Abstract
Amide hydrogen-deuterium exchange mass spectrometry is powerful for describing combinatorial coupling effects of a cooperative ligand pair binding at noncontiguous sites: adenosine at the ATP-pocket and a docking peptide (PIFtide) at the PIF-pocket, on a model protein kinase PDK1. Binding of two ligands to PDK1 reveal multiple hotspots of synergistic allostery with cumulative effects greater than the sum of individual effects mediated by each ligand. We quantified this synergism and ranked these hotspots using a difference in deuteration-based approach, which showed that the strongest synergistic effects were observed at three of the critical catalytic loci of kinases: the αB-αC helices, and HRD-motif loop, and DFG-motif. Additionally, we observed weaker synergistic effects at a distal GHI-subdomain locus. Synergistic changes in deuterium exchange observed at a distal site but not at the intermediate sites of the large lobe of the kinase reveals allosteric propagation in proteins to operate through two modes. Direct electrostatic interactions between polar and charged amino acids that mediate targeted relay of allosteric signals, and diffused relay of allosteric signals through soft matter-like hydrophobic core amino acids. Furthermore, we provide evidence that the conserved β-3 strand lysine of protein kinases (Lys111 of PDK1) functions as an integrator node to coordinate allosteric coupling of the two ligand-binding sites. It maintains indirect interactions with the ATP-pocket and mediates a critical salt bridge with a glutamate (Glu130) of αC helix, which is conserved across all kinases. In summary, allosteric propagation in cooperative, dual-liganded enzyme targets is bidirectional and synergistic and offers a strategy for combinatorial drug development.
Significance
Rational drug design has traditionally targeted orthosteric sites for common biological ligands such as nucleotides. Loss of specificity is a major disadvantage in such an approach. Allosteric site(s) offer an alternative, and a combination of orthosteric targeting and allosteric drugs offer greater potency and specificity. However, in vitro quantitation of the combinatorial effects of two or more drugs targeting a common protein has remained a challenge. We present a simple method to quantify combinatorial effects in a dual-liganded kinase model by hydrogen-deuterium exchange mass spectrometry. This method is easily scalable for proteins with more than two ligands, as well as for rapid pairwise screening of candidate molecules.
Introduction
Allostery is a fundamental property of all proteins enabling protein-ligand-protein interactions at a primary binding site to alter the binding properties of a second ligand at a distal, noncontiguous site (1), thereby playing a central role in executing and regulating cellular functions (2). All long-range conformational changes accompanying protein-ligand interactions and distal to the primary interface constitute allosteric changes (1,3,4). Dynamics is inherently linked to protein allostery (5,6) and a complete thermodynamic description of allostery requires measurement of both enthalpic (protein-ligand interface) as well as entropic (conformational dynamics) components (7). A molecular understanding of allostery is achieved by combining high-resolution structural maps from static crystallographic and cryogenic electron microscopy models with conformational dynamics measurements across multiple timescales. Mapping combinatorial allostery in enzymes interacting with more than one ligand across space requires a deconvolution of multiple allosteric networks, making it a bigger challenge (8, 9, 10, 11, 12).
Amide hydrogen-deuterium exchange mass spectrometry (HDXMS) is a powerful technique for mapping allostery in single proteins and large multiprotein assemblies (13, 14, 15, 16, 17). Deuterium exchange is initiated by dilution of an aqueous sample protein in an equivalent buffer constituted in heavy water (D2O) (seconds to minutes), followed by lowering of pHread ∼2.5 and endoprotease-mediated proteolysis followed by mass spectrometry (13,18,19). A major advantage of HDXMS in mapping protein-ligand interactions is that it offers both a direct readout of interacting residues at the interface and accompanying conformational changes. Allostery in single-ligand systems is readily identifiable when increases in deuterium exchange are observed in loci upon ligand binding (20). In the absence of mutagenesis or other biophysical measurements, it is harder to distinguish between orthosteric and allosteric effects when predominantly decreased exchange is observed across the entire protein in the liganded state. In these instances, allosteric sites can be distinguished from changes at orthosteric sites by integrating HDXMS and x-ray crystallography or cryogenic electron microscopy maps. Although structures provide high-resolution maps of the protein-ligand interface, HDXMS changes distal to the interface are attributable to allostery (14). For biological ligands with multiple functional groups interacting with the target protein, relative contributions to the overall strength of the interaction can be deconstructed with HDXMS carried out in variable concentrations of urea (21). Characterizing allostery in multiliganded systems (combinatorial allostery) represents a bigger challenge because of knowledge gaps in ligand cooperativity, in which studies of individual ligand-bound states are insufficient to fully describe allosteric effects in multiply-liganded states. Here, we describe combinatorial allostery from two ligands interacting at noncontiguous sites in a model protein kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) by HDXMS.
Protein kinases are a large important family of enzymes that catalyze phosphotransfer to protein substrates. Kinases have conserved structural scaffolds, consisting of a smaller N-terminal and a larger C-terminal lobe. The N-lobe harbors a characteristic ATP-binding pocket with conserved residues that catalyze the transfer of ATP’s γ-phosphate to the acceptor residues in substrates (22, 23, 24). Protein kinases are key mediators in numerous cellular processes and their dysfunction has been linked to cancer, diabetes, and neurological and inflammatory diseases (25). Allostery is fundamental for kinase regulation (26,27). Some kinases, such as PDK1 and Aurora A kinase, harbor an allosteric pocket within the kinase domain (28). Other kinases regulate their activities either through dimerization, e.g. EGFR kinase (29), or through complex formation with binding partner proteins, such as the independently expressed regulatory subunits of the protein kinase A (PKA) holoenzyme (30). Because of their crucial role in multiple diseases, kinases are attractive targets for inhibitor design (31) and small molecules targeting allosteric regulatory sites offer improved specificity in comparison to compounds solely targeting conserved ATP-binding sites. Consequently, a number of recent drug discovery efforts toward kinases focus on the unique allosteric modalities of kinases (28). Thus, there is a need for an effective strategy to quantitate combinatorial allosteric effects in kinases in a physiologically relevant context in which ligands occupy the ATP-binding site and, additionally, a substrate-binding or anchoring site. This study describes such a strategy.
PDK1 is a master kinase downstream of the PI3-kinase and the growth factor signaling pathway. It is responsible for phosphorylation of other AGC-family kinases, including Akt/PKB, S6K, PKA at their activation loops, resulting in kinase activation and stimulation of their individual downstream pathways (32, 33, 34, 35). PDK1 contains two ligand-binding sites in the N-terminal lobe: the orthosteric ATP-binding pocket and a regulatory allosteric site called the PIF-pocket (“O” and “A,” respectively, Fig. 1, A and B). As the primary interaction site for ATP, “O” is the critical orthosteric site fundamental to kinase catalysis. PDK1 activity is further regulated by the docking of C-terminal hydrophobic motif of a subset of its substrates such as S6K and PKC at the PIF-pocket, which elicits allosteric changes across both lobes of the kinase. A peptide ligand (PIFtide), as well as a number of small-molecule activators have been developed that bind at the PIF-pocket and enhance PDK1 kinase activity (36, 37, 38, 39, 40). These PIF-pocket ligands can be designed to function as selective allosteric modulators/inhibitors for substrates requiring docking at the PIF-pocket. Additionally, a reverse allosteric effect upon adenosine binding at the ATP-pocket of PDK1 was found to enhance binding of PIFtide (41). PDK1 and its sets of ligands thus offer a unique set of two ligands binding to a single target protein to monitor and quantitate allosteric coupling. (Fig. 1 A).
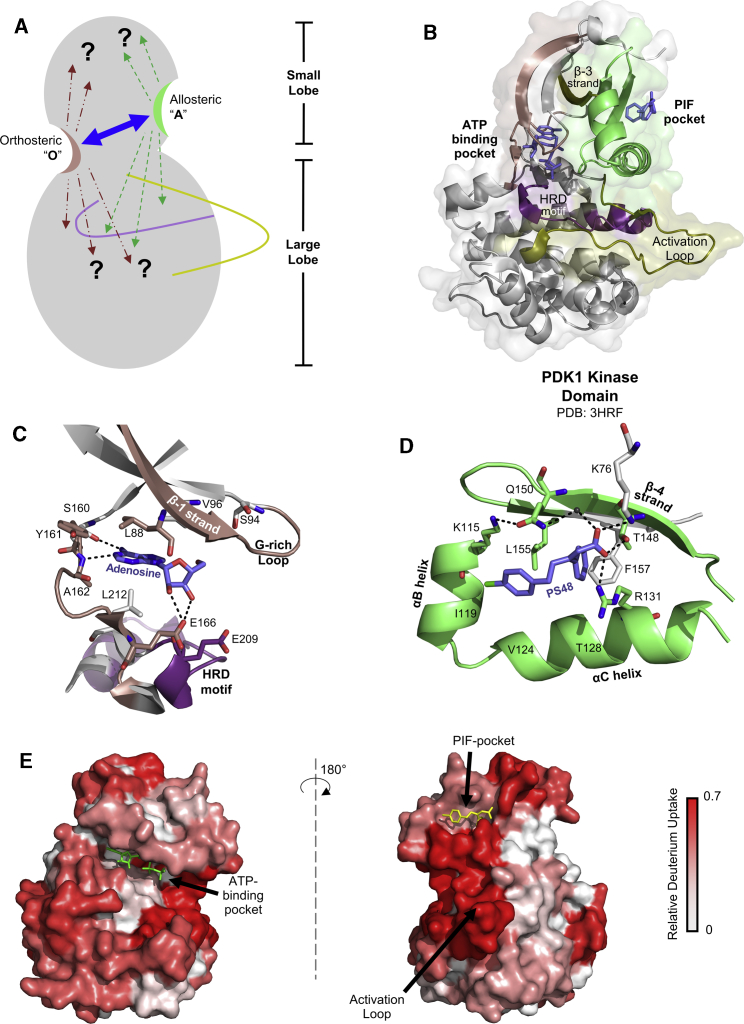
Figure 1.
Basis for allosteric crosstalk between the two noncontiguous ligand sites of PDK1 kinase. (A) Kinase domain of PDK1 in cartoon representation showing the N- and C-terminal lobes and loops critical for catalysis of phosphotransfer activity: HRD motif loop (purple) and activation loop (olive). Two ligand binding sites at the small lobe are marked “O” for the orthosteric ATP-binding pocket and “A” for its corresponding allosteric PIF-pocket. A double-headed arrow between them indicates bidirectional allostery. The interplay between PDK1 dynamics and the allosteric effects of ligand binding at either site is unknown. (B) Structure of PDK1 bound to ATP and PS48 (PDB: 3HRF (38)) showing ATP- (salmon) and PIF-pocket sites (green). Binding ligands are blue. Important secondary structure elements of the kinase are highlighted: β-1 strand, G-rich loop and the hinge region at the ATP-binding pocket (salmon), the HRD motif loop (purple), β-3 strand and activation loop (olive), and PIF-pocket (αB, αC, and β-4) (green). Shown is a close-up of ATP- (C) and PIF-pocket (D), with important orthosteric contacts at the ATP-pocket (using adenosine-bound PDK1 (PDB: 5LVN)) and PIF-pocket (using PDK1-ATP-PS48 complex structure (PDB: 3HRF)). (E) Deuterium exchange heatmaps showing relative fractional deuterium uptake (data uncorrected for back exchange) in apo-PDK1 ranging from 0 to a maximum of 0.7 (PDB: 3HRF). Shown is a 180° view of apo-PDK1 heatmap at deuterium exchange time (t = 1 min). Ligands are shown only to demarcate the two pockets. To see this figure in color, go online.
In this study, we have mapped the allosteric coupling between ATP- and PIF-pockets in PDK1. Our results reveal that HDXMS offers a quantitative readout of synergism in intraprotein allosteric networks, i.e., enhanced changes in conformation and dynamics arising from concurrent binding of ligands at two sites. Furthermore, we uncover a kinase-specific allosteric mechanism in which a conserved lysine on β-3 strand (Lys72 in protein kinase A (42)) functions as an integrator node for convergence and integration of allosteric signals from distinct ligand-binding sites.
Materials and Methods
Key materials
Ni-NTA Sepharose resin was from GE Healthcare (Chicago, IL). Insect cell expression system and all the insect-cell-related material were from Invitrogen (Thermo Fisher Scientific, Waltham, MA). Site-directed mutagenesis was performed using a QuikChange strategy (Agilent Technologies, Santa Clara, CA). PIFtide (REPRILSEEEQEMFRDFDYIADWC) is a peptide derived from the hydrophobic motif of PRK2 (a PDK1 substrate) synthesized by Pepscan (Lelystad, the Netherlands). Deuterium oxide (D2O) for HDXMS was from Cambridge Isotope Laboratories (Tewksbury, MA). Water, acetonitrile, and methanol for liquid chromatography coupled to mass spectrometry (LC-MS) were from Thermo Fisher Scientific. Trifluoroacetic acid was from Fluka Biochemica (Buchs, Switzerland). All other reagents and chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Purification of PDK1 protein kinase
Although dimerization of full length PDK1 has been described (43, 44, 45), there is no report suggesting dimerization or oligomerization of the PDK1 catalytic domain by itself. We have previously purified PDK1 catalytic domain (residues 50–359) and reported on its properties extensively (37,38,46), and it elutes as a single Gaussian peak corresponding to the estimated molecular weight of the monomer. However, it crystallizes in crystal packing I, with the PIF-pocket occupied by Tyr288 of a neighboring molecule (46). To avoid this potential crystallization artifact, the PDK1 protein used in this study is PDK1 50–359 [Y288G,Q292A], which elutes as a single monomer with an unambiguous Gaussian elution profile in size-exclusion chromatography and crystallizes in a different packing (38). Adenosine, PIFtide and other compounds tested produced the same effects on wildtype and PDK1 [Y288G,Q292A] mutant (38,41), indicating that this motif does not impact the effects described in this study.
PDK1 50–359 [Y288G,Q292A] was purified as previously described (38). In brief, the His-tagged recombinant protein was purified through Ni-NTA affinity chromatography, followed by a 4-h dialysis in buffer A (50 mM Tris-HCl (pH 7.4), 500 mM NaCl, PMSF, and 0.02% β-mercaptoethanol) to remove excess imidazole. The hexahistidine tag was then cleaved by overnight incubation with TEV protease at 4°C and affinity purified through a second Ni-NTA resin to recover the flowthrough consisting of cleaved PDK1 50–359 alone. After a final gel filtration chromatography step using buffer B (20 mM Tris-HCl (pH 7.4), 500 mM NaCl, and 1 mM DTT), the purified protein fractions were pooled and concentrated to 1.1 mg/mL (31 μM), aliquoted, flash frozen in liquid nitrogen, and stored at −80°C until HDXMS analysis. The gel filtration chromatography profile for purification of PDK1 is shown in Fig. S1.
Amide hydrogen-deuterium exchange
Amide hydrogen-deuterium exchange reactions were initiated by diluting apo PDK1 (unliganded) and under various ligand-bound conditions in buffer B reconstituted in D2O (pHread ∼7.4). Ligand concentrations used ensured >98% ligand bound to PDK1. PIFtide stock solution (400 μM) was prepared in 50 mM Tris (pH 7.4), adenosine stock solution (20 mM) was prepared in water. Of these stocks, 1 μL PIFtide and 2 μL adenosine were used per deuterium exchange reaction to achieve a final reaction concentration of 10 μM and 1 mM, respectively. Deuterium exchange was initiated by dilution of 1 μL of PDK1 with 39 μL of dilution buffer, prepared using buffer B reconstituted in 99% D2O (36 μL), 3 μL ligand stock solution in buffer B in H2O, to achieve a final D2O concentration of 90% in all the reactions. This premixing of ligands with buffer B in D2O ensured protein was not exposed to high concentrations of ligands before dilution in deuterium exchange buffer.
Deuterium exchange reactions were carried out for apo, single-ligand-bound (adenosine, PIFtide) and two-ligand-bound (adenosine and PIFtide) PDK1 for times, t = 0.5, 1, 5, 10, and 100 min as they span both fast and slow deuterium exchange regimes (47). All reactions were carried out at 30°C, and quenched using 10 μL prechilled solution (0.5% trifluoroacetic acid, 5 M GnCl) to achieve a pHread of 2.5. All deuterium exchange reactions were carried out in triplicates, along with 10 undeuterated reactions for peptide identification.
LC-MS
Quenched deuterium exchange reactions were immediately injected into nanoUPLC HDX Sample Manager (Waters Corporation, Milford, MA) for online pepsin digestion using Poroszyme prepacked pepsin column (Applied Biosystems, Foster City, CA), using 0.1% formic acid in LC-MS water at 100 μL/min. The eluted peptides were then trapped and desalted by a VanGuard C-18 column (Waters Corporation), followed by reverse-phase separation using ACQUITY 2.1 × 5 mm BEH C-18 column (Waters Corporation) using a 8–40% gradient of 0.1% formic acid in acetonitrile flowing at 40 μL/min using nano-ACQUITY binary solvent manager (Waters Corporation).
Eluted peptides were analyzed by Synapt G2 Si mass spectrometer (Waters Corporation) operating in positive ion mode using an MSE acquisition method (48,49). The mass spectrometer was continuously calibrated using 200 fmol/μL Glu-Fibrinopeptide B standard flowing at 1 μL/min. All other parameters for pepsin digestion and LC-MS were same as described (50).
HDXMS data analysis
Peptides of PDK1 in mass spectrometry data were identified from 10 separate pepsin digests of aqueous PDK1 (undeuterated) by ProteinLynx Global Server 3.0 software (Waters Corporation), searching against a human PDK1 sequence database from UniProt (UniProt: O15530) residues 50–359 along with substitution of two residues to account for Y288G and Q292A mutations. We then applied the following filters for PDK1 reporter peptides for HDXMS: minimal signal intensity, 5000; maximal sequence length, 25 residues; minimal fragmentation products per amino acid, 0.2; mass tolerance, 10 ppm; and present in a minimum of three out of 10 replicate digests. Spectral peak assignment and calculation of deuterium uptake was performed using DynamX 3.0 software (Waters Corporation, Milford, MA). Only 35 peptides with high signal/noise ratios were analyzed yielding a total 89% sequence coverage and an average redundancy of 1.56. Deuterium uptake was calculated by DynamX 3.0 as the shift in centroids of individual spectral profiles relative to that of the undeuterated peptide. This difference in centroid in each deuterium-exchanged peptide represents the average increase in deuterium exchange with time. The deuterium uptake values reported in this study are uncorrected for back exchange, with ∼30% average back exchange for our experimental setup (51). A summary of the data analysis statistics is described in Table S2. The list of peptides, charge states and deuterium exchange values for the peptides in apo, individually ligand-bound and dual-ligand-bound states are shown in Tables S3 and S4.
Deuterium exchange time and difference plots were generated by DynamX 3.0. Deuterium difference plots compare deuterium uptake values for each peptide from one state to the other as mentioned and are plotted against individual peptides listed from N- to C-terminus along the x axis. Because deuterium uptake values for each peptide are an average of three replicates, the maximal standard errors for all peptides are shown in gray in all deuterium exchange difference plots. All difference plots follow the convention of difference between ligand-bound minus ligand-free states, respectively, in which negative difference at the site for ligand indicated in bold (O for adenosine, A for PIFtide) represents ligand binding induced protection from deuterium exchange.
Structure-based statistical mechanical model of allostery
The structure-based statistical mechanical model of allostery (SBSMMA), which accounts for the causality of allosteric communication upon perturbations such as ligand binding and/or mutations (10,12,52), was used in this study. Here, allosteric signaling caused by binding of adenosine PIFtide and, upon perturbation at Lys111 (identified from HDXMS mapping of PDK1 allostery), was quantified in terms of the energetics using the AlloSigMA web-server (53).
The crystal structure of PDK1 kinase domain complexed with both ATP and PS48 (Protein Data Bank, PDB: 3HRF) was used to construct the Cα-harmonic models for both unperturbed (0) and perturbed (P) states of the protein domain. The energy function for the unperturbed state is given by
| (1) |
where represents the 3N-dimensional vector of displacements of Cα-atoms relative to the reference structure. The distance between Cα-atoms i and j and the corresponding distance in the reference structure are and , respectively. The distance-dependent force constant ∼ stipulated a global distance cutoff at 25 Å (54). Perturbation mimicking ligand binding to a site S is modeled by an additional harmonic restraining term to the force constants of all pairs of residues constituting S. Similarly, alteration of the local interactions between a single residue m and its neighbors j is simulated by increasing or decreasing the force constants, which gives
| (2) |
Next, two sets of low-frequency normal modes and , characterizing the configurational ensembles of the corresponding states are obtained. The normal modes are then used to calculate the per-residue allosteric potential for each state, which evaluates the total elastic work experienced by a residue i as a result of changes in the displacements of the neighbors within dc = 11 Å in each state, yielding
| (3) |
where represents a configurational state and . In the last step, briefly, integrating over the ensemble of all configuration states of a residue i allows calculation of the corresponding free energies, and therefore the change in free energy upon a perturbation P is
| (4) |
More detailed descriptions can be found in previous work (12,52). To estimate the background-free effect, the per-residue allosteric modulation is calculated by the deviation of the free energy change from its average value over all residues of the protein chain. A positive allosteric modulation indicates an increase of configurational work exerted on residue i due to a perturbation P, potentially leading to conformational changes, whereas a negative one may prevent conformational changes via stabilization of the residue. As mentioned above, the allosteric response due to the weakening or a strengthening of the local interactions of single residues can be quantified using the SBSMMA. To evaluate the allosteric effect emanating from Lys111, the modulation range , which provides a generic description of the maximal strength of allosteric signaling (12), was obtained from the difference in modulations caused by the opposite scenario. These per-residue Δhi-values were used to generate plots of configurational work against residue numbers arranged along the x axis (Figs. 6 E and S1, A and B). These values were also used to generate SBSMMA heat maps (Fig. 6 D and S1, C and D). Allosteric responses on the level of sites can be obtained by averaging the modulation values of all residues belonging to a site of interest (Table S1).
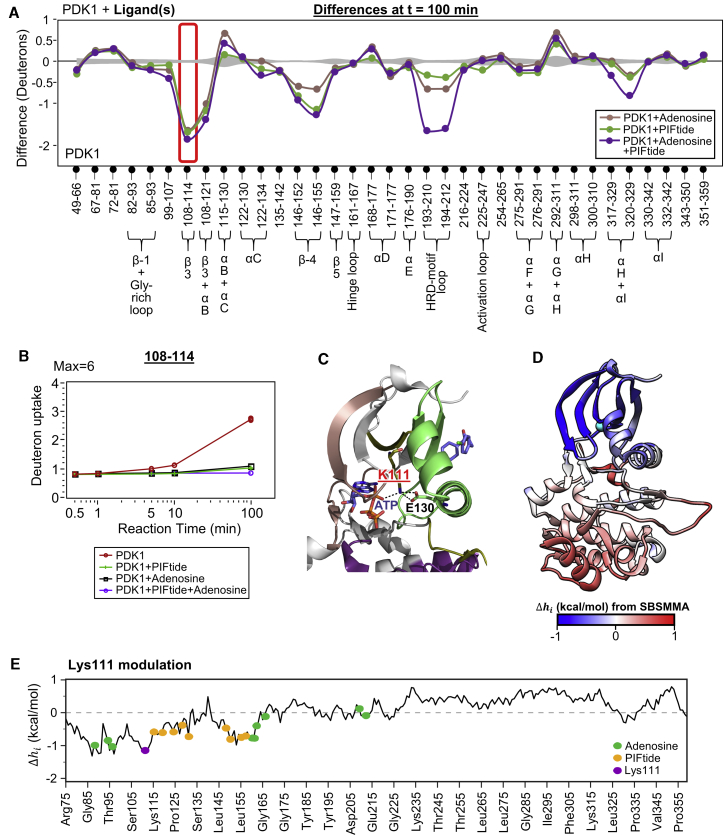
Figure 6.
Lys111 integrates allosteric signals from both ATP- and PIF-pockets. (A) Difference plot comparing protein-wide differences in deuterium exchange between ligand-bound and apo PDK1 at t = 100 min, with indicated ligand(s) bound as per key. Axes and error curves are similar to the difference plots in Fig. 2. Peptide 108–114, highlighted in red box, spans the β-3 strand and shows significant protection only at 100 min. (B) Semilog deuterium exchange plot of the peptide 108–114 in apo, adenosine-, and PIFtide-bound states, as shown in key below. Presence of either ligand decreases exchange in the peptide after 100 min. (C) A network of interactions between the conserved Lys111 on β-3 strand and conserved Glu130 of αC helix of the PIF-pocket, as well as the α-phosphate of ATP in PDK1 (PDB: 3HRF). (D) Heatmap of the allosteric configurational work due to Lys111 modulation, as calculated using SBSMMA, with blue indicating negative , whereas red is positive. (E) A plot of Δhi output from SBSMMA for Lys111 perturbation, with along y axis and PDK1 residues along x axis. See also Tables S3 and S4. To see this figure in color, go online.
Results
Mapping allosteric signatures of adenosine and PIFtide binding to PDK1
PDK1 contains two spatially distinct ligand-binding sites (38) on the small N-lobe of the kinase (Fig. 1, A and B), labeled “O” for orthosteric ATP/adenosine binding site and “A” for an allosterically coupled distal site (PIF-pocket). In turn, “A” corresponds to the orthosteric site for PIFtide and “O” is its known allosterically coupled distal site. Orthosteric contacts for ligand binding at either pocket (Fig. 1, C and D) are defined based upon distance constraints (typically ≤5 Å) obtained from structural coordinates. Such distance constraints do not reflect the true energetic contributions of individual residues for maintenance of protein-ligand complexes in solution. The energetic contributions to protein-ligand and protein-protein interfaces are mediated by smaller subsets of the total number of proximal contacts, as shown by alanine scanning mutagenesis (55) or HDXMS (20,56). To map key PDK1 residues that contribute to the interface with adenosine or PIFtide at their respective noncontiguous binding sites, we carried out comparative HDXMS of apo and ligand-bound PDK1.
HDXMS of apo PDK1 at deuteration times, t = 0.5–100 min were carried out as described in Materials and Methods. Relative deuterium exchange in all peptides were mapped onto the structure of PDK1-ATP-PS48 complex (PDB: 3HRF (38)) to generate a deuterium exchange heatmap (Fig. 1 E). Loop regions expectedly showed greater relative deuterium exchange in apo PDK1, whereas ATP- and PIF-pockets showed both high and low relative deuterium exchange at all time points of deuterium exchange.
A deuterium exchange difference plot (Fig. 2 A) shows changes in deuterium exchange across PDK1 upon adenosine binding with decreased deuterium exchange broadly across all identified orthosteric contact residues (“O”). Deuterium exchange time plots for representative peptides are shown (Fig. 2, B–D).
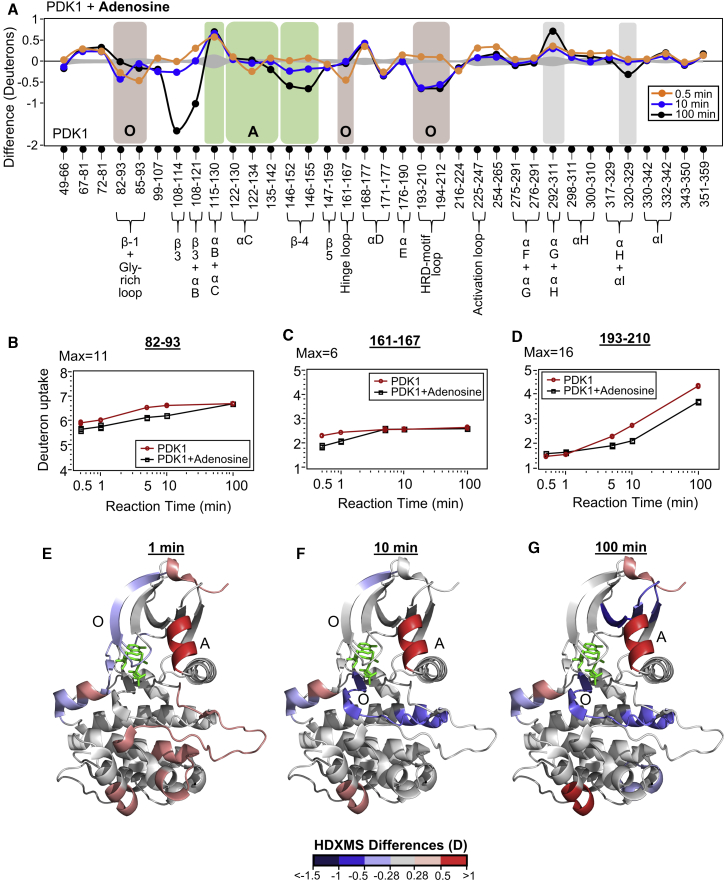
Figure 2.
Orthosteric and allosteric changes upon adenosine binding. (A) Deuterium exchange difference plot (deuterons versus peptic cleavage peptides represented from the N- to C-terminus) comparing differences in deuterium exchange between adenosine-bound and apo PDK1 at different deuterium exchange time points, as indicated in key. Each difference value is an average of three replicates, with standard errors for each peptide plotted as a gray curve along the y = 0 line. Peptides spanning residues lining the ATP-pocket (“O”) and PIF-pocket (“A”) are boxed brown and green, respectively. (B–D) Semilog deuterium uptake plots (deuterons exchanged with time (minutes)) for selected peptides spanning residues lining the ATP-pocket for apo (red) and adenosine-bound (black) PDK1. (E–G) Deuterium exchange differences between adenosine-bound and apo-PDK1 at t = 1, 10, and 100 min, respectively, mapped to the structure of PDK1-ATP-PS48 complex (PDB: 3HRF). Decreased and increased exchange are indicated in gradients of blue and red, as per key. See also Fig. S2; Tables S1 and S3. To see this figure in color, go online.
Allostery upon adenosine binding was detected in the distal “A” site, with increased deuterium exchange in peptide 115–130 (αB + αC helix), but no changes in 122–134 (αC helix, Fig. 2 A). Increased deuterium exchange seen at αB helix is consistent with molecular dynamics simulations (41). The β-4 strand peptide 146–155 and part of the “A” site showed decreased exchange upon adenosine binding (Fig. 2, A and E–G). Importantly, a large decrease in HDXMS at peptide 108–114 was observed after 100 min deuterium exchange, and its significance is discussed at the end of this section. Increased deuterium exchange was also observed at additional distal loci in peptides 67–81, 168–177, activation loop (225–247), and 254–265. The C-terminal GHI-subdomain of the kinase also presented two allosteric loci with altered deuterium exchange in peptides 292–311 and 317–329 (Fig. 2 A, boxed gray). Overall, five peptides spanning the orthosteric ATP-binding site showed decreased deuterium exchange due to adenosine binding, one peptide of PIF-pocket showed increased and two peptides showed decreased deuterium exchange. At sites distal to both pockets, one peptide each showed increased and decreased exchange in the adenosine-bound state. Importantly, adenosine binding resulted in changes in deuterium exchange in its known “A” site and in catalytically important regions of the kinase including the β3 strand. The C-lobe GHI-subdomain was additionally identified as a new allosteric hotspot for adenosine.
Interactions of PIFtide with the PIF-pocket of PDK1 were similarly mapped by HDXMS. PIFtide-bound PDK1 showed deuterium exchange protection at all three subsites of PIF-pocket (“A,” Fig. 3 A). The αB-αC helices with high intrinsic deuterium exchange, as well as the β-4 strand showed decreased deuterium exchange in PIFtide-bound PDK1 (Fig. 3, B–D). PIFtide binding also resulted in decreased deuterium exchange at the HRD-motif loop (193–212) of the ATP-binding pocket, with no changes at other subsites forming the ATP-pocket (Fig. 3, A and E–G). This decrease in deuterium exchange at ATP-pocket by PIFtide binding is an opposite effect to the increased exchange in the αB helix of PIF-pocket elicited by adenosine binding and highlight an asymmetry in the allosteric effects observed with these two cooperative ligands. Together, these results indicated that for adenosine and PIFtide, all subsites of the two ligand-binding pockets were not equivalently allosterically coupled as measured by deuterium exchange. For adenosine and PIFtide, only the HRD-motif loop of the ATP-pocket, αB helix, and β-4 strand of the PIF-pocket showed corresponding allosteric changes in deuterium exchange.
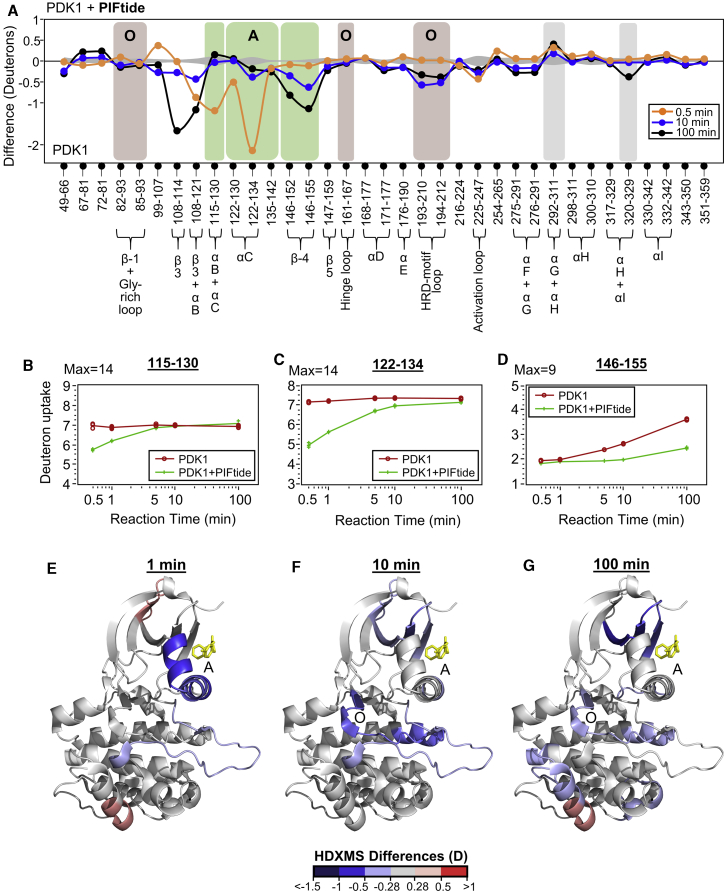
Figure 3.
Orthosteric and allosteric changes upon PIFtide binding. (A) Deuterium exchange difference plot (deuterons versus peptic cleavage peptides displayed from the N- to C-terminus) comparing differences in deuterium exchange between PIFtide-bound and apo PDK1 at different deuterium exchange time points, as indicated in key. Axes, time points, error curves, and highlighted regions of interest are similar to Fig. 2A. (B–D) Semilog deuterium uptake plots showing deuterons exchanged with time (minutes) for selected peptides spanning residues lining the PIF-pocket for apo (red) and PIFtide-bound (green) PDK1. (E–G) Deuterium exchange differences between PIFtide-bound and apo-PDK1 at t = 1, 10, and 100 min, respectively, mapped to structure of PDK1-ATP-PS48 complex (PDB: 3HRF). Regions showing decreased exchange are indicated in shades of blue, whereas those showing increased exchange (positive differences) in shades of red, as per key. See also Fig. S2; Tables S1, S3, and S4. To see this figure in color, go online.
Changes in deuterium exchange due to PIFtide binding were also observed in β-3 strand (108–114, discussed in a later section), the activation loop (225–247), as well as the two GHI-subdomain loci (292–311 and 320–329). Interestingly, both adenosine and PIFtide binding elicited similar changes in deuterium exchange at the two GHI-subdomain loci. Overall, PIFtide binding resulted in decreased deuterium exchange at five peptides of the PIF-pocket and two peptides of the ATP-pocket. At GHI-subdomain sites distal to both pockets, one peptide each showed increased and decreased deuterium exchange in PIFtide-bound PDK1.
To model the energetics of allosteric coupling between the two noncontiguous ligand-binding sites of PDK1, we applied the SBSMMA, which describes allosteric communication generated by perturbations such as ligand binding and/or mutations (12,52). The SBSMMA models the protein as a network of spheres positioned at the C α-atoms and analyzes the normal modes of their harmonics using a distance-dependent force constant, without taking into consideration residue type or any specific interactions. The readout of allosteric response from SBSMMA is in the form of configurational work, (kcal/mol), calculated for each residue i of the protein. Positive modulation, i.e., , is indicative of conformational changes in response to the allosteric perturbation. Indeed, conformational changes taking place as a result of the allosteric signaling can lead to changes in the structural dynamics and in the ensemble distribution of sampled conformations. In the case of a shift in the conformational ensemble toward more closed structures at the site of HDXMS measurement, decreased deuterium exchange can be expected, whereas a shift toward more open structures would result in a localized increase in deuterium exchange. On the other hand, a negative is an indicator of decrease in configurational work, i.e., allosteric stabilization that may result in either a similar or reduced deuterium exchange.
We performed SBSMMA analysis for adenosine and PIFtide binding to PDK1 as described in the Materials and Methods, and observed that adenosine binding generated negative -values for β-4 strand (−0.23 kcal/mol), with only minor changes at αB and αC helices (0.05 and −0.11 kcal/mol, respectively) (Fig. S2 A; Table S1). PIFtide binding resulted in a negative for β-1 strand and Gly-rich loop (−0.65 kcal/mol) and a positive at the hinge loop and the HRD-motif loop (0.11 and 0.27 kcal/mol respectively) (Fig. S2 B; Table S1). Furthermore, both ligands resulted in strong allosteric effects at the large lobe of PDK1, including the GHI-subdomain loci (Table S1). Importantly, results from SBSMMA aligned well with HDXMS results. Heat maps of these -values (Fig. S2, C and D) confirm these loci as common hotspots of allostery by these ligands.
αB and αC helices, HRD-motif loop, and GHI-subdomain are hotspots of combinatorial allostery
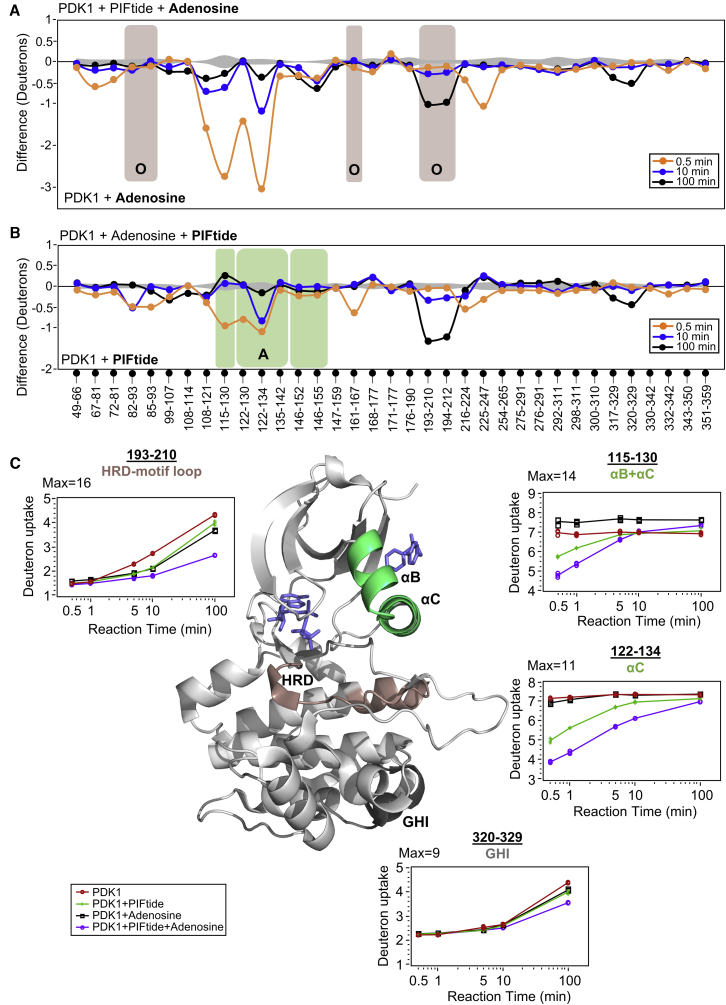
We next mapped orthosteric and allosteric effects in dual-ligand-bound PDK1 (adenosine and PIFtide) by HDXMS. A deuterium exchange difference plot of PDK1-adenosine-PIFtide ternary complex with apo-PDK1 (Fig. S3) showed deuterium exchange protection at both ligand pockets as well as at one of the GHI-subdomain loci. A deuterium exchange difference plot of PDK1-adenosine-PIFtide ternary complex with PDK1-adenosine complex (Fig. 4 A) showed decreased exchange at the HRD-motif loop (193–212) alone, whereas the other “O” subsites β-1 strand and Gly-rich loop (82–93) and hinge loop (161–167) showed no change. Difference plot comparing exchange in the ternary complex with PDK1-PIFtide complex (Fig. 4 B) showed decreased exchange at αB-αC helices (115–134) of the PIF-pocket (“A”), whereas β-4 strand peptides (146–155) showed no difference.
Figure 4.
Combinatorial allostery in PDK1 upon ligand binding at both ATP- and PIF-pockets. (A–B) Deuterium exchange difference plots of PDK1 bound to both PIFtide and adenosine relative to singly liganded PDK1. Axes, time points, error curves, and highlighted regions of interest are as per Fig. 2A. Decreased deuterium exchange at 193–212 (ATP-pocket, salmon) in (A) and 115–134 (PIF-pocket, green) in (B) reflect cooperatively enhanced binding of adenosine and PIFtide, respectively, in the presence of the corresponding cooperative ligand. (C) Semilog deuterium uptake plots for representative peptides that show enhanced protection from deuterium exchange in presence of both ligands. Red, green, black, and purple plots represent deuterium exchange in apo-, PIFtide-bound, adenosine-bound, and both (adenosine + PIFtide)-bound PDK1, respectively, as indicated in the key. Structure of PDK1-ATP-PS48 complex (PDB: 3HRF) in the center highlights the loci revealing combinatorial allosteric effects. See also Fig. S3; Tables S3 and S4. To see this figure in color, go online.
Furthermore, one of the two GHI-subdomain loci (320–329) showed enhanced deuterium exchange protection due to concurrently bound ligands relative to individual-ligand-bound states, indicating an integrative combinatorial effect in the adenosine and PIFtide-bound state (Fig. 4 C).
Overall, a comparison of deuterium exchange in the dual-liganded complex relative to adenosine-bound PDK1 indicated additional decreases in exchange in two peptides alone of “O” site. In contrast, all peptides at the “A” site showed a decreased exchange due to PIFtide binding. At sites distal to both pockets, five peptides showed larger decreases in exchange. Similarly, for ternary complex relative to PIFtide-bound PDK1, three peptides of “A” site showed decreased exchange, and five peptides of “O” site showing decreased exchange due to adenosine binding. However, only three peptides distal to both pockets showed a negative difference.
Allosteric coupling factor for quantitation of synergistic allosteric effects
A comparison of deuterium exchange differences between doubly liganded versus singly liganded PDK1 showed certain loci where effects of both ligands were more pronounced than at other spots. To map and quantitate synergism in the allosteric effects of the two cooperative ligands (adenosine and PIFtide), we assessed any added deuterium exchange protection observed with both ligands bound, relative to that seen in singly liganded states. We postulate that deuterium exchange in the doubly liganded state offers a direct readout of the synergistic allosteric effects because of combined effects of both ligands. We define a term, allosteric coupling factor (ACF), to denote the deuterium exchange difference in the doubly liganded state (relative to the apo state) (C) minus the sum of deuterium exchange differences in singly bound states (relative to the apo state):
ACF offers a readout of allosteric synergism across different PDK1 peptides with differences in deuterium exchange (O, A, and C) relative to apo-PDK1. Negative and positive ACF, in turn, denote synergistic stabilization or destabilization at any peptide.
Positive ACF-values were not observed for any PDK1 peptide indicating no synergistic destabilization in doubly liganded PDK1. Peptides of PDK1 that showed synergistic allosteric stabilization with PIFtide and adenosine are summarized in Table 1, ranked in decreasing ACF for all deuterium exchange times. Broadly, ACF-values followed the same pattern with time as in singly bound states where, in high-exchanging peptides, the magnitude of ACF was most negative at early time points and low-exchanging peptides showed greatest magnitude ACF-values at longer deuterium exchange times.
Table 1.
Allosteric Synergism across PDK1-Adenosine-PIFtide Complex
| Peptidea | Siteb | MEAc | Allosteric Coupling Factord C – (O + A) |
||||
|---|---|---|---|---|---|---|---|
| 0.5 min | 1 min | 5 min | 10 min | 100 min | |||
| 115–130 | αB | 14 | −1.5∗ | −1.4 | −0.9 | −0.6 | −0.4 |
| 122–134 | αC | 11 | −0.9 | −1.2∗ | −1.1 | −0.8 | −0.1 |
| 122–130 | αC | 7 | −0.9∗ | −0.8 | −0.1 | 0.0 | −0.1 |
| 108–121 | β3 + αB | 13 | −0.7∗ | −0.7 | −0.6 | −0.3 | 0.8 |
| 193–210 | HRD | 16 | −0.2 | −0.2 | 0.2 | 0.3 | −0.7∗ |
| 194–212 | HRD | 17 | −0.1 | −0.1 | 0.2 | 0.3 | −0.6∗ |
| 67–81 | N-term | 9 | −0.5∗ | −0.5∗ | −0.2 | −0.3 | −0.3 |
| 72–81 | N-term | 6 | −0.4 | −0.5∗ | −0.3 | −0.2 | −0.3 |
| 216–224 | DFG | 8 | −0.3 | −0.4∗ | −0.1 | −0.1 | 0.1 |
| 317–329 | GHI | 12 | −0.1 | −0.2 | −0.1 | −0.1 | −0.3∗ |
| 146–155 | β4 | 9 | −0.3∗ | −0.1 | 0.2 | 0.2 | 0.5 |
| 146–152 | β4 | 6 | −0.2∗ | −0.1 | 0.1 | 0.2 | 0.5 |
| 320–329 | GHI | 9 | −0.1∗ | −0.1 | 0.1 | −0.1 | −0.1∗ |
N-term, N-terminus.
PDK1 peptides showing synergistic allostery ranked in increasing order of negative ACF. The lowest ACF-values for each peptide across deuterium exchange time points are marked by asterisks.
Secondary structural loci on PDK1 kinase
Number of maximal exchangeable amide (MEA) hydrogens in the peptide, calculated by subtracting the number of Prolines and 1 (for the N-terminal residue) from the total number of residues in the peptide.
ACF, defined as the composite difference in deuterium exchange protection in doubly-bound PDK1 (C) minus the added decreased exchange in singly adenosine (O) and PIFtide (A) bound PDK1 relative to apo-PDK1. Negative values reflect stabilizing effects due to concurrent binding of two ligands.
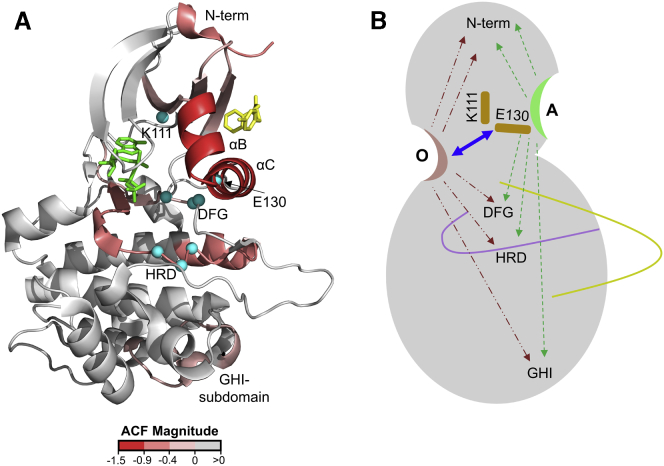
Overall, peptides spanning αB and αC helices of PIF-pocket showed the highest magnitude of negative ACF-values, indicative of strong synergism (−1.5 for peptide 115–130 and −1.2 for 122–134), followed by catalytic/HRD-motif loop of the ATP-pocket (−0.7 for 193–210). N-terminal peptides (−0.5 for peptide 67–81) and loop containing the catalytic DFG-motif (−0.4 for peptide 216–224) were additional hotspots of synergistic allostery based on magnitude of ACF. Furthermore, we found that β-4 strand of PIF-pocket (−0.3 for peptide 146–155) and the GHI-subdomain locus (−0.3 for 317–329, −0.1 for peptide 320–329) showed ACF-values of smallest magnitude, and therefore represent regions with smallest synergistic effects. Importantly, ACF magnitudes correlated with the most catalytically critical regions of the kinase: αB-αC helices, β-3 strand, HRD and DFG motifs (Fig. 5, A and B; (57,58)).
Figure 5.
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.bpj.2020.09.019
Hotspots of synergistic allostery detected by ACF span crucial kinase motifs. (A) Heat map of synergistic allosteric effects due to dual adenosine and PIFtide binding to PDK1 as calculated by ACF mapped onto PDK1-ATP-PS48 complex (PDB: 3HRF), with ligands in green and yellow. The extent of synergism is denoted by shades of red as per key (more negative ACF equals greater synergism). Residues spanning catalytically crucial motifs of the kinase that showed synergistic effects are shown as cyan spheres. (B) Cartoon representation of PDK1 showing the observed combinatorial bidirectional allosteric effects upon dual ligand binding at the two pockets, catalytic sites, and distal GHI-subdomain locus. See also Tables S3 and S4. To see this figure in color, go online.
Lys111, a critical lynchpin residue for integration of allosteric signals
Deuterium exchange plots for adenosine and PIFtide singly bound to PDK1 showed a sharp drop in deuterium exchange for the β-3 strand peptide (108–114) at t = 100 min alone (Figs. 2 A and 3 A). A combined difference plot, showing deuterium exchange differences at t = 100 min for adenosine, PIFtide, and both ligand-bound PDK1 relative to apo-PDK1 (Fig. 6 A), highlights this reduction in HDX. A deuterium uptake plot for this peptide (Fig. 6 B) shows slow exchange up to t = 10 min (∼1.1 D) with a jump in exchange after 100 min (∼2.7 D). After 100 min exchange, protection was observed with both singly bound adenosine and PIFtide (1.1 and 1.0 D, respectively). In the doubly liganded state, a bigger decrease in exchange was seen (0.8 D).
The β-3 strand, an important element in all kinases, is sandwiched between the two ligand-binding sites within the small lobe of the kinase. It contains a conserved lysine (Lys111) that mediates an important salt bridge interaction with the α-phosphate of ATP on one side and a conserved glutamate (Glu130) of the αC helix of PIF-pocket on the other (Fig. 6 C). This salt bridge has been clearly described in PKA (59). Our results suggest that even in the absence of the phosphate moieties of ATP, e.g. with adenosine, this conserved lysine and its salt bridge with Glu130 connected bidirectional allosteric relays between the “O” and “A” sites, functioning as an integrator node.
To further analyze the allosteric links between Lys111 and the two noncontiguous ligand-binding sites, we performed single-residue perturbation analysis for Lys111 using the AlloSigMA web-server as described in Materials and Methods. The results showed that perturbation of Lys111 induced strong allosteric effects at both ATP- and PIF-pockets (Fig. 6, D and E) in the form of negative modulation ( and ). This indicated that allosteric communication via Lys111 was capable of promoting overall stabilization of both pockets. Furthermore, the model predicted strong allosteric effects at the distal GHI-subdomain locus originated by Lys111 perturbation were similar to those observed with both PIFtide and adenosine. These results strongly support the role of Lys111 as an integrator node for allosteric signaling across the kinase domain of PDK1.
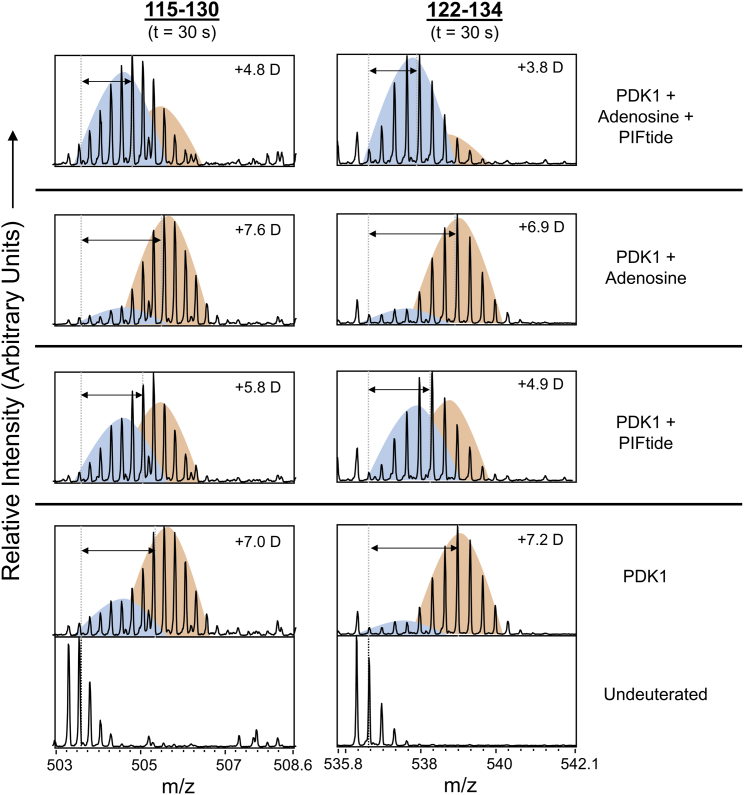
Disorder-to-order transitions at αB-αC helices for kinase activation
A closer inspection of mass spectral profiles of deuterium-exchanged peptides spanning the αB and αC helices (115–130 and 122–134) of the PIF-pocket revealed a characteristic bimodal isotopic distribution with a low-exchanging and a high-exchanging subpopulation (Figs. 7, S4, and S5). This is reflective of EX1 deuterium exchange kinetics and indicative of coordinated folding or unfolding events (13,60). These helices exhibit disordered-like behavior in solution in the apo state (38). Therefore, these two subpopulations correspond to distinct well-folded and disordered states within the conformational ensemble, with a high-exchanging disordered state predominating in apo-PDK1 (Figs. 7, S4, and S5). Orthosteric binding of PIFtide induced a shift in the ensemble toward a more ordered conformation. Adenosine binding resulted in an ensemble shift toward a more disordered state for the peptide 115–130, but not for 122–134. Thus, the allosterically induced disorder due to adenosine binding was confined to αB helix alone. Furthermore, we observed the biggest shift toward the ordered state in the dual-ligand state (Fig. 7). Through these shifts, it is apparent that PIFtide preferentially binds adenosine-bound PDK1, as allosteric destabilization at these helices caused by adenosine enhances PIFtide binding. Together, these results show that the allosteric activation of kinase is driven by a disorder-to-order switch in αB and αC helices, coordinated via the Lys111 integrator node. Increased helical order in αC helix would also stabilize the critical Glu130-Lys111 salt bridge.
Figure 7.
Disorder-order transitions in PDK1 αB and αC helices function as conformational switches for kinase activation. Stacked mass spectral envelope plots for two overlapping peptides, 115–130 and 122–134, spanning αB and αC helices in different states at deuterium exchange time t = 0.5 min. Shift in centroids are relative to the undeuterated (aqueous) peptide. Bimodal isotopic distributions can be resolved into low-exchanging (blue) and high-exchanging states (orange). See also Figs. S4 and S5. To see this figure in color, go online.
Discussion
Although a general understanding of allostery has greatly improved with recent experimental and theoretical advances (7), rational development of allosteric drugs has lagged (61). Further, allostery has been examined in singly ligand-bound states but has not been explored as a base for screening second ligands for more targeted drug discovery. However, doubly liganded states are especially relevant for pharmacological targets such as kinases, which share a conserved ATP-binding pocket and diverse allosteric pockets. Synergistic allosteric modulation using a combination of two ligands presents a potential new starting point for 2-compound combinatorial drug development. HDXMS is ideal for mapping protein-ligand interfaces and dynamics of protein-ligand interactions because it simultaneously offers a peptide-level readout of bidirectional allostery in protein kinases. Using this approach, we have mapped combinatorial effects of adenosine and PIFtide binding at corresponding ligand-binding sites, identified a common distal allosteric GHI-subdomain site that shows synergistic effects and developed a method to quantify the composite allosteric effect across PDK1 to rank the hotspots of allosteric synergism.
Protein kinases are important pharmacological targets (31) and contain conserved ATP/ADP binding pockets regulated by allosteric switches to fine-tune their enzymatic activity (62). Most drug candidates are screened against the unliganded apo protein and compounds identified target the conserved ATP-binding pocket making them prone to cross talk interactions with other kinases causing reduced specificity and potency (31). Synergistic ligands to ATP-mimetics can be used to improve both these attributes (63,64). Synergy or antagonism between the ligands binding at allosterically linked sites can also be utilized to modulate interactions between protein kinases and regulatory effector proteins (65).
PDK1 with its extensive repertoire of site-specific ligands is an excellent model system for studying combinatorial allostery (37,38,41). Ligands targeting noncontiguous sites of PDK1 afforded a reduction in deuterium exchange greater than the sum of reduction generated by individual ligands. ACF derived from these measurements enabled ranking of hotspots of synergistic allostery in PDK1. The greatest synergism was observed at αB helix, αC helix, and the HRD-motif loop sites of the two ligand pockets. A distal locus in the C-terminal lobe at the GHI-subdomain (317–329) also showed allosteric synergism (Table 1), revealing long-range propagation of synergistic allosteric effects. GHI-subdomain plays an important role in interactions with protein substrates and allosteric regulators (58), and synergism at this locus shows that ligands at ATP- and PIF-pockets exert a combined allosteric control at the GHI-subdomain. Furthermore, we identified two additional loci of synergism: regions 67–81 (N-terminus) and 216–224 (DFG-motif loop), that were not readily identifiable from deuterium exchange difference plots. Importantly, sites with negative values of ACF correlated with the catalytically most important elements of kinases (57), highlighting a strong allosteric coupling between the two ligands (Fig. 5).
The analysis presented here has major implications for our fundamental understanding of allostery. Our results show that partially liganded states of an enzyme are partially primed states, and full functional activity requires binding of two ligands at allosterically coupled sites making a singly ligand-bound state, and not the apo state, a preferred starting point for allosteric inhibitor screening/design. Rather than a discrete inactive-active state binary, our results posit allostery in proteins operates through a continuum of conformational substates within a broad ensemble (66). Thus, current theoretical frameworks describing combinatorial allostery, such as one based on discrete Boolean logic and end-states (67), would need to account for synergistically enhanced changes in protein conformation and dynamics.
Cooperative allostery through asymmetric conformational changes
It has been theorized that all cooperative allosteric effects must be bidirectional and symmetrical (68), based on the principle of microscopic reversibility. Bidirectional allosteric coupling between ATP- and PIF-pockets of PDK1 is well established (38,41). Interestingly, our results revealed that cooperative ligands binding at these sites induced asymmetrical changes in dynamics at their corresponding allosteric hotspots: PIFtide induced stabilization at HRD-motif loop of the ATP-pocket, and adenosine induced increased disorder at αB helix of the PIF-pocket (Figs. 2, 3, and 6) necessary for enhanced PIFtide binding through optimal positioning of residues from PIFtide. Moreover, these results also indicated that not all orthosteric subsites are allosterically coupled. Our results therefore provide the first experimental evidence of asymmetric changes in structural dynamics between allosterically linked sites.
Proteins as soft matter for allosteric propagation
A more nuanced view of allostery incorporating both conformational as well as dynamics changes has come to the forefront in the last two decades (6,7). It has been proposed that multiple “pathways” of interactions connect two allosterically coupled sites in a protein (69), and binding of ligands at these sites alters the relative contribution of each of these pathways to the allosteric link (70). Our results indicate allosteric coupling between small lobe (ATP- and PIF-pockets) and large lobe (GHI-subdomain) of PDK1 may not be connected entirely by well-defined pathways, as we observed no differences in deuterium exchange across many regions in the large lobe, but only at specific loci in GHI-subdomain (Figs. 2 and 3). Additionally, we observed synergism only at a single GHI-subdomain locus (Figs. 4 and 5). This suggests a hybrid model for allosteric propagation, comprising well-defined directional relays via charged amino acid side chains, as well as more fluid, diffused interactions through the C-lobe’s hydrophobic core. Propagation of allostery through the “bulk” hydrophobic protein core would resemble energy dissipation through soft matter such as hydrogels (71).
Integrator nodes in kinase allostery networks
In addition to its known orthosteric interactions with the α-phosphate of ATP, our results showed that Lys111 plays a critical role in integrating bidirectional allosteric signals in singly adenosine-bound PDK1 (Fig. 6) and doubly liganded states (Fig. 7). This has been independently derived by SBSMMA analysis in which Lys111 perturbation caused nonzero at both ligand sites as well as the GHI-subdomain (Fig. 6 D). In a closely related kinase from the AGC superfamily, PKA, substituting the β-3 strand lysine (Lys72) with histidine, generated a classical “kinase-dead” mutant with altered deuterium exchange kinetics (42). Substitution of this critical lysine with any other amino acid was found to abolish allosteric relays arising from ATP binding. Additionally, we have previously described a compound that binds at the PIF-pocket of another AGC kinase, atypical PKC, and inhibits its kinase activity by affecting the equivalent β-3 strand lysine (72). We therefore postulate that this conserved lysine functions as an integrator node for transduction of coordinated allosteric signals across all protein kinases, as kinases retain a number of conserved elements that allow extraction of fundamental features such as the underlying allosteric networks (26). Both the β-3 strand lysine as well as the αC helix glutamate are conserved among all protein kinases (22), as well as the salt bridge between them (73). Furthermore, the disorder-to-order conformational switch of αC helix (Fig. 7) can be modulated by the Lys111-Glu130 salt bridge for allosteric kinase activation (59).
In summary, our results describe for the first time, to our knowledge, a framework for quantitation of synergistic allostery in doubly liganded kinases, and show that the combinatorial allosteric effects rely on integrator nodes and nonuniform distribution of the hotspots of synergistic allostery. This strategy will enable rational development as well as screening of synergistic pairs of ligands for enhanced therapeutic effects with lower cross-reactivity.
Author Contributions
Conceptualization, A.G. and G.S.A.; methodology, A.G. and G.S.A.; resources, L.Z.F.G.; investigation, A.G.; software, W.V.T. and E.G.; formal analysis, A.G., W.V.T., and E.G.; visualization, A.G. and W.V.T.; writing (original draft), A.G. and G.S.A.; writing (review and editing), A.G., L.Z.F.G., W.V.T., I.N.B., R.M.B., and G.S.A.; supervision: I.N.B., R.M.B., and G.S.A.
Acknowledgments
We are thankful to Dr. Nikhil Kumar Tulsian for help with additional HDXMS experiments at 20°C.
This work was supported by a Competitive Research Proposal grant (R-154-000-B12-281) funded by the National Research Foundation, Singapore awarded to G.S.A. A.G. was supported by an RSB-funded Research Fellowship from Singapore Ministry of Education Academic Research Fund (MOE2019-T1-B24-114) awarded to G.S.A.
Editor: Elizabeth Komives.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.09.019.
Supporting Material
References
- 1.Monod J., Changeux J.P., Jacob F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 2.Nussinov R., Tsai C.J., Ma B. The underappreciated role of allostery in the cellular network. Annu. Rev. Biophys. 2013;42:169–189. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perutz M.F. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 4.Koshland D.E., Jr., Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 5.Cooper A., Dryden D.T. Allostery without conformational change. A plausible model. Eur. Biophys. J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 6.Gunasekaran K., Ma B., Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 7.Motlagh H.N., Wrabl J.O., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iturriaga-Vásquez P., Alzate-Morales J., Reyes-Parada M. Multiple binding sites in the nicotinic acetylcholine receptors: an opportunity for polypharmacolgy. Pharmacol. Res. 2015;101:9–17. doi: 10.1016/j.phrs.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Sieghart W. Allosteric modulation of GABAA receptors via multiple drug-binding sites. Adv. Pharmacol. 2015;72:53–96. doi: 10.1016/bs.apha.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Tee W.V., Guarnera E., Berezovsky I.N. Reversing allosteric communication: from detecting allosteric sites to inducing and tuning targeted allosteric response. PLoS Comput. Biol. 2018;14:e1006228. doi: 10.1371/journal.pcbi.1006228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarnera E., Berezovsky I.N. On the perturbation nature of allostery: sites, mutations, and signal modulation. Curr. Opin. Struct. Biol. 2019;56:18–27. doi: 10.1016/j.sbi.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Guarnera E., Berezovsky I.N. Toward comprehensive allosteric control over protein activity. Structure. 2019;27:866–878.e1. doi: 10.1016/j.str.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Hoofnagle A.N., Resing K.A., Ahn N.G. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 14.Chandramohan A., Krishnamurthy S., Anand G.S. Predicting allosteric effects from orthosteric binding in hsp90-ligand interactions: implications for fragment-based drug design. PLoS Comput. Biol. 2016;12:e1004840. doi: 10.1371/journal.pcbi.1004840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamurthy S., Veesler D., Anand G.S. Distinguishing direct binding interactions from allosteric effects in the protease-HK97 prohead I δ domain complex by amide H/D exchange mass spectrometry. Bacteriophage. 2014;4:e959816. doi: 10.4161/21597073.2014.959816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson B., McConnell P., Cooper J.A. Allosteric coupling of CARMIL and V-1 binding to capping protein revealed by hydrogen-deuterium exchange. Cell Rep. 2018;23:2795–2804. doi: 10.1016/j.celrep.2018.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badireddy S., Yunfeng G., Anand G.S. Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004390. M110 004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Smith D.L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson R.S., Walsh K.A. Mass spectrometric measurement of protein amide hydrogen exchange rates of apo- and holo-myoglobin. Protein Sci. 1994;3:2411–2418. doi: 10.1002/pro.5560031224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand G.S., Hughes C.A., Komives E.A. Amide H/2H exchange reveals communication between the cAMP and catalytic subunit-binding sites in the R(I)alpha subunit of protein kinase A. J. Mol. Biol. 2002;323:377–386. doi: 10.1016/s0022-2836(02)00919-1. [DOI] [PubMed] [Google Scholar]
- 21.Chandramohan A., Tulsian N.K., Anand G.S. Dissecting orthosteric contacts for a reverse-fragment-based ligand design. Anal. Chem. 2017;89:7876–7885. doi: 10.1021/acs.analchem.7b00587. [DOI] [PubMed] [Google Scholar]
- 22.Hanks S.K., Quinn A.M., Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 23.Manning G., Whyte D.B., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 24.Taylor S.S., Radzio-Andzelm E., Narayana N. Catalytic subunit of cyclic AMP-dependent protein kinase: structure and dynamics of the active site cleft. Pharmacol. Ther. 1999;82:133–141. doi: 10.1016/s0163-7258(99)00007-8. [DOI] [PubMed] [Google Scholar]
- 25.Arencibia J.M., Pastor-Flores D., Biondi R.M. AGC protein kinases: from structural mechanism of regulation to allosteric drug development for the treatment of human diseases. Biochim. Biophys. Acta. 2013;1834:1302–1321. doi: 10.1016/j.bbapap.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Kornev A.P., Taylor S.S. Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 2015;40:628–647. doi: 10.1016/j.tibs.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Z., Resing K.A., Ahn N.G. Networks for the allosteric control of protein kinases. Curr. Opin. Struct. Biol. 2006;16:686–692. doi: 10.1016/j.sbi.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Leroux A.E., Schulze J.O., Biondi R.M. AGC kinases, mechanisms of regulation and innovative drug development. Semin. Cancer Biol. 2018;48:1–17. doi: 10.1016/j.semcancer.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Lavoie H., Li J.J., Sicheri F. Dimerization-induced allostery in protein kinase regulation. Trends Biochem. Sci. 2014;39:475–486. doi: 10.1016/j.tibs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Taylor S.S., Ilouz R., Kornev A.P. Assembly of allosteric macromolecular switches: lessons from PKA. Nat. Rev. Mol. Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhullar K.S., Lagarón N.O., Rupasinghe H.P.V. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol. Cancer. 2018;17:48. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alessi D.R., James S.R., Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 33.Stokoe D., Stephens L.R., Hawkins P.T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 34.Dennis P.B., Pullen N., Thomas G. Phosphorylation sites in the autoinhibitory domain participate in p70(s6k) activation loop phosphorylation. J. Biol. Chem. 1998;273:14845–14852. doi: 10.1074/jbc.273.24.14845. [DOI] [PubMed] [Google Scholar]
- 35.Cheng X., Ma Y., Taylor S.S. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busschots K., Lopez-Garcia L.A., Biondi R.M. Substrate-selective inhibition of protein kinase PDK1 by small compounds that bind to the PIF-pocket allosteric docking site. Chem. Biol. 2012;19:1152–1163. doi: 10.1016/j.chembiol.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Engel M., Hindie V., Biondi R.M. Allosteric activation of the protein kinase PDK1 with low molecular weight compounds. EMBO J. 2006;25:5469–5480. doi: 10.1038/sj.emboj.7601416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hindie V., Stroba A., Biondi R.M. Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1. Nat. Chem. Biol. 2009;5:758–764. doi: 10.1038/nchembio.208. [DOI] [PubMed] [Google Scholar]
- 39.Sadowsky J.D., Burlingame M.A., Wells J.A. Turning a protein kinase on or off from a single allosteric site via disulfide trapping. Proc. Natl. Acad. Sci. USA. 2011;108:6056–6061. doi: 10.1073/pnas.1102376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rettenmaier T.J., Sadowsky J.D., Wells J.A. A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc. Natl. Acad. Sci. USA. 2014;111:18590–18595. doi: 10.1073/pnas.1415365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze J.O., Saladino G., Biondi R.M. Bidirectional allosteric communication between the ATP-binding site and the regulatory PIF pocket in PDK1 protein kinase. Cell Chem. Biol. 2016;23:1193–1205. doi: 10.1016/j.chembiol.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Iyer G.H., Garrod S., Taylor S.S. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J. Mol. Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Kang J.A., Jeong S.P., Park S.G. Transition from heterotypic to homotypic PDK1 homodimerization is essential for TCR-mediated NF-κB activation. J. Immunol. 2013;190:4508–4515. doi: 10.4049/jimmunol.1202923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masters T.A., Calleja V., Larijani B. Regulation of 3-phosphoinositide-dependent protein kinase 1 activity by homodimerization in live cells. Sci. Signal. 2010;3:ra78. doi: 10.1126/scisignal.2000738. [DOI] [PubMed] [Google Scholar]
- 45.Heras-Martínez G.L., Calleja V., Requejo-Isidro J. A complex interplay of anionic phospholipid binding regulates 3′-phosphoinositide-dependent-kinase-1 homodimer activation. Sci. Rep. 2019;9:14527. doi: 10.1038/s41598-019-50742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biondi R.M., Komander D., van Aalten D.M. High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J. 2002;21:4219–4228. doi: 10.1093/emboj/cdf437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markwick P.R.L., Peacock R.B., Komives E.A. Accurate prediction of amide exchange in the fast limit reveals thrombin allostery. Biophys. J. 2019;116:49–56. doi: 10.1016/j.bpj.2018.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bateman R.H., Carruthers R., Vissers J.P. A novel precursor ion discovery method on a hybrid quadrupole orthogonal acceleration time-of-flight (Q-TOF) mass spectrometer for studying protein phosphorylation. J. Am. Soc. Mass Spectrom. 2002;13:792–803. doi: 10.1016/S1044-0305(02)00420-8. [DOI] [PubMed] [Google Scholar]
- 49.Silva J.C., Denny R., Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 50.Tulsian N.K., Krishnamurthy S., Anand G.S. Channeling of cAMP in PDE-PKA complexes promotes signal adaptation. Biophys. J. 2017;112:2552–2566. doi: 10.1016/j.bpj.2017.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moorthy B.S., Gao Y., Anand G.S. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002295. M110 002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guarnera E., Berezovsky I.N. Structure-based statistical mechanical model accounts for the causality and energetics of allosteric communication. PLoS Comput. Biol. 2016;12:e1004678. doi: 10.1371/journal.pcbi.1004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guarnera E., Tan Z.W., Berezovsky I.N. AlloSigMA: allosteric signaling and mutation analysis server. Bioinformatics. 2017;33:3996–3998. doi: 10.1093/bioinformatics/btx430. [DOI] [PubMed] [Google Scholar]
- 54.Hinsen K., Petrescu A.-J., Kneller G.R. Harmonicity in slow protein dynamics. Chem. Phys. 2000;261:25–37. [Google Scholar]
- 55.Cunningham B.C., Wells J.A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 56.Hughes C.A., Mandell J.G., Komives E.A. Phosphorylation causes subtle changes in solvent accessibility at the interdomain interface of methylesterase CheB. J. Mol. Biol. 2001;307:967–976. doi: 10.1006/jmbi.2001.4523. [DOI] [PubMed] [Google Scholar]
- 57.Fabbro D., Cowan-Jacob S.W., Moebitz H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015;172:2675–2700. doi: 10.1111/bph.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor S.S., Kornev A.P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith C.M., Radzio-Andzelm E., Taylor S.S. The catalytic subunit of cAMP-dependent protein kinase: prototype for an extended network of communication. Prog. Biophys. Mol. Biol. 1999;71:313–341. doi: 10.1016/s0079-6107(98)00059-5. [DOI] [PubMed] [Google Scholar]
- 60.Weis D.D., Wales T.E., Ten Eyck L.F. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 61.Nussinov R., Tsai C.J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 62.Leroux A.E., Gross L.Z.F., Biondi R.M. Allosteric regulation of protein kinases downstream of PI3-kinase signalling. Adv. Exp. Med. Biol. 2019;1163:279–311. doi: 10.1007/978-981-13-8719-7_12. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J., Adrián F.J., Gray N.S. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazzoletti M., Bortolin F., Broggini M. Combination of PI3K/mTOR inhibitors: antitumor activity and molecular correlates. Cancer Res. 2011;71:4573–4584. doi: 10.1158/0008-5472.CAN-10-4322. [DOI] [PubMed] [Google Scholar]
- 65.Leroux A.E., Biondi R.M. Renaissance of allostery to disrupt protein kinase interactions. Trends Biochem. Sci. 2020;45:27–41. doi: 10.1016/j.tibs.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galstyan V., Funk L., Phillips R. Combinatorial control through allostery. J. Phys. Chem. B. 2019;123:2792–2800. doi: 10.1021/acs.jpcb.8b12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hertig S., Latorraca N.R., Dror R.O. Revealing atomic-level mechanisms of protein allostery with molecular dynamics simulations. PLoS Comput. Biol. 2016;12:e1004746. doi: 10.1371/journal.pcbi.1004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moorthy B.S., Anand G.S. Multistate allostery in response regulators: phosphorylation and mutagenesis activate RegA via alternate modes. J. Mol. Biol. 2012;417:468–487. doi: 10.1016/j.jmb.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 70.del Sol A., Tsai C.J., Nussinov R. The origin of allosteric functional modulation: multiple pre-existing pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cui K., Sun T.L., Gong J.P. Multiscale energy dissipation mechanism in tough and self-healing hydrogels. Phys. Rev. Lett. 2018;121:185501. doi: 10.1103/PhysRevLett.121.185501. [DOI] [PubMed] [Google Scholar]
- 72.Zhang H., Neimanis S., Biondi R.M. Molecular mechanism of regulation of the atypical protein kinase C by N-terminal domains and an allosteric small compound. Chem. Biol. 2014;21:754–765. doi: 10.1016/j.chembiol.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Scheeff E.D., Bourne P.E. Structural evolution of the protein kinase-like superfamily. PLoS Comput. Biol. 2005;1:e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.