The Centers for Disease Control and Prevention estimates that VRE causes an estimated 54,000 infections and 539 million dollars in attributable health care costs annually. Despite improvements in hand washing, environmental cleaning, and antibiotic use, VRE is still prevalent in many hospitals. There is a pressing need to better understand the processes by which patients acquire VRE. Multiple lines of evidence suggest that intestinal microbiota may help some patients resist VRE acquisition. In this large case-control study, we compared the 16S profile of intestinal microbiota on admission in patients that did and did not subsequently acquire VRE. The 16S profile did not predict subsequent VRE acquisition, in part due to rapid and dramatic change in the gut microbiome following hospitalization. However, Blautia spp. present on admission predicted decreased Enterococcus abundance after VRE acquisition, and Lactobacillus spp. present on admission predicted Enterococcus dominance after VRE acquisition. Thus, VRE acquisition and domination may be distinct processes.
KEYWORDS: microbiome, vancomycin-resistant Enterococcus, colonization resistance, hospital-acquired infection
ABSTRACT
Vancomycin-resistant Enterococcus (VRE) is a leading cause of hospital-acquired infections and continues to spread despite widespread implementation of pathogen-targeted control guidelines. Commensal gut microbiota provide colonization resistance to VRE, but the role of gut microbiota in VRE acquisition in at-risk patients is unknown. To address this gap in our understanding, we performed a case-control study of gut microbiota in hospitalized patients who did (cases) and did not (controls) acquire VRE. We matched case subjects to control subjects by known risk factors and “time at risk,” defined as the time elapsed between admission until positive VRE screen. We characterized gut bacterial communities using 16S rRNA gene amplicon sequencing of rectal swab specimens. We analyzed 236 samples from 59 matched case-control pairs. At baseline, case and control subjects did not differ in gut microbiota when measured by community diversity (P = 0.33) or composition (P = 0.30). After hospitalization, gut communities of cases and controls differed only in the abundance of the Enterococcus-containing operational taxonomic unit (OTU), with the gut microbiota of case subjects having more of this OTU than time-matched control subjects (P = 0.01). Otherwise, case and control communities after the time at risk did not differ in diversity (P = 0.33) or community structure (P = 0.12). Among patients who became VRE colonized, those having the Blautia-containing OTU on admission had lower Enterococcus relative abundance once colonized (P = 0.004). Our results demonstrate that the 16S profile of the gut microbiome does not predict VRE acquisition in hospitalized patients, likely due to rapid and profound microbiota change. The gut microbiome does not predict VRE acquisition, but it may be associated with Enterococcus expansion, suggesting that these should be considered two distinct processes.
IMPORTANCE The Centers for Disease Control and Prevention estimates that VRE causes an estimated 54,000 infections and 539 million dollars in attributable health care costs annually. Despite improvements in hand washing, environmental cleaning, and antibiotic use, VRE is still prevalent in many hospitals. There is a pressing need to better understand the processes by which patients acquire VRE. Multiple lines of evidence suggest that intestinal microbiota may help some patients resist VRE acquisition. In this large case-control study, we compared the 16S profile of intestinal microbiota on admission in patients that did and did not subsequently acquire VRE. The 16S profile did not predict subsequent VRE acquisition, in part due to rapid and dramatic change in the gut microbiome following hospitalization. However, Blautia spp. present on admission predicted decreased Enterococcus abundance after VRE acquisition, and Lactobacillus spp. present on admission predicted Enterococcus dominance after VRE acquisition. Thus, VRE acquisition and domination may be distinct processes.
INTRODUCTION
Vancomycin-resistant Enterococcus (VRE) species are highly antibiotic-resistant bacteria, are a leading cause of health care-associated infections, and are classified as a serious public health threat by the Centers for Disease Control and Prevention (1, 2). Colonization with VRE precedes infection (3, 4), and molecular epidemiologic analyses show patient-to-patient hospital transmission is the primary means of spread (5). Preventing transmission between hospitalized patients is a significant challenge, and despite the widespread application of pathogen-targeted control measures (6), VRE remains prevalent in many hospitals (1, 2).
Both indirect human evidence and animal experimentation demonstrate that gut microbiota prevent VRE colonization when a patient is exposed, a phenomenon termed “colonization resistance” (7–9). Colonization resistance may entail competition for resources, secretion of bactericidal factors (10, 11), and indirect stimulation of host immune defense mechanisms that target VRE (12, 13). Though colonization resistance plays a crucial role in suppressing VRE expansion and preventing VRE infection (14, 15), to date, no study has evaluated whether variation in intestinal microbiota can explain variation in VRE acquisition among at-risk patients.
To address this gap in our understanding of VRE transmission, we investigated whether the gut microbiome of at-risk patients predicts VRE colonization in a hospitalized patient population. We hypothesized that if the gut microbiome can confer colonization resistance for VRE acquisition, variation in baseline microbiota would explain variation in patient susceptibility to VRE acquisition. To test this hypothesis, we designed a case-control study using 16S rRNA gene amplicon sequencing of rectal swabs acquired from hospitalized patients.
RESULTS
Study population and medication exposures.
We studied gut microbiome communities in 236 rectal swab samples from 59 matched pairs of case and control subjects (Table 1). Cases and controls did not differ in demographics (age, sex, ethnicity) or in the relative frequency of common comorbidities (e.g., immunosuppression, malignancy, or gastrointestinal disease). Antibiotic use was widespread among all subjects and was nearly equal across groups (Table 2). Vancomycin, cefepime, metronidazole, and piperacillin-tazobactam were the most commonly used antibiotics in the study population. Cases and controls did not differ significantly in their exposure to any specific antibiotics prior to initial sampling. More cases received proton pump inhibitors prior to initial sampling (P = 0.04). During time at risk (between initial and subsequent sampling), case and controls did not differ in their exposure to antibiotics or proton pump inhibitors.
TABLE 1.
Demographics and comorbidities of matched cohortsa
| Demographic or clinical characteristicb |
No. of individuals (proportion) with characteristic or value specified |
P value | |
|---|---|---|---|
| Controls (n = 59) | Cases (n = 59) | ||
| Demographics | |||
| Age (mean ± SE) | 57.19 ± 1.62 | 60.2 ± 1.95 | 0.23 |
| Female | 23 (0.39) | 22 (0.38) | 0.56 |
| Nonwhite race | 9 (0.15) | 9 (0.15) | 0.28 |
| Diagnoses and comorbidities | |||
| C. difficile infection | 4 (0.07) | 11 (0.18) | 0.07 |
| Leukemia | 21 (0.36) | 17 (0.29) | 0.38 |
| Lymphoma | 12 (0.21) | 10 (0.17) | 0.49 |
| Bone marrow transplant | 15 (0.25) | 14 (0.24) | 0.64 |
| Solid organ malignancy | 35 (0.6) | 40 (0.67) | 0.33 |
| Metastatic malignancy | 29 (0.49) | 35 (0.59) | 0.10 |
| Diabetes | 27 (0.46) | 23 (0.39) | 0.59 |
| Coronary artery disease | 6 (0.11) | 10 (0.17) | 0.72 |
| Congestive heart failure | 19 (0.32) | 18 (0.3) | 0.60 |
| COPD | 21 (0.35) | 36 (0.61) | 0.02 |
| Peripheral vascular disease | 6 (0.1) | 2 (0.03) | 0.31 |
| End-stage renal disease | 18 (0.31) | 26 (0.44) | 0.07 |
| Connective tissue disease | 1 (0.01) | 4 (0.06) | 0.35 |
| Peptic ulcer disease | 9 (0.15) | 6 (0.11) | 0.50 |
| Cirrhosis | 2 (0.04) | 9 (0.15) | 0.06 |
| Cerebrovascular disease | 12 (0.21) | 17 (0.29) | 0.54 |
| Hemiplegia | 4 (0.06) | 12 (0.2) | 0.07 |
| Dementia | 1 (0.01) | 3 (0.05) | 0.34 |
| Charlson score (mean ± SE) | 3.71 ± 0.22 | 4.45 ± 0.25 | 0.05 |
Cases and controls were matched by “time at risk” and unit or ward.
C. difficile, Clostridium difficile; COPD, chronic obstructive pulmonary disease.
TABLE 2.
Medication exposure of matched cohorts
| Sampling time and medication | Prevalence of exposurea
|
P value | Duration of exposureb
|
P value | ||
|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | |||
| Prior to admission swab | ||||||
| Antibiotics | ||||||
| Any antibiotics | 29 (0.49) | 40 (0.68) | 0.05 | 1.72 ± 0.62 | 2.32 ± 0.47 | 0.44 |
| Vancomycin | 14 (0.24) | 21 (0.36) | 0.17 | 0.52 ± 0.3 | 0.36 ± 0.06 | 0.61 |
| Metronidazole | 8 (0.14) | 14 (0.24) | 0.17 | 0.14 ± 0.04 | 0.4 ± 0.16 | 0.16 |
| Piperacillin-tazobactam | 8 (0.14) | 12 (0.2) | 0.29 | 0.14 ± 0.04 | 0.21 ± 0.06 | 0.25 |
| Cefepime | 7 (0.12) | 11 (0.19) | 0.32 | 0.4 ± 0.3 | 0.32 ± 0.12 | 0.81 |
| Proton pump inhibitors | 9 (0.15) | 19 (0.32) | 0.04 | 0.2 ± 0.07 | 0.61 ± 0.19 | 0.07 |
| Between admission and “time at risk” swab |
||||||
| Antibiotics | ||||||
| Any antibiotics | 56 (0.95) | 52 (0.88) | 0.18 | 21.66 ± 4.98 | 22.36 ± 3.66 | 0.82 |
| Vancomycin | 37 (0.63) | 39 (0.66) | 0.66 | 3.55 ± 0.82 | 3.33 ± 0.82 | 0.73 |
| Metronidazole | 20 (0.34) | 24 (0.41) | 0.43 | 2.01 ± 0.72 | 2.29 ± 0.6 | 0.75 |
| Piperacillin-tazobactam | 26 (0.44) | 25 (0.42) | 0.83 | 3.66 ± 1.13 | 2.8 ± 0.64 | 0.42 |
| Cefepime | 21 (0.36) | 24 (0.41) | 0.56 | 2.92 ± 1.01 | 3.1 ± 0.82 | 0.85 |
| Proton pump inhibitors | 33 (0.56) | 39 (0.66) | 0.23 | 5.15 ± 1.48 | 6.75 ± 1.32 | 0.13 |
Prevalence values are reported as number of case or control individuals (proportion).
Duration values are reported as numbers of days of therapy ± standard deviation (SD).
Admission gut microbiota do not predict VRE acquisition.
We first compared baseline microbiota across patients who did (cases) and did not (controls) subsequently acquire VRE. Baseline gut communities of cases and controls did not differ in their community composition, determined either via permutation testing (P = 0.30 by permutational multivariate analysis of variance [PERMANOVA]) or via visualization (principal-component analysis; Fig. 1, left). Similarly, baseline gut communities of cases and controls did not differ in their species diversity as measured by the Shannon diversity index (mean of 2.72 ± 0.90 for controls, mean of 2.71 ± 0.76 for cases, P = 0.96 for all matched case-control pairs) (Fig. 1, right). We noted that Enterococcus (OTU0004) was among the top 10 operational taxonomic units (OTUs) identified in both cases and controls at the time of admission. Despite the high abundance of the genus Enterococcus on admission, none of these swabs had vancomycin-resistant Enterococcus at the time of admission when evaluated with VRESelect chromogenic medium. Enterococcus (OTU0002) colonization did not imply VRE colonization, as OTU0004 captured both resistant and sensitive strains. We concluded that the gut microbiota, as represented by the 16S profile of these samples of hospitalized patients, do not predict subsequent VRE acquisition.
FIG 1.
In hospitalized patients, admission gut microbiota do not predict subsequent VRE acquisition. We used 16S rRNA sequencing to characterize gut bacterial communities in 118 hospitalized patients who tested negative for VRE at admission. We compared admission gut microbiota across 59 matched cases (patients who acquired VRE) and controls (patients who did not acquire VRE). (Left) Visualization of admission gut microbial communities using principal-component analysis. The admission gut communities of cases and controls did not differ in their community composition, either visually or via permutation testing (P = 0.3 by PERMANOVA). (Right) Comparison of average species diversity as measured by Shannon diversity index in admission gut communities. The admission gut communities of cases and controls did not differ in their community Shannon diversity index (P = 0.96 by conditional logistic regression).
At the time of VRE detection, the gut communities of cases and controls differ only in the abundance of Enterococcus.
We next compared gut communities across matched cases and controls after time at risk: after cases had been colonized and time-matched controls had not. After time at risk, gut microbiota did differ across cases and controls (P < 0.001 by PERMANOVA), though Shannon diversity index did not (mean of 2.38 ± 0.11 for controls, mean of 2.22 ± 0.12 for cases, P = 0.33 for all matched case-control pairs). The difference in gut microbiota was driven by the increased relative abundance of a single OTU, the Enterococcus-classified taxonomic group (OTU0004), which was greater in cases than controls (P = 0.01 via mvabund, P < 0.001 via random forest) (Fig. 2). When this Enterococcus OTU was excluded from the analysis, we found no significant difference in communities across cases and controls (P = 0.12 by PERMANOVA) (Fig. 3). We thus concluded that at the time of VRE acquisition, the gut microbiota of VRE-infected and uninfected patients differ only in the relative abundance of Enterococcus and do not consistently differ in their non-Enterococcus microbiota.
FIG 2.
After the time at risk, the gut microbiota of cases and controls differ primarily in their relative abundance of Enterococcus. The 10 most abundant bacterial taxa are shown in controls (top) and cases (bottom), at the time of admission (left), and following matched time at risk (right). Cases and controls did not differ in their admission microbiota (left). After the time at risk, the gut microbiota of cases were enriched with Enterococcus relative to control (P < 0.01, mvabund), but otherwise did not differ in their community composition (P > 0.05 for all other taxa, mvabund).
FIG 3.
With the exception of Enterococcus, gut communities of VRE-infected and uninfected patients do not differ. When we excluded Enterococcus OTU enriched in VRE-infected patients, we found no remaining difference in bacterial community composition, either visually (principal-component analysis) or via permutation testing (P = 0.12 by PERMANOVA).
Gut microbiota change rapidly and profoundly in hospitalized patients.
Given the lack of differentiation of gut communities across cases and controls at admission and at the time of VRE colonization, we then asked whether the temporal change in gut microbiota could distinguish the two groups. We did this by calculating the relative dissimilarity of admission and index (time-at-risk) communities for each subject using Jaccard distance, a metric of dissimilarity between gut microbial communities measured on a scale of 0 (complete similarity) to 1 (complete dissimilarity) (Fig. 4). The gut communities of both groups underwent a rapid, profound change in composition. Within several days of admission, gut communities of both cases and controls bore little similarity to the communities detected at the time of admission. The size of the change in gut communities did not differ across cohorts (0.87 ± 0.02 for cases, 0.86 ± 0.02 controls, P = 0.84). Cases and controls also had similar decreases in Shannon diversity (−0.48 ± 0.10 for cases, −0.40 ± 0.08 for controls, P = 0.93 for all matched case-control pairs). We found that Jaccard distance was significantly correlated with time (Spearman’s rank correlation coefficient ρ = 0.32, P = 0.0006) and determined that a negative exponential model best fit the data, with gut microbiota approaching complete dissimilarity at an exponential rate of 0.47 × e−0.47t+32 (t representing the time between swabs). We found no significant difference in the rate of change between the two groups. We noted that the predicted mean Jaccard distance for two rectal swab samples taken on the same day (t = 0) was 0.79 ± 0.058, implying a substantial amount of variation in community structure within the same day of admission.
FIG 4.
Rapid and dramatic change in gut microbiota among hospitalized patients. We calculated the dissimilarity between admission and subsequent (index, time at risk) gut communities in both cohorts with Jaccard distance. Both cases and controls exhibited rapid changes in gut communities during hospitalization, with Jaccard distance changing at an exponential rate. Cases and controls did not differ from each other in temporal disruption of gut microbiota. Dashed lines in the figure represent the 95% confidence interval for predicted mean Jaccard distance (inner ribbon) and predicted Jaccard distance for an individual subject (outer ribbon).
Gut microbiota on admission are associated with Enterococcus expansion.
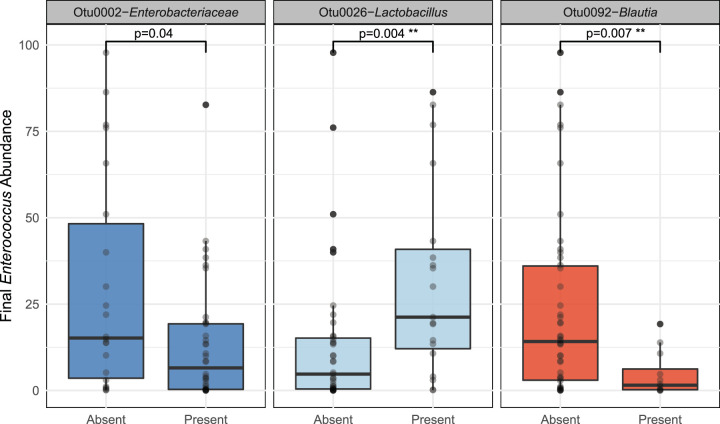
Finding no difference in the community composition, diversity, or temporal rate of change across patients who did (cases) and did not (controls) acquire VRE during their hospitalization, we asked whether gut microbiota on admission could predict the relative abundance of Enterococcus in VRE-colonized patients. We built a random forest regression model to identify taxa present on admission that were predictive of final Enterococcus relative abundance. In cases, only Blautia and Lactobacillus were significant after correcting for multiple testing and feature importance bias (Fig. 5; see Tables S1 and S2 and Fig. S2 in the supplemental material). In cases, Blautia spp. (OTU 0092) on admission was predictive of decreased Enterococcus (−10.3% relative-abundance-adjusted P [relative abundance P] = 0.004 by Mann-Whitney U test), and Lactobacillus spp. (OTU 0026) was predictive with an increased abundance of Enterococcus after the time at risk (+12.5% relative abundance P = 0.007 by Mann-Whitney U test). A random forest regression model applied to the control population identified the same Lactobacillus and Blautia taxa as predictive of Enterococcus abundance after the time at risk (–3.7% relative abundance P = 2.4 × 10−8 and +3.6% relative abundance for Lactobacillus P = 0.003 by Mann-Whitney U test). In controls, Lactobacillus and Blautia were not the only predictive taxa, as Phascolarctobacterium, Prevotella, and Bifidiobacterium were also predictive of decreased Enterococcus abundance. While more taxa were predictive of final Enterococcus abundance in controls, the effect size of these taxa was smaller, as controls had a lower abundance of Enterococcus after the time at risk (Table S2). Thus, we found that the presence of specific anaerobes previously implicated in Enterococcus colonization resistance (8, 15, 16) is predictive of decreased Enterococcus abundance in both VRE-colonized and uncolonized patients. These findings suggest that VRE acquisition and Enterococcus expansion are two distinct processes with different risk factors and pathophysiology.
FIG 5.
Presence of Blautia species on admission is predictive of decreased Enterococcus abundance at the time of VRE acquisition. A random forest regression model identified seven OTUs present on admission that predicted subsequent relative abundance of Enterococcus spp. Of these, the presence of Enterobacteriaceae spp., Lactobacillus spp., and Blautia spp. were significant predictors of the final relative abundance of Enterococcus spp.. Only Lactobacillus spp. and Blautia spp. remained significant after correcting for multiple testing. Significance was determined using the Mann-Whitney U test controlled for multiple comparisons.
Most important taxa identified by random forest regression model in cases. We built a random forest regression model to identify taxa present on admission that were predictive of final Enterococcus relative abundance. In cases, only Blautia and Lactobacillus were significant after correcting for multiple testing and feature importance bias. Download Table S1, DOCX file, 0.01 MB (14.2KB, docx) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Most important taxa identified by random forest regression model in cases. We built a random forest regression model to identify taxa present on admission that were predictive of final Enterococcus relative abundance. In cases, only Blautia and Lactobacillus were significant after correcting for multiple testing and feature importance bias. Download Table S2, DOCX file, 0.01 MB (14.6KB, docx) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, gut microbiota did not predict VRE acquisition in hospitalized patients. Secondary analysis identified individual members of the gut microbiota that do predict Enterococcus abundance at the time of VRE acquisition, implying that acquisition and expansion of VRE may be distinct processes. The community composition, diversity, and temporal rate of change did not differ across patients who did (cases) and did not (controls) acquire VRE during their hospitalization. As expected based on the study design, gut communities of cases had a greater abundance of Enterococcus than controls after the time at risk.
Gut communities of all subjects demonstrated a rapid and dramatic change during hospitalization that was time dependent. In this population, antibiotic use was prevalent (Table 2), gut microbial communities were remarkably dynamic (Fig. 5), and admission gut microbiota provided very little information about microbiota after the time at risk. Our model of Jaccard distance over time estimated a mean Jaccard distance of 0.79 between two rectal swabs taken on the same day of admission, implying that only 21% of gut microbiota remain constant with resampling within 24 h. Given the significant correlation between Jaccard distance and time, some of this change is likely due to the disruptive pressures that face gut microbiota upon hospitalization (i.e., antibiotics). However, a large portion of this change may represent stochasticity and noise introduced by variation in sample collection and storage. Other studies have found that gut microbiota change dramatically during hospitalization (17–19), but to our knowledge, ours is the largest study to examine this change, the only study to look at the rate of change, and the only study show that most of the change occurs very early (within 72 h). These results have important implications for the clinical use of gut microbiota for therapy, prediction, and risk stratification. Given the rapid change of gut communities upon hospitalization, a single static 16S analysis of gut microbiota may miss subtle dynamics important for VRE acquisition and is subject to a large amount of noise that may obscure a true biologically meaningful association. Future study of the gut microbiota in VRE acquisition may need to move beyond traditional 16S analysis, which can be time-consuming and miss important species-level information (20, 21). Real-time metagenomics and rapid, ultrasensitive quantification technologies hold promise as tools with better resolution to evaluate these processes (22, 23).
Despite the dramatic change in community structure, we did find some evidence of colonization resistance, as admission microbiota were predictive of Enterococcus abundance at the time of VRE detection. VRE-colonized subjects with Blautia had less Enterococcus expansion, consistent with prior studies (8, 15, 16). We hypothesize that there may be a distinction between VRE acquisition and VRE expansion. In conjunction with earlier studies (8, 16, 24), our findings suggest that commensal anaerobes may play a significant role in suppressing VRE once colonized. In this context, our results further support the possibility of microbiome manipulation to reduce VRE burden even in patients already colonized to prevent progression to VRE infection in the individual patient (15, 24) or transmission into the surrounding environment and other hospitalized patients (7, 8, 16).
We noted that more cases received proton pump inhibitor (PPI) therapy prior to initial sampling than controls (Table 2), consistent with prior studies showing that PPI use is a risk factor for VRE colonization (25, 26). Despite this difference in treatment, we found no meaningful difference in the community structure of cases and controls on admission (Fig. 1). This may imply that the increased risk of VRE colonization from PPI therapy is not mediated through changes in gut microbiota, but by elimination of the gut acid barrier to ingested bacteria (27). We believe these findings are hypothesis generating for future studies of the role of PPI therapy in VRE acquisition.
In this retrospective case-control study, we controlled for multiple confounders with our time- and unit-matched design. We used machine learning algorithms robust to multicollinearity and overfitting , and applied permutation heuristics to correct for feature importance bias and decrease our false discovery rate. This study reveals an opportunity for future studies to delineate key differences in pathophysiology between VRE acquisition and domination.
In summary, VRE acquisition and expansion may be two distinct processes, and efforts to manipulate the microbiome to prevent the spread of VRE may be more beneficial in reducing VRE domination in colonized patients than in preventing VRE acquisition in uncolonized patients. Future studies of the role of the gut microbiota in VRE acquisition may need to move beyond single time point 16S analyses and address the role of temporal dynamics and stochasticity of gut microbiota in the acquisition and expansion of VRE.
MATERIALS AND METHODS
Study setting and design.
We previously conducted a retrospective case-control study of clinical risk factors for VRE acquisition among patients who did (cases) and did not (controls) acquire VRE during their admissions at the University of Michigan Healthcare System from January 2013 until June 2016 (25). We studied gut microbiome communities in 236 rectal swab samples from 59 matched pairs of case and control subjects from patients admitted to the University of Michigan Hospital in 2016. Sixty-four out of 118 subjects in this study (54%) were a part of our previous clinical risk factor analysis. The remaining subjects were admitted from June to December 2016 (outside the time frame of the previous study by 6 months). The University of Michigan Healthcare system consists of ∼1,000 inpatient beds and serves as a tertiary referral hospital for southeastern Michigan. The institutional review board at the University of Michigan approved the study before its initiation.
VRE case definition.
The infection control practice throughout the study period was to perform routine surveillance for VRE on eight adult units, including intensive care units, the hematology and oncology ward, and the bone marrow transplant ward. All patients were routinely screened on admission and weekly thereafter with rectal swabs that were tested by Bio-Rad VRESelect chromogenic medium to detect VRE. Cases were defined as subjects with an initial negative swab followed by a positive swab when evaluated by this selective culture. We further identified the “time at risk” for each case patient, here defined as the time elapsed between admission and positive VRE screen. We matched each case subject to a control subject with an initial negative swab followed by repeat negative swab within the same time at risk (±5%). An additional matching factor was the unit from which the first positive VRE was recovered for cases or the matched swab after the time at risk for controls.
Bacterial DNA isolation.
Genomic DNA was extracted from rectal swabs resuspended in 360 μl ATL buffer (Qiagen DNeasy blood and tissue kit) and homogenized in fecal DNA bead tubes using a modified protocol previously demonstrated to isolate bacterial DNA (28, 29). Sterile laboratory water and AE buffer (10 mM Tris-Cl, 0.5 mM EDTA; pH 9.0) used in DNA isolation were collected and analyzed as potential sources of contamination. ZymoBIOMICS microbial community DNA standard (Zymo Research catalog no. D6306) was sequenced for error analysis.
16S rRNA gene sequencing.
The V4 region of the 16S rRNA gene was amplified using published primers and the dual-indexing sequencing strategy developed previously (28). Sequencing was performed using the Illumina MiSeq platform (San Diego, CA) and a MiSeq reagent kit V2 (500 cycles) according to the manufacturer’s instructions with modifications found in the standard operating procedure of the laboratory of Patrick Schloss (28, 30). Accuprime high-fidelity Taq was used in place of Accuprime Pfx SuperMix (31). Primary PCR cycling conditions were 95°C for 2 min, followed by 20 cycles of touchdown PCR (1 cycle consisting of 95°C for 20 s, 60°C for 20 s and decreasing 0.3 degrees each cycle, 72°C for 5 min), and then 20 cycles of standard PCR (1 cycle consisting of 95°C for 20 s, 55°C for 15 s, and 72°C for 5 min), and finished with 72°C for 10 min.
Statistical analyses.
Sequence data were processed and analyzed using the software mothur v.1.43.0 (32) according to the standard operating procedure for MiSeq sequence data using a minimum sequence length of 250 bp (28, 33). To summarize, the SILVA rRNA database (34) was used as a reference for sequence alignment and taxonomic classification. K-mer searching with 8-mers was used to assign raw sequences to their closest matching template in the reference database, and pairwise alignment was performed with the Needleman-Wunsch (35) and NAST algorithms (36). A k-mer-based naive Bayesian classifier (37) was used to assign sequences to their correct taxonomy with a bootstrap confidence score threshold of 80. Pairwise distances between aligned sequences were calculated by the method employed by Sogin et al. (38), where pairwise distance equals mismatches, including indels, divided by sequence length. A distance matrix was passed to the OptiCLUST clustering algorithm (39) to cluster sequences into “operational taxonomic units” (OTUs) by maximizing the Matthews correlation coefficient with a dissimilarity threshold of 3% (40).
After clustering and classification of raw sequencing data, we evaluated differences in community structure with permutational multivariate analysis of variance (PERMANOVA) in the vegan package (v 2.0-4) (41) in R (v 3.6.4) (42). We performed resampling of multiple generalized linear models with the mvabund (43) package in R to look for individual OTU differences between communities. We set a significance threshold of 0.01 after adjusting for multiple comparisons using a stepdown resampling procedure to reduce the type I error rate (44). We confirmed individual OTU differences with random forest classification and regression models built with the ranger package in R (v 0.11.2) (45). We used the caret (v 6.0-84) (46) package in R for cross-validation and to optimize the hyperparameters of the number of decision trees in the model and the number of features considered by each tree when splitting a node. We corrected for feature importance bias in random forest models with a permutation importance (PIMP) heuristic developed by Altmann et al. (47). All OTUs were included in diversity and abundance analyses. We performed bivariate analysis with conditional logistic regression using the survival (v 3.1-8) package in R (48, 49). Differences in means of final Enterococcus abundance were compared with the nonparametric Mann-Whitney U test. We used the vegan package in R to calculate both the average species diversity in an individual rectal swab (Shannon diversity) and the change in community structure between the initial swab and second swab for each subject (Jaccard distance). We used Spearman’s rank correlation coefficient to determine whether Jaccard distance was significantly correlated with the time between swabs, and we used nonlinear least-squares regression to fit a model of Jaccard distance over time for cases and controls.
Adequacy of sequencing.
We performed 16S rRNA gene amplicon sequencing on 236 rectal swab specimens and 15 negative-control specimens, which identified 1,188 unique operational taxonomic units (genus-level bacterial taxa) at a dissimilarity threshold of 3%. After bioinformatics processing, the mean number of reads per sample was 71,484 ± 2,684. No specimens were excluded from the analysis. Rectal swab specimens had clear differences in community structure compared to control specimens, which was confirmed as statistically significant using multiple methods of hypothesis testing (mvabund and PERMANOVA [adonis], P < 0.01 for both) (see Fig. S1 in the supplemental material). Sequences generated from negative-control specimens were dominated by a single Pseudomonas-classified OTU (OTU001). This OTU was included in all reported analyses, though the exclusion of this OTU did not affect any of the reported results.
Comparison of rectal swab communities with negative sequencing controls. A total of 236 rectal swab specimens and 13 negative-control specimens were studied using 16S rRNA gene amplicon sequencing. Bacterial communities in rectal swabs were distinct from those detected in negative controls, both visually and via permutation testing (P < 0.001 by PERMANOVA). Download FIG S1, EPS file, 1.6 MB (1.6MB, eps) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Random forest identifies OTUs on admission is predictive of final Enterococcus abundance. We built a random forest regression model using admission microbiota and antimicrobial exposures to predict the abundance of Enterococcus at the time of VRE acquisition. Seven individual OTUs were significant features at P < 0.01 after correcting for feature importance bias. Download FIG S2, EPS file, 1.4 MB (1.4MB, eps) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
Sequences are available via the NCBI Sequence Read Archive (accession number PRJNA633879). OTU tables, taxonomy classification tables, and metadata tables are available at https://github.com/rishichanderraj/Microbiota_Predictors_VRE_Acquisition. We have excluded protected health information (PHI) attached with this metadata. Potential collaborators are welcome to contact our group with reasonable requests that guarantee patient safety and privacy.
ACKNOWLEDGMENTS
We thank Aline Penkevich for assistance with acquiring rectal swabs for sequencing.
This work was supported by the National Institutes of Health (grant R01 HL144599 to R.P.D., R01 AI143852 to R.J.W., and 5T32 HL007749-27 to R.C.).
We declare that we have no conflicts of interest to disclose.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Antibiotic Resistance Coordination and Strategy Unit, Division of Healthcare Quality Promotion, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA.
- 2.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288−1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zirakzadeh A, Gastineau DA, Mandrekar JN, Burke JP, Johnston PB, Patel R. 2008. Vancomycin-resistant enterococcal colonization appears associated with increased mortality among allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant 41:385–392. doi: 10.1038/sj.bmt.1705912. [DOI] [PubMed] [Google Scholar]
- 4.Zirakzadeh A, Patel R. 2006. Vancomycin-resistant enterococci: colonization, infection, detection, and treatment. Mayo Clin Proc 81:529–536. doi: 10.4065/81.4.529. [DOI] [PubMed] [Google Scholar]
- 5.Bonten MJ, Hayden MK, Nathan C, van Voorhis J, Matushek M, Slaughter S, Rice T, Weinstein RA. 1996. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet 348:1615–1619. doi: 10.1016/S0140-6736(96)02331-8. [DOI] [PubMed] [Google Scholar]
- 6.Society for Healthcare Epidemiology of America, Infectious Diseases Society of America, Pediatric Infectious Diseases Society. 2012. Policy statement on antimicrobial stewardship by the Society for Healthcare Epidemiology of America (SHEA), the Infectious Diseases Society of America (IDSA), and the Pediatric Infectious Diseases Society (PIDS). Infect Control Hosp Epidemiol 33:322–327. doi: 10.1086/665010. [DOI] [PubMed] [Google Scholar]
- 7.Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, Taur Y, Jenq RR, van den Brink MRM, Xavier JB, Pamer EG. 2013. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, Kim GJ, Ling L, Pamer EG. 2017. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21:592–602.e4. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Der Waaij D, Berghuis-de Vries JM, Lekkerkerk-Van Der Wees JEC. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SG, Becattini S, Moody TU, Shliaha PV, Littmann ER, Seok R, Gjonbalaj M, Eaton V, Fontana E, Amoretti L, Wright R, Caballero S, Wang ZMX, Jung HJ, Morjaria SM, Leiner IM, Qin W, Ramos RJJF, Cross JR, Narushima S, Honda K, Peled JU, Hendrickson RC, Taur Y, van den Brink MRM, Pamer EG. 2019. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572:665–669. doi: 10.1038/s41586-019-1501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cash HL, Whitham CV, Behrendt CL, Hooper LV. 2006. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. 2005. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 14.Bhalla A, Pultz NJ, Ray AJ, Hoyen CK, Eckstein EC, Donskey CJ. 2003. Antianaerobic antibiotic therapy promotes overgrowth of antibiotic-resistant, Gram-negative bacilli and vancomycin-resistant enterococci in the stool of colonized patients. Infect Control Hosp Epidemiol 24:644–649. doi: 10.1086/502267. [DOI] [PubMed] [Google Scholar]
- 15.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M-A, Jenq RR, van den Brink MRM, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald D, Ackermann G, Khailova L, Baird C, Heyland D, Kozar R, Lemieux M, Derenski K, King J, Vis-Kampen C, Knight R, Wischmeyer PE. 2016. Extreme dysbiosis of the microbiome in critical illness. mSphere 1:199–215. doi: 10.1128/mSphere.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravi A, Halstead FD, Bamford A, Casey A, Thomson NM, van Schaik W, Snelson C, Goulden R, Foster-Nyarko E, Savva GM, Whitehouse T, Pallen MJ, Oppenheim BA. 2019. Loss of microbial diversity and pathogen domination of the gut microbiota in critically ill patients. Microb Genom 5:e000293. doi: 10.1099/mgen.0.000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livanos AE, Snider EJ, Whittier S, Chong DH, Wang TC, Abrams JA, Freedberg DE. 2018. Rapid gastrointestinal loss of clostridial clusters IV and XIVa in the ICU associates with an expansion of gut pathogens. PLoS One 13:e0200322. doi: 10.1371/journal.pone.0200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojima M, Motooka D, Shimizu K, Gotoh K, Shintani A, Yoshiya K, Nakamura S, Ogura H, Iida T, Shimazu T. 2016. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci 61:1628–1634. doi: 10.1007/s10620-015-4011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubin KA, Mathur D, McKenney PT, Taylor BP, Littmann ER, Peled JU, Van Den Brink MRM, Taur Y, Pamer EG, Xavier JB. 2019. Diversification and evolution of vancomycin-resistant Enterococcus faecium during intestinal domination. Infect Immun 87:e00102-19. doi: 10.1128/IAI.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pendleton KM, Erb-Downward JR, Bao Y, Branton WR, Falkowski NR, Newton DW, Huffnagle GB, Dickson RP. 2017. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am J Respir Crit Care Med 196:1610–1612. doi: 10.1164/rccm.201703-0537LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanderraj R, Dickson RP. 2018. Rethinking pneumonia: a paradigm shift with practical utility. Proc Natl Acad Sci U S A 115:13148–13150. doi: 10.1073/pnas.1819024116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chanderraj R, Millar JA, Patel TS, Read AF, Washer L, Kaye KS, Woods RJ. 2019. VRE acquisition in a tertiary care hospital: testing the roles of antibiotic use, proton pump inhibitor use, and colonization pressure. Open Forum Infect Dis 6:ofz139. doi: 10.1093/ofid/ofz139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willems RPJ, Van Dijk K, Ket JCF, Vandenbroucke-Grauls CMJE. 2020. Evaluation of the association between gastric acid suppression and risk of intestinal colonization with multidrug-resistant microorganisms: a systematic review and meta-analysis. JAMA Intern Med 180:561. doi: 10.1001/jamainternmed.2020.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannella RA, Broitman SA, Zamcheck N. 1972. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut 13:251–256. doi: 10.1136/gut.13.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. 2012. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 80:3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss PD. 2019. SchlossLab/MiSeq_WetLab_SOP. GitHub. https://github.com/SchlossLab/MiSeq_WetLab_SOP.
- 31.Korbie DJ, Mattick JS. 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc 3:1452–1456. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- 32.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss PD. 2019. MiSeq SOP:mothur. https://mothur.org/wiki/miseq_sop/.
- 34.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590−D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westcott SL, Schloss PD. 2017. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2:e00073-17. doi: 10.1128/mSphereDirect.00073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews BW. 1975. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 41.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara RB, Simpson G, Solymos P, Stevenes MHH, Wagner H. 2012. Vegan: community ecology package. R package version 2.0-2.
- 42.R Development Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 43.Wang Y, Naumann U, Wright ST, Warton DI. 2012. mvabund− an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol 3:471–474. doi: 10.1111/j.2041-210X.2012.00190.x. [DOI] [Google Scholar]
- 44.ter Braak CJF, Peres-Neto P, Dray S. 2017. A critical issue in model-based inference for studying trait-based community assembly and a solution. PeerJ 2017 5:e2885. doi: 10.7717/peerj.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright MN, Ziegler A. 2017. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw 77. doi: 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 46.Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Softw 28:1–26. doi: 10.18637/jss.v077.i01.27774042 [DOI] [Google Scholar]
- 47.Altmann A, Toloşi L, Sander O, Lengauer T. 2010. Permutation importance: a corrected feature importance measure. Bioinformatics 26:1340–1347. doi: 10.1093/bioinformatics/btq134. [DOI] [PubMed] [Google Scholar]
- 48.Therneau T. 2015. A package for survival analysis in S. 2.38.
- 49.Therneau T, Grambsh P. 2000. Modeling survival data: extending the Cox model. Springer-Verlag, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Most important taxa identified by random forest regression model in cases. We built a random forest regression model to identify taxa present on admission that were predictive of final Enterococcus relative abundance. In cases, only Blautia and Lactobacillus were significant after correcting for multiple testing and feature importance bias. Download Table S1, DOCX file, 0.01 MB (14.2KB, docx) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Most important taxa identified by random forest regression model in cases. We built a random forest regression model to identify taxa present on admission that were predictive of final Enterococcus relative abundance. In cases, only Blautia and Lactobacillus were significant after correcting for multiple testing and feature importance bias. Download Table S2, DOCX file, 0.01 MB (14.6KB, docx) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of rectal swab communities with negative sequencing controls. A total of 236 rectal swab specimens and 13 negative-control specimens were studied using 16S rRNA gene amplicon sequencing. Bacterial communities in rectal swabs were distinct from those detected in negative controls, both visually and via permutation testing (P < 0.001 by PERMANOVA). Download FIG S1, EPS file, 1.6 MB (1.6MB, eps) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Random forest identifies OTUs on admission is predictive of final Enterococcus abundance. We built a random forest regression model using admission microbiota and antimicrobial exposures to predict the abundance of Enterococcus at the time of VRE acquisition. Seven individual OTUs were significant features at P < 0.01 after correcting for feature importance bias. Download FIG S2, EPS file, 1.4 MB (1.4MB, eps) .
Copyright © 2020 Chanderraj et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequences are available via the NCBI Sequence Read Archive (accession number PRJNA633879). OTU tables, taxonomy classification tables, and metadata tables are available at https://github.com/rishichanderraj/Microbiota_Predictors_VRE_Acquisition. We have excluded protected health information (PHI) attached with this metadata. Potential collaborators are welcome to contact our group with reasonable requests that guarantee patient safety and privacy.