Abstract
Among the fish of the genus Oryzias, two species are frequently used as model animals in biological research. In Thailand, Oryzias mekongensis is usually found in natural freshwater near the Mekong Basin in the northeast region, while O. songkhramensis inhabits the Songkhram Basin. For differential morphological identification, the coloured bands on the dorsal and ventral margins of the caudal fin are used to distinguish O. mekongensis from O. songkhramensis. However, these characteristics are insufficient to justify species differentiation, and little molecular evidence is available to supplement them. This study aimed to investigate the molecular population and transcriptome profiles of adult O. mekongensis and O. songkhramensis. In the molecular tree based on cytochrome b sequences, O. mekongensis exhibited four clades that were clearly distinguished from O. songkhramensis. Clade 1 of the O. mekongensis population was close to the Mekong River and lived in the eastern portion of the upper northeast region. Clade 2 was far from the Mekong River and inhabited the middle region of the Songkhram River. Clade 3 was positioned to the west of the Songkhram River, and clade 4 was to the south of the Songkhram River Basin. After RNA sequencing using an Illumina HiSeq 2500 platform, the gene category annotations hardly differentiated the species and were discussed in the text. Based on the present findings, population dispersal of these Oryzias species might be associated with geographic variations of the upper northeast region. Molecular genetics and transcriptome profiling might advance our understanding of the evolution of teleost fish.
Introduction
In the teleost genus Oryzias, commonly called “medaka” or “ricefish”, there are more than 25 recognized species in East Asia and Southeast Asia [1, 2]. Two species, Japanese medaka (O. latipes) and Java medaka (O. javanicus), have been utilized as nonmammalian vertebrate models in many molecular biology studies [3, 4]. Experiments using medaka have been performed in fields including molecular physiology, endocrinology, genetics and evolution [3, 5–7]. In Thailand, five species of Oryzias have been reported: O. minutillus, O. javanicus, O. dancena, O. mekongensis and O. songkhramensis [1, 8, 9]. Oryzias minutillus is found throughout freshwater environments in all regions [10], while O. javanicus and O. dancena have a wide range in brackish water [1].
In the northeast region of Thailand, O. mekongensis is widely found in natural freshwater, such as paddy fields and shallow ponds, in the Mekong River Basin, while O. songkhramensis is commonly found in the middle area of the upper northeast region based on the Songkhram River Basin [8, 11]. For morphological identification, orange-red and black bands on the dorsal and ventral portions of the caudal fin are predominant in O. mekongensis [11]. In contrast, light yellow and thin black bands appear on the dorsal and ventral margins of the caudal fin of O. songkhramensis [8]. However, these characteristics are too obscure or insufficient to justify the differentiation of O. mekongensis from O. songkhramensis in the overlapping habitats of both species [12].
Cytochrome b (cytb) is a mitochondrial DNA-encoded polypeptide that has been used for molecular genetic variation in several studies of bony fish [13–15]. Takehana et al. [16] previously clarified the natural distribution of wild Japanese medaka in association with geographic variations based on the cytochrome b gene. The cytb gene is also a potential molecular genetics tool to evaluate fish populations, including medaka [13–16]. In Thailand, only O. minutillus has been investigated at the population genetics level in association with geographic variation [12]. In contrast, the genetic diversity of O. mekongensis and O. songkhramensis in the northeast region of Thailand remains unclear [12].
Transcriptome sequencing is an approach for the analysis of mRNAs that allows the investigation of a set of RNA transcripts to obtain information about gene prediction, gene pathways and gene function [17, 18]. This approach has provided profiles that improved our understanding of molecular processes in many organisms [19–22].
RNA sequencing (RNA-Seq) has been used to achieve transcriptome profiling in teleost fish research [23]. For instance, previous works included transcriptome analysis of Selincuo naked carp (Gymnocypris selincuoensis) [24], comparative transcriptome analysis of four Percidae [25] and transcriptomic characterization of the response to ectoparasitic infection in mangrove rivulus (Kryptolebias marmoratus) [26]. In fish species of Oryzias, transcriptome sequencing data sets have been reported for O. javanicus, O. melastigma, O. latipes and O. woworae [3, 27–30]. Recently, Ngamniyom et al. [12] provided transcriptome profiling data for male and female O. minutillus.
Therefore, the aims of this study were to investigate the molecular populations and transcriptome profiles of wild adult fish of O. mekongensis and O. songkhramensis from Thailand. Furthermore, we emphasized the RNA-Seq data of both species of Oryzias to improve the genetic resources for freshwater teleosts.
Materials and methods
Fish were randomly captured from shallow ponds, ditches and paddy fields in the northeast region of Thailand by using a hand net (Fig 1A) and (Table 1). All field sites for fish sampling were in public areas that did not require any special permit. To confirm the species of Oryzias, adult fish individuals with a standard length (>18 mm) were anaesthetized with 100 mg/L tricaine methanesulfonate solution (MS-222) (Sigma-Aldrich, MO, USA). After anaesthesia, O. mekongensis and O. songkhramensis were identified according to the key ricefish species in Thailand described by Magtoon [8] and Termvidchakorna and Magtoon [9]. The caudal fins of O. mekongensis and O. songkhramensis were dissected from the fish bodies and stored in absolute ethanol at −20°C prior to DNA extraction. The procedure used for the sacrifice of fish was consistent with the Canadian Council on Animal Care Guidelines on the Care and Use of Fish in Research, Teaching and Testing, 2005 (https://www.ccac.ca/Documents/Standards/Guidelines/Fish.pdf) and the National and Institutional Guidelines for Animal Care and Use for Vertebrates from the Institute for Animals for Scientific Purpose Development (IAD) National Research Council of Thailand (NRCT). The Animal Care and Use Committee of Srinakharinwirot University provided ethics approval for all animal experiments in this study, and the approval license was SWU-A-026_2562.
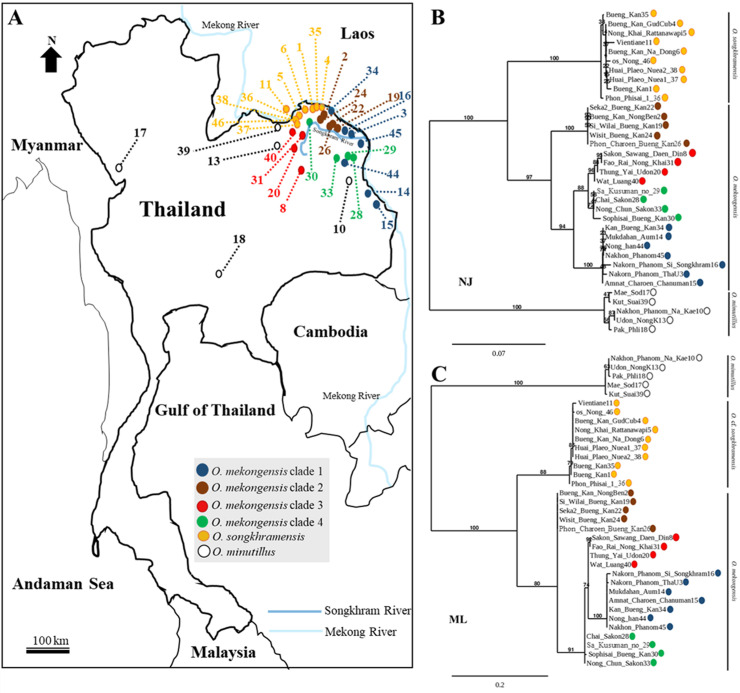
Fig 1. Map drawing of fish collections in the northeast region of Thailand.
(A) Neighbour joining (NJ) method (B) and maximum likelihood (ML) method (C) for the molecular trees of O. mekongensis and O. songkhramensis based on the mitochondrial cytb gene. Brown, red, blue and green circles indicate the individual sample of O. mekongensis for each clade. Yellow circles represent the samples of O. songkhramensis. White circles indicate O. minutillus as an outgroup.
Table 1. Geographic coordinates of collection sites and accession numbers of cytb sequences.
| specimen | geographic coordinate | species | accession number |
|---|---|---|---|
| Bueng Kan1 | 18°23'45.2"N 103°25'58.4"E | O. songkhramensis | MN657279 |
| Bueng Kan35 | 18°24'29.2"N 103°27'40.4"E | O. songkhramensis | MN657280 |
| NongBen2 | 18°20'34.1"N 103°39'50.4"E | O. mekongensis | MN657281 |
| Nakorn Phanom ThaU3 | 17°38'22.0"N 104°26'02.5"E | O. mekongensis | MN657282 |
| GudChap4 | 18°24'00.6"N 103°32'48.9"E | O. songkhramensis | MN657283 |
| Nong Khai Rattanawapi5 | 18°14'20.8"N 103°11'13.6"E | O. songkhramensis | MN657284 |
| Na Dong6 | 18°16'11.2"N 103°17'32.1"E | O. songkhramensis | MN657285 |
| Sawang Daen Din8 | 17°24'00.3"N 103°21'25.5"E | O. mekongensis | MN657286 |
| Na Kae10 | 16°57'45.8"N 104°27'48.8"E | O. minutillus | MN657287 |
| Vientiane11 | 18°12'17.0"N 102°49'31.8"E | O. songkhramensis | MN657288 |
| Udon NongK13 | 17°43'04.3"N 102°50'50.8"E | O. minutillus | MN657289 |
| Mukdahan Aum14 | 16°21'55.5"N 104°33'24.8"E | O. mekongensis | MN657290 |
| Chanuman15 | 16°13'48.3"N 104°59'09.7"E | O. mekongensis | MN657291 |
| Si Songkhram16 | 17°43'12.0"N 104°19'06.2"E | O. mekongensis | MN657292 |
| Mae Sod17 | 16°42'27.2"N 98°36'43.8"E | O. minutillus | MN657293 |
| Pak Phli18 | 14°05'37.1"N 101°16'37.0"E | O. minutillus | MN657294 |
| Si Wilai19 | 17°57'31.1"N 104°02'15.5"E | O. mekongensis | MN657295 |
| Thung Yai20 | 17°30'24.8"N 103°12'03.2"E | O. mekongensis | MN657296 |
| Seka22 | 18°11'46.3"N 103°44'57.1"E | O. mekongensis | MN657297 |
| Wisit24 | 18°22'17.0"N 103°37'08.8"E | O. mekongensis | MN657298 |
| Phon Charoen26 | 18°01'32.0"N 103°37'31.1"E | O. mekongensis | MN657299 |
| Chai Sakon28 | 17°20'48.2"N 104°15'54.7"E | O. mekongensis | MN657300 |
| SKusuman29 | 17°21'47.7"N 104°10'29.5"E | O. mekongensis | MN657301 |
| Sophisai30 | 18°08'25.4"N 103°31'03.0"E | O. mekongensis | MN657302 |
| Fao Rai31 | 17°59'30.5"N 103°23'16.4"E | O. mekongensis | MN657303 |
| Nong Chun33 | 17°35'46.7"N 104°00'23.4"E | O. mekongensis | MN657304 |
| Kan Bueng Kan34 | 18°15'24.6"N 103°52'10.9"E | O. mekongensis | MN657305 |
| Phon Phisai36 | 18°05'18.2"N 103°04'57.7"E | O. songkhramensis | MN657306 |
| Huai Plaeo Nuea37 | 18°03'06.7"N 103°08'07.3"E | O. songkhramensis | MN657307 |
| Huai Plaeo Nuea38 | 18°03'41.0"N 103°08'56.4"E | O. songkhramensis | MN657308 |
| Kut Suai39 | 17°54'19.6"N 103°03'12.3"E | O. minutillus | MN657309 |
| Wat Luang40 | 17°55'25.3"N 103°03'28.6"E | O. mekongensis | MN657310 |
| Nong han44 | 17°15'39.3"N 104°09'16.8"E | O. mekongensis | MN657311 |
| Nakhon Phanom45 | 17°36'42.3"N 104°26'12.1"E | O. mekongensis | MN657312 |
| os Nong46 | 18°03'26.9"N 103°05'25.4"E | O. songkhramensis | MN657313 |
The genomic DNA of the medaka fin was extracted by using the DNeasy Blood & Tissue Kit (Qiagen, Germany) according to the manufacturer's protocol. The paired primers used for DNA target amplification of cytb were 5´–ggACgCTCCgCTgCTAgCCC–3´ and 5´–CCTggTTTgggAgTCAggg–3´ (~1394 bp) with Ex Taq polymerase (Takara, Japan). The PCR thermal cycling steps consisted of an initial denaturation at 95°C for 3 min; 33 cycles of denaturation for 30 s at 94°C, annealing for 40 s at 55°C, and extension for 2 min at 72°C; and a final extension for 5 min at 72°C. The PCR products were confirmed on 1% agarose gels stained with SmartGlow™ Safe Green Pre-Stain (Accuris Instruments, US) under a Blue light DNA transilluminator, and they were extracted from the gels by using the QIAquick Gel Extraction Kit (Qiagen, Germany) according to the manufacturer's instructions. DNA sequencing was conducted using an automated DNA analyser ABI 3730xl system (Applied Biosystems, US). Nucleotide sequences were deposited in GenBank from the National Center for Biotechnology Information (NCBI) with the following accession numbers: MN657279-MN657313 for cytb. The nucleotide sequences of all samples were aligned and trimmed with MUltiple Sequence Comparison by Log-Expectation (MUSCLE) software [31] and curated using Gblocks [32]. Molecular trees with neighbour-joining and maximum likelihood were generated and rendered using PhyML with 1000 bootstraps and TreeDyn [33]. Bootstrap support was justified at > 70%.
For RNA-Seq analysis, twenty males and twenty females of O. mekongensis (n = 40) sampled evenly from all sites and twenty males and twenty females of O. songkhramensis (n = 40) sampled equally from all sites were separated by sex and species and maintained in aquariums containing freshwater without chlorine. The environments of the aquaria were established, and the fish were allowed to acclimatize for 1 week under the following conditions: pH, 6.9–7.3; salinity, 0.05–0.06 ppt; 27–29°C; dissolved oxygen, 6.0–6.5 mg/L. The fish were fed ad libitum 2 times per day with Kyorin Hikari food for medaka (Kyorin Food Industries, Hyogo, Japan) under a photoperiod of 12 hr light:12 hr darkness. The water in the fish aquaria was changed every day.
Total RNA extractions were performed by using an RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. During RNA purification, the nucleic solutions were treated with RNase-free DNase I (Qiagen, Germany). The quality of the total RNA was evaluated by gel electrophoresis in 1% agarose gels. The concentrations were quantified by using Thermo Scientific™ NanoDrop 2000 and 2000c (Thermo Fisher Scientific, MA, USA) and confirmed via Qubit 2.0 fluorometric quantitation (Thermo Fisher Scientific, MA, USA). Equal concentrations of RNA of both species were preserved at –80°C.
After the quality control procedures, the mRNAs were enriched from the total RNA using oligo(dT)25 beads (200 ng RNA per sample), and fragments were randomly placed in RNA fragmentation buffer (Illumina, CA, USA). The rRNA was removed using the Illumina Ribo-Zero Plus rRNA Depletion Kit (Illumina, WI, USA). For first-strand synthesis, complementary DNA (cDNA) synthesis was performed using a TruSeq mRNA kit (Illumina, CA, USA) with random hexamers and a SuperScript® III First-Strand Synthesis System (Invitrogen, CA, USA). Second-strand cDNA was synthesized in a buffer (mRNAseq Illumina, CA, USA) supplemented with E. coli polymerase I, RNase H and dNTPs, and the cDNAs were purified by AMPure XP beads. The cDNA library quantification was measured by using a Qubit 2.0 fluorometer, and the insert size was verified with an Agilent 2100 Bioanalyzer with an Agilent RNA 6000 Nano Kit (Agilent Technologies, MD, USA). The sequencing was processed on a HiSeq 2500 Sequencing System (Illumina, CA, USA) according to Dillies et al. [34]. The accession numbers of the transcriptome data were GSE142602, GPL27950, GSM4232740, PRJNA635451 and SAMN15041390, which were deposited in a public functional genomics database, the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra). For transcriptome reconstruction, SOAPnuke v1.5.2 software was used to filter raw reads, low-quality reads, noisy reads, adaptor-polluted reads and reads with a high content of unknown bases to obtain clean reads. Trinity software (r20140413p1) composed of Inchworm, Chrysalis and Butterfly was used to analyse the assembly data, and Tgicl v2.0.6 was applied to cluster transcripts [35]. Nt (nucleotide) databases were annotated by using NCBI blast v2.2.23. Diamond software v0.8.31 was used for Nr (NCBI non-redundant protein sequences), KOG (euKaryotic Orthologous Groups) and KAAS r140224 for KEGG (Kyoto Encyclopedia of Genes and Genome). Pfam and protein prediction were analysed by the HMMER 3.0 package hmmscan and Blast2GO v2.5.0. NR was used to carry out Gene Ontology (GO) annotation. KEGG annotations were assigned by using the KEGG Automatic Annotation Server. KEGG enrichment was performed with GOSeq 1.10.0, topGO 2.10.0 for GO and KOBAS v2.0.12 for KEGG. SwissProt (UniProtKB/Swiss-Prot) was used for the manually annotated and reviewed database of the UniProt Knowledgebase (UniProtKB) (http://ftp.ebi.ac.uk/pub/databases/swissprot). Unigenes encoding predicted transcription factors (TFs) were mapped to the AnimalTFDB2.0 database to obtain the TF family using getorf EMBOSS:6.5.7.0 with–minsize 150 parameters and hmmsearch v3.0. Differentially expressed genes (DEGs) were identified according to the gene expression levels of both groups by using DEGseq, DEseq2, EBseq, NOIseq and PossionDis to detect the DEGs. The DEG identification methods used the log2-fold change ratios between samples and the P value (cut off 0.05) and q-value for PossionDis or DEseq2. Heatmaps of TF expression and DEGs were created with pheatmap and the appropriate function of R.
Results
The O. songkhramensis populations were found to be limited to the upper west part of the Songkhram River Basin, while the O. mekongensis populations were spread throughout the upper northeast region based on the Mekong River Basin (Fig 1A). Regarding molecular variation based on the cytb gene, O. mekongensis (n = 20) was clearly isolated from O. songkhramensis (n = 10) with bootstrap support (≥ 80%) and from O. minutillus (n = 5) (≥ 100%) for the neighbour-joining and maximum likelihood methods (Fig 1B and 1C).
All specimens of O. mekongensis were monophyletic and were a sister group to the O. songkhramensis populations. Among the three Oryzias species, the O. mekongensis populations were more closely related to O. songkhramensis than were the O. minutillus population. Four clades of wild fish populations were represented at the molecular level in O. mekongensis according to neighbour-joining and maximum likelihood (≥ 70%). In contrast, there were no subgroups of the O. songkhramensis population, which had only a single clade. For the clades of the O. mekongensis population, clade 1 approached the Mekong River and lived in the eastern part of the upper northeast region (n = 7). The fish in clade 2 spread out from the Mekong River and were distributed across the middle part of the Songkhram River (n = 5). Clade 3 of the O. mekongensis population naturally inhabited the western part of the Songkhram River Basin (n = 4). Conversely, clade 4 comprised the fish population in the southern part of the Songkhram River (n = 3). However, one sample of this clade was biased to the west (Fig 1B and 1C).
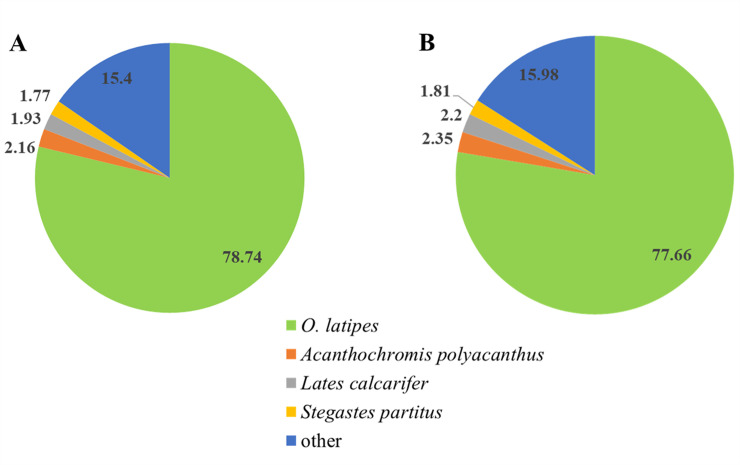
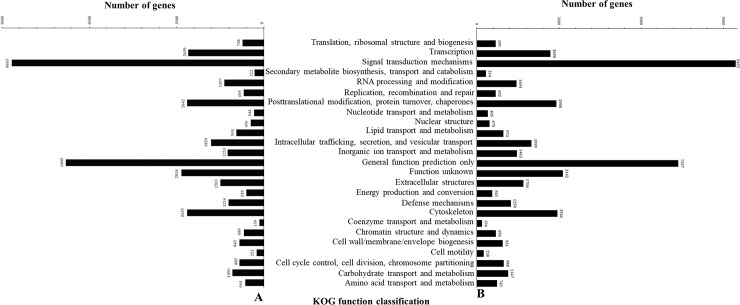
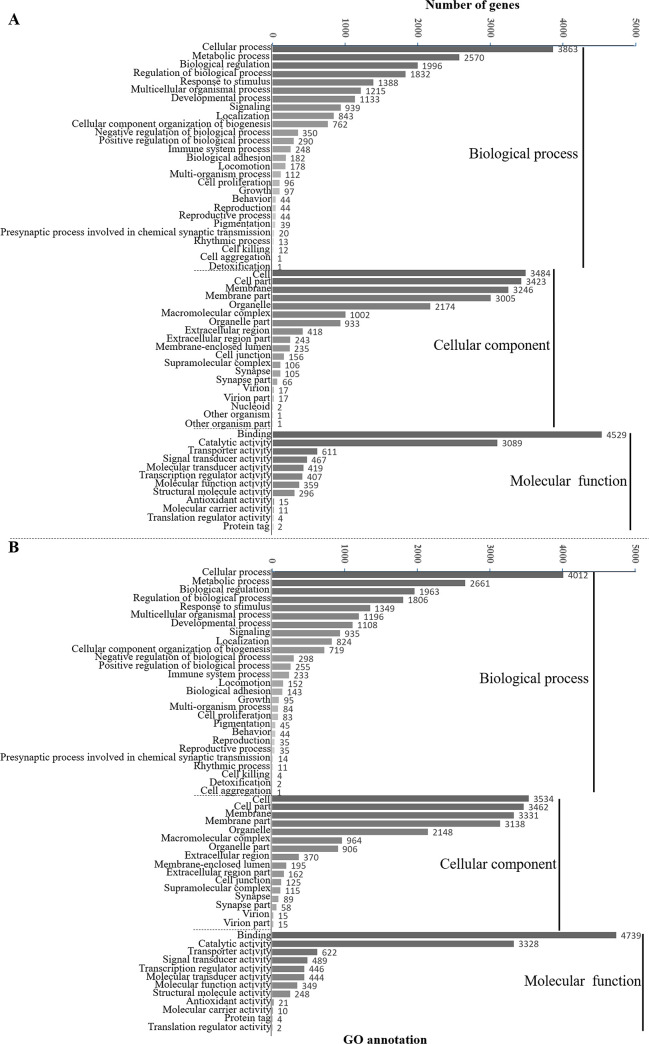
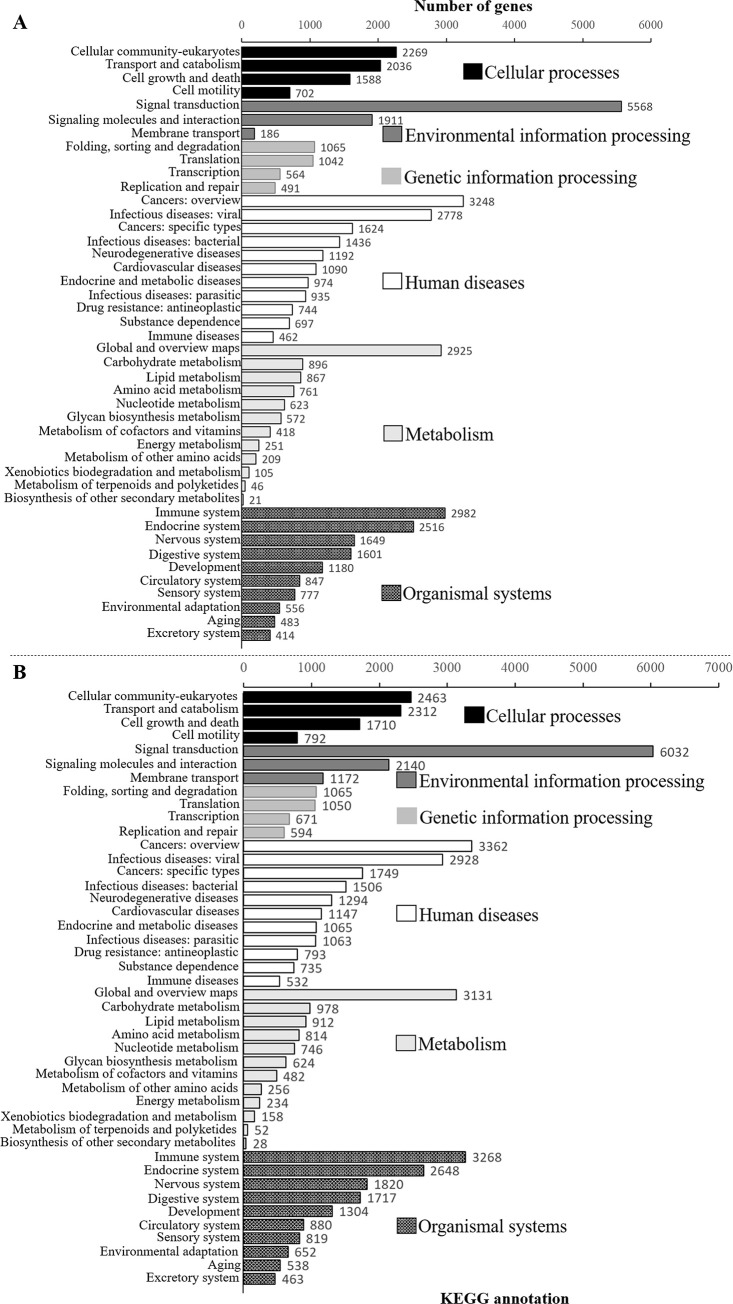
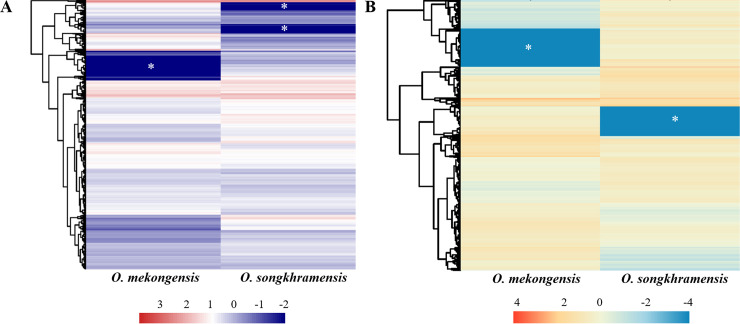
In the transcriptomic analysis of the adult fish, the distributions of the non-redundant database (NR) annotated species of O. mekongensis and O. songkhramensis were similarly matched to O. latipes at 78.74 and 77.66%, respectively (Fig 2A and 2B). In KOG (euKaryotic Orthologous Groups), 25 functions were contained in O. mekongensis and O. songkhramensis, and “signal transduction mechanisms” and “general function prediction only” dominated in both species of medaka fish. In contrast, “coenzyme transport and metabolism” was the least represented among the gene function types in both O. mekongensis and O. songkhramensis (Fig 3A and 3B). In the gene ontology (GO) annotation, the three main categories of gene or gene products were represented, including “biological process”, “cellular component” and “molecular function”. Twenty-seven gene processes of “biological process” and twelve gene functions of “molecular function” were detected in O. mekongensis and O. songkhramensis. However, “cellular component” accounted for 19 and 16 components in O. mekongensis and O. songkhramensis, respectively. The categories “components of nucleoids”, “other organisms” and “other organism parts” were lacking in O. songkhramensis. Cellular process, cell and binding accounted for the highest gene numbers for their categories in both species (Fig 4A and 4B). In the KEGG (Kyoto Encyclopedia of Genes and Genome), the six major classes consisted of “cellular processes”, “environmental information processing”, “genetic information processing”, “human diseases”, “metabolism” and “organismal systems”. Cellular community-eukaryotes, signal transduction, folding, sorting and degradation, cancers: overview, global and overview maps and immune system showed the highest gene numbers among their classes for both O. mekongensis and O. songkhramensis (Fig 5A and 5B). In the SwissProt annotation, the unigenes of O. mekongensis approached sp|Q5T197|DCST1_HUMAN with expectation values (e-values) of 1.3e-27 and 1.0e-20 and sp|Q68F72|S15A4_XENLA with e-values of 1.2e-218 and 2.1e-55 in the SwissProt database. In O. songkhramensis, the unigenes were annotated to sp|P58875|S14L2_BOVIN with e-values of 1.3e-108 and 5.6e-169, sp|Q91WM3|U3IP2_MOUSE with an e-value of 1.6e-158, and sp|Q14416|GRM2_HUMAN with an e-value of 1.7e-148. Details of the annotation list format are provided in the S1 Table for O. mekongensis and O. songkhramensis. Regarding TF expression, one cluster showed quite low expression in O. mekongensis compared with O. songkhramensis. In contrast, two clusters showed clearly lower TF expression in O. songkhramensis than in O. mekongensis (Fig 6A). In the DEGs, there was one gene cluster of differentially expressed factors who expression quite low in O. mekongensis compared with O. songkhramensis. The low gene expression of O. mekongensis (14.58–12.46 log2-fold) consisted of the microtubule-associated protein TAU (Mapt), integral membrane protein 2B (Itm2b or Bri2), calpactin-1 light chain, interferon regulatory factor 2 binding protein 2 (Irf2bp2), heterogeneous nuclear ribonucleoprotein A1 (Hnrnpa1), microsomal glutathione S-transferase 3 (Mgst3), lysophosphatidic acid receptor 6-like (Lpar6), histone-lysine N-methyltransferase EHMT1-like, Wiskott-Aldrich syndrome protein (WASp) and 26S protease regulatory subunit 8 (Psmc5). Similarly, for the DEGs of O. mekongensis, one cluster showed obviously lower gene expression in O. songkhramensis (Fig 6B). Gene expression of O. songkhramensis was detected at low levels (16.16–12.57 log2-fold) compared with that of O. mekongensis, followed by heat shock protein family A member 8 (Hspa8), tyrosine-protein phosphatase non-receptor (Ptpn1), sister chromatid cohesion protein PDS5 homolog B (Pds5b), cyclin-dependent kinase 8 (Cdk8), TBCC domain-containing protein 1 (Tbccd1), protein disulfide-isomerase A3 (Pdia3), calpain-1, coatomer subunit alpha, junction plakoglobin-like, plakophilin 2 and apolipoprotein A-IV-like (ApoA4).
Fig 2.
Distribution of non-redundant database annotated species (%) of O. Mekongensis (A) and O. Songkhramensis (B). The image refers to O. latipes, A. polyacanthus, L. calcarifer and S. partitus.
Fig 3.
EuKaryotic Orthologous Groups (KOG) classification and gene number in O. Mekongensis (A) and O. Songkhramensis (B). Twenty-five classes of KOG were observed between the two species.
Fig 4.
Gene Ontology (GO) for annotation of gene number in O. Mekongensis (A) and O. Songkhramensis (B). Three major criteria for GO between both species.
Fig 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) in O. Mekongensis (A) and O. Songkhramensis (B). They represented the six main classifications and gene numbers.
Fig 6.
Unigenes encoding predictions of transcription factor expression (A) and differentially expressed genes (B) between O. Mekongensis and O. Songkhramensis from the heatmap representations. Asterisks indicate the substantially lower expression of gene clusters in both species.
Discussion
In this study, the populations of O. mekongensis could be divided into four groups based on the mitochondrial cytb gene. This result was consistent with reports of fish populations that examined genetic variation during succession by using the cytb gene [14, 15, 36]. In Oryzias, Takehana et al. [16] reported the use of the cytb gene to understand the diversity of wild Japanese medaka populations that correlated with their geographic variation in Japan. Therefore, the cytb gene might be suitable for evaluating the genetic variation of O. mekongensis populations in the northeastern region of Thailand. Moreover, this mitochondrial gene is also a potential tool to phylogenetically differentiate among O. mekongensis, O. songkhramensis and O. minutillus. It is well known that the cytb gene is one of the standard DNA barcodes for fishes [37, 38]. The cytb gene might converted to a molecular barcode and utilized in the further identification of freshwater ricefish in Thailand. Ngamniyom et al. [12] reported a molecular diversity destitution of wild Thai medaka with biogeographic variation throughout Thailand. In the present study, the wild populations of O. mekongensis showed genetic variation but the populations of O. songkhramensis did not. The physical geography of northeastern Thailand is a plateau, and the upper part forms the structure of the Sakon Nakorn Basin involving the Mekong River, Songkhram River and complexed canals [39, 40]. Thus, the genetic diversity of O. mekongensis might depend on the Mekong and Songkhram River Basins. Magtoon [8] described the natural habitat of O. songkhramensis, which mainly lives in the upper part of the Mekong River Basin in northeastern Thailand. The distribution of O. songkhramensis was not wider than that of O. mekongensis. The population of O. songkhramensis in this study was similar to the original findings for this fish population in the report of Magtoon [8]. These results suggest that the range distribution of O. songkhramensis may be narrow, which may have resulted in the single group within the O. songkhramensis population in this study.
Morphologically, O. mekongensis and O. songkhramensis have different coloured lines on their caudal fins, but there are no colored lines on the caudal fin of O. minutillus [1, 8]. Among the three species in this study, O. mekongensis was more closely related to O. songkhramensis than O. minutillus based on the cytb partial sequences. This molecular biology result is congruent with the morphology of the caudal fin, which may be considered both with and without coloured lines.
In RNA-Seq analysis, the species distributions of O. mekongensis and O. songkhramensis were closer to those of Japanese medaka than to those of the spiny chromis (Acanthochromis polyacanthus), barramundi (Lates calcarifer), and bicolour damselfish (Stegastes partitus). Investigators have already provided genome data for these fish species from transcriptome profiling, genome information and biosystem databases of the NCBI [41]. Moreover, the species distributions of both of the fish in this study were more similar to those of Japanese medaka than to those of Thai medaka in a previous report by Ngamniyom et al. [12]. RNA-Seq of O. mekongensis and O. songkhramensis confirmed that transcriptomic sequences of both species were conserved with medaka species close to Japanese medaka. Furthermore, the results of this study add to the molecular genetics resources for teleost fish.
In the databases of annotated genes, the KOG, GO and KEGG catalogue groups were quite similar for the gene functions of O. mekongensis and O. songkhramensis. The predominant gene functions and processes of O. mekongensis and O. songkhramensis are presented. In addition, the general gene functions of both fish species in this study were consistent with several reports of freshwater fish, such as Thai medaka, pond loach (Misgurnus anguillicaudatus), Yellow River scaleless carp (Gymnocypris eckloni) and goldfish (Carassius auratus) [12, 42–44]. These results suggested that the represented gene predictions of O. mekongensis and O. songkhramensis might conserve the main gene functions not only among Oryzias fish but also among other freshwater fish. It is well known that unigene annotations against the databases of KOG, GO and KEGG are important for understanding the candidate genes, gene functions and biological pathways in various organisms [45, 46]. The gene predictions of O. mekongensis and O. songkhramensis were dominated by “signal transduction, signal mechanism, cellular processes, cell and binding”. Therefore, these gene functional groups might be necessary for important roles in the cell biology of O. mekongensis and O. songkhramensis, as annotated by KOG, GO and KEGG. Regarding TF expression and DEGs, TFs have been investigated as an important step in gene regulatory networks [47], and DEGs are utilized as potential markers to observe different patterns of gene expression [48]. In this study, there were polymorphisms of gene expression clusters between O. mekongensis and O. songkhramensis. These findings might be species-specific gene expression patterns that differ between O. mekongensis and O. songkhramensis.
The analysis of DEGs can be performed through transcriptomic profiles to understand alterations in genes or genes in different species [49–51]. For DEG screening, the transcriptomic analyses showed that some gene expression levels were different between O. mekongensis and O. songkhramensis. One cluster of genes was highly expressed in O. mekongensis compared with O. songkhramensis. In contrast, one cluster showed higher gene expression in O. songkhramensis than in O. mekongensis.
In general pathways or functions of gene, the Mapt is microtubules associated protein regulated by phosphorylation [52], while the Itm2b plays a role of neurite outgrowth [53]. Calpactin-1 light chain regulate cytoskeletal proteins [54], and WASp is regulator of the actin cytoskeleton [55]. In vertebrates, Irf2bp2 is known as a transcriptional corepressor [56]. Hnrnpa1 is important regulation of RNA synthesis [57], but Histone-lysine N-methyltransferase EHMT1 exhibit functions in silencing of gene expression [58]. The Lpar6 is G protein-coupled receptor of lipid signaling [59], and the Mgst3 is essential gene in metabolize of endogenous and exogenous substrates [60]. In addition, Psmc5 plays the important processes for the maintenance of protein homeostasis [61].
Hspa8 functions as a chaperone in a cellular folding of translation, and Ptpn1works as a regulator of unfolding [62, 63]. It has known that Pds5b plays a role during meiosis and DNA repair associated with chromatin [64], while Cdk8 is a coactivator in the regulated transcription of genes [65]. Gonçalves et al [66] demonstrated that Tbccd1 was required for a centrosome positioning. Pdia3 played the crucial role for catalysing formation of disulfide bonds in proteins [67]. In neurons, the calpain-1is calcium-dependent cysteine protease for neurodegeneration [68]. For cytosolic protein complex, the coatomer subunit alpha binds to dilysine motifs between the endoplasmic reticulum and golgi body [69]. Plakophilin 2 is known for a role in cell–cell adhesion [70]. In major component of high density lipoprotein, ApoA4 plays an important function in lipoprotein metabolism [71]. Considering into all above mentions of gene functions and pathways, they might therefore be differences in such predominant level-specific gene expression profiles between the two fish.
To distribute the present knowledge, we provided the molecular populations of O. mekongensis and O. songkhramensis and the transcriptomic patterns of these species from Thailand in publicly available databases. These results may enhance the available genetic resources to further understand the evolution of fish in the genus Oryzias.
Supporting information
Tables show the information from the SwissProt database for both species.
(XLS)
Data Availability
All nucleotide sequences are available from GenBank database (accession numbers MN657279, MN657280, MN657281, MN657282, MN657283, MN657284, MN657285, MN657286, MN657287, MN657288, MN657289, MN657290, MN657291, MN657292, MN657293, MN657294, MN657295, MN657296, MN657297, MN657298,MN657299, MN657300, MN657301, MN657302, MN657303, MN657304, MN657305, MN657306, MN657307, MN657308, MN657309, MN657310, MN657311, MN657312, MN657313. Gene Expression Omnibus data is available from NCBI (accession number GSE142602, GPL27950). Sequence Read Archive (SRA) database is available from NCBI (accession numbers SRS6721712, SRP265012).
Funding Statement
The author received no specific funding for this work.
References
- 1.Parenti LR. A phylogenetic analysis and taxonomic revision of ricefishes, Oryzias and relatives (Beloniformes, Adrianichthyidae). Zool J Linn Soc. 2008;154: 494–610. [Google Scholar]
- 2.Parenti L, Hadiaty R, Lumbantobing D, Herder F. Two new ricefishes of the genus Oryzias (Atherinomorpha: Beloniformes: Adrianichthyidae) augment the endemic freshwater fish fauna of Southeastern Sulawesi, Indonesia. Copeia. 2013;2013: 403–414. [Google Scholar]
- 3.Takehana Y, Zahm M, Cabau C, Klopp C, Roques C, Bouchez O, et al. Genome sequence of the Euryhaline Javafish medaka, Oryzias javanicus: a small aquarium fish model for studies on adaptation to salinity. G3. 2020;10: 907–915. 10.1534/g3.119.400725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilgers L, Schwarzer J. The untapped potential of medaka and its wild relatives. Elife. 2019;8: e46994 10.7554/eLife.46994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inoue K, Takei Y. Asian medaka fishes offer new models for studying mechanisms of seawater adaptation. Comp Biochem Physiol B Biochem Mol Biol. 2003;136: 635–645. 10.1016/s1096-4959(03)00204-5 [DOI] [PubMed] [Google Scholar]
- 6.Setiamarga DH, Miya M, Yamanoue Y, Azuma Y, Inoue JG, Ishiguro NB, et al. Divergence time of the two regional medaka populations in Japan as a new time scale for comparative genomics of vertebrates. Biol Lett. 2009;5: 812–816. 10.1098/rsbl.2009.0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai S, Koyama J, Fujii K. Effects of 17β-estradiol on the reproduction of Java-medaka (Oryzias javanicus), a new test fish species. Mar Pollut Bull. 2005;51: 708–714. 10.1016/j.marpolbul.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 8.Magtoon W. Oryzias songkhramensis, a new species of ricefish (Beloniformes; Adrianichthyidae) from Northeast Thailand and Central Laos. Trop Nat Hist. 2010;10: 107–129. [Google Scholar]
- 9.Termvidchakorn A, Magtoon W. Development and identification of the ricefish Oryzias in Thailand. ScienceAsia. 2008;34: 416–423. [Google Scholar]
- 10.Ngamniyom A, Magtoon W, Nagahama Y, Sasayama Y. Expression levels of hormone receptors and bone morphogenic protein in fins of medaka. Zoolog Sci. 2009;26: 74–79. 10.2108/zsj.26.74 [DOI] [PubMed] [Google Scholar]
- 11.Uwa H, Magtoon W. Description and karyotype of a new ricefish, Oryzias mekongensis, from Thailand. Copeia. 1986;1986: 473–478. [Google Scholar]
- 12.Ngamniyom A, Sriyapai T, Sriyapai P. Molecular analysis of population and de novo transcriptome sequencing of Thai medaka, Oryzias minutillus (Teleostei: Adrianichthyidae). Heliyon. 2020;6: e03079 10.1016/j.heliyon.2019.e03079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker CS, Perry A, Chambers GK, Smith PJ. Population variation in the mitochondrial cytochrome b gene of the orange roughy Hoplostethus atlanticus and the hoki Macruronus novaezelandiae. Mar Biol. 1995;122: 503–509. [Google Scholar]
- 14.Habib M, Lakra WS, Mohindra V, Khare P, Barman AS, Singh A, et al. Evaluation of cytochrome b mtDNA sequences in genetic diversity studies of Channa marulius (Channidae: Perciformes). Mol Biol Rep. 2011;38: 841–846. 10.1007/s11033-010-0175-2 [DOI] [PubMed] [Google Scholar]
- 15.Çiftci Y, Eroğlu O, Firidin Ş. Mitochondrial cytochrome b sequence variation in three Sturgeon species (A. stellatus Pallas, 1771, A. gueldenstaedtii Brandt, 1833, H. huso Linnaeus, 1758) from the Black Sea Coasts of Turkey. Turk J Fish Aquat Sci. 2013;13: 291–303. [Google Scholar]
- 16.Takehana Y, Nagai N, Matsuda M, Tsuchiya K, Sakaizumi M. Geographic variation and diversity of the cytochrome b gene in Japanese wild populations of medaka, Oryzias latipes. Zoolog Sci. 2003;20: 1279–1291. 10.2108/zsj.20.1279 [DOI] [PubMed] [Google Scholar]
- 17.Kukurba KR, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015;2015: 951–969. 10.1101/pdb.top084970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, He L, Cai L. Transcriptome sequencing: RNA-seq. Methods Mol Biol. 2018;1754: 15–27. 10.1007/978-1-4939-7717-8_2 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10: 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrne A, Cole C, Volden R, Vollmers C. Realizing the potential of full-length transcriptome sequencing. Philos Trans R Soc Lond B Biol Sci. 2019;374: 20190097 10.1098/rstb.2019.0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins LJ, Biggs PJ, Voelckel C, Joly S. An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Inform. 2008;21: 3–14. [PubMed] [Google Scholar]
- 22.Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA. 2017;8 10.1002/wrna.1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian X, Ba Y, Zhuang Q, Zhong G. RNA-Seq technology and its application in fish transcriptomics. OMICS. 2014;18: 98–110. 10.1089/omi.2013.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng X, Jia Y, Zhu R, Chen K, Chen Y. Characterization and analysis of the transcriptome in Gymnocypris selincuoensis on the Qinghai-Tibetan Plateau using single-molecule long-read sequencing and RNA-seq. DNA Res. 2019;26: 353–363. 10.1093/dnares/dsz014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie P, Yi SK, Yao H, Chi W, Guo Y, Ma XF, et al. Comparative transcriptome analysis reveals potential evolutionary differences in adaptation of temperature and body shape among four Percidae species. PLoS One. 2019;14: e0215933 10.1371/journal.pone.0215933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawluk RJ, Uren Webster TM, Cable J, Garcia de Leaniz C, Consuegra S. Immune-related transcriptional responses to parasitic infection in a naturally inbred fish: roles of genotype and individual variation. Genome Biol Evol. 2018;10: 319–327. 10.1093/gbe/evx274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YK, Lee SY, Kim BS, Kim DS, Nam YK. Isolation and mRNA expression analysis of aquaporin isoforms in marine medaka Oryzias dancena, a euryhaline teleost. Comp Biochem Physiol A Mol Integr Physiol. 2014;171: 1–8. 10.1016/j.cbpa.2014.01.012 [DOI] [PubMed] [Google Scholar]
- 28.Kim B-M, Choi B-S, Kim H-S, Rhee J-S, Au DWT, Wu RSS, et al. Transcriptome profiling of larvae of the marine medaka Oryzias melastigma by Illumina RNA-seq. Mar Genomics. 2015;24: 255–258. 10.1016/j.margen.2015.07.013 [DOI] [PubMed] [Google Scholar]
- 29.Schartl M, Shen Y, Maurus K, Walter R, Tomlinson C, Wilson RK, et al. Whole body melanoma transcriptome response in medaka. PLoS One. 2015;10: e0143057 10.1371/journal.pone.0143057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mokodongan DF, Montenegro J, Mochida K, Fujimoto S, Ishikawa A, Kakioka R, et al. Phylogenomics reveals habitat-associated body shape divergence in Oryzias woworae species group (Teleostei: Adrianichthyidae). Mol Phylogenet Evol. 2018;118: 194–203. 10.1016/j.ympev.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56: 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- 33.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36: W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14: 671–683. 10.1093/bib/bbs046 [DOI] [PubMed] [Google Scholar]
- 35.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phadphon P, Amontailak T, Kotchantuek N, Srithawong S, Kutanan W, Suwannapoom C. Genetic diversity of the endangered mekong giant catfish, striped catfish, and their hybrids from Thailand. Trop Conserv Sci. 2019;12: 1940082919869487. [Google Scholar]
- 37.Hänfling B, Lawson Handley L, Read DS, Hahn C, Li J, Nichols P, et al. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol Ecol. 2016;25: 3101–3119. 10.1111/mec.13660 [DOI] [PubMed] [Google Scholar]
- 38.Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989;86: 6196–6200. 10.1073/pnas.86.16.6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kutanan W, Ghirotto S, Bertorelle G, Srithawong S, Srithongdaeng K, Pontham N, et al. Geography has more influence than language on maternal genetic structure of various northeastern Thai ethnicities. J Hum Genet. 2014;59: 512–520. 10.1038/jhg.2014.64 [DOI] [PubMed] [Google Scholar]
- 40.Shrestha S, Bhatta B, Shrestha M, Shrestha PK. Integrated assessment of the climate and landuse change impact on hydrology and water quality in the Songkhram River Basin, Thailand. Sci Total Environ. 2018;643: 1610–1622. 10.1016/j.scitotenv.2018.06.306 [DOI] [PubMed] [Google Scholar]
- 41.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38: D492–D496. 10.1093/nar/gkp858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo W, Cao X, Xu X, Huang S, Liu C, Tomljanovic T. Developmental transcriptome analysis and identification of genes involved in formation of intestinal air-breathing function of Dojo loach, Misgurnus anguillicaudatus. Sci Rep. 2016;6: 31845 10.1038/srep31845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi D, Chao Y, Wu R, Xia M, Chen Q, Zheng Z. Transcriptome analysis provides insights into the adaptive responses to hypoxia of a schizothoracine fish (Gymnocypris eckloni). Front Physiol. 2018;9: 1326 10.3389/fphys.2018.01326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Tan H, Zhang M, Zhao R, Wang S, Qin Q, et al. The hybrid genome of a new goldfish-like fish lineage provides insights into the origin of the goldfish. Front Genet. 2020;11: 122 10.3389/fgene.2020.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14: 482–517. 10.1038/s41596-018-0103-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakaya A, Katayama T, Itoh M, Hiranuka K, Kawashima S, Moriya Y, et al. KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2013;41: D353–357. 10.1093/nar/gks1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47: W212–W224. 10.1093/nar/gkz446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang S, Zhou X, Pei Y, Wang H, He K, Zhao A. Identification of differentially expressed genes in porcine ovaries at proestrus and estrus stages using RNA-Seq technique. Biomed Res Int. 2018;2018: 9150723 10.1155/2018/9150723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernal MA, Schunter C, Lehmann R, Lightfoot DJ, Allan BJ, Veilleux HD, et al. Species-specific molecular responses of wild coral reef fishes during a marine heatwave. Sci Adv. 2020;6: eaay3423 10.1126/sciadv.aay3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marra NJ, Stanhope MJ, Jue NK, Wang M, Sun Q, Pavinski Bitar P, et al. White shark genome reveals ancient elasmobranch adaptations associated with wound healing and the maintenance of genome stability. Proc Natl Acad Sci U S A. 2019;116: 4446–4455. 10.1073/pnas.1819778116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Fors L, Slotte T, Theopold U, Binzer-Panchal M, Wheat CW, et al. Differential expression of immune genes between two closely related beetle species with different immunocompetence following attack by asecodes parviclava. Genome Biol Evol. 2020;12: 522–534. 10.1093/gbe/evaa075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbier P, Zejneli O, Martinho M, Lasorsa A, Belle V, Smet-Nocca C, et al. Role of Tau as a Microtubule-Associated Protein: Structural and Functional Aspects. Front Aging Neurosci. 2019;11: 204 10.3389/fnagi.2019.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin L, Fluhrer R, Reiss K, Kremmer E, Saftig P, Haass C. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J Biol Chem. 2008;283(3): 1644–1652. 10.1074/jbc.M706661200 [DOI] [PubMed] [Google Scholar]
- 54.Zokas L, Glenney JR Jr. The calpactin light chain is tightly linked to the cytoskeletal form of calpactin I: studies using monoclonal antibodies to calpactin subunits. J Cell Biol. 1987; 105(5): 2111–2121. 10.1083/jcb.105.5.2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouma G, Burns SO, Thrasher AJ. Wiskott-Aldrich Syndrome: Immunodeficiency resulting from defective cell migration and impaired immunostimulatory activation. Immunobiology. 2009;214(9–10): 778–790. 10.1016/j.imbio.2009.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carneiro FR, Ramalho-Oliveira R, Mognol GP, Viola JP. Interferon regulatory factor 2 binding protein 2 is a new NFAT1 partner and represses its transcriptional activity. Mol Cell Biol. 2011;31(14): 2889–2901. 10.1128/MCB.00974-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi ST, Huang P, Li HP, Lai MM. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 2000;19(17): 4701–4711. 10.1093/emboj/19.17.4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson ZL, Yamamoto TM, McMellen A, Kim H, Hughes CJ, Wheeler LJ, et al. Histone methyltransferases EHMT1 and EHMT2 (GLP/G9A) maintain PARP inhibitor resistance in high-grade serous ovarian carcinoma. Clin Epigenetics. 2019;11(1): 165 10.1186/s13148-019-0758-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tao K, Guo S, Chen R, Yang C, Jian L, Yu H, et al. Lysophosphatidic Acid Receptor 6 (LPAR6) Expression and Prospective Signaling Pathway Analysis in Breast Cancer. Mol Diagn Ther. 2019;23(1): 127–138. 10.1007/s40291-019-00384-3 [DOI] [PubMed] [Google Scholar]
- 60.Uno Y, Murayama N, Kunori M, Yamazaki H. Characterization of microsomal glutathione S-transferases MGST1, MGST2, and MGST3 in cynomolgus macaque. Drug Metab Dispos. 2013;41(9): 1621–1625. 10.1124/dmd.113.052977 [DOI] [PubMed] [Google Scholar]
- 61.Kanayama HO, Tamura T, Ugai S, Kagawa S, Tanahashi N, Yoshimura T, et al. Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms. Eur J Biochem. 1992;206(2): 567–578. 10.1111/j.1432-1033.1992.tb16961.x [DOI] [PubMed] [Google Scholar]
- 62.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6): 670–684. 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sangwan V, Paliouras GN, Abella JV, Dubé N, Monast A, Tremblay ML, et al. Regulation of the Met receptor-tyrosine kinase by the protein-tyrosine phosphatase 1B and T-cell phosphatase. J Biol Chem. 2008;283(49): 34374–34383. 10.1074/jbc.M805916200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118(Pt 10): 2133–2141. 10.1242/jcs.02355 [DOI] [PubMed] [Google Scholar]
- 65.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16(4): 509–520. 10.1016/j.molcel.2004.10.014 [DOI] [PubMed] [Google Scholar]
- 66.Gonçalves J, Nolasco S, Nascimento R, Fanarraga M, Zabala JC, Soares H. TBCCD1, a new centrosomal protein, is required for centrosome and Golgi apparatus positioning. EMBO Rep. 2010;11(3): 194–200. 10.1038/embor.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chamberlain N, Korwin-Mihavics BR, Nakada EM, Bruno SR, Heppner DE, Chapman DG, et al. Lung epithelial protein disulfide isomerase A3 (PDIA3) plays an important role in influenza infection, inflammation, and airway mechanics. Redox Biol. 2019;22: 101129 10.1016/j.redox.2019.101129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baudry M, Bi X. Calpain-1 and Calpain-2: The Yin and Yang of Synaptic Plasticity and Neurodegeneration. Trends Neurosci. 2016;39(4): 235–245. 10.1016/j.tins.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19(15): 3905–39017. 10.1093/emboj/19.15.3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cerrone M, Montnach J, Lin X, Zhao YT, Zhang M, Agullo-Pascual E, et al. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nat Commun. 2017;8(1): 106 10.1038/s41467-017-00127-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qu J, Ko CW, Tso P, Bhargava A. Apolipoprotein A-IV: A Multifunctional Protein Involved in Protection against Atherosclerosis and Diabetes. Cells. 2019;8(4): 319 10.3390/cells8040319 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables show the information from the SwissProt database for both species.
(XLS)
Data Availability Statement
All nucleotide sequences are available from GenBank database (accession numbers MN657279, MN657280, MN657281, MN657282, MN657283, MN657284, MN657285, MN657286, MN657287, MN657288, MN657289, MN657290, MN657291, MN657292, MN657293, MN657294, MN657295, MN657296, MN657297, MN657298,MN657299, MN657300, MN657301, MN657302, MN657303, MN657304, MN657305, MN657306, MN657307, MN657308, MN657309, MN657310, MN657311, MN657312, MN657313. Gene Expression Omnibus data is available from NCBI (accession number GSE142602, GPL27950). Sequence Read Archive (SRA) database is available from NCBI (accession numbers SRS6721712, SRP265012).