Abstract
Previously limited to symptomatic patients, our hospital introduced a universal admission screening strategy for coronavirus disease 2019 on 25 April 2020. All patients were tested by RT-PCR. We observed decreased viral loads linked to increased screening of asymptomatic patients highlighting the fact that viral load values could guide infection control decisions.
Keywords: Asymptomatic patients, coronavirus disease 2019, infection control, pre-hospitalization, RT-PCR, screening, severe acute respiratory disease coronavirus 2, viral load
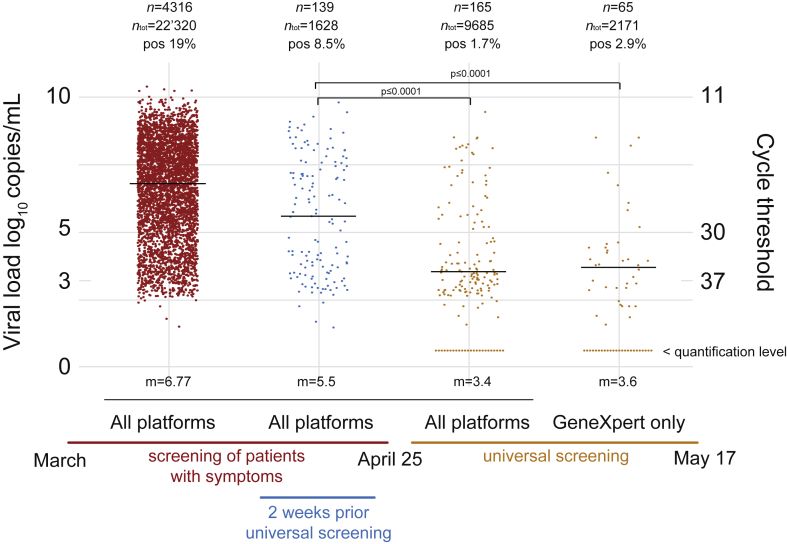
Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) quantitative RT-PCR has been used as a crucial diagnostic tool [1]. Between the beginning of March and 17 May 2020, our molecular diagnostic laboratory located in a tertiary care university hospital (Lausanne, Switzerland), performed more than 34 000 SARS-CoV-2 RT-PCR tests using three different platforms: our high throughput automated molecular diagnostic platform (MDx platform) [2], the cobas SARS-CoV-2 test (Roche, Rotkreuz, Switzerland) [4] and the GeneXpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA, USA). In addition to delivering a qualitative yes/no answer, RT-PCR can provide quantitative values based on cycle threshold (Ct). To provide precise and reliable quantitative or semi-quantitative information, laboratories must transform the Ct values into viral loads using positive controls obtained from viral culture and/or from calibrated positive plasmid controls. In our laboratory, we used these two types of positive controls to determine the correlation between the Ct value and the viral load and this calibration was performed for all our instruments [3,4]. Reporting viral load values can be used (a) by the laboratory as an internal quality assessment tool, (b) by clinicians to evaluate the progression of the infection (in lower respiratory tract specimens or across time) or (c) to address patient contagiousness and hence to guide infection control decisions [[5], [6], [7], [8], [9]]. Regarding the latter application and during the first deconfinement phase after the lockdown in Switzerland, the benefit of reporting the viral load value appeared of utmost importance. From 25 April 2020, a universal admission or pre-intervention screening strategy including asymptomatic patients was introduced in our hospital. We observed an abrupt decrease of viral load in patients screened after 25 April compared with patients screened during the epidemic period, when the screening strategy focused mainly on symptomatic patients (Fig. 1). This shift is explained by an increase in screening of asymptomatic patients with low viral load compared with symptomatic individuals, who are more likely to have high viral loads. This abrupt change was confirmed by looking only at a shorter period of 2 weeks just before the shift to the universal screening strategy. The GeneXpert SARS-CoV-2 test was broadly used during this period, detecting the SARS-CoV-2-specific N2 region (encoding for viral nucleoprotein N2) and the E-gene (encoding for a protein of the envelope). Viral load calculation was based on the E-gene as the Ct correlated with the other two platforms and could therefore be compared directly. Among the very high Cts, we obtained several results positive only for N2, suggesting a very low viral load at the detection limit of the GeneXpert assay [10,11]. By retesting these specimens with other RT-PCR platforms and reviewing clinical data, we could demonstrate that these N2-only positive results corresponded to true detection of viral RNA. When only the N2 gene was positive, the reported result was ‘positive result, low viral load, quantification impossible’.
Fig. 1.
Median viral load value of positive SARS-CoV-2 RT-PCR was compared across two periods: the epidemic period (left), during which mainly symptomatic patients were screened, and the post-epidemic period (right) when all patients were tested on hospital admission. A decrease of median viral load (with an increased number of specimens with viral loads <1000 copies/mL) was observed during the universal screening period when many individuals tested were asymptomatic. Patient samples analysed using the GeneXpert test and showing only an N-positive PCR are displayed below the quantification limit. Cycle threshold values (Ct) were converted to viral loads using the formula –0.27Ct + 13.04 [3,4] generated using purified viral RNA and synthetic plasmids, kindly provided by the Institute of Virology of the University of Berlin, la Charité. Significance of viral load decrease was assessed using the non-parametric Wilcoxon–Mann–Whitney test with p ≤ 0.0001. m, viral load median value; n, number of positive samples; ntot, total number of tests; pos, percentage of positive tests.
Since April 2020, we have reported all SARS-CoV-2 RT-PCR results quantitatively. This is important because it provides some information regarding the robustness of the result and about the contagiousness of the patients. As an individual with a viral load less than 1000 copies/mL is probably exhibiting negligible contagiousness. However, to assess contagiousness, it is important to also consider the presence or absence of symptoms of respiratory tract infection as proposed by the Centers for Disease Control and Prevention regarding transmission-based precautions, that moved from a test-based strategy to a symptom-based strategy [12]. Even after complete resolution of symptoms, some patients can have a prolonged positive test result [8]. We observed as many as 32 patients with positive results as long as 30 days after the first documented positive results [13]. Although most of them exhibited very high Cts, corresponding to less than 1000 copies/mL, we also observed an asymptomatic patient with sustained high viral loads up to 5 weeks after infection. Hence, we think it important to systematically consider both the viral load and the presence of symptoms, because clear data on the potential relationship between virus load and contagiousness are still missing [9]. On the other hand, detection of low viral load in the upper respiratory tract of asymptomatic patients may occur during the onset of infection. However, a person with a low viral low tested early in the disease course might become highly infectious within the 24 hours following the first test and with a strong increase of viral loads. Some suggest repeated testing of the same patients over a period of 24 hours to monitor the viral load but we only recommend re-testing when the interval is 72 hours or more, because daily retesting might become rapidly problematic as a result of the pending world shortage of reagents and the high workload in most laboratories. Furthermore, a nasopharyngeal SARS-CoV-2 swab result could be very dependent on the quality of the sampling. Moreover, a very high viral load may be present in the lung in a patient with a COVID-19 pneumonia when the viral load in the nasopharyngeal swab may be much lower, even negative. Finally, serological investigations might help to address the disease timeline in a patient.
In conclusion, although many pre-analytical issues can affect the result of any respiratory virus RT-PCR tests, our data also highlight the importance of viral load quantification and their interpretation in a clinical context for the interpretation of SARS-CoV-2 RT-PCR tests for the care of patients with positive results.
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The data were obtained during a quality enhancement project at our institution. According to national law, the performance and publishing of the results of such a project can be done without asking the permission of the competent research ethics committee.
Acknowledgments
We thank all the staff of the Laboratory of Molecular Diagnostic of the Institute of Microbiology of the University of Lausanne.
Contributor Information
G. Greub, Email: Gilbert.Greub@chuv.ch.
O. Opota, Email: Onya.Opota@chuv.ch.
References
- 1.Deng S.Q., Peng H.J. Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. J Clin Med. 2020;9(2) doi: 10.3390/jcm9020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greub G., Sahli R., Brouillet R., Jaton K. Ten years of R&D and full automation in molecular diagnosis. Future Microbiol. 2016;11:403–425. doi: 10.2217/fmb.15.152. [DOI] [PubMed] [Google Scholar]
- 3.Jacot D., Greub G., Jaton K., Opota O. Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.08.004. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Opota O., Brouillet R., Greub G., Jaton K. Comparison of SARS-CoV-2 RT-PCR on a high-throughput molecular diagnostic platform and the cobas SARS-CoV-2 test for the diagnostic of COVID-19 on various clinical samples. Pathog Dis. 2020 doi: 10.1093/femspd/ftaa061. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones T.C., Mühlemann B., Veith T., Biele G., Zuchowski M., Hoffmann J. An analysis of SARS-CoV-2 viral load by patient age. medRxiv. 2020:2020. 06.08.20125484. [Google Scholar]
- 6.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa345. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao A.T., Tong Y.X., Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa460. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tom M.R., Mina M.J. To interpret the SARS-CoV-2 test, consider the cycle threshold value. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa619. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V. The detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 assays. J Clin Microbiol. 2020 doi: 10.1128/JCM.00772-20. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 test. J Clin Microbiol. 2020 doi: 10.1128/JCM.00926-20. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC . CDC; Atlanta, GA: 2020. Discontinuation of isolation for persons with COVID-19 not in healthcare settings. epub ahead of print. [Google Scholar]
- 12.Mueller L., Scherz V., Greub G., Jaton K., Opota O. Computer-aided medical microbiology monitoring tool: a strategy to adapt to the SARS-CoV-2 epidemic and that highlights RT-PCR consistency. medRxiv. 2020 doi: 10.3389/fcimb.2021.594577. 2020.07.27.20162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]