Abstract
Recent studies in mice suggested that KLF5 (Kruppel like factor 5), a zinc-finger transcription factor, plays an important role in skeletal muscle development and regeneration. As an important factor in the process of muscle development, KLF5 participates in the regulation of the cell cycle, cell survival, and cell dryness under different environmental conditions, but it is not clear whether KLF5 participates in muscle atrophy. Therefore, we investigated whether KLF5 can regulate the atrophy of chicken satellite cells in vitro and examined its mechanism of action. qPCR showed that KLF5 gene knockdown promoted the expression of key genes in muscle atrophy. Subsequently, we sequenced and analyzed the transcriptomes of KLF5 silenced and control cells, and we showed that the differentially expressed genes were mainly enriched in 10 signaling pathways (P<0.05), with differential gene and enrichment analyses indicating that the Wnt signaling pathways are extremely important. In conclusion, our results indicate that KLF5 may regulate the atrophy of chicken skeletal muscle through the Wnt/β-catenin signaling pathway.
Keywords: atrophy, KLF5, satellite cell, skeletal muscle, Wnt/β-catenin
Introduction
Skeletal muscle, also known as striated muscle, is a significant motor organ in organisms. It is not only an important motor organ of livestock and poultry but also an important source of human intake of high-quality protein. Post-hatch muscle growth occurs by a process called hypertrophy [14]. Muscle growth is due to the growth of muscle fibers, resulting in an increase in myofibrils and an increase in the number of myofibrillar nuclei, but the number of muscle fibers remains basically constant after an animal’s birth [3,8]. Skeletal muscle satellite cells (SMSCs) are skeletal muscle progenitor cells responsible for postnatal growth and repair. The function of SMSCs are diverse,including functions in myonuclear production and synthesis and muscle adaptation and regeneration [31]. These processes are principally coordinated by the MyoD family (MyoD, Myf5, myogenin, and MRF4) and MEF2 family (MEF2A-D) transcription factors [27]. Furthermore, the Sp1 transcription factor links ERK5 to Kruppel-like factor (KLF) 2/4, and nephronectin, a KLF transcriptional target, is involved in myocyte fusion [32]. Recently, some new roles of KLF in skeletal muscle biology have been gradually discovered.
KLFs are a subfamily of the zinc-finger class of DNA-binding transcriptional regulators [13]. A total of 18 KLFs have been found in the human genome, many of which are involved in the regulation of the development and function of the human and animal myocardium, smooth muscle, and skeletal muscle. Like other members of the KLF family, KLF5 contains three C2H2 zinc finger domains at its carbon end, which are used to identify and bind DNA. Studies have shown that KLF5 can identify and bind to the Sp1 locus in the promoter region of a target gene, GC and CACCC box, and the most of its target gene promoter regions contain one or more GC enrichment sites [19]. Fascinating research in Dr. Ryozo Nagai’s lab has shown that KLF5 is an important regulator of muscle lipid metabolism and system energy balance [28]. In the heart, published evidence to date indicates that KLF5 is mainly expressed in cardiac fibroblasts [13]; KLF5 is an important transcription factor that not only causes changes in smooth muscle phenotype but also causes cardiac hypertrophy and fibrosis [25]. Recently, Hayashi and coworkers clearly pointed out in an article in elife that klf5 is an essential regulator of skeletal muscle differentiation, acting in concert with myogenic transcription factors such as MyoD and Mef2. In poultry [16], the KLF5 gene does not significantly regulate proliferation of chicken skeletal muscle satellite cells, but it does promote differentiation and inhibit apoptosis [35]. On the other hand, KLF15 participates in muscle catabolism through transcriptional regulation of FoxO1, terragin-1, and MuRF1 and upregulates BCAT2, negatively affecting mTOR and thereby inducing degradation of branched chain amino acids [6]; it also upregulates the gene expression of the E3 ubiquitin ligases Atrogin1 and MuRF1 and negatively modulates myofiber size [22]. Compared with KLF5 in terms of promoting of proliferation, overexpression of KLF15 in cultured SMCs (skeletal muscle cells) could effectively inhibit SMC proliferation [25]. What’s more, skeletal muscle atrophy was inhibited by inhibiting Atrogin1/mafbx-mediated MyoD proteolysis [18], and this suggests that there may be an inverse relationship between differentiation and atrophy.
Previous research has suggested that muscle differentiation and atrophy are closely related in human-related diseases [7, 21]. Even more interestingly is that MyoD interacts with Atrogin1, and MyoD and Atrogin1 are key regulators of muscle differentiation and atrophy, respectively [34]. In addition, KLF15 acts contrary to KLF5 in muscle differentiation, possibly suggesting that KLF5 acts as an inhibitor of the muscular atrophy process. Although numerous studies have shown that KLF5 is involved in the regulation of cell proliferation, the cell cycle, cell survival, cell migration, cell dryness, and cell differentiation under different environmental conditions, the correlation between KLF5 and cell atrophy is unclear. In this study, we proved that KLF5 regulates chicken skeletal muscle atrophy may via the canonical Wnt/β-catenin signaling pathway by determining the effects of KLF5 knockdown by KLF5-specifc small interfering RNA (siRNA).
Materials and Methods
Animal procedures
These experiments were approved by the Committee on the Care and Use of Laboratory Animals of the State-level Animal Experimental Teaching Demonstration Center of Sichuan Agricultural University (No. 20192020061). All research work was conducted in strict accordance with the Sichuan Agricultural University (SAU) Laboratory Animal Welfare and Ethics guidelines.
Isolation and culture of chicken skeletal muscle satellite cells
In this study, tissue samples and primary skeletal muscle satellite cells were collected or obtained from six 5-day-old male ROSS-308 white-feathered brock chickens, which were purchased from a Yunda poultry cooperative in Chengdu, Sichuan province China.The pectoralis muscle was removed from six different chickens and used for preparation of primary myogenic cultures. About 5 g of the treated pectoralis muscle was digested with 3 times the volume of 0.1% collagenase I (SIGMA, Santa Clara, CA, USA) and 0.25% trypsin (Gibco, Beijing, China). The cell suspension was centrifuged and fresh medium was added, then the cells were blown evenly and seeded on a 12-well plate for culture. Chicken SMSCs were initially cultured in growth medium composed of 1% penicillin–streptomycin, 10% gestational horse serum, 10% fetal bovine serum (FBS), and Dulbecco’s Modified Eagle Medium (DMEM) with 0.5% chicken embryo extract at 37°C under 5% CO2 with saturating humidity for about 2–4 days, and the medium was refreshed every 24 h. In order to induce cell differentiation, DMEM containing 2% gestational horse serum, 5% fetal bovine serum, and 1% penicillin streptomycin was, was used instead of the original growth medium.
Knockdown of KLF5 mRNA in chicken skeletal satellite cells
Small interfering RNA-mediated knockdown was conducted in 6-well plates. When the cells reached approximately 70% confluence, the cells in each well were transfected with KLF5 siRNAs (Treatment group) and negative siRNA (control group); the cells had not yet begun to differentiate. Based on previous research, three siRNAs for KLF5 were chemically synthesized in the desalted, preannealed duplex form (Sangon Biotech, Shanghai, China) (Table 1). Cell transfection was performed using the Lipofectamine 3000 reagent protocol (Invitrogen). The Lipofectamine 3000 and siRNA were diluted with optim-MEM culture medium. The diluted siRNA and Lipofectamine 3000 were mixed evenly and placed at room temperature for 10 min. The composite was added to the cell culture plate and mixed in the culture plate. Knockdown efficiency was estimated by quantitative qPCR of KLF5 mRNA.
Table 1. Design of siRNA targets on chicken KLF5 CDS.
| siRNA | Sense strands (5’→3’) | Anti-sense strands (5’→3’) |
|---|---|---|
| KLF5 siRNA1 | GAAGUACAGAAGAGACAGUTT | ACUGUCUCUUCUGUACUUCTT |
| KLF5 siRNA2 | UUCACAACCCGAAUUUACCTT | GGUAAAUUCGGGUUGUGAATT |
| KLF5 siRNA3 | GUAACCCAGAUUUGGAGAATT | UUCUCCAAAUCUGGGUUACTT |
| Negative siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
RNA extraction and Quantitative real-time PCR (qPCR)
Total RNA from tissue or cells was isolated with Total RNA Isolation Kit reagent (ROREGENE, Shanghai, China), and its concentration and purity were determined based on the A260/280 absorbance ratio (values from 1.8 to 2.0 are preferred). The integrity of the 18S and 28S rRNA bands was determined using 2% agarose gel.
qPCR primers (Table 2) were designed with the Primer Premier 5 software. A 15 µl reaction containing 6.5 µl of SYBR® Premix Ex TaqTM (Takara Bio, Kusatsu, Japan), 1.5 µl of cDNA, 0.3 µl of forward primer, 0.3 µl of reverse primer, and 6.4 µl of RNase-free H2O (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China) were used for qPCR. The reaction was carried out with the following amplification conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, the primer-specific annealing temperature for 10 s, and 72°C for 10 s. The qPCR data were normalized relative to the expression of β-actin (endogenous control), and the 2−∆∆Ct method was applied to quantify mRNA expression levels. Three replicates were performed for qPCR analysis.
Table 2. Primers used in this study.
| Gene | Forward primer (5’→3’) | Reverse primer(5’→3’) |
|---|---|---|
| KLF5 | F: CGCGCTCGGATGAATTAACG | R: GATACAGAACAGCCTCGGCA |
| Atrogin 1 | F: TCAACGGGTCGGCAAGTCT | R: TCCCTCCCATCGCTCAGTC |
| MuRF1 | F: GGCAGCAGCATCATCTCGG | R: CCTCGCAGGTGACGCAGTAG |
| β-actin | F: GTCCACCGCAAATGCTTCTAA | R: TGCGCATTTATGGGTTTTGTT |
| LEF1 | F: CATCAAGTCCTCGCTGGTCAACG | R: TAGGAATCCTGCGCTGGAGTCTG |
| CYP1A1 | F: ACGGACTCATTGATTGGGCA | R: GGGTACAAGGCAGCGTACAT |
| MYOM1 | F: GCAGTCAACCGTAAGCCAGTACC | R: CTGTAGGCTGCTGACTCAAGGTTG |
| NOB1 | F: AGGTGTTGGCGCTCACG | R: GCATCTCCTGGAGCTCATCC |
| MYF6 | F: AGCGGAGGAGGCTGAAGAAGATC | R: TGATGGCGCTCCTCAGGATCTC |
| Wnt3a | F: GTGGCTTTTGCAGTGACCAG | R: GTTGTGCCTGTTCATGGCTG |
| Wnt5a | F: TGGCTTCTCAGTACCTCGTAGTGG | R: TAGAGACCACCAAGAGCTGGCTTC |
| Wnt5b | F: GTGCCATCAAGGAGTGCCAGTAC | R: GCCGAAGACGGACGTGTTGTC |
| Wnt7a | F: CTCCGCCGACATCAGATACG | R: CCCGGAATTTCGGCAGAGTT |
| Wnt10a | F: GCCAACACGGTGTGCCTGAC | R: CGTGGATGGCGATCTGGATGC |
| β-catenin | F: TACTGCAGGCACACTACACA | R: TTTGTCTTGTTGAGCAGGGC |
Library construction and transcriptome sequencing
The isolated mRNA was enriched, and eukaryotic mRNA was enriched by Oligo (dT) magnetic beads binding to the polyA tail of mRNA by A-T complementary pairing. Fragmentation buffer was then added to break the mRNA into short fragments. The mRNA was used as a template to synthesize one strand of cDNA with random hexamers. Then buffer, dNTPs, and DNA polymerase were added to synthesize the second strand of cDNA. The purified double-stranded cDNA was then repaired at the end, a tail was added, and the sequencing joint was connected. Next AMPure XP beads were used to select the fragment size, and PCR enrichment was carried out to obtain the final cDNA library. Finally, the quality of the libraries was evaluated with a Qubit 2.0 fluorometer, Agilent 2100 Bioanalyzer instrument, and qPCR, and then the libraries were sequenced using the Illumina HiSeq platform.
Enrichment analysis
Gene Ontology (GO) analyses were performed using Genomatix (https://www.genomatix.de/) and the biological process (BP), cellular component (CC), and molecular function (MF) aspect of GO. Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) pathway analysis was carried out for differentially expressed genes with a probability (P)-value <0.05 and a false discovery rate (FDR) <0.05.
Statistical analyses
Comparisons between two groups were analyzed using a two-tailed Student’s t-test. Differences among more than two groups were analyzed using one-way ANOVA followed by Tukey-Kramer post hoc tests. All these analyses were performed using the JMP Pro software (SAS Institute, Cary, NC, USA). Values of P<0.05 were considered statistically significant. All data are shown as the mean ± SEM.
Results
Knockdown of KLF5 expression in chicken SMSCs
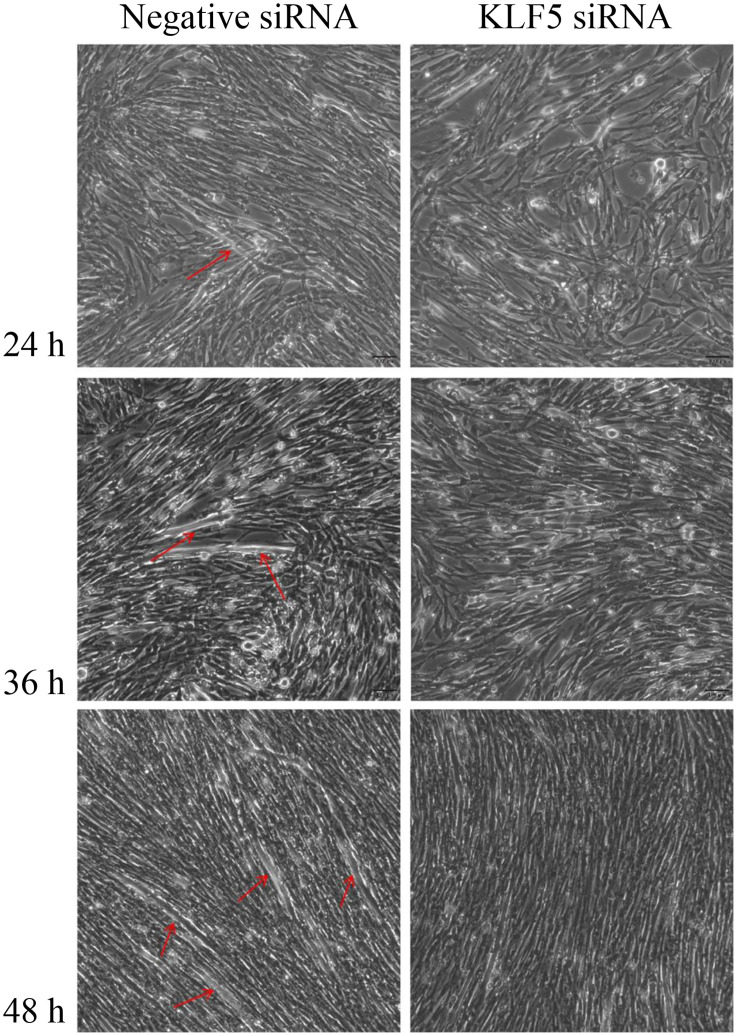
In previous studies, it was been found that KLF5 silencing causes damage to myotube formation [35]. To investigate the effect of KLF5 gene knockdown on cell growth and differentiation, negative siRNA and KLF5 siRNA were transfected with SMSCs after 70–80% of the cells reached confluence. Morphologically, SMSCs began to differentiate into myotubes 24 h after transfection with negative siRNA, as shown in Fig. 1. Further, the SMSCs differentiated into larger and more numerous myotubes at 48 h, and the number of myotubes increased significantly at 72 h. On the other hand, the cells transfected with KLF5 siRNA did not form myotubes. This showed shows that the differentiation of SMSCs transfected with KLF5 siRNA at 24 h, 48 h and 72 h after confluence was significantly lower than that of the negative siRNA group.
Fig. 1.
The effect of KLF5 knockdown on the differentiation of chicken SMSCs. The SMSCs were transfected with KLF5 siRNA or negative siRNA and cultured in differentiation medium for subsequent experiments. The myotube formation of cells transfected at 24 h, 48 h, and 72 h was observed under a light microscope. Red arrows indicate myotubes. Black scale bar=100 µm.
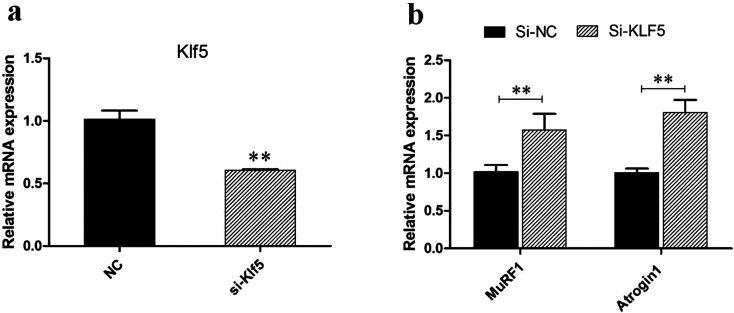
In order to further investigate the function of KLF5 and explore whether KLF5 plays a role in the process of chicken skeletal muscle atrophy, we designed a siRNA interference fragment for KLF5 according to its gene sequence. After 36 h of transfection of chicken satellite cells, samples were collected, and the interference efficiency was detected by qPCR. The results showed that the expression of KLF5 gene could be significantly reduced by transfection of the siRNA interference fragment (P<0.01) (Fig. 2a).
Fig. 2.
Interference with the KLF5 gene promotes the expression of muscular atrophy genes. a The expression of mRNA in the KLF5 interference group (si-KLF5) and NC after 36 h. b Expression of apoptosis-promoting genes after KLF5 interference.
Effects of KLF5 gene knockdown on the atrophy of chicken SMSCs cells
In order to further verify the effect of KLF5 gene on chicken skeletal atrophy, we detected the mRNA expression of MuRF1 and Atrogin1 in the skeletal muscle satellite cell interference group and control group. The results showed that after the interfering with KLF5 expression, the mRNA expression of MuRF1 and Atrogin1 increased significantly within 36 h compared with the control group (P<0.01) (Fig. 2b). Atrogin1 and MuRF1 are E3 ubiquitin ligases and which are important regulators of ubiquitin-mediated protein degradation in skeletal muscle and key markers of muscle atrophy [17]. Therefore, we speculate that KLF5 has a negative regulatory effect on chicken muscle atrophy.
Differential expression analysis
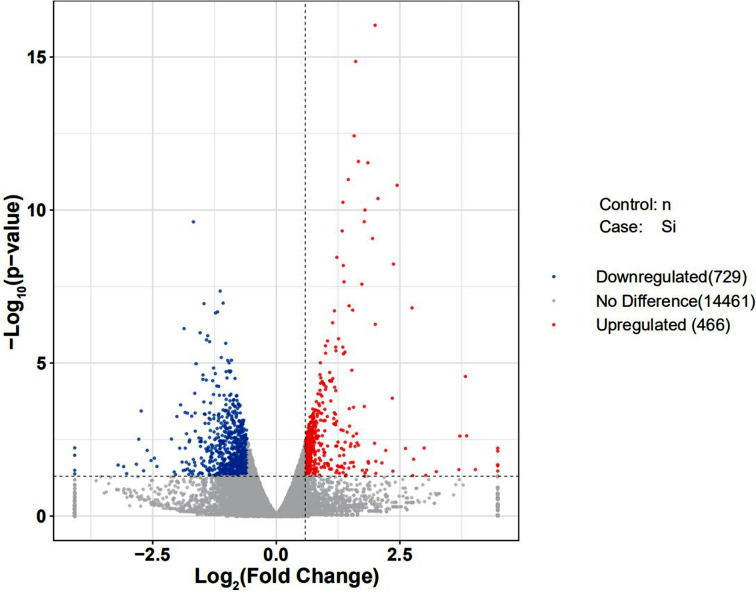
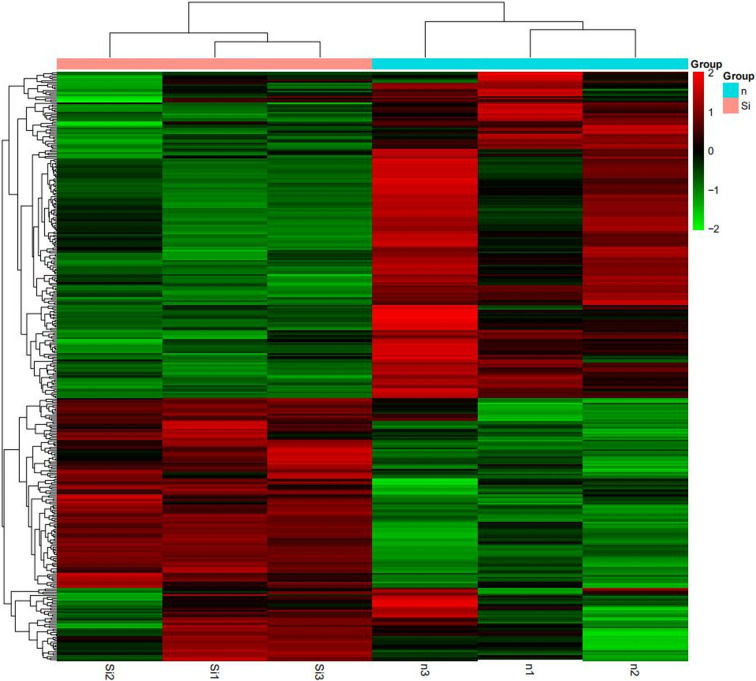
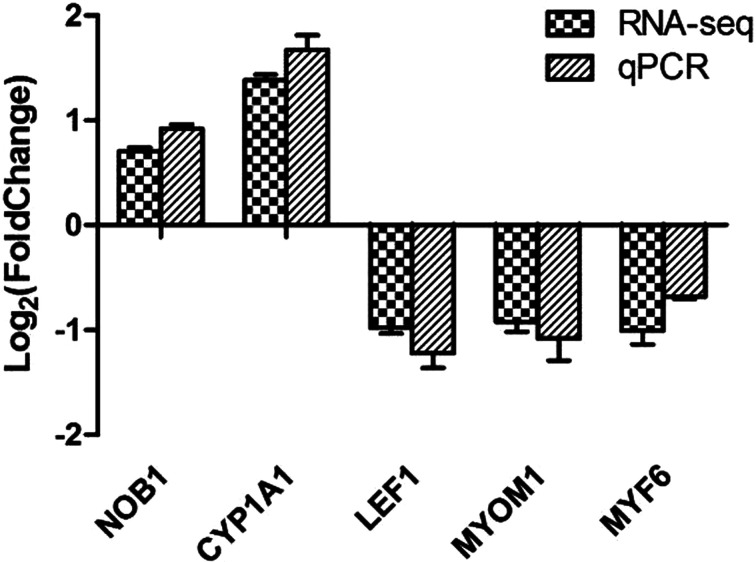
In order to discover how KLF5 gene affects chicken skeletal muscle atrophy, we sequenced and analyzed the transcriptomes of the KLF5 interference group and control group cells. We obtained the expression pattern of the gene in a sample (Fig. 3) and used DEseq to analyze the differentially expressed genes from transcriptome sequencing, the screening conditions: the expression difference multiple log2FoldChange >1 and significance set at P-value <0.05. The results showed that there were 1195 differentially expressed genes in the interference group and the control group, of which 729 and 466 genes were significantly downregulated and upregulated, respectively (Fig. 4). This includes a large number of genes related to muscle development, including MYL6, MYH1B, MYO16, and LEF1. We further screened and clustered the differentially expressed genes genes related to muscle development (Fig. 5). In order to verify the transcriptome sequencing results, we randomly selected six differentially expressed genes from the transcriptome sequencing results, which were verified by qPCR, including MYF6, MYOM1, NOB1, LEF1 and CYP1A1 (Fig. 6). The results of qPCR were consistent with the transcriptome sequencing results, indicating that the transcriptome sequencing data were reliable and could be further analyzed.
Fig. 3.
Expression patterns of genes in samples. (a) Boxplot: the upper and lower limits are 90%, the upper and lower edges are 75%, and the horizontal line is the median. (b) FPKM density distribution map: the abscissa is the log10 (FPKM) value of the gene, and the ordinate is the gene distribution density of the corresponding expression amount.
Fig. 4.
Volcano map of differentially expressed genes. The two dashed vertical lines are the thresholds of 2 times, the dashed horizontal line is the threshold of P-value=0.05, the red dots are the upregulated genes, the blue dots are the downregulated genes, and gray dots represent nonsignificantly differentially expressed genes (n v.s. Si).
Fig. 5.
Cluster diagram of differentially expressed genes. Red indicates genes with higher expression, and green indicated genes with lower expression genes (n v.s. Si).
Fig. 6.
Verification of RNA-seq results by qPCR.
GO enrichment analysis
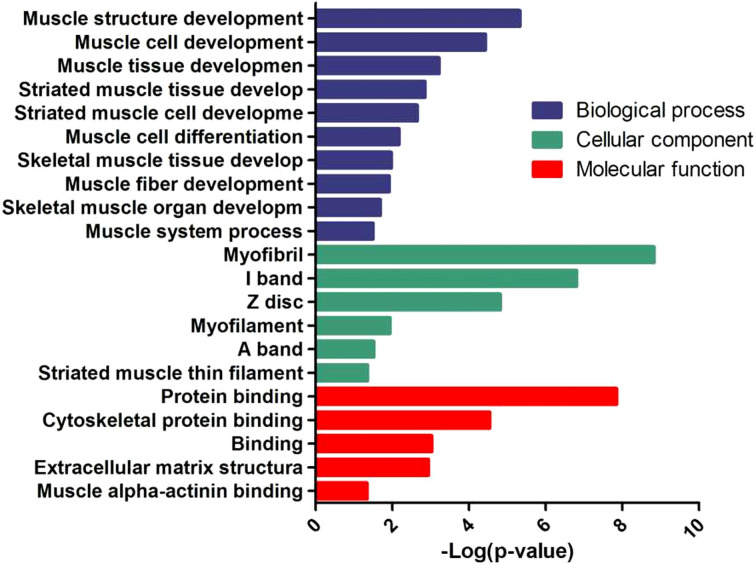
Analysis of Go enrichment enables us to understand which GO function items show significant enrichment in differentially expressed genes and speculate about biological functions related to the differentially expressed genes. After interfering with the expression of KLF5 in chicken skeletal muscle satellite cells, we sequenced the transcriptome of the control group to obtain the differentially expressed genes. The main Go enrichment items are shown in Fig. 7. Differential genes are mainly enriched growth and development of muscle tissue and differentiation of muscle cells in the BPs and the composition of muscle tissue and the characteristics of muscle fiber in CCs; in MFs, they were mainly related to protein binding and energy metabolism.
Fig. 7.
GO analysis of differentially expressed genes.
KEGG enrichment analysis
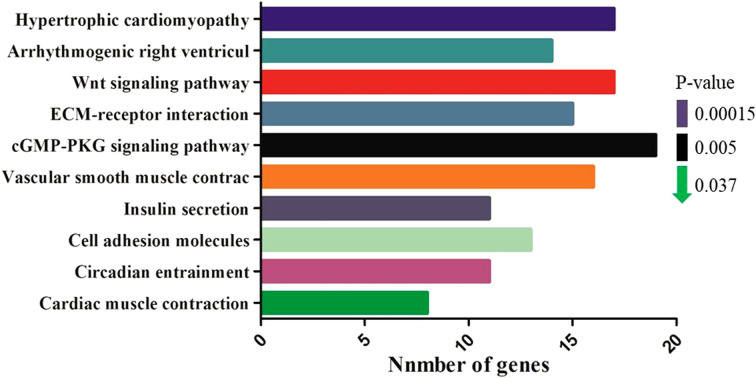
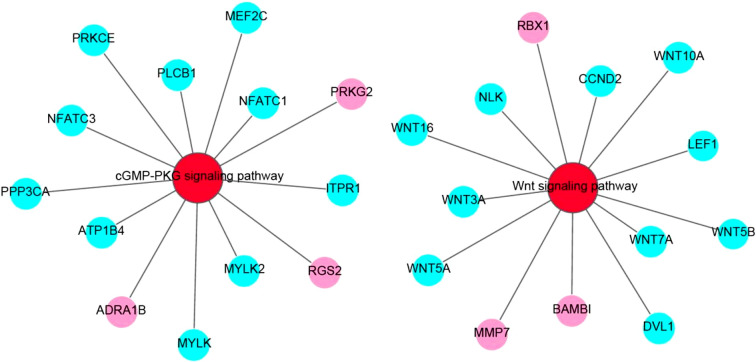
Our analysis showed that the differentially expressed genes were mainly enriched in 10 signaling pathways (P<0.05), which were mainly related to intercellular regulation, muscle development, energy metabolism, and diseases (Fig. 8), and among them, the cGMP-PKG signaling pathway and Wnt signaling pathway are two pivotal pathways for muscle development. The differentially expressed genes in these two pathways through network diagram (Fig. 9). Wnt signaling pathway is an important signaling pathway that regulates the development of skeletal muscle. It mainly regulates the growth, development, and regeneration of skeletal muscle by inducing the development of skeletal muscle in the embryonic stage and activating skeletal muscle satellite cells.
Fig. 8.
KEGG pathway analysis of differentially expressed genes. The top ten pathways of significance are shown in the figure, and the abscissa represents the number of enriched genes.
Fig. 9.
Network diagram of two pivotal pathways for muscle development in KEGG enrichment analysis. Red circles represent pathway, aqua circles represent downregulated differentially expressed genes, and pink circles represent upregulated differentially expressed gene.
Verification of Wnt signal pathway-related gene expression
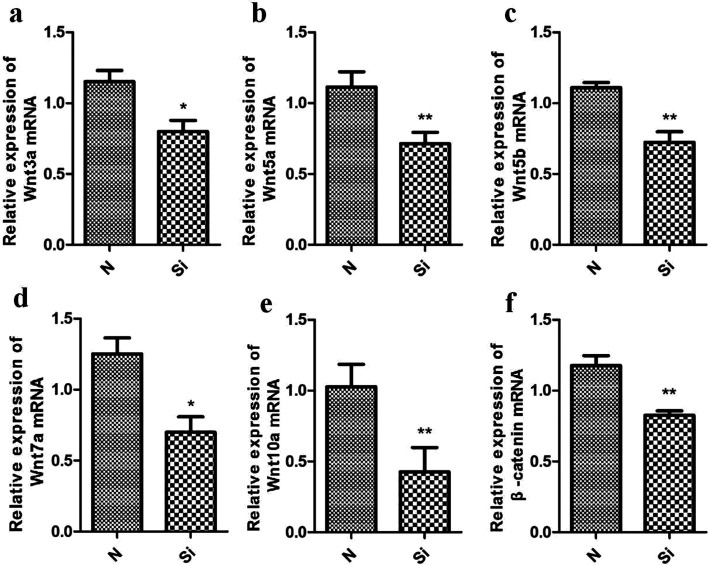
By interfering with the expression of KLF5 in chicken skeletal muscle satellite cells and sequencing the transcriptome, the Wnt signal pathway, an important signal pathway regulating skeletal muscle development, was screened out by differential gene analysis. Previous genetic studies have shown that the Wnt/β-catenin signaling pathway regulates the normal development of skeletal muscle [5]. We validated the expression of Wnt signaling pathway-related genes after KLF5 interference in chicken skeletal muscle satellite cells by qPCR. The expression of Wnt3a, Wnt5a, Wnt5b, Wnt7a, Wnt10a, and β-catenin in the KLF5 interference group and control group decreased significantly (P<0.05) (Fig. 10). The results suggested that KLF5 may regulates the development of chicken skeletal muscle satellite cells through the Wnt signaling pathway.
Fig. 10.
The expression of Wnt signaling pathway-related genes. *P<0.05; **P<0.01. Data are expressed as the mean ± SEM.
Discussion
Loss of muscle mass and fiber volume in adults is called atrophy and is characterized by increased protein degradation [2, 15]. KLF5 transcription factors play a crucial role in the muscle regeneration, myoblast differentiation, and satellite cell differentiation in mice, but it is unknown if it regulates muscle atrophy, especially in poultry [16]. However, it has been confirmed that KLF15 can degrade mammalian target of rapamycin (mTOR) through branched chain amino acids (BCAA) and negatively regulate the size of muscle fibers [33]. For SMCs, the response of KLF15 to stimulation of proliferation was strongly weakened after vascular injury; this contrast with the induction of KLF4 and KLF5 that occurs after vascular injury [25]. Therefore, KLF15 and KLF5 which are members of the same family of transcription factors have opposite effects on muscle differentiation, so it is possible that they might have the opposite effects on muscle atrophy. In addition, previous studies have identified MyoD as the second MAFbx target for skeletal muscle atrophy. It also relies on atrophic signaling pathways, and its expression appears to be inhibited at both the RNA and protein levels, suggesting that the downregulation of MyoD in vivo is essential for muscle loss [12, 20, 34]. In this study, we explored whether KLF5 plays a role in skeletal muscle atrophy and its potential regulatory mechanism by knocking out KLF5. After interfering with KLF5 expression, MuRF1 and Atrogin1 mRNA expression increased significantly (P<0.01) compared with the control group within 36 h, indicating that KLF5 has some interaction with MuRF1 and Atrogin1 that promotes muscle atrophy.
Based on the results of the present study, we speculate that KLF5 can regulate the process of atrophy of chicken skeletal muscle, but the specific regulatory mechanism remains unclear. The regulation of muscle fiber development is a complex and delicate process that requires many regulatory factors to cooperate with each other. Here, we interfered with the expression of KLF5 in chicken skeletal muscle satellite cells, and sequenced the transcriptome of the control group to explore the way that KLF5 regulates the development of chicken skeletal muscle satellite cells. A total of 1,195 differentially expressed genes were obtained by transcriptome sequencing, including a large number of genes related to skeletal muscle development, such as Myf5, MYF6, and MyoG in the myogenic regulatory factors (MRFs) family. The MRFs gene family is involved in the whole process of muscle development, from the initial myoprogenitor cell formation to the formation of muscle fibers and subsequently to muscle maturity after birth, which runs through all the links of muscle development. MyoD regulates the differentiation of myocytes, while Myf5 regulates the regeneration of myocytes and keeps the number of myocytes constant [11]. According to the functional analysis of different genes screened by transcriptome sequencing, KLF5 can affect the development of chicken skeletal muscle satellite cells by regulating the expression of members of the myogenic factor family, multiple members of the myosin family, and other myogenic factors.
The differentially expressed genes were obtained by transcriptome sequencing and GO cluster analysis. The differentially expressed genes mainly enriched items related to muscle development, such as myofibril, myofilament, and muscle cell development, skeletal muscle development, muscle organ development, and muscle system processes. Furthermore, muscle mass and fiber size are essentially determined by protein turn over, that is, the balanc of protein synthesis and degradation in muscle fibers [30]; when the degradation rate is greater than the synthetic speed, muscle atrophy occurs. An enrichment analysis of the KEGG pathway of the differentially expressed genes from the transcriptome sequencing revealed that the differentially expressed genes were mainly concentrate in muscle development, intercellular regulation, energy metabolism, and disease-related signaling pathways. For example, in terms of muscle development and disease pathways, excessive degradation of skeletal muscle protein is very harmful to the human body and may lead to death [29]. Furthermore, the Wnt signaling pathway is a very important signaling pathway in skeletal muscle development. It can affect the development of skeletal muscle in the embryo and the maintenance of skeletal muscle homeostasis in adult [1], and mainly through the regulation of (MRFs) that induce muscle development [24].
In order to verify whether KLF5 regulates the development of chicken skeletal muscle satellite cells through the Wnt signaling pathway, we interfered with the expression of KLF5 in chicken skeletal muscle satellite cells and detected the expression of related genes in the Wnt signaling pathway. The results showed that the expression of Wnt3a, Wnt5a, Wnt5b, Wnt7a, Wnt10a, and β-catenin decreased significantly after KLF5 expression was disturbed. The classical Wnt/β-catenin signaling pathway promotes the regeneration of skeletal muscle after injury by inducing muscle satellite cells to switch from proliferation to differentiation [9]. In the classical Wnt signaling pathway, β-catenin plays a central regulatory role. Some studies have shown that β-catenin can directly interact with MyoD, enhance the connection between MyoD and E-box, and thus start the myogenic process [26]. In addition, β-catenin can promote the self-renewal of satellite cells and inhibit the process of satellite cells immediately entering myogenic differentiation. It has been pointed out that the activity of spinal muscular atrophy protein is regulated in the process of development and cell differentiation and that SMN activity and snRNP synthesis are strongly downregulated in neurons and myogenic differentiation [10]. Taken together,the above indicate that there may be a close relationship between muscle differentiation and atrophy. Furthermore, non-canonical Wnt signaling is also needed to complete the regeneration of skeletal muscle. Wnt7a can regulate the differentiation of injured skeletal muscle cells through multiple non-canonical Wnt signaling pathways, such as by activating the Akt/mTOR signaling pathway to induce muscle fiber hypertrophy and promote muscle development [4]. Muscle atrophy occurs when the rate of protein degradation exceeds the rate of protein synthesis, and a previous study showed that Wnt7a activated the PI3K/Akt/mTOR pathway through its receptor, fzd7, and increased the number of muscle fibers [23]. In our study, after interfering with KLF5, levels of expression of the pivotal signal molecules β-catenin, Wnt3a, Wnt5a, Wnt5b, and Wnt7a in the Wnt signaling pathway were significantly reduced. Combined with the findings of previous studies, the present study suggests that downregulation of these genes would hinder the differentiation and confluence of skeletal muscle satellite cells, hinder the development of muscle fibers, and cause muscle atrophy, which indicates that KLF5 may regulates the atrophy of chicken skeletal muscle through the Wnt/β-catenin signaling pathway.
Author Contributions
Data curation, H.-D.Y. and X.-S.J.; formal analysis, D.-H.Z. and Y.-D.H.; funding acquisition, Y.W. and Y.-P.L.; investigation, H.-D.Y. and H.-R.D.; methodology, C.-L.Y. and H.-R.D.; project administration, Y.W.; resources, Y.-D.H.; software, C.-W.Y. and C.-L.Y.; supervision, Q.-Y.L.; validation, Q.-Y.L.; Visualization, C.-W.Y.; writing – original draft, D.-H.Z.; writing – review & editing, X.-S.J. and Y.-P.L.
Conflicts of Interest
All authors have no conflict of interest to declare.
Acknowledgments
This study was supported by the Sichuan Science and Technology Program, the Open Fund of Farm Animal Genetic Resources Exploration (Grant NO. 2016NYZ0043), and the Innovation Key Laboratory of Sichuan Province (Grant NO. 2017JZ0033).
References
- 1.Abu-Elmagd M., Robson L., Sweetman D., Hadley J., Francis-West P., Münsterberg A.2010. Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. Dev. Biol. 337: 211–219. doi: 10.1016/j.ydbio.2009.10.023 [DOI] [PubMed] [Google Scholar]
- 2.Jagoe R.T., Goldberg A.L.2001. What do we really know about the ubiquitin-proteasome pathway in muscle atrophy? Curr. Opin. Clin. Nutr. Metab. Care 4: 183–190. doi: 10.1097/00075197-200105000-00003 [DOI] [PubMed] [Google Scholar]
- 3.Allen R.E., Merkel R.A., Young R.B.1979. Cellular aspects of muscle growth: myogenic cell proliferation. J. Anim. Sci. 49: 115–127. doi: 10.2527/jas1979.491115x [DOI] [PubMed] [Google Scholar]
- 4.Bentzinger C.F., von Maltzahn J., Dumont N.A., Stark D.A., Wang Y.X., Nhan K., Frenette J., Cornelison D.D., Rudnicki M.A.2014. Wnt7a stimulates myogenic stem cell motility and engraftment resulting in improved muscle strength. J. Cell Biol. 205: 97–111. doi: 10.1083/jcb.201310035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Amerongen R., Berns A.2006. Knockout mouse models to study Wnt signal transduction. Trends Genet. 22: 678–689. doi: 10.1016/j.tig.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 6.Bonaldo P., Sandri M.2013. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 6: 25–39. doi: 10.1242/dmm.010389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong J.T., Chen C.2009. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell. Mol. Life Sci. 66: 2691–2706. doi: 10.1007/s00018-009-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgerton V.R., Roy R.R.1991. Regulation of skeletal muscle fiber size, shape and function. J. Biomech. 24:(Suppl 1): 123–133. doi: 10.1016/0021-9290(91)90383-X [DOI] [PubMed] [Google Scholar]
- 9.Figeac N., Zammit P.S.2015. Coordinated action of Axin1 and Axin2 suppresses β-catenin to regulate muscle stem cell function. Cell. Signal. 27: 1652–1665. doi: 10.1016/j.cellsig.2015.03.025 [DOI] [PubMed] [Google Scholar]
- 10.Gabanella F., Carissimi C., Usiello A., Pellizzoni L.2005. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 14: 3629–3642. doi: 10.1093/hmg/ddi390 [DOI] [PubMed] [Google Scholar]
- 11.Gayraud-Morel B., Chrétien F., Flamant P., Gomès D., Zammit P.S., Tajbakhsh S.2007. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 312: 13–28. doi: 10.1016/j.ydbio.2007.08.059 [DOI] [PubMed] [Google Scholar]
- 12.Guttridge D.C., Mayo M.W., Madrid L.V., Wang C.Y., Baldwin A.S., Jr.2000. NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science 289: 2363–2366. doi: 10.1126/science.289.5488.2363 [DOI] [PubMed] [Google Scholar]
- 13.Haldar S.M., Ibrahim O.A., Jain M.K.2007. Kruppel-like Factors (KLFs) in muscle biology. J. Mol. Cell. Cardiol. 43: 1–10. doi: 10.1016/j.yjmcc.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding R.L., Halevy O., Yahav S., Velleman S.G.2016. The effect of temperature on proliferation and differentiation of chicken skeletal muscle satellite cells isolated from different muscle types. Physiol. Rep. 4: e12770. doi: 10.14814/phy2.12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasselgren P.O.1999. Glucocorticoids and muscle catabolism. Curr. Opin. Clin. Nutr. Metab. Care 2: 201–205. doi: 10.1097/00075197-199905000-00002 [DOI] [PubMed] [Google Scholar]
- 16.Hayashi S., Manabe I., Suzuki Y., Relaix F., Oishi Y.2016. Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. eLife 5: e17462. doi: 10.7554/eLife.17462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumucio J.P., Mendias C.L.2013. Atrogin-1, MuRF-1, and sarcopenia. Endocrine 43: 12–21. doi: 10.1007/s12020-012-9751-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagirand-Cantaloube J., Cornille K., Csibi A., Batonnet-Pichon S., Leibovitch M.P., Leibovitch S.A., Gianni P.2009. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLoS One 4: e4973. doi: 10.1371/journal.pone.0004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima S., Kobayashi A., Gotoh O., Ohkuma Y., Fujii-Kuriyama Y., Sogawa K.1997. Transcriptional activation domain of human BTEB2, a GC box-binding factor. J. Biochem. 121: 389–396. doi: 10.1093/oxfordjournals.jbchem.a021600 [DOI] [PubMed] [Google Scholar]
- 20.Langen R.C.J., Van Der Velden J.L., Schols A.M.W.J., Kelders M.C.J.M., Wouters E.F., Janssen-Heininger Y.M.2004. Tumor necrosis factor-alpha inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. 18: 227–237. doi: 10.1096/fj.03-0251com [DOI] [PubMed] [Google Scholar]
- 21.Li H., Malhotra S., Kumar A.2008. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J. Mol. Med. (Berl.) 86: 1113–1126. doi: 10.1007/s00109-008-0373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y.P., Chen Y., John J., Moylan J., Jin B., Mann D.L., Reid M.B.2005. TNF-α acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 19: 362–370. doi: 10.1096/fj.04-2364com [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Maltzahn J., Renaud J.M., Parise G., Rudnicki M.A.2012. Wnt7a treatment ameliorates muscular dystrophy. Proc. Natl. Acad. Sci. USA 109: 20614–20619. doi: 10.1073/pnas.1215765109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy M.M., Keefe A.C., Lawson J.A., Flygare S.D., Yandell M., Kardon G.2014. Transiently active Wnt/β-catenin signaling is not required but must be silenced for stem cell function during muscle regeneration. Stem Cell Reports 3: 475–488. doi: 10.1016/j.stemcr.2014.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai R., Shindo T., Manabe I., Suzuki T., Kurabayashi M.2003. KLF5/BTEB2, a Krüppel-like zinc-finger type transcription factor, mediates both smooth muscle cell activation and cardiac hypertrophy. Adv. Exp. Med. Biol. 538: 57–65, discussion 66. doi: 10.1007/978-1-4419-9029-7_5 [DOI] [PubMed] [Google Scholar]
- 26.Nelson W.J., Nusse R.2004. Convergence of Wnt, β-catenin, and cadherin pathways. Science 303: 1483–1487. doi: 10.1126/science.1094291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molkentin J.D., Olson E.N.1996. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 93: 9366–9373. doi: 10.1073/pnas.93.18.9366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oishi Y., Manabe I., Tobe K., Ohsugi M., Kubota T., Fujiu K., Maemura K., Kubota N., Kadowaki T., Nagai R.2008. SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ. Nat. Med. 14: 656–666. doi: 10.1038/nm1756 [DOI] [PubMed] [Google Scholar]
- 29.Sandri M.2010. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am. J. Physiol. Cell Physiol. 298: C1291–C1297. doi: 10.1152/ajpcell.00531.2009 [DOI] [PubMed] [Google Scholar]
- 30.Schiaffino S., Dyar K.A., Ciciliot S., Blaauw B., Sandri M.2013. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 280: 4294–4314. doi: 10.1111/febs.12253 [DOI] [PubMed] [Google Scholar]
- 31.Schultz E., McCormick K.M.1994. Skeletal muscle satellite cells. Rev. Physiol. Biochem. Pharmacol. 123: 213–257. doi: 10.1007/BFb0030904 [DOI] [PubMed] [Google Scholar]
- 32.Sunadome K., Yamamoto T., Ebisuya M., Kondoh K., Sehara-Fujisawa A., Nishida E.2011. ERK5 regulates muscle cell fusion through Klf transcription factors. Dev. Cell 20: 192–205. doi: 10.1016/j.devcel.2010.12.005 [DOI] [PubMed] [Google Scholar]
- 33.Shimizu N., Yoshikawa N., Ito N., Maruyama T., Suzuki Y., Takeda S., Nakae J., Tagata Y., Nishitani S., Takehana K., Sano M., Fukuda K., Suematsu M., Morimoto C., Tanaka H.2011. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 13: 170–182. doi: 10.1016/j.cmet.2011.01.001 [DOI] [PubMed] [Google Scholar]
- 34.Tintignac L.A., Lagirand J., Batonnet S., Sirri V., Leibovitch M.P., Leibovitch S.A.2005. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 280: 2847–2856. doi: 10.1074/jbc.M411346200 [DOI] [PubMed] [Google Scholar]
- 35.Zhang X.X., Lian T., Ran J.S., Li Z.Q., Han S.S., Liu Y.P.2019. KLF5 functions in proliferation, differentiation, and apoptosis of chicken satellite cells. 3 Biotechnol. 9: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]