Extremophiles, organisms that thrive in extreme environments, are key models for research on biological adaption. They can provide hints for the origin and evolution of life, as well as improve the understanding of biogeochemical cycling of elements. Extremely acidophilic bacteria such as Acidiphilium are widespread in acid mine drainage (AMD) systems, but the metabolic potential, ecological functions, and evolutionary history of this genus are still ambiguous. Here, we sequenced the genomes of three new Acidiphilium strains and performed comparative genomic analysis on this extremely acidophilic bacterial genus. We found in the genomes of Acidiphilium an abundant repertoire of horizontally transferred genes (HTGs) contributing to environmental adaption and metabolic ability expansion, as indicated by phylogenetic reconstruction and gene context comparison. This study has advanced our understanding of microbial evolution and biogeochemical cycling in extreme niches.
KEYWORDS: acid mine drainage, evolution, horizontal gene transfer, comparative genomics, Acidiphilium
ABSTRACT
Here, we report three new Acidiphilium genomes, reclassified existing Acidiphilium species, and performed the first comparative genomic analysis on Acidiphilium in an attempt to address the metabolic potential, ecological functions, and evolutionary history of the genus Acidiphilium. In the genomes of Acidiphilium, we found an abundant repertoire of horizontally transferred genes (HTGs) contributing to environmental adaption and metabolic expansion, including genes conferring photosynthesis (puf, puh), CO2 assimilation (rbc), capacity for methane metabolism (mmo, mdh, frm), nitrogen source utilization (nar, cyn, hmp), sulfur compound utilization (sox, psr, sqr), and multiple metal and osmotic stress resistance capacities (czc, cop, ect). Additionally, the predicted donors of horizontal gene transfer were present in a cooccurrence network of Acidiphilium. Genome-scale positive selection analysis revealed that 15 genes contained adaptive mutations, most of which were multifunctional and played critical roles in the survival of extreme conditions. We proposed that Acidiphilium originated in mild conditions and adapted to extreme environments such as acidic mineral sites after the acquisition of many essential functions.
IMPORTANCE Extremophiles, organisms that thrive in extreme environments, are key models for research on biological adaption. They can provide hints for the origin and evolution of life, as well as improve the understanding of biogeochemical cycling of elements. Extremely acidophilic bacteria such as Acidiphilium are widespread in acid mine drainage (AMD) systems, but the metabolic potential, ecological functions, and evolutionary history of this genus are still ambiguous. Here, we sequenced the genomes of three new Acidiphilium strains and performed comparative genomic analysis on this extremely acidophilic bacterial genus. We found in the genomes of Acidiphilium an abundant repertoire of horizontally transferred genes (HTGs) contributing to environmental adaption and metabolic ability expansion, as indicated by phylogenetic reconstruction and gene context comparison. This study has advanced our understanding of microbial evolution and biogeochemical cycling in extreme niches.
INTRODUCTION
Prokaryotes occupy almost all environmental niches and have dominated the majority of Earth’s evolutionary history. Extremophiles that thrive in extreme environments represent a key research field in many disciplines, ranging from the adaption to extreme conditions to the cycling of elements in biogeochemistry. Extremophiles also have important implications for the research on the origin of life and the search for life on other planets (1, 2). Acid mine drainage (AMD), characterized by extreme acidity and high concentrations of metals and sulfate, represents an extreme ecological condition and a major global challenge (3). The primary microbial taxa in AMD include Acidiphilium, Acidisphaera, Acidithiobacillus, and Leptospirillum (4). The biological factors that contribute to the formation of this hyperacidic environment as well as the adaptive mechanisms of the organisms inhabiting it are hot topics in current research (3, 5). The genus Acidiphilium belongs to the family Acetobacteraceae, class Rhodospirillales, and appears frequently in AMD environments (6, 7). Members of this genus are Gram-negative, photosynthetic, aerobic and facultative anaerobic, metal-respiring, acidophilic heterotrophs (8–10). They grow at pH 1.5 to 7.5, are able to utilize a wide range of organic and inorganic substrates, and synthesize poly-β-hydroxybutyrate (PHB) for carbon storage (11–13). Acidiphilium can also resist multiple harmful stressors such as toxic metals (e.g., Cd, Ni, Cr) and osmotic pressure (14–16). There has also been increased interest in the application of Acidiphilium spp. for microbial fuel cells (MFCs) (17), as well as in metal mobilization from minerals or waste both in pure culture and in coculture (18–20). Nevertheless, little is known about whether there are divergences in the functional potential and niche partitioning among Acidiphilium-affiliated species. In addition, the evolutionary history of many notable properties such as carbon assimilation and metal resistance in Acidiphilium is still elusive. Acidiphilium is one of only four genera in the family Acetobacteraceae found in acidic mineral sites, with the other three genera being Acidicaldus, Acidisphaera, and Acidocella (21–24). Evolutionary drivers such as horizontal gene transfer (HGT) and selection pressure might have played their parts in the adaptive evolution of Acidiphilium that survives in harsh acidic mineral conditions. However, their relative contributions are still ambiguous. Horizontal gene transfer refers to the acquisition of genetic elements from distant lineages for genetic and phenotypic innovations, a process contributing significantly to evolution within challenging environments and during global geologic and/or climatic events (25, 26). Positive selection, on the other hand, mediating survival fitness by adaptive mutations, has also been an indispensable driving force in microbial evolution, and recent investigations have shifted from testing selection on individual genes to the entire genomes (27–30).
To assess the differences in metabolic capacity and niche adaption potential among Acidiphilium species, and to unravel the evolutionary history of many fundamental genetic properties of Acidiphilium, we performed whole-genome sequencing of three novel strains of Acidiphilium isolated from two different AMD sites. Comparative genome analysis was carried out, focusing on understanding the roles of evolutionary processes in shaping the genomes of Acidiphilium. For this purpose, we conducted a detailed comparison of Acidiphilium species. We performed ancestral genomic reconstruction, cooccurrence analysis, and extensive phylogenetic analyses and explored the genomic arrangements of pathways of interest. We also focused on discovering genes under positive selection.
RESULTS
Genomic features and reclassification of Acidiphilium.
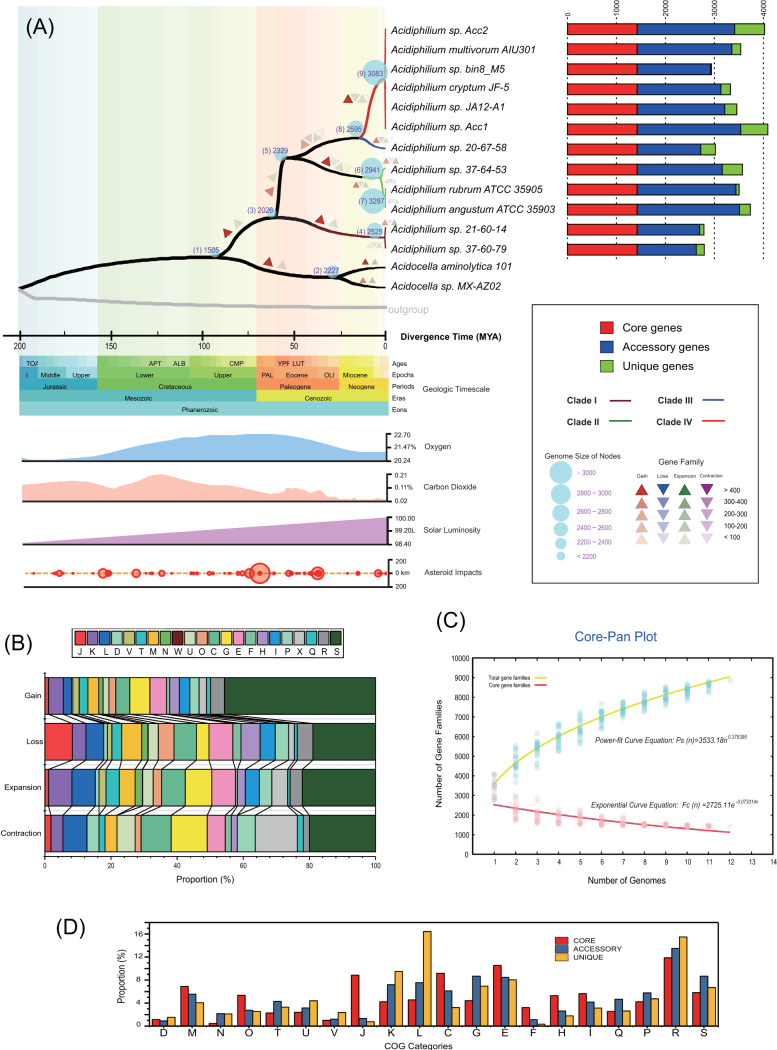
Three Acidiphilium genomes (AccI, AccII, and ZJSH63) were sequenced, resulting in a complete genome of strain AccI (a single chromosome and seven plasmids) and high-quality drafts of strains AccII and ZJSH63, according to the MISAG standards (31). The characteristics of these genomes and the other publicly available genomes of Acidiphilium spp. used in this study are shown in Table 1. The visualization of strain AccI chromosome (applying colors based on clusters of orthologous group [COG] classes) and comparative analysis of our three genomes were performed (see Fig. S1 at https://doi.org/10.6084/m9.figshare.12892016.v1). The genome size of Acidiphilium was about 4 Mbp. Although strains AccI and AccII were isolated from the same site, strain AccI shared more gene families with strain ZJSH63 than AccII. Strain AccII contained the most unique gene families among our three strains. COG annotations showed that AccII contained more unique genes with adaptive functions than ZJSH63 and AccI, especially those related to COG category L (replication, recombination, and repair) and COG N (cell motility). State-of-the-art whole-genome average nucleotide identity (ANI) analysis (32) classified all Acidiphilium genomes into four clades (species) based on an ANI cutoff of 95% (see Fig. S2 at https://doi.org/10.6084/m9.figshare.12892016.v1). We found some disagreement between our ANI results and previous nomenclatures of the Acidiphilium strains (mainly based on 16S rRNA sequences) in GenBank/JGI-IMG databases (33–35). For example, our ANI results showed that strains of Acidiphilium cryptum and Acidiphilium multivorum, as well as Acidiphilium angustum and Acidiphilium rubrum, should be classified as the same species (ANI > 95%). The major problem with previous species classification, based on 16S rRNA gene sequencing, was the low resolution, as shown by the low bootstrap values of the phylogenetic tree constructed with 16S rRNA sequences (see Fig. S3B at https://doi.org/10.6084/m9.figshare.12892016.v1). This shortage might be overcome by ANI analysis (32). Thus, the genomes of Acidiphilium spp. were thereafter referred to as clades I to IV according to their new classification based on ANI (Table 1; see Fig. S2 at https://doi.org/10.6084/m9.figshare.12892016.v1). Strain CAG727 (GCA_000437515.1) was determined not to be a member of Acidiphilium, given that it was phylogenetically distant from other Acidiphilium strains (see Fig. S2 and S3 at https://doi.org/10.6084/m9.figshare.12892016.v1), and was therefore excluded from further analyses. The clustering of Acidiphilium strains based on ANI values was mostly congruent with their geographic locations. For example, strains of clades I, II, and III were isolated from North America, while strains of clade IV were isolated from Europe and East Asia (see Fig. S2 at https://doi.org/10.6084/m9.figshare.12892016.v1). Whole-genome synteny analysis of all available complete sequences of Acidiphilium (strains AccI, JF-5, and AIU301, which belong to the same clade) showed that nine conserved locally colinear blocks (LCBs) were present in these strains, but they differed in their order of arrangement and similarity (see Fig. S4A in https://doi.org/10.6084/m9.figshare.12892016.v1). A similarity-based whole-genome comparison of Acidiphilium spp. with strain AccI as the reference showed that many genomic regions were not common to all isolates, many of which harbored hypothetical proteins and mobile genetic elements (see Fig. S5 at https://doi.org/10.6084/m9.figshare.12892016.v1).
TABLE 1.
General features of bacterial genomes used in this study
| Organism and strain | GenBank/IMG-ER accession no. | Level | Contig | N50 (bp) | No. of plasmids | Completeness (%) | Size (Mb) | Coding density (%) | GC (%) | Clade assigned | No. of genes | No. of proteins | Source | Geographic location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acidiphilium sp. strain ZJSH63 | 2828882166 | Draft | 301 | 124,801 | 95.3 | 4.39 | 90.6 | 66.5 | IV | 4,330 | 4,245 | Acid mine drainage | Fujian, China | |
| Acidiphilium sp. strain AccI | 2824045439 | Complete | 1 | 4,058,204 | 7 | 100 | 4.18 | 90.6 | 66.7 | IV | 4,304 | 4,224 | Acid mine drainage | Guangdong, China |
| Acidiphilium sp. strain AccII | 2824049744 | Draft | 716 | 93,659 | 98.0 | 4.69 | 89.4 | 65.5 | IV | 4,352 | 4,347 | Acid mine drainage | Guangdong, China | |
| Acidiphilium angustum ATCC 35903 | GCA_000701585.1/2561511102 | Draft | 206 | 71,968 | 98.5 | 4.07 | 89.6 | 63.6 | II | 3,851 | 3,731 | Acid mine | ||
| Acidiphilium cryptum JF-5 | GCA_000016725.1/640427101 | Complete | 1 | 3,389,227 | 8 | 100 | 3.96 | 90.8 | 67.1 | IV | 3,747 | 3,574 | Acid mine | |
| Acidiphilium multivorum AIU301 | GCA_000202835.1 | Complete | 1 | 3,749,411 | 8 | 100 | 4.21 | 89.8 | 67.0 | IV | 3,991 | 3,803 | Acid mine water | Iwate, Matsuo, Japan |
| Acidiphilium rubrum ATCC 35905 | GCA_900156265.1/2681812815 | Draft | 78 | 136,338 | 98.6 | 3.98 | 89.9 | 63.7 | II | 3,752 | 3,692 | Acidic coal mine drainage | Pennsylvania, USA | |
| Acidiphilium sp. strain 20-67-58 | GCA_002255515.1 | Draft | 119 | 80,126 | 98.6 | 3.41 | 89.4 | 66.6 | III | 3,199 | 3,073 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 21-60-14 | GCA_002255745.1 | Draft | 132 | 63,048 | 98.6 | 3.06 | 90.8 | 60.2 | I | 2,940 | 2,828 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 21-62-4 | GCA_002255545.1 | Draft | 433 | 1,584 | 16.2 | 0.69 | 85.0 | 61.7 | II | 910 | 832 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 21-66-27 | GCA_002255645.1 | Draft | 542 | 1,752 | 11.5 | 1.02 | 82.6 | 65.7 | III | 1,316 | 1,189 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 21-68-69 | GCA_002279225.1 | Draft | 884 | 2,201 | 44.6 | 1.81 | 82.3 | 67.9 | III | 2,291 | 1,999 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 34-60-192 | GCA_002282645.1 | Draft | 89 | 65,514 | 83.8 | 3.11 | 90.6 | 60.1 | I | 3,029 | 2,695 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 34-64-41 | GCA_002282635.1 | Draft | 180 | 38,599 | 93.9 | 3.86 | 88.6 | 63.7 | II | 3,623 | 3,413 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 37-60-79 | GCA_002279355.1 | Draft | 98 | 72,002 | 96.6 | 3.07 | 90.9 | 60.0 | I | 2,916 | 2,791 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 37-64-53 | GCA_002279345.1 | Draft | 211 | 43,062 | 97.3 | 4.03 | 88.3 | 63.5 | II | 3,799 | 3,620 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain 37-67-22 | GCA_002279335.1 | Draft | 937 | 3,121 | 62.2 | 2.48 | 89.9 | 67.0 | III | 2,942 | 2,632 | Mine wastewater | Ontario, Canada | |
| Acidiphilium sp. strain bin8_M5 | 2734482270 | Draft | 74 | 76,901 | 97.3 | 3.08 | 92.7 | 68.8 | IV | 3,042 | 2,992 | Acid mine drainage | Guangdong, China | |
| Acidiphilium sp. strain CAG727 | GCA_000437515.1 | Draft | 116 | 9,562 | 82.5 | 1.73 | —a | 45.4 | — | 1,509 | 1,479 | Gut microbiota | ||
| Acidiphilium sp. strain JA12-A1 | GCA_000724705.2/2571042905 | Draft | 296 | 44,500 | 98.6 | 4.18 | 88.4 | 66.9 | IV | 4,059 | 3,719 | Acid mine drainage | Lusatia, Germany, Europe | |
| Acidiphilium sp. strain PM | GCA_000219295.2 | Draft | 627 | 12,446 | 91.2 | 3.93 | 86.4 | 66.4 | IV | 3,908 | 3,859 | Acidic, metal-rich water | Rio Tinto, Spain, Europe |
—, not available.
Core genome and pangenome of Acidiphilium.
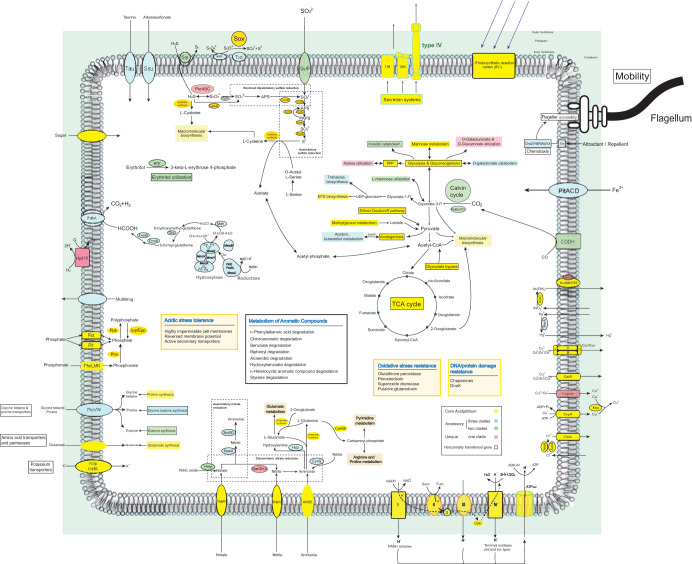
Twelve genomes of the genus Acidiphilium, with estimated completeness over 97.0%, were carefully chosen for further pangenome analysis and genomic content reconstruction. The phylogenetic trees based on the concatenated alignment of 133 core genes inferred with neighbor-joining (NJ) methods were congruent with that based on whole-genome sequences (Fig. 1A; see Fig. S3A at https://doi.org/10.6084/m9.figshare.12892016.v1). The pangenome of the 12 Acidiphilium strains possessed 8,845 gene families, while the core genome possessed 1,422 gene families accounting for only 16.1% of the pangenome (Fig. 1C). Core and pangenome analyses of the 12 Acidiphilium genomes revealed an ‘‘open’’ pangenome fitted into a power-law regression function [Ps (n) = 3,533.18n0.375395], while the core genome was fitted into an exponential regression [Fc (n) = 2,725.11e −0.073314n] (Fig. 1A). The open pangenome suggested that species have undergone considerable gene exchanging to extend their functional profiles (36). Functional COG annotation revealed that the core genome had a higher proportion of genes classified in COG categories J (translation, ribosomal structure, and biogenesis), C (energy production and conversion), O (posttranslational modification, protein turnover, chaperones), F (nucleotide transport and metabolism), and H (coenzyme transport and metabolism), all associated with basic biological functions. The accessory genome and strain-specific genes were biased toward COG categories G (carbohydrate transport and metabolism), L (replication, recombination, and repair), P (inorganic ion transport and metabolism), and N (cell motility) (Fig. 1D), which were probably related to the adaption of Acidiphilium to oligotrophic, metal-laden, and acidic environments that often cause DNA damage. Gene ontology (GO) enrichment analyses produced similar results (see Table S1 in the supplemental material). Detailed metabolic reconstruction of Acidiphilium was also performed; the core/specific metabolic features are shown in Fig. 2 using different colors, and pathways containing predicted horizontally transferred genes are marked with black rectangles.
FIG 1.
(A) The evolutionary timeline of Acidiphilium was estimated (left) using RelTime on top of the rooted NJ tree based on the concatenated alignment of 133 core genes. Ancestral genome content reconstruction of Acidiphilium was performed with Count software, and the color depth represents the numbers of reconstructed gain, loss, expansion, and contraction events of each lineage. Data of asteroid impacts, solar luminosity, and fluctuations in atmospheric oxygen and carbon dioxide concentrations are displayed synchronously with divergence times in the form of time panels (left). A stack bar diagram (right) shows the number of genes shared by all strains (i.e., the core genome), the number of genes shared by partial strains (i.e., the accessory genome), and the number of strain-specific genes (i.e., the unique gene) in the tested strains. (B) Stack bar chart showing functional proportions of Acidiphilium gene families undergoing gain, loss, expansion, and contraction events as based on COG classes. (C) Mathematical modeling of the pangenome and core genome of Acidiphilium. (D) Bar chart showing functional proportions (based on COG categories) of different parts of the Acidiphilium pangenome (i.e., core, accessory, unique). Detailed descriptions of the COG categories are provided in Text S1 in the supplemental material.
FIG 2.
Overview of metabolic potentials in Acidiphilium as predicted from genome annotation; core/specific metabolic features are shown with different colors, and pathways containing predicted horizontally transferred genes are marked with black rectangles.
GO enrichment analyses (P value < 0.05) of Acidiphilium gene families. Download Table S1, XLSX file, 0.02 MB (16.4KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed description for abbreviations of COG categories. Download Text S1, DOCX file, 0.01 MB (11.9KB, docx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MGEs and CRISPR-Cas systems.
Mobile genetic elements (MGEs), such as insertion sequences, transposases, genomic islands (GIs), plasmids, and phages, are known signals of HGT events, and the number of MGEs correlates positively with the frequency of HGT (37). MGEs in the genomes of Acidiphilium were identified in this study (Table S2). The average number of transposon sequences per genome was 307, with A. multivorum AIU301 harboring the greatest number (841). Members of the ISAli7, ISGalb1, ISMex27, ISAan1, and ISAcr4 families were most common. The average number of sequences located in GIs per genome was 527, with Acidiphilium sp. strain ZJSH63 containing the most (1,055). The average number of prophages and/or prophage remnants per genome was 23, with Acidiphilium sp. strain AccII harboring the most (58, with a total size of 69.9 kb). The number of plasmids in Acidiphilium could reach eight (A. cryptum JF-5 and A. multivorum AIU301). The functional gene profiles of plasmids from these three completely sequenced strains were also compared (see Fig. S4B to D at https://doi.org/10.6084/m9.figshare.12892016.v1). Approximately 66 gene families were shared among plasmids from these three strains, and certain degrees of collinearity were observed. COG L (replication, recombination, and repair) functions were enriched in the plasmid genomes. Type I-C/E/V and II-C CRISPR-Cas systems were also found in Acidiphilium spp., with Acidiphilium sp. strain PM containing the most CRISPR-Cas-related genes or spacers (38). The abundant MGEs present in genomes of Acidiphilium indicated that HGT might have contributed significantly to the genomic evolution of Acidiphilium species during niche adaption, while the CRISPR-Cas system would also help protect the genomes of Acidiphilium by eliminating harmful genomic intrusion events, balancing genomic stability and functional investments (39). A recent study also revealed that spacer sequences of the CRISPR-Cas system could not only specify the targets of Cas nucleases but also facilitate HGT (40).
Statistics of mobile genetic elements (MGEs) present in the genomes of Acidiphilium spp. Download Table S2, XLSX file, 0.5 MB (539KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evolutionary analyses of Acidiphilium.
The evolutionary timeline of Acidiphilium was also estimated on the rooted core gene tree (Fig. 1A). Overall, gene families undergoing gain events outnumbered those undergoing loss events by approximately three times (6,319 versus 2,231), and gene families undergoing expansion events outnumbered those undergoing contraction events by approximately 20 times (1,173 versus 55) in the genomes of Acidiphilium. Our analyses suggested that there has been an ongoing increase in genomic content throughout the evolutionary history of this genus, from an estimated 2,026 gene families in the common ancestor to over 3,000 gene families. Predicted gain events of over 400 gene families occurred at nodes 3, 4, 6, and 9, accounting for approximately 14% to 25% of gene families at the corresponding nodes. Of all gene families undergoing gain events, about half encoded hypothetical proteins. A considerable proportion of gain events were related to COG category G (carbohydrate transport and metabolism, 6.1%) and COG category E (amino acid transport and metabolism, 5.0%), and a notable proportion of gene families undergoing expansion events were also related to COG categories G (carbohydrate transport and metabolism, 8.0%), E (amino acid transport and metabolism, 7.2%), C (energy production and conversion, 7.2%), K (transcription, 7.0%), and L (replication, recombination, and repair, 7.0%) (Fig. 1B). It seems that the COG categories involved are carbohydrate metabolism and transport as well as amino acid metabolism and transport, which reflect the adaptive strategies of Acidiphilium, including the expansion of metabolic abilities to utilize a variety of potential nutrients, while the acquisition of efficient repair mechanisms is in response to damage of biological molecules possibly caused by harsh environments such as AMD sites. This was in line with previous work which showed that larger genomes preferentially accumulated genes associated with metabolism, regulation, and energy conversion (41). We also found that 8.3% of gene families undergoing loss events belonged to COG category J (translation, ribosomal structure, and biogenesis) and that 12.7% of gene families undergoing contraction events belonged to COG category X (mobilome: prophages, transposons) (Fig. 1B), which were probably related to a holistic adjustment toward a more efficient operational and survival mode of these heterotrophs. The most recent common ancestor (MRCA) of Acidiphilium spp. was estimated to have emerged around 60.3 million years ago (Mya) (Fig. 1A, left), not long after a recorded strong asteroid impact, which we postulated to be one of the possible courses of significant changes to the Earth’s atmosphere, since it coincided with a decrease in atmospheric O2 and increase in atmospheric CO2.
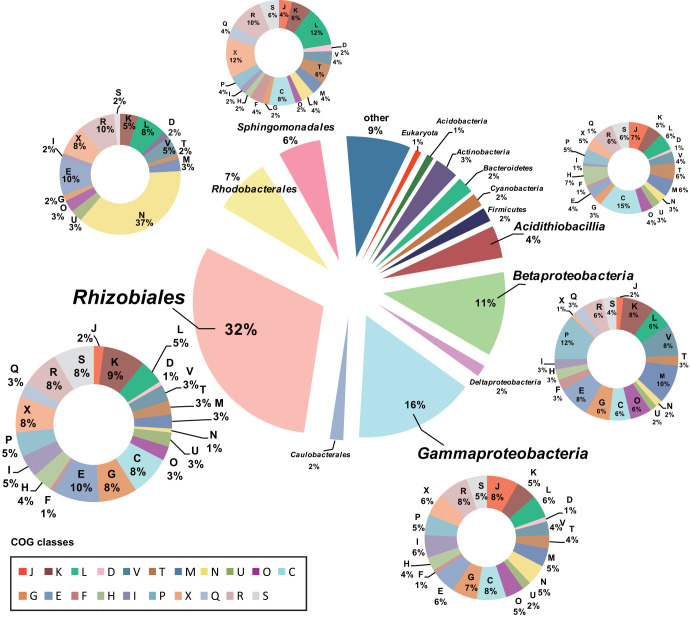
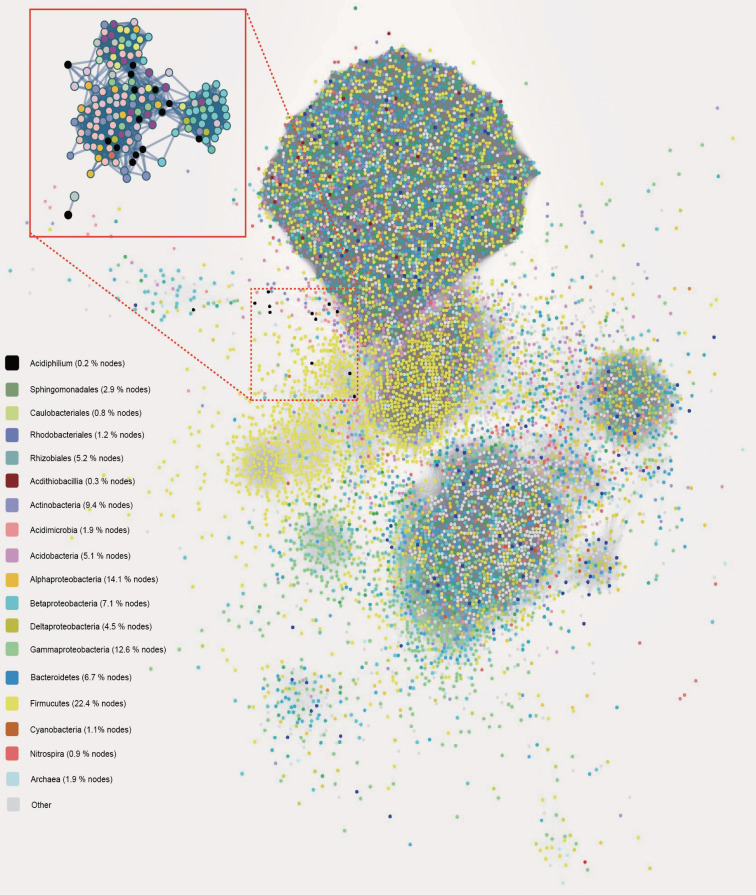
We extracted HGT events predicted with the IMG Annotation Pipeline (Table S3). Results showed that a notable set of genes were identified as being acquired via HGT, accounting for up to 18.9% of genes among tested Acidiphilium genomes, indicating the chimeric nature of these genomes. Cross-order HGT events from Rhizobiales were the most frequent, accounting for ∼32% of total HGT events, followed by cross-class HGT events from Gammaproteobacteria (∼16%) and Betaproteobacteria (∼11%) and cross-order HGT events from Rhodobacterales (∼7%) and Sphingomonadales (∼6%). A considerable proportion (∼4%) of genes were derived via cross-class HGT from the typical AMD autotroph Acidithiobacillia (Fig. 3). The above-mentioned HGT donors were almost all consistently present together with Acidiphilium in the cooccurrence network based on 16S rRNA gene amplicon data sets generated from AMD samples (Fig. 4). In the cooccurrence network, Acidiphilium accounted for 0.2% of the nodes, while the HGT donors occupied 0.3 to 22.4% of the nodes. Furthermore, most of these HGT donors were present in the first-neighbor network of Acidiphilium. Function annotations of putative horizontally transferred genes (HTGs) based on COG classes showed that these genes were biased toward COG categories E (amino acid transport and metabolism), K (transcription), C (energy production and conversion), and G (carbohydrate transport and metabolism), which are associated with metabolic and energy production processes, COG categories L (replication, recombination, and repair) and V (defense mechanisms), all associated with defense and repair mechanism, COG category X (mobilome: prophages, transposons), involved in the mobilization of genome fragments, and COG category M (cell wall/membrane/envelope biogenesis), related to enhanced cellular barriers against external disturbance. COG category N (cell motility) was found to account for up to 37% of Rhodobacterales-derived HGT (Fig. 3), which might facilitate Acidiphilium to swim away from harmful environments and/or toward nutrients. We further performed detailed analyses of gene synteny and phylogeny for examination and quantification of the cumulative impact of HGT on Acidiphilium evolution.
FIG 3.
Pie charts showing donors that putatively transferred genes to Acidiphilium spp., their relative contributions to total HGT events, and the functional proportions of the HGT functions derived from respective donors based on COG classes. Detailed descriptions of the COG categories are provided in Text S1 in the supplemental material.
FIG 4.
Cooccurrence network based on correlation analysis of 16S rRNA amplicon sequencing data sets of AMD samples (n = 205). Each node denotes a microbial OTU at a 97% cutoff. The first neighbors of Acidiphilium nodes (highlighted by a red rectangle) were selected using the tool “first neighbors of selected nodes” in Cytoscape.
Extracted information of putative horizontally transferred genes from the results of IMG annotation. Download Table S3, XLSX file, 0.04 MB (45.8KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Environmental stress adaption.
Acidiphilium was predicted to originate at a time when the O2 concentration present in the atmosphere reached its peak (22.7%) (Fig. 1A, left). As Acidiphilium evolved, the atmospheric O2 concentration decreased to about 20.6% in a consistent manner. In contrast, solar luminosity gradually increased to current levels (100 L), and the atmospheric CO2 concentrations first increased by approximately 0.1%, followed by a decrease of about 0.05% (Fig. 1A, left). The bd-type oxidase encoded by cydAB for oxygen-reducing energy production was present in all clades of Acidiphilium but was not uniform with gene synteny and discrepancy of phylogeny compared with the species tree (see Fig. S3, S6, and S7 at https://doi.org/10.6084/m9.figshare.12892016.v1). This suggested that independent HGT events contributed to the acquisitions of cydAB after the speciation of clades I and II but before the divergence of clades III and IV. The acquisitions of cydAB genes were probably in adaption to decreasing atmospheric O2 concentrations, considering the high affinity of bd-type oxidase even at low O2 concentrations (42). Additionally, cytochrome bo3-type ubiquinol oxidase (CyoABCD), which facilitates growth at low pH and low O2 concentrations (43), was found in Acidiphilium clades I and IV, which were probably acquired from species sharing a habitat with Acidiphilium, such as Acidithiobacillus, Acidihalobacter, and Acidiferrobacter species (see Fig. S8 at https://doi.org/10.6084/m9.figshare.12892016.v1). The nuo gene cluster in Acidiphilium that encodes NADH:ubiquinone oxidoreductase (complex I), and functions preferentially under aerobic conditions (44–46), was also HGT derived (see Fig. S9 at https://doi.org/10.6084/m9.figshare.12892016.v1). These acquisitions might explain the facultative anaerobic ability of Acidiphilium. The HGT-derived Calvin cycle-related gene cluster prk-rbcLS-cbbX was present in clades II and IV of Acidiphilium (see Fig. S10 to S14 at https://doi.org/10.6084/m9.figshare.12892016.v1), which might help Acidiphilium overcome oligotrophic AMD conditions through CO2 assimilation. Carbon monoxide dehydrogenase HTGs (CoxLMS) were detected in clades III and IV (see Fig. S15 and S16 at https://doi.org/10.6084/m9.figshare.12892016.v1). This suggested that Acidiphilium might utilize CO that was present in mine areas (47) as an energy supplement and source of CO2, since the atmospheric CO2 concentration dropped to a lower level upon the diversification of Acidiphilium (Fig. 1A, left). Photosystem II-type photosynthetic reaction centers (PufBALMC and PuhA) were found in all clades of Acidiphilium, probably transferred to Acidiphilium after the speciation of clade III (see Fig. S17 to S19 at https://doi.org/10.6084/m9.figshare.12892016.v1), coinciding with the acquisition of gene clusters prk-rbcLS-cbbX and coxLMS. Ni/Fe hydrogenase HTGs were detected only in clade IV (see Fig. S20 at https://doi.org/10.6084/m9.figshare.12892016.v1). AMD sites that Acidiphilium inhabits are hyperosmotic and rich in various metals due to the corrosion of minerals by sulfuric acid and chemoautotrophic microbes (48). We discovered a set of HGT-derived heavy metal resistance genes as well as osmotic pressure resistance genes in the genomes of Acidiphilium (see Fig. S21 to S46 at https://doi.org/10.6084/m9.figshare.12892016.v1). For example, apcA (49), arsH (50), which encodes an NADPH-dependent metal-reducing cytochrome/protein, merA, encoding mercuric reductase, chrA, encoding a chromate transporter, and arsRCB, which confers arsenic resistance, were present in all clades of Acidiphilium. It was notable that two of our strains, AccI and AccII, harbored more copies of ferric ion-reducing apcA genes (49) than strain ZJSH63, probably due to the higher ferric ion concentration in the sites that AccI and AccII inhabited. HGT-derived toxic divalent cation (e.g., cadmium, zinc, cobalt, and copper) resistance genes czcABC, czcD, copA, and mco and regulatory genes czrRS were also present in all clades of Acidiphilium, while HGT-derived Cu homeostasis genes copCD (51) were detected only in some strains of clade IV. Chelation of metals with polyphosphate (polyP) is also an effective metal resistance mechanism of acidophiles (52). HGT-derived alkaline phosphatases (Alp), which release phosphate groups from various compounds, and phosphate import systems encoded by pstSACB and pitA were present in all clades of Acidiphilium. Compatible solute uptake and biosynthesis as well as potassium uptake are known as common strategies to counteract osmotic stress (53, 54). The HGT-derived biosynthetic pathway of the compatible solute hydroxyectoine conferred by the gene cluster ectRABCD-ask (55) was detected in the genomes of clades III and IV. HGT-derived kdpABCDE, which confer resistance to osmotic stress by uptake of K+, were present in all clades of Acidiphilium, likely acquired via individual HGT. HGT-derived gene cluster proXWV-betBA-soxBDAG, involved in the uptake and biosynthesis of glycine betaine, is present in all clades of Acidiphilium except for clade I. This gene cluster was likely gained before diversification of clades III and IV but after speciation of clade II. A standalone HTG, betC, also involved in glycine betaine synthesis, was present only in clades III and IV. Motility and chemotaxis conferred by flagellar and sensing proteins might help microbes swim away from harmful environments and toward favorable chemicals or other nutrients. We found operons involved in flagellar biosynthesis such as flg, flh, and fli located in identified genomic islands (GIs) (Table S2C), and the HGT-derived chemotaxis operon cheDYBRWAYX was also found in all clades of Acidiphilium.
Metabolic potential expansion through HGT.
Many parts of the sulfur, nitrogen, and carbon metabolic pathways in Acidiphilium were also acquired through HGT. The Sox multienzyme complex (encoded by soxCDXYZAB and the regulatory soxH and soxR genes), which oxidizes thiosulfate to sulfate (56), was present in all clades of Acidiphilium and was likely derived via independent HGT events in clade I, clade II, and the MRCA of clades III and IV (see Fig. S47 and S48 at https://doi.org/10.6084/m9.figshare.12892016.v1). HTG sqr, encoding sulfide:quinone oxidoreductase, was found in clades III and IV (see Fig. S49 at https://doi.org/10.6084/m9.figshare.12892016.v1). HGT-derived polysulfide reductase (PsrABC) was found in clade IV, while thiosulfate sulfurtransferase (Tst) was detected only in clades II and IV (see Fig. S50 to S52 at https://doi.org/10.6084/m9.figshare.12892016.v1). Homologues of Sdo1 in Acidithiobacillus caldus MTH-04 (A5904_0421), a new sulfur dioxygenase associated with tetrathionate oxidation (57), were detected in all clades of Acidiphilium except clade II, likely acquired after the diversification of clades III and IV (see Fig. S53 and S54 at https://doi.org/10.6084/m9.figshare.12892016.v1). Thiosulfate dehydrogenase/tetrathionate reductase (encoded by tsdA), which mediates the flow of electrons into respiratory or photosynthetic electron chains (58), was also detected in Acidiphilium outside clade I (see Fig. S55 at https://doi.org/10.6084/m9.figshare.12892016.v1), which was likely acquired after the emergence of clade III (see Fig. S56 at https://doi.org/10.6084/m9.figshare.12892016.v1). Reversible assimilatory sulfate reduction (conferred by cysND and cysJHI), found in all clades of Acidiphilium, was inferred to be acquired before diversification of clades III and IV but after the speciation of clade II (see Fig. S57 to S59 at https://doi.org/10.6084/m9.figshare.12892016.v1). Nitrogen metabolism enzymes for nitrate/nitrite transporter (NarK), dissimilatory nitrate reductase (NarGHJI), nitric oxide dioxygenases (Hmp), cyanate lyase (CynS), hydroxylamine reductase (Hcp), assimilatory nitrate reductase (NasA), and nitrite reductase (NirBD) were all derived via HGT. However, they were only sparsely present in Acidiphilium (mostly clade IV) (see Fig. S60 to S64 at https://doi.org/10.6084/m9.figshare.12892016.v1).
Abundant HTGs involved in carbon metabolism were also detected, including those coding for a complete methane catabolic pathway and many key genes involved in hydrocarbon utilization. An HGT-derived methane catabolism pathway (conferred by genes mmoXCYB, fdhA, mdh, gfa, frmA, and frmB) was found in almost all clades of Acidiphilium (see Fig. S65 to S70 at https://doi.org/10.6084/m9.figshare.12892016.v1). Soluble methane monooxygenase (sMMO) converts methane to methanol, which could be further converted to formaldehyde by methanol dehydrogenase (Mdh). Formaldehyde is eventually converted to CO2 via enzymes encoded by gfa, frmA, and frmB (59, 60). Multiple genes related to glycolysis and gluconeogenesis (fba, fbp, GNL), the pentose phosphate pathway (tkt, tal, pgd, rpe, G6PD, prsA, and xfp), the Entner-Doudoroff pathway (gdh, ddgk, and pglD), methylglyoxal metabolism (megR), tricarboxylic acid cycle or glyoxylate bypass (fumC, mdh, mdh, mls, icl), acetogenesis (spxB, pta, PDHA1, actP), and the previously mentioned prk-rbcLS-cbbX involved in the Calvin cycle were proposed to have been acquired via HGT (see Fig. S71 to S92 at https://doi.org/10.6084/m9.figshare.12892016.v1). In addition, the rha operon, involved in metabolism of l-rhamnose, was present in the genomes of clades II and IV, likely acquired by HGT (see Fig. S93 at https://doi.org/10.6084/m9.figshare.12892016.v1). For sugar alcohol metabolism, genes involved in the metabolism of erythritol and inositol were found only in clades II and IV. The HGT-derived cluster iolBDC-inoKEF-iolIG1HEG2G3, conferring inositol catabolic ability, was detected in clade IV, while a similar but differently arranged gene cluster, iolDEG1G2-inoEKF-iolBCG3, was detected in clade II (see Fig. S94 at https://doi.org/10.6084/m9.figshare.12892016.v1). In addition, the ery operon, involved in erythritol utilization, was presented in clades II and IV of Acidiphilium, likely acquired via cross-order HGT (see Fig. S95 at https://doi.org/10.6084/m9.figshare.12892016.v1).
Multiple genes involved in the biodegradation and metabolism of aromatic compounds showed patchy distribution throughout the genus Acidiphilium. Many were gained via HGT (see Fig. S96 to S103 at https://doi.org/10.6084/m9.figshare.12892016.v1), including genes fadA, fadB, fadD (n-phenylalkanoic acid degradation), gene cluster catFIJ (chloroaromatic degradation), gene cluster benABCD-catBCA (benzoate degradation), and genes pcaGH, pcaB, pcaC, pcaD, pcaT, pcaQ, and pcaIJ (beta-ketoadipate pathway), together with pobA and pcaK (hydroxybenzoate degradation), iqoAB (n-heterocyclic aromatic compound degradation), and fahA and faaH (styrene degradation).
Positive selection analyses.
Genome-scale positive selection analyses were exhaustively performed on all 21 genomes of Acidiphilium in this study in order to expand the input data set. The complete genome of Acidiphilium sp. AccI was used as the anchor genome. Results showed that 15 genes were identified as being under positive selection, including Ap_37 (exoribonuclease), Ap_279 (acyl coenzyme A [acyl-CoA] dehydrogenase), Ap_346 (H+/Cl− exchange transporter), Ap_374 (membrane component of nitrite reductase), Ap_696 (hypothetical protein), Ap_835 (N-acetyltransferase), Ap_1029 (hypothetical protein), Ap_1038 (Holliday junction resolvase), Ap_1473 (3-hydroxy acid dehydrogenase), Ap_1766 (hypothetical protein), Ap_1855 (asparagine synthetase), Ap_2034 (transposase), Ap_2355 (pyridoxine/pyridoxamine 5′-phosphate oxidase), Ap_2619 (cysteine synthase), and Ap_2636 (thioredoxin) (Table S4).
Genes in Acidiphilium identified as being under positive selection using the PosiGene pipeline. Download Table S4, DOCX file, 0.01 MB (14.2KB, docx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, we present a detailed analysis of the metabolic capabilities and evolutionary history of the genus Acidiphilium. Abundant HGT events were found to contribute substantially to the genomic contents of Acidiphilium, providing this genus with unprecedented elasticity to counteract harsh conditions such as those found in AMD. HGT may also have had a great impact on the diversity of Acidiphilium gene repertoires. Genome size dynamics (“Why are some genomes really big and others quite small?”) and the occurrence of horizontal gene transfer (“Why does lateral transfer occur in so many species and how?”) were listed as two world-class scientific questions that awaited answers by the editorial of the journal Science entitled “So much more to know” (61), and we believe that the present study might provide some clues for these questions. Our results showed that the genome size of Acidiphilium was relatively large (∼4 Mbp), with overwhelming gene family gains predicted across its evolution. This is in sharp comparison with its endosymbiotic Acetobacteraceae relative (genome size of ∼2 Mbp) that underwent genome reduction (62, 63). Microbes tend to evolve relatively large genomes with higher nutrient uptake and metabolic potential as a means to compensate for fluctuating and inhospitable environments (64–66). This theory could be applied to Acidiphilium spp. that inhabit hyperacidic, metal-laden, nutrient-depleted AMD environments, which are quite different from the stable environments (with plentiful easy-to-metabolize resources) in which their endosymbiotic Acetobacteraceae relatives dwell (67). The gene repertoire of microbes might evolve rapidly, with HGT being a major source of gene acquisitions in microbial genomes, and a number of genomic analyses have shown that microorganisms adopted new a “lifestyle” via HGT in the colonization of new niches (68). In addition, HGT occurs mainly in the form of horizontal operon transfer (HOT) (69), since many functional modules require a contiguous gene cluster, as exemplified by the acquisition of the photosynthetic operon in Rhodobacteraceae (70). Consistent with this, our results showed that HGT of functional genes (cluster) might have conferred to Acidiphilium better environmental adaptations as well as the expansion of a wide range of metabolic abilities. For example, the coincided acquisitions of the photosystem II-type photosynthetic reaction center (RC), Calvin cycle enzymes, and carbon monoxide dehydrogenases might have conferred adaptive benefits to Acidiphilium by taking advantage of the high CO levels (∼50 ppm) in mining areas as an energy source (47, 71) and the increasing solar luminosity for enhanced CO2 assimilation and/or energy production (72). The methane (CH4), metal sulfides, hydrogen (H2), and hydrogen sulfide (H2S) that are present in mining areas (47, 73) are also potential energy sources for Acidiphilium. Hydrogen (H2) might be formed in AMD areas through the acid dissolution of metals and minerals, and Ni/Fe hydrogenases might exploit this as an electron donor to support chemolithotrophic growth (74). Correspondingly, Acidiphilium acquired a nearly complete repertoire of methane and sulfur metabolic genes, as well as genes encoding Ni/Fe hydrogenase. Evidence of expression of the above-mentioned pathways has been shown in a Rhodospirillales relative (75). This evidence together with previously observed related phenotypes of Acidiphilium (see the introduction) suggests that these HGT-derived genes may also be functional in Acidiphilium. Numerous HGT-derived resistance genes for AMD adaption were present in Acidiphilium, similar to other AMD inhabitants (76, 77), reflecting diverse strategies of Acidiphilium to avert the deleterious effects of toxic metals and osmotic pressure in AMD environments. The acquired metabolic capacity of organic compounds and hydrocarbon in Acidiphilium suggested a mutualistic interaction of autotrophic acidophiles in AMD. For instance, metabolotoxic organic byproducts excreted by autotrophs (e.g., Acidithiobacillus) might be utilized by chemoorganotrophs (e.g., Acidiphilium), a process that in turn might stimulate the metabolic processes of the autotrophs (78–80). AMD microbial communities tend to form biofilms on mineral substrates for better metabolic cooperation and enhanced resistance against harsh environments (80–82). The microbes in biofilms are usually active, and the high community density, with increased proximity of microbes encapsulated in biofilm, might create more numerous opportunities for the efficient occurrence of HGT. In addition, MGEs, such as plasmids, might contribute to the development, stabilization, and expansion of biofilm (83–85). Consistent with this, our results showed that the predicted donors of HGT were also present in the cooccurrence network of Acidiphilium. Cooccurrence networks in microbial communities may provide hints for ecological interactions between species, on which HGT might have an influence (86, 87). Positive selection was also found to be an important driving force for adaptive evolution of Acidiphilium. Genes can be changed by positive selection for fixation of beneficial variants in a population/species over time if they increase survival fitness, which might help fine-tune gene expression in adaption to changing environmental conditions (27–30). Those genes under positive selection in Acidiphilium were prone to play key multifunctional roles, of which even small adaptive changes in their coding sequences might influence multiple pathways, bringing considerable benefits for survival of microbes during evolution in response to changing global conditions and shifting of niches. For example, the positively selected gene (PSG) cysK (Ap_2619, encoding cysteine synthase) might perform functions related to sulfide utilization, tellurite resistance, and growth inhibition (88–90); the PSG pdxH (Ap_2355), encoding pyridoxine/pyridoxamine 5′-phosphate oxidase, might act as a potent quencher of reactive oxygen intermediates and as an essential cofactor in amino acid metabolism (91, 92); and, finally, the PSG trxA (Ap_2636) encodes thioredoxin, a small redox protein that may play important roles in electron transfer, transcriptional regulation, immune response, and oxidative stress defense (93–96). However, further experiments are required to confirm their actual functions in Acidiphilium. The genus Acidiphilium is one of only four genera in the family Acetobacteraceae reported to colonize metal-rich AMD sites thus far, with the other genera of Acetobacteraceae found primarily in more moderate environments such as vinegar production environments and breweries (38, 97, 98). We suggest that the ancestor of Acidiphilium may have originated in mild or moderate conditions but then adapted to extreme environments, such as AMD niches, with the help of HGT and probable positive selection on the genes, similar to what has been found in acidophilic archaeal lineages such as Thermoplasmatales and Sulfolobales, which seemed to have evolved independently from moderately acidophilic ancestors (99). It is foreseeable that as more Acidiphilium strains are isolated and sequenced, the panorama of Acidiphilium evolution will gradually unfold before us.
Conclusions.
Extremophiles that thrive in extremely acidic environments are research model organisms for microbial adaption and evolution. In this study, we provided evidence that Acidiphilium is characterized by a complex lifestyle granted by HGT. By way of gene acquisitions, Acidiphilium has greatly expanded its genetic diversity, resulting in functional divergence. Acidiphilium has acquired multiple abilities via HGT, such as photosynthesis, CO2 assimilation, metal resistance, and organic compound metabolism, which would facilitate beneficial interactions with cohabitant autotrophs. In addition, the predicted donors of HGT were present in the cooccurrence network of Acidiphilium. Positive selection on new mutations was also an important driving force in the evolution of Acidiphilium. We further proposed that microorganisms originating under mild conditions can adapt to extreme environments such as AMD sites after the acquisition of multiple adaptive functions. Taken together, this study has shed light on the ecological roles and evolutionary scenario of Acidiphilium and is a good example of research on the adaption and evolution of extremophiles.
MATERIALS AND METHODS
DNA extraction, genome sequencing, and assembly.
Strains AccI and AccII were isolated from an acid mine drainage (AMD) water sample obtained in the Mangzi mining area (formed by oxidizing dissolution of pyrite, characterized by a high ferric ion concentration), Yunnan Province, China (long 103.5, lat 23.3, altitude 1,847 m). Strain ZJSH63 was isolated from an AMD water sample obtained in the heap leaching area for copper ore in the Zijinshan Gold and Copper Mine, Fujian Province (lat 25.2, long 116.4, altitude 282.6 m), China. Genomic DNA of strains AccI, AccII, and ZJSH63 were extracted using the Qiagen genomic DNA extraction kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions. After the DNA sample passed quality testing, the large fragments were subjected to agarose recovery using a BluePippin automatic nucleic acid recovery instrument (SAGE Science). The DNA was damaged and repaired; after purification, the DNA fragments were end repaired and linked with adenine. After a purification and ligation reaction, Qubit was used to accurately quantify the constructed DNA library by following official protocol (http://cshprotocols.cshlp.org/content/2017/6/pdb.prot094730). The DNA library was subjected to the PacBio Sequel platform for sequencing at Guangdong Magigene Biotechnology Co. Ltd. (Guangzhou, China). After sequencing, SMRT Link v5.1.0 (https://www.pacb.com/support/software-downloads/) was utilized for correction and assembly.
ANI and whole-genome alignments.
JSpecies v1.2.1 was used to calculate average nucleotide identity (ANI) based on the BLASTN algorithm with default parameters (100). BLASTN-based whole-genome comparison of Acidiphilium strains (completeness > 97%) was performed and represented with BRIG-0.95 (101). We utilized Circos (102) for construction and visualization of the multiple genome alignments of strains with completely sequenced genomes, including AccI, JF-5, and AIU301.
Pangenome analyses and gene family evolution analyses of Acidiphilium.
A summary of features for the Acidiphilium genomes involved in this study are listed in Table 1. BUSCO (103) was used to estimate the completeness of each genome against its bacterial core gene set. Gene family clustering of 12 Acidiphilium genomes (completeness > 97%) together with UniProt search, GO Slim annotation, and GO enrichment analyses (default cutoff P value, 0.05) was performed via OrthoVenn2 (104) with default parameters. The BPGA pipeline (105) was used to perform model extrapolations of the Acidiphilium pangenome/core genome by applying default parameters. We applied COUNT (106) under the Wagner parsimony algorithm for ancestor genome size estimation and for detecting the gain, loss, expansion, and contraction events of gene families with the penalty ratio set to 1.
Phylogenic analyses and divergence time estimation.
Phylogenetic trees based on 133 concatenated core genes and 16S rRNA gene sequences of Acidiphilium were constructed with the neighbor-joining (NJ) method using MEGA-X (107) with 1,000 bootstrap replicates. A chronogram for Acidiphilium species with branch lengths reflecting divergence times was inferred on the core gene tree of Acidiphilium using the RelTime method (108) implemented in MEGA-X with the JTT matrix-based model as described previously (109). The TimeTree reference data (110) that integrated data of asteroid impacts (Earth Impact Database, http://www.impact-structures.com/database-of-earth-impact-structures/), solar luminosity (111), and fluctuations in atmospheric O2 (112) and CO2 (113–116) amount were displayed synchronously with divergence times in the form of time panels. Phylogenetic trees based on protein sequences of functional genes were constructed using PhyML (117) with the maximum likelihood (ML) method and 1,000 bootstrap replicates, followed by visualization with iTOL (118). Sequences were aligned with Muscle (119) and trimmed with Gblocks (120) before tree construction.
Genome annotation and horizontally transferred gene prediction.
We applied RAST (121), KEGG (122), and COG (123) databases (BLASTP cutoff, E value < 10−5) for genome annotation. We also extracted information of putative horizontally transferred genes from IMG Annotation results. Genome neighbor (context) visualizations were conducted with the EFI-GNT tool (124). Identification of putative horizontally transferred genes (HTGs) in the genomes of Acidiphilium was performed via the Integrated Microbial Genomes (IMG) system (125), which defined genes as being putative lateral transfers by the following principle: genes that have their best BLAST hits (best bit scores) or >90% of the best hits outside the taxonomic lineage of the genome (i.e., to genomes from another phylum, class, etc.) but with lower-scoring hits or no hits within the lineage.
Prediction of mobile genetic elements.
We applied the ISfinder (126) to predict and classify insertion sequences (IS) and transposases within Acidiphilium genomes with BLASTP (cutoff E value, 1e−5). IslandViewer 4 (127) was used to detect putative genomic islands (GIs) distributed within Acidiphilium genomes. PHASTER (128) was applied to detect prophage and prophage remnant sequences within Acidiphilium genomes. We also applied CRISPRCasFinder (129) for detection of CRISPRs and Cas within Acidiphilium genomes.
Construction of cooccurrence network.
To identify the associations between Acidiphilium and other microbes in AMD environments, 16S rRNA amplicon sequencing data sets of AMD samples (n = 205) were collected from the Sequence Read Archive (SRA) database (see Table S5 in the supplemental material). The QIIME (130) pipeline was applied to analyze these data sets. Sequences were clustered into operational taxonomic units (OTUs) at the 97% similarity level with the “closed reference OTU picking” strategy against the QIIME formatted Greengenes v.13.8 reference database (http://greengenes.lbl.gov). Rare OTUs, with fewer than five occurrences, were removed before network construction. The cooccurrence network was constructed using CoNet (131), which was implemented in Cytoscape v.3.6.1 based on the OTU occurrence frequency. Pairwise scores between OTUs were calculated using Spearman rank correlations applying a threshold rho of >0.6 and a P value of <0.01. The cooccurrence network was visualized with Organic layout in Cytoscape v. 3.6.1 (132).
16S rRNA amplicon sequencing datasets of acid mine drainage (AMD) samples collected from the Sequence Read Archive (SRA) database for identification of the associations between Acidiphilium and other microbes in AMD environments. Download Table S5, XLSX file, 0.01 MB (12KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome-wide detection of positively selected genes.
We used the PosiGene pipeline (133) for genome-wide detection of positively selected genes in the above-mentioned strains of Acidiphilium spp., in which Acidiphilium sp. AccI was used as the anchor species, reference, and target species. Genes were considered under positive selection if the branch-wide test resulted in false discovery rates (FDR) of <0.05 and adjusted P values of <0.05.
Data availability.
The genome sequences of Acidiphilium strains AccI, AccII, and ZJSH63 have been deposited in the JGI IMG-ER database under ER Genome IDs 2824045439, 2824049744, and 2828882166, respectively.
ACKNOWLEDGMENTS
We thank Han Zhou for assistance in data processing and figure creation and the NCBI database and IMG-ER database for providing the genome sequences of Acidiphilium spp.
This work was funded by the National Natural Science Foundation of China (grant no. 91851206 and 41877345), by the Ministry of Science and Technology of China (project no. 2018YFE0110200), by open funding from the State Key Laboratory of Comprehensive Utilization of Low-Grade Refractory Gold Ores (Zijin Mining Group Co., Ltd.) (grant no. 738010212), and by Fundamental Research Funds for the Central Universities of Central South University (grant no. 2019zzts996).
We also thank the Hunan International Scientific and Technological Cooperation Base of Environmental Microbiology and Application, China.
We declare no conflicts of interest.
REFERENCES
- 1.Merino N, Aronson HS, Bojanova DP, Feyhl-Buska J, Wong ML, Zhang S, Giovannelli D. 2019. Living at the extremes: extremophiles and the limits of life in a planetary context. Front Microbiol 10:780. doi: 10.3389/fmicb.2019.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature 409:1092–1101. doi: 10.1038/35059215. [DOI] [PubMed] [Google Scholar]
- 3.Hua ZS, Han YJ, Chen LX, Liu J, Hu M, Li SJ, Kuang JL, Chain PS, Huang LN, Shu WS. 2015. Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. ISME J 9:1280–1294. doi: 10.1038/ismej.2014.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadnikov VV, Ivasenko DA, Beletsky AV, Mardanov AV, Danilova EV, Pimenov NV, Karnachuk OV, Ravin NV. 2016. Effect of metal concentration on the microbial community in acid mine drainage of a polysulfide ore deposit. Microbiology 85:745–751. doi: 10.1134/S0026261716060126. [DOI] [PubMed] [Google Scholar]
- 5.Kuang J, Huang L, He Z, Chen L, Hua Z, Jia P, Li S, Liu J, Li J, Zhou J, Shu W. 2016. Predicting taxonomic and functional structure of microbial communities in acid mine drainage. ISME J 10:1527–1539. doi: 10.1038/ismej.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuang JL, Huang L, Chen L, Hua Z, Li S, Hu M, Li J, Shu W. 2013. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050. doi: 10.1038/ismej.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sievers M, Ludwig W, Teuber M. 1994. Phylogenetic positioning of Acetobacter, Gluconobacter, Rhodopila and Acidiphilium species as a branch of acidophilic bacteria in the α-subclass of Proteobacteria based on 16S ribosomal DNA sequences. Syst Appl Microbiol 17:189–196. doi: 10.1016/S0723-2020(11)80006-8. [DOI] [Google Scholar]
- 8.Cummings DE, Fendorf S, Singh N, Sani RK, Peyton BM, Magnuson TS. 2007. Reduction of Cr(VI) under acidic conditions by the facultative Fe(III)-reducing bacterium Acidiphilium cryptum. Environ Sci Technol 41:146–152. doi: 10.1021/es061333k. [DOI] [PubMed] [Google Scholar]
- 9.Küsel K, Dorsch T, Acker G, Stackebrandt E. 1999. Microbial reduction of Fe(III) in acidic sediments: isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. Appl Environ Microbiol 65:3633–3640. doi: 10.1128/AEM.65.8.3633-3640.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomi T, Shibata Y, Ikeda Y, Taniguchi S, Haik C, Mataga N, Shimada K, Itoh S. 2007. Energy and electron transfer in the photosynthetic reaction center complex of Acidiphilium rubrum containing Zn-bacteriochlorophyll a studied by femtosecond up-conversion spectroscopy. Biochim Biophys Acta 1767:22–30. doi: 10.1016/j.bbabio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Okamura K, Kawai A, Wakao N, Yamada T, Hiraishi A. 2015. Acidiphilium iwatense sp. nov., isolated from an acid mine drainage treatment plant, and emendation of the genus Acidiphilium. Int J Syst Evol Microbiol 65:42–48. doi: 10.1099/ijs.0.065052-0. [DOI] [PubMed] [Google Scholar]
- 12.Rohwerder T, Sand W. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology (Reading) 149:1699–1710. doi: 10.1099/mic.0.26212-0. [DOI] [PubMed] [Google Scholar]
- 13.Xu A-l, Xia J-l, Song Z-w, Jiang P, Xia Y, Wan M-x, Zhang R-y, Yang Y, Liu K-k. 2013. The effect of energy substrates on PHB accumulation of Acidiphilium cryptum DX1-1. Curr Microbiol 67:379–387. doi: 10.1007/s00284-013-0373-y. [DOI] [PubMed] [Google Scholar]
- 14.Fischer JR, Quentmeier A, Gansel S, Sabados V, Friedrich CG. 2002. Inducible aluminum resistance of Acidiphilium cryptum and aluminum tolerance of other acidophilic bacteria. Arch Microbiol 178:554–558. doi: 10.1007/s00203-002-0482-7. [DOI] [PubMed] [Google Scholar]
- 15.Mahapatra NR, Banerjee PC. 1996. Extreme tolerance to cadmium and high resistance to copper, nickel and zinc in different Acidiphilium strains. Lett Appl Microbiol 23:393–397. doi: 10.1111/j.1472-765X.1996.tb01344.x. [DOI] [Google Scholar]
- 16.San Martin-Uriz P, Mirete S, Alcolea PJ, Gomez MJ, Amils R, Gonzalez-Pastor JE. 2014. Nickel-resistance determinants in Acidiphilium sp. PM identified by genome-wide functional screening. PLoS One 9:e95041. doi: 10.1371/journal.pone.0095041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borole AP, O’Neill H, Tsouris C, Cesar S. 2008. A microbial fuel cell operating at low pH using the acidophile Acidiphilium cryptum. Biotechnol Lett 30:1367–1372. doi: 10.1007/s10529-008-9700-y. [DOI] [PubMed] [Google Scholar]
- 18.Xu A-l, Xia J-l, Zhang S, Yang Y, Nie Z-y, Qiu G-z. 2010. Bioleaching of chalcopyrite by UV-induced mutagenized Acidiphilium cryptum and Acidithiobacillus ferrooxidans. Trans Nonferrous Metals Soc China 20:315–321. doi: 10.1016/S1003-6326(09)60140-0. [DOI] [Google Scholar]
- 19.González E, Rodríguez JM, Muñoz JÁ, Blázquez ML, Ballester A, González F. 2018. The contribution of Acidiphilium cryptum to the dissolution of low-grade manganese ores. Hydrometallurgy 175:312–318. doi: 10.1016/j.hydromet.2017.12.008. [DOI] [Google Scholar]
- 20.Priya A, Hait S. 2017. Feasibility of bioleaching of selected metals from electronic; Waste by Acidiphilium acidophilum. Waste Biomass Valorization 9:871–877. [Google Scholar]
- 21.Almeida WI, Vieira RP, Cardoso AM, Silveira CB, Costa RG, Gonzalez AM, Paranhos R, Medeiros JA, Freitas FA, Albano RM, Martins OB. 2009. Archaeal and bacterial communities of heavy metal contaminated acidic waters from zinc mine residues in Sepetiba Bay. Extremophiles 13:263–271. doi: 10.1007/s00792-008-0214-2. [DOI] [PubMed] [Google Scholar]
- 22.Hiraishi A, Matsuzawa Y, Kanbe T, Wakao N. 2000. Acidisphaera rubrifaciens gen. nov., sp. nov., an aerobic bacteriochlorophyll-containing bacterium isolated from acidic environments. Int J Syst Evol Microbiol 50:1539–1546. doi: 10.1099/00207713-50-4-1539. [DOI] [PubMed] [Google Scholar]
- 23.Jones RM, Hedrich S, Johnson DB. 2013. Acidocella aromatica sp. nov.: an acidophilic heterotrophic alphaproteobacterium with unusual phenotypic traits. Extremophiles 17:841–850. doi: 10.1007/s00792-013-0566-0. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Yang H, Zhang X, Xiao Y, Guo X, Liu X. 2016. Genomic analysis unravels reduced inorganic sulfur compound oxidation of heterotrophic acidophilic Acidicaldus sp. strain DX-1. BioMed Res Int 2016:8137012. doi: 10.1155/2016/8137012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren M, Feng X, Huang Y, Wang H, Hu Z, Clingenpeel S, Swan BK, Fonseca MM, Posada D, Stepanauskas R, Hollibaugh JT, Foster PG, Woyke T, Luo H. 2019. Phylogenomics suggests oxygen availability as a driving force in Thaumarchaeota evolution. ISME J 13:2150–2161. doi: 10.1038/s41396-019-0418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salcher MM, Schaefle D, Kaspar M, Neuenschwander SM, Ghai R. 2019. Evolution in action: habitat transition from sediment to the pelagial leads to genome streamlining in Methylophilaceae. ISME J 13:2764–2777. doi: 10.1038/s41396-019-0471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viana MVC, Sahm A, Goes Neto A, Figueiredo HCP, Wattam AR, Azevedo V. 2018. Rapidly evolving changes and gene loss associated with host switching in Corynebacterium pseudotuberculosis. PLoS One 13:e0207304. doi: 10.1371/journal.pone.0207304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. 2007. Genes under positive selection in Escherichia coli. Genome Res 17:1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Props R, Monsieurs P, Vandamme P, Leys N, Denef VJ, Boon N. 2019. Gene expansion and positive selection as bacterial adaptations to oligotrophic conditions. mSphere 4:e00011-19. doi: 10.1128/mSphereDirect.00011-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Jalan N, Zhou X, Goss E, Jones JB, Setubal JC, Deng X, Wang N. 2015. Positive selection is the main driving force for evolution of citrus canker-causing Xanthomonas. ISME J 9:2128–2138. doi: 10.1038/ismej.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, The Genome Standards Consortium , et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison AP. 1981. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int J Syst Bacteriol 31:327–332. doi: 10.1099/00207713-31-3-327. [DOI] [Google Scholar]
- 34.Wakao N, Nagasawa N, Matsuura T, Matsukura H, Matsumoto T, Hiraishi A, Sakurai Y, Shiota H. 1994. Acidiphilium multivorum sp. nov., an acidophilic chemoorganotrophic bacterium from pyritic acid mine drainage. J Gen Appl Microbiol 40:143–159. doi: 10.2323/jgam.40.143. [DOI] [Google Scholar]
- 35.Wichlacz PL, Unz RF, Langworthy TA. 1986. Acidiphilium angustum sp. nov., Acidiphilium facilis sp. nov., and Acidiphilium rubrum sp. nov.: acidophilic heterotrophic bacteria isolated from acidic coal mine drainage. Int J Syst Bacteriol 36:197–201. doi: 10.1099/00207713-36-2-197. [DOI] [Google Scholar]
- 36.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Springael D, Top EM. 2004. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol 12:53–58. doi: 10.1016/j.tim.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 38.De Roos J, Verce M, Aerts M, Vandamme P, De Vuyst L. 2018. Temporal and spatial distribution of the acetic acid bacterium communities throughout the wooden casks used for the fermentation and maturation of lambic beer underlines their functional role. Appl Environ Microbiol 11:e02846-17. doi: 10.1128/AEM.02846-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 40.Varble A, Meaden S, Barrangou R, Westra ER, Marraffini LA. 2019. Recombination between phages and CRISPR-cas loci facilitates horizontal gene transfer in staphylococci. Nat Microbiol 4:956–963. doi: 10.1038/s41564-019-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konstantinidis KT, Tiedje JM. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci U S A 101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Govantes F, Albrecht JA, Gunsalus RP. 2000. Oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon: roles of multiple promoters and the Fnr‐1 and Fnr‐2 binding sites. Mol Microbiol 37:1456–1469. doi: 10.1046/j.1365-2958.2000.02100.x. [DOI] [PubMed] [Google Scholar]
- 43.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. 2012. The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim Biophys Acta 1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Chadwick GL, Hemp J, Fischer WW, Orphan VJ. 2018. Convergent evolution of unusual complex I homologs with increased proton pumping capacity: energetic and ecological implications. ISME J 12:2668–2680. doi: 10.1038/s41396-018-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontã T, Smets BF. 2018. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J 12:1779–1793. doi: 10.1038/s41396-018-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma P, Mattos MJTD, Hellingwerf KJ, Bekker M. 2012. On the function of the various quinone species in Escherichia coli. FEBS J 279:3364–3373. doi: 10.1111/j.1742-4658.2012.08608.x. [DOI] [PubMed] [Google Scholar]
- 47.Ozmen I, Aksoy E. 2015. Respiratory emergencies and management of mining accidents. Turk Thorac J 16(Suppl 1):S18–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie J, He Z, Liu X, Liu X, Van Nostrand JD, Deng Y, Wu L, Zhou J, Qiu G. 2011. GeoChip-based analysis of the functional gene diversity and metabolic potential of microbial communities in acid mine drainage. Appl Environ Microbiol 77:991–999. doi: 10.1128/AEM.01798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magnuson TS, Swenson MW, Paszczynski AJ, Deobald LA, Kerk D, Cummings DE. 2010. Proteogenomic and functional analysis of chromate reduction in Acidiphilium cryptum JF-5, an Fe(III)-respiring acidophile. Biometals 23:1129–1138. doi: 10.1007/s10534-010-9360-y. [DOI] [PubMed] [Google Scholar]
- 50.Xue XM, Yan Y, Xu HJ, Wang N, Zhang X, Ye J. 2014. ArsH from Synechocystis sp. PCC 6803 reduces chromate and ferric iron. FEMS Microbiol Lett 356:105–112. doi: 10.1111/1574-6968.12481. [DOI] [PubMed] [Google Scholar]
- 51.Rehan M, Furnholm T, Finethy RH, Chu F, El-Fadly G, Tisa LS. 2014. Copper tolerance in Frankia sp. strain EuI1c involves surface binding and copper transport. Appl Microbiol Biotechnol 98:8005–8015. doi: 10.1007/s00253-014-5849-6. [DOI] [PubMed] [Google Scholar]
- 52.Renninger N, Knopp R, Nitsche H, Clark DS, Keasling JD. 2004. Uranyl precipitation by Pseudomonas aeruginosa via controlled polyphosphate metabolism. Appl Environ Microbiol 70:7404–7412. doi: 10.1128/AEM.70.12.7404-7412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khaleque HN, Gonzalez C, Shafique R, Kaksonen AH, Holmes DS, Watkin ELJ. 2019. Uncovering the mechanisms of halotolerance in the extremely acidophilic members of the Acidihalobacter genus through comparative genome analysis. Front Microbiol 10:155. doi: 10.3389/fmicb.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roesser M, Müller V. 2010. Osmoadaptation in bacteria and archaea: common principles and differences. Environ Microbiol 3:743–754. doi: 10.1046/j.1462-2920.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- 55.Moritz KD, Amendt B, Witt EMHJ, Galinski EA. 2015. The hydroxyectoine gene cluster of the non-halophilic acidophile Acidiphilium cryptum. Extremophiles 19:87–99. doi: 10.1007/s00792-014-0687-0. [DOI] [PubMed] [Google Scholar]
- 56.Welte C, Hafner S, Krätzer C, Quentmeier A, Friedrich CG, Dahl C. 2009. Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett 583:1281–1286. doi: 10.1016/j.febslet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 57.Wu W, Pang X, Lin J, Liu X, Wang R, Lin J, Chen L. 2017. Discovery of a new subgroup of sulfur dioxygenases and characterization of sulfur dioxygenases in the sulfur metabolic network of Acidithiobacillus caldus. PLoS One 12:e0183668. doi: 10.1371/journal.pone.0183668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IAC, Dahl C. 2012. Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ Microbiol 14:2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x. [DOI] [PubMed] [Google Scholar]
- 59.Murrell JC, Smith TJ. 2010. Biochemistry and molecular biology of methane monooxygenase In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 60.Wang VC, Maji S, Chen PP, Lee HK, Yu SS, Chan SI. 2017. Alkane oxidation: methane monooxygenases, related enzymes, and their biomimetics. Chem Rev 117:8574–8621. doi: 10.1021/acs.chemrev.6b00624. [DOI] [PubMed] [Google Scholar]
- 61.AAAS. 2005. So much more to know. Science 309:78–102. doi: 10.1126/science.309.5731.78b. [DOI] [PubMed] [Google Scholar]
- 62.Chouaia B, Gaiarsa S, Crotti E, Comandatore F, Esposti MD, Ricci I, Alma A, Favia G, Bandi C, Daffonchio D. 2014. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol Evol 6:912–920. doi: 10.1093/gbe/evu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L, Illeghems K, Van Kerrebroeck S, Borremans W, Cleenwerck I, Smagghe G, De Vuyst L, Vandamme P. 2016. Whole-genome sequence analysis of Bombella intestini LMG 28161T, a novel acetic acid bacterium isolated from the crop of a red-tailed bumble bee, Bombus lapidarius. PLoS One 11:e0165611. doi: 10.1371/journal.pone.0165611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bentkowski P, Van Oosterhout C, Mock T. 2015. A model of genome size evolution for prokaryotes in stable and fluctuating environments. Genome Biol Evol 7:2344–2351. doi: 10.1093/gbe/evv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guieysse B, Wuertz S. 2012. Metabolically versatile large-genome prokaryotes. Curr Opin Biotechnol 23:467–473. doi: 10.1016/j.copbio.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 66.Bentkowski P, van Oosterhout C, Ashby B, Mock T. 2017. The effect of extrinsic mortality on genome size evolution in prokaryotes. ISME J 11:1011–1018. doi: 10.1038/ismej.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown BP, Wernegreen JJ. 2019. Genomic erosion and extensive horizontal gene transfer in gut-associated Acetobacteraceae. BMC Genomics 20:472. doi: 10.1186/s12864-019-5844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vos M, Hesselman MC, Te Beek TA, van Passel MWJ, Eyre-Walker A. 2015. Rates of lateral gene transfer in prokaryotes: high but why? Trends Microbiol 23:598–605. doi: 10.1016/j.tim.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Lindsey ARI, Newton ILG. 2019. Some like it HOT: horizontal operon transfer. Cell 176:1243–1245. doi: 10.1016/j.cell.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 70.Brinkmann H, Göker M, Koblížek M, Wagner-Döbler I, Petersen J. 2018. Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J 12:1994–2010. doi: 10.1038/s41396-018-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yelton AP, Comolli LR, Justice NB, Castelle C, Denef VJ, Thomas BC, Banfield JF. 2013. Comparative genomics in acid mine drainage biofilm communities reveals metabolic and structural differentiation of co-occurring archaea. BMC Genomics 14:485. doi: 10.1186/1471-2164-14-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kishimoto N, Fukaya F, Inagaki K, Sugio T, Tanaka H, Tano T. 1995. Distribution of bacteriochlorophyll a among aerobic and acidophilic bacteria and light-enhanced CO2-incorporation in Acidiphilium rubrum. FEMS Microbiol Ecol 16:291–296. doi: 10.1111/j.1574-6941.1995.tb00293.x. [DOI] [Google Scholar]
- 73.Ly T, Wright JR, Weit N, McLimans CJ, Ulrich N, Tokarev V, Valkanas MM, Trun N, Rummel S, Grant CJ, Lamendella R. 2019. Microbial communities associated with passive acidic abandoned coal mine remediation. Front Microbiol 10:1955. doi: 10.3389/fmicb.2019.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hedrich S, Johnson DB. 2013. Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol Lett 349:40–45. doi: 10.1111/1574-6968.12290. [DOI] [PubMed] [Google Scholar]
- 75.Orlova MV, Tarlachkov SV, Dubinina GA, Belousova EV, Tutukina MN, Grabovich MY. 2016. Genomic insights into metabolic versatility of a lithotrophic sulfur-oxidizing diazotrophic alphaproteobacterium Azospirillum thiophilum. FEMS Microbiol Ecol 92:fiw199. doi: 10.1093/femsec/fiw199. [DOI] [PubMed] [Google Scholar]
- 76.Li L, Liu Z, Meng D, Liu X, Li X, Zhang M, Tao J, Gu Y, Zhong S, Yin H. 2019. Comparative genomic analysis reveals distribution, organization, and evolution of metal resistance genes in the genus Acidithiobacillus. Appl Environ Microbiol 85:e02153-18. doi: 10.1128/AEM.02153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Navarro CA, Von Bernath D, Jerez CA. 2013. Heavy metal resistance strategies of acidophilic bacteria and their acquisition: importance for biomining and bioremediation. Biol Res 46:363–371. doi: 10.4067/S0716-97602013000400008. [DOI] [PubMed] [Google Scholar]
- 78.Liu H, Yin H, Dai Y, Dai Z, Liu Y, Li Q, Jiang H, Liu X. 2011. The co-culture of Acidithiobacillus ferrooxidans and Acidiphilium acidophilum enhances the growth, iron oxidation, and CO2 fixation. Arch Microbiol 193:857–866. doi: 10.1007/s00203-011-0723-8. [DOI] [PubMed] [Google Scholar]
- 79.Muravyov M, Panyushkina A. 2020. Distinct roles of acidophiles in complete oxidation of high-sulfur ferric leach product of zinc sulfide concentrate. Microorganisms 8:386. doi: 10.3390/microorganisms8030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y, Diao M, Liu K, Qian L, Nguyen AV, Qiu G. 2013. Column bioleaching of low-grade copper ore by Acidithiobacillus ferrooxidans in pure and mixed cultures with a heterotrophic acidophile Acidiphilium sp. Hydrometallurgy 131–132:93–98. doi: 10.1016/j.hydromet.2012.09.003. [DOI] [Google Scholar]
- 81.Denef VJ, Mueller RS, Banfield JF. 2010. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J 4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- 82.Goltsman DSA, Comolli LR, Thomas BC, Banfield JF. 2015. Community transcriptomics reveals unexpected high microbial diversity in acidophilic biofilm communities. ISME J 9:1014–1023. doi: 10.1038/ismej.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 84.Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 85.Reisner A, Holler BM, Molin S, Zechner EL. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J Bacteriol 188:3582–3588. doi: 10.1128/JB.188.10.3582-3588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fang P, Peng F, Gao X, Xiao P, Yang J. 2019. Decoupling the dynamics of bacterial taxonomy and antibiotic resistance function in a subtropical urban reservoir as revealed by high-frequency sampling. Front Microbiol 10:1448. doi: 10.3389/fmicb.2019.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow C-ET, Sachdeva R, Jones AC, Schwalbach MS, Rose JM, Hewson I, Patel A, Sun F, Caron DA, Fuhrman JA. 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramirez A, Castañeda M, Xiqui ML, Sosa A, Baca BE. 2010. Identification, cloning and characterization of cysK, the gene encoding O-acetylserine (thiol)-lyase from Azospirillum brasilense, which is involved in tellurite resistance. FEMS Microbiol Lett 261:272–279. doi: 10.1111/j.1574-6968.2006.00369.x. [DOI] [PubMed] [Google Scholar]
- 89.Johnson PM, Beck CM, Morse RP, Garza-Sánchez F, Low DA, Hayes CS, Goulding CW. 2016. Unraveling the essential role of CysK in CDI toxin activation. Proc Natl Acad Sci U S A 113:9792–9797. doi: 10.1073/pnas.1607112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaundal S, Uttam M, Thakur KG. 2016. Dual role of a biosynthetic enzyme, CysK, in contact dependent growth inhibition in bacteria. PLoS One 11:e0159844. doi: 10.1371/journal.pone.0159844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ankisettypalli K, Cheng JJ, Baker EN, Bashiri G. 2016. PdxH proteins of mycobacteria are typical members of the classical pyridoxine/pyridoxamine 5'-phosphate oxidase family. FEBS Lett 590:453–460. doi: 10.1002/1873-3468.12080. [DOI] [PubMed] [Google Scholar]
- 92.Matxain JM, Padro D, Ristilä M, Strid A, Eriksson LA. 2009. Evidence of high *OH radical quenching efficiency by vitamin B6. J Phys Chem B 113:9629–9632. doi: 10.1021/jp903023c. [DOI] [PubMed] [Google Scholar]
- 93.Mosterz J, Hochgräfe F, Jürgen B, Schweder T, Hecker M. 2010. The role of thioredoxin TrxA in Bacillus subtilis: a proteomics and transcriptomics approach. Proteomics 8:2676–2690. doi: 10.1002/pmic.200701015. [DOI] [PubMed] [Google Scholar]
- 94.Lu J, Holmgren A. 2014. The thioredoxin antioxidant system. Free Radic Biol Med 66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 95.Möller MC, Hederstedt L. 2008. Extracytoplasmic processes impaired by inactivation of trxA (thioredoxin gene) in Bacillus subtilis. J Bacteriol 190:4660–4665. doi: 10.1128/JB.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun JS, Li YX, Li S. 2012. Cynoglossus semilaevis thioredoxin: a reductase and an antioxidant with immunostimulatory property. Cell Stress Chaperones 17:445–455. doi: 10.1007/s12192-012-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Roos J, De Vuyst L. 2018. Acetic acid bacteria in fermented foods and beverages. Curr Opin Biotechnol 49:115–119. doi: 10.1016/j.copbio.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 98.Azuma Y, Hosoyama A, Matsutani M, Furuya N, Horikawa H, Harada T, Hirakawa H, Kuhara S, Matsushita K, Fujita N, Shirai M. 2009. Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res 37:5768–5783. doi: 10.1093/nar/gkp612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Colman DR, Poudel S, Hamilton TL, Havig JR, Selensky MJ, Shock EL, Boyd ES. 2018. Geobiological feedbacks and the evolution of thermoacidophiles. ISME J 12:225–236. doi: 10.1038/ismej.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alikhan NF, Petty NK, Zakour NLB, Beatson SAA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 104.Yi W, Coleman-Derr D, Chen G, Yong QG. 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43:W78–84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chaudhari NM, Gupta VK, Dutta C. 2016. BPGA—an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miklós CS. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 107.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tamura K, Tao Q, Kumar S. 2018. Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol Biol Evol 35:1770–1782. doi: 10.1093/molbev/msy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beatriz M. 2018. Estimating timetrees with MEGA and the TimeTree resource. Mol Biol Evol 35:2334–2342. doi: 10.1093/molbev/msy133. [DOI] [PubMed] [Google Scholar]
- 110.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 111.Gough DO. 1981. Solar interior structure and luminosity variations. Sol Phys 74:21–34. doi: 10.1007/BF00151270. [DOI] [Google Scholar]
- 112.Holland HD. 2006. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci 361:903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beerling DJ, Royer DL. 2011. Convergent cenozoic CO2 history. Nat Geosci 4:418–420. doi: 10.1038/ngeo1186. [DOI] [Google Scholar]
- 114.Berner AR. 1990. Atmospheric carbon dioxide levels over phanerozoic time. Science 249:1382–1386. doi: 10.1126/science.249.4975.1382. [DOI] [PubMed] [Google Scholar]
- 115.Hessler AM, Lowe DR, Jones RL, Bird DK. 2004. A lower limit for atmospheric carbon dioxide levels 3.2 billion years ago. Nature 428:736–738. doi: 10.1038/nature02471. [DOI] [PubMed] [Google Scholar]
- 116.Petit JR, Jouzel J, Raynaud D, Barkov NI, Barnola J-M, Basile I, Bender M, Chappellaz J, Davis M, Delaygue G, Delmotte M, Kotlyakov VM, Legrand M, Lipenkov VY, Lorius C, PÉpin L, Ritz C, Saltzman E, Stievenard M. 1999. Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature 399:429–436. doi: 10.1038/20859. [DOI] [Google Scholar]
- 117.Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 118.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 121.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75–75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. 2019. New approach for understanding genome variations in KEGG. Nucleic Acids Res 47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gerlt AJ. 2017. Genomic enzymology: Web tools for leveraging protein family sequence-function space and genome context to discover novel functions. Biochemistry 56:4293–4308. doi: 10.1021/acs.biochem.7b00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Anderson I, Lykidis A, Mavromatis K. 2009. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res 38:D382–D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FS, Simon Fraser University Research Computing Group . 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Faust K, Raes J. 2016. CoNet app: inference of biological association networks using Cytoscape. F1000Res 5:1519. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang P-L, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sahm A, Bens M, Platzer M, Szafranski K. 2017. PosiGene: automated and easy-to-use pipeline for genome-wide detection of positively selected genes. Nucleic Acids Res 45:e100. doi: 10.1093/nar/gkx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO enrichment analyses (P value < 0.05) of Acidiphilium gene families. Download Table S1, XLSX file, 0.02 MB (16.4KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed description for abbreviations of COG categories. Download Text S1, DOCX file, 0.01 MB (11.9KB, docx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistics of mobile genetic elements (MGEs) present in the genomes of Acidiphilium spp. Download Table S2, XLSX file, 0.5 MB (539KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extracted information of putative horizontally transferred genes from the results of IMG annotation. Download Table S3, XLSX file, 0.04 MB (45.8KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes in Acidiphilium identified as being under positive selection using the PosiGene pipeline. Download Table S4, DOCX file, 0.01 MB (14.2KB, docx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA amplicon sequencing datasets of acid mine drainage (AMD) samples collected from the Sequence Read Archive (SRA) database for identification of the associations between Acidiphilium and other microbes in AMD environments. Download Table S5, XLSX file, 0.01 MB (12KB, xlsx) .
Copyright © 2020 Li et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The genome sequences of Acidiphilium strains AccI, AccII, and ZJSH63 have been deposited in the JGI IMG-ER database under ER Genome IDs 2824045439, 2824049744, and 2828882166, respectively.