytic leukemia to transcription factor networks. Mol Syst Biol 2019, 15:e8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearl JR, Colantuoni C, Bergey DE, Funk CC, Shannon P, Basu B, Casella AM, Oshone RT, Hood L, Price ND, et al. : Genome-scale transcriptional regulatory network models of psychiatric and neurodegenerative disorders. Cell Syst 2019, 8 122–135.e7. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Cho D-Y, Lee H, Fear J, Oliver B, Przytycka TM: Reprogramming of regulatory network using expression uncovers sex-specific gene regulation in Drosophila. Nat Commun 2018, 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Zhu W, Wang Q, Gu J, Huang LF, Sun X: Differential regulatory network-based quantification and prioritization of key genes underlying cancer drug resistance based on time-course RNA-seq data. PLoS Comput Biol 2019, 15:e1007435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.••.Zhou X, Cai X: Inference of differential gene regulatory networks based on gene expression and genetic perturbation data. Bioinformatics 2020, 36:197–204.FSSEM learns directed edges by integrating gene expression and natural perturbation data (eQTL, FSSEM learns directed edges by integrating gene expression and natural perturbation data (eQTL, CNV, etc) for a pair of conditions. The method imposes sparsity of the individual GRNs and the sparsity of the differences between them. FFSEM was shown to outperform single condition SEM.
- 30.Bhuva DD, Cursons J, Smyth GK, Davis MJ: Differential coexpression-based detection of conditional relationships in transcriptional data: comparative analysis and application to breast cancer. Genome Biol 2019, 20:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.•.Tu J-J, Ou Yang L, Yan H, Zhang X-F, Qin H: Joint reconstruction of multiple gene networks by simultaneously capturing inter-tumor and intra-tumor heterogeneity. Bioinformatics 2020, 36:2755–2762.NETI2 is a sparse Gaussian Graphical Model approach to learn context-specific networks. NETI2 can model within sample heterogeneity as well as between sample heterogeneity by learning context-specific and shared networks. NETI2 was applied to breast cancer subtype data and inferred cancer subtype specific networks as well as a shared network that was common to all subtypes representing noncancerous cells.
- 32.••.Osmanbeyoglu HU, Shimizu F, Rynne Vidal A, Alonso Curbelo D, Chen H-A, Wen HY, Yeung T-L, Jelinic P, Razavi P, Lowe SW, et al. : Chromatin-informed inference of transcriptional programs in gynecologic and basal breast cancers. Nat Commun 2019, 10:1–12.PSIONIC uses a multi-task learning approach to integrate TF motifs, ATAC-seq and tumor expression data to learn patient-specific regression weights that reflect the activities of TFs in multiple cancers. PSIONIC can integrate information from multiple cancer types and was shown to outperform single task approaches.
- 33.Caruana R: Multitask learning. Mach Learn 1997, 28:41–75. [Google Scholar]
- 34.Cancer Genome Atlas Network: Comprehensive molecular portraits of human breast tumours. Nature 2012, 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.•.Castro DM, Veaux NR de, Miraldi ER, Bonneau R: Multi-study inference of regulatory networks for more accurate models of gene regulation. PLoS Comput Biol 2019, 15:e1006591.Inferelator-AMuSR is a dependency network learning approach that uses multi-task learning with structured sparsity to encourage similarity between networks from each context. Inferelator-AMuSR can incorporate additional data as context-specific priors. This approach was applied to learn species-specific regulatory networks in S. cerevisiae and B. subtilis and was shown to outperform ensemble-based or batch correction-based approaches to integrate data for network inference.
- 36.Kumar A, Daumé H: Learning task grouping and overlap in multi-task learning. In proceedings of the 29th international coference on international conference on machine learning Omnipress 2012:1723–1730. [Google Scholar]
- 37.Wang J, Zibetti C, Shang P, Sripathi SR, Zhang P, Cano M, Hoang T, Xia S, Ji H, Merbs SL, et al. : ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat Commun 2018, 9:1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Zhang S, YI X, Plewczynski D, Li MJ: Exploring 3D chromatin contacts in gene regulation: the evolution of approaches for the identification of functional enhancer-promoter interaction. Comput Struct Biotechnol J 2020, 18: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, et al. : Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet 2019, 51:1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijayabaskar MS, Goode DK, Obier N, Lichtinger M, Emmett AML, Abidin FNZ, Shar N, Hannah R, Assi SA, Lie-A-Ling M, et al. : Identification of gene specific cis-regulatory elements during differentiation of mouse embryonic stem cells: an integrative approach using high-throughput datasets. PLoS Comput Biol 2019, 15:e1007337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong Y, Sun J, Wong CF, Kang Q, Ru B, Wong CN, Chan AS, Leung SY, Zhang J: MICMIC: identification of DNA methylation of distal regulatory regions with causal effects on tumorigenesis. Genome Biol 2018, 19:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S, Chasman D, Knaack S, Roy S: In silico prediction of high-resolution Hi-C interaction matrices. Nat Commun 2019, 10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belokopytova PS, Nuriddinov MA, Mozheiko EA, Fishman D, Fishman V: Quantitative prediction of enhancer–promoter interactions. Genome Res 2020, 30:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hait TA, Amar D, Shamir R, Elkon R: FOCS: a novel method for analyzing enhancer and gene activity patterns infers an extensive enhancer–promoter map. Genome Biol 2018, 19:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinstein JN, Collisson EA, Mills GB, Shaw KM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM: The cancer genome atlas pan-cancer analysis project. Nat Genet 2013, 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh S, Yang Y, Póczos B, Ma J: Predicting enhancer-promoter interaction from genomic sequence with deep neural networks. Quant Biol 2019, 7:122–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W, Wong WH, Jiang R: DeepTACT: predicting 3D chromatin contacts via bootstrapping deep learning. Nucleic Acids Res 2019, 47 e60–e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talukder A, Saadat S, Li X, Hu H: EPIP: a novel approach for condition-specific enhancer–promoter interaction prediction. Bioinformatics 2019, 35:3877–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanay A, Regev A: Single cell genomics: from phenomenology to mechanism. Nature 2017, 541:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shema E, Bernstein BE, Buenrostro JD: Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat Genet 2019, 51:19–25. [DOI] [PubMed] [Google Scholar]
- 51.Karemaker ID, Vermeulen M: Single-cell DNA methylation profiling: technologies and biological applications. Trends Biotechnol 2018, 36:952–965. [DOI] [PubMed] [Google Scholar]
- 52.Ramani V, Deng X, Qiu R, Lee C, Disteche CM, Noble WS, Shendure J, Duan Z: Sci-Hi-C: a single-cell Hi-C method for mapping 3D genome organization in large number of single cells. Methods 2020, 170:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonnaffoux A, Herbach U, Richard A, Guillemin A, Gonin-Giraud S, Gros P-A, Gandrillon O: WASABI: a dynamic iterative framework for gene regulatory network inference. BMC Bioinf 2019, 20:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali TM: Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods 2020, 17:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.•.Bravo González Blas C, Quan X, Duran-Romaña R, Taskiran II, Koldere D, Davie K, Christiaens V, Makhzami S, Hulselmans G, Waegeneer M, et al. : Identification of genomic enhancers through spatial integration of single-cell transcriptomics and epigenomics. Mol Syst Biol 2020:16.ScoMAP is an approach that first defines scATAC-seq and scRNA-seq cell clusters and then uses this information and pseudo time to map single cells with scATAC-seq and scRNA-seq measurements to virtual cells that are organized to mimic a 2D representation of the tissue. Using the virtual map, enhancers are linked to target genes with regression trees that predict gene expression based on candidate region accessibility. When applied to the Drosophilia eye-antennal disc, most regions regulating gene expression were in non-promoter regions. For genes that showed cell-type specific expression, promoter accessibility correlated poorly with gene expression indicating the importance of more distal enhancers.
- 56.•.Jansen C, Ramirez RN, El-Ali NC, Gomez-Cabrero D, Tegner J, Merkenschlager M, Conesa A, Mortazavi A: Building gene regulatory networks from scATAC-seq and scRNA-seq using Linked Self Organizing Maps. PLoS Comput Biol 2019, 15: e1006555.SOMatic leverages a self-organizing maps (SOMs) to cluster scRNA-seq and scATAC-seq separately. Each cluster represents genes and genomic regions with similar scRNA-seq or scATAC-seq profiles. A linking function based on the presence of regulatory regions in gene promoters is used to link the scRNA-seq clusters to the scATAC-seq clusters. When applied to mouse pre-B cell differentiation with Ikaros over-expression (a regulator of lymphocyte differentiation), the method was able to recover known and potentially novel targets of Ikaros.
- 57.••.Zeng W, Chen X, Duren Z, Wang Y, Jiang R, Wong WH: DC3 is a method for deconvolution and coupled clustering from bulk and single-cell genomics data. Nat Commun 2019, 10:1–11.A non-negative matrix factorization approach to integrate scATAC-seq and scRNA-seq and HiChIP data to infer cell-type specific regulatory programs. HiChIP data is used to link scATAC-seq clusters to scRNA-seq clusters in a cell-type specific manner. The algorithm is versatile in that can be applied for the integration of any combinations of these three data types whether they originate from single cell or bulk samples.
- 58.Chakraborty A, Ay F: The role of 3D genome organization in disease: from compartments to single nucleotides. Semin Cell Dev Biol 2018, 10.1016/j.semcdb.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Park J, Susztak K, Zhang NR, Li M: Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun 2019, 10:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixit A, Parnas O, Li B, Chen J, Fulco C, Jerby-Arnon L, Marjanovic N, Dionne D, Burks T, Raychowdhury R, et al. : Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 2016:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, Mumbach MR, Ji AL, Kim DS, Cho SW, et al. : Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. e17 Cell 2019, 176:361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.