Abstract
Endophytic fungi play an important role in plant growth. The composition and structure of endophytes vary in different plant tissues, which are specific habitats for endophyte colonization. To analyze the diversity and structural composition of endophytic fungi from toothed clubmoss (Huperzia serrata) that was artificially cultivated for 3 years, we investigated endophytic fungi from the roots, stems and leaves using comparative sequence analysis of the ITS2 region of the fungal rRNA genes sequenced with high-throughput sequencing technology. Seven fungal phyla were identified, and fungal diversity and structure varied across different tissues, with the most distinctive community features found in the roots. A total of 555 operational taxonomic units (OTUs) were detected, and 198 were common to all samples, and 43, 16, 16 OTUs were unique to the root, stem, leaf samples, respectively. Taxonomic classification showed that Ascomycota and Basidiomycota were dominant phyla, and Cladosporium, Oidiodendron, Phyllosticta, Sebacina and Ilyonectria were dominant genera. The relative abundance heat map at the genus level suggested that H. serrata had characteristic endophytic fungal microbiomes. Line discriminant analysis effect size analysis and principal coordinate analysis demonstrated that fungal communities were tissue-type and tissue-site specific. Overall, our study provides new insights into the complex composition of endophytic fungi in H. serrata.
Introduction
Huperzia serrata, a traditional medicinal plant in China, has bioactive properties favorable for treating fever, schizophrenia, and myasthenia gravis [1, 2]. Huperzine A isolated from H. serrata has a potent anti-acetylcholinesterase (AChE) activity [3], which has been approved in China as a drug to treat Alzheimer’s disease [4] and is currently used as a supplement for preventing further memory degeneration in USA [5]. However, wild H. serrata shows a low huperzine A content (ca. 0.007%), a limited geographic distribution, and an extremely slow growth [6]. Meanwhile, the excessive mining of wild H. serrata has degraded their habitat [2]. Wild H. serrata populations are insufficient to meet market demand. Therefore, it is very important and necessary to develop artificial cultivation of H. serrata, however there are still many challenges to growing H. serrata in a non-wild setting.
Along with emerging research on plant-microbe interactions, accumulating evidences suggest that endophytic fungi play important roles in plant growth [7, 8]. Some endophytic fungi can benefit plants by producing plant hormones [7], improving stress resistance [8], protecting plants from phytopathogens [9], and enabling nutrient uptake [10]. Previous studies examining the endophytic fungi of H. serrata and their secondary metabolic products revealed that they exhibited various biological activities, including antimicrobial activity, acetylcholinesterase inhibitory activity, nematocidal activity, and inhibition of nitric oxide production, among others [11–14]. In addition, some of endophytic fungi in H. serrata can produce huperzine A [15–17]. The distribution of some endophytic fungi in the host plants exhibited tissue specificity, which is an important influencing factor for accumulation of bioactive substances in different tissues [18, 19]. Although there are many studies focusing on endophytes of H. serrata, the understanding of the endophytic community in different tissues associated with H. serrata is still limited. Therefore, it is necessary to study the diversity and composition of endophytes in different tissues within H. serrata.
The Illumina-based high-throughput sequencing technology can comprehensively reveal the diversity and composition of plant-associated endophytes [20]. Lee et al. used high-throughput sequencing technology to reveal diversity of endophytic bacterial, archaeal and fungal communities inhabiting different rhizocompartments of tomato plants in real-world environments [21]. Chen et al. used high-throughput sequencing technology to analyze and compare the endophytic fungal community structures associated with stems and roots of Dendrobium huoshanense [22]. However, few researches on the diversity and composition of endophytes in H. serrata based on high-throughput sequencing have been conducted.
In the present study, the Illumina-based high-throughput sequencing analysis of the ITS2 region of fungal ribosomal RNA (rRNA) genes was conducted to describe the diversity and composition of endophytic fungi in the various tissues of H. serrata. As far as we know, this is the first time that the high-throughput amplicon sequencing has been used to study fungal community structure and diversity in H. serrata. Our results provide new insights into the fungal communities and lay a foundation for further study of H. serrata.
Materials and methods
Plant materials and treatments
Healthy three-year-old artificially cultivated H. serrata plants were randomly collected in July 2019 from the medicinal plant plantation in Huayuan County, Xiangxi Tujia and Miao Autonomous Prefecture, Hunan Province, China. These collected plants were developed from the mature spores of H. Serrata in the plantation, which guaranteed the exact growth age of them, and were properly managed, including watering, weeding and deworming. All plant samples were placed in aseptic bags that were placed on ice and immediately transported back to our laboratory, and three plants were randomly selected for investigation. After removing all sporangia, the root (marked as R1, R2, R3), stem (marked as S1, S2, S3) and leaf (marked as L1, L2, L3) samples of all three plants were separately collected using a sterile scissors and surface-sterilized using a series of washing steps: 70% (v/v) ethanol for 1 min, 3% (v/v) sodium hypochlorite solution for 3 min, 2.5% (w/v) sodium thiosulfate for 5 min, and rinsing the samples five times with sterile water [21]. Subsequently, nine samples from the root, stem and leaf tissues of three H. Serrata plants were used separately to extract the fungal genome DNA within them.
DNA extraction, PCR amplification, and ITS clone library construction
The root, stem, and leaf samples were rapidly ground to fine powder with liquid nitrogen in a sterilized and pre-cooled mortar. The resulting powder was then transferred to a bead tube for total DNA extraction using a MN NucleoSpin 96 Soil DNA kit (Macherey-Nagel, Dueren, Germany) according to the manufacturer instructions. DNA was stored at −20°C until subsequent analysis. The target-specific primers ITS1F (5'-CCTGGTCATTTAGAGGAAGTAA-3'), ITS4 (5'-TCCTCCGCTTATTGATATGC-3'), and fITS7 (5'-GTGARTCATCGAATCTTTG -3'), which do not amplify the chloroplast or mitochondrial rRNA genes of H. serrata, were used to amplify the ITS region of the fungal rRNA genes. PCRs (50 μL) were assembled and conducted using a reaction program as follows: 5 min at 94°C, 35 cycles of 1 min at 94°C, 50 sec at 54°C, and 60 sec at 68°C, then 10 min at 68°C. The PCR products were examined on a 1.8% agarose gel and purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States). Then, the construction and sequencing of the ITS clone libraries were performed by Beijing Biomarker Biotechnology Co., Ltd (Biomarker Biotechnology, Beijing, China) using an Illumina HiSeq 2500 platform.
Sequence processing and data analysis
FLASH software (v1.2.11) [23] was used to splice the reads of each sample through overlap, and the resulting spliced sequences were used as raw tags. Trimmomatic software (v0.33) [24] was used to filter the spliced raw tags to obtain high-quality tags (clean tags). UCHIME software (v8.1) [25] was used to identify and remove chimeric sequences to obtain the final effective tags, which were further clustered into operational taxonomic units (OTUs) with 97% pairwise identity.
The representative sequences of OTUs were used to perform taxonomic analysis through aligned to the UNITE database using QIIME software (v1.7.0) [26]. QIIME software was also used to select the OTU sequence with the highest abundance at the taxonomic level of the genus, carry out multiple sequence alignment, construct the phylogenetic tree, and then create a graph with the Python language tool. R software was used to obtain Venn diagrams and microbial community bar plots to characterize the richness of a specific fungal community.
Alpha diversity indexes, including Chao1, Ace, Shannon and Simpson, were evaluated using Mothur software (version v.1.30) [27]. In addition, OTU coverage was also counted to determine whether the sequencing results were representative of the actual microbial communities in our samples. Rarefaction curves, reflecting the sequencing depth, were calculated and constructed using QIIME software.
Beta diversity indexes were evaluated using QIIME software to compare the similarity of different samples in species diversity. Principal Coordinate Analyses (PCoA) utilizing the Bray-Curtis distances were used to observe the relationships between fungal community structures in different tissues. Analysis of Unweighted Pair-Group Method with Arithmetic Means (UPGMA) was carried out to determine whether the samples had significant microbial community differences in a UPGMA tree. The heat map of genera differences between groups was drawn based on the OTU-Table to understand the fungal community composition at genus level among different tissues. Furthermore, Line discriminant analysis effect size (LEfSe) analysis [28] using Galaxy online (http://huttenhower.sph.harvard.edu/galaxy/) was used to identify differentially abundant features among samples for biomarker discovery.
Results
Characteristics of sample sequences
The quality of the sequencing data was evaluated mainly through the statistics of sequence number, sequence length, GC content, Q20 and Q30 quality values, effective ratio, and other parameters in each stage (S1 Table). The number of effective tags per sample ranged from 230,078 to 231,811, and the average length of sequences from the root, stem and leaf samples was 304, 303 and 299 bp, respectively, however the length of sequencing tags in the three tissue samples mainly fell within the range of 280–370 bp (S1 Fig). The rarefaction curves (Fig 1) tended to be flat, indicating that our sequencing depth was sufficient. Similarly, more than 0.999 coverage suggested that the ITS libraries were large enough to capture most of the fungal diversity in the samples used in this study (Table 1).
Fig 1. Rarefaction curves based on the ITS2 sequences of endophytic fungi from the root (R), stem (S) and leaf (L) samples associated with H. serrata.
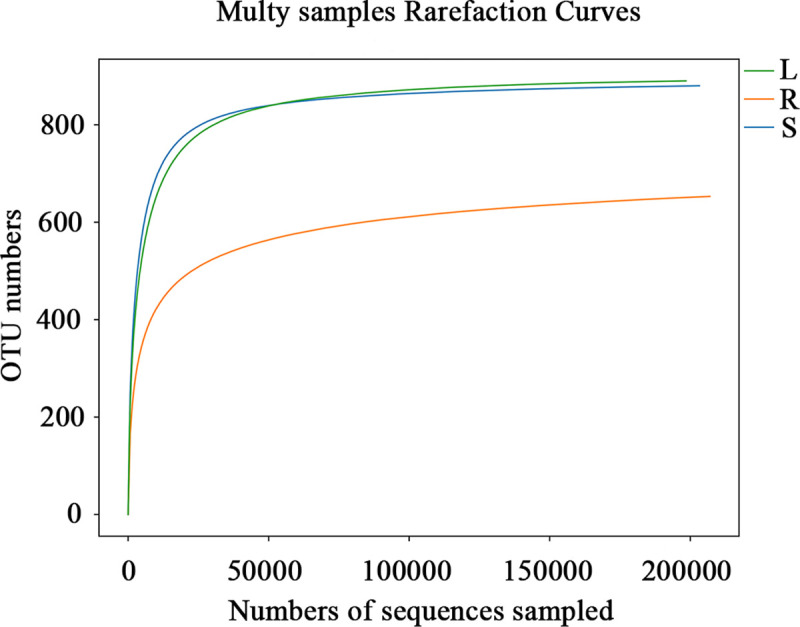
Table 1. The richness and diversity indexes of endophytic fungi from the root, stem and leaf samples associated with H. serrata.
| Sample origin | OTUs | Total OTUs | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|---|
| Roots | 284 | 555 | 314.5561 | 350.1111 | 0.051 | 3.4706 | 0.9998 |
| Stems | 465 | 467.8932 | 466.5 | 0.0321 | 4.1734 | 1 | |
| Leaves | 484 | 501.5221 | 507.25 | 0.0475 | 3.8822 | 0.9999 |
Richness and diversity of endophytic fungi
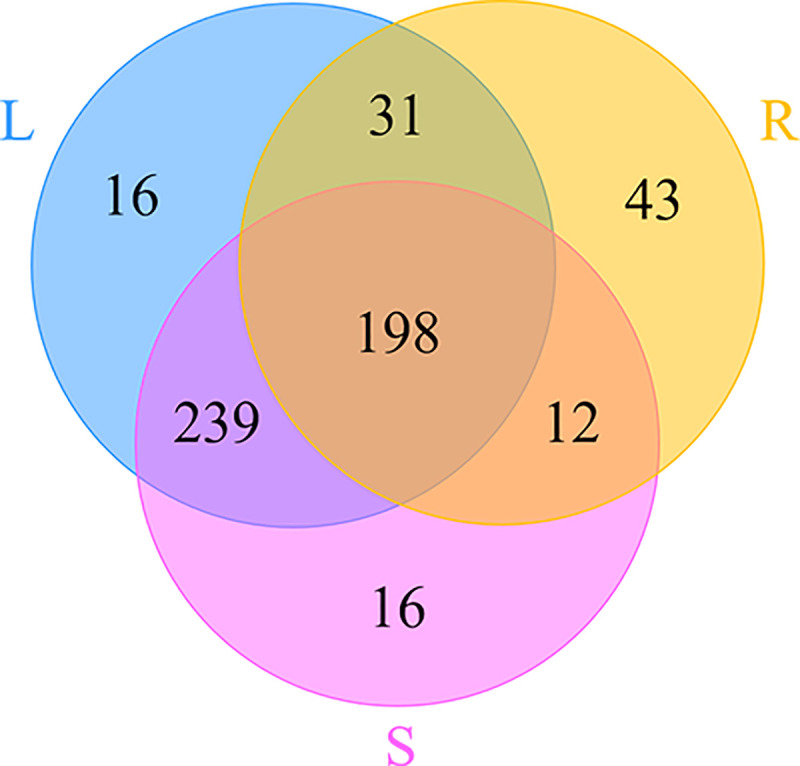
A total of 555 OTUs were detected across all of the ITS libraries, including 284 OTUs in the root samples, 465 OTUs in the stem samples and 484 OTUs in the leaf samples (Table 1). Among them, 198 OTUs were common to all samples, and 43, 16 and 16 OTUs were exclusive to the root, stem and leaf samples, respectively (Fig 2). The Chao1 and Ace indexes showed that the community richness of the endophytic fungi in the leaves was higher than that in the stems and roots (Table 1). However, the Shannon and Simpson indexes showed that the community diversity of the endophytic fungi in the stems was highest, followed by the leaves and roots (Table 1).
Fig 2. Venn diagram showing the OTUs shared among the root (R), stem (S) and leaf (L) samples associated with H. serrata.
Taxonomic distribution of endophytic fungi
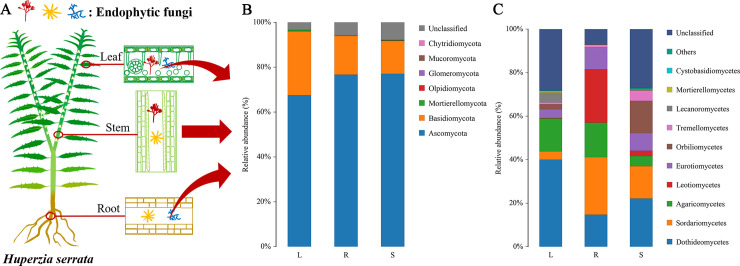
The taxonomic distribution of endophytic fungi in the roots, stems and leaves of H. serrata was displayed in Fig 3. After screening out rare OTUs, the remaining OTUs represented 7 phyla, 21 classes, 50 orders, 95 families, and 120 genera (Table 2). The 7 identified phyla were Ascomycota, Basidiomycota, Chytridiomycota, Glomeromycota, Mortierellomycota, Mucoromycota, and Olpidiomycota, and the relative abundances of these phyla varied across the three tissues (Fig 3B). Among them, Ascomycota and Basidiomycota were predominant phyla, accounting for 71.9% and 13.2% of sequences, respectively, while the other phyla were all below 1% of sequences (Table 3). In addition, all the phyla were found in the leaf samples, but Chytridiomycota and Glomeromycota were absent in the root samples and the stem samples, respectively.
Fig 3.
Distribution of endophytic fungi in H. serrata (A) and relative abundances of endophytic fungi at the phylum (B) level and class (C) level in the root (R), stem (S) and leaf (L) samples of H. serrata.
Table 2. Numbers of taxa of endophytic fungi in the root, stem and leaf samples of H. serrata.
| Sample origin | Phylum | Class | Order | Family | Genus |
| Roots | 6 | 14 | 30 | 61 | 80 |
| Stems | 6 | 20 | 48 | 91 | 113 |
| Leaves | 7 | 20 | 49 | 90 | 110 |
| Total | 7 | 21 | 50 | 95 | 120 |
Table 3. Distribution of fungal microbiome at phylum level in the root, stem and leaf samples of H. serrata.
| Taxon | Relative abundance (%) | ||
|---|---|---|---|
| Roots | Stems | Leaves | |
| Ascomycota | 76.67 | 76.31 | 65.07 |
| Basidiomycota | 17.29 | 15.05 | 28.48 |
| Chytridiomycota | 0 | 0.01 | 0.004 |
| Glomeromycota | 0.07 | 0 | 0.0005 |
| Mortierellomycota | 0.01 | 0.18 | 0.60 |
| Mucoromycota | 0.0004 | 0.03 | 0.02 |
| Olpidiomycota | 0.001 | 0.16 | 0.02 |
| Unclassified | 5.95 | 8.26 | 5.80 |
In detail, 21 classes belonging to the 7 phyla were identified (S2 Table). Among them, 20 classes were identified in the leaf and stem samples, respectively, while only 14 classes were identified in the root samples. The predominant classes (top 10) were Dothideomycetes, Sordariomycetes, Eurotiomycetes, Tremellomycetes, Leotiomycetes, Agaricomycetes, Mortierellomycetes, Cystobasidiomycetes, Ustilaginomycetes, and Orbiliomycetes in all samples (Fig 3C). However, class distributions differed greatly across the three tissues (S2 Table). For example, GS18 was not found in the leaf samples, Glomeromycetes was not found in the stem samples, and Pezizomycetes, Agaricostilbomycetes, Cystobasidiomycetes, Exobasidiomycetes, Utilaginomycetes, Spizellomycetes, GS17 were not found in the root samples (S2 Table).
The top 26 genera (i.e., those with relative abundance > 0.2%) were selected to make a heatmap clustering (S2 Fig and Table 4), which further indicated that species distributions differed greatly across the three tissue samples. Among them, 20 genera belonged to Ascomycota, while 5 belonged to Basidiomycota and 1 belonged to Mortierellomycota. The genera of Oidiodendron, Ilyonectria, Chloridium, Russula, Sebacina, Cladophialophora, Periconia, Pezicula, Roussoella, Scytalidium, Dactylonectria, Papiliotrema, Pochonia, and Verticillium were mainly distributed in the roots. The genera of Cladosporium, Saitozyma, Pyrenochaetopsis, Claviceps, Cyphellophora, and Purpureocillium were mainly distributed in the stems, while the dominant genera in the leaves included Phyllosticta, Serendipita, Devriesia, and Mortierella. In addition, Botryosphaeria, Scytalidium, and Idriella were the exclusive genera in the root samples, while Phialophora, Lecophagus, Clavaria, and Peniophora were exclusive to the stem samples, and no genus was exclusive to the leaf samples (S3 Table).
Table 4. Distribution of fungal microbiome at genus level (relative abundance > 0.2%) in the root, stem and leaf samples of H. serrata.
| Phyla | Class | Genus | Relative abundance (%) | ||
|---|---|---|---|---|---|
| Roots | Stems | Leaves | |||
| Ascomycota | Dothideomycetes | Phyllosticta | 0.02 | 0.07 | 11.64 |
| Cladosporium | 2.55 | 6.52 | 5.74 | ||
| Devriesia | 0.001 | 0.20 | 0.66 | ||
| Pyrenochaetopsis | 0.10 | 2.29 | 1.51 | ||
| Periconia | 2.77 | 0.02 | 0.02 | ||
| Roussoella | 1.85 | 1.23 | 0.08 | ||
| Eurotiomycetes | Cyphellophora | 0.001 | 1.03 | 0.89 | |
| Cladophialophora | 6.54 | 0.50 | 2.14 | ||
| Leotiomycetes | Pezicula | 2.70 | 0.001 | 0 | |
| Scytalidium | 1.73 | 0 | 0.001 | ||
| Oidiodendron | 11.55 | 1.97 | 0.06 | ||
| Sordariomycetes | Chloridium | 8.38 | 0.10 | 0.01 | |
| Verticillium | 0.47 | 0.09 | 0.29 | ||
| Clonostachys | 0.28 | 0.27 | 0.20 | ||
| Claviceps | 0.0004 | 1.56 | 0.12 | ||
| Pochonia | 0.48 | 0.13 | 0.02 | ||
| Trichoderma | 0.50 | 0.72 | 0.02 | ||
| Dactylonectria | 1.59 | 0.004 | 0.001 | ||
| Ilyonectria | 9.70 | 0.02 | 0.04 | ||
| Purpureocillium | 0.02 | 0.41 | 0.16 | ||
| Basidiomycota | Agaricomycetes | Russula | 8.29 | 0 | 0.001 |
| Sebacina | 7.27 | 4.12 | 0.31 | ||
| Serendipita | 0.0004 | 0.003 | 1.38 | ||
| Tremellomycetes | Papiliotrema | 0.50 | 0.09 | 0.07 | |
| Saitozyma | 0.73 | 3.30 | 0.12 | ||
| Mortierellomycota | Mortierellomycetes | Mortierella | 0.01 | 0.18 | 0.60 |
In addition, the endophytic fungi of the root samples possessed a better taxonomic annotation of their OTUs and harbored a minimum proportion of unclassified OTUs. For examples, the percentage of classified taxa at the class level in the roots were 85.46%, which were higher than that in the stems (72.13%) and leaves (59.04%), while 61.65% classified taxa at genus level in the roots were higher than 29.97% in the stems and 28.84% in the leaves (Fig 3C and S3 Fig).
Comparative analysis of endophytic fungi
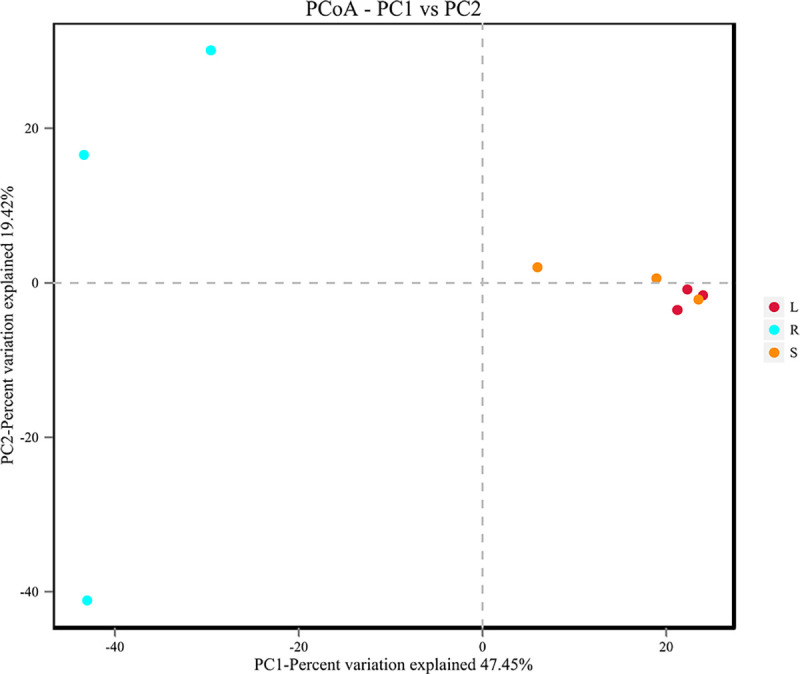
Important distinctions were found in the composition of fungal communities in the root, stem and leaf samples. Two different clusters were observed at the genus level in the UPGMA tree: the fungal microbiota from H. serrata leaves and stems clustered together, but the roots formed their own cluster and distinctly separated from those of the stems and leaves (S4 Fig), suggesting that the endophyte microbiomes of the leaf samples were more similar to that of the stem samples than to that of the root samples, and that the endophytic fungi of the leaf and stem samples might share a same origin. PCoA revealed the main variations in fungal community composition among the three tissues (Fig 4), and the highest variations in the microbiota of different samples were 25.99% (PC1) and 17.25% (PC2), representing a strong separation based on the plant tissues.
Fig 4. Comparison of microbial communities in the root (R), stem (S) and leaf (L) samples of H. serrata based on Bray-Curtis.
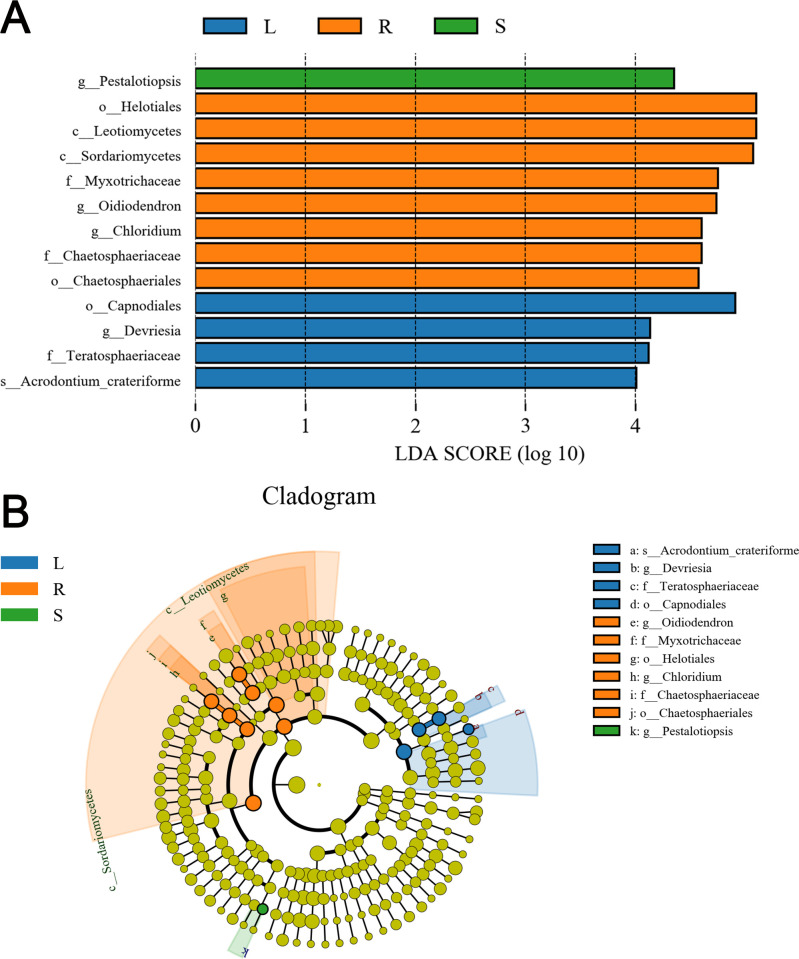
Significantly different taxon abundances of endophytic fungi were found among the three tissues, as determined by LEfSe (Fig 5). At the genus level, Pestalotiopsis was significantly enriched in the stems, while Oidiodendron and Chloridium were more abundant in the roots. Hannaella exhibited relatively higher abundance in the leaves than in the roots and stems. These differentially abundant taxa can be considered as potential biomarkers (line discriminant analysis (LDA) score > 4, P < 0.05).
Fig 5. Groups from the phylum-to-species level determined to be significant representatives of their sample type based on LEfSe analysis.
(A) The figure shows the taxa with an LDA score greater than 4.0. The length of the histogram represents the influence of different taxa (LDA score), and different colors represent different grouped taxa. (B) The cladogram represents the taxonomic hierarchical structure of the identified habitat biomarkers generated using LEfSe. The circles radiating from the inside to the outside of the branching diagram represent the taxonomic levels from the phylum to the species; each small circle at different classification levels represents a classification at this level, and the diameter of the small circle is proportional to the relative abundance. Taxa with no significant differences are shown in yellow, while taxa with significant differences are colored according to the grouping of the most abundant taxa.
Discussion
In this study, the endophytic fungi associated with the root, stem and leaf tissues of three-year-old H. serrata were characterized and compared. Alpha diversity analysis (Chao1, Ace, Shannon, Simpson) indicated that richness and diversity of endophytic fungi in the H. serrata root samples were lowest (Table 1), which were different from those of the D. huoshanense roots [22], that have a higher richness and diversity of endophytic fungi. A total of 555 fungal OTUs were detected from the three tissue samples of H. serrata, and were further classified into 7 phyla, 21 classes, 50 orders, 95 families and 120 genera. However, there were still some OTUs sequences were not taxonomically annotated at different taxon level, such as 38.35% unclassified taxa (OTUs) at genus level in the root samples (Fig 3C, S3 Fig), indicating that we still lacked the full knowledges of endophytic fungi of the investigated H. serrata in nature.
The distribution of endophytic fungal taxa varied greatly among different plant tissues, which might be related to host genotype [29], growing environment [30], and plant age [31]. The root, stem, and leaf tissues of the investigated H. serrata were colonized by some of the same fungal communities, but in different proportions. Ascomycota and Basidiomycota were the most abundant phyla in all of the three tissues of H. serrata, which were consistent with the dominant phyla in the roots of Sinopodophyllum hexandrum [32] and Pennisetum sinese [30], in the barks of Eucommia ulmoides [33], and the culturable fungal endophytes of H. serrata [18].
The dominant genera of endophytic fungi differed among the three tissues of the investigated H. serrata in our study. The genera of Oidiodendron and Cladosporium were dominant in the root and stem samples of the investigated H. serrata, respectively. Some strains of Oidiodendron were reported to promote nitrogen uptake and plant growth [34], and exhibit metal tolerance [35]. Some strains of Cladosporium exhibited an antimicrobial activity against Bacillus cereus IIIM 25 (Gram positive) and Escherichia coli ATCC 25922 (Gram negative) [36], and effectively reduced the infection of nematodes in some plant roots [37]. The genera of Oidiodendron and Cladosporium might confer similar benefit upon the host H. serrata. Some of endophytic fungi can produce antifungal active substances [38], which can improve their competitiveness and prevent colonization by other fungi. The tetranorlabdane diterpenoids from the extract of endophytic fungus Botryosphaeria sp. P483 isolated from H. serrata exhibited obviously antifungal activities against to Gaeumannomyces graminis, Fusarium moniliforme, F. solani, F. oxysporum and Pyricularia oryzae [11]. Interestingly, the genus of Botryosphaeria mainly existed in the root samples in our study, which might partly explain why the richness and diversity of endophytic fungi in the root samples were lower than that in the leaf and stem samples.
Endophytic fungi are considered as the fungi that live inside healthy plant tissues at a certain or whole stage of life cycle and do not cause obvious plant diseases [39]. Some endophytic fungi in the investigated H. serrata were identified as plant pathogenic fungi, such as the dominant genus Phyllosticta (11.56%) in the leaf samples. Phyllosticta was an important group of plant pathogenic fungi distributed worldwide that causes serious diseases, e.g., citrus and grapevine black spots [40, 41]. An abundant Phyllosticta in the leaf samples might be attributed to the investigated H. serrata leaves invaded by this pathogenic fungus, but that did not cause obvious lesions of the host plant temporarily. Another possible explanation was the limitation of the surface sterilization method used in this study, which was responsible for these identified plant pathogenic fungi as endophytes, because this surface sterilization method could not entirely remove the microorganisms adhered to the tissue surfaces.
Currently, about 12 fungal genera have been reported to produce huperzine A [15–17, 42–47], of which 7 genera were found in H. serrata in this study. Among them, Cladosporium, Trichoderma, and Fusarium with the relative abundance > 0.1% had the highest distribution in the stems, followed by the leaves and the roots, indicating that the endophytic fungi in the stems and the leaves were more involved in the synthesis of some secondary metabolites than those in the roots. Five of the reported fungal genera, that can produce huperzine A, were not detected in our study, which might be attributed to the investigated H. serrata, because the planting environment and age of host plant were important factors affecting the composition of its endophytic fungi. Therefore, it is necessary to further investigate the endophytic microorganisms of H. serrata from different growing environments and years, which were beneficial to obtain a comprehensive understanding of the endophyte community in H. serrata. Besides these endophytic fungi of the reported genera with an ability to produce huperzine, our results uncovered that H. serrata contained diverse endophytic fungi of yet unexplored potential importance, which a reservoir for developing endophyte resources for artificial cultivation H. serrata and production of useful bioactive compounds.
Conclusion
In conclusion, this study reveals the community composition and structure of the endophytic fungi in the roots, stems, and leaves of H. serrata, and found endophytic fungal communities varying across the three tissues, and uncovered some dominant endophytic fungi in the three tissues, which benefit further scientific understanding of fungal community ecology in this medicinally important plant and better to tap functionally important endophytes.
Supporting information
(TIF)
The dendrogram represents complete-linkage agglomerative clustering based on Euclidean dissimilarities.
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the Biomarker Technologies Corporation for technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
National Natural Science Foundation of China (Grant Numbers: 31770134 and 31370118): P Z http://www.nsfc.gov.cn/english/site_1/ Fundamental Research Funds for the Central Universities, SCUEC (Grant Numbers: CZT18005, 18006, 18007): FJ S, AH L, P Z http://www.polymer.cn/ss/projects/show.aspx?id=1738 Fund for Key Laboratory Construction of Hubei Province (Grant Number: 2018BFC360): FJ S, AH L, P Z Development Project of Science and Technology Innovation Capability of Shanxi University of Chinese Medicine (Grant Number: 2019PY-132): LY M Conceived and designed the research: Peng Zhang, Liyun Miao, Fajun Song. Performed the experiments, analysis and interpretation of data: Shipeng Fan, Liyun Miao, Haodong Li, Aihua Lin. Wrote the manuscript: Shipeng Fan, Liyun Miao, Peng Zhang.
References
- 1.Skolnick AA. Old Chinese herbal medicine used for fever yields possible new Alzheimer disease therapy. JAMA. 1997;277(10):776 10.1001/jama.1997.03540340010004 [DOI] [PubMed] [Google Scholar]
- 2.Ma X, Tan C, Zhu D, Gang DR, Xiao P. Huperzine A from Huperzia species—an ethnopharmacolgical review. J Ethnopharmacol. 2007;113(1):15–34. 10.1016/j.jep.2007.05.030 [DOI] [PubMed] [Google Scholar]
- 3.Wang YE, Yue DX, Tang XC. [Anti-cholinesterase activity of huperzine A]. Zhongguo Yao Li Xue Bao. 1986;7(2):110–3. 10.1016/0024-3205(86)90602-8 [DOI] [PubMed] [Google Scholar]
- 4.Liu MY, Liu HC. [Intelligence promoting Chinese materia medica]. Zhongguo Zhong Xi Yi Jie He Za Zhi. 1995;15(1):59–61. [PubMed] [Google Scholar]
- 5.Qian ZM, Ke Y. Huperzine A: Is it an effective disease-modifying drug for Alzheimer's disease? Front Aging Neurosci. 2014;6:216 10.3389/fnagi.2014.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X, Tan C, Zhu D, Gang DR. Is there a better source of huperzine A than Huperzia serrata? Huperzine A content of Huperziaceae species in China. J Agric Food Chem. 2005;53(5):1393–8. 10.1021/jf048193n [DOI] [PubMed] [Google Scholar]
- 7.Wani ZA, Mirza DN, Arora P, Riyaz-Ul-Hassan S. Molecular phylogeny, diversity, community structure, and plant growth promoting properties of fungal endophytes associated with the corms of saffron plant: An insight into the microbiome of Crocus sativus Linn. Fungal Biol. 2016;120(12):1509–24. 10.1016/j.funbio.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 8.Khan AL, Hamayun M, Kang SM, Kim YH, Jung HY, Lee JH, et al. Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiol. 2012;12:3 10.1186/1471-2180-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Gao BL, Li XX, Zhang ZB, Yan RM, Yang HL, et al. Phylogenetic diversity of culturable endophytic fungi in Dongxiang wild rice (Oryza rufipogon Griff), detection of polyketide synthase gene and their antagonistic activity analysis. Fungal Biol. 2015;119(11):1032–45. 10.1016/j.funbio.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 10.Zhao XL, Yang JZ, Liu S, Chen CL, Zhu HY, Cao JX. The colonization patterns of different fungi on roots of Cymbidium hybridum plantlets and their respective inoculation effects on growth and nutrient uptake of orchid plantlets. World J Microbiol Biotechnol. 2014;30(7):1993–2003. 10.1007/s11274-014-1623-2 [DOI] [PubMed] [Google Scholar]
- 11.Chen YM, Yang YH, Li XN, Zou C, Zhao PJ. Diterpenoids from the endophytic fungus Botryosphaeria sp. P483 of the Chinese herbal medicine Huperzia serrata. Molecules. 2015;20(9):16924–32. 10.3390/molecules200916924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang YH, Yang DS, Li GH, Pu XJ, Mo MH, Zhao PJ. Antibacterial diketopiperazines from an endophytic fungus Bionectria sp. Y1085. J Antibiot (Tokyo). 2019;72(10):752–8. 10.1038/s41429-019-0209-5 [DOI] [PubMed] [Google Scholar]
- 13.Yu FX, Li Z, Chen Y, Yang YH, Li GH, Zhao PJ. Four new steroids from the endophytic fungus Chaetomium sp. M453 derived of Chinese herbal medicine Huperzia serrata. Fitoterapia. 2017;117:41–6. 10.1016/j.fitote.2016.12.012 [DOI] [PubMed] [Google Scholar]
- 14.Qi B, Liu X, Mo T, Li SS, Wang J, Shi XP, et al. Nitric oxide inhibitory polyketides from Penicillium chrysogenum MT-12, an endophytic fungus isolated from Huperzia serrata. Fitoterapia. 2017;123:35–43. 10.1016/j.fitote.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 15.Zaki AG, El-Shatoury EH, Ahmed AS, Al-Hagar OEA. Production and enhancement of the acetylcholinesterase inhibitor, huperzine A, from an endophytic Alternaria brassicae AGF041. Appl Microbiol Biotechnol. 2019;103(14):5867–78. 10.1007/s00253-019-09897-7 [DOI] [PubMed] [Google Scholar]
- 16.Thi Minh Le T, Thi Hong Hoang A, Thi Bich Le T, Thi Bich Vo T, Van Quyen D, Hoang Chu H. Isolation of endophytic fungi and screening of huperzine A-producing fungus from Huperzia serrata in Vietnam. Sci Rep. 2019;9(1):16152 10.1038/s41598-019-52481-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz-Miranda OL, Folch-Mallol J, Martinez-Morales F, Gesto-Borroto R, Villarreal ML, Taketa AC. Identification of a huperzine A-producing endophytic fungus from Phlegmariurus taxifolius. Mol Biol Rep. 2020;47(1):489–95. 10.1007/s11033-019-05155-1 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Lai Z, Li XX, Yan RM, Zhang ZB, Yang HL, et al. Isolation, diversity and acetylcholinesterase inhibitory activity of the culturable endophytic fungi harboured in Huperzia serrata from Jinggang Mountain, China. World J Microbiol Biotechnol. 2016;32(2):20 10.1007/s11274-015-1966-3 [DOI] [PubMed] [Google Scholar]
- 19.Gong A, Zhou T, Xiao C, Jiang W, Zhou Y, Zhang J, et al. Association between dipsacus saponin VI level and diversity of endophytic fungi in roots of Dipsacus asperoides. World J Microbiol Biotechnol. 2019;35(3):42 10.1007/s11274-019-2616-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu WZ Zhuoyan; Yuhan Liu; Xiufang Hu; Ying Guo; Li Junfeng. Application of high-throughput internal transcribed spacer rRNA metagenomics analysis in deciphering endophytic fungi diversity of Dendrobium Officinale. J Biobsed Mater Bio. 2017;11:106–18. 10.1166/jbmb.2017.1647 [DOI] [Google Scholar]
- 21.Lee SA, Kim Y, Kim JM, Chu B, Joa JH, Sang MK, et al. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci Rep. 2019;9(1):9300 10.1038/s41598-019-45660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Dai J, Song X, Jiang X, Zhao Q, Sun C, et al. Endophytic microbiota comparison of Dendrobium huoshanense root and stem in different growth years. Planta Med. 2020;86(13–14):967–75. 10.1055/a-1046-1022 [DOI] [PubMed] [Google Scholar]
- 23.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albrectsen BR, Siddique AB, Decker VHG, Unterseher M, Robinson KM. Both plant genotype and herbivory shape aspen endophyte communities. Oecologia. 2018;187(2):535–45. 10.1007/s00442-018-4097-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng ZS, Liu XD, Zhang BC, Jiao S, Qi XY, Sun ZH, et al. The root endophytic fungi community structure of Pennisetum sinese from four representative provinces in China. Microorganisms. 2019;7(9). 10.3390/microorganisms7090332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs Benjamin, Krischke Markus, Mueller Martin J., Krauss Jochen. Plant age and seasonal timing determine endophyte growth and alkaloid biosynthesis. Fungal Ecol. 2017;29:52–58. 10.1016/j.funeco.2017.06.003 [DOI] [Google Scholar]
- 32.Ning Y, Li YL, Zhou GY, Yang LC, Xu WH. [Community composition and diversity of endophytic fungi from roots of Sinopodophyllum hexandrum in forest of Upper-north mountain of Qinghai province]. Zhongguo Zhong Yao Za Zhi. 2016;41(7):1227–34. 10.4268/cjcmm20160712 [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Dong CB, Chen WH, Liang JD, Han YF, Liang ZQ. [Community composition and ecological functional structural analysis of endophytic fungi in bark of Eucommia ulmoides in different areas]. Zhongguo Zhong Yao Za Zhi. 2019;44(6):1126–34. 10.19540/j.cnki.cjcmm.20181226.005 [DOI] [PubMed] [Google Scholar]
- 34.Wei X, Chen J, Zhang C, Pan D. A new Oidiodendron maius strain isolated from Rhododendron fortunei and its effects on nitrogen uptake and plant growth. Front Microbiol. 2016;7:1327 10.3389/fmicb.2016.01327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiapello M, Martino E, Perotto S. Common and metal-specific proteomic responses to cadmium and zinc in the metal tolerant ericoid mycorrhizal fungus Oidiodendron maius Zn. Metallomics. 2015;7(5):805–15. 10.1039/c5mt00024f [DOI] [PubMed] [Google Scholar]
- 36.Syed Naseer, Khursheed A. Bhat, Masroor Qadri, Syed Riyaz‐Ul‐Hassan, Fayaz A. Malik, Mohammad A. Khuroo. Bioactivity‐guided isolation, antimicrobial and cytotoxic evaluation of secondary metabolites from Cladosporium tenuissimum associated with Pinus wallichiana. ChemistrySelect 2017;2(3):1311–4. 10.1002/slct.201601942 [DOI] [Google Scholar]
- 37.Zhou WQ, Vijay C. Verma, Terry A. Wheeler, Jason E. Woodward, James L. Starr, Gregory Alan Sword. Tapping into the cotton fungal phytobiome for novel nematode biological control tools. Phytobiomes Journal 2020;4(1). 10.1094/PBIOMES-08-19-0043-SC [DOI] [Google Scholar]
- 38.Gupta S, Chaturvedi P, Kulkarni MG, Van Staden J. A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol Adv. 2020;39:107462 10.1016/j.biotechadv.2019.107462 [DOI] [PubMed] [Google Scholar]
- 39.Clay K. Fungal endophytes of grasses: A defensive mutualism between plants and fungi. Ecology. 1988;69(1):10–6. 10.2307/1943155 [DOI] [Google Scholar]
- 40.Tran NT, Miles AK, Dietzgen RG, Dewdney MM, Zhang K, Rollins JA, et al. Sexual reproduction in the citrus black spot pathogen, Phyllosticta citricarpa. Phytopathology. 2017;107(6):732–9. 10.1094/PHYTO-11-16-0419-R [DOI] [PubMed] [Google Scholar]
- 41.Rinaldi PA, Paffetti D, Comparini C, Broggini GAL, Gessler C, Mugnai L. Genetic variability of Phyllosticta ampelicida, the agent of black rot disease of grapevine. Phytopathology. 2017;107(11):1406–16. 10.1094/PHYTO-11-16-0404-R [DOI] [PubMed] [Google Scholar]
- 42.Le TTM, Hoang ATH, Nguyen NP, Le TTB, Trinh HTT, Vo TTB, et al. A novel huperzine A-producing endophytic fungus Fusarium sp. Rsp5.2 isolated from Huperzia serrate. Biotechnol Lett. 2020;42(6):987–95. 10.1007/s10529-020-02836-x [DOI] [PubMed] [Google Scholar]
- 43.Shu SH, Zhao XM, Wang WJ, Zhang GW, Cosoveanu A, Ahn Y, et al. Identification of a novel endophytic fungus from Huperzia serrata which produces huperzine A. World J Microbiol Biotechnol. 2014;30(12):3101–9. 10.1007/s11274-014-1737-6 [DOI] [PubMed] [Google Scholar]
- 44.Su J, Yang M. Huperzine A production by Paecilomyces tenuis YS-13, an endophytic fungus isolated from Huperzia serrata. Nat Prod Res. 2015;29(11):1035–41. 10.1080/14786419.2014.980245 [DOI] [PubMed] [Google Scholar]
- 45.Dong LH, Fan SW, Ling QZ, Huang BB, Wei ZJ. Indentification of huperzine A-producing endophytic fungi isolated from Huperzia serrata. World J Microbiol Biotechnol. 2014;30(3):1011–7. 10.1007/s11274-013-1519-6 [DOI] [PubMed] [Google Scholar]
- 46.Zhu D, Wang J, Zeng Q, Zhang Z, Yan R. A novel endophytic huperzine A-producing fungus, Shiraia sp. Slf14, isolated from Huperzia serrata. J Appl Microbiol. 2010;109(4):1469–78. 10.1111/j.1365-2672.2010.04777.x [DOI] [PubMed] [Google Scholar]
- 47.Zhang ZB, Zeng QG, Yan RM, Wang Y, Zou ZR, Zhu D. Endophytic fungus Cladosporium cladosporioides LF70 from Huperzia serrata produces huperzine A. World J Microbiol Biotechnol 2011;27:479–86. 10.1007/s11274-010-0476-6 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
The dendrogram represents complete-linkage agglomerative clustering based on Euclidean dissimilarities.
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.